Brassinosteroid and glucose signals interact extensively during the development of etiolated seedlings.
Abstract
Brassinosteroid (BR) and glucose (Glc) regulate many common responses in plants. Here, we demonstrate that under etiolated growth conditions, extensive interdependence/overlap occurs between BR- and Glc-regulated gene expression as well as physiological responses. Glc could regulate the transcript level of 72% of BR-regulated genes at the whole-genome level, of which 58% of genes were affected synergistically and 42% of genes were regulated antagonistically. Presence of Glc along with BR in medium could affect BR induction/repression of 85% of BR-regulated genes. Glc could also regulate several genes involved in BR metabolism and signaling. Both BR and Glc coregulate a large number of genes involved in abiotic/biotic stress responses and growth and development. Physiologically, Glc and BR interact to regulate hypocotyl elongation growth of etiolated Arabidopsis (Arabidopsis thaliana) seedlings in a dose-dependent manner. Glc may interact with BR via a HEXOKINASE1 (HXK1)-mediated pathway to regulate etiolated hypocotyl elongation. BRASSINOSTEROID INSENSITIVE1 (BRI1) is epistatic to HXK1, as the Glc insensitive2bri1-6 double mutant displayed severe defects in hypocotyl elongation growth similar to its bri1-6 parent. Analysis of Glc and BR sensitivity in mutants defective in auxin response/signaling further suggested that Glc and BR signals may converge at S-phase kinase-associated protein1-Cullin-F-box-TRANSPORT INHIBITOR RESPONSE1/AUXIN-RELATED F-BOX-AUXIN/INDOLE-3-ACETIC ACID-mediated auxin-signaling machinery to regulate etiolated hypocotyl elongation growth in Arabidopsis.
Plants constantly sense the changes in their environment and transmit these signals as part of normal development. For optimal growth and development, plants need to coordinate complex developmental processes and, at the same time, sense and respond to endogenous physiological factors and external environmental stimuli. Many factors such as light, nutrients, and phytohormones are known to regulate these developmental processes. All these factors probably form a complex signal response network to bring about optimum growth changes to enable better fitness in plants. Among the phytohormones, brassinosteroids (BRs) are very important for plant growth and development. BRs are a class of polyhydroxylated sterol derivatives that are small growth-promoting molecules found at low concentrations throughout the plant kingdom. The BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) heterodimerizes with BRI1-ASSOCIATED KINASE1 (BAK1) after binding to BR. BRI1 and BAK1 subsequently act together to inhibit a Glycogen synthase kinase3-like kinase BRASSINOSTEROID INSENSITIVE2 (BIN2; Li et al., 2001). In absence of BR, BIN2 phosphorylates BRASSINAZOLE RESISTANT1 (BZR1). BIN2-mediated phosphorylation inhibits DNA binding capacity of BZR1 and promotes its binding to 14-3-3 proteins, which ultimately leads to its cytoplasmic retention or degradation (He et al., 2002; Gampala et al., 2007; Ryu et al., 2007). Signaling by BRI1/BAK1 removes this inhibition, and unphosphorylated BZR1 translocates to the nucleus, where it acts together with the transcription factor BRI1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1) to regulate expression of BR-inducible genes (Wang et al., 2002; Yin et al., 2002, 2005). BZR1 not only activates BR-induced genes and promotes cell elongation, but also suppresses BR biosynthetic genes such as CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD), leading to feedback inhibition of BR biosynthesis (He et al., 2002). BRs govern many processes involved in morphogenetic changes. BRs stimulate cell elongation by increasing cell wall plasticity and can also affect cell shape and expansion via regulation of microtubule dynamics. BRs control a vast number of responses in plants, such as seed germination, hypocotyl elongation, and senescence. BRs are also involved in controlling stomatal development and physiology. They have also been implicated to play a role in both biotic and abiotic stresses (Dhaubhadel et al., 1999, 2002; Kagale et al., 2007; Koh et al., 2007; Xia et al., 2009). BRs also interact with auxin to regulate both root elongation and tropic responses in plants (Bao et al., 2004; Li et al., 2005; Kim et al., 2007).
In plants, the availability of nutrient resources regulates many physiological and developmental processes profoundly. Apart from their metabolic functions, sugars such as Suc and Glc have also been reported to act as signals that can trigger changes in gene expression in plants (Price et al., 2004). Glc as a signaling molecule can also influence almost every aspect of plant growth and development such as cell proliferation and death, cell expansion and elongation, seed germination, seedling growth and development, primary root length, shoot and root gravitropism, lateral roots, root hairs, shoot meristem maintenance, reproduction, senescence, photosynthetic gene expression, crop yield and product quality, carbon and nitrogen metabolism, and stress responses (Rolland et al., 2002, 2006; Rolland and Sheen, 2005; Chen et al., 2006; Ramon et al., 2008; Mishra et al., 2009; Smeekens et al., 2010; Eveland and Jackson, 2012; Gupta et al., 2012; Singh et al., 2014). There are three distinct Glc-signaling pathways in plants: the Arabidopsis (Arabidopsis thaliana) HEXOKINASE1 (AtHXK1) signaling function-dependent pathway, the AtHXK1 catalytic function-dependent pathway, and the AtHXK1-independent pathway (Xiao et al., 2000). In the AtHXK1-dependent pathway, AtHXK1 acts as a Glc sensor (Moore et al., 2003). The AtHXK1-independent pathway involves G-protein signaling (Chen and Jones, 2004). The G-protein complex is represented by single Gα (G-PROTEIN α SUBUNIT1 [GPA1]), Gβ (G-PROTEIN β SUBUNIT1), and Gγ (G-PROTEIN γ SUBUNIT1 [AGG1] and AGG2) subunits. The REGULATOR OF G-PROTEIN SIGNALING1 (AtRGS1) contained a seven-transmembrane domain and RGS box and accelerates intrinsic guanosine triphosphatase activity of G-protein. The THYLAKOID FORMATION1 (THF1) protein is located on the outer membrane and stroma of plastids. The heterotrimeric G-protein complex via GPA1 interacts with the AtRGS1 and THF1 (Huang et al., 2006).
Sugar signaling has been reported to exhibit cross talk with several different response pathways, such as those involved in nitrogen responses, environmental responses, light responses, phytohormones, and stress responses. However, not much is known about Glc and BR signaling interaction. High level of sugar causes repression of CPD, an important gene involved in brassinolide biosynthesis (Szekeres et al., 1996; Smeekens, 1998). The CPD-antisense lines as well as cabbage1 (dwarf1-6 [dwf1-6]) mutant Arabidopsis plants display a clear reduction in starch content and assimilatory capacity (Schlüter et al., 2002). BRs are reported to regulate the process of sugar uptake in tomato (Lycopersicon peruvianum; Goetz et al., 2000). The dx mutation leads to production of a nonfunctional DWARF enzyme in tomato. In dx fruits, levels of starch and various sugars are reduced, which could be partially normalized by BR application (Lisso et al., 2006). The sugar hypersensitivity of a brassinosteroid, light, sugar mutant could be rescued by exogenous BR application, suggesting an interaction of sugar with BR (Laxmi et al., 2004). In rice (Oryza sativa), overexpression of a sterol C-22 hydroxylase, which controls BR levels, increased seed weight, which was associated with more allocation of sugars to seeds (Wu et al., 2008). These results suggest a role of BR in sugar allocation during grain filling in rice. In Saccharum spp., the LEUCINE-RICH REPEAT RECEPTOR-LIKE PROTEIN KINASE ScBAK1 is found to be expressed predominantly in bundle sheath cells of the mature leaf and potentially involved in cellular signaling cascades mediated by high levels of sugar in this organ (Vicentini et al., 2009). UDP-glycosyltransferases UGT73C5 and UGT73C6 function in conjugation of Glc to BR, which ultimately leads to reduced BR activity (Poppenberger et al., 2005; Husar et al., 2011). Recently, our group has also shown a dual role for BR-Glc cross talk in modulating shoot and root gravitropic responses of Arabidopsis seedlings (Gupta et al., 2012; Singh et al., 2014). Although a number of developmental responses of early seedlings are simultaneously controlled by BR and sugar, no systematic study has been performed to explore the interaction between Glc and BR signaling on global gene expression profiles and, in turn, on plant growth and development in the dark. In this study, whole-genome transcript profiling along with physiological analysis has been performed to find out the interdependence/overlap between Glc and BR response pathways in the etiolated seedlings of the model plant system Arabidopsis.
