Metabolomics analysis of P450 tandem gene double mutants leads to a new model for the defense-related tryptophan metabolic network.
Abstract
In Arabidopsis (Arabidopsis thaliana), a number of defense-related metabolites are synthesized via indole-3-acetonitrile (IAN), including camalexin and indole-3-carboxylic acid (ICOOH) derivatives. Cytochrome P450 71A13 (CYP71A13) is a key enzyme for camalexin biosynthesis and catalyzes the conversion of indole-3-acetaldoxime (IAOx) to IAN. The CYP71A13 gene is located in tandem with its close homolog CYP71A12, also encoding an IAOx dehydratase. However, for CYP71A12, indole-3-carbaldehyde and cyanide were identified as major reaction products. To clarify CYP71A12 function in vivo and to better understand IAN metabolism, we generated two cyp71a12 cyp71a13 double knockout mutant lines. CYP71A12-specific transcription activator-like effector nucleases were introduced into the cyp71a13 background, and very efficient somatic mutagenesis was achieved. We observed stable transmission of the cyp71a12 mutation to the following generations, which is a major challenge for targeted mutagenesis in Arabidopsis. In contrast to cyp71a13 plants, in which camalexin accumulation is partially reduced, double mutants synthesized only traces of camalexin, demonstrating that CYP71A12 contributes to camalexin biosynthesis in leaf tissue. A major role of CYP71A12 was identified for the inducible biosynthesis of ICOOH. Specifically, the ICOOH methyl ester was reduced to 12% of the wild-type level in AgNO3-challenged cyp71a12 leaves. In contrast, indole-3-carbaldehyde derivatives apparently are synthesized via alternative pathways, such as the degradation of indole glucosinolates. Based on these results, we present a model for this surprisingly complex metabolic network with multiple IAN sources and channeling of IAOx-derived IAN into camalexin biosynthesis. In conclusion, transcription activator-like effector nuclease-mediated mutation is a powerful tool for functional analysis of tandem genes in secondary metabolism.
In response to pathogens, cruciferous plants synthesize a large variety of Trp-derived phytoalexins, which are metabolically related to indole glucosinolates (Rauhut and Glawischnig, 2009; Pedras et al., 2011). Both classes of metabolites are important for defense against fungal pathogens (Bednarek et al., 2009; Clay et al., 2009; Pedras et al., 2011). In the biosynthesis of camalexin, the characteristic phytoalexin in Arabidopsis (Arabidopsis thaliana), Trp is converted to indole-3-acetaldoxime (IAOx) by CYP79B2 and CYP79B3 (Glawischnig et al., 2004). IAOx is dehydrated to indole-3-acetonitrile (IAN), oxidized, and conjugated with glutathione (Nafisi et al., 2007; Parisy et al., 2007; Böttcher et al., 2009; Su et al., 2011). From this glutathione conjugate (GS-IAN), a Cys conjugate [Cys(IAN)] is formed, involving γ-GLUTAMYL PEPTIDASE1 (GGP1) and GGP3 (Geu-Flores et al., 2011). Cys(IAN) is then converted to camalexin by the unique bifunctional cytochrome P450 CYP71B15/PAD3 (Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009). The cyp71b15/pad3 mutant synthesizes only traces (typically 1% of the wild-type level) of camalexin (Glazebrook and Ausubel, 1994), largely independent of the applied stimulus triggering its biosynthesis.
CYP71A13 is highly transcriptionally coregulated with CYP71B15/PAD3 and, therefore, was a clear candidate for involvement in camalexin biosynthesis. A cyp71a13 mutant showed strong reduction in camalexin formation in response to AgNO3, Alternaria brassicicola, and Pseudomonas syringae in leaves, but in all cases, a significant amount of camalexin was still synthesized (Nafisi et al., 2007). CYP71A13 shows 89% identity on the amino acid level compared with CYP71A12, and the corresponding genes are located as tandem copies on chromosome 2. CYP71A13, expressed in Escherichia coli, converted IAOx to IAN in vitro (Nafisi et al., 2007). CYP71A12 could partially functionally replace CYP71A13 in a Nicotiana benthamiana expression system (Møldrup et al., 2013) and also catalyzed the formation of IAN and indole-3-carbaldehyde (ICHO) from IAOx in vitro (Klein et al., 2013). It is involved in camalexin biosynthesis in roots in response to Flg22 treatment (Millet et al., 2010). The extent to which CYP71A12 also plays a role in camalexin biosynthesis in leaves remained unclear. A third homolog, CYP71A18, shares 87% and 85% identity on the amino acid level to CYP71A12 and CYP71A13, respectively. It is expressed very weakly in leaf tissue also in response to pathogen infection (http://bbc.botany.utoronto.ca/efp). Its biological function is unclear.
In addition to camalexin, derivatives of ICHO and indole-3-carboxylic acid (ICOOH) are synthesized in Arabidopsis in substantial quantities (Hagemeier et al., 2001; Bednarek et al., 2005; Böttcher et al., 2009, 2014; Iven et al., 2012). In response to AgNO3 treatment, the total molar amount of these derivatives was similar to that of camalexin. Analysis of cyp79b2 cyp79b3 mutants demonstrated that, similar to camalexin, ICHO/ICOOH derivatives are synthesized from IAOx (Böttcher et al., 2009, 2014). In addition, incorporation studies suggest IAN as a putative biosynthetic precursor. Consequently, here, we address the extent to which CYP71A12 and/or CYP71A13 are important for the biosynthesis of these metabolites in leaves.
Reverse genetics in Arabidopsis typically relies on transfer DNA- or transposon-induced mutant alleles, and mutational events from distinct plants can be merged into plant lines carrying multiple mutations following Mendelian genetics (Bolle et al., 2011). The genetic versatility of plant secondary metabolism has been accelerated by gene duplication events, resulting in functionally redundant tandemly arranged gene copies (Hofberger et al., 2013). However, due to the lack of recombination, it is very unlikely that distinct mutational events of tandemly arranged genes are combined into a higher order mutant plant. Recently developed approaches for targeted genome editing offer a solution to this problem but have thus far not been applied to unravel plant secondary metabolic pathways. We used TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASEs (TALENs; Christian et al., 2010; Joung and Sander, 2013) to create stable cyp71a12 cyp71a13 double knockout lines. These double mutants synthesized only traces of camalexin, demonstrating that, in addition to CYP71A13, CYP71A12 is involved in camalexin biosynthesis in leaves. Based on a detailed metabolite analysis and on the characteristics of the corresponding enzymes, a differential function for CYP71A12 and CYP71A13 in the biosynthesis of other IAN-derived metabolites was established. This work demonstrates that targeted genome editing eliminates the limitations of classical genetic approaches and breaks ground for the elucidation of plant secondary metabolic pathways by reverse genetic approaches.
