SUMMARY
A precise regulation of flowering time is central to plant species survival. Therefore, mechanisms have evolved in plants to integrate different environmental cues to optimize flowering time. In this study we show that the wheat gene TaFT, which integrates photoperiod and vernalization signals promoting flowering, interacts with bZIP proteins TaFDL2 and TaFDL6. We also show that TaFDL2 can interact in vitro with five ACGT elements in the promoter of the meristem identity gene VRN1, suggesting that TaFDL2 is a functional homologue of Arabidopsis FD. No direct interactions between the TaFT protein and the VRN1 promoter were detected. Transgenic wheat plants overexpressing TaFT showed parallel increases in VRN1 transcripts, suggesting that TaFT provides transcriptional activation to VRN1, possibly through interactions with the TaFDL2 protein. The same transgenic plants also showed increased transcript levels of TaFT2 (a TaFT paralogue) indicating that TaFT2 acts downstream of TaFT. The fact that TaFT2 interacts with the different bZIP protein TaFDL13, which lacks the ability to interact with the VRN1 promoter, suggests that TaFT and TaFT2 have different gene targets. This observation agrees with the functional divergence observed for the TaFT and TaFT2 orthologous genes in rice. The temperate cereals analyzed so far show VRN1 transcripts in the leaves, a characteristic not observed in Arabidopsis or rice. The high levels of TaFDL2 transcripts observed in wheat leaves provide a simple explanation for this difference. We present a hypothesis to explain the conservation of VRN1 expression in the leaves of temperate cereals.
Keywords: wheat, flowering, FT, FD, VRN1, AP1
INTRODUCTION
The precise control of flowering is central to plant reproductive success and species survival. Optimization of flowering time to maximize grain yield is also an important target in cereal breeding programs. Plants have evolved mechanisms to integrate different environmental signals, including photoperiod and vernalization, to flower under conditions that optimize seed production. These different environmental signals are perceived by separate parts of the plant and their integration requires a precise spatial and temporal coordination. Photoperiod, for example, is perceived by the leaves, whereas, cold temperature is perceived directly by the shoot apical meristem (SAM) (Bernier, 1988).
The photoperiod signal perceived in the leaves is transmitted to the SAM to induce flowering, as demonstrated by grafting experiment (reviewed in Zeevaart, 1976). This transmissible substance named “florigen” (Chailakhyan, 1937) was not identified until recently (Corbesier and Coupland, 2006). Studies in Arabidopsis and rice have now demonstrated that this flowering signal is the FLOWERING LOCUS T (FT) (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin, et al., 2007; Mathieu, et al., 2007; Tamaki et al., 2007), a small globular protein similar to the RAF kinase inhibitors found in animals (Kardailsky et al., 1999; Kobayashi et al., 1999). Over-expression of FT is associated with early flowering in a wide range of plant species suggesting a highly conserved function (Böhlenius et al., 2006; Kardailsky et al., 1999; Kobayashi et al., 1999; Kojima et al., 2002; Lifschitz et al., 2006; Lin et al., 2007; Yan et al., 2006).
FT is the primary target of CONSTANS (CO), a B-box zinc finger CCT protein that plays a central role in the photoperiod pathway (Putterill et al., 1995; Robson et al., 2001; Wigge et al., 2005). When Arabidopsis plants are exposed to long days (LD), CO activates FT in the leaf vascular tissue (phloem) (An et al., 2004). The FT protein moves then through the phloem to the SAM, where it interacts with the bZIP transcription factor FD to activate the expression of the MADS-box meristem identity gene APETALA 1 (AP1) (Abe et al., 2005; Corbesier et al., 2007; Wigge et al., 2005). The FT-FD protein interaction is spatially regulated by the preferential expression of FD in the SAM, and temporally by the integration of different environmental signals that converge to regulate FT (Wigge et al., 2005). According to this model, FD provides specificity in the recognition of the DNA target and FT acts in concert with FD to transcriptionally activate AP1 (Wigge et al., 2005).
In the temperate cereals, the role of the FT homologues seems to be similar to the one described above for Arabidopsis. The over-expression of TaFT (Ta indicates Triticum aestivum L.) in transgenic wheat plants significantly accelerates flowering relative to the non-transgenic controls, suggesting a conserved role as flowering promoter (Yan et al., 2006). In addition, both TaFT and HvFT (Hv indicates Hordeum vulgare L.) are up-regulated by long days, and increased transcript levels correlate with accelerated flowering times (Turner et al., 2005; Yan et al., 2006). Arabidopsis and the temperate cereals differ in the spatial transcription profile of the main target of FT, AP1 in Arabidopsis and the homologous VRN1 in temperate cereals. In wheat and barley VRN1 is normally expressed in the leaves at high levels (Schmitz et al., 2000; Yan et al., 2003) whereas in Arabidopsis, AP1 transcripts are either not detected in the leaves or present at very low levels (e.g. in the vascular tissues of cotyledons, Abe et al., 2005). Interestingly, the ectopic expression of FD in 35S::FD transgenic Arabidopsis plants results in high expression of AP1 in the leaves (Wigge et al., 2005). Based on this result we hypothesized that a wheat functional homologue of Arabidopsis FD, designated as TaFD-like (TaFDL, hereafter), should be normally expressed in leaves.
The understanding of the role of TaFT in the regulation of flowering in the temperate cereals is complicated by the existence of a similar (78% identical) paralogous copy in both wheat (TaFT2) and barley (HvFT2) (Faure et al., 2007; Yan et al., 2006). Since the TaFT-TaFT2 duplication occurred after the divergence between the grasses and Arabidopsis, this duplication is independent of the FT-TSF (Twin Sister of FT) duplication in Arabidopsis.
In this study we provide evidence that TaFT2 is regulated by TaFT and that these two genes interact with different TaFDL partners. We also show that only one of the TaFDL proteins that interact with TaFT was able to interact with the VRN1 promoter. The expression of this particular TaFDL gene in the leaves provides a simple explanation for the presence of VRN1 transcripts in the leaves of temperate cereals. Finally, we discuss a hypothesis for the putative role of VRN1 expression in the leaves of temperate cereals.
RESULTS
FD-like genes are transcribed in wheat leaves
Using cDNA samples from leaves of hexaploid wheat Chinese Spring at the 5th-leaf stage and primer pairs for 16 TaFDL genes (Table S1) we were able to amplify five different FDL genes. The cDNA sequences of these genes and their predicted protein sequences were deposited in GenBank: TaFDL2 (EU307112), TaFDL3 (EU307113), TaFDL6 (EU307114), TaFDL13 (EU307115) and TaFDL15 (EU307116). This result confirmed that at least some TaFDL genes are transcribed in the leaves.