RESULTS
Genomewide Analysis of Glc and BR Interactions in Dark-Grown Arabidopsis Seedlings
Effect of Glc on BR-Regulated Gene Profiles
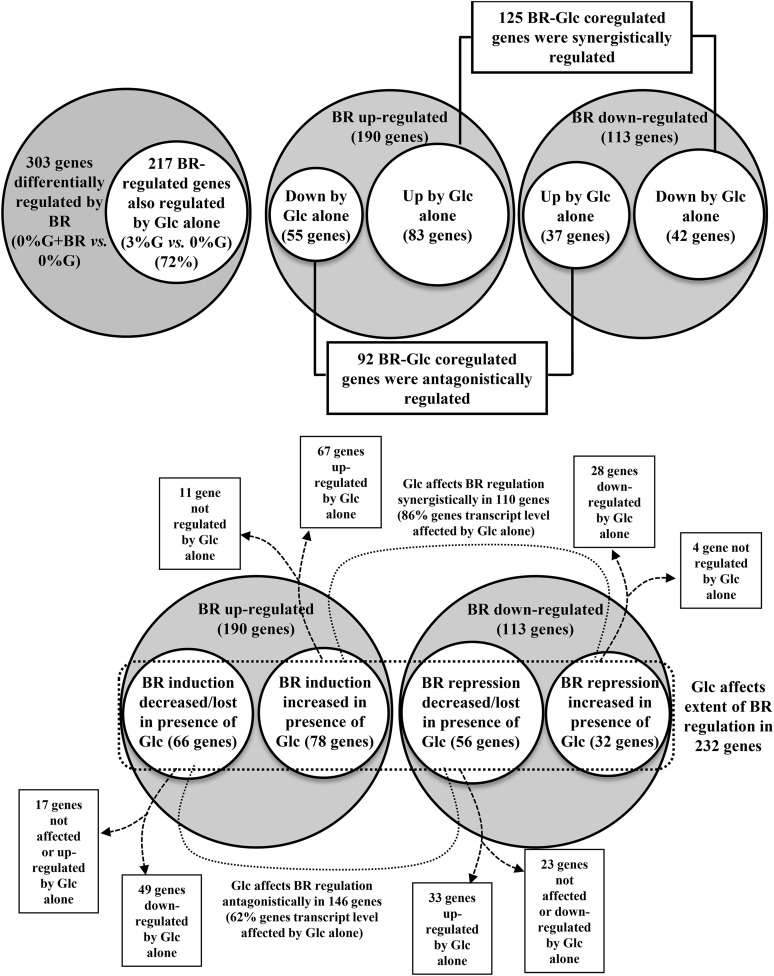
To determine the global effect of Glc on BR-regulated gene profiles during skotomorphogenic development, whole-genome transcript profiling of 6-d-old, etiolated wild-type (ecotype Columbia-0 [Col-0]) seedlings treated with BR and/or Glc was performed by using Agilent’s Arabidopsis gene expression microarray (4x44K). Our results show that a significant number of genes were being simultaneously regulated by Glc and BR. BR could regulate 303 genes (fold change ≤ 2.0), which include 190 (63%) up-regulated and 113 (37%) down-regulated genes (Fig. 1, A and B; Supplemental Table S1). Glc and BR could simultaneously regulate transcript level of several genes. Glc alone could regulate 72% (217) of BR-regulated genes (Fig. 1A; Supplemental Table S2). Fifty-eight percent (125) of them were regulated synergistically, and the remaining 92 (42%) were regulated antagonistically (Fig. 1B; Supplemental Table S2). Thus, at whole-genome level, Glc and BR could interact both synergistically and antagonistically.
Figure 1.
Effect of Glc on BR-regulated genes in dark-grown seedlings. A, In etiolated seedlings, BR can altogether up- or down-regulate a total of 303 genes in Glc-free medium (cutoff, 2-fold). Glc alone can independently affect 217 (72%) genes out of a total of 303 BR-regulated genes. B, Out of these 217 genes, 125 (58%) genes were synergistically, and the remaining 92 (42%) genes were antagonistically regulated by BR and Glc treatment alone. C, Presence of Glc can also change the extent of BR induction or repression by more than 2-fold for almost 85% of BR-affected genes (i.e. 256 genes out of a total of 303 BR-regulated genes). Out of these 256 genes, the extent of 110 (43%) genes was affected synergistically, and the extent of 146 (57%) genes was affected antagonistically in the presence of Glc. Glc can also affect BR regulation of those genes that are themselves not regulated by Glc alone.
Out of 303 BR-regulated genes (0% [w/v] Glc versus 0% [w/v] Glc + BR), BR-mediated up-regulation and down-regulation of 256 (85%) genes (0% [w/v] Glc versus 0% [w/v] Glc + BR compared with 0% [w/v] Glc versus 3% [w/v] Glc + BR) was significantly affected (2-fold or more/less or lost) in the presence of Glc (Fig. 1C; Supplemental Table S3). One hundred ten (43%) of them were affected synergistically (BR up-regulation and down-regulation increased), while 146 (57%) were antagonistically (BR up-regulation and down-regulation decreased or lost) affected (Fig. 1C; Supplemental Table S3). Interestingly, for 14% from synergistically regulated and 30% from antagonistically affected genes, Glc treatment alone could not cause any change in the transcript level (Fig. 1C; Supplemental Table S3). Regulation of signaling outcomes has often been correlated with posttranslational regulatory mechanisms such as protein phosphorylation-dephosphorylation, proteasomal degradation, etc. Glc might utilize these regulatory mechanisms to affect the BR induction/repression of such genes.
Effect of Glc on Transcription of Genes Involved in BR Physiology
When the fold change cutoff was relaxed to 1.5 (±), many BR-related genes could show differential expression patterns in one or more of our experimental conditions (Supplemental Table S4). These genes include BR biosynthetic genes such as CPD, DWF4, BRASSINOSTEROID-6-OXIDASE1 (BR6OX1), BR6OX2, and CYTOCHROME P450, FAMILY 90, SUBFAMILY D, POLYPEPTIDE1 (CYP90D1), BR perception- and signaling-related genes such as BRI1-LIKE1 (BRL1), BRI1 SUPPRESSOR1, and BES1-INTERACTING MYC-LIKE1 (BIM1), early BR-responsive genes such as BR ENHANCED EXPRESSION1 (BEE1), BEE2, and TOUCH4 (TCH4), and BR homeostasis-related genes such as PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1), etc. Glc was able to regulate the expression of most of these BR-related genes. The transcript levels of CYP90D1, BRL1, BIM1, and BES1/BZR1 HOMOLOG1 were induced, whereas CPD, BR6OX2, BEE1, TCH4, BAS1, etc. were repressed upon Glc treatment. All these results together suggest that Glc significantly affects most steps of BR biosynthesis, perception, signaling, and homeostasis.
Quantitative Real-Time PCR Validation of Microarray Results
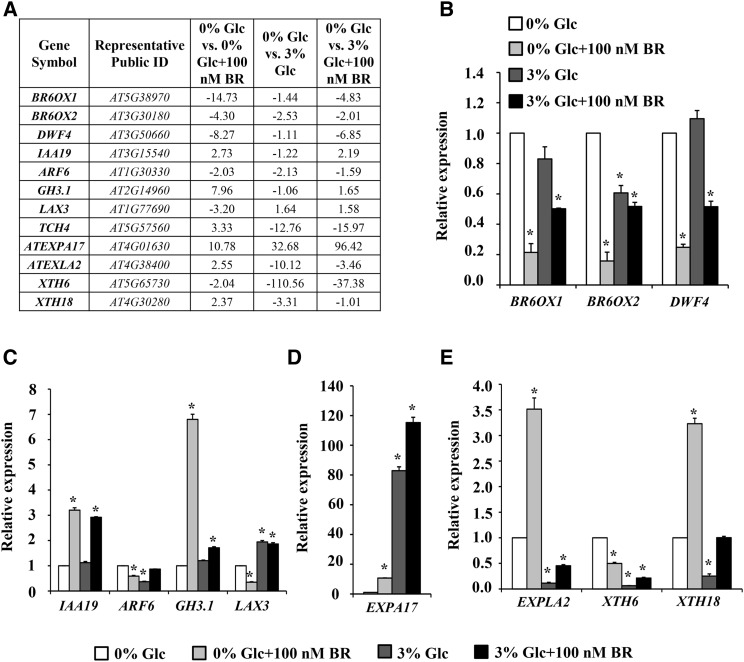
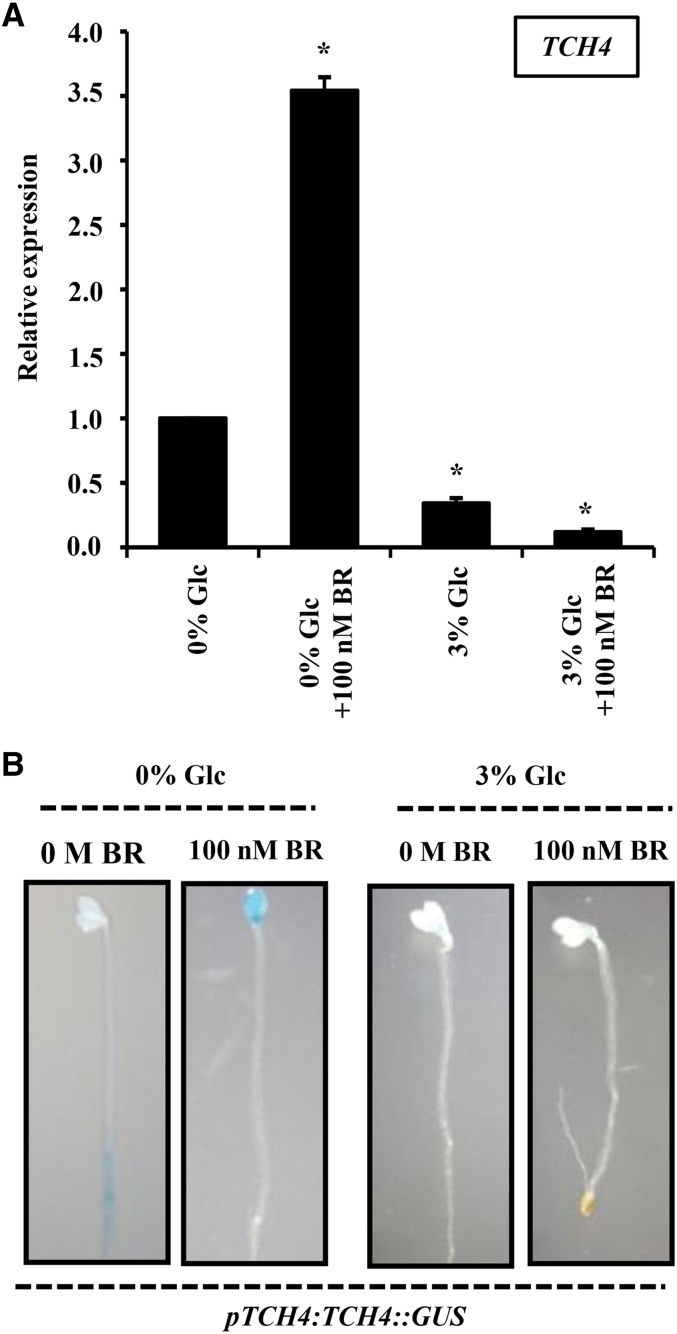
Validation of microarray data was done by conducting quantitative real-time PCR of some well-known BR-regulated genes and an additional set of selected genes. The expression pattern of some auxin-regulated genes and a few cell wall remodelling-related genes was also checked (Fig. 2). The real-time expression of these representative genes followed the similar pattern as obtained from microarray. The microarray data were further supported by pTCH4:TCH4::GUS expression pattern. TCH4 was up-regulated by BR and down-regulated by Glc (Fig. 3A). At increased Glc concentration (3% [w/v]), a significant reduction was observed in pTCH4:TCH4::GUS expression, while BR was able to induce the GUS expression in Glc-free or low-Glc-containing medium. However, at higher Glc concentrations, BR induction of GUS expression was significantly inhibited (Fig. 3B).
