Abstract
Ralstonia solanacearum, the causal agent of bacterial wilt, exerts its pathogenicity through more than a hundred secreted proteins, many of them depending directly on the functionality of a type 3 secretion system. To date, only few type 3 effectors have been identified as required for bacterial pathogenicity, notably because of redundancy among the large R. solanacearum effector repertoire. In order to identify groups of effectors collectively promoting disease on susceptible hosts, we investigated the role of putative post-translational regulators in the control of type 3 secretion. A shotgun secretome analysis with label-free quantification using tandem mass spectrometry was performed on the R. solanacearum GMI1000 strain. There were 228 proteins identified, among which a large proportion of type 3 effectors, called Rip (Ralstonia injected proteins). Thanks to this proteomic approach, RipBJ was identified as a new effector specifically secreted through type 3 secretion system and translocated into plant cells. A focused Rip secretome analysis using hpa (hypersensitive response and pathogenicity associated) mutants revealed a fine secretion regulation and specific subsets of Rips with different secretion patterns. We showed that a set of Rips (RipF1, RipW, RipX, RipAB, and RipAM) are secreted in an Hpa-independent manner. We hypothesize that these Rips could be preferentially involved in the first stages of type 3 secretion. In addition, the secretion of about thirty other Rips is controlled by HpaB and HpaG. HpaB, a candidate chaperone was shown to positively control secretion of numerous Rips, whereas HpaG was shown to act as a negative regulator of secretion. To evaluate the impact of altered type 3 effectors secretion on plant pathogenesis, the hpa mutants were assayed on several host plants. HpaB was required for bacterial pathogenicity on multiple hosts whereas HpaG was found to be specifically required for full R. solanacearum pathogenicity on the legume plant Medicago truncatula.
The soil-borne vascular bacterium R. solanacearum is described as one of the most destructive plant pathogenic bacterium worldwide (1), mainly because of its broad host range and wide geographic distribution. Indeed, R. solanacearum attacks more than 250 plant species, distributed in more than 50 botanical families (2). R. solanacearum causes dramatic crop losses, more specifically in the tropics, notably affecting emerging countries. Durable protection strategies against this bacterium are lacking. R. solanacearum penetrates into the plant via the roots, and then colonizes the xylem vessels. The bacterium reaches the aerial parts of the plant, causes wilting symptoms leading to the death of the plant, and will eventually return to the soil, completing the cycle (3). To achieve these first colonization steps, the bacterium uses a molecular syringe called the type 3 secretion system (T3SS),1 which delivers virulence factors, the type 3 effectors (T3Es) into the host cells. Collectively, these effectors constitute one of the main weapons of the pathogenicity arsenal of R. solanacearum, as it is the case for many other Gram-negative phytopathogenic bacteria (i.e. Pseudomonas spp., Xanthomonas spp., and Erwinia spp.) (4). Many early substrates of the secretion apparatus are highly conserved among these bacteria, including the cytoplasmic, inner and outer membrane ring components, and the needle. The translocators (intermediate substrates) and the translocated effectors (late substrates) show more diversity, which may contribute to the host specificities of each pathogenic bacterium (4–6). The core component proteins are encoded by the hrp (hypersensitive response and pathogenicity) and hrc (hrp-conserved) genes, located on the hrp gene cluster, whereas the T3Es are distributed throughout the genomes of these bacteria (7–10).
R. solanacearum strains possess large T3E repertoires, with 72 Rips (Ralstonia injected proteins) identified in the model strain GMI1000 (11). Rip delivery through the T3SS is under a fine transcriptional control, orchestrated by the regulatory protein HrpB. This transcription factor activates both the expression of the T3SS encoding genes and of the T3E genes (12). The R. solanacearum T3SS transcriptional regulatory system is well described (13), whereas little is known about the post-translational control of Rip delivery. There were 94 Rip genes identified among strains of the R. solanacearum species complex sequenced to date (11). This large repertoire of virulence factors may explain the wide host range of this bacterium. However, because of Rip redundancy (14–16), single Rip are often dispensable for bacterial pathogenicity on a given host, except for RipG7 (formerly named GALA7) on the legume plant Medicago truncatula (17). To date, many questions remain, we still do not know whether there could be a specific regulation of Rip delivery, depending on the host plant, the host tissues, or on the stage of bacterial colonization. Are all Rips delivered into the host at the same time, in equal quantities, or is there a hierarchical and quantitative control?
To better understand this complex mechanism, we studied the delivery of Rips via the T3SS through in vitro Rip secretion. We used optimized secretion conditions (18) in which the bacterium is in an HrpB-inducing environment, mimicking the in planta conditions and leading to Rip secretion. With this standardized and controlled system, we aimed at identifying sets of Rips with conserved and typical secretion patterns. Like it has been shown for other pathogenic bacteria (19, 20), we can hypothesize that Rip delivery is post-translationally controlled by helper proteins. These helper proteins are described in both plant and animal pathogenic bacteria, but are not orthologs. They are known to regulate T3E transit through the T3SS at post-transcriptional and post-translational levels, stabilizing T3Es, preventing their degradation or ordering and mediating their recognition by the T3SS (19, 21, 22). Some secretion helper proteins have been well described, like the T3Cs (type 3 chaperones), or the T3S4 (type 3 secretion substrate specificity switch) proteins (6, 22). Our previous work on R. solanacearum HpaP, a putative T3S4 protein, revealed that this regulatory protein could act as a modulator of the secretion level, targeting some T3Es, notably via direct protein interactions (23).
To advance our knowledge on the R. solanacearum T3S and on its post-translational control, we explored the complete secretome of R. solanacearum, taking advantage of the most recent tandem MS approach. In this work, we describe the most complete secretome (228 proteins including Rips) of a plant pathogenic bacterium using a MS-based shotgun approach with a label-free quantification (24). This allowed us to compare secretomes between hpa (hrp-associated) mutants and evaluate their respective involvement in T3S. In parallel, the phenotypic analysis of these hpa mutants on different host plants was carried out. This first in-depth proteomic approach on R. solanacearum enhances our knowledge on the T3S regulation and is a new mean to identify sets of Rips potentially coregulated, likely to be relevant for host specificity.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains used in this work are listed in Table I. Escherichia coli strains were grown at 37 °C in Luria-Bertani medium. Ralstonia solanacearum strains were grown in complete B medium or minimal medium, as previously described (25). When needed, antibiotics were added at the following final concentrations (mg/L): kanamycin (50), spectinomycin (40), gentamycin (10), tetracycline (10) for R. solanacearum and kanamycin (25), spectinomycin (40), gentamycin (10), tetracycline (10), ampicillin (50), chloramphenicol (25) for E. coli. The plasmids used in this study, listed in Table I, were constructed by Gateway technology (Life Technologies, Carlsbad, CA) following the instructions of the manufacturer. Genes cloned in the pDONR207 plasmid were amplified in two steps. The first PCR was performed using the specific primers indicated in Supplemental Experimental Procedures. The second PCR was performed using 1 μl of the first PCR as matrix and attB universal primers (oNP291 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATG-3′ and oNP292 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTC-3′).
Table I. Strains and plasmids used in this study. *Cmr, Gmr, Kmr, Tcr and Spr, resistances to chloramphenicol, gentamycin, kanamycin, tetracycline and spectinomycin, respectively.
| Characteristics | Source | |
|---|---|---|
| R. solanacearum strains | ||
| GMI1000 | Wild-type strain | (8) |
| GMI1694 | GMI1000 hrcV::Ω mutant, Spr | (37) |
| GRS230 | GMI1000 ΔhpaG mutant, Gmr | (37) |
| GRS266 | GMI1000 hpaD::Ω mutant, Spr | (37) |
| GRS474 | GMI1000 hpaB::Ω mutant, Spr | This study |
| GRS503 | GMI1000 Rsp0213::Ω mutant, Spr | This study |
| Plasmids used for cloning | ||
| pENTR/D-TOPO | Gateway™ entry vector, Kmr | Life Technologies |
| pENTR/SD/D-TOPO | Gateway™ entry vector, Shine–Dalgarno sequence, Kmr | Life Technologies |
| pDONR207 | Gateway™ entry vector, Gmr Cmr | Life Technologies |
| pAA3 | pENTR/D-TOPO derivating carrying ripD, Kmr | This study |
| pACC109 | pDONR207 derivating carrying ripH2, Gmr | This study |
| pACC220 | pDONR207 derivating carrying ripAN, Gmr | This study |
| pACC351 | pDONR207 derivating carrying ripH3, Gmr | This study |
| pACC387 | pDONR207 derivating carrying ripH1, Gmr | This study |
| pACC478 | pDONR207 derivative carrying ripW, Gmr | This study |
| pACC646 | pENTR/SD/D-TOPO derivating hpaG, Kmr | This study |
| pACC702 | pENTR/SD/D-TOPO derivating carrying ripAB, Kmr | This study |
| pACC750 | pENTR/SD/D-TOPO derivating carrying RSp0213, Kmr | This study |
| pFL12 | pMT1 derivatives carrying a KpnI-HindIII fragment of pNP224 containing the ripG7 promoter Kmr | This study |
| pFL13 | pLAFR6 carrying pFL12 Tcr Kmr | This study |
| pLAFR6 | pLAFR1 with trp terminators Tcr | (25) |
| pLBy5 | pDONR207 derivating carrying hpaB, Gmr | This study |
| pLBy14 | pDONR207 derivating carrying ripAV, Gmr | This study |
| pMT1 | pSC205 derivative carrying CyaA4–1197 N-terminal part, Kmr | (23) |
| pNP224 | pSC205 derivative carrying ripG7 promoter (from RSc1445146 to RSc1445497) upstream of the Gateway™ cassette Kmr | This study |
| pNP329 | pRCG derivative (63) with the RipG7 promoter followed by a Gateway™ destination cassette and a triple HA epitope tag, Gmr Kmr | (23) |
| pSC205 | pET26b(+) (Clontech) carrying a Gateway™ cassette Kmr | (23) |
| Plasmids for transformation in R. solanacearum | ||
| pACC618 | pNP329 derivative carrying ripH2, Gmr Kmr | This study |
| pACC619 | pNP329 derivative carrying ripH3, Gmr Kmr | This study |
| pACC636 | pNP329 derivative carrying ripH1, Gmr Kmr | This study |
| pAM5 | pLAFR3 carrying 2-kb fragment containing hrpB, Tcr | (18) |
| pEG3 | pNP329 derivative carrying ripAV, Gmr Kmr | This study |
| pEG4 | pNP329 derivative carrying ripW, Gmr Kmr | This study |
| pEG5 | pNP329 derivative carrying ripAN, Gmr Kmr | This study |
| pEG7 | pNP329 derivative carrying ripD, Gmr Kmr | This study |
| pEG8 | pNP329 derivative carrying hpaG, Gmr Kmr | This study |
| pEG10 | pNP329 derivative carrying ripAB, Gmr Kmr | This study |
| pEG11 | pNP329 derivative carrying hpaB, Gmr Kmr | This study |
| pFL11 | pNP329 derivative carrying RSp0213, Gmr Kmr | This study |
| pFL14 | pFL13 derivative carrying RSp0213, Tcr Kmr | This study |
Generation of R. solanacearum Mutants
R. solanacearum mutants are listed in Table I. hrcV, hpaG, and hpaD mutants were previously generated (12). In this study we constructed hpaB and ripBJ (RSp0213) mutants. To generate hpaB mutation in the GMI1000 wild-type strain, a 1.6 kb DNA fragment encompassing hpaB was PCR-amplified using the following primers G-UPLNG 5′-AAGCTTCCCGGACGGCTCAACGC-3′ and HPAB-rev 5′-AAGCTTGTAGATGTGCTGCTCGA-3′ and then cloned into the pUC18 vector. The Ω -spectinomycin interposon (26) was cloned into the unique NotI restriction site within the hpaB open reading frame to generate plasmid pSG424. pSG424 was linearized by XbaI and used to transform R. solanacearum as previously described (27). Strain GRS474 was obtained by selecting a double recombination event using spectinomycin resistance and hpaB gene disruption was then checked by PCR. The same strategy was used to generate the strain GRS503 (RSp0213 mutant). p213fw 5′-TCTAGATCCAGGTGGCGCTGCAG-3′ and p213rev 5′-GTTCGACGTGGATGCGGGC-3′ primers were used to amplify and clone a 1.6 kb fragment containing RSp0213 into the pUC vector. The Ω -spectinomycin interposon (26) was cloned into the SmaI restriction site of RSp0213 (pSG493). Transformation into R. solanacearum was performed and checked as previously described for generation of GRS474 strain.
