Abstract
Background
Accumulating evidence links colorectal cancer (CRC) with the intestinal microbiota. However, the disturbance of intestinal microbiota and the role of Fusobacterium nucleatum during the colorectal adenoma-carcinoma sequence have not yet been evaluated.
Methods
454 FLX pyrosequencing was used to evaluate the disturbance of intestinal microbiota during the adenoma-carcinoma sequence pathway of CRC. Intestinal microbiota and mucosa tumor-immune cytokines were detected in mice after introducing 1,2-dimethylhydrazine (DMH), F. nucleatum or Berberine (BBR), using pyrosequencing and Bio-Plex Pro™ cytokine assays, respectively. Protein expressions were detected by western blotting.
Results
The levels of opportunistic pathogens, such as Fusobacterium, Streptococcus and Enterococcus spp. gradually increased during the colorectal adenoma-carcinoma sequence in human fecal and mucosal samples. F. nucleatum treatment significantly altered lumen microbial structures, with increased Tenericutes and Verrucomicrobia (opportunistic pathogens) (P < 0.05 = in wild-type C57BL/6 and mice with DMH treatment). BBR intervention reversed the F. nucleatum-mediated increase in opportunistic pathogens, and the secretion of IL-21/22/31, CD40L and the expression of p-STAT3, p-STAT5 and p-ERK1/2 in mice, compared with mice fed with F. nucleatum alone.
Conclusions
F. nucleatum colonization in the intestine may prompt colorectal tumorigenesis. BBR could rescue F. nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment and blocking the activation of tumorigenesis-related pathways.
Keywords: colorectal tumorigenesis, intestinal microbiota, fusobacterium nucleatum, berberine, tumor-immune cytokine
INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer-related mortality worldwide, and its incidence has increased rapidly in recent years in China [1, 2]. The etiology of CRC includes genetic factors; external environment factors, such as dietary structure, smoking, drinking and other unhealthy lifestyles; and internal environment factors, characterized by intestinal microbiota disturbance, immunological derangement and the activation of tumor-related signaling pathways. Accumulating evidence links CRC with the intestinal microbiota [3–5].
There are about 100 trillion bacteria in the human intestine, which constitute the intestinal microbiome [6]. This large group of bacteria is termed intestinal symbiotic bacteria, because of their mutualistic and interdependent relationship with the human body during the long period of co-evolution [7]. The intestinal microbiota community is closely related to the development of CRC via their influence on the physiological functions of the colorectum and even the entire digestive system [7, 8].
Our previous research indicated a structural imbalance in the gut microbiota in patients with advanced colorectal adenoma (CRA), which was represented by a reduction of butyrate-producing bacteria and an increase of opportunistic pathogens compared with healthy controls [2]. However, the exact role of the intestinal microbiota during the occurrence and development of CRC requires further exploration. In addition, whether the microbiota imbalance could be restored by certain chemicals or factors remains unknown.
Berberine (BBR) is an isoquinoline alkaloid and a pharmacological component of the Chinese herb Coptis chinensis, which has been used to treat intestinal infections, particularly bacterial diarrhea, for thousands of years in China [9]. Recently, BBR has been reported to modulate microbiota structures, which contributed to improving obesity and insulin resistance in mice [10, 11]. However, BBR has a poor oral bioavailability and is difficult to absorb into the bloodstream from the intestines [12, 13], which has contributed to the lack of data concerning the mechanism of BBR's action. Here, we hypothesized that BBR might modulate intestinal microbiota to prevent colorectal carcinogenesis.
The current study was undertaken to evaluate the intestinal microbiota disturbance during the colorectal adenoma-carcinoma sequence, using pyrosequencing of the 16S ribosome RNA (rRNA) genome from fecal samples of healthy controls and patients with CRA or CRC. We also attempted to identify the effect on colonic tumorigenesis of the bacteria Fusobacterium nucleatum (F. nucleatum) and of BBR on F. nucleatum-induced tumorigenesis in vivo. These findings may provide a more comprehensive understanding of the role of intestinal microbiota during the occurrence and development of CRC and have important implications for the prevention and treatment of CRC.