RESULTS
Enzymatic Parameters of CYP71A12 and CYP71A13
Previously, it was shown that recombinant CYP71A13 and CYP71A12 convert IAOx to IAN (Nafisi et al., 2007; Klein et al., 2013). In the presence of thiol donors such as Cys or glutathione, CYP71A13 produces Cys(IAN) or GS-IAN in vitro as a side product, while CYP71A12 generates ICHO as a side product (Klein et al., 2013). Here, we studied the enzymatic characteristics of CYP71A12 and CYP71A13, expressed in yeast (Saccharomyces cerevisiae), to understand their functional differences in more detail (Fig. 1). When CYP71A13-containing microsomes were incubated with IAOx and NADPH, IAN was synthesized (Supplemental Fig. S1). In the vector control, no IAN formation was observed. When glutathione was added, formation of GS-IAN and traces of ICHO were observed, as reported previously by Klein et al. (2013). The amount of GS-IAN synthesized varied strongly between enzyme preparations (2.7%–21.5% [molar] GS-IAN of total product; n = 3), indicating the influence of yeast proteins on the product spectrum. For CYP71A12, after 30 min of enzymatic reaction, ICHO was the major product of the IAOx turnover independent of reduced glutathione addition (60% ± 10% [molar] ICHO of total product; n = 4). In addition, NADPH-dependent turnover of IAN to ICHO accompanied by a release of cyanide was detected for CYP71A12 (Supplemental Fig. S1), as observed for CYP71B6 (Böttcher et al., 2014). In conclusion, CYP71A12 consecutively catalyzes a CYP71A13-type and a CYP71B6-type reaction.
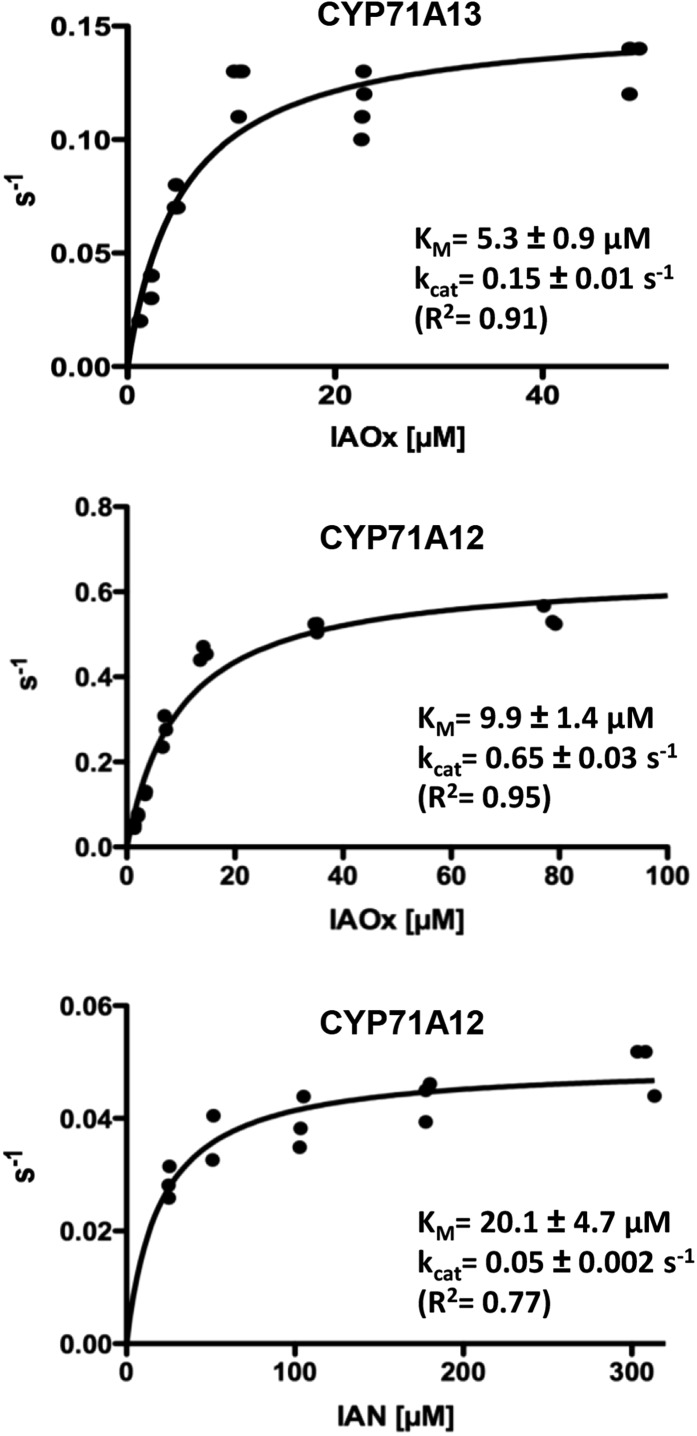
Figure 1.
Enzymatic parameters of CYP71A12 with IAOx and IAN as substrates and of CYP71A13 with IAOx as substrate. All turnover rates (s−1) were calculated based on P450 quantification (Supplemental Fig. S1). Data points represent turnover rates, calculated based on product quantification after individual enzymatic conversions.
Concentration of active cytochrome P450 was determined by carbon monoxide differential spectroscopy (Supplemental Fig. S1), and the kinetic parameters of CYP71A12 and CYP71A13 were determined (Fig. 1). For the reaction with IAOx, catalytic efficiency was approximately 0.066 µm−1 s−1 for CYP71A12 and approximately 0.029 µm−1 s−1 for CYP71A13. Besides the limitations regarding comparisons between catalytic efficiencies of different enzymes (Eisenthal et al., 2007), it is clear that both enzymes are efficiently dehydrating IAOx. The catalytic efficiency for the turnover of IAN by CYP71A12 was approximately 0.001 µm−1 s−1, indicating that IAOx is preferred over IAN as a substrate of CYP71A12.