Construction of Strains with Effector-HA Tag and Complementations
Effector-3HA fusions were done using pNP329 (23) with the Gateway technology (Life Technologies) to allow stable chromosomic insertion. pNP329 derivatives carrying ripD, ripH1, ripH2, ripH3, ripW, ripAB, ripAN, and ripAV-3HA tags were linearized using ScaI enzyme, and then introduced into five strains, i.e. the wild-type (GMI1000) (8), and the hrcV, hpaB, hpaD, and hpaG mutants (GMI1694, GRS474, GRS266, and GRS230) by natural transformation as previously described (27). Stable chromosomal insertions in the defined bacterial chromosome site were checked by PCR using oNP611 5′-GAAAGCACGCTGTTTCCGCTATTT-3′, oNP612 5′-GCGTAGTGCGCAAGACGAACAA-3′ and oNP613 5′-GGCTCAAGGAGAAGAGCCTTCAGA-3′ primers. The hpaB and hpaG mutants were complemented with their corresponding wild-type alleles, using pNP329 and the same procedure.
Secretome Sample Preparation
Five R. solanacearum strains were used for MS secretome analysis: GMI1000 wild-type strain, GMI1694 hrcV mutant, GRS474 hpaB mutant, GRS266 hpaD mutant and GRS230 hpaG mutant (Table I). These R. solanacearum strains carrying the pAM5 plasmid (18), that leads to a higher transcriptional activity of T3SS-regulated genes, were first cultivated in B medium overnight, pelleted for 5 min at 4000 rpm and then resuspended in 15 ml of minimal medium (supplemented with 0.1 g/L congo red and 10 mm glucose and 10 mm glutamate, with 10 μg/ml tetracycline) at OD600 nm = 0.2. Bacterial cultures were then harvested after 8 h of culture, set at the same concentration, according to the OD600 nm and pelleted for 10 min at 5000 rpm. One milliliter of supernatant was filter-sterilized (0.22 μm) and the proteins were precipitated using 1:1 (v/v) TCA 25%. The protein pellet was washed twice with acetone 90% and resuspended in 40 μl of Laemmli buffer with 0.2 m of Tris-HCl pH 7.4. Concentrated supernatants were then loaded onto a SDS-PAGE 10% bisacrylamid gel for a short migration. Portions of gels that contain the proteins (1 cm of sample migration) were cut for shotgun MS analysis.
Protein In-Gel Digestion
Each lane was cut and washed for 15 min with an acetonitrile/100 mm ammonium bicarbonate mixture (1:1). Digestion was performed in 50 mm ammonium bicarbonate pH 8.0 and the quantity of modified trypsin (Promega, sequencing grade, Madison, WI) was 0.1 μg per sample. Digestion was achieved for 6 h at 37 °C. The supernatant was conserved. Peptides were extracted by 5% formic acid in water/acetonitrile (v/v). Supernatant and extracted tryptic peptides were dried and resuspended in 50 μl of 0.1% (v/v) formic acid and 2% (v/v) acetonitrile.
Nano Liquid Chromatography Coupled to Mass Spectrometry in Tandem
A Q Exactive (Thermo Fisher Scientific) coupled to Eksigent 2Dnano LC (AB-Sciex, Framingham, MA) were used for the nano-LC-MS/MS analysis. Four microliters of sample were injected on the nanoLc-Ultra system (AB-Sciex) chain. Sample was loaded at 7.5 μl/min on the precolumn cartridge (C18, 5 μm, 120 Å, 20 mm Nanoseparations) and desalted with 0.1% formic acid. Then, peptides were separated with a gradient of acetonitrile on the reverse phase column C18 (stationary phase: C18 Biosphere, 3 μm; column: 75 μm i.d., 150 mm; Nanoseparations). Buffers were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The peptide separation was achieved with a linear gradient from 5 to 30% B for 40 min at 300 nL/min−1. Eluted peptides were analyzed on-line with a Q Exactive mass spectrometer (Thermo Ficher Scientific, San Jose, CA) using a nanoelectrospray interface. Ionization (1.8 kV ionization potential) was performed with stainless steel emitters (30 μm i.d.; (Thermo Ficher Scientific). Peptide ions were analyzed using Xcalibur 2.1 with the following data-dependent acquisition steps: (1) full MS scan (mass-to-charge ratio (m/z) 400 to 1400) and (2) MS/MS. Step two was repeated for the eight major ions detected in step one. Dynamic exclusion was set to 40 s. Lock mass option was chosen “best,” MS resolution 70,000 at m/z 400, auto gain control was 1e6, maximum injection time 250 ms. For MS2 the resolution was 17,500 at m/z 400, auto gain control was 2e5, maximum injection time 120 ms, isolation window m/z = 3, normalized collision energy: 30, underfill ratio 0.5%, intensity threshold 8.3e3. Charge state: 2, 3, 4; dynamic exclusion 40 s.
Data Processing and Bioinformatics Analysis
The R. solanacearum strain GMI1000 database was downloaded from Database site (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi, October 2012, 5122 protein entries). This database was merged and in conjunction with reverse and contaminant databases, were searched by X!Tandem (Sledge Hammer version 2013.09.01.1, http://www.thegpm.org/tandem/) using X!TandemPipeline (version 3.3.3) developed by PAPPSO platform (http://pappso.inra.fr/bioinfo/). Enzymatic cleavage was declared as a trypsin digestion with one possible miss-cleavage. Cys carboxyamidomethylation and Met oxidation were set to static and possible modifications, respectively. Precursor mass was 10 ppm and fragment mass tolerance was 0.02 Th. A refinement search was added with similar parameters except for semitrypsic peptides and possible N-ter proteins. For data of proteomic, only peptides with an E value smaller than 0.1 were reported. Identified proteins were filtered and grouped using X!TandemPipeline (http://pappso.inra.fr/bioinfo/xtandempipeline/) according to: (1) a minimum of two different peptides was required with an E value smaller than 0.05, (2) a protein log (E value) (calculated as the product of unique peptide E values) smaller than 10−3. These criteria led to a false discovery rate (FDR) of 0.08% for the wild-type strain and of 0.07% for the five strains (wild-type, hrcV and hpa mutants) for peptide and protein identification. To take redundancy into account, proteins with at least one peptide in common were grouped. This allowed to group proteins of similar function. Within each group, proteins with at least one specific peptide relatively to other members of the group were reported as subgroups. Only proteins detected on at least two samples on at least one strain were considered. Label-free quantification of proteins was achieved in two steps. The first one is the spectral counting (SC) (28–30), which is a strategy to determine a relative quantification of protein from their number of spectra obtained with tryptic peptides in MS. This quantification is based on the fact that the more of a particular protein is present in a sample, the more MS spectra are detected for peptides of that protein. The second step is performed using protein abundance index (PAI) for every protein and was obtained by dividing SC data of the observed protein by the number of theoretically observable tryptic peptides for that protein (31).
Experimental Design and Statistical Rationale
Concerning the secretome samples used for MS experiments, four independent biological replicates were generated for each of the five strains studied (wild-type strain, hrcV, hpaB, hpaD, and hpaG mutants). We found that four replicates were the best possible compromise in order to duly handle experimental variability and to do quantitative analysis. The hrcV mutant was used as a negative control of T3S, allowing discrimination of type 3 proteins. In order to be compared, samples were normalized dividing each PAI by the total sum of PAIs on the corresponding sample. Thus normalized PAI values were log-scaled to be able to adjust a linear model for every protein. With the aim of better handling multiple comparisons, general linear hypothesis tests (glht) (32) were subsequently run on the model to discriminate proteins differentially quantified between bacterial strains. All statistical procedures were carried out under [R] v3.1.1 environment (33) using base packages as well as “multcomp” (32), “limma” (34), and “venneuler” (35) packages. The R source code created for this study is available upon request.
