The crystal structure of the catalytic domain of MARK4 was determined and compared with those of other proteins belonging to the MARK/PAR-1 protein kinase family.
Keywords: MARK4, PAR-1 serine/threonine protein kinase, transferase
Abstract
Microtubule-associated protein/microtubule affinity-regulating kinase 4 (MARK4) is a serine/threonine kinase involved in the phosphorylation of MAP proteins that regulate microtubule dynamics. Abnormal activity of MARK4 has been proposed to contribute to neurofibrillary tangle formation in Alzheimer’s disease. The crystal structure of the catalytic and ubiquitin-associated domains of MARK4 with a potent pyrazolopyrimidine inhibitor has been determined to 2.8 Å resolution with an R work of 22.8%. The overall structure of MARK4 is similar to those of the other known MARK isoforms. The inhibitor is located in the ATP-binding site, with the pyrazolopyrimidine group interacting with the inter-lobe hinge region while the aminocyclohexane moiety interacts with the catalytic loop and the DFG motif, forcing the activation loop out of the ATP-binding pocket.
1. Introduction
The MARK/PAR-1 family protein kinases have been widely studied because of their importance in microtubule assembly and in cell-cycle progression (for a review see Matenia & Mandelkow, 2009 ▸). Orthologs of this kinase family have been found in species ranging from yeast to mammals (Tassan & Le Goff, 2004 ▸). In humans, the four PAR-1 paralogs have been shown to phosphorylate microtubule-associated proteins (tau, MAP2 and MAP4) and to regulate the transition between stable and dynamic microtubules, and thus were named MAP/microtubule affinity-regulating kinases or MARKs (Drewes et al., 1995 ▸). Members of the MARK family are highly conserved and are associated with the centrosome throughout mitosis (Trinczek et al., 2004 ▸). Thus, it is likely that these kinases are important in cell-cycle control and other cellular processes. They have also been implicated in a number of other functions such as glucose metabolism and immune-system pathways (see Tassan & LeGoff, 2004 ▸). Of particular interest is the role of MARK in the phosphorylation of tau protein in neuronal microtubules, which causes the microtubules to destabilize and the tau proteins to aggregate, a symptom of Alzheimer’s disease (Chin et al., 2000 ▸).
In mammals, there are four MARK family members (MARK1–MARK4), each with a molecular weight of approximately 85 kDa, with high sequence homology (88–90% identity) and a similar multidomain structure (Drewes et al., 1998 ▸). While the function and interplay of the domains is not well understood, the conservation of many of these features across species suggests the modulation of MARK kinase activity by interactions with other regulatory proteins (see Matenia & Mandelkow, 2009 ▸).
While all of the MARK family members are believed to be involved in microtubule stability, the MARK4 isoform is found mainly in brain tissue. This suggests that MARK4 may be the primary homolog responsible for microtubule destabilization in neuronal cells and in the tau-protein phosphorylation seen in Alzheimer’s disease (Trinczek et al., 2004 ▸; Lund et al., 2014 ▸). Thus, it is hoped that the inhibition of MARK4 might be a means to prevent the hyperphosphorylation of tau and slow the progression of the disease.
The crystal structures of the catalytic and the ubiquitin-associated domains of three of these isoforms have previously been reported: MARK1 (Marx et al., 2006 ▸), MARK2 (Panneerselvam et al., 2006 ▸) and MARK3 (Murphy et al., 2007 ▸). To better understand the mechanism of small-molecule inhibition of MARK kinases, and the structural differences among the MARK family members, we have determined the 2.8 Å resolution crystal structure of the catalytic and ubiquitin-associated domains of the fourth member of this family, MARK4, in complex with a potent small-molecule pyrazolopyrimidine-based inhibitor (Fig. 1 ▸) selected for synthesis from the patent literature (Lim et al., 2011 ▸, example 7). Knowledge of the binding mode of this inhibitor might serve as a model in the design of inhibitors of MARK4. Moreover, while the structure of MARK4 is very similar to those of the other MARK isoforms, its structure may help to locate areas of differentiation to be used to add protein specificity to our inhibitors for the treatment of Alzheimer’s disease.