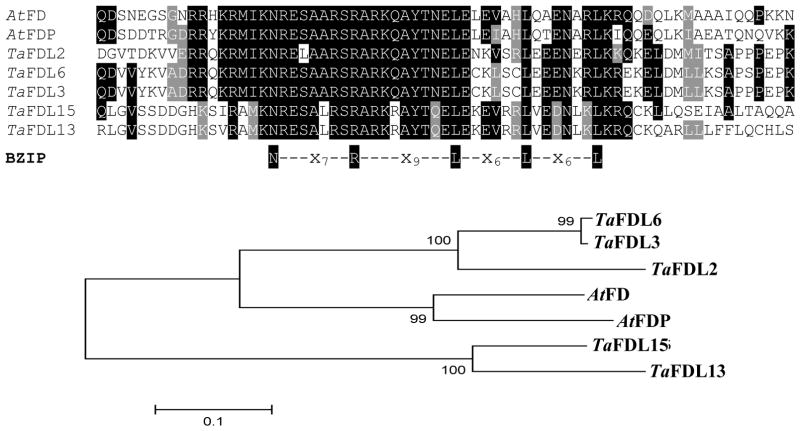
A multiple sequence alignment of the five predicted wheat FD-like proteins and the Arabidopsis proteins FD and FDP is presented in Fig. 1. The alignment shows a conserved region of 69 amino acids without gaps including perfectly conserved bZIP domains (N-x7-R-x9-L-x6-L-x6-L). The regions outside this 69 amino acids region were not well conserved among the different proteins and were excluded for the tree construction (Fig. 1).
Figure 1.
Comparison among wheat and Arabidopsis proteins using MEGA4 (Tamura et al. 2007). Top: ClustalW multiple sequence alignment of Arabidopsis FD and FDP with five wheat FD-like protein sequences based on the bZIP domain and conserved un-gapped flanking regions. Sequences used in the alignment: AtFD (At4g35900), AtFDP (At2g17770), TaFD-like protein sequences TaFDL2 (EU307112), TaFDL3 (EU307113), TaFDL6 (EU307114), TaFDL13 (EU307115) and TaFDL15 (EU307116). Bottom: Neighbor joining tree based on the previous alignments. Numbers in the nodes indicate bootstrap confidence values based on 1000 iterations.
The TaFT protein interacts with the bZIP proteins TaFDL2 and TaFDL6
Yeast two-hybrid assays were used to test the interactions between TaFT and the different TaFDL proteins. We first performed auto-activation tests and confirmed that TaFT was not able to activate the reporter genes when used alone as bait or prey. The TaFT bait construct was then co-transformed into yeast with individual prey constructs for TaFDL2, TaFDL3, TaFDL6, TaFDL13, and TaFDL15.
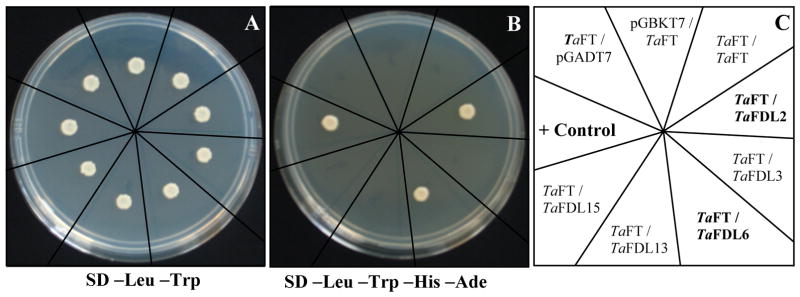
The two proteins used as positive controls, Murine p53 and SV40 large T-antigen (Iwabuchi et al., 1993; Li and Fields, 1993), showed the expected interaction on the SD –Leu –Trp –His –Ade medium (Figure 2B). Under the same selective conditions, TaFDL2 and TaFDL6 also showed very strong interactions with the TaFT bait (Figure 2B). A TaFDL6 truncation construct carrying only the bZIP domain showed a weaker interaction with the TaFT bait than the full-length protein (data not shown). These interactions were validated using reverse bait / prey constructs. When used as bait, both TaFDL2 and TaFDL6 showed strong interactions with co-transformed TaFT prey construct (data not shown).
Figure 2.
Yeast two-hybrid assays using TaFT bait. A) Yeast cells plated on SD-LEU-TRP; B) Equal amount of yeast cells as in A, but plated on SD-LEU-TRP-HIS-ADE; C) Diagram indicating the different bait / prey combinations used in the assay (baits are indicated first in each pair): Positive control: Murine P53 / SV40 T-antigen.
TaFT2 protein interacts with TaFDL13
We also investigated the interactions between TaFDL proteins with TaFT2. Auto-activation tests confirmed that TaFT2 was not able to activate the reporter genes when used alone as bait or prey or in combination with TaFT (Fig. 3).
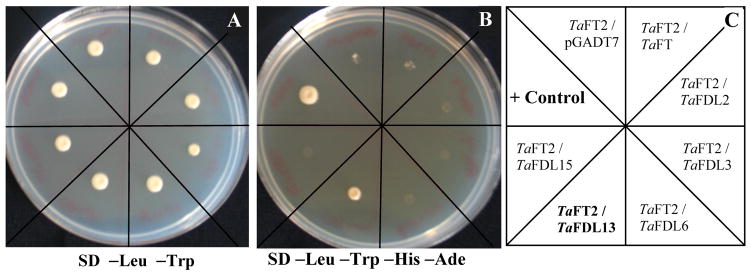
Figure 3.
Yeast two-hybrid assays using TaFT2 as bait. A) Yeast cells plated on SD-LEU-TRP; B) Equal amount of yeast cells as in A, but plated on SD-LEU-TRP-HIS-ADE; C) Diagram indicating the different bait / prey combinations used in the assay (baits are indicated first in each pair): Positive control: Murine P53 / SV40 T-antigen.
Interestingly, co-transformation of TaFT2 bait with TaFDL2, TaFDL3, TaFDL6, TaFDL13, and TaFDL15 prey constructs showed different results from those observed with TaFT. Whereas TaFT showed strong interactions with TaFDL2 and TaFDL6, TaFT2 interacted only with TaFDL13 among these five FD-like proteins (Fig. 3). This interaction was also validated using the reverse bait / prey combination, in which TaFDL13 was used as a bait construct and TaFT2 as a prey (data not shown).
TaFDL2 protein binds to bZip binding sites in the VRN1 promoter in vitro
Members of the bZIP transcription factor family exhibit DNA binding specificity to DNA motifs with an ACGT core sequence including the A box (TACGTA), G box (CACGTG), C box (GACGTC) (Foster et al., 1994; Izawa et al., 1993), and hybrid C-box/G-box motifs (Martinez-Garcia et al., 1998). In Arabidopsis, a C-box present in the AP1 promoter has been suggested as the binding site for the interaction between the FT-FD protein complex and the AP1 promoter (Wigge et al., 2005). Since this is a critical step in the initiation of the flowering cascade, we investigated if the same interaction can occur in wheat.