Figure 2.
Validation of microarray results using real-time PCR. A, The relative transcript levels of a few selected genes from microarray data. Real-time PCR results of selected BR (B), auxin (C), and cell wall-related genes (D and E). Values represent the average from two biological replicates, and error bars represent se (Student’s t test, P < 0.05; *, control versus treatment). ARF6, AUXIN RESPONSE FACTOR6; GH3.1, GRETCHEN HAGEN3; LAX3, LIKE AUXIN RESISTANT3; ATEXPA17, EXPANSIN A17; ATEXLA2, EXPANSIN-LIKE A2; XTH6, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE6.
Figure 3.
Glc interacts antagonistically with BR to regulate TCH4 expression. A, The relative abundance of TCH4 transcript upon BR and Glc treatments in 7-d-old etiolated wild-type (Col-0) seedlings. B, GUS expression analysis in hypocotyls and cotyledons of 7-d-old dark-grown pTCH4:TCH4::GUS seedlings in the absence or presence of Glc (0% and 3% [w/v]) and/or BR (0 m and 100 nm). BR regulation of TCH4 expression was lost in presence of Glc, suggesting, mainly, an antagonistic interaction between BR and Glc response. Values represent the average from two biological replicates, and error bars represent se (Student’s t test, P < 0.05; *, control versus treatment).
BR and Glc Mainly Regulate Genes Involved in Morphogenesis, Stress Responses, and Developmental Pathways
BR-regulated genes include members belonging to different gene families. Although the largest groups of genes regulated by BR and Glc were of unknown function, many genes fell into gene family groups with assigned functions (Supplemental Table S1). BR-regulated genes include members of the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) gene family, GDSL-motif lipase/hydrolase family proteins, glutathione S-transferases, the Leu-rich repeat family, the UDP-glucoronosyl/UDP-glucosyl transferase family, xyloglucan endotransglycosylases, plant U-box proteins, class III peroxidases, the cytochrome P450 family, etc. BR and Glc together could also regulate a significant numbers of genes known to be involved in different hormone responses, such as auxin, cytokinin, ethylene, abscisic acid (ABA), and jasmonic acid responses (Supplemental Table S5). We also categorized the differentially regulated gene sets under various treatment conditions using the Gene Ontology (GO; https://www.arabidopsis.org/tools/bulk/go/index.jsp) search program available at The Arabidopsis Information Resource (Rhee et al., 2003; Berardini et al., 2004). Subcategories of molecular functions, cellular components, and biological processes were prepared using these GO assigned terms. GO analysis also shows that there is a significant overlap between BR- and Glc-regulated genes in terms of cellular components, molecular functions, and biological processes (Supplemental Fig. S1). Biological process predictions revealed that for all differentially regulated gene lists, GO terms stress responses, response to abiotic or biotic stimulus, and signal transduction were significantly enriched compared with the whole-genome functional categorization (Supplemental Fig. S1). A large number of BR-regulated genes were found to encode for genes belonging to abiotic and biotic stress categories, and most of them were also regulated by Glc alone (Supplemental Table S6). These genes include several peroxidases, glutathione S-transferases, resistance genes, pathogenesis-related genes, genes involved in regulating redox state, etc.
Minute changes in transcript levels of transcription factors (TFs; i.e. <±2-fold) can cause significant alterations in the signaling outputs during key biological processes. Due to stringent statistical significance criteria selection, these minor changes are often left undetected. Upon relaxing the fold change cutoff to ≥1.5, we found that 9.2% (75 genes) of BR-regulated genes (811) were encoding for TFs. Glc alone was also able to regulate transcript levels in 80% (60) of BR-regulated TF-encoding genes (Supplemental Fig. S2). We also identified statistically overrepresented (P ≤ 10–3) TF-binding sites present in promoters (1,000 bp upstream of the translational start site) of BR- and Glc-regulated genes using the ATHENA (http://www.bioinformatics2.wsu.edu/Athena) promoter sequence analysis tool (O’Connor et al., 2005). It was found that a majority of the enriched cis-regulatory elements present in promoters of BR and Glc-coregulated genes were previously reported to be either ABA- or biotic/abiotic stress-responsive elements (Supplemental Table S7), further suggesting that Glc and BR may together control various stress responses in plants.
Physiological Significance of BR-Glc Interaction in Etiolated Seedling Growth
BR-Glc Interaction in Controlling Hypocotyl Growth
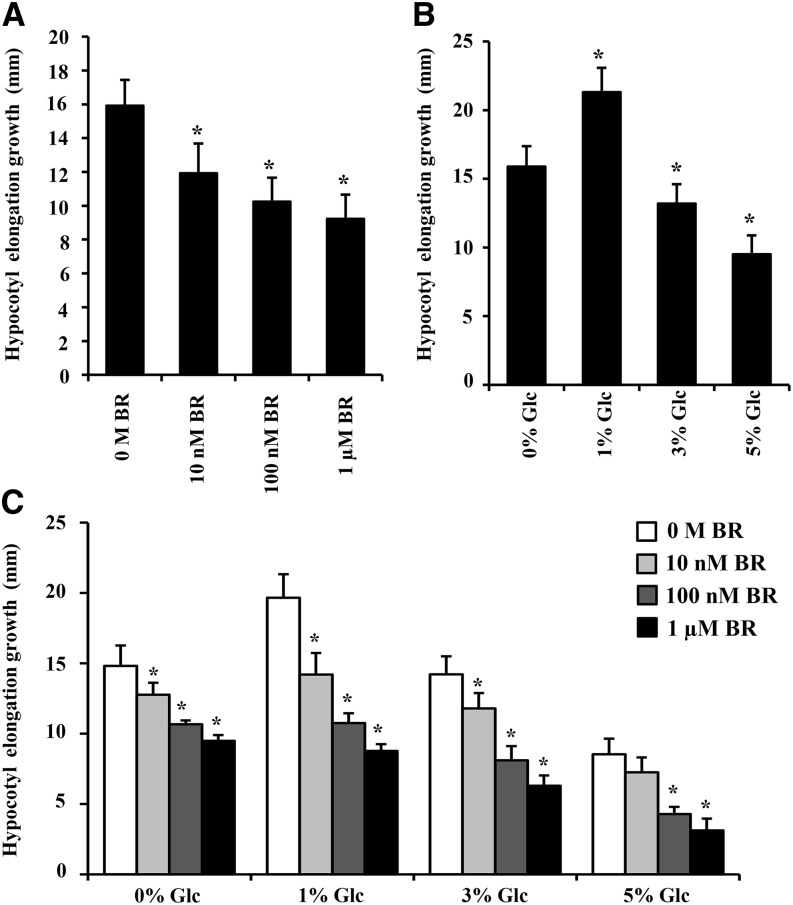
In microarray, both BR and Glc were found to regulate many key genes involved in hypocotyl growth and development (Supplemental Table S8). So, we checked the Glc and BR sensitivity of the wild type for regulation of hypocotyl elongation growth. Imbibed wild-type (Col-0) seeds were directly sown on one-half-strength Murashige and Skoog (MS) medium supplemented with independent and/or combined treatments of increasing Glc (0%, 1%, 3%, and 5% [w/v]) and BR (0 m, 10 nm, 100 nm, and 1 µm) concentrations for 7 d either in dark or long-day conditions (Fig. 4, A–C; Supplemental Figs. S3 and S4, A–C), and changes in hypocotyl elongation growth were quantified. In dark conditions, exogenous BR could reduce etiolated hypocotyl elongation in a dose-dependent manner (Fig. 4A), whereas, in long-day condition or light-grown seedlings, BR alone was able to increase hypocotyl elongation growth at all concentrations tested (Supplemental Fig. S4A). Glc at lower concentration (1% [w/v] Glc) increased and, at higher concentration (5% [w/v] Glc), decreased the hypocotyl elongation growth in both dark- and light-grown seedlings (Fig. 4B; Supplemental Fig. S4B). In light-grown seedlings, BR-induced hypocotyl elongation growth was significantly inhibited upon cotreatment with higher Glc concentrations (Supplemental Fig. S4C). In etiolated seedlings, the inhibitory effect of BR (10 nm) was more efficient in presence of lower Glc concentrations (1% [w/v] Glc) compared with cotreatment with higher Glc concentrations (5% [w/v] Glc; Fig. 4C; Supplemental Fig. S3). These results suggest that Glc cotreatment at lower doses supports and, at higher concentrations, antagonizes BR response in terms of hypocotyl elongation growth regulation.
Figure 4.
Glc and BR regulation of hypocotyl elongation growth in etiolated seedlings. Quantification of hypocotyl elongation in 7-d-old etiolated wild-type (Col-0) seedlings treated without or with increasing concentrations of BR (A), increasing concentrations of Glc (B), and Glc-free medium or increased Glc concentration containing one-half-strength MS medium supplemented without or with increasing concentrations of BR (C). Glc and BR work antagonistically at lower Glc concentrations but act synergistically at higher Glc concentrations for regulation of hypocotyl elongation growth. Values represent the average from two biological replicates, each having 30 seedlings, and error bars represent se (Student’s t test, P < 0.001; *, control versus treatment).