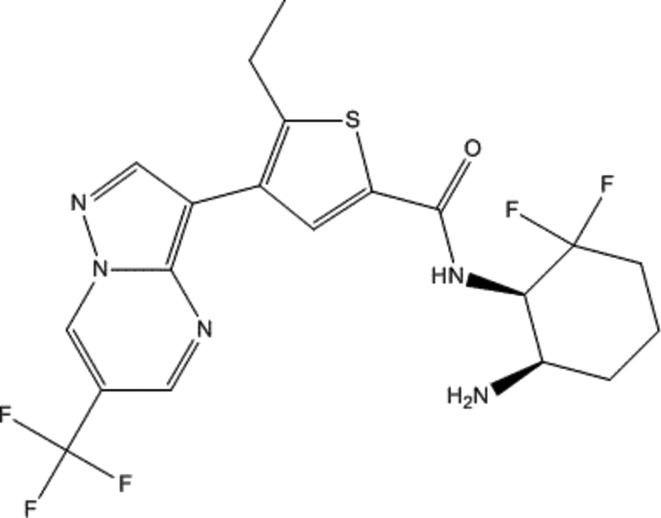
Figure 1.
The structure of [(1R,6R)-6-amino-2,2-difluorocyclohexyl]-5-ethyl-4-{6-(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-3-yl}-2-thiophenecarboxamide (compound 1).
2. Materials and methods
2.1. Protein expression and purification
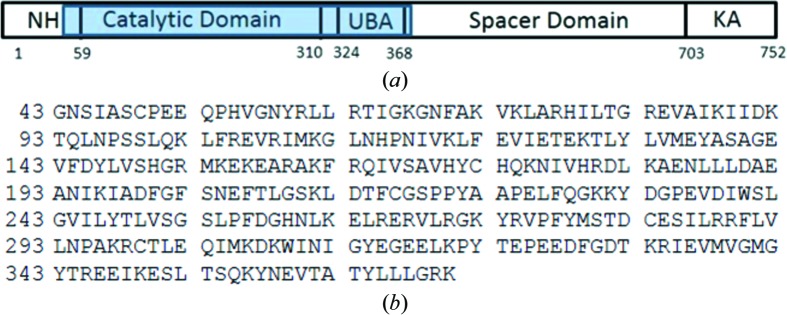
cDNA encoding the kinase and UAB domains of MARK4 (Asn44–Lys370; Fig. 2 ▸) was cloned into pET-28b vector with an N-terminal six-histidine tag followed by a TVMV protease cleavage site. The recombinant pET-28 plasmid containing the MARK4 gene of interest with an N-terminal His tag was transformed into Escherichia coli BL21(DE3) cells for expression. For expression, the transformed cells were grown at 37°C with shaking at 250 rev min−1 in LB medium until the OD600 nm reached 1.3, when the temperature was reduced to 18°C and the cells were allowed to equilibrate. IPTG was then added to a final concentration of 0.5 mM and the cells were allowed to grow overnight at 18°C for 20 h with shaking at 250 rev min−1. For the purification of MARK4, all steps were performed at 277 K unless noted otherwise. The frozen pellet from 1 l of culture was suspended in 100 ml lysis buffer [25 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 30 mM imidazole, two tablets of cOmplete EDTA-free protease inhibitor (Sigma–Aldrich), 2 U ml−1 Benzonase nuclease (EMD Milllipore), 0.2 mg ml−1 lysozyme, 5 mM DTT, 1 mM TCEP]. The cells were lysed by sonication (Branson Digital Sonifier with a microtip probe). The lysate was clarified by sedimentation at 120 000g for 45 min (Thermo F40L-8 × 100 rotor) and the supernatant was loaded onto a 5 ml HisTrap FF Crude column (GE Healthcare Life Sciences) pre-equilibrated with buffer A (25 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 5 mM DTT, 1 mM TCEP). The column was washed with 20 column volumes of buffer A and then eluted with 20 column volumes of buffer A containing 250 mM imidazole pH 7.5. The eluted protein was concentrated by centrifugal ultrafiltration to 6 ml using Amicon Ultra-15 units with a 10 000 Da cutoff. The retentate was loaded onto a HiLoad 16/600 Superdex 200 pg column (GE Healthcare Life Sciences) pre-equilibrated with buffer B (25 mM HEPES pH 7.5, 350 mM NaCl, 5 mM DTT, 1 mM TCEP, 5% glycerol). The fractions containing pure monomeric His-TVMV-MARK4 (Asn44–Lys370) protein were pooled. The His tag was removed by digestion with purified recombinant His-TVMV protease for 16 h at 4°C [1:10 ratio of TVMV protease to MARK4 (Asn44–Lys370)]. The His-TVMV protease and residual His tag were removed by loading the reaction mixture onto a 2 ml column of Ni Sepharose 6 Fast Flow (GE Healthcare Life Sciences) pre-equilibrated with buffer A. The column-flowthrough fraction was concentrated to 6 ml and loaded again onto a HiLoad 16/600 Superdex 200 pg column (GE Healthcare Life Sciences) pre-equilibrated with buffer B (25 mM HEPES pH 7.5, 350 mM NaCl, 5 mM DTT, 1 mM TCEP, 5% glycerol). The final protein purity was greater than 90% as determined by SDS–PAGE with Coomassie Brilliant Blue staining. The final protein has a molecular weight of 37 562 Da as measured by mass spectometry (compared with a calculated mass of 37 560.52 Da for the 328 residues).