Generation and Analysis of cyp71a13 Plants Harboring CYP71A12-Specific TALENs
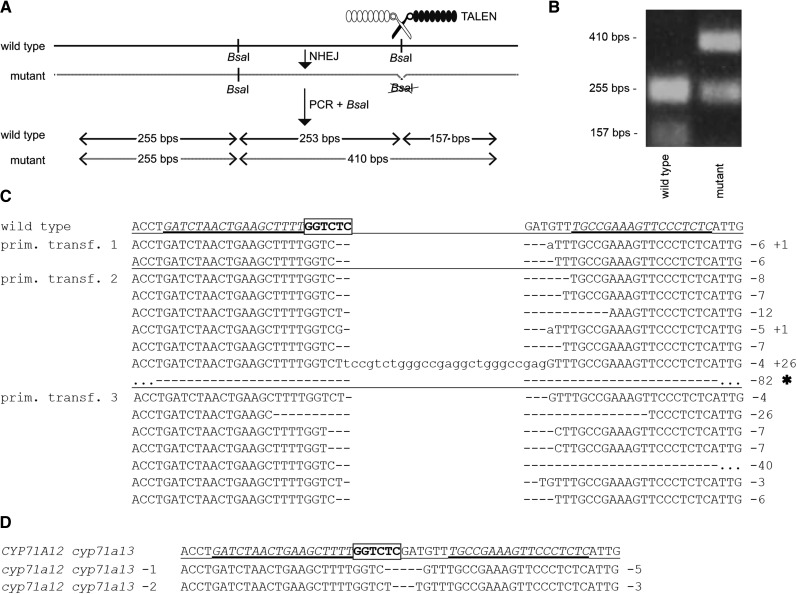
In TALEN-mediated genome editing, two distinct TALEN proteins, each containing a DNA-binding domain and a FokI cleavage domain, bind on opposite sites of a given cleavage site. Upon TALEN-induced cleavage, the strands are rejoined by nonhomologous end joining repair, which typically results in small deletions (Joung and Sander, 2013). To simplify the identification of plants with mutations, the cleavage sites are positioned in such a way that the TALEN-induced deletions cause the loss of endonuclease recognition sites (Hoshaw et al., 2010). We aimed to induce mutations at the CYP71A12 gene and generated two TALENs that bind on opposite sites of a BsaI endonuclease recognition site that is present within the CYP71A12 coding sequence. Loss of this site was monitored by BsaI-based cleaved-amplified polymorphic sequence (CAPS) marker analysis (Fig. 2, A and B).
Figure 2.
Somatic mutagenesis and inheritance of TALEN-mediated mutations in CYP71A12. A, Schematic representation of the analysis of targeted mutagenesis; the TALEN-binding site targeting one of two BsaI sites is indicated. NHEJ, Nonhomologous end joining. B, Representative CAPS analysis for a wild-type plant and a homozygous mutant plant. C and D, Analysis of TALEN-induced mutations in CYP71A12. The sequence of the CYP71A12 wild-type allele with TALEN-binding sites is in italic letters, and the targeted BsaI restriction site is in boldface; insertions are in lowercase letters, and deletions are indicated as dashes. C, Somatic events detected. D, Stable lines generated. prim. transf., Primary transformant; *, of the 82-bp deletion event, only 56 deleted bp are indicated.
These CYP71A12-specific TALENs were transformed into cyp71a13-1 knockout mutants with the aim to generate cyp71a12 cyp71a13-1 double mutants. Using primers specific for the FokI domain, PCR was carried out on seven primary transformants and confirmed the presence of the TALEN coding sequence. A CYP71A12-specific CAPS marker assay indicated that three of the seven tested plants indeed contain the desired mutation at the CYP71A12 locus. For each of these three primary transformants, the PCR product was cloned, and random clones were analyzed via BsaI digestion. A variety of mutant alleles in CYP71A12 were detected by sequence analysis (Fig. 2C). Notably, among 10 random clones for primary transformant 3, no wild-type sequence was detected, whereas for primary transformants 2 and 1, 20% and 50% of the clones, respectively, contained a wild-type sequence. This demonstrates efficient TALEN activity especially in plant 3.
Inheritance of TALEN-Induced cyp71a12 Mutations
T1, T2, and T3 plants of primary transformant 3 were analyzed for the presence of TALEN constructs using FokI-specific primers as indicators for a transgenic line. The presence of TALEN sequence segregated 44:4 for T2 and 9:1 for T3 (transgenic to wild-type plants).
A total of 150 T2 plants of each line were screened for TALEN-induced mutation in the targeted BsaI restriction site in the CYP71A12 coding sequence. For plants 1 and 2, none of the progeny showed evidence for mutation of this BsaI site in CYP71A12, indicating that all progeny plants are homozygous for the CYP71A12 wild-type allele. In contrast, for line 3, 19% of the T2 plants showed amplicons that lack the targeted BsaI site, indicating that they have homozygous or transheterozygous cyp71a12 mutant alleles. In 21% of the T2 plants, the amplicons were only partially cleaved by BsaI, indicating that the plants are probably heterozygous. In 60% of the T2 plants, the informative BsaI restriction site was cleaved, indicating the presence of a CYP71A12 wild-type allele.
Sequence analysis determined two distinct CYP71A12 mutant alleles, carrying a 5-bp and a 3-bp deletion, respectively. The latter one causes a loss of Asp-488. Interestingly, only this 3-bp deletion allele was among the 16 characterized somatic events (Fig. 2). One T2 plant lacking the TALEN gene and homozygous for the 5-bp deletion allele was identified (cyp71a12 cyp71a13-1). In the third generation, also a nontransgenic homozygous plant carrying the 3-bp deletion was obtained (cyp71a12 cyp71a13-2). The genotype was confirmed in the T3 and T4 generations, which were analyzed for metabolic phenotypes.
With respect to potential off targets of the TALEN pair and resulting unwanted mutations, the genomic sequences of CYP71A18 (with coding sequence 91% identical to CYP71A12) and CYP71B15/PAD3 (related with respect to the metabolic phenotype) were checked for their integrity, and no change in sequence was observed.