Type 3 Secretion Assays and Immunoblot Analysis
The secretion pattern of thirteen T3Es was followed in five R. solanacearum backgrounds. First, the same five R. solanacearum strains as for MS experiments were used for secretion assays with available T3E specific antibodies. Second, for the other T3Es studied, T3E-HA fusions were generated (ripD, ripH1, ripH2, ripH3, ripW, ripAB, ripAN, and ripAV) and introduced into the five strains (see above). pAM5 (18) was added by electroporation into these 40 strains. Then, secretion assays were performed as previously described (23). Proteins from an equal amount of cultures (normalized by OD600) were loaded in each lane. Total proteins were stained with Silver Stain Plus kit (Bio-Rad, Hercules, CA) (supplemental Fig. S1) or transferred to nitrocellulose membrane for Western blotting. Antibodies used were RipG7 (23), RipP1 (formerly PopP1) (36), RipP2 (formerly PopP2) (courtesy of L. Deslandes, INRA/CNRS, Castanet Tolosan, France), RipX (formerly PopA) (18), RipAA (formerly AvrA) (25), and an antihemagglutinin coupled with horseradish peroxidase (anti-HA) (Santa Cruz Biotechnology, Santa Cruz, CA). When needed, goat anti-rabbit antibody conjugated with horseradish peroxidase was used as secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Biologically independent experiments were performed twice.
Adenylate Cyclase (CyaA) Translocation Assays
A KpnI-HindIII fragment of pNP224 containing the ripG7 promoter was cloned into pMT1 that carries CyaA 4–1197 N-terminal part (pFL12). Then pFL12 was cloned into the KpnI site of pLAFR6 to allow plasmid replication in R. solanacearum (pFL13). RSp0213 was then integrated into pFL13 by gateway cloning. RSp0213-CyaA' (pFL14) construct was transformed into GMI1000 (wild-type strain) and GMI1694 (hrcV mutant). Translocation assays were performed as previously described on Nicotiana tabacum (37). The cAMP production was measured using the cAMP Biotrak enzyme immunoassay (EIA) System kit by GE Healthcare (Buckinghamshire, UK). Four independent biological replicates were performed.
Plant Assays and Statistical Analysis
For Arabidopsis thaliana assay, 16 Col-0 ecotype plants were inoculated as previously described (38) using 108 bacteria/ml, without cutting the roots. For M. truncatula (ecotype A17), two types of inoculation were performed. For soil inoculations, 16 M. truncatula plants with cut roots were soaked with 800 ml of a bacterial solution at 106 bacteria/ml. In vitro inoculations were done as previously described (39) with 107 bacteria/ml. Disease development was monitored every day, and plants with at least 50% of wilting were considered as dead for the statistical survival analysis. To compare the disease development of two given strains, we used the Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon test. A p value smaller than 0.05 was considered significant, indicating that the Ho hypothesis of similarity of the survival experience of the tested strains can be rejected. Statistical analyses were done with Prism version 5.00 (GraphPad Software). Each experiment was repeated at least three times.
Internal Growth Curve
Internal Growth curve was determined mostly as previously described (39). Fourteen days post-inoculation, at least three pools of three plants were harvested, sterilized in 70% ethanol and rinsed three times in water. Plants were weighted, grinded and then resuspended in sterile water. Bacterial concentrations were determined by plating dilutions on B medium with antibiotic if necessary. Experiments were repeated three times.
RESULTS
Secretome of R. solanacearum GMI1000 Grown in Type 3 Secretion Inducing Conditions
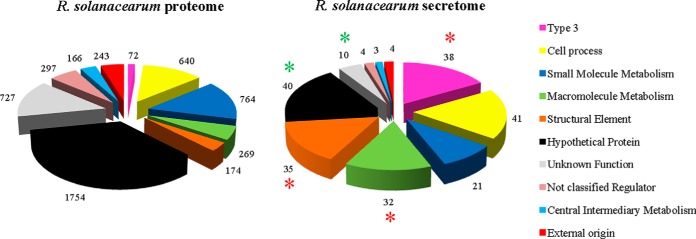
Using LC-MS/MS, we analyzed the secretome of the R. solanacearum GMI1000 wild-type strain. Bacteria were grown in secretion inducing media, and four independent biological secretome samples were generated by a shotgun approach with label-free quantification. The experimental procedure scheme is shown in supplemental Fig. S2A. Identification of peptides was done using X!TandemPipeline developed by PAPPSO platform (http://pappso.inra.fr/). Protein identification was done using the R. solanacearum genome database (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi) and a contaminant database (trypsin, keratins, etc). A protein was considered as present in the supernatant when spectra were detected in at least two independent replicates. The number of spectra corresponding to each detected protein, the normalized protein abundance index (PAI), and all the peptide sequences are provided in supplemental Table S1. The analysis of strain GMI1000 secretome identified 228 proteins which were classified into functional groups according to the R. solanacearum proteome database (see supplemental Table S1; Fig. 1). A Bonferroni-corrected hypergeometric test (with a p value < 0.001) revealed a significant enrichment for three categories corresponding to macromolecule metabolism, structural elements, and type 3-associated proteins (Fig. 1). Among the 32 proteins belonging to the “macromolecule metabolism” class many previously characterized R. solanacearum secreted proteins were found, e.g. plant cell wall degrading enzymes (endo- and exoglucanases, polygalacturonases) (supplemental Table S1). The category of type 3-associated proteins showed the highest enrichment (p value = 5.72.10−34). The 38 detected type 3-associated proteins included three structural elements of the syringe, the HrpY pilin (the major component of the Hrp pilus) and the two translocators RipF1_1 and RipF1_2 (formerly named PopF1 and PopF2), which are thought to form a pore in the plant cell (supplemental Fig. S2B). The 35 other type 3 proteins correspond to Rip protein effectors.
Fig. 1.
R. solanacearum GMI1000 wild-type secretome shows enrichment in T3-proteins compared with the wild-type proteome. Distribution of total R. solanacearum proteins according to the predicted functions ((https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi) (left pie) compared with detected proteins by LC-MS/MS (right pie). Bonferroni-corrected hypergeometric tests (p values < 0.001) reveal three significantly enriched categories in the secretome (red asterisks), i.e. “Type 3” (p = 5.72.10−34), “Macromolecule Metabolism” (p = 1.79.10−6) and “Structural Element” (p = 6.05.10−13); as well as two significantly under-represented categories (green asterisks), i.e. “Hypothetical Protein” (p = 1.06.10−8) and “Unknown” (p = 2.06.10−5).
Type 3 Secretome of R. solanacearum
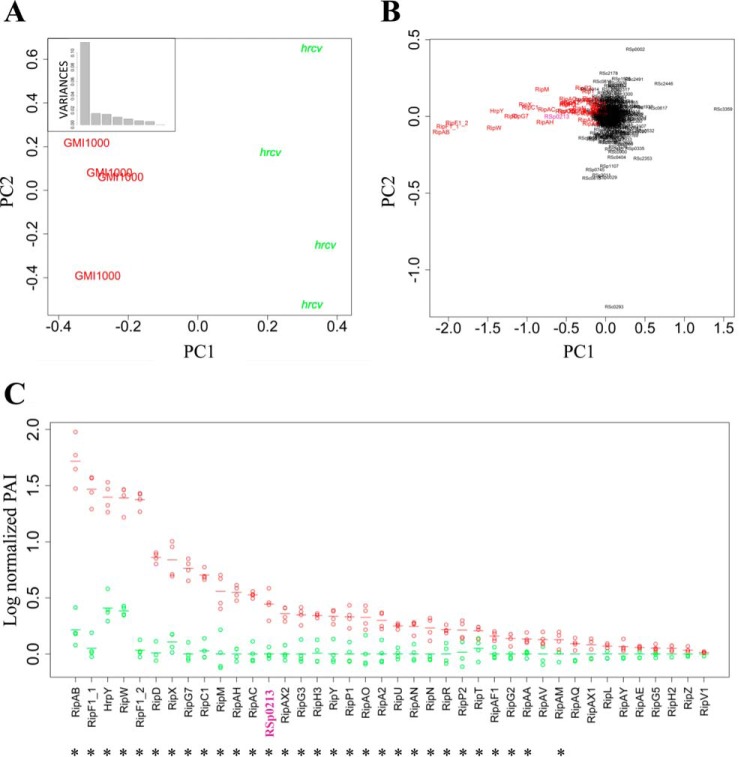
In order to define the set of T3-secreted proteins, we compared the GMI1000 wild-type secretome with the T3SS-defective hrcV mutant. There were 224 proteins detected in the hrcV mutant culture supernatant (supplemental Fig. S3). A principal component analysis (PCA) on log-normalized PAI data was carried out to identify and visualize the sources of variability between all eight analyzed samples (four for each bacterial strain). The PCA on Fig. 2A clearly shows that the “strain” effect defining the first principal component, explains 66% of the overall variation. The positioning of the individual proteins on the PCA highlights that variance on this first component is quasi-exclusively because of T3-associated proteins (Fig. 2B). A similar analysis conducted excluding the annotated T3-associated proteins, shows that the strain effect is still present (supplemental Fig. S4A), but in this case the first principal component only explains 30% of the overall variance (supplemental Fig. S4B), with very few discriminating proteins being responsible for this component (supplemental Fig. S4C). A linear model was fitted and the subsequent glht post-hoc tests were performed to discriminate the proteins differentially secreted between GMI1000 and the hrcV mutant, revealing 63 proteins differentially secreted (p value < 0.05) (supplemental Table S2). Twenty-nine out of these 63 proteins were found to be T3-associated proteins (HrpY and 28 Rips). These 29 proteins are among the most significantly different between both strains with the lowest p values (supplemental Table S2). The secretion pattern of all the T3-proteins detected in both strains is shown in Fig. 2C. A noticeable point is that none or, at best, very few spectra were detected for any of the Rip T3Es (except RipW) in the hrcV mutant (supplemental Table S3, Fig. 2C). The barely detected T3S SC data on the hrcV replicates suggest a minimal bacterial lysis in our secretome samples. Overall, these first data can be considered as a good control for the whole process, from the protein extraction protocol in the supernatants to the statistical analysis of the MS detection.
Fig. 2.
R. solanacearum LC-MS/MS secretomes of GMI1000 wild-type strain and of hrcV mutant (a hrp defective mutant) bring specific and quantitative data on Rip secretion. A, PCA with the detected proteins in the wild-type strain and in the hrcV mutant highlights the strain effect on the first component, which presents a standard deviation of 0.34 and explains 66% of the total variance (box). B, protein distribution on the same PCA shows that the most discriminating proteins are T3-associated proteins (in red), and reveals a new Rip candidate: RSp0213 (in purple). C, log-normalized PAI values of T3-associated proteins detected in the wild-type (red) and in the hrcV mutant (green) secretomes. The proteins were classified according to their level of secretion in the wild-type strain (from the strongest to the lowest log-normalized PAI values). Four biological replicates were analyzed for each strain. Each dot represents a replicate; dashes represent the mean values of all four replicates. Asterisks highlight proteins presenting significantly different secretion levels between both strains (glht post-hoc test, p value < 0.05).