MATERIALS AND METHODS
Human specimen collection
Fecal samples and left colonic tissues were collected from consecutive patients who had undergone colonoscopy or colorectal carcinoma surgery in the Renji Hospital between 1 January 2012 and 30 July 2012. Patients must have met the following inclusion criteria to enter the study: 1) patients were over 50 years of age, because current US, European and Asian guidelines define the age threshold for endoscopy at 50 y [2]; 2) patients had a normal bowel frequency (minimum of once every 2 day and a maximum of twice per day) [14]; and 3) patients underwent colonoscopies with adequate withdrawal time [15] by well-trained gastroenterologists using standard colonoscopy equipment. The exclusion criteria were the same as our previous study [2]. Patients with CRA or CRC confirmed by both colonoscopy and pathological examination were included in the CRA group or CRC group, respectively. Patients without obvious abnormalities were enrolled in the NC (Negative control) group. We enrolled 52 cases in the NC group, 47 patients in the CRA group and 42 patients in the CRC group. All subjects were asked to provide fresh stool samples, which were immediately stored at −80°C for further analysis. The samples were collected and preserved according to our previous study [2].
All procedures were undertaken in accordance with the Declaration of Helsinki. The Ethics Committees in the Renji Hospital at each participating center approved the study protocol. Informed consent was obtained from all of the subjects. An independent data and safety committee monitored the trial and reviewed the results.
Bacterial strains and culturing
Fusobacterium nucleatum subsp. nucleatum ATCC 25586 [16] and Escherichia coli MG1655 ATCC 47076 [17] were purchased from American type culture collection (ATCC). F. nucleatum were cultured overnight at 37°C under anaerobic conditions (DG250, Don Whitley Scientific, West Yorkshire, UK) in brain heart infusion (BHI) broth supplemented with hemin, K2HPO4, vitamin K1, and L-Cysteine [18]. The commensal E. coli strain MG1655 was used as the non-pathogenic control and was cultured in Luria-Bertani (LB) medium [17]. Bacteria were centrifuged after culturing, and suspended in phosphate buffered solution (PBS) for animal experiments.
Chemicals
We injected 1,2-dimethylhydrazine (DMH) into mice to model the occurrence of a colonic tumor [19]. DMH is one of the two isomers of dimethylhydrazine. DMH is a potent carcinogen that acts as a DNA methylating agent and it is used to induce colon tumors in experimental animals. DMH and BBR were both obtained from Sigma Chemical Co. (St. Louis, MO, USA) and were prepared by dissolving them in PBS.
Animal experiments
All mice were maintained in specific pathogen-free (SPF) conditions at the Animal Experimental Center of Tongji University. Fifty male C57BL/6-APCMin/+ mice (The APCMin/+ (adenomatous polyposis coli, APC; multiple intestinal neoplasia, Min) mouse is a popular animal model for studies of human colon cancer) were randomly and equally separated into five groups: Control (Ctr), F. nucleatum (Fn), E. coli (Ec), BBR and Fn+BBR. Ninety male wild-type C57BL/6 were separated into nine groups: Control (Ctr), DMH, Fn, Fn+DMH, Ec, Ec+DMH, BBR, BBR+DMH and BBR+Fn+DMH. Experiments were performed after adaptive breeding for 1 week. Bacteria were fed at 109 colony forming units (CFU) suspended with 0.1 ml PBS per day. DMH was injected subcutaneous at a dose of 20 mg/kg once weekly. BBR was administrated by gavage at a dose of 100 mg/kg two hours after bacterial feeding. PBS was used as the control treatment. All treatments were performed for 8 weeks to ensure aberrant crypt foci (ACF) formation or 20 weeks to ensure tumor formation. ACF are clusters of abnormal tube-like glands in the lining of the colon and rectum. ACF form before colorectal polyps and are one of the earliest changes seen in the colon that may lead to cancer. Study end points included the occurrence of colonic ACF or tumors, and changes in the lumen microbial structures and the expression of mucosa tumor immune cytokines in mice of different groups. The ACF and colon tumors were identified by methylene blue staining and hematoxylin-eosin staining.
DNA preparation
The E.Z.N.A. Stool DNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) was used to extract DNA from 200 mg of fecal samples for wild-type C57BL/6 mice, C57BL/6-APCMin/+ mice and humans, according to the manufacturer's instructions. The QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) was used to isolate DNA from colonic tissues, with additional bead-beating steps on a FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA), as previously described [20, 21].
454 FLX pyrosequencing
To investigate the microbiota community composition in fecal or colon tissues, V1~V3 hypervariable regions of the 16S rRNA gene were amplified by polymerase chain reaction (PCR) using universal primers (27F 5′- AGAGTTTGATCCTGGCTCAG - 3′, 533R 5′ - TTACCGC GGCTGCTGGCAC - 3′) incorporating the FLX Titanium adaptors and a sample barcode sequence [2, 22]. The prepared DNA was pyrosequenced by using a Roche 454 GS FLX, in accordance with the manufacturer's instructions.