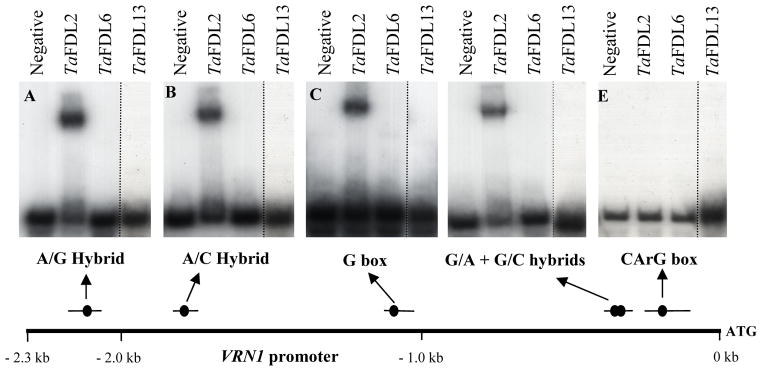
The VRN1 (the AP1 homologue) promoter region in T. monococcum most likely does not extend beyond 2.3 kb upstream from its start codon, since an un-interrupted 67-kb stretch of nested retroelements was found upstream from this region (AY188331). Within this 2.3 kb region we found a G box and four putative hybrid boxes (Fig. 4).
Figure 4.
Electrophoresis Mobility Shift Assays (EMSA). Potential bZIP binding sites present in the VRN1 promoter were radiolabeled and used as probes in the binding reactions with purified recombinant proteins TaFDL2, TaFDL6, and TaFDL13. The first lane in each panel labeled as ‘Negative’ contains the probe alone. TaFDL13 was tested in a separate experiment. The four probes included the following potential bZIP binding sites: A) A/G hybrid of A- and G-boxes (TACGTG), B) A/C hybrid of A and C-boxes (TACGTC), C) G box (CACGTG), D) G/A hybrid of G and A-boxes (CACGTA) plus a G/C hybrid of G and C-boxes (CACGTC), E) CArG box. The bottom scheme indicates the position and size of the different probes and potential bZIP binding sites.
Four segments of the VRN1 promoter, each including one or two of the five putative binding sites (presented at scale in Fig. 4) were radiolabeled as DNA probes. The purified bZIP proteins TaFDL2, TaFDL6 and TaFDL13, which showed interactions with either TaFT or TaFT2 in the yeast two-hybrid assays, were used with the radiolabeled DNA probes in the EMSA experiments. Among these three bZIP proteins, only the TaFDL2 protein was able to bind to the putative bZIP binding sites in the TaVRN1 promoter, and none of them was able to bind to the CArG box located approximately 160 bp upstream from the VRN1 start codon (Fig. 4). To determine the binding specificity of TaFDL2 oligonucleotides including either four copies of the wild type bZIP binding sequence or a mutant version in which the ACGT core sequences were replaced by AATT (Table S3) were tested in binding assays and competition experiments. As shown in Fig. S1A, TaFLD2 binds specifically to the wild-type bZIP sequence in a concentration dependent manner, and is out competed by an excess of un-labeled cold oligonucleotides. Furthermore, the mutation of ACGT to AATT in the bZIP binding sequence completely abolishes the interactions (Fig. S1B).
In a separate EMSA experiment, five overlapping segments covering the complete (2.3-kb) VRN1 promoter region were used as probes in binding reactions with TaFT protein. No interactions were observed (data not shown).
Spatial and temporal localization of TaFDL2 transcripts
Our yeast two-hybrid assays and EMSA experiments confirmed that the wheat protein TaFDL2 interacts with TaFT and with the VRN1 promoter in a similar way as described for the homologous FT and FD proteins in Arabidopsis. Therefore, to understand the spatial and temporal regulation of VRN1 in wheat it is important to know the spatial and temporal transcription profiles of TaFDL2. For comparative purposes we also tested TaFDL6 and TaFDL13 transcript levels.
Abundant TaFDL2 transcripts were detected by RT-PCR in the RNA samples extracted from leaves of vernalized and unvernalized plants of common winter wheat variety Jagger (Fig. 5). Two RNA samples from vegetative and early reproductive SAM (including a small portion of the crown) extracted from Chinese Spring plants, also showed abundant TaFDL2 transcripts. RT-PCR analyses for the other TaFDL genes showed that TaFDL6 transcript levels were even more abundant than TaFDL2 in all these tissues; whereas TaFDL13 transcripts were more abundant in the apices than in the leaves (Fig. 5).
Figure 5.
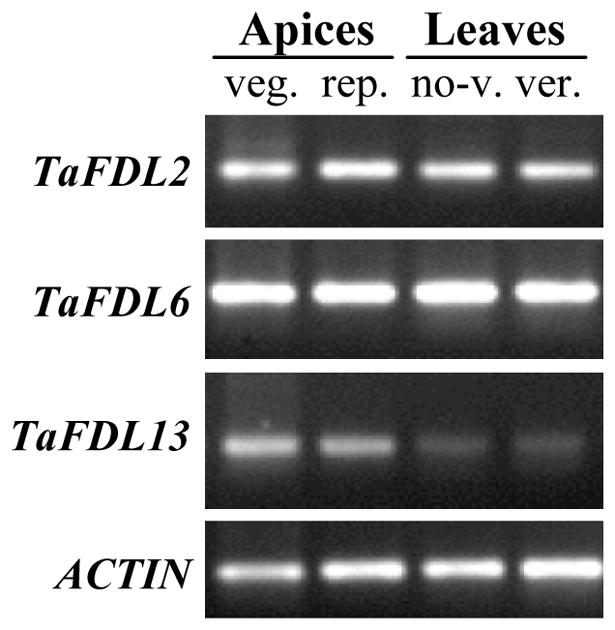
Transcript levels of TaFDL2, TaFDL6, and TaFDL13 in apices (spring wheat Chinese Spring) and leaves (winter wheat Jagger). ACTIN is included as endogenous control. Abbreviations: veg.= vegetative apex, rep.= reproductive apex, no-v= non vernalized leaves, ver.= vernalized leaves.