cyp71a12 cyp71a13 Double Mutants Are Camalexin Deficient
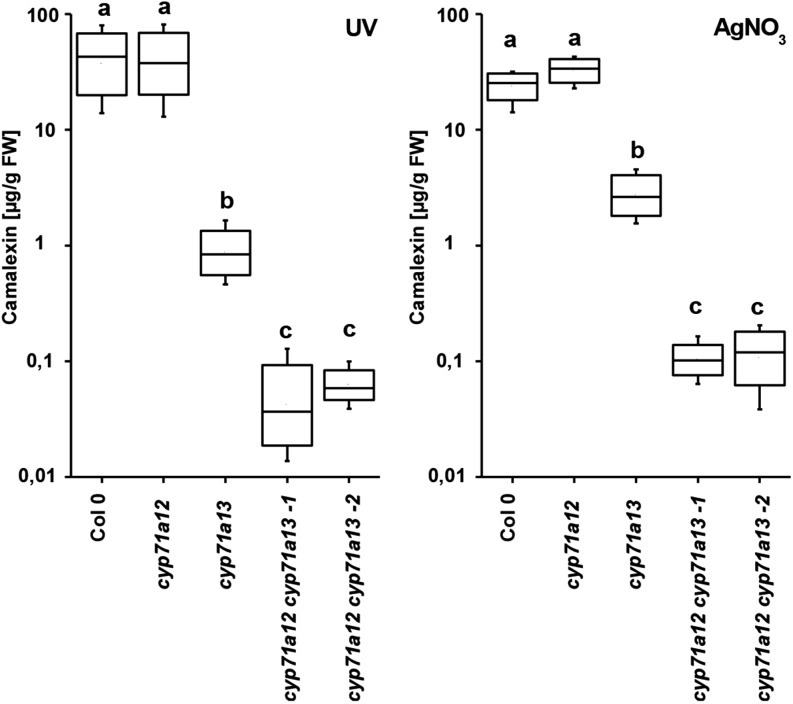
We analyzed camalexin levels in rosette leaves of 6-week-old plants of cyp71a12 and cyp71a13 single knockout and cyp71a12 cyp71a13 double knockout mutants in response to AgNO3 and UV irradiation (Fig. 3). For cyp71a12, no significant differences in comparison with the wild type were detected. In cyp71a13, we determined approximately 2.2% of wild-type camalexin level in response to UV irradiation and approximately 12% in response to AgNO3, similar to previous observations (Nafisi et al., 2007). Camalexin concentration in the cyp71a12 cyp71a13 double mutant was less than approximately 0.15% and 0.5% of the wild-type level for UV irradiation and AgNO3 treatment, respectively. In comparison with cyp71a13, camalexin concentrations were significantly reduced in cyp71a12 cyp71a13 double mutants for both treatments. In summary, the strongly reduced camalexin level in the cyp71a12 cyp71a13 double mutant as compared with the cyp71a13 single mutant suggests that CYP71A12 contributes to camalexin biosynthesis in leaf tissue.
Figure 3.
Camalexin quantification. Camalexin concentrations in response to AgNO3 and UV light were determined in single and double mutants of cyp71a12 and cyp71a13 (n = 10). Different letters indicate significant differences according to ANOVA (Scheffé’s test; P < 0.05). FW, Fresh weight.
Metabolite Profiling of cyp71a12 cyp71a13 Indicates Multiple Sources for IAN
Previously, we identified the spectrum of compounds that derive from IAOx provided by CYP79B2 and CYP79B3 (Böttcher et al., 2009). Here, we performed targeted metabolite profiling for these compounds in a cyp79b2 cyp79b3 double mutant and in single and double mutants of cyp71a12 and cyp71a13 (Fig. 4; Supplemental Table S1). In accordance with previous findings (Glawischnig et al., 2004; Böttcher et al., 2009), cyp79b2 cyp79b3 leaves were deficient in camalexin and ICHO/ICOOH derivatives. Interestingly, irrespective of the treatment, we observed strong accumulation of Trp in cyp79b2 cyp79b3 in comparison with the wild type, consistent with the lack of major Trp sinks in this line (Fig. 4E).
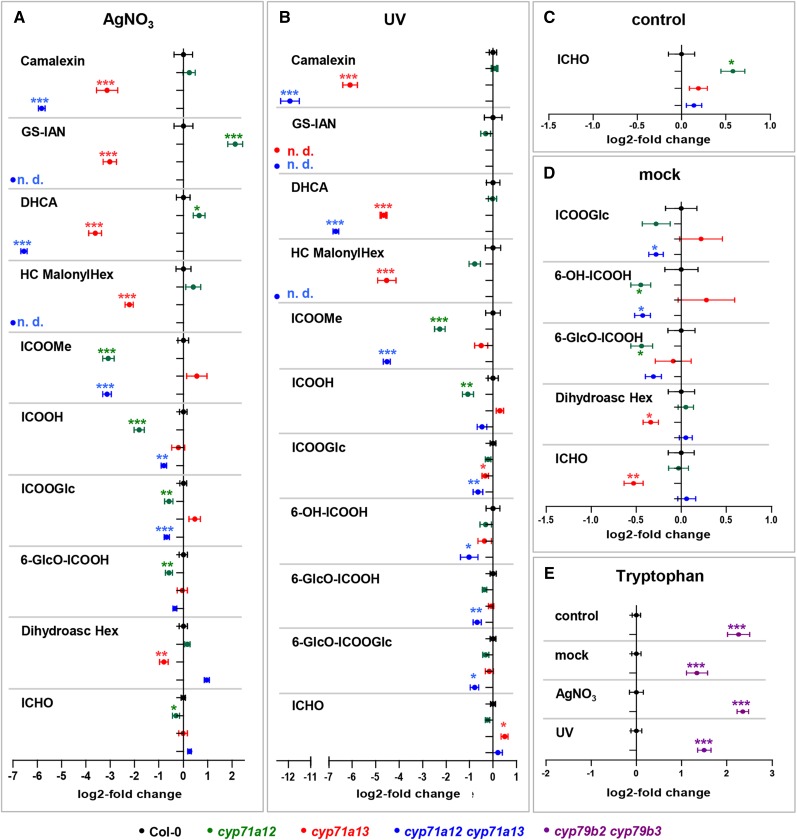
Figure 4.
Metabolomics analysis. A to D, Metabolites in rosette leaves of wild-type (Columbia-0 [Col-0]; black), cyp71a12 (green), cyp71a13 (red), and cyp71a12 cyp71a13 (blue) plants, 24 h after detachment and spraying with AgNO3 (A), 22 h after detachment and 2 h of UV irradiation (B), untreated (C), or 24 h after detachment and spraying with water (D). Characterized metabolites for which significant differences in cyp71a12, cyp71a13, or cyp71a12 cyp71a13 in comparison with the wild type were observed. The full data set is presented in Supplemental Table S1. Error bars indicate se (n = 9–11, except n = 20 for cyp71a12 cyp71a13 in A and D). DHCA, Dihydrocamalexic acid; HC MalonylHex, hydroxycamalexin malonylhexoside; ICOOGlc, Glc ester of ICOOH; 6-OH-ICOOH, 6-hydroxyindole-3-carboxylic acid; 6-GlcO-ICOOH, 6-hydroxyindole-3-carboxylic acid 6-O-β-d-glucoside. E, Relative quantification of Trp in Col-0 (black) and cyp79b2 cyp79b3 (violet) in the data sets denoted. Significance analysis of differences between the wild type and mutant was performed by two-tailed Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001).