Figure 2.
(a) Schematic diagram of human MARK4: N-terminal header (NH), serine/threonine kinase or catalytic domain (CD), short linker, ubiquitin-associated domain (UBA), spacer domain and globular kinase-associated domain (KA). The range of residues in the construct used in this study is highlighted in blue. (b) The amino-acid sequence of the human MARK4 construct used in the crystallization trials. The residue numbering is based on the UniProt sequence numbering.
2.2. Crystallization
The protein was complexed with compound 1 (5.5 µl 0.1 M stock solution), incubated on ice for 2 h and then concentrated to 225 µl using an Amicon Ultra 10 000 Da molecular-weight cutoff spin-filtration unit. The protein concentration was determined to be 23.7 mg ml−1 via the A 280. The concentrated complex was clarified by centrifugation (11 000 rev min−1 for 7 min). Crystals of MARK4 were grown by vapor diffusion by mixing 1 µl protein solution with 1 µl reservoir solution consisting of 20%(w/v) PEG 6000, 0.2 M MgCl2, 0.1 M Tris pH 8.5. Crystallization trays were set up at room temperature and then placed at 19°C to incubate. Crystals were observed under several conditions in 5–14 d.
2.3. Data collection and processing
A crystal was harvested, transferred into a drop of crystallization solution (15 µl well solution + 5 µl 100% ethylene glycol) and then flash-cooled in liquid nitrogen. A 2.8 Å resolution data set was collected on a Rayonix MX-300 detector at a temperature of ∼100 K on beamline 08ID-1 at the Canadian Light Source (CLS). The data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▸). The reduced structure-factor data file was 99.8% complete to 2.8 Å resolution. MARK4 crystallized in space group P62 (unit-cell parameters a = b = 111.48, c = 69.78 Å) with one molecule in the asymmetric unit (Matthews coefficient of 3.33 Å3 Da−1; solvent content 63.0%; Table 1 ▸)
Table 1. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 08ID-1, CLS |
| Wavelength (Å) | 0.980 |
| Temperature (K) | 100.0 |
| Detector | Rayonix MX-300 CCD |
| Crystal-to-detector distance (mm) | 347.4 |
| Rotation range per image (°) | 0.8 |
| Total rotation range (°) | 160 |
| Exposure time per image (s) | 0.8 |
| Space group | P62 |
| a, b, c (Å) | 111.48, 111.48, 69.78 |
| α, β, γ (°) | 90, 90, 120 |
| Mosaicity (°) | 1.28 |
| Resolution range (Å) | 50.000–2.800 (2.900–2.800) |
| Total No. of reflections | 107134 (11634) |
| No. of unique reflections | 12441 (1227) |
| Completeness (%) | 99.800 (100.000) |
| Multiplicity | 8.600 (9.400) |
| 〈I/σ(I)〉 | 15.800 (3.900) |
| R r.i.m. † | 0.104 (0.809) |
| Overall B factor from Wilson plot (Å2) | 101.800 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
The structure of MARK3 (PDB entry 3fe3; Panneerselvam et al., 2006 ▸), with waters and ligand removed, was used as the starting structure in the molecular-replacement search with Phaser (McCoy et al., 2007 ▸) without any modifications. The search gave a single solution which was used as the starting point for autoBUSTER (Bricogne et al., 2011 ▸) refinement. The first cycle of autoBUSTER refinement gave an R work of 33.45% (R free = 36.20%). The model and electron-density maps were examined with Coot (Emsley et al., 2010 ▸). There was clear electron density in the binding site. The model was carefully examined and the amino-acid differences between MARK3 and MARK4 were applied. A total of four autoBUSTER refinement cycles were run prior to fitting of the ligand.