We also analyzed the independent as well as combined effects of Glc and BR on apical hook formation and maintenance. Imbibed wild-type (Col-0) seeds were directly sown on one-half-strength MS medium supplemented with independent and/or combined treatments of increasing Glc (0% and 3% [w/v]) and BR (0 m, 10 nm, 100 nm, and 1 µm) concentrations in the dark for 3 d, and apical hook phenotypes of seedlings were observed. In absence of Glc, BR caused a considerable apical hook opening, while in presence of Glc, BR could not execute the similar effect (Supplemental Fig. S5).
Involvement of Glc-Signaling Components in Controlling Hypocotyl Growth
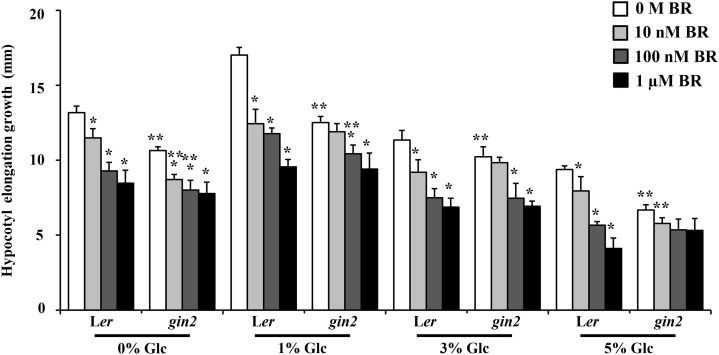
BR sensitivity of AtHXK1-dependent and -independent Glc-signaling mutants was examined in terms of hypocotyl elongation growth regulation. AtHXK1-dependent pathway mutant glucose insensitive2-1 (gin2-1), in addition to being resistant toward exogenous Glc application, also showed attenuated response toward exogenous BR in terms of hypocotyl elongation growth inhibition compared with the wild type (Fig. 5; Supplemental Fig. S6). At the same time, AtHXK1-independent pathway mutants rgs1-1, rgs1-2, thf1-1, gpa1-1, gpa1-2, and gpa1-3 were found to have similar sensitivity toward BR for hypocotyl length control compared with their respective wild types (Supplemental Figs. S7 and S8). Although gin2-1 mutant was not completely resistant toward Glc- and BR-regulated etiolated hypocotyl elongation growth, it showed significantly reduced response, suggesting that the AtHXK1-dependent pathway is required for a proper BR-Glc regulation of etiolated hypocotyl elongation, and any perturbation in the same leads to altered response toward BR.
Figure 5.
Involvement of AtHXK1-dependent Glc signaling during BR regulation of hypocotyl elongation growth. Quantification of hypocotyl elongation in 7-d-old etiolated seedlings of the wild type (Ler) and AtHXK1-dependent Glc sensor mutant gin2-1 growing on Glc-free (0%) medium or increasing concentrations of Glc (1%, 3%, and 5% [w/v]) containing one-half-strength MS medium supplemented without or with increasing BR concentrations (10 nm, 100 nm, and 1 µm). The gin2-1 mutant showed less response to BR in terms of hypocotyl elongation growth compared with the wild type. Values represent the average from two biological replicates, each having 30 seedlings, and error bar represents se (Student’s t test, P < 0.001; *, control versus treatment; and **, wild type versus mutant).
Involvement of BR-Signaling Components in Controlling Hypocotyl Growth
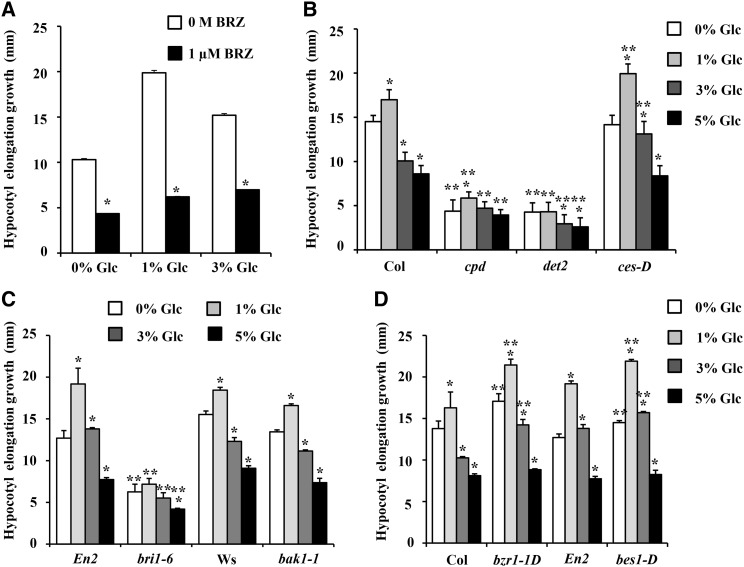
Inhibition of endogenous BR biosynthesis by brassinazole (BRZ; 1 µm) could perturb the Glc regulation of hypocotyl elongation growth, suggesting that BR is essential for Glc response (Fig. 6A). The BR biosynthesis and signaling mutants were treated with increasing concentrations of Glc, and changes in hypocotyl elongation growth were quantified (Fig. 6, B–D; Supplemental Fig. S9). BR biosynthesis-defective mutants cpd and deetiolated2 (det2) were found to be resistant, whereas cesta-D (ces-D) mutant having constitutively higher BR biosynthesis displayed increased hypocotyl elongation growth at all Glc concentrations tested compared with the wild type (Fig. 6B). BR perception-defective mutant bri1-6 was found to be resistant for Glc treatment for hypocotyl elongation growth, while another BR perception-defective mutant bak1-1 showed wild-type-like sensitivity toward Glc regulation of hypocotyl elongation (Fig. 6C), suggesting the Glc-BR control of hypocotyl elongation growth to be BRI1 mediated. The bzr1-1D and bes1-D mutants that have endogenously high BR signaling were also found to show more hypocotyl elongation growth at all Glc concentrations tested compared with the wild type (Fig. 6D). These results further suggest the involvement of BR biosynthesis, perception, and signaling components in Glc-BR interaction controlling hypocotyl elongation growth in the dark.
Figure 6.
Dependence of Glc regulation of hypocotyl elongation growth upon BR-signaling components. A, Quantification of hypocotyl elongation growth in 7-d-old etiolated seedlings of the wild type (Col-0) growing on Glc-free medium or increasing concentrations of Glc containing one-half-strength MS medium supplemented without or with 1 µm BRZ. B to D, Quantification of hypocotyl elongation growth in the wild type and BR-signaling mutants. BR biosynthesis mutants cpd and det2 and BR receptor mutant bri1-6 were found to be resistant toward Glc-mediated hypocotyl elongation, while BR overproducer mutant ces-D and mutants with endogenously high BR signaling (bzr1-1D and bes1-D) showed more hypocotyl elongation at all Glc concentrations tested compared with their respective wild types. Values represent the average from two biological replicates, each having at least 20 seedlings, and error bar represents se (Student’s t test, P < 0.001; *, control versus treatment; and **, wild type versus mutant).
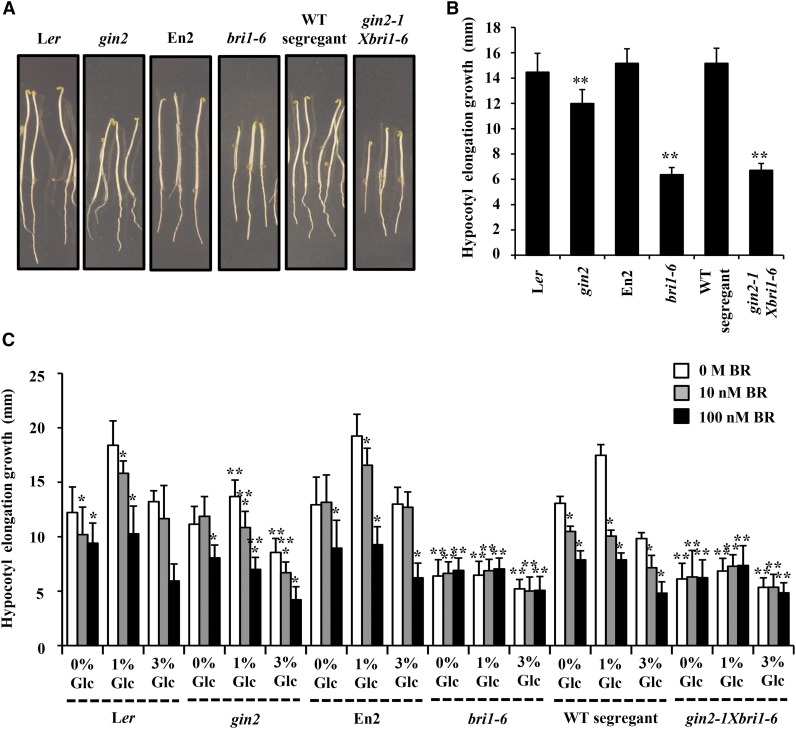
BRI1 Is Epistatic to HXK1 for the Regulation of Etiolated Hypocotyl Elongation Growth
To study the integration of BR and Glc signals at the genetic level, we used gin2-1bri1-6 double mutant (Gupta et al., 2015). As the ecotype background for gin2-1 mutant is Landsberg erecta (Ler) and for bri1-6 mutant is Enkheim (En2), wild-type segregants were screened and used as control for gin2-1bri1-6 double mutant. Phenotypically, the etiolated seedling of gin2-1bri1-6 double mutant was found to have compromised hypocotyl elongation growth similar to that of bri1-6 rather than the gin2 parent. This observation suggested BRI1 to be epistatic to HXK1 in regulation of etiolated hypocotyl elongation growth (Fig. 7, A and B). We then analyzed the Glc-BR sensitivity of gin2-1bri1-6 double mutant in terms of hypocotyl elongation growth regulation. In gin2-1 mutant, exogenous application of Glc and BR could still affect hypocotyl elongation, albeit to a significantly lesser extent than the wild type (Ler); however, the complete resistance of bri1-6 mutant and gin2-1bri1-6 double mutant toward Glc-BR regulation of hypocotyl elongation growth further proved that BRI1 is epistatic to HXK1 during BR-Glc regulation of etiolated hypocotyl elongation growth (Fig. 7C).