RipBJ, a New R. solanacearum Type 3 Effector
The global comparison of the GMI1000 wild-type secretome with the hrcV mutant secretome by LC MS/MS allowed us to highlight proteins specifically secreted by the T3SS (Fig. 2B). We noticed that the RSp0213 hypothetical protein clustered within the T3-proteins in the PCA (Fig. 2B) and RSp0213 appeared to be differentially secreted between both strains, with a p value = 1.50.10−5 (Fig. 2C; supplemental Table S2). In order to test the hypothesis that RSp0213 encodes a previously overlooked Rip effector protein, we performed in vitro secretion assay using GMI1000 and hrcV mutant strains expressing an RSp0213-HA fusion construct. Immunoblotting using an anti-HA antibody revealed that RSp0213-HA fusion proteins were present in both strains, but the RSp0213-HA secretion was only detected from the wild-type strain (Fig. 3A). To definitely show that RSp0213 is a bona fide Rip T3E, we performed a translocation assay based on the adenylate cyclase reporter system (40). GMI1000 and hrcV mutant strains carrying RSp0213-CyaA' fusion constructs were generated, and infiltrated in tobacco leaves. Seven hours after infiltration, tobacco leaves were harvested and cAMP levels were measured using cAMP Biotrak competitive enzyme immunoassay (37). High amounts of cAMP were detected for the wild-type strain but not for the hrcV T3-defective mutant, indicating that RSp0213 was translocated into plant cells through the T3SS (Fig. 3B). These experiments showed that RSp0213 encodes a T3E, which was therefore renamed RipBJ following the proposed nomenclature for R. solanacearum T3Es (11). A ripBJ disruption mutant was generated and used to evaluate the contribution of this effector to pathogenicity on A. thaliana and M. truncatula, and to hypersensitive response (HR) elicitation after infiltration of tobacco leaves. No differences could be observed between the ripBJ mutant and the wild-type strain for none of these bioassays (supplemental Fig. S5).
Fig. 3.
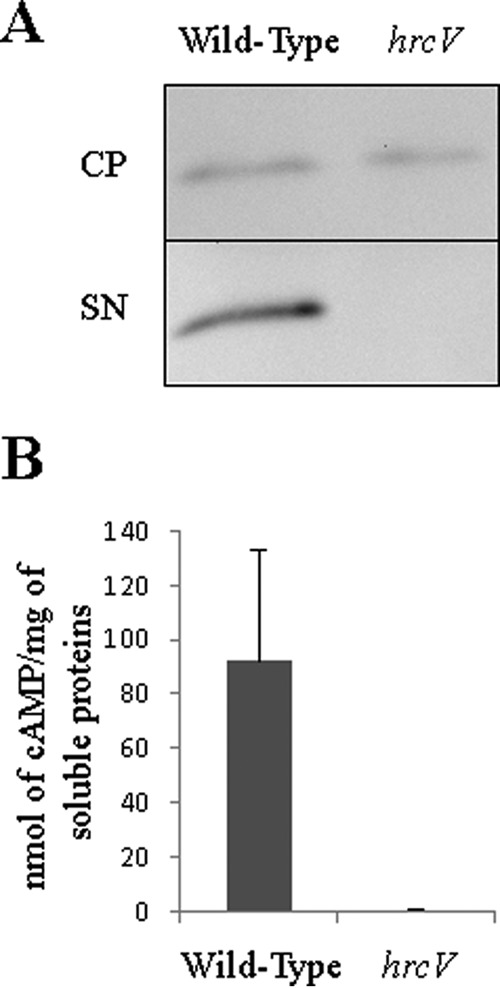
RSp0213 is secreted and translocated through the T3SS. A, the wild-type strain and the hrcv mutant were transformed to express RSp0213-HA fusion. Secretion assays were performed and total proteins from bacterial cell pellet (CP) and proteins in the supernatant (SN) were detected by Western-Blot. Detected signal size corresponds to the RSp0213-HA size (18 kDa). B, translocation assay of RSp0213 protein in N. tabacum leaves using RSp0213-CyaA' fusion protein. Cyclic adenosine monophosphate (cAMP) levels were detected to determine the level of translocation of RSp0213-CyaA' protein in tobacco leaves for each strain. Four independent biological replicates were made.
Insights into Type 3 Secretion Regulation from the Secretome Analysis of hrp-Associated Mutants
The comparative secretome analysis between the wild-type strain and a hrp deficient mutant showed that our analysis enables the fine characterization of T3-secretome, but also allows for the comparative quantification of detected proteins between strains. The sensitivity of the method incited us to explore and compare the secretomes of potential T3S regulators. Some of them were characterized in phytopathogenic bacteria for their participation to T3E delivery, by promoting or repressing secretion through protein–protein interactions (21, 23). In R. solanacearum strain GMI1000, four potential Hpa proteins were identified: HpaB (RSp0853), HpaD (RSp0848), HpaG (RSp0842), and HpaP (RSp0862). Hpa genes are hrpB-regulated (41), physically associated to the hrp-cluster and not secreted. HpaB and HpaD have features of molecular chaperones (20, 42) with small sizes (16.8 kDa and 13.9 kDa, respectively) and acidic pI (4.4 and 4.2, respectively). HpaP was recently shown to modulate T3S (23) and HpaG (also known as LrpE) is a leucine-rich repeats (LRR) containing protein that negatively regulates the production of Hrp pili (43). We conducted secretome analyses of the hpaB, hpaD, and hpaG mutants using a similar experimental procedure as described above (supplemental Fig. S2C). We detected 260, 279, and 262 proteins in the secretomes of the hpaB, hpaD, and hpaG mutants, respectively (supplemental Table S3; supplemental Fig. S3). The first two components of the PCA conducted on the secreted proteins detected in the five strains (wild-type, hrcV and hpa mutants) clearly identified the strain effect, discriminating all strains and particularly singling out the hrcV mutant (Fig. 4A). The proteins responsible for this sample distribution are highlighted on Fig. 4B. Group 1 contains the RipF1 translocon proteins (44) and RipAB (formerly named PopB) (18). Group 2 consists of the HrpY pilin and the harpins RipX and RipW (formerly PopA and PopW) (45, 46). Group 3 contains RipC1, RipD, RipG7, RipM, and RipAC, which are among the most abundant T3Es in the wild-type strain supernatant (see supplemental Table S1). Group 4 contains proteins with a lower secretion level in the wild-type strain (supplemental Table S1) but with a high discriminating value upon the glht post-hoc test (supplemental Table S2), whereas group 5 corresponds to the bulk of the remaining proteins. PCA performed on the five strain secretomes, excluding T3-associated proteins, shows no highly discriminating component (supplemental Fig. S6).
Fig. 4.
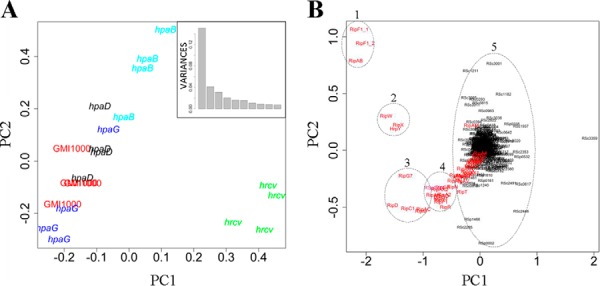
R. solanacearum LC-MS/MS secretomes of T3-mutants highlight specific secretion patterns for each mutant. A, PCA with all the secreted proteins in the wild-type strain, in the hrcV mutant and in three hpa mutants highlights the strain effect on the first component, which represents a standard deviation of 0.39 and explains 44% of the total variance (box). B, Same PCA as in A shows that main discriminating proteins belong to T3-associated proteins (in red) and different protein groups may be identified (dotted line circles).
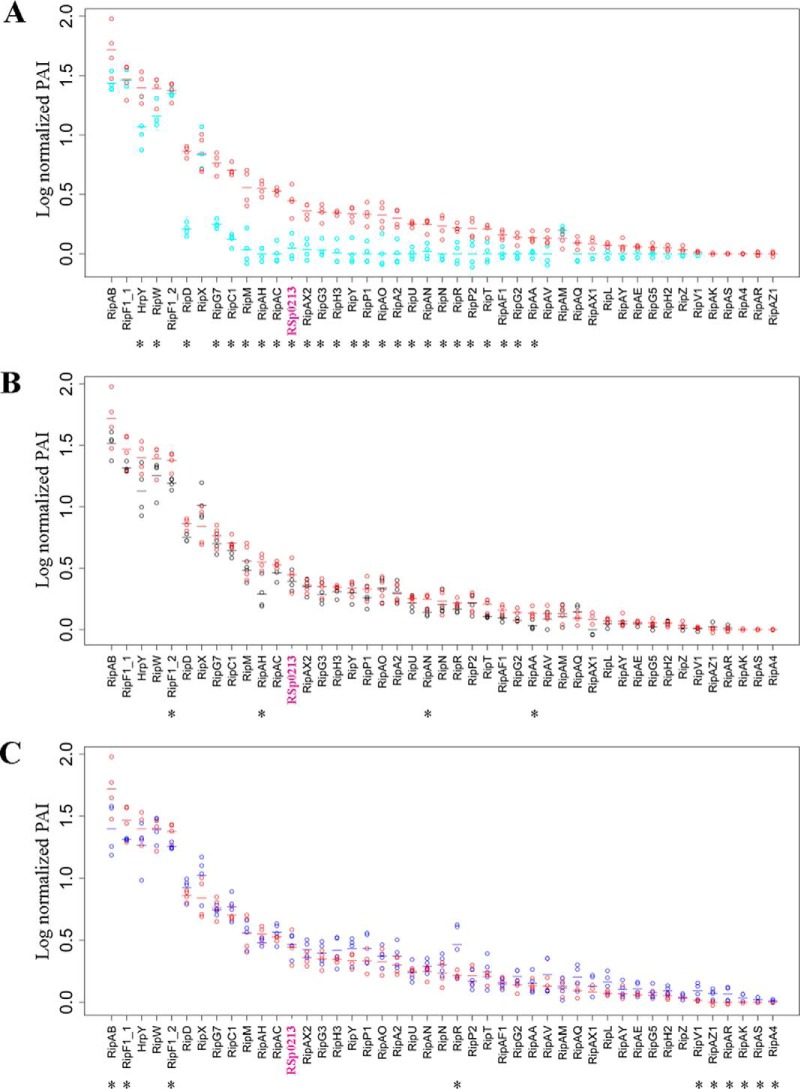
To more accurately evaluate the role of the hpa genes in T3S, we compared the detected T3-proteins in the wild-type strain versus each hpa mutant using a glht post-hoc test (Fig. 5; supplemental Table S3). The hpaB mutant shows three classes of secretion patterns: (1) a class of proteins with a high level of secretion, but mainly not differentially secreted as compared with GMI1000, which corresponds to HrpY, RipF1, and RipAB proteins, along with the RipW and RipX harpins; (2) a class of 25 proteins with a reduced level of secretion in the hpaB mutant; and (3) a class of five effectors with a low level of detection, which are not detected in the hpaB mutant (Fig. 5A). The hpaB mutant appears therefore strongly altered in its secretion of effectors but not of the components of the T3SS machinery. The hpaD mutant shows a secretion pattern close to the wild-type strain, with only four Rip significantly differentially detected (Fig. 5B). In contrast, the hpaG mutant secretome reveals a global over-secretion pattern. Indeed, even if few Rips are differently secreted in the mutant compared with the wild-type (RipAB, RipF1_1, RipF1_2, and RipR), we can observe that numerous Rips are more secreted by the hpaG mutant than the wild-type (Fig. 5C). Six Rips were detected only in the hpaG mutant (RipV1, RipAK, RipAS, RipA4, RipAR, and RipAZ1).