A quantitative RT-PCR analysis of cDNA samples from leaves further confirmed the high transcript levels of TaFDL2. We used available cDNA samples from spring wheat lines Chinese Spring (CS) and CS (Hope 7B) chromosome substitution line (dominant TaFT allele), previously characterized for TaFT and VRN1 (Yan et al., 2006). In all the samples, we found high levels of TaFDL2 transcripts (close to those of ACTIN). We detected no significant differences in TaFDL2 transcript levels between the two lines, or during the six weeks of vernalization at 4°C (Fig. S2). Plants kept at room temperature showed an approximately 2-fold increase in TaFDL2 transcript levels during the six weeks, but the differences were not significant (Fig. S2). In our previous study, these same samples showed significant increases in TaFT and VRN1 transcript levels with development (in both vernalized and unvernalized plants), and higher transcript levels in CS (Hope 7B) than in CS (Yan et al., 2006)
Transcript levels of TaFT and VRN1 are correlated
We have shown that TaFDL2 transcripts are abundant in both leaves and apices, and do not seem to be a limiting factor in the induction of VRN1. Therefore, if the interactions described above between TaFDL2, TaFT and the VRN1 promoter also occur in planta, modifications of the transcription levels of TaFT should be paralleled by the VRN1 transcript levels. To test this prediction we used two independent transgenic hexaploid wheat lines Tr-1 and Tr-2, generated in a previous study in the winter variety Jagger (Yan, et al. 2006). These transgenic plants exhibit increased levels of TaFT as a result of the insertion of a dominant TaFT allele for spring growth habit from the variety Hope (Yan, et al. 2006).
TaFT transcript levels in the leaves of unvernalized plants from both transgenic lines (LD photoperiod) were significantly higher than in the non-transgenic sister-lines or Jagger, at the two developmental stages tested (Figure 6). Transcript levels at the 5th leaf stage were approximately 5-fold higher than at the 1st leaf stage and more than 8-fold higher than ACTIN. The VRN1 transcript levels paralleled the TaFT ones, with significantly higher VRN1 transcript levels in transgenic plants than in the non-transgenic controls (Figure 6), and higher transcript levels at the 5th leaf-stage than at the 1st leaf stage. In this experiment, non-transgenic control plants took approximately 124 days from sowing to heading, whereas Tr-1 and Tr-2 plants headed on average 46 and 44 days after sowing, approximately 80 days earlier than the non-transgenic control.
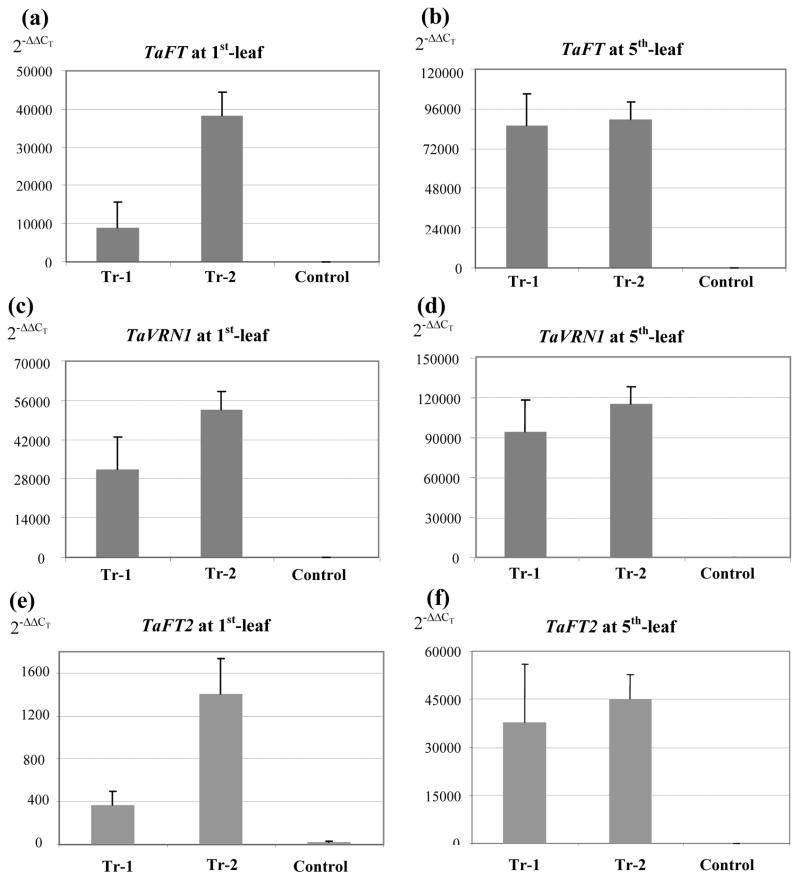
Figure 6.
Q-PCR of TaFT, VRN1 and TaFT2 in transgenic plants overexpressing TaFT. Tr-1 (average of 6 plants) and Tr-2 (average of 13 plants) are two independent transgenic lines. The controls include 9 non-transgenic sister plants and 10 Jagger plants. Plants were grown at 20–24 °C under LD (16light / 8 dark). Transcript levels for TaFT (A–B), VRN1 (C–D), and TaFT2 (E–F) were examined at the 1st-leaf stage (A, C, E) and the 5th-leaf stage (B, D, F). The same calibrator was used for TaFT and VRN1, so their units are comparable. A different corrector was used for TaFT2 because of its low transcript levels relative to TaFT and VRN1, therefore their scales are not comparable. When using the same calibrator, TaFT2 transcript levels were 40–70 fold lower than TaFT in the non-transgenic control plants.
A 40-fold (1st leaf stage) to 500-fold (5th leaf-stage) increase in TaFT2 transcript levels was observed in the TaFT transgenic lines relative to the controls, suggesting that TaFT2 may be regulated directly or indirectly by TaFT. In the non-transgenic plants the transcript levels of TaFT2 were 20- to 100-fold lower than those of TaFT.
DISCUSSION
bZIP transcription factors
The bZIP transcription factors are characterized by a basic region that binds DNA and a leucine zipper dimerization motif. This family is found in all eukaryotes but has more members in plants than in humans (Homo sapiens) or worm (Caenorhabditis elegans) (Riechmann et al., 2000). Approximately 75 different bZIP transcription factors involved in pathogen resistance, light and stress signaling, flower development, and seed maturation have been described in Arabidopsis (Jakoby et al., 2002).
The bZIP transcription factor FD plays a central role in the regulation of flowering in Arabidopsis (Wigge et al. 2005, Abe et al. 2005). FD is expressed in the SAM before floral induction, and its transcript levels increase with time after germination under short or long day photoperiods (Abe et al. 2005). Under inductive photoperiod, the FT protein is expressed in the phloem and travels to the shoot apex where it interacts with FD (Corbesier et al., 2007; Jaeger, et al., 2007; Mathieu, et al., 2007; Tamaki et al., 2007). The interaction between these two proteins in planta has been demonstrated in Arabidopsis by both bimolecular fluorescent complementation (Abe et al., 2005) and chromatin immunoprecipitation (ChIP) using FT antibodies in plants overexpressing FD (Wigge et al., 2005).