Figure 7.
BRI1 is epistatic to HXK1. A, Pictures showing phenotypic differences between 7-d-old etiolated seedlings of ecotypes (Ler and En2), wild-type (WT) segregant, Glc sensor mutant gin2-1, BR perception mutant bri1-6, and heterozygous double mutant gin2-1bri1-6 growing on one-half-strength MS medium. B, Difference between hypocotyl elongation growth of the 7-d-old etiolated wild type (Ler and En2), wild-type segregant, Glc sensor mutant gin2-1, BR perception mutant bri1-6, and homozygous double mutant gin2-1bri1-6 seedlings growing on one-half-strength MS medium. C, Analysis of Glc and BR sensitivity of the wild type (Ler and En2), wild-type segregant, Glc sensor mutant gin2-1, BR perception mutant bri1-6, and homozygous double mutant gin2-1bri1-6 seedlings in terms of hypocotyl elongation growth in the dark. The gin2-1bri1-6 double mutant displayed compromised etiolated hypocotyl growth as well as Glc and BR sensitivity similar to its bri1-6 parent rather than gin2-1 parent, suggesting that BRI1 is epistatic to HXK1 during BR-Glc regulation of etiolated seedling hypocotyl elongation growth. Data represent the average of values from 20 seedlings, and error bar represents se (Student’s t test, P < 0.001; *, control versus treatment; and **, wild type versus mutant).
Involvement of Auxin-Signaling Components in Controlling Hypocotyl Growth
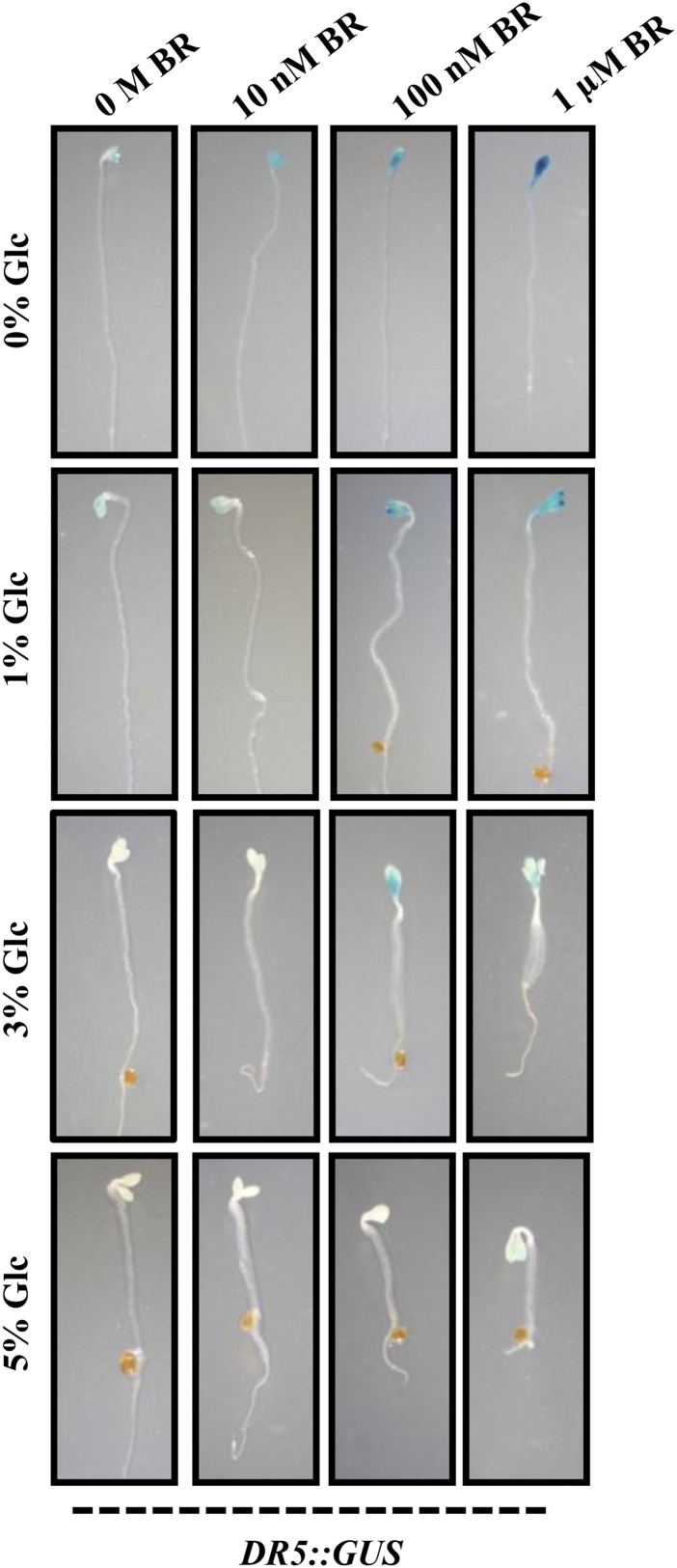
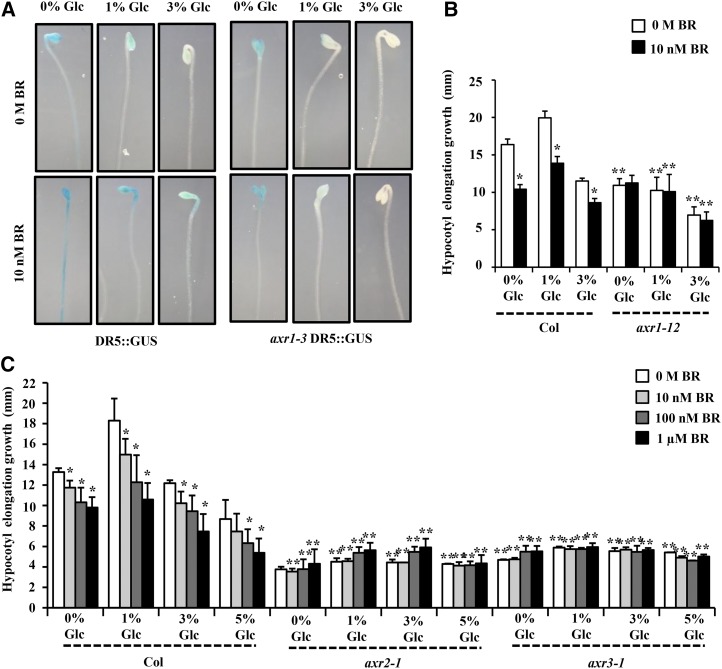
BR interacts with other hormones, especially with GA and auxin, to control early seedling growth and development (Nemhauser et al., 2004; Mouchel et al., 2006; Hardtke et al., 2007; Vert et al., 2008; Kuppusamy et al., 2009; Song et al., 2009). There are also various reports for sugar and auxin interaction during plant growth and development (Moore et al., 2003; Mishra et al., 2009; Mudgil et al., 2009; Booker et al., 2010). To determine any involvement of auxin machinery in Glc-BR control of etiolated hypocotyl elongation growth, we used well-established auxin reporter auxin responsive promoter DR5::GUS. Imbibed seeds were germinated and grown vertically in Glc-free medium or increasing concentrations of Glc (1%, 3%, and 5% [w/v]) containing one-half-strength MS medium supplemented with different concentrations of BR (0 m, 10 nm, 100 nm, and 1 µm) in the dark for 7 d, and the GUS activity was checked. BR was able to induce the DR5::GUS expression in Glc-free or low-Glc-containing medium (Fig. 8). However, at higher Glc concentrations (3% and 5% [w/v]), BR induction of GUS expression was significantly inhibited (Fig. 8). These results suggest that in etiolated seedlings, Glc and BR might involve an auxin-mediated mechanism to regulate hypocotyl elongation.
Figure 8.
BR-Glc regulation of DR5::GUS expression in etiolated seedlings. GUS expression analysis in hypocotyls and cotyledons of 7-d-old dark-grown DR5::GUS seedlings in absence or presence of different concentrations of Glc (0%, 1%, 3%, and 5% [w/v]) and/or BR (0 m, 10 nm, 100 nm, and 1 µm). BR induction of DR5::GUS expression was lost in presence of increased Glc concentration (3% and 5% [w/v]), suggesting, mainly, an antagonistic interaction between BR and Glc.