In order to elucidate the role of CYP71A12 and CYP71A13 in the biosynthesis of camalexin precursors and of soluble ICHO/ICOOH derivatives, their accumulation levels were determined relative to the wild type under four different conditions: detached leaves sprayed with AgNO3 or water (mock) and incubated for 24 h, detached leaves irradiated with UV light for 2 h and incubated for 22 h, and untreated leaves (control). Irrespective of the treatment, the camalexin level was strongly reduced in cyp71a13 and was nearly absent in cyp71a12 cyp71a13 (Fig. 4, A and B), consistent with our targeted absolute quantification (Fig. 3). Similarly, significantly reduced levels were detected for the biosynthetic intermediates GS-IAN and dihydrocamalexic acid as well as for a hydroxycamalexin malonylhexoside representing the major camalexin metabolite (Fig. 4). This demonstrates that the synthesis of GS-IAN as a biosynthetic precursor of camalexin is dependent on CYP71A12/A13.
Among the analyzed ICOOH derivatives, the methyl ester of ICOOH (ICOOMe) was strongly and significantly reduced in cyp71a12 in response to UV irradiation (21% of the wild-type level; P = 3.6E−4) and AgNO3 (12% of the wild-type level; P = 2.3E−5; Fig. 4, A and B). For cyp71a12 cyp71a13, we detected 4.3% (P = 5.2E−5) and 11% (P = 1.8E−9) of the wild-type level for UV and AgNO3 treatment, respectively. In conclusion, CYP71A12 is important for the biosynthesis of this ICOOH derivative, which is de novo formed in response to both stress applications but below the detection limit in mock-treated and control leaves. For a number of other ICOOH derivatives, which in contrast to ICOOMe constitutively accumulate already in nontreated leaf tissue, we observed significantly reduced levels (Fig. 4), although to a much lower degree and not necessarily in all data sets. For example, the level of the Glc ester of ICOOH, which is a major compound, was reduced to 64% (P = 5.4E−3) and 68% (P = 1.1E−4) of the wild-type level in cyp71a12 cyp71a13, respectively, in UV- and AgNO3-challenged leaves.
In contrast, we did not observe any significant reduction of ICHO derivatives in cyp71a12 cyp71a13, although exogenously applied IAN is converted to ICHO derivatives in vivo (Böttcher et al., 2009, 2014). IAN synthesized by CYP71A12/A13 is not a major precursor of these metabolites.
DISCUSSION
TALEN-Induced Mutations Are Somatically Frequent But Rarely Inherited
For the applied TALEN pair, introduction of a mutation into CYP71A12 was very efficient in leaf tissue. For several primary transformants, we have observed somatic apparently transheterozygous mutations, with the wild-type allele being underrepresented. We detected 12 different mutated alleles: nine of them were deletions and three were insertion/deletion combinations.
In leaves of the primary transformant from which stable mutant alleles were transmitted, we did not detect a wild-type allele, indicating that very efficient somatic TALEN activity is a prerequisite for generating heritable lines, as observed by Christian et al. (2013). Most of the somatic events were not inherited, in accordance with other attempts to generate stable mutant lines via TALEN technology (Christian et al., 2013). This indicated that TALENs are mostly inactive in germline tissue. Whether this is due to them being expressed under the control of the 35S promoter remains to be investigated. Once a novel cyp71a12 allele was transmitted to the T2 generation, it was inherited in a Mendelian fashion, freely segregating from the TALEN transgene. Consequently, homozygous cyp71a12 cyp71a13 T2 plants were identified that did not carry the TALEN construct.
CYP71A12 Functionally Overlaps with CYP71A13 But Plays a Role in the Formation of ICOOH Derivatives in Leaf Tissue
CYP71A12 catalyzed two consecutive reactions, the dehydration of IAOx to IAN, which is then further converted to ICHO and cyanide (Klein et al., 2013; this study). These two activities were shown for CYP71A13 and CYP71B6, respectively (Nafisi et al., 2007; Böttcher et al., 2014), suggesting some genetic redundancy for these steps. Based on the comparison of cyp71a13 and cyp71a12 cyp71a13 phenotypes, the contribution of CYP71A12 to camalexin formation in leaves is significant but minor in relation to CYP71A13. In contrast, we observed a strong reduction of ICOOMe in cyp71a12, which was not further enhanced in the double knockout, demonstrating that CYP71A12 is important for the biosynthesis of this compound (Fig. 4, A and B). To a lesser extent, we also detected a contribution of CYP71A12 to the biosynthesis of ICOOH, Glc ester of ICOOH, 6-hydroxyindole-3-carboxylic acid, and 6-hydroxyindole-3-carboxylic acid 6-O-β-d-glucoside (Fig. 4, A, B, and D). We conclude that the biosynthesis of inducible ICOOH derivatives is the major biological function of CYP71A12 in leaves.
Metabolite Profiles of cyp71a12 cyp71a13 Double Mutants Suggest Multiple IAN Sources and Metabolite Channeling in Camalexin Biosynthesis
In cyp71a12 cyp71a13 double mutants, only traces of camalexin are synthesized, showing that dehydration of IAOx by CYP71A12 and CYP71A13 is essential for IAN synthesis as a camalexin precursor. This phenotype was observed when a 5-bp deletion allele of cyp71a12 was present, but also for a 3-bp deletion allele, resulting in the deletion of Asp-488. This amino acid, conserved in the CYP71A12/A13/A18 branch, might be essential for enzymatic function, or its deletion might destroy protein structure.
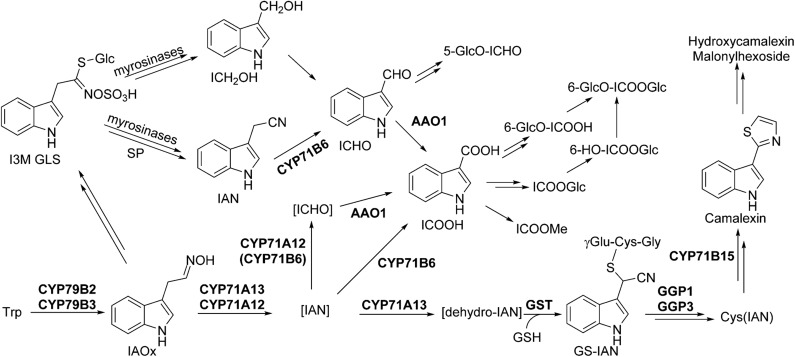
For ICOOH derivatives, smaller changes than for camalexin have been observed in comparison with the wild type, and the level of ICHO derivatives was essentially unaffected. The known ICHO/ICOOH biosynthetic genes CYP71B6 and Aldehyde Oxidase1 (AAO1) are to some extent coregulated with the camalexin biosynthetic genes CYP71A13 and CYP71B15 (Böttcher et al., 2014), indicating that different timing of protein expression and different leaf cell types for the two biosynthetic routes are unlikely to explain this observation. Moreover, the known enzymes of camalexin and ICHO/ICOOH derivative biosynthesis are cytosolic or endoplasmic reticulum bound with catalytic activity on the cytosolic side, so both processes occur in the same subcellular compartment. Therefore, for camalexin, ICOOMe, and other ICOOH derivative biosynthesis, we propose a different degree of exchange of IAN synthesized as product/intermediate of the CYP71A12/A13 reaction, with an IAN pool derived from glucosinolate degradation (Fig. 5). Possibly, the glucosinolate degradation product indole-3-carbinol (Agerbirk et al., 2008) is the major source for ICHO derivatives.