The ChIP experiment showed enrichment for the C-box region in the AP1 promoter only under conditions promoting the expression of FT, suggesting that a stable FT-FD-AP1 complex was formed in Arabidopsis (Wigge et al 2005). The presence of a similar interaction in wheat is indirectly supported by the in vitro interactions (TaFT –TaFDL2 and TaFDL2-VRN1 promoter) described before and by the spatial correlation between the expression of TaFDL2 and VRN1. High levels of TaFDL2 and VRN1 are observed in the leaves of wheat plants expressing TaFT, paralleling the induction of AP1 in the leaves of Arabidopsis when FD is expressed ectopically in this tissue (Wigge et al., 2005).
To explore further if a similar TaFT-TaFDL2-VRN1 complex could be also detected in wheat we included both TaFT and TaFDL2 proteins in the same binding reaction with the VRN1 promoter segments (SOM). Unfortunately, we failed to observe any supershift relative to the TaFDL2 protein used alone (SOM). These results suggest that either there is no stable TaFT-TaFDL2-VRN1 complex in wheat, or more likely that the conditions used in the binding assays were not conductive to the formation of a stable complex. It is also possible that additional proteins are necessary to stabilize this complex. ChIP studies will be necessary to confirm the existence of a TaFT-TaFDL2-VRN1 complex in wheat.
The identification of wheat FD homologue was not a trivial task due to the limited sequence conservation among bZIP transcription factors, which is generally limited to the bZIP motif (Fig. 1). Therefore, we relied on the simultaneous interaction of the TaFDL candidates with TaFT in yeast-two-hybrid tests and with the VRN1 promoter in EMSA experiments to select the best candidate. Only TaFDL2 fulfilled both selection criteria among the five TaFDL genes identified in wheat leaves, making it the best candidate for a functional homologue of Arabidopsis FD.
Expression results showed that TaFDL2 transcripts accumulate in the vegetative and reproductive apices (Fig. 5) suggesting that the TaFDL2 protein is present in the correct tissue and developmental stage to be involved in flowering induction once TaFT becomes available.
Duplication and functional differentiation of FT and FT2 genes
Phylogenetic and comparative mapping analyses have shown that orthologues of wheat TaFT and TaFT2 are present in barley and rice, indicating that the duplication that originated these paralogous genes predated the divergence of the grasses (Faure et al., 2007; Yan et al., 2006). The same phylogenetic analyses also showed that the TaFT-TaFT2 duplication occurred after the divergence of the grass species from the dicots. Therefore, this duplication is independent from the FT-TSF (Twin Sister of FT) duplication in the Arabidopsis lineage (Faure et al., 2007; Yan et al., 2006). Arabidopsis tsf mutations delay flowering and enhance the phenotype of ft mutants; whereas TSF over-expression causes precocious flowering under short day (Michaels et al., 2005; Yamaguchi et al., 2005).
Unfortunately, similarities and differences between Arabidopsis FT and TSF (Michaels et al., 2005; Sung et al., 2006; Yamaguchi et al., 2005) cannot be used to infer the functions of TaFT and TaFT2 in the grasses because independently duplicated genes may have different sub-functionalization. Therefore, the specific roles of the TaFT and TaFT2 genes need to be studied directly in the grass species. Detailed microcolinearity studies between barley HvFT gene region on chromosome arm 7HS and the rice region on rice chromosome arm 6S including Hd3a (the functional orthologue of Arabidopsis FT) have confirmed that these genes are true orthologues (Yan et al., 2006). However, this relationship is complicated by a recent tandem duplication that occurred only in the rice lineage resulting in closely linked genes Hd3a (=OsFTL2) and RFT1 (=OsFTL3) (Izawa et al., 2002; Kojima et al., 2002; Yan et al., 2006). Comparative mapping studies also confirmed that HvFT2, located in the short arm of barley chromosome 3H, is orthologous to rice OsFTL1, which is located on the colinear region of the short arm of rice chromosome 1 (Faure et al., 2007).
In rice, a short day plant, Hd3a transcript levels are rapidly induced under SD, but this effect can be suppressed by short exposures to light in the middle of the night (night breaks). These characteristics are not observed for other rice FT-like genes (including OsFTL1 and OsFTL3) suggesting that Hd3a is the central gene in the photoperiod pathway in rice (Ishikawa et al., 2005). Recent studies have confirmed that the Hd3a protein moves from the leaves to the apices where it induces flowering, confirming that it is the functional homologue of Arabidopsis FT (Tamaki et al., 2007).
Whereas over-expression of Hd3a results in early flowering in rice (Kojima et al., 2002), over-expression of OsFTL1 produces more complex phenotypes including elongation of internodes, loss of apical dominance, and a terminal tissue at the apical meristem (Izawa et al., 2002). This terminal tissue is composed of multiple glumes, and occasionally a terminal floret at the tip instead of a panicle, suggesting that OsFTL1 may be involved in the regulation of panicle and floral development rather than in the regulation of flowering initiation, which is mainly regulated by Hd3a (Izawa et al., 2002).
The different functions described above for Hd3a and OsFTL1 may also apply to the orthologous wheat TaFT and TaFT2 genes. The finding that these wheat paralogues interact with different FD-like proteins provides a putative molecular explanation for a functional differentiation. TaFT interacts with the TaFDL2 and TaFDL6 proteins, whereas TaFT2 interacts with TaFDL13, which belongs to a different clade of FD-like proteins (Fig. 1). We showed here that the interaction between TaFT and TaFDL2 likely confers TaFT the ability to regulate VRN1 transcription and therefore, to affect the fate of the SAM. On the contrary, TaFDL13 (the TaFT2 partner) showed no interactions with the VRN1 promoter. Since TaFDL13 is mainly expressed in the apical region, it may provide TaFT2 specificity for targets expressed in this tissue.
A role of TaFT2 in processes occurring after the SAM reproductive differentiation is also supported by the transcription profiles of HvFT2. The proteins coded by TaFT2 and HvFT2 are 98% identical (excluding 7 initial amino acids) suggesting that they may also share similar structural and functional characteristics. Faure et al. (2007) found that HvFT2 is upregulated by LD, but this up-regulation occurs two to three weeks after the differentiation of the SAM. Our results in wheat exhibit a similar trend. The transcript levels of TaFT in the transgenic plants overexpressing TaFT are very high from the first leaf, whereas TaFT2 transcript levels were low at the first-leaf stage and reached high levels at the fifth–leaf stage (Fig. 6, E–F), suggesting a delayed induction relative to TaFT. In the non-transgenic control plants the transcript levels of TaFT were 40–70 fold higher than those of TaFT2, also suggesting a more central role of TaFT. More importantly, the strong upregulation of TaFT2 in the two TaFT overexpressing transgenic wheat lines relative to the control suggests that TaFT2 may be regulated directly or indirectly by TaFT (Fig. 6, E–F).