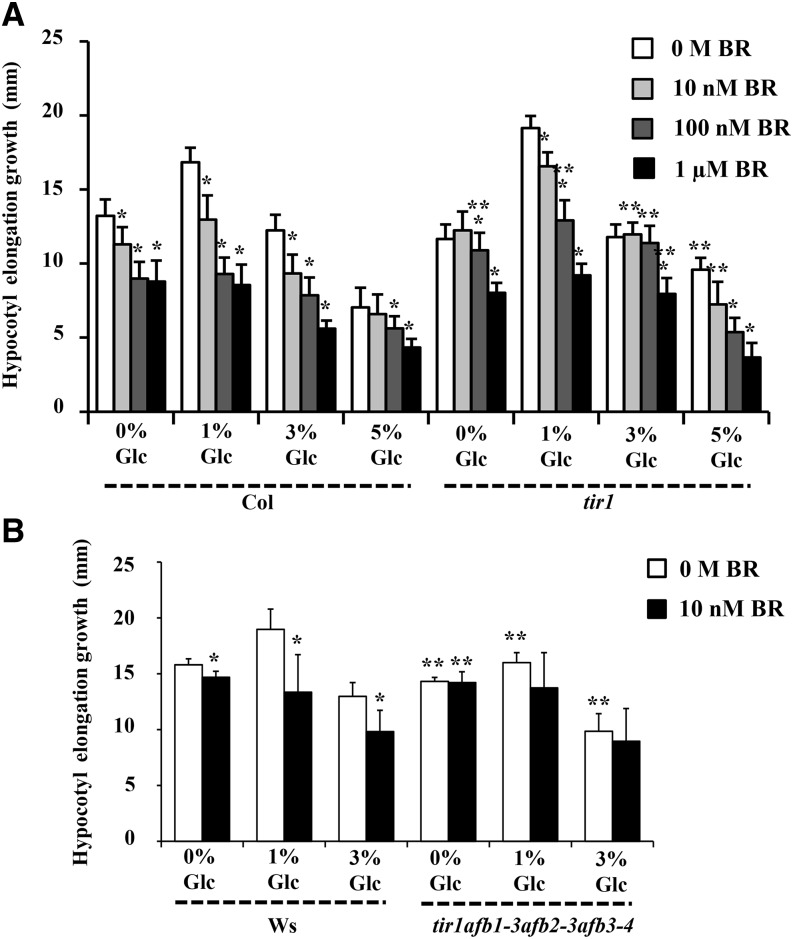
To further check the participation of a conventional auxin-signaling cascade during BR-Glc regulation of etiolated hypocotyl elongation growth, we analyzed BR/Glc sensitivity of mutants with altered auxin signaling or transport. The auxin-signaling mutants carrying null allele in transport inhibitor response1 (tir1; encoding the F-box protein TIR1) and all the tested auxin influx/efflux carrier mutants showed a nearly normal response toward Glc and BR for hypocotyl elongation growth control (Fig. 9A; Supplemental Fig. S10, A–C). However, combining loss-of- function alleles of tir1 with other members of this F-box gene family by using transport inhibitor resistant1 (tir1)/auxin-signaling f-box1 (afb1-3)/afb2-3/afb3-4 quadruple mutant (described as class-III mutants by Dharmasiri et al., 2005) could significantly reduce the BR sensitivity with respect to etiolated hypocotyl elongation growth (Fig. 9B; Supplemental Fig. S11). All these results suggest involvement of TIR1/AFB-dependent signaling during BR/Glc cross talk.
Figure 9.
Involvement of TIR1/AFB mediated auxin perception during Glc-BR for regulation of hypocotyl elongation in etiolated seedlings. Quantification of hypocotyl elongation growth in the 7-d-old etiolated wild type (Col-0 and Wassilewskija), the auxin-signaling mutant tir1 (A), and tir1afb1-3afb2-3afb3-4 quadruple mutant seedlings (B) growing on one-half-strength MS medium supplemented with independent as well as combined treatments of Glc and BR. The tir1 mutants displayed wild-type-like sensitivity for Glc and higher concentrations of BR treatments. However, the tir1afb1-3afb2-3afb3-4 quadruple mutant showed resistance for Glc and BR compared with the wild type in terms of etiolated hypocotyl elongation growth. These results indicate the involvement of TIR1/AFB-dependent auxin perception during BR-Glc cross talk. Values represent the average from two independent biological replicates, each having 20 seedlings, and error bars represent se. (Student’s t test, P < 0.001; *, control versus treatment; and **, wild type versus mutant).
A weak allele in auxin-resistant1-3 (axr1-3) showed a nearly normal response toward Glc and BR for hypocotyl elongation growth control (Supplemental Fig. S10D). However, the BR induction of DR5::GUS was strongly inhibited in axr1-3 mutant background (Fig. 10A). We also tested a strong axr1-12 mutant allele for Glc-BR response, and it showed compromised response in terms of hypocotyl elongation growth (Fig. 10B). Further, we used axr2-1/iaa7 and axr3-1/iaa17 gain-of-function mutants, and both axr2-1/iaa7 and axr3-1/iaa17 mutants were found highly resistant for different concentrations of Glc and BR for hypocotyl elongation growth control compared with the wild type (Fig. 10C). The mutation in these AUX/IAA repressors makes them stable such that they are not recognized by proteasomal machinery and become constitutively active. Collectively, our results suggest that an intact auxin-signaling pathway acts as a nodal point downstream to both Glc and BR and is required for optimal hypocotyl elongation growth of Arabidopsis etiolated seedlings.
Figure 10.
Involvement of auxin-signaling components in Glc-BR regulation of hypocotyl elongation in etiolated seedlings. A, The BR induction of DR5::GUS expression was inhibited in axr1-3 mutation background. Also, in axr1-3 mutant background, even lower concentrations of Glc were able to inhibit the DR5::GUS expression, suggesting the involvement of AXR1-mediated mechanisms for BR-Glc cross talk. B and C, Quantification of hypocotyl elongation growth in the 7-d-old etiolated wild type (Col-0) and auxin-signaling mutants axr1-12, axr2-1, and axr3-1 seedlings growing on one-half-strength MS medium supplemented with independent as well as combined treatments of Glc and BR in the dark. The auxin-signaling mutant axr1-12 seedlings were found to have significantly reduced sensitivity, whereas axr2-1 and axr3-1 mutants were found highly resistant for different concentrations of Glc and BR for regulation of etiolated hypocotyl elongation growth compared with the wild type. These results indicate the dependence of both BR and Glc action upon these auxin-signaling components for regulation of etiolated hypocotyl growth. Values represent the average from two independent biological replicates, each having at least 15 seedlings (except for axr1-12; data from two biological replicates, 10 seedlings each), and error bars represent se (Student’s t test, P < 0.001; *, control versus treatment; and **, wild type versus mutant).
DISCUSSION
Both sugars and BRs are fundamental to plants and regulate a number of similar processes. In the literature, only a few reports describe a possible sugar and BR interaction in plants (Szekeres et al., 1996; Smeekens, 1998; Goetz et al., 2000; Laxmi et al., 2004; Vicentini et al., 2009). Sugars and phytohormones regulate a wide range of genes at the transcriptional level. High levels of sugar induce the expression of genes involved in many key physiological processes, such as respiration, synthesis of polysaccharides, storage proteins, and pigments. Sugars are also known to induce the expression of defense response-related genes in plants. By contrast, sugar deprivation enhances the expression of genes involved in photosynthesis and resource remobilization, such as starch, lipid, and protein degradation (Yu et al., 1991; Koch, 1996; Ho et al., 2001). Some global gene expression studies in Arabidopsis seedlings have previously shown that sugar signaling could interact with hormones and other nutrient signaling networks at the genetic level to regulate expression of genes (Mishra et al., 2009). Similarly, BR has also been reported to regulate genes involved in various key processes in plants, such as photomorphogenesis, flowering, biotic and abiotic stress responses, etc. (Goda et al., 2002, 2004; Luo et al., 2010; Sun et al., 2010; Bai et al., 2012). Interactions of sugars with phytohormones such as ethylene, ABA, and auxin have already been established (Gibson, 2004; Mishra et al., 2009). To reveal the overlap between BR and Glc signals, an extensive whole-genome transcription profiling approach was employed to elucidate the extent of dependence between these two signaling pathways.
At the whole-genome level, Glc can broadly affect BR-regulated gene expression. BR alone could regulate 303 genes in dark-grown seedlings. A large number of genes have been found to be simultaneously regulated by both Glc and BR, with Glc alone being able to affect the expression of 71% of BR-regulated genes. Out of these BR and Glc-coregulated genes, 58% of genes were synergistic, while 42% of genes showed antagonistic regulation. Interestingly, Glc could also significantly affect induction or suppression of 85% of the BR-regulated genes. Fifty-seven percent of genes were regulated antagonistically, whereas 43% of genes were regulated synergistically in presence of Glc. Interestingly, presence of Glc can even affect those BR-regulated genes that showed no significant expression upon treatment with Glc alone. This indicates that Glc may also employ other mechanisms to regulate BR-regulated gene expression or a BR-dependent factor is required for sensitivity of these genes to Glc.
In microarray, Glc alone was found to affect expression of many important genes involved in BR biosynthesis, metabolism, perception, and signaling. Consequently, a number of BR-related and -regulated genes were found to be differentially regulated in presence of Glc. These observations altogether suggest that Glc may regulate BR-related responses by modulating both BR biosynthesis as well as BR signaling at a global scale. Similarly, BR induction of pTCH4:TCH4::GUS was abolished in presence of Glc in the medium.
A large number of genes regulated by BR and Glc encoded for proteins whose molecular and biological functions were unknown; however, many genes fell into groups with assigned functions. GO analysis revealed that a majority of BR-regulated genes were involved in developmental processes and stress responses. The BR-regulated genes include a large number of important gene families that include members of the AUX/IAA gene family, cytochrome p450, glucosyl hydrolase family proteins, GDSL-motif lipase/hydrolase family proteins, glutathione S-transferases, the Leu-rich repeat family, U-box domain-containing proteins, UDP-glucoronosyl/UDP-glucosyl transferase family proteins, xyloglucan endotransglycosylases, and peroxidase gene families. Collectively, all these genes are known to be involved in controlling morphogenesis, response to stress, response to abiotic or biotic stimulus, and developmental processes. Similarly, our microarray data also showed that BR significantly changed transcript abundance of members of various TF families. The enriched TF binding sites in promoters (1 kb) of BR- and Glc-regulated genes are either ABA responsive or biotic/abiotic stress responsive, further strengthening that Glc and BR may together control stress response in plants. Altogether, GO analysis, gene family enrichment analysis, TF analysis, and cis-regulatory element analysis of BR-regulated genes suggest that both BR and Glc regulate genes mainly belonging to either plant growth and development or the stress response category.