The ligand was fitted using the rhofit routine. A single solution (CC = 0.70) was found for the ligand fitting. Four additional refinement cycles were run to complete the refinement. The final model has an R work of 22.9% (R free = 29.5%) for the 2326 atoms including 32 ligand atoms and five solvent molecules (see Table 2 ▸).
Table 2. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 36.4900–2.7800 (3.0400–2.7800) |
| Completeness (%) | 99.0 |
| σ Cutoff | F > 0.000σ(F) |
| No. of reflections, working set | 12426 (2716) |
| No. of reflections, test set | 657 (156) |
| Final R cryst | 0.239 (0.271) |
| Final R free | 0.311 (0.317) |
| Cruickshank DPI | 0.5730 |
| No. of non-H atoms | |
| Protein | 2285 |
| Ligand | 32 |
| Solvent | 8 |
| Total | 2325 |
| R.m.s. deviations | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.190 |
| Average B factors (Å2) | |
| Protein | 99 |
| Ligand | 78 |
3. Results and discussion
3.1. Overall structure of MARK4
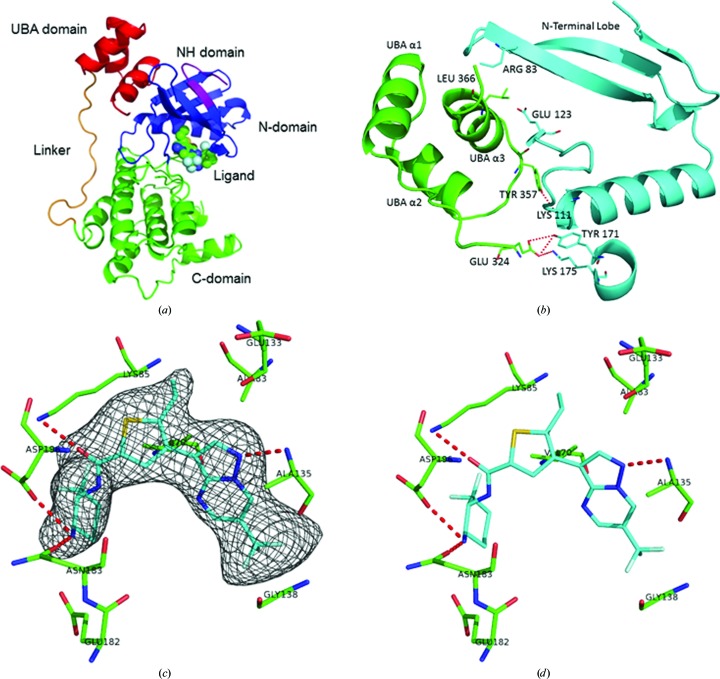
The fragment of human MARK4 used in this study incorporates the kinase domain (Tyr59–Ile310), a short linker sequence (Asn311–Thr323) and the UBA domain (Glu324–Gly368). With this crystal structure of MARK4, we now have the structures of the catalytic domains of all four MARK isoforms. Given the high degree of sequence homology, it is not surprising that all four structures have the same overall topology and arrangement of domains: the catalytic domain with the characteristic kinase bilobal fold, the linker sequence running up the face of the catalytic domain, and the UBA domain which binds to the N-terminal lobe of the catalytic domain via hydrophobic contacts (Fig. 3 ▸ a).
Figure 3.
Structure of the MARK4 complex. (a) Cartoon diagram of the MARK4 structure showing the NH domain (purple), N-domain (blue), C-domain (green), linker (orange) and UBA domain (red). (b) Cartoon diagram of the interaction between the CD (blue) and UBA (green) domains in MARK4. (c) The final F o − F c OMIT electron-density map contoured at 2.5 r.m.s.d. is shown with the final model of compound 1 in the ATP-binding site of MARK4. (d) Stick figure of the binding of compound 1 showing the hydrogen-bonding scheme (the drawings were generated using PyMOL).