Similarities between the Arabidopsis and wheat flowering pathways
FT transcripts in Arabidopsis and wheat are rapidly upregulated upon transfer of plants to LD (Imaizumi et al., 2003; Faure et al., 2007; Turner et al., 2005; Yan et al., 2006). In addition over-expression of FT in transgenic plants results in precocious flowering confirming the conserved role of this gene as a promoter of flowering (Kobayashi, et al., 1999; Yan et al., 2006). The observed protein-protein interaction between TaFT and TaFDL2, and protein-DNA interaction between TaFDL2 and the VRN1 promoter suggest that the molecular interactions downstream of FT are also conserved between wheat and Arabidopsis. These interactions are incorporated into our current model of flowering regulation in temperate cereals (Fig. 7).
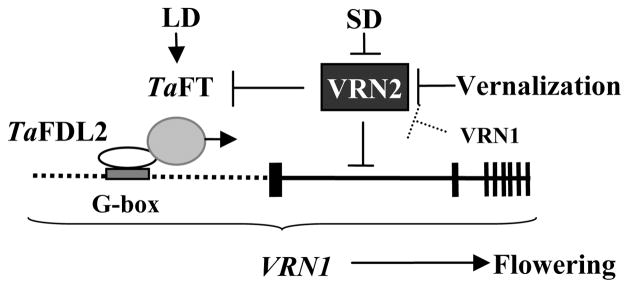
Figure 7.
Model for the regulation of flowering time in temperate cereals. The horizontal dotted line represents the VRN1 promoter and the vertical boxes indicate the eight exons of VRN1. The white oval represents the TaFDL2 protein interacting with the G-box (or hybrid boxes, grey rectangle) in the VRN1 promoter. The grey circle represents the TaFT protein interacting with TaFDL2. “→” indicates induction and “⊥” indicates repression. The doted line from VRN1 to VRN2 indicates a negative regulatory feedback loop. LD= long days, SD= short days.
The model presented in Fig. 7 shows the interaction between TaFDL2 and the G-box and hybrid boxes in the VRN1 promoter (Fig. 4). A similar protein-DNA interaction has been previously identified in Arabidopsis, where FD can bind to a 130-bp region of the AP1 promoter including a C-box motif (Wigge et al., 2005).
In this study, we observed a close correlation between VRN1 and TaFT transcript levels in transgenic wheat plants overexpressing TaFT. Lower transcript levels of TaFT in the transgenic plant Tr-1 relative to Tr-2, and in the 1st leaf stage relative to the 5th leaf stage (Fig. 6, A–B) were paralleled by similar differences in VRN1 transcript levels (Fig. 6, C–D). More importantly, VRN1 transcript levels were 500- to 1000-fold higher in the transgenic wheat plants over-expressing TaFT than in the non-transgenic control plants. Additional experiments in Arabidopsis showed that the addition of a strong transcriptional activation domain to FT increases its flowering promoting activity (Wigge et al., 2005). Taken together, these results suggest that TaFT is a limiting factor in the activation of VRN1.
Since we detected no binding between the TaFT protein and the VRN1 promoter by EMSA (data not shown), it is unlikely that TaFT can regulate the transcription of VRN1 through a direct protein-DNA interaction. These results, combined with the observed yeast two-hybrid interaction between TaFDL2 and TaFT (Figure 2) and the TaFDL2 interaction with the VRN1 promoter, suggest a model in which TaFT and TaFDL2 act in concert to regulate the transcription of VRN1 (Figure 7).
Differences between the Arabidopsis and wheat flowering pathways
Although the conservation of the last steps of the photoperiod pathway suggests that their origin predates the divergence of the monocot and dicot lineages, the differences in the vernalization pathways between Arabidopsis and the temperate cereals suggest independent origins. This is consistent with the view that the temperate Pooideae grasses are a specialized monophyletic group that evolved from subtropical bambusoid-like species (Clayton and Renvoize, 1986; Preston and Kellogg, 2008).
The central repressor in the vernalization pathway of Arabidopsis, the MADS-box gene FLOWERING LOCUS C (FLC) (Michaels and Amasino, 1999; Sheldon et al., 1999), has not been found in the grass species; whereas VRN2, the central repressor of flowering in the vernalization pathway of the temperate cereals, has not been found in Arabidopsis (Yan et al., 2004b). FLC delays flowering by interacting with regulatory regions of FT in the leaves and SOC1 in the meristems (Helliwell et al., 2006; Searle et al., 2006; Wigge et al., 2005). Vernalization permanently down-regulates FLC releasing FT and SOC1 to promote the transcription of AP1 (Michaels and Amasino, 1999).
In the temperate cereals, VRN2 is also down-regulated by vernalization (Yan et al., 2004b), but in contrast to FLC, it also can be down-regulated by short days (Dubcovsky et al., 2006; Trevaskis et al., 2006) (Fig. 7). The position of VRN2 upstream of VRN1 and TaFT in the model is supported by results from transgenic winter wheat plants with reduced VRN2 transcript levels (Yan et al., 2004b) and isogenic lines for VRN2 (Yan et al., 2006). Plants with reduced or non-functional VRN2 transcripts showed up-regulation of TaFT and VRN1 and a significant acceleration of flowering (Yan et al., 2004b, 2006). The increase in VRN1 transcript levels is then followed by down-regulation of VRN2 (Fig. 7) suggesting the existence of a regulatory feedback loop that has not been described in Arabidopsis (Loukoianov et al., 2005).
Arabidopsis and the temperate cereals also differ in the genes that show natural variation in vernalization requirement. In Arabidopsis, most of the natural mutants with reduced or non-vernalization requirement are concentrated in FRI and FLC (Gazzani et al., 2003; Michaels, et al., 2003); whereas in the temperate cereals they were detected in VRN1 (Fu et al., 2005; Yan et al., 2004a; Yan et al., 2003), VRN2 (Cockram et al., 2007; vonZitzewitz et al., 2005; Yan et al., 2004b), and TaFT (Yan et al., 2006). The known natural mutations for TaFT and VRN1 are dominant for spring growth habit and are located within regulatory, rather than coding regions of these genes, suggesting the disruption of recognition sites for a flowering repressor, likely VRN2.