Glc and BR act antagonistically at low Glc concentration and synergistically at higher Glc concentrations for hypocotyl elongation growth regulation in dark-grown seedlings. Further, both BR and Glc were also found to regulate many key genes involved in hypocotyl growth and development in microarray. Glc receptor mutant gin2-1 was found to be less sensitive toward BR compared with the wild type at all the Glc concentrations tested. HXK1-independent pathway mutants have similar sensitivity toward Glc and BR for hypocotyl length regulation compared with the wild type. Therefore, for regulation of hypocotyl elongation, BR and Glc can interact via the AtHXK1-dependent pathway. However, we cannot deny the possible involvement of other factors, as the gin2-1 mutant did not show complete resistance and showed a compromised sensitivity toward Glc and BR regulation of etiolated hypocotyl elongation growth. BR biosynthesis and signaling mutant analysis suggested that both BR biosynthesis and signaling components are involved in Glc control of hypocotyl length. In BR biosynthesis mutants cpd and det2 and BR receptor mutant bri1-6, in which endogenous BR levels and/or signaling is low, Glc sensitivity for hypocotyl elongation growth was highly compromised. In ces-D, bzr1-1D, and bes1-D mutant seedlings in which endogenous BR levels and/or signaling is high, hypocotyl elongation growth was constitutively more at all Glc concentrations tested compared with the wild type. Further analysis of etiolated hypocotyl growth phenotypes in gin2-1bri1-6 double mutant confirmed the interaction between Glc and BR signals genetically and also proved that BRI1 is epistatic to HXK1 during BR-Glc regulation of etiolated hypocotyl elongation growth.
The functional importance of auxin signaling and transport interaction with both Glc and BR independently in controlling several developmental processes has already been documented before. Glc was able to inhibit BR induction of DR5::GUS expression, suggesting that BR and Glc work antagonistically to regulate auxin-related gene expression during dark-growth conditions. The SKP-Cullin-F-box (SCF) TIR1/AFB, AUX-IAA, and AUXIN-BINDING PROTEIN1 are the major receptor/coreceptor systems studied in context of auxin signaling. AXR1, a regulatory protein for SCF-mediated protein degradation, has previously been reported to confer altered sensitivity toward auxin, ethylene, methyl jasmonate, and cytokinin (Timpte et al., 1995; Tiryaki and Staswick, 2002). AUX/IAA genes are auxin-inducible genes, which encode for transcription regulators that repress the auxin-signaling pathway (Gray et al., 2001). Presence of auxin decreases the AUX/IAA protein levels through degradation via the TIR1/AFB-mediated proteasomal pathway. The phenotype analysis of auxin perception-defective mutants and gene expression results suggested involvement of TIR1/AFB-dependent signaling during BR/Glc cross talk. Analysis of Glc and BR sensitivity in mutants defective in auxin response/signaling further suggested that Glc and BR may converge at SCF-TIR1/AFB-AUX/IAA-mediated auxin-signaling machinery for controlling etiolated hypocotyl elongation growth. Altogether, our studies suggest that Glc and BR might interact via auxin-mediated mechanisms to regulate etiolated seedling development. Further, dissecting how precisely Glc and BR signals integrate with each other and with other environmental signals at tissue-specific developmental and temporal levels will enable us to better understand the mechanism of plant growth regulation in broader context.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The following seed stocks were obtained from the Arabidopsis Biological Resource Center at Ohio State University: gin2-1 (AT4G29130, CS6383); det2 (AT2G38050, CS6159); bri1-6 (AT4G39400, CS399); bak1-1 (AT4G33430, CS6125); bzr1-1D (AT1G75080, CS65987); bes1-D (AT1G19350, CS65988); tir1-1 (AT3G62980, CS3798); axr1-3 (AT1G05180, CS3075); axr1-3 DR5::GUS (AT1G05180, CS16705); axr1-12 (AT1G05180, CS3076); axr3-1 (AT1G04250, CS57504); and ethylene insensitive root1-1 (AT5G57090, CS8058). The axr2-1 (AT3G23050, CS3077) seed stock was obtained from the European Arabidopsis Stock Centre. The following lines were obtained from published sources: rgs1-1 and rgs1-2 (AT3G26090; Chen et al., 2003); gpa1-1, gpa1-2, and gpa1-3 (AT2G26300; Ullah et al., 2001); thf1-1 (AT2G20890; Huang et al., 2006); cpd (AT5G05690; Szekeres et al., 1996); ces-D (Poppenberger et al., 2011); pTCH4:TCH4::GUS (Xu et al., 1995); DR5::GUS (Ulmasov et al., 1997); pin-formed3-4 (pin3-4; AT1G70940; Friml et al., 2002); pin7-2 (AT1G23080; Friml et al., 2003); multidrug resistant1-1 (mdr1-1; AT3G28860; Noh et al., 2001); P-glycoprotein1-100 (AT2G36910; Lin and Wang, 2005); tir1/afb1-3/afb2-3/afb3-4 (AT3G62980/AT4G03190/AT3G26810/AT1G12820; Dharmasiri et al., 2005); and gin2-1bri1-6 (AT4G29130/AT4G39400; Gupta et al., 2015). All mutant lines were in Col-0 background, except the following: the gin2-1 mutant was in the Ler background; bri1-6 and bes1-D mutants were in the En2 background; the bak1-1, gpa1-1, gpa1-2, mdr1-1, mdr1-101, and tir1/afb1-3/afb2-3/afb3-4 mutants were derived from the Wassilewskija background. All chemicals were purchased from Sigma, except agar, which was purchased from Himedia, and BRZ, which was purchased from TCI Chemicals. Epibrassinolide was prepared as 10–2 m stock solution in 50% (v/v) ethanol; BRZ was prepared as 10–2 m stock solution in dimethyl sulfoxide; and 5-bromo-4-chloro-3-indolyl-β-d-GlcA was prepared as 100 mg L–1 stock solution in N,N-dimethylformamide (DMF). Seeds were surface sterilized and imbibed at 4°C for 48 h. Seed germination was carried out in a climate-controlled growth room under long-day conditions (16 h of light and 8 h of darkness, 80 µmol m–2 s–1 light intensity) at 22°C ± 2°C temperature. For the dark-grown seedlings, seeds on plates were first exposed to 12-h light (80 µmol m–2 s–1 light intensity) to stimulate germination; the plates were wrapped with two layers of aluminum foil and placed in the growth chamber. For the study of hypocotyl elongation, seeds were germinated and grown vertically on one-half-strength MS and 0.8% (w/v) agar containing medium with different concentrations of Glc and/or BR and grown vertically in a climate-controlled growth room (22°C ± 2°C) in the dark. In all experiments, plates were sealed with gas-permeable tape to avoid ethylene accumulation. All end point analyses were taken on day 7 unless otherwise specified, although the experiments were observed for a longer period.
Gene Expression Analysis
Plant tissue sample preparation.
Imbibed seeds were sown on square (120 × 120 mm) petri dishes containing one-half-strength MS medium supplemented with 1% (w/v) Suc and 0.8% (w/v) agar and grown vertically. For etiolated growth, seeds on plates were first exposed to 12 h of light to stimulate germination; the plates were wrapped with two layers of aluminum foil and placed in the growth chamber for all the treatments. Once the plant material was uniformly germinated, the experimental conditions were applied. Five-day-old uniformly grown etiolated seedlings were washed seven times with sterile water with last wash given by one-half-strength MS liquid medium without Suc to remove residual exogenous sugar. To deplete internal sugars, seedlings were further kept in sugar-free liquid one-half-strength MS medium for 24 h in the dark. All subsequent steps were performed in the dark, and the cultures were shaken at 140 rpm at 22°C. Briefly, seedlings were treated with 0% (w/v) Glc, 0% (w/v) Glc + 100 nm BR, 3% (w/v) Glc, and 3% (w/v) Glc + 100 nm BR containing liquid one-half-strength MS medium for 3 h in the dark. Afterward, seedlings were flash frozen in liquid nitrogen and stored at –80°C. The RNA was prepared from frozen tissue using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. The RNA was quantified and tested for quality before it was used for subsequent analyses.
Microarray Analysis
Three biological replicates were performed. After initial processing and quality assessments, total RNA from each sample was amplified and cyanine dye-labeled using Agilent’s One-Color Quick Amp Labeling Kit following the manufacturer’s protocols (version 6.5). After the labeling, the complementary RNA was cleaned and examined with the Nanodrop ND-2000 (Thermo Scientific). Equal amounts of cyanine dye-labeled complementary RNA (1.65 μg; for the one-color protocol) was hybridized to Arabidopsis (Arabidopsis thaliana) microarray slides (4x44K; Agilent) for 18 h at 65°C using Agilent’s Gene Expression hybridization kit. Washes were conducted as recommended by the manufacturer using Agilent’s gene expression wash pack. Arrays were scanned with an Agilent scanner (model no. G2505B). Spot intensities and other quality control features were extracted with Agilent’s Feature Extraction software (version 10.7.3.1). GeneSpring 11.5.1 software was used for the analysis of the expression data. The raw data from the biological replicate samples was normalized using the percentile shift summarization algorithm, and the signature lists of the significantly altered genes (P ≤ 0.005; fold change, ≥2) for each condition were generated using one-way ANOVA with Benjamini Hochberg False Discovery Rate in GeneSpring 11.5.1. Additional microarray data presentation and manipulation were assessed using Microsoft Excel. All data are MIAME compliant, and the raw data has been deposited in the ArrayExpress database through MIAMExpress (accession no. E–MEXP–3545).
Quantitative Real-Time PCR
For real-time PCR, first-strand complementary DNA (cDNA) was synthesized by reverse transcription using 2 µg of total RNA in 20 µL of reaction volume using high-capacity cDNA reverse transcription kit (Applied Biosystems). Diluted cDNA samples (5 µm) were used for quantitative reverse transcription-PCR analysis, and 5 µm of each primer was mixed with SYBR Green PCR master mix as per the manufacturer’s instructions. Primers for all the candidate genes were designed preferentially from the 3′ end of the gene using PRIMER EXPRESS (version 3.0; PE Applied Biosystems) with default parameters. The reaction was carried out in 96-well optical reaction plates (Applied Biosystems) using the ABI Prism 7900 HP Fast Real-Time PCR System (Applied Biosystems). To normalize the variance among samples, 18S ribosomal RNA was used as the internal control. The mRNA levels for each candidate gene in different samples were determined using the Delta Delta cycle threshold method (Livak and Schmittgen, 2001). Relative expression values were calculated after normalizing against the maximum expression value. The values represent the average of the two biological replicates (each with three technical replicates), and error bars present se. For all experiments, a Student’s t test with paired two-tailed distribution was used for statistical analysis. Primers used for real-time PCR are described in Supplemental Figure S12.