Fig. 5.
Log-normalized PAI values of T3-associated proteins detected in the secretomes of the wild-type (red) and of hpa mutants show different T3S patterns. A, hpaB mutant in cyan. B, hpaD mutant in black. C, hpaG mutant in blue. Four biological replicates were analyzed for each bacterial strain. Each dot represents a replicate; dashes represent the mean value for all four replicates. The proteins were classified according to their level of secretion in the wild-type strain (from the strongest to the lowest log-normalized PAI detection). Asterisks highlight proteins presenting significantly different secretion levels between strains (glht post-hoc test, p value < 0.05).
Three Main Classes of Type 3 Effectors Based on Secretion Pattern Specificities
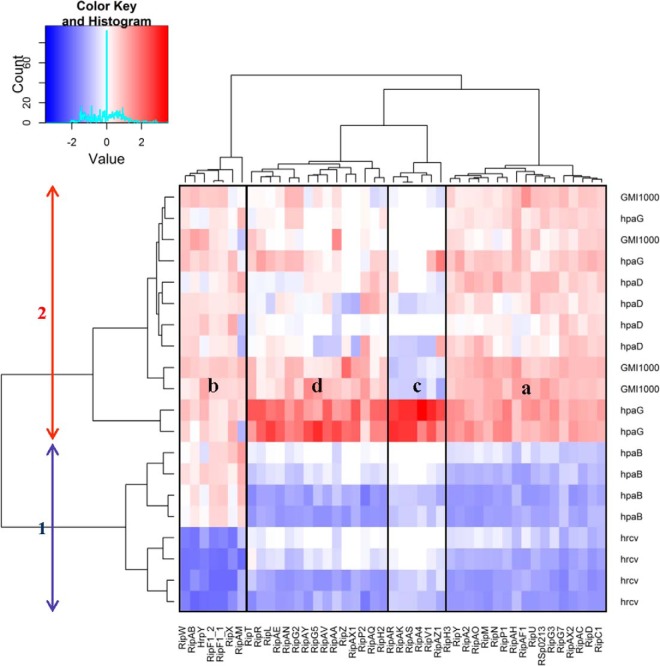
We used the log-normalized PAI data to perform a double hierarchical clustering in order to classify both the secreted Rips and the bacterial samples, according to the secretion of those Rip proteins. This analysis revealed two main groups of strains: hrcV and hpaB mutants on one hand, and hpaD, hpaG mutants, and GMI1000 on the other hand (Fig. 6). The separation of these two groups is caused by the differences in the quantification of 17 proteins from group “a” (including the newly identified RipBJ (RSp0213). These proteins are more abundant in the GMI1000/hpaD/hpaG secretomes than in the hpaB/hrcV ones. Likewise, the “b” group differentiates the hrcV from the other strains on the study, because of the absence of the Hrp pilus component and of the harpins in the hrcV mutant, thus revealing that regulation of RipAB and RipAM secretion is tightly linked at the post translational level. The “c” group corresponds to the six Rips only detected in the hpaG mutant secretome. The “d” subgroup clustered the 15 remaining Rips, which are also quantified in higher amount in the hpaG mutant.
Fig. 6.
Double hierarchical clustering of bacterial strains and T3-proteins. Bacterial strains thus produced two main clusters (arrows). Different protein subgroups according to their secretion patterns could be identified (a to d). White color represents a Z-score of 0 whereas red and blue shades represent higher and lower log-normalized PAI values, respectively.
In order to validate these specific secretion patterns obtained by MS, secretion assays were performed on several effectors showing (according to Fig. 2C) either high levels of secretion (RipAB, RipW, RipD, RipX, and RipG7), intermediate levels of secretion (RipH3, RipP1, RipAN, and RipP2), weak levels of secretion (RipAA, RipAV, and RipH2), or not detected by MS (RipH1). The abundance of these selected proteins was determined by immunoblotting protein extracts of cell pellets and of culture supernatants produced in the same conditions as the MS samples. In pellets, all the Rips were produced by the different strains in equal amount, except for hpaG mutant, as a slight overproduction can be observed for some Rips (Fig. 7). In supernatants, a different pattern of secretion can be observed: RipW, RipX, and RipAB were secreted in equal quantity by the three hpa mutants when compared with the wild-type strain (Fig. 7A). Combining MS data and Western blot experiments confirmed that the harpins RipW and RipX, as well as RipAB, share a similar secretion pattern. These Rips are thus secreted (in high amounts) independently of the three Hpa proteins. In Fig. 7B, we can observe that RipG7 is equally secreted in the hpaD and hpaG mutants compared with the wild-type, and less secreted in the hpaB mutant. This reflects the partial involvement of HpaB for strongly secreted Rips. Fig. 7C and 7D show the proteins (RipH3, RipP1, RipP2, RipAA, RipD, RipH2, RipAN, and RipAV) that are not secreted by hrcV nor hpaB mutants. This data confirms the secretion patterns observed by MS and highlights the specific requirement of hpaB for their secretion. Additionally, we observe an increased secretion for all these proteins in the hpaG mutant, which also validate the original over-secreting MS pattern of this mutant (Fig. 5C). We can notice that this over-secretion can be independent (Fig. 7C) or not (Fig. 7D) of an over-production of these proteins in the hpaG mutant. This reflects the involvement of HpaG in the control of T3E synthesis, but also potentially in transcriptional regulations or in protein stabilization. Finally, Fig. 7E shows that Western blot detection is more sensitive that MS detection as we can observe an hpaB-dependent pattern of secretion for RipH1, not detected in our MS analysis. However, the immunoblot analyses confirmed the specific patterns observed with MS analysis.
Fig. 7.
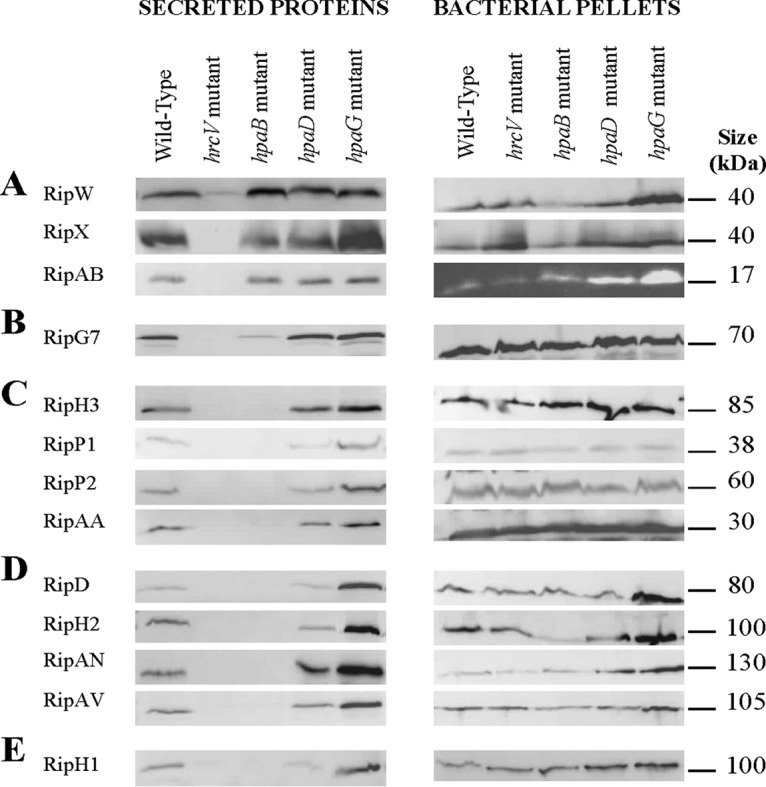
Western blot validations of Rip secretion patterns. Secretion assays were performed in T3-inducing conditions, for the GMI1000 wild-type strain, and the hrcV, hpaB, hpaD, and hpaG mutants. Total proteins from bacterial pellets and supernatants were analyzed by Western-blot. Two to three biological replicates were done for each set of five strains.