3.2. The catalytic domain
The overall conformation of the catalytic domain is analogous to the other MARK serine/threonine protein kinases. The first 11 residues (Gly43–Gln53) are insufficiently ordered to model in the MARK4 crystal structure. The first visible residue, Pro54 of the N-terminal header, forms a short β-strand in line with the N-terminal lobe β-sheet. The P-loop (residues Gly66–Val73), located between β-strands 1 and 2, is conserved in all of the MARK isoforms and has been shown to be rather flexible in the apo structures but is stabilized by substrate interactions in the nucleotide-bound structures (Timm et al., 2008 ▸). In the case of the MARK4–inhibitor complex it is very well ordered in the structure, perhaps owing to the proximity of the difluorocyclohexane of the ligand to the side chain of Val73.
The most variable region in the MARK structures is in the catalytic loop (Lys175–Ala184). This may be owing to the difference in the residue at the start of the loop (Tyr170 in MARK1, Phe170 in MARK2, Arg173 in MARK3 and Asn176 in MARK4) that causes variations in the orientation of the side chain and may be responsible for many of the differences seen between the isoforms in the catalytic domain (Marx et al., 2006 ▸). None of the other nonconserved amino-acid differences in the catalytic domain of MARK4 make a significant alteration to the secondary structure.
As is typical for a kinase in an inactive conformation, most of the activation loop of MARK4 is not seen in the crystal structure. Asp199 of the DFG motif is involved in interactions with both the ligand and Asn186 (Fig. 3 ▸ a). This interaction stabilizes the start of the activation loop and positions the aspartate towards the OH group of the substrate. The middle of the activation loop (Glu206–Gly217) is disordered and is not seen in the MARK4 structure. The C-terminal end of the loop (Ser218–Glu225) is distant from the DGF motif, which is consistent with an open, inactive form of the activation loop.
3.3. The linker and ubiquitin-associated domains
The ubiquitin-associated (UBA) domain (Glu324–Gly368) is connected to the end of the catalytic domain by means of a short linker region (Asn311–Thr323). Surprisingly, the linker, which is clearly seen in the electron-density map, follows a similar extended conformation along the back face of the catalytic domain in all of the MARK isoforms without making many direct interactions with the catalytic domain. This positions the UBA domain alongside the N-terminal lobe β-sheet. The interaction between the N-terminal lobe and UBA domain is possible because of an unusual fold in which one of the three short α-helices is inverted relative to the canonical UBA-domain fold (Murphy et al., 2007 ▸). The UBA-domain α3 helix is involved in hydrophobic contacts with the catalytic domain N-terminal lobe in all four MARK crystal structures. In the MARK4 structure the primary interaction involves Asn58, His78, Arg83 and Glu123 on the N-terminal lobe with Glu359, Thr363 and Leu366 on the UBA domain (Fig. 3 ▸ b). While little sequence homology is apparent among the UBA domains, the main-chain conformation of the UBA domain is very similar in all isoforms (r.m.s.d. on Cα atoms of 0.41 Å), with most of the amino-acid differences located on the exterior of the domain, away from the N-lobe interface.
3.4. Ligand-binding interactions
As predicted, the inhibitor binds in the putative ATP-binding site (Fig. 3 ▸ c). The only direct interaction with the hinge is from the pyrazole N atom to the amino group of Ala138 (2.9 Å). While one of the F atoms is relatively close to the carboxyl of Ala138 (3.3 Å), it does not appear to make a direct interaction. The other end of the ligand binds in the phosphate pocket, with the O atom making a hydrogen bond to the side chain of Lys88 (2.8 Å), and the terminal N atom interacts with the side chains of Glu185 (3.0 Å), Asn186 (3.2 Å) and Asp199 (2.8 Å). As noted, residue Val73 is in close proximity to one of the F atoms of the cyclohexane (3.2 Å) and may play a role in stabilizing the catalytic loop. Since all of these residues are conserved among the MARK isoforms, it is not surprising that this compound shows pan-MARK activity (IC50 values of 4.1 nM for MARK1, 2.5 nM for MARK2, 3.9 nM for MARK3 and 4.6 nM for MARK4).