Figure 5.
Model for the biosynthetic pathways of Trp-derived secondary metabolites in Arabidopsis. We propose pools of free IAN and ICHO derived from glucosinolate degradation and, in addition, IAN and ICHO as channeled intermediates of camalexin and ICOOH biosynthesis. Enzyme functions are denoted. Square brackets indicate proposed channeling of intermediates, and double arrows indicate multiple steps. ICOOGlc, Glc ester of ICOOH; I3M GLS, indole-3-methyl glucosinolate; SP, specifier protein; 6-OH-ICOOH, 6-hydroxyindole-3-carboxylic acid; 6-GlcO-ICOOH, 6-hydroxyindole-3-carboxylic acid 6-O-β-d-glucoside.
Most likely, IAN synthesized by CYP71A12 and CYP71A13 during the course of camalexin biosynthesis is directly channeled. In the presence of glutathione, GS-IAN is a minor product of IAN turnover by CYP71A13 in vitro and was not detected after turnover by CYP71A12. However, in planta, specific interaction with glutathione S-transferases, GGP1 (Geu-Flores et al., 2011), and CYP71B15 (Glazebrook and Ausubel, 1994; Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009) might drive efficient IAN conversion. Also, other cytochrome P450 enzymes could play a role in activating IAN in the camalexin biosynthetic pathway, functionally overlapping with CYP71A13, as indicated by the complementation of camalexin deficiency in cyp71a13 by the addition of IAN (Nafisi et al., 2007).
In addition, we propose a free cellular IAN pool, which could be fed by the degradation of indole glucosinolates (de Vos et al., 2008), from which ICHO derivatives and subsequently ICOOH derivatives are synthesized, involving CYP71B6. The binding constant of IAN and CYP71B6 is very low (Böttcher et al., 2014), consistent with the fact that IAN did not accumulate in leaves in response to AgNO3, UV irradiation, or Phytophthora spp. infection (Böttcher et al., 2009, 2014; this study). As IAN can act as an auxin (indole-3-acetic acid) precursor (Bartling et al., 1992; Normanly et al., 1997; Kriechbaumer et al., 2007), avoiding IAN accumulation ensures that the production of IAN-derived defense compounds can be induced without effects on the indole-3-acetic acid pool, which could counteract defense responses.
MATERIALS AND METHODS
Yeast Expression and Enzymatic Analysis of CYP71A12 and CYP71A13
IAOx was synthesized according to Ahmad et al. (1960) with the following modifications: 5 mg of indole-3-acetaldehyde (Sigma-Aldrich) was suspended in 400 µL of 1 m sodium carbonate. The suspension was extracted three times with 500 µL of ethyl acetate. The organic phases were combined, 500 µL of 0.1 m hydroxylamine was added, and after 1 h of shaking, the organic phase was removed, dried over Na2SO4, and evaporated to dryness.
The CYP71A12 coding sequence was amplified from the complementary DNA clone R21987 (Arabidopsis Biological Resource Center) using the primers 5′-GGATTAAUAATGATGTCTAATATTCAAGAAATGGAAATGGATATTG-3′ and 5′-GGGTTAAUTTAAATAACGGAAGATGGAAATG-3′. CYP71A13 was amplified from total Arabidopsis (Arabidopsis thaliana) Col-0 leaf complementary DNA using the primers 5′-GGATTAAUAATGATGTCTAATATTCAAGAAATGGAAATGGATATTG-3′ and 5′-GGGTTAAUTTACACAACCGAAGATGGAAATG-3′. The PCR fragments were cloned into pYEDP60u via USER technology (Nour-Eldin et al., 2006) and transformed into yeast (Saccharomyces cerevisiae) WAT11 (Pompon et al., 1996). Yeast microsomes were prepared as described by Schuhegger et al. (2006). Cyanide derivatization and HPLC analysis were performed as described previously (Böttcher et al., 2009). The concentration of active cytochrome P450 was determined by carbon monoxide differential spectroscopy (Omura and Sato, 1964). For analysis of the enzymatic parameters, reactions were performed with 18 µg of microsomal protein (representing approximately 27 ng of CYP71A12 or 59 ng of CYP71A13) in 100 µL of potassium phosphate buffer (20 mm, pH 7.5) for 30 min and then stopped by adding 200 µL of methanol. IAOx, IAN, and ICHO were analyzed by reverse-phase HPLC (MultoHigh 100 RP18, 5-µm particle size; Göhler Analytik) as follows: flow rate of 1 mL min−1; solvents, 0.3% (v/v) formic acid in water (A) and acetonitrile (B); and gradient: 0 to 2 min, isocratic, 23% B; 2 to 16 min, linear from 23% to 48% B; 16 to 16.5 min, linear from 48% to 100% B; 16.5 to 18.5 min, isocratic, 100% B. The HPLC device was equipped with a photodiode array detector (Dionex). Retention time values were as follows: IAOx, 11.8/12.6 min; IAN, 15.6 min; ICHO, 9.5 min; and GS-IAN, 8.2 min. Quantification was based on calibration curves with authentic standards. Turnover rates were calculated for individual enzymatic conversions, and the data were fitted to Michaelis-Menten kinetics using GraphPad Prism 4 software.