The Abundances of the Detected Effectors in the Bacterial Supernatants and That of Their Corresponding Gene Transcripts are Positively Correlated
In order to identify critical parameters determining the MS detection threshold, we investigated whether MS detection of the 44 secreted Rip effectors could be correlated either with structural features such as the isoelectric point (pI) or molecular weight (MW), or with transcription levels (as deduced from RNA sequencing approaches) (supplemental Fig. S7). The pI values of the detected Rips ranges from 4.2 (RipX) to 11.8 (RipAO), displaying a homogenous distribution between all the ranges of pI (supplemental Fig. S7A and S7B). The MW of the Rips ranges from 10 kDa (RipAG and RipAH) to 278 kDa (RipS4), with half of the Rips between 20 kDa to 80 kDa. The distributions observed in supplemental S7C and S7D could not clearly attribute a lower or higher detection rate according to the MW, since this detection ranged from RipAH (10 kDa) to RipR (186 kDa). However, the RipA and RipS protein families (formerly named AWR and SKWP families, respectively) having the highest molecular weight among R. solanacearum T3Es were less efficiently detected. Indeed, among the five paralogs of the RipA family (ranging from 113 kDa to 141 kDa), only RipA2 is detected and none of the six RipS paralogs (>250 kDa, except for RipS6) could be detected (Fig. 2; see supplemental Table S4 for exhaustive pI and MW Rip data). We also looked for a possible correlation between RNA abundance (RNA-Seq data) and MS detection. This was done by taking advantage of RNA-Seq experiments performed in the same inducing medium (Guidot et al., in preparation). We observed a positive correlation between the Rip RNA abundance and the cognate protein detection in the supernatant (supplemental Fig. S7E and S7F). We fitted a linear model to explain the log-normalized PAI values by the squared rpkm values from the RNA-Seq dataset. We obtained the following formula log(normalized PAI) = 0.1509 + 0.0093*rpkm2 where the p values for intercept (0.1509) and slope (0.0093) are 0.07 and 6.10−4, respectively (supplemental Fig. S7G). Therefore, except for the RipA and RipS family which correspond to very large proteins, no apparent differences could be attributed between detected and not-detected Rips in term of pI or MW, the threshold of Rip detection being very probably dependent on gene transcription, and thus to the amount of intracellular proteins produced.
Altered Secretion of Rip Proteins in the hpaB and hpaG Mutants is Associated with Impaired Virulence on M. truncatula
We then wanted to evaluate the impact of these modulations of Rip secretion on R. solanacearum pathogenicity on several host plants. We first validated that the hpaB, hpaD, and hpaG mutants were not affected in their growth in complete or minimal medium (data not shown). The three hpa mutant strains were leaf-infiltrated on N. tabacum, and root-inoculated on A. thaliana and M. truncatula. The hpaD mutant triggered an HR similar to the wild-type strain on tobacco plants, and virulence on A. thaliana and M. truncatula plants were undistinguishable from the wild-type strain (supplemental Fig. S8). The hpaB mutant was strongly affected in pathogenicity (Fig. 8). Indeed, the hpaB mutant is no more able to trigger an HR on tobacco, a phenotype complemented by the wild-type hpaB allele (Fig. 8A). The hpaB mutant was also unable to induce disease on A. thaliana (Figs. 8B and 9C), or on M. truncatula (Fig. 8D). Finally, HpaG was found to be dispensable for HR induction on tobacco (supplemental Fig. S8A) as well as for R. solanacearum pathogenicity on A. thaliana (Fig. 9A). However, disease development was delayed and significantly reduced on M. truncatula compared with the wild-type strain (Fig. 9B and 9C). M. truncatula in planta bacterial growth showed that HpaG is also required for bacterial multiplication, with a decrease of more than 100-fold for the hpaG mutant compared with the wild-type strain. Bacterial multiplication was restored in the hpaG-complemented strain (Fig. 9C).
Fig. 8.
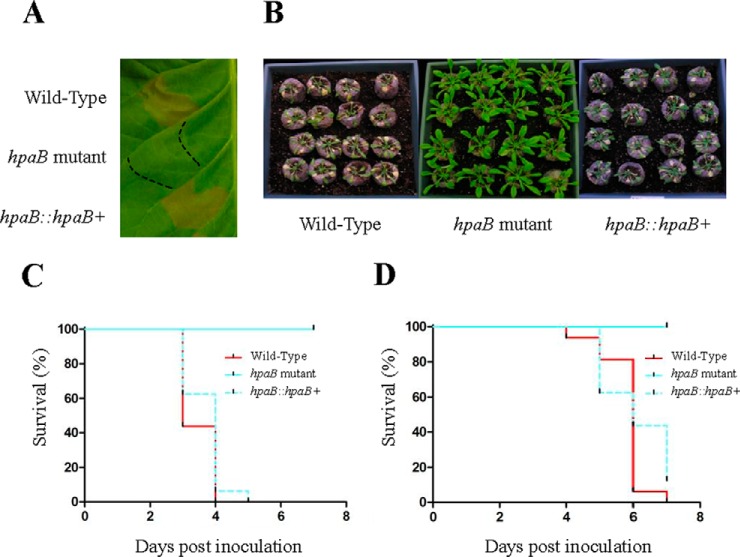
HpaB is required to induce HR on tobacco and disease on A. thaliana and M. truncatula. A, GMI1000 wild-type strain, hpaB mutant and complemented hpaB mutant inoculated on N. tabacum. HR is observed only with GMI1000 and with the complemented hpaB mutant 24 h after leaf infiltration. B, pictures of A. thaliana Col-0 plants 6 days post inoculation. C, Kaplan-Meier survival analysis of 16 A. thaliana inoculated plants. Gehan-Breslow-Wilcoxon test indicates that the wild-type strain curve (red) is significantly different from the hpaB mutant curve (cyan) (p value < 0.001), contrary to the complemented hpaB curve (dotted cyan) (p value = 0.8702). D, Kaplan-Meier survival analysis of 16 M. truncatula inoculated plants. Gehan-Breslow-Wilcoxon test indicates that the wild-type strain curve (red) is significantly different from the hpaB curve (cyan) (p value < 0.001), contrary to the complemented hpaB curve (dotted cyan) (p value = 0.5416). All these experiments were repeated at least three times.
Fig. 9.
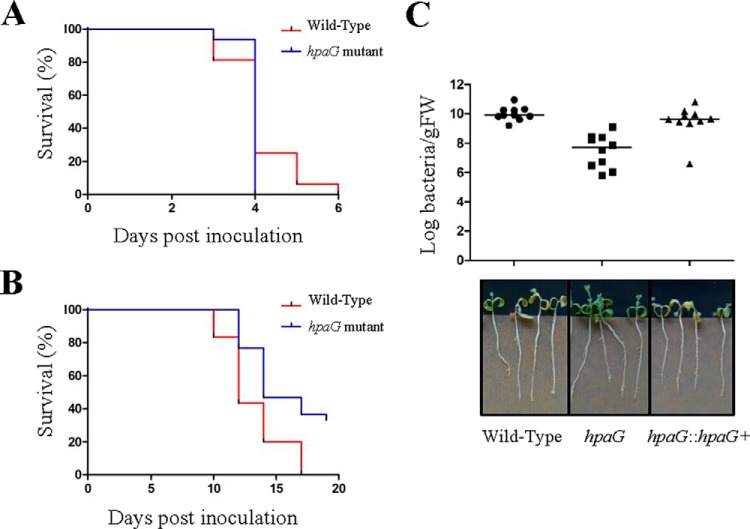
hpaG mutant is strongly reduced in virulence on M. truncatula. A, Kaplan-Meier survival analysis of 16 A. thaliana plants inoculated with R. solanacearum wild-type strain (red) and hpaG mutant (blue). Gehan-Breslow-Wilcoxon test indicates that the two curves are not significantly different (p value = 0.4845). B, Kaplan-Meier survival analysis of 16 M. truncatula plants inoculated with R. solanacearum wild-type strain (red) and hpaG mutant (blue). Gehan-Breslow-Wilcoxon test indicates that the two curves are significantly different (p value = 0.002). Three biological replicates were performed (A–B). C, measurement of in planta bacterial growth using in vitro M. truncatula plants, 14 days post inoculation. Each dot represents a replicate of three plants, the black line specifies the median of the ten replicates. Mann-Whitney-Wilcoxon tests were performed and indicate that the in planta bacterial multiplication of the hpaG mutant is different from the wild-type strain (p value < 0.001), but not the complemented hpaG mutant (p value = 0.1655). Representative pictures were taken 17 days post inoculation.
DISCUSSION
Phytopathogenic bacteria have evolved multiple strategies to infect plants. A major determinant of bacterial pathogenicity is the T3SS that delivers effector proteins directly into the host cells. Despite increasing knowledge on the control of the T3SS through transcriptional regulatory networks, only limited information on post-translational regulations is available. This study provides the first characterization of the T3-dependent secretome of R. solanacearum using the recent and advanced Q Exactive technology. To date, some T3-secretomes of animal pathogenic bacteria have been characterized (47–49), but only partial secretome and proteome analyses are available from plant pathogenic bacteria since these studies mainly used two-dimensional gel electrophoresis followed by MALDI-TOF-MS or nano-LC-MS/MS approaches (50–55). With a total of 228 proteins detected in the wild-type strain supernatant (supplemental Table S1), this study therefore represents the most exhaustive secretome inventory to date among bacterial plant pathogens. In addition, the analysis of hpa mutants revealed specific T3-secretome patterns, showing both the sensitivity of the methodology and its efficiency to decrypt T3-secretomes and to describe the impact of T3-post-translational regulators in the secretion process. Combining the data obtained for the four biological replicates of the five strains analyzed in this study, we could detect 320 secreted proteins. Among them, 66% were predicted to be secreted using SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/), SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP-4.0/), and MaGe (Magnifying Genomes Microbial Genome Annotation System) (http://www.genoscope.cns.fr/agc/microscope/mage/index.php). We also showed with Western blot experiments that the non-detection of some Rips using MS is not because of a non-secretion in our experimental conditions, but is because of the MS detection threshold. Detection of 60% of the R. solanacearum GMI1000 known effectors (43 out of 72 (11)) is satisfactory compared with other T3-secretome studies (51–55) and considering that several Rip effectors may certainly be low-abundant proteins produced at the host cell contact. We observed a direct correlation between rip gene transcription and the quantitative detection of the corresponding proteins through MS analysis. Quantitative MS data were also supported by Western blot experiments. Absence or residual spectral counting for Rips in the hrcV mutant was confirmed by the absence of detection of Rips in bacterial supernatants after immunoblot experiments whereas these proteins were well produced in the hrcV bacterial pellets. Contrary to all the other Rips tested, RipW was detected as slightly secreted by the hrcV mutant as shown by MS (Fig. 2C) or Western blot experiments (Fig. 7A). The contrasting secretion patterns of the hpaB and hpaG mutants, with specific proteins either less or more secreted than the wild-type, were also validated by specific immuno-detections. All these observations confirm the overall quality and sensitivity (reproducibility of detection of differentially secreted proteins) of the MS experimental process we used.