Measurement of Hypocotyl Elongation Growth and Apical Hook Phenotype Analysis
All end-point analyses were performed on day 7. For all the experiments, digital images of seedlings were captured using a Nikon Coolpix digital camera. Hypocotyl length of etiolated seedlings 7 d after treatment was measured using the ImageJ (http://rsb.info.nih.gov/ij/) program from National Institutes of Health. The apical hook phenotypes were observed under a Nikon SMZ1500 Stereo-Zoom microscope, and digital images of the seedlings were captured using a Nikon Coolpix digital camera connected with a Nikon SMZ1500 Stereo-Zoom microscope 3 d after treatment.
GUS Histochemical Staining
Imbibed seeds of pTCH4:TCH4::GUS, DR5::GUS, and axr1-3DR5::GUS were germinated and grown directly in Glc-free (0%) medium or increased Glc (1%, 3%, and 5% [w/v]) medium containing one-half-strength MS supplemented with or without BR (10 nm/100 nm/1 µm) and solidified with 0.8% (w/v) agar in a climate-controlled growth room for 7 d in darkness. GUS activities were then determined by incubating the seedlings at 37°C in a GUS staining solution (sodium phosphate buffer, pH 7, 0.1 m; K3Fe(CN)6, 0.5 mm; K4Fe(CN)6, 0.5 mm; EDTA, 50 mm; and 5-bromo-4-chloro-3-indolyl-β-d-GlcA, 1 mg mL–1) for 3 to 4 h. The seedlings were then kept in 70% (v/v) ethanol for the removal of chlorophyll. The seedlings were then observed under a Nikon SMZ1500 Stereo-Zoom microscope, and photographs were taken by a Nikon Coolpix digital camera connected with a Nikon SMZ1500 Stereo-Zoom microscope. The experiments were repeated at least twice, with each replicate having 10 seedlings, yielding similar results.
Statistical Analyses
All experiments reported in this study were performed at least three times, yielding similar results. All values reported in this work are averages from two independent biological replicates, each having at least 30 seedlings unless otherwise specified. For quantitative reverse transcription-PCR results, the values represent the average of the two biological replicates (each with three technical replicates). Error bars represent se. Statistical significance for all the experiments was evaluated using Microsoft Excel. For all experiments, statistical differences between both control/treatment and wild-type/mutant pairs were analyzed using Student’s t test evaluation with paired two-tailed distribution. P value cutoff was taken at P < 0.001, except where stated otherwise.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Functional categorization of BR- and Glc-regulated genes on the basis of GO cellular component, GO molecular function, and GO biological process.
Supplemental Figure S2. TF family enrichment analysis of BR- and Glc-regulated genes (1.5-fold, ±) in etiolated seedlings.
Supplemental Figure S3. Regulation of hypocotyl growth by Glc and BR in etiolated seedlings.
Supplemental Figure S4. Regulation of hypocotyl growth by Glc and BR in light-grown seedlings.
Supplemental Figure S5. Regulation of apical hook growth and development by Glc and BR in etiolated seedlings.
Supplemental Figure S6. Glc-BR regulation of hypocotyl growth in AtHXK1-dependennt Glc-signaling mutant gin2-1.
Supplemental Figure S7. Glc-BR control of hypocotyl length in AtHXK1-independent pathway mutants rgs1 and thf1.
Supplemental Figure S8. Glc-BR control of hypocotyl length in AtHXK1-independent pathway mutant gpa1.
Supplemental Figure S9. Glc regulation of hypocotyl growth in BR-signaling mutants.
Supplemental Figure S10. Glc-BR control of hypocotyl elongation growth in auxin transport/signaling-defective mutants.
Supplemental Figure S11. Glc-BR regulation of etiolated hypocotyl elongation growth in auxin perception mutant.
Supplemental Figure S12. List of primers used in this study.
Supplemental Table S1. BR-regulated genes.
Supplemental Table S2. BR-regulated genes up- and down-regulated by Glc alone.
Supplemental Table S3. Genes in which BR regulation is affected (2-fold more/less or lost) significantly in presence of Glc.
Supplemental Table S4. Genes involved in BR physiology.
Supplemental Table S5. BR- and Glc-regulated genes involved in various phytohormone responses.
Supplemental Table S6. BR-regulated genes involved in stress responses.
Supplemental Table S7. Cis-regulatory element analysis of promoters of BR-regulated genes.
Supplemental Table S8. BR-regulated genes involved in hypocotyl growth and development.
Supplementary Material
Acknowledgments
We thank the National Institute of Plant Genome Research Central Instrument facility (Real-Time PCR division) and Microarray facility for assistance.
Glossary
- BR
brassinosteroid
- Col-0
ecotype Columbia-0
- ABA
abscisic acid
- GO
Gene Ontology
- TF
transcription factor
- MS
Murashige and Skoog
- BRZ
brassinazole
- Ler
Landsberg erecta
- cDNA
complementary DNA
Footnotes
This work was supported by the Department of Science and Technology, Government of India (grant no. SR/FT/LS–102/2008), a National Institute of Plant Genome Research core grant, and the Council of Scientific and Industrial Research, India (research fellowships to A.G. and M.S.).
References
- Bai MY, Fan M, Oh E, Wang ZY (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker KS, Schwarz J, Garrett MB, Jones AM (2010) Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS ONE 5: e12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Gao Y, Jones AM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141: 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389: 338–350 [DOI] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Browning KS, Gallie DR, Krishna P (2002) Brassinosteroid functions to protect the translational machinery and heat-shock protein synthesis following thermal stress. Plant J 29: 681–691 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Chaudhary S, Dobinson KF, Krishna P (1999) Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol Biol 40: 333–342 [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP (2012) Sugars, signalling, and plant development. J Exp Bot 63: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. (2004) Sugar and phytohormone response pathways: navigating a signalling network. J Exp Bot 55: 253–264 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Roitsch T (2000) Tissue-specific induction of mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role of steroid hormones in assimilate partitioning. Plant J 22: 515–522 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gupta A, Singh M, Jones AM, Laxmi A (2012) Hypocotyl directional growth in Arabidopsis: a complex trait. Plant Physiol 159: 1463–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A (2015) Interaction between glucose and Brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol 168: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Dorcey E, Osmont KS, Sibout R (2007) Phytohormone collaboration: zooming in on auxin-brassinosteroid interactions. Trends Cell Biol 17: 485–492 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S, Chao Y, Tong W, Yu S (2001) Sugar coordinately and differentially regulates growth- and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husar S, Berthiller F, Fujioka S, Rozhon W, Khan M, Kalaivanan F, Elias L, Higgins GS, Li Y, Schuhmacher R, et al. (2011) Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364 [DOI] [PubMed] [Google Scholar]
- Kim TW, Lee SM, Joo SH, Yun HS, Lee Y, Kaufman PB, Kirakosyan A, Kim SH, Nam KH, Lee JS, et al. (2007) Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrate modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Koh S, Lee SC, Kim MK, Koh JH, Lee S, An G, Choe S, Kim SR (2007) T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65: 453–466 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Walcher CL, Nemhauser JL (2009) Cross-regulatory mechanisms in hormone signaling. Plant Mol Biol 69: 375–381 [DOI] [PubMed] [Google Scholar]
- Laxmi A, Paul LK, Peters JL, Khurana JP (2004) Arabidopsis constitutive photomorphogenic mutant, bls1, displays altered brassinosteroid response and sugar sensitivity. Plant Mol Biol 56: 185–201 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW (2005) Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H (2005) Two homologous ABC transport proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138: 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J, Altmann T, Müssig C (2006) Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67: 2232–2238 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M, et al. (2010) Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BS, Singh M, Aggrawal P, Laxmi A (2009) Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS ONE 4: e4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS (2006) BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM (2009) Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 21: 3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, et al. (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, Luschnig C, Fujioka S, Sieberer T (2011) CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J 30: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J (2008) Sugar sensing and signaling. The Arabidopsis Book 6: e0117, doi/10.1199/tab.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Sheen J (2005) Sugar sensing and signalling networks in plants. Biochem Soc Trans 33: 269–271 [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U, Köpke D, Altmann T, Müssig C (2002) Analysis of carbohydrate metabolism of CPD antisense plants and the brassinosteroid deficient cbb1 mutant. Plant Cell Environ 25: 783–791 [Google Scholar]
- Singh M, Gupta A, Laxmi A (2014) Glucose control of root growth direction in Arabidopsis thaliana. J Exp Bot 65: 2981–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. (1998) Sugar regulation of gene expression in plants. Curr Opin Plant Biol 1: 230–234 [DOI] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW (2009) Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol Plant 2: 755–772 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M (1995) The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J 8: 561–569 [DOI] [PubMed] [Google Scholar]
- Tiryaki I, Staswick PE (2002) An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol 130: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini R, Felix JdeM, Dornelas MC, Menossi M (2009) Characterization of a sugarcane (Saccharum spp.) gene homolog to the brassinosteroid insensitive1-associated receptor kinase 1 that is associated to sugar content. Plant Cell Rep 28: 481–491 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ (2009) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150: 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) Multiple sugar signal transduction pathways in plants. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu SM, Kuo YH, Sheu G, Sheu YJ, Liu LF (1991) Metabolic derepression of α-amylase gene expression in suspension-cultured cells of rice. J Biol Chem 266: 21131–21137 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.