TALEN Design and Cloning
TALEN effector-binding elements proceeded by a T, 18 bp long, separated by a 12-bp spacer sequence, were identified manually. Potential TALEN-binding sites on CYP71A12 exon sequences flanking restriction sites were screened manually. Low off-target probability was estimated using The Arabidopsis Information Resource Patmatch with the single effector-binding elements (mismatches, three; mismatch type, substitutions). TALEN-encoding modules lacking the repeats were assembled with BsaI site-flanked modules containing short 35S promoter (plCH51277; Weber et al., 2011), HA-Nuclear Localization Signal (de Lange et al., 2014), truncated TALEN N- and C-terminal sequences, FokI (Mussolino et al., 2011), and octopine synthase terminator (plCH41432; Weber et al., 2011) into pICH47732 (Weber et al., 2011) and pICH47742 (Weber et al., 2011). The repeat domains of TALEN211 and TALEN212 were created using a previously described method (Morbitzer et al., 2011) and cloned via BpiI into pICH47732 TALEN∆Rep and pICH47742 TALEN∆Rep. BpiI-flanked TALEN modules with repeats were assembled together with pICH47751 kanamycin, conferring in planta resistance against kanamycin, and pICH47766 (Weber et al., 2011) into pICH50505 (Weber et al., 2011).
Generation of Stably Transformed Arabidopsis Lines
The transfer DNA expression vector was transformed into Agrobacterium tumefaciens strain GV3101 MP90. cyp71a13-1 (SALK_105136) plants were transformed by the floral dip method (Clough and Bent, 1998). The progeny were selected on solid one-half-strength Murashige and Skoog medium (Duchefa) containing 50 µg mL−1 kanamycin (Duchefa). Seven plants were singled out, and the presence of the TALEN construct was confirmed by PCR on the FokI gene with the primer pair 5′-GTGAAATCTGAATTGGAAGAG-3′ and 5′-TATCTCACCGTTATTAAATTTCC-3′.
Screening for Mutation Events
From leaves of primary transformants, genomic DNA was isolated and a sequence of 665 bp in the target region of the TALEN pair was amplified with the primer pair 5′-AAGCCGTGATTAAAGAGGTG-3′ and 5′-AAATTGTAGGATATGCTTATTTTCT-3′. A total of 5 µL of the PCR product was used directly for digestion with BsaI (New England Biolabs). The amplicon sequence contains two cleavage sites for BsaI, one targeted by the TALEN pair, resulting in different digestion patterns of the wild-type sequence and mutated or partially mutated CYP71A12 (the wild type, 255, 253, and 157 bp; TALEN mutated, 255 and 410 bp). PCR products representing plants carrying somatic mutations were cloned into pGEM T-Easy (Promega). Corresponding Escherichia coli XL1 Blue clones were randomly picked, and the plasmids harbored were sequenced.
For screening of T2 and T3 plants, PCR was conducted with the CYP71A12 TALEN primer set using small leaf discs as template (annealing, 54°C; Phire Plant Direct PCR Kit; Thermo Scientific). Of each PCR product, 5 µL was used directly for digestion with BsaI.
Plant Lines, Growth Conditions, and Stress Treatments
Plants were grown on a 3:1 mixture of soil (Einheitserde) and sand in a growth chamber in a 12-h photoperiod at a light intensity of 80 to 100 µmol m−2 s−1 at 21°C. AgNO3 challenge was conferred by spraying 5 mm AgNO3 on detached leaves and incubating for 24 h in the growth chamber. For UV treatment, leaves were detached and placed under a UV lamp (Desaga; λ = 254 nm, 8 W) at a distance of 20 cm, irradiated for 2 h, and incubated for an additional 22 h in the growth chamber. cyp79b2 cyp79b3 and cyp71a13-1 (SALK_105136) knockout lines have been described previously (Zhao et al., 2002; Nafisi et al., 2007). cyp71a12 (Millet et al., 2010; GABI-Kat 127 H03) was provided by the European Arabidopsis Stock Centre.
Camalexin Quantification
Rosette leaves of 6-week-old cyp71a12 cyp71a13-1, cyp71a12 cyp71a13-2, cyp71a12, cyp71a13-1, and Col-0 plants were analyzed untreated or after 5 mm AgNO3 or UV exposure. Leaves were weighed, and camalexin was extracted using 400 µL of methanol:water (80:20, v/v) for 1 h in a thermoshaker at 65°C. Extracts were cleaned twice by centrifugation and analyzed by reverse-phase HPLC (MultoHigh 100 RP18, 5-µm particle size; Göhler Analytik) as follows: flow rate of 1 mL min−1; solvents, 0.3% (v/v) formic acid in water (A) and acetonitrile (B); and gradient, 0 to 1 min, isocratic, 20% B; 1 to 7 min, linear from 20% to 80% B; 7 to 7.5 min, linear from 80% to 100% B; 7.5 to 9 min, isocratic, 100% B. Camalexin (retention time of 8.4 min) was quantified using a fluorescence detector (318-nm excitation and 370-nm emission) based on calibration with an authentic standard (Schuhegger et al., 2006).
Metabolomic Analysis
Rosette leaves (50–150 mg) of 6-week-old cyp71a12 cyp71a13-1, cyp79b2 cyp79b3, cyp71a12, cyp71a13-1, and Col-0 (AgNO3/UV treated and untreated/mock control) plants were analyzed. Individual leaves were weighed, transferred into 2-mL tubes, and frozen in liquid nitrogen. Samples were homogenized in a ball mill using steel balls (3 mm) and placed in a precooled (−70°C) rack. After 400 µL of precooled (−70°C) methanol:water (80:20, v/v) was added, samples were immediately vortexed and slowly thawed under periodic vortexing. At room temperature, 400 pmol of biochanin A (Sigma-Aldrich) dissolved in methanol:water (1:1, v/v), per 100 mg fresh weight, was added, and the samples were extracted for 1 h in a thermoshaker at room temperature. Samples were centrifuged for 10 min at 16,000g, and after the supernatant was collected, the pellet was extracted for a second time by adding 400 µL of methanol:water (80:20, v/v) and shaking for another 1 h at room temperature. Samples were centrifuged, and supernatants were combined and evaporated to dryness in a vacuum centrifuge (less than 10 mbar, 30°C).