Our study identified a novel effector, RipBJ, which contains an N-terminal sequence homologous to RipAG and RipAH, but also to AvrRpm1 and AvrPto P. syringae effectors (supplemental Fig. S5D). This N-terminal sequence domain contains putative myristoylation/acylation signals which were shown to be required for the targeting of AvrRpm1 to the plasma membrane (56). However, no difference could be observed between the wild-type strain and the ripBJ mutant in pathogenicity assays, probably because of functional redundancy among Rip effectors (15–17). In a former study (23), we provided evidence of a post-translational control of T3E secretion in R. solanacearum by the HpaP protein. In this work, we combined proteomic and phenotypic analyses studying three additional hpa mutants, hpaB, hpaD, and hpaG. Our work showed that these hpa mutants have three distinct secretion patterns, with specific proteomic signatures highlighting Rip classes of T3S.
hpaB mutant secretome analysis showed the strongest Rip secretory phenotype. Indeed, secretion of almost all the Rips detected in the wild-type strain supernatant was partly or fully altered in the hpaB mutant. This global defect in Rip secretion is likely to be the cause of the nonpathogenicity phenotype of the hpaB mutant. This corroborates previous works on hpaB homologs in several phytopathogenic bacteria. In another strain of R. solanacearum, HpaB was shown to be required for the translocation of at least 66 T3Es (57, 58). Studies in Xanthomonas campestris pv. vesicatoria (Xcv) showed that HpaB had a central role in Xcv efficient T3S, notably interacting with various control proteins and T3Es (6, 22). HpaB homologs were shown to be required in bacterial pathogenicity but with different specificities according to the pathosystems studied (57, 59, 60). In our study, we show that the hpaB mutant is still capable of secreting HrpY, furthermore there is also efficient secretion of RipX and RipW harpins (45, 46), altogether suggesting that the Hrp pilus is functional in this mutant. Interestingly, RipAB appears to be the most abundantly secreted Rip, and is still efficiently secreted in the hpaB mutant. Additionally, the hierarchical clustering obtained taking into account the five strains clustered the HrpY pilin, the RipF1_1 and RipF1_2 translocators, the RipX and RipW harpins with RipAB and RipAM. The RipF1 paralogs are required for efficient Rip translocation (11), but a direct interaction between RipF1_1 and HrpY could not be identified (44). Even if RipAM has a lower secretion level compared with the other proteins of this group, similar pattern is observed. All together, these data led us to hypothesize that RipX, RipW, RipAB, and RipAM could be involved, in an Hpa protein-independent manner, in the first stages of Rip translocation process. This hypothesis is in accordance with a polymutant approach in Pst DC3000 which highlighted a consortium of redundant translocators required to promote Pst T3E injection into plant cells (61, 62). RipF1, RipW, RipX, RipAB, and RipAM all belong to the R. solanacearum core effectome (11). It will be of particular interest to study more precisely how they take part in the formation of the Hrp translocation apparatus as accessory cotranslocators, notably by looking for interactions between them and the HrpY pilin.
The hpaG mutant secretome analysis showed a deregulation of the hrp-secretion with an unexpected MS-over-secretion pattern, numerous Rips being more detected compared with the wild-type strain. Although still able to induce an HR on tobacco plants and the full disease on A. thaliana, the hpaG mutant is specifically impaired in its pathogenicity on M. truncatula. In addition, RipG7, which is strictly required for R. solanacearum pathogenicity on M. truncatula (17), is still efficiently secreted by the hpaG mutant. A study with another strain of R. solanacearum on a HpaG homolog protein (LrpE) showed a slight virulence reduction of the lrpE mutant on eggplant (43). Thus, we hypothesize that these host phenotypes depend more on altered secretion of some effectors directly involved on certain hosts to confer disease. One or several Rips could be directly involved in this phenotype on the model legume plant, letting us hypothesize a potential HpaG function as a secretion suppressor of Rips involved in the induction of plant immunity. We also observed that over-secretion of Rips by the hpaG mutant can also be associated with specific over-production of Rips, suggesting a role for HpaG in transcriptional control or on protein stabilization for a subset of Rips.
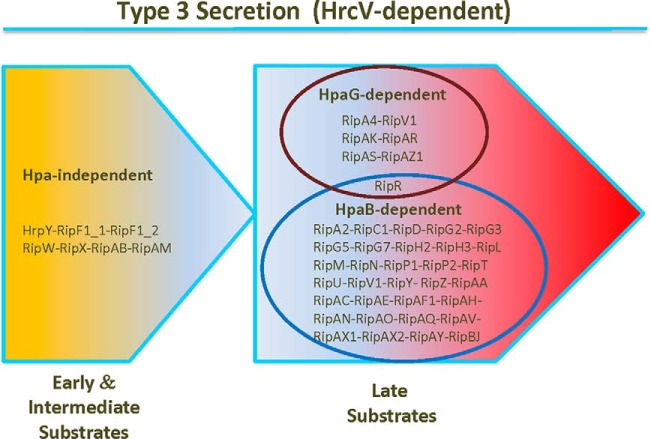
This article constitutes another piece of the puzzle of T3S post-translational control, and even if many questions remain, this first R. solanacearum secretome study supports the existence of a T3S hierarchy with specific subsets of effectors having differential secretion patterns (Fig. 10). The study of three hpa mutants revealed previously unknown complex regulations of T3S, and Rips with similar regulation profiles. This work also showed that HpaB and HpaG proteins are post-translational regulators of T3S, with opposite roles on T3S. Indeed, although there is no evidence for a role of HpaD in T3S post-translational control, HpaB clearly promotes secretion of numerous Rips and acts as a positive regulator of T3S, whereas HpaG seems to negatively regulate secretion of other Rips. A recent review on orchestration of T3S from animal and plant pathogenic bacteria classified the pilus as an “early” T3S substrate, translocon proteins as “intermediate” substrates and effector proteins as “late” substrates (6). Here, we identified a first set of Rips whose secretion is Hpa-independent which we propose to qualify as putative “early/intermediate” substrates as they include secreted syringe components HrpY and RipF1 proteins. We highlighted a second set of Rips whose secretion is Hpa-dependent, which we hypothesize as putative “late” substrates, with either a strict, or a quantitative Hpa-secretion control. This also suggests a sequential secretion in R. solanacearum, between “early/intermediate” and “late” substrates, helping to define new subsets of Rips under secretion process (Fig. 10). This work illustrates how the Rip secretion is finely tuned and how this control can impact host specificity. We showed that we were able to monitor secretion of more than 40 Rips per assay. A polymutant approach could help to identify T3Es collectively involved on different host plants.
Fig. 10.
Involvement of Hpa proteins in Rip secretion. This illustration proposes subsets of Rips for which T3S is coregulated, in an Hpa-independent or Hpa-dependent way. The model is based on the T3-specific secretion patterns of the GMI1000 wild-type strain and of the hpa mutants. Data obtained from the statistical analysis on MS assays, from the following hierarchical clustering, and from the Western blot validation experiments were combined. The two groups represent “early/intermediate” and “late” substrates of T3SS, respectively. Among the late substrates, the blue circle contains the Rips whose secretion is positively regulated by HpaB, whereas the red circle contains the Rips whose secretion is negatively regulated by HpaG.
Supplementary Material
Acknowledgments
We thank Jean Luc Pariente and Claudette Icher for tobacco plant preparation. We thank Elise Gay for technical assistance for construction of strains with effector-HA tag, and Lenaïck Belliot for LBy5 and LBy14 plasmids. We thank Alice Guidot and Laure Plener for generously providing transcription data of Rip genes. We thank Laurent Deslandes for the gift of the PopP2 antibody. We thank Anne Aubert Frambourg and Michel Zivy for fruitful discussions.
Footnotes
Author contributions: F.L., M.T., C.H. and F.V. Designed Research; F.L., M.T., C.H., D.L., Q.K. and F.V Performed Research; F.L., M.T., C.H., D.R., A.C.C., N.P., S.G. and F.V. Contributed new reagents or analytic tools; F.L., C.H., D.R. and F.V. Analyzed data; F.L., C.H., D.R., N.P., S.G. and F.V. Wrote or contributed to the writing of the paper.
* This work was supported by a French Agence Nationale de la Recherche grant (ANR-2010-JCJC-1710-01) to F.V. This work benefited from interactions promoted by COST Action FA 1208 https://www.cost-sustain.org. Our work is performed at the Laboratoire des Interactions Plantes-Microorganismes that is part of the Laboratoire d'Excellence (LABEX) entitled TULIP (ANR-10-LABX-41). F.L and D.L. were funded by a grant from the French Ministry of National Education and Research.
 This article contains supplemental Procedures, Figs. S1 to S8, and Tables S1 to S4.
This article contains supplemental Procedures, Figs. S1 to S8, and Tables S1 to S4.
1 The abbreviations used are:
- T3SS
- Type 3 secretion system
- cAMP
- cyclic adenosine monophosphate
- glht
- general linear hypothesis test
- hrc
- hypersensitive response and pathogenicity conserved
- hrp
- hypersensitive response and pathogenicity
- hpa
- hypersensitive response and pathogenicity associated
- HA
- human influenza hemagglutinin
- kDa
- kilo dalton
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- m/z
- mass-to-charge ratio
- MS
- mass spectra from precursor ion
- MS2
- fragmentation mass spectra
- MW
- molecular weight
- OD
- optical density
- PAI
- protein abundance index
- PCA
- principal component analysis
- ppm
- part per million
- Rip
- Ralstonia injected protein
- rpkm
- reads per kilobase per million mapped reads
- rpm
- rotation per minute
- SC
- spectral counting
- T3
- type 3
- T3C
- type 3 chaperone
- T3E
- type 3 effector
- T3S
- type 3 secretion
- T3S4
- type 3 secretion substrate switch specificity
- TCA
- trichloroacetic acid
- Th
- thomson unit
- v/v
- volume to volume ratio.