An additional characteristic that seems to be unique for the temperate cereals is the high level of expression of VRN1 in the leaves (Schmitz et al., 2000; Yan et al., 2003). In contrast, AP1 transcripts are abundant in the induced SAM and floral primordia in Arabidopsis, but are undetectable or present at much lower levels in some vegetative tissues (e.g. vascular tissues of cotyledons, Abe et al., 2005). Interestingly, transgenic 35S:FD Arabidopsis plants with ectopic expression of FD show high levels of AP1 transcripts in the leaves (Wigge et al., 2005), suggesting that the spatial differences between AP1 and VRN1 transcription profiles are the result of differences between the spatial transcription profiles of Arabidopsis FD and its wheat homologue (TaFDL2). The results presented here support this hypothesis. We first identified TaFDL2 as the functional homologue of Arabidopsis FD by its ability to interact with TaFT and the VRN1 promoter, and then showed that TaFDL2 exhibits high transcript levels in the leaves of both vernalized and unvernalized winter wheat plants from very early developmental stages (Fig. 5). The simultaneous presence of TaFDL2 and TaFT in the leaves of spring or vernalized winter wheat plants provides a simple explanation for the simultaneous presence of VRN1. The regulation of TaFT by different environmental cues provides temporal specificity to this interaction.
Although the results presented above explain the molecular mechanism by which VRN1 transcription is regulated in the leaves, they do not explain why such a complex regulation of a meristem identity gene has been conserved in the leaves of the temperate grasses analyzed so far. Upregulation of VRN1 transcripts in the leaves of winter genotypes has been confirmed in wheat (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003), barley (Trevaskis et al., 2003), Lolium (Petersen et al., 2004), and oats (Preston and Kellogg, 2008) suggesting conservation across several tribes of temperate cereals (Triticeae, Poeae, and Aveneae).
An interesting observation is that VRN1 transcript levels are negatively associated with the accumulation of COR (cold responsive) genes and with the degree of frost tolerance (Danyluk et al., 2003). Frost tolerance increases during the acclimation of wheat plants to cold but non-freezing temperatures, but decreases after the transition between the vegetative and reproductive apices. Near isogenic lines for the VRN1 gene carrying the recessive vrn1 allele (winter growth habit) can tolerate 11°C lower freezing temperatures than lines carrying the dominant Vrn1 allele (spring growth habit) (Limin and Fowler, 2006). Similarly, spring lines grown under SD, which down-regulates VRN1 transcript levels, can tolerate 8.5°C lower temperatures than the same lines under LD (Limin and Fowler, 2006). A similar result has been reported for barley. In a double-haploid population segregating for VRN-H1 lines carrying the recessive vrn-H1 allele showed higher CBF (C-repeat binding factors) transcript levels than those carrying the dominant Vrn-H1 allele (Stockinger et al., 2007). In addition, lines grown under SD (reduced VRN-H1 levels) showed higher CBF transcript levels than lines grown under LD. CBF transcription factors are rapidly upregulated by cold temperatures, inducing the expression of COR genes and playing a critical role in cold acclimation and frost tolerance in temperate cereals (Francia et al., 2004; Knox et al., 2008; Miller et al., 2006; Skinner et al., 2005; Vágújfalvi et al., 2003). Based on these results, it is tempting to speculate that the temperate cereals have developed the ability to use the presence of VRN1 in the leaves as a signal to down-regulate the cold-tolerance regulatory network. Since VRN1 is upregulated upon the arrival of the spring, its presence would prevent the upregulation of the frost tolerance genes in the spring but not in the fall, when cold temperatures are an indication of future frost events.
CONCLUSION
In summary, we have identified the wheat functional homologue of Arabidopsis FD, TaFDL2 and confirmed that the protein coded by this gene interacts with TaFT and the VRN1 promoter. Transcription of TaFDL2 in the leaves provides a simple explanation for the characteristic expression of VRN1 in the leaves in temperate cereals. We also showed here that TaFT2 is partially regulated by TaFT and that the proteins coded by these two genes interact with different bZIP proteins providing the molecular basis for the sub-functionalization of these paralogous genes.
EXPERIMENTAL PROCEDURES
Wheat FD-like genes
The database used for the searches is available at The Gene Index (TGI) website (http://compbio.dfci.harvard.edu/tgi/). Starting with Arabidopsis FD protein sequence At4g35900, a TBLASTN search was carried out against wheat (Triticum aestivum) TGI Gene Index release 10.0 to search for TaFD-like genes in wheat. The top 16 gene sequences producing high-scoring segment pairs were chosen and further investigated. Primers were designed to amplify cDNAs for each of the top 16 TaFD-like genes (Table S1). Restriction sites (underlined) were included in the oligos to facilitate the cloning of the PCR products into yeast vectors.
A multiple sequence alignment and a Neighbor Joining phylogenetic tree were constructed using Mega v.4 (Tamura et al., 2007). The tree was based on a conserved un-gapped 69 amino acid region including the bZIP domain (Fig. 1). Bootstrap confidence values for the nodes were based on 1000 iterations.
Yeast two-hybrid assays
The yeast cloning vectors pGBKT7 and pGADT7, control vectors pGADT7-T and PGBKT7-53, and the yeast strain AH109 used in the yeast two-hybrid assays were obtained from Clontech (Mountain View, CA, USA). The yeast two-hybrid assays were performed according to the manufacturer’s instructions. Wheat full-length TaFT and TaFT2 cDNAs were fused to GAL4 DNA binding domain of pGBKT7 or the GAL4 activation domain of pGADT7 to generate bait and prey constructs, respectively. Full-length coding regions of TaFDL2, TaFDL6, TaFDL13 and TaFDL15, and a 375bp cDNA fragment of TaFDL3 (including the putative bZIP binding domain) were cloned in frame with GAL4 activation domain into the prey vector pGADT7.
Table S2 describes the primers and restriction sites used to generate yeast bait and prey constructs. The appropriate plasmids were transformed into yeast strain AH109 using the lithium acetate method and selected on SD medium lacking leucine (Leu) and tryptophan (Trp). After 4 days of incubation at 30°C, yeast cells were re-plated out on selection plates containing SD medium lacking Leu, Trp, histidine (His) and adenine (Ade) for the interaction test.
For validation, coding regions of the three FDL proteins that showed interactions with either TaFT or TaFT2 bait constructs were switched from prey constructs to the bait vector pGBKT7 using restriction sites EcoRI and PstI for TaFDL2, BamHI and PstI for TaFDL6, EcoRI and BamHI for TaFDL13. The new constructs were retested by yeast two-hybrid assays with TaFT or TaFT2 prey constructs.
Electrophoretic Mobility Shift Assays (EMSA)
TaFDL2, TaFDL6 and TaFDL13 cDNA fragments (Table S3) were cloned in frame with GST coding region into the corresponding sites of pGEX-6p-1 (GE Healthcare, Piscataway, NJ http://www4.gelifesciences.com/) to generate TaFDL2-GST, TaFDL6-GST and TaFDL13-GST fusion constructs. Four VRN1 promoter fragments containing one or two putative bZIP binding sites were generated by PCR (Table S3) and cloned in pGEM-T easy vector (Promega, Madison, WI, USA). Detailed EMSA protocols are described in the SOM.