The binding of ligand to MARK4 does not cause a significant hinge movement of the N- and C-terminal lobes compared with the apo structures of the other MARK isoforms. This may be owing to the anchoring of the lobes by the linker and UBA domain.
4. Summary
The structure of MARK4 completes the crystallographic structures of the four human MARK isoforms. Given the high degree of homology among this family, one would not expect any major differences in the structure, which is what we find. For a drug-design effort, however, the structure of MARK4 might provide insight into how to target MARK4 over the other isoforms. MARK4 plays a key role in microtubule organization in neuronal cells and may contribute to the abnormal aggregation of tau seen in Alzheimer’s disease. If this specificity can be achieved then the inhibition of MARK4 might be a means to prevent the hyperphosphorylation of tau and slow the progression of Alzheimer’s disease.
Supplementary Material
PDB reference: MARK4 catalytic domain in complex with a pyrazolopyrimidine inhibitor, 5es1
Supporting Information.. DOI: 10.1107/S2053230X15024747/tt5077sup1.pdf
Acknowledgments
We would like to thank Shamrock Structures and the staff of the Canadian Light Source for data collection. We would also like to thank Weixu Zhai (Department of Chemistry, Bristol-Myers Squibb), Arundutt Silamkoti and Sushmita Nandi (Biocon Bristol-Myers Squibb Research and Development Center) for providing compound 1, Leads Discovery and Optimization (Bristol-Myers Squibb) for providing the binding data and the Bristol-Myers Squibb MARK4 drug discovery team lead by Angela Cacace and Lorin Thompson for their support of the crystallography project. We also thank Steven Sheriff for assistance in the preparation of the manuscript.
References
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). AutoBUSTER. Cambridge: Global Phasing Ltd.
- Chin, J. Y., Knowles, R. B., Schneider, A., Drewes, G., Mandelkow, E.-M. & Hyman, B. T. (2000). J. Neuropathol. Exp. Neurol. 59, 966–971. [DOI] [PubMed]
- Drewes, G., Ebneth, A. & Mandelkow, E.-M. (1998). Trends Biochem. Sci. 23, 307–311. [DOI] [PubMed]
- Drewes, G., Trinczek, B., Illenberger, S., Biernat, J., Schmitt-Ulms, G., Meyer, H. E., Mandelkow, E.-M. & Mandelkow, E. (1995). J. Biol. Chem. 270, 7679–7688. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Lim, J., Taoka, B. M., Lee, S., Northrup, A., Altman, M. D., Sloman, D. L., Stanton, M. G. & Noucti, N. (2011). Patent WO/2011/087999.
- Lund, H., Gustafsson, E., Svensson, A., Nilsson, M., Berg, M., Sunnemark, D. & von Euler, G. (2014). Acta Neuropathol. Commun. 2, 22–36. [DOI] [PMC free article] [PubMed]
- Marx, A., Nugoor, C., Muller, J., Panneerselvam, S., Timm, T., Bilang, M., Mylonas, E., Svergun, D. I., Mandelkow, E.-M. & Mandelkow, E. (2006). J. Biol. Chem. 281, 27586–27599. [DOI] [PubMed]
- Matenia, D. & Mandelkow, E.-M. (2009). Trends Biochem. Sci. 34, 332–342. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murphy, J. M., Korzhnev, D. M., Ceccarelli, D. F., Briant, D. J., Zarrine-Afsar, A., Sicheri, F., Kay, L. E. & Pawson, T. (2007). Proc. Natl Acad. Sci. USA, 104, 14336–14341. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Panneerselvam, S., Marx, A., Mandelkow, E.-M. & Mandelkow, E. (2006). Structure, 14, 173–183. [DOI] [PubMed]
- Tassan, J.-P. & Le Goff, X. (2004). Biol. Chem. 96, 193–199. [DOI] [PubMed]
- Timm, T., Marx, A., Paneerselvam, S., Mandelkow, E. & Madelkow, E.-M. (2007). BMC Neurosci. 9, S9. [DOI] [PMC free article] [PubMed]
- Trinczek, B., Brajenovic, M., Ebneth, A. & Drewes, G. (2004). J. Biol. Chem. 279, 5915–5923. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: MARK4 catalytic domain in complex with a pyrazolopyrimidine inhibitor, 5es1
Supporting Information.. DOI: 10.1107/S2053230X15024747/tt5077sup1.pdf