The residue was reconstituted in 50% methanol (400 µL per 100 mg fresh weight), sonicated for 10 min at 20°C, and centrifuged for 10 min at 12,000g. One microliter of the supernatant was separated on an Agilent Infinity 1290 UHPLC System (1290 binary pump, 1290 autosampler with 20-µL loop, 1290 thermostatted column compartment, and 1260 diode array detector) equipped with a Zorbax RRHD Eclipse Plus C18 column (100 × 2.1 mm, 1.8-µm particle size; Agilent). The following binary gradient was applied at a flow rate of 400 µL min−1: 0 to 12 min, linear from 95% A (0.1% [v/v] formic acid in water) and 5% B (0.1% [v/v] formic acid in acetonitrile) to 65% B; 12 to 15 min, isocratic, 95% B; and 15 to 17 min, isocratic, 5% B. The column temperature was maintained at 40°C. Eluting compounds were detected from a mass-to-charge ratio (m/z) of 100 to 1,100 using an Agilent 6550 iFunnel Q-TOF LC/MS-System equipped with a Dual Agilent Jet Stream electrospray ion source in positive and negative ion modes. The following instrument settings were applied for positive (negative) ion mode: nebulizer gas, nitrogen, 35 pounds-force per square inch gauge; dry gas, nitrogen, 200°C, 18 L min−1; sheath gas, nitrogen, 300°C, 12 L min−1; capillary voltage, 3,000 V; nozzle voltage, 0 V; fragmentor voltage, 300 V; high-pressure funnel, voltage drop 150 V, radio frequency (RF) voltage 100 V; low-pressure funnel, voltage drop 100 V, RF voltage 60 V; funnel exit direct current 40 V; octopole RF voltage, 750 V; collision gas, nitrogen; collision energy, 0 V; and acquisition rate, 3 Hz. The mass spectrometer was operated in extended dynamic range (2 GHz) mode, and the slicer mode was set to high sensitivity. The mass resolution (Resolution [full width at half maximum]) within the analyzed m/z range was 12,000 to 27,000. For reference mass correction, a solution of purine (5 µm) and hexakis-(2,2,3,3-tetrafluoropropoxy)phosphazine (0.5 µm) in acetonitrile:water (95:5, v/v) was continuously introduced through the second sprayer of the dual ion source at a flow rate of 15 µL min−1 using an external HPLC pump equipped with a 1:100 splitting device. Collision-induced dissociation mass spectra were acquired in targeted tandem mass spectrometry mode using a medium isolation width of 4 m/z and applying collision energies in the range of 5 to 25 V. MassHunter software packages were used for data acquisition (version B.05.01) as well as qualitative (version B.06.00) and quantitative (version B.06.00) analyses.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Enzymatic properties of CYP71A12 and CYP71A13.
Supplemental Table S1. Analytical data and relative quantifications for all indolic compounds analyzed.
Supplementary Material
Acknowledgments
We thank Franziska Fellermeier for improvement of the enzyme assays, Heidi Miller-Mommerskamp for propagation of plant lines, and Alfons Gierl for hosting the E.G. laboratory.
Glossary
- IAOx
indole-3-acetaldoxime
- IAN
indole-3-acetonitrile
- GS-IAN
glutathione conjugate of indole-3-acetonitrile
- Cys(IAN)
cysteine conjugate of indole-3-acetonitrile
- ICHO
indole-3-carbaldehyde
- ICOOH
indole-3-carboxylic acid
- CAPS
cleaved-amplified polymorphic sequence
- ICOOMe
methyl ester of indole-3-carboxylic acid
- Col-0
Columbia-0
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. GL346/5 [Heisenberg Fellowship to E.G.] and LA1338/5 [to T.L.]), the Hans-Fischer-Gesellschaft, and the TUM Junior Fellow Fund.
References
- Agerbirk N, De Vos M, Kim JH, Jander G (2008) Indole glucosinolate breakdown and its biological effects. Phytochem Rev 8: 101–120 [Google Scholar]
- Ahmad A, Eelnurme I, Spenser I (1960) Indolyl-3-acetaldoxime. Can J Chem 38: 2523 [Google Scholar]
- Bartling D, Seedorf M, Mithöfer A, Weiler EW (1992) Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur J Biochem 205: 417–424 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Schneider B, Svatos A, Oldham NJ, Hahlbrock K (2005) Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol 138: 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Schneider A, Leister D (2011) Perspectives on systematic analyses of gene function in Arabidopsis thaliana: new tools, topics and trends. Curr Genomics 12: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Chapman A, Fellermeier F, Choudhary M, Scheel D, Glawischnig E (2014) The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol 165: 841–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Qi Y, Zhang Y, Voytas DF (2013) Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda) 3: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Lange O, Wolf C, Dietze J, Elsaesser J, Morbitzer R, Lahaye T (2014) Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res 42: 7436–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos M, Kriksunov KL, Jander G (2008) Indole-3-acetonitrile production from indole glucosinolates deters oviposition by Pieris rapae. Plant Physiol 146: 916–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal R, Danson MJ, Hough DW (2007) Catalytic efficiency and kcat/KM: a useful comparator? Trends Biotechnol 25: 247–249 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F, Møldrup ME, Böttcher C, Olsen CE, Scheel D, Halkier BA (2011) Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23: 2456–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier J, Schneider B, Oldham NJ, Hahlbrock K (2001) Accumulation of soluble and wall-bound indolic metabolites in Arabidopsis thaliana leaves infected with virulent or avirulent Pseudomonas syringae pathovar tomato strains. Proc Natl Acad Sci USA 98: 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofberger JA, Lyons E, Edger PP, Pires JC, Schranz ME (2013) Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol Evol 5: 2155–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshaw JP, Unger-Wallace E, Zhang F, Voytas DF (2010) A transient assay for monitoring zinc finger nuclease activity at endogenous plant gene targets. Methods Mol Biol 649: 299–313 [DOI] [PubMed] [Google Scholar]
- Iven T, König S, Singh S, Braus-Stromeyer SA, Bischoff M, Tietze LF, Braus GH, Lipka V, Feussner I, Dröge-Laser W (2012) Transcriptional activation and production of tryptophan-derived secondary metabolites in Arabidopsis roots contributes to the defense against the fungal vascular pathogen Verticillium longisporum. Mol Plant 5: 1389–1402 [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AP, Anarat-Cappillino G, Sattely ES (2013) Minimum set of cytochromes P450 for reconstituting the biosynthesis of camalexin, a major Arabidopsis antibiotic. Angew Chem Int Ed Engl 52: 13625–13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2007) Maize nitrilases have a dual role in auxin homeostasis and beta-cyanoalanine hydrolysis. J Exp Bot 58: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møldrup ME, Salomonsen B, Geu-Flores F, Olsen CE, Halkier BA (2013) De novo genetic engineering of the camalexin biosynthetic pathway. J Biotechnol 167: 296–301 [DOI] [PubMed] [Google Scholar]
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T (2011) Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res 39: 5790–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B (1997) Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239: 2370–2378 [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F (2007) Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Pedras MS, Yaya EE, Glawischnig E (2011) The phytoalexins from cultivated and wild crucifers: chemistry and biology. Nat Prod Rep 28: 1381–1405 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Rauhut T, Glawischnig E (2009) Evolution of camalexin and structurally related indolic compounds. Phytochemistry 70: 1638–1644 [DOI] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E (2006) CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Xu J, Li Y, Lei L, Zhao L, Yang H, Feng J, Liu G, Ren D (2011) Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23: 364–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.