Quantitative PCR and RT-PCR analyses
RNA samples were extracted from leaves and apices using the TRIZOL method (INVITROGEN). ACTIN was used as endogenous control for both the RT-PCR and the quantitative PCR SYBR GREEN® Applied Biosystems, http://www3.appliedbiosystems.com experiments, with primers described before (Dubcovsky et al., 2006). SYBR GREEN® systems for VRN1 and TaFT were developed in the previous study (Yan et al., 2006). The primers for the new SYBR GREEN® systems and RT-PCR experiments for TaFT2, TaFDL2, TaFDL6, and TaFDL13 are listed in Table S4. Quantitative PCR experiments were performed in an ABI7000. The 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to normalize and calibrate transcript values relative to the endogenous controls. For each experiment, the same calibrator was used across replications, genotypes and environmental conditions to make units comparable. However, different calibrators were used for different experiments and therefore, their units are not comparable.
RNA samples for the RT-PCR analysis of the different TaFDL transcript levels were extracted from leaves of unvernalized and vernalized (6 weeks at 4°C and LD) winter wheat variety Jagger. The apical region samples were extracted from the spring wheat variety Chinese Spring at the vegetative and early reproductive stages. Thirty plants were pooled to generate sufficient tissue. The apical region includes the SAM and a small portion of the crown.
Supplementary Material
Figure S1. TaFDL2 protein binds to bZIP binding sites in a concentration-dependent manner, but not to bZIP mutant sequences.
Figure S2. Transcript levels of TaFDL2 in the leaves of vernalized and unvernalized plants of Chinese Spring and the Chinese Spring Hope substitution line 7B carrying the dominant Vrn-B3 allele.
Table S1. Primers used to amplify FD-like cDNA fragments from wheat leaves.
Table S2. Primers and restriction sites used in the construction of yeast plasmids.
Table S3. Primers used in the EMSA experiment.
Table S4. RT-PCR and SYBR GREEN® quantitative PCR systems for TaFT2, TaFDL2, TaFDL6 and TaFDL13
Acknowledgments
This research was supported by the United States Department of Agriculture Cooperative State Research, Education, and Extension Services National Research Initiative competitive grant 2007-35301-17737 and 2007-35301-18188. The authors thank Ann Blechl (USDA-Agricultural Research Service Albany) for the transgenic wheat plants.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- An HL, Roussot C, Suarez-Lopez P, Corbesler L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Bernier G. The control of floral evocation and morphogenesis. Annu Rev Pl Phys Plant Mol Biol. 1988;39:175–219. [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK. Connecting the hormonal nature of plant development processes. Doklady Akad Nauk SSSR. 1937;16:227–230. [Google Scholar]
- Clayton WD, Renvoize SA. Grasses of the world. London: Royal Botanic Gardens: Kew; 1986. Genera Graminum. [Google Scholar]
- Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O’Sullivan DM. Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor Appl Genet. 2007;115:993–1001. doi: 10.1007/s00122-007-0626-x. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Coupland G. The quest for florigen: a review of recent progress. J Exp Bot. 2006;57:3395–3403. doi: 10.1093/jxb/erl095. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH. Plant bZIP proteins gather at ACGT elements. Faseb J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N. Two loci on chromosome 5H determine low-temperature tolerance in a Nure (winter) × Tremois (spring) barley map. Theor Appl Genet. 2004;108:670–680. doi: 10.1007/s00122-003-1468-9. [DOI] [PubMed] [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner J, Hayes P, Dubcovsky J. Large deletions in the first intron of the VRN-1 vernalization gene are associated with spring growth habit in barley and polyploid wheat. Mol Gen Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, Peacock WJ, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signaling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K. Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell. 2005;17:3326–3336. doi: 10.1105/tpc.105.037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K, Li B, Bartel P, Fields S. Use of the 2-hybrid system to identify the domain of P53 involved in oligomerization. Oncogene. 1993;8:1693–1696. [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. Plant bZIP protein-DNA binding-specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Gene Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–11. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Knox AK, Li C, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J. Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol Biol. 2008;67:257–270. doi: 10.1007/s11103-008-9316-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Li B, Fields S. Identification of mutations in P53 that affect its binding to Sv40 large T-antigen by using the yeast 2-hybrid system. Faseb J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limin AE, Fowler DB. Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta. 2006;224:360–366. doi: 10.1007/s00425-006-0219-y. [DOI] [PubMed] [Google Scholar]
- Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka KI, Miura E, Xoconostle-Cazares B, Gendler K, Jorgensene RA, Phinney B, Lough TJ, Lucas WJ. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Moyano E, Alcocer MJC, Martin C. Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J. 1998;13:489–505. doi: 10.1046/j.1365-313x.1998.00050.x. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17:1055–60. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He YH, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, Galiba G, Dubcovsky J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol Gen Genomics. 2006;275:193–203. doi: 10.1007/s00438-005-0076-6. [DOI] [PubMed] [Google Scholar]
- Petersen K, Didion T, Andersen CH, Nielsen KK. MADS-box genes from perennial ryegrass differentially expressed during transition from vegetative to reproductive growth. J Pl Physiol. 2004;161:439–447. doi: 10.1078/0176-1617-01212. [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. Discrete developmental roles for temperate cereal grass VRN1/FUL-like genes in flowering competency and the transition to flowering. Plant Physiol. 2008;146:265–276. doi: 10.1104/pp.107.109561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MMR, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol. 2000;42:899–913. doi: 10.1023/a:1006425619953. [DOI] [PubMed] [Google Scholar]
- Searle I, He YH, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Gene Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007;51:308–321. doi: 10.1111/j.1365-313X.2007.0141.x. [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Gene Dev. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis software (MEGA) Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. The cold regulated transcriptional activator Cbf3 is linked to the frost-tolerance gene Fr-A2 on wheat chromosome 5A. Mol Gen Genomics. 2003;269:60–67. doi: 10.1007/s00438-003-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonZitzewitz J, Szûcs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Mol Biol. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet. 2004a;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004b;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. Physiology of flower formation. Annu Rev Pl Phys Plant Mol Biol. 1976;27:321–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. TaFDL2 protein binds to bZIP binding sites in a concentration-dependent manner, but not to bZIP mutant sequences.
Figure S2. Transcript levels of TaFDL2 in the leaves of vernalized and unvernalized plants of Chinese Spring and the Chinese Spring Hope substitution line 7B carrying the dominant Vrn-B3 allele.
Table S1. Primers used to amplify FD-like cDNA fragments from wheat leaves.
Table S2. Primers and restriction sites used in the construction of yeast plasmids.
Table S3. Primers used in the EMSA experiment.
Table S4. RT-PCR and SYBR GREEN® quantitative PCR systems for TaFT2, TaFDL2, TaFDL6 and TaFDL13