REFERENCES
- 1.Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., Dow M., Verdier V., Beer S. V., Machado M. A., Toth I., Salmond G., and Foster G. D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayward A. C. (1991) Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87 [DOI] [PubMed] [Google Scholar]
- 3.Genin S. (2010) Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187, 920–928 [DOI] [PubMed] [Google Scholar]
- 4.Tampakaki A. P., Skandalis N., Gazi A. D., Bastaki M. N., Sarris P. F., Charova S. N., Kokkinidis M., and Panopoulos N. J. (2010) Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370 [DOI] [PubMed] [Google Scholar]
- 5.Büttner D., and He S. Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Büttner D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petnicki-Ocwieja T., Schneider D. J., Tam V. C., Chancey S. T., Shan L., Jamir Y., Schechter L. M., Janes M. D., Buell C. R., Tang X., Collmer A., and Alfano J. R. (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 99, 7652–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., Billault A., Brottier P., Camus J. C., Cattolico L., Chandler M., Choisne N., Claudel-Renard C., Cunnac S., Demange N., Gaspin C., Lavie M., Moisan A., Robert C., Saurin W., Schiex T., Siguier P., Thebault P., Whalen M., Wincker P., Levy M., Weissenbach J., and Boucher C. A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502 [DOI] [PubMed] [Google Scholar]
- 9.Oh C.-S., and Beer S. V. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192 [DOI] [PubMed] [Google Scholar]
- 10.Thieme F., Koebnik R., Bekel T., Berger C., Boch J., Büttner D., Caldana C., Gaigalat L., Goesmann A., Kay S., Kirchner O., Lanz C., Linke B., McHardy A. C., Meyer F., Mittenhuber G., Nies D. H., Niesbach-Klösgen U., Patschkowski T., Rückert C., Rupp O., Schneiker S., Schuster S. C., Vorhölter F.-J., Weber E., Pühler A., Bonas U., Bartels D., and Kaiser O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peeters N., Carrère S., Anisimova M., Plener L., Cazalé A.-C., and Genin S. (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunnac S., Boucher C., and Genin S. (2004) Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186, 2309–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genin S., and Denny T. P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 [DOI] [PubMed] [Google Scholar]
- 14.Remigi P., Anisimova M., Guidot A., Genin S., and Peeters N. (2011) Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solé M., Popa C., Mith O., Sohn K. H., Jones J. D. G., Deslandes L., and Valls M. (2012) The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant-Microbe Interact. 25, 941–953 [DOI] [PubMed] [Google Scholar]
- 16.Chen L., Shirota M., Zhang Y., Kiba A., Hikichi Y., and Ohnishi K. (2013) Involvement of HLK effectors in Ralstonia solanacearum disease development in tomato. J. Gen. Plant Pathol. 80, 79–84 [Google Scholar]
- 17.Angot A., Peeters N., Lechner E., Vailleau F., Baud C., Gentzbittel L., Sartorel E., Genschik P., Boucher C., and Genin S. (2006) Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103, 14620–14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guéneron M., Timmers A. C., Boucher C., and Arlat M. (2000) Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the hrp-secretion apparatus of Ralstonia solanacearum. Mol. Microbiol. 36, 261–277 [DOI] [PubMed] [Google Scholar]
- 19.Feldman M. F., and Cornelis G. R. (2003) The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219, 151–158 [DOI] [PubMed] [Google Scholar]
- 20.Parsot C., Hamiaux C., and Page A. L. (2003) The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14 [DOI] [PubMed] [Google Scholar]
- 21.Büttner D., and Bonas U. (2006) Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr. Opin. Microbiol. 9, 193–200 [DOI] [PubMed] [Google Scholar]
- 22.Lohou D., Lonjon F., Genin S., and Vailleau F. (2013) Type III chaperones & Co in bacterial plant pathogens: a set of specialized bodyguards mediating effector delivery. Front. Plant Sci. 4, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohou D., Turner M., Lonjon F., Cazalé A.-C., Peeters N., Genin S., and Vailleau F. (2014) HpaP modulates type III effector secretion in Ralstonia solanacearum and harbors a substrate specificity switch domain essential for virulence. Mol. Plant Pathol. 15, 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirmoradian M., Budamgunta H., Chingin K., Zhang B., Astorga-Wells J., and Zubarev R. A. (2013) Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol. Cell. Proteomics 12, 3330–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poueymiro M., Cunnac S., Barberis P., Deslandes L., Peeters N., Cazale-Noel A.-C., Boucher C., and Genin S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant-Microbe Interact. 22, 538–550 [DOI] [PubMed] [Google Scholar]
- 26.Prentki P., and Krisch H. M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313 [DOI] [PubMed] [Google Scholar]
- 27.González A., Plener L., Restrepo S., Boucher C., and Genin S. (2011) Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ. Microbiol. 13, 3172–3185 [DOI] [PubMed] [Google Scholar]
- 28.Washburn M. P., Wolters D., and Yates J. R. (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 29.Sadygov R. G., Liu H., and Yates J. R. (2004) Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal. Chem. 76, 1664–1671 [DOI] [PubMed] [Google Scholar]
- 30.Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J. C., Hallett M., Paiement J., Kearney R. E., Nilsson T., and Bergeron J. J. M. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 31.Rappsilber J., and Mann M. (2002) What does it mean to identify a protein in proteomics? Trends Biochem. Sci. 27, 74–78 [DOI] [PubMed] [Google Scholar]
- 32.Hothorn T., Bretz F., and Westfall P. (2008) Simultaneous inference in general parametric models. Biom. J. Biom. Z. 50, 346–363 [DOI] [PubMed] [Google Scholar]
- 33.R Core Team (2014) R: A language and environment for statistical computing.R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 34.Smyth G. K. (2005) in Bioinformatics and computational biology solutions using R and Bioconductor Springer, NY, pp 397–420 [Google Scholar]
- 35.Wilkinson L. (2012) Exact and approximate area-proportional circular Venn and Euler diagrams. IEEE Trans. Vis. Comput. Graph. 18, 321–331 [DOI] [PubMed] [Google Scholar]
- 36.Lavie M., Shillington E., Eguiluz C., Grimsley N., and Boucher C. (2002) PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant. Microbe Interact. 15, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 37.Cunnac S., Occhialini A., Barberis P., Boucher C., and Genin S. (2004) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128 [DOI] [PubMed] [Google Scholar]
- 38.Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., Somssich I., Genin S., and Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vailleau F., Sartorel E., Jardinaud M.-F., Chardon F., Genin S., Huguet T., Gentzbittel L., and Petitprez M. (2007) Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula. Mol. Plant-Microbe Interact. 20, 159–167 [DOI] [PubMed] [Google Scholar]
- 40.Sory M. P., and Cornelis G. R. (1994) Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14, 583–594 [DOI] [PubMed] [Google Scholar]
- 41.Occhialini A., Cunnac S., Reymond N., Genin S., and Boucher C. (2005) Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18, 938–949 [DOI] [PubMed] [Google Scholar]
- 42.Thomas N. A., Ma I., Prasad M. E., and Rafuse C. (2012) Expanded roles for multicargo and class 1B effector chaperones in type III secretion. J. Bacteriol. 194, 3767–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murata Y., Tamura N., Nakaho K., and Mukaihara T. (2006) Mutations in the lrpE gene of Ralstonia solanacearum affects Hrp pili production and virulence. Mol. Plant-Microbe Interact. 19, 884–895 [DOI] [PubMed] [Google Scholar]
- 44.Meyer D., Cunnac S., Guéneron M., Declercq C., Van Gijsegem F., Lauber E., Boucher C., and Arlat M. (2006) PopF1 and PopF2, two proteins secreted by the type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 188, 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arlat M., Van Gijsegem F., Huet J. C., Pernollet J. C., and Boucher C. A. (1994) PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J.-G., Liu H.-X., Cao J., Chen L.-F., Gu C., Allen C., and Guo J.-H. (2010) PopW of Ralstonia solanacearum, a new two-domain harpin targeting the plant cell wall. Mol. Plant Pathol. 11, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng W., de Hoog C. L., Yu H. B., Li Y., Croxen M. A., Thomas N. A., Puente J. L., Foster L. J., and Finlay B. B. (2010) A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J. Biol. Chem. 285, 6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niemann G. S., Brown R. N., Gustin J. K., Stufkens A., Shaikh-Kidwai A. S., Li J., McDermott J. E., Brewer H. M., Schepmoes A., Smith R. D., Adkins J. N., and Heffron F. (2011) Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng W., Yu H. B., de Hoog C. L., Stoynov N., Li Y., Foster L. J., and Finlay B. B. (2012) Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11, 692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazemi-Pour N., Condemine G., and Hugouvieux-Cotte-Pattat N. (2004) The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 4, 3177–3186 [DOI] [PubMed] [Google Scholar]
- 51.Nissinen R. M., Ytterberg A. J., Bogdanove A. J., VAN Wijk K. J., and Beer S. V. (2007) Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8, 55–67 [DOI] [PubMed] [Google Scholar]
- 52.Robin G. P., Ortiz E., Szurek B., Brizard J.-P., and Koebnik R. (2014) Comparative proteomics reveal new HrpX-regulated proteins of Xanthomonas oryzae pv. oryzae. J. Proteomics 97, 256–264 [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., Kim S. G., Wu J., Huh H.-H., Lee S.-J., Rakwal R., Agrawal G. K., Park Z.-Y., Young Kang K., and Kim S. T. (2013) Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics 13, 1901–1912 [DOI] [PubMed] [Google Scholar]
- 54.Haapalainen M., Mosorin H., Dorati F., Wu R.-F., Roine E., Taira S., Nissinen R., Mattinen L., Jackson R., Pirhonen M., and Lin N.-C. (2012) Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194, 4810–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schumacher J., Waite C. J., Bennett M. H., Perez M. F., Shethi K., and Buck M. (2014) Differential secretome analysis of Pseudomonas syringae pv tomato using gel-free MS proteomics. Front. Plant Sci. 5, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nimchuk Z., Marois E., Kjemtrup S., Leister R. T., Katagiri F., and Dangl J. L. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363 [DOI] [PubMed] [Google Scholar]
- 57.Mukaihara T., Tamura N., Murata Y., and Iwabuchi M. (2004) Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54, 863–875 [DOI] [PubMed] [Google Scholar]
- 58.Mukaihara T., Tamura N., and Iwabuchi M. (2010) Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant-Microbe Interact. 23, 251–262 [DOI] [PubMed] [Google Scholar]
- 59.Kim J.-G., Park B. K., Yoo C.-H., Jeon E., Oh J., and Hwang I. (2003) Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Büttner D., Gürlebeck D., Noël L. D., and Bonas U. (2004) HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54, 755–768 [DOI] [PubMed] [Google Scholar]
- 61.Kvitko B. H., Ramos A. R., Morello J. E., Oh H.-S., and Collmer A. (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189, 8059–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi M.-S., Kim W., Lee C., and Oh C.-S. (2013) Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol. Plant-Microbe Interact. 26, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 63.Monteiro F., Solé M., van Dijk I., and Valls M. (2012) A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum. Mol. Plant-Microbe Interact. 25, 557–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.