Taxonomic analysis
The obtained sequences were analyzed using MOTHUR software (version 1.14). The quality control and specific taxonomic procedures were performed similarly to our previous study [2]. According to the taxonomy information, differences among specimens or among clinical groups were analyzed using Metastats and principal component analysis (PCA).
Real-time quantitative PCR assay
DNA from each specimen was subjected to real-time quantitative PCR (qPCR) assays to determine the amounts of total bacteria and F. nucleatum by detecting the 16S genes. The qPCR assay was performed in triplicate with a SYBR Premix Ex Taq (Takara) on an ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Amplifications were performed under the following reaction conditions: 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at the required temperature for 40 sec and at 60°C for 1 min. Cycle threshold (CT) values were calculated using the automated settings for SDS 2.2 (Applied Biosystems). The primer sequences of 16S and F. nucleatum for each assay were the same as those used in our previous studies [2, 16]. Relative abundance was calculated by the ΔΔCT method.
Bio-Plex Pro™ mice mucosa cytokine assays
Bio-Plex Pro™ cytokine assays (BIO-RAD, Hercules, CA, USA) are magnetic bead-based multiplex assays designed to measure multiple members of this diverse group of proteins in a minimal volume of matrix, such as serum, plasma, tissue supernatant and other biological fluids [23]. The use of magnetic beads allows researchers to automate the wash steps on a Bio-Plex Pro wash station. Magnetic separation offers greater convenience, productivity and reproducibility compared with vacuum filtration.
We used mice colonic tissue lysates extracted using RIPA with PMSF (1:100), Protease Inhibitor Cocktail (1:100) and Phosphatase Inhibitor Cocktail (1:100). The Bio-Plex Pro™ suspension array system is operated around three core elements of xMAP technology fluorescently dyed microspheres to permit discrimination of individual tests within a multiplex suspension; a dedicated flow-cytometer with two lasers and associated optics to measure the different molecules bound to the surface of the beads; and a high-speed digital signal processor to manage the fluorescence data efficiently.
Western blot analysis
Proteins were extracted from mice colonic tissue and quantified using a Pierce™ BCA Protein Assay Kit (Thermo Scientific™). Proteins were subjected to 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes, and blocked with 5% bovine serum Albumin in PBS containing 0.1% Tween-20. Equal protein loading was controlled by Ponceau Red staining of the membranes. Blots were probed using primary antibodies. A GAPDH antibody was used as a control for whole-cell lysates. Antibodies were purchased from Cell Signaling Technology Inc., except for GAPDH (Kangchen), and AKT2 and p-AKT2 (Abcam). After washes, membranes were incubated with the appropriate HRP-conjugated secondary antibodies (Kangchen), and blots were detected using the Chemiluminescent Western Blot Detection (Thermo Scientific™).
Statistical analysis
The Mann-Whitney U and One-Way ANOVA tests were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) and MOTHUR, when comparing the intestinal microbiota structures among the different human subject groups or mice groups with various treatments. Student's t-test was used to describe colon ACF or tumors, and the expression of mucosa tumor-immune cytokines, compared with corresponding mice groups, according to the study design. Two-sided P-values <0.05 were considered statistically significant.
RESULTS
Structural transformation of lumen microbiota during the colorectal adenoma-carcinoma sequence
We first analyzed the fecal microbial community among negative control subjects, CRA and CRC patients. A total of 2,296,326 optimized sequences were obtained with an average of 437 base pairs per sequence. The sequences were clustered using 3% dissimilarity as an indicator of an operational taxonomic unit (OTU). There were no significant differences among the relative microbiota abundance of the fecal samples from negative subjects group (FN, n = 52), CRA group (FA, n = 47) and CRC group (FC, n = 42) when comparing the richness estimators (Ace and Chao) and the diversity estimators (Shannon and Simpson). The rarefaction curve showed the ends of the curve per sample tending towards stability (Supplementary Figure 1), which indicated that the pyrosequencing data generated in this study were sufficient to reflect the diversity of the intestinal microbiota.
Principal component analysis (PCA), which is based on unweighted Unifrac metrics, showed alteration of fecal intestinal microbiota communities in patients with CRA or CRC compared with the negative controls (Figure 1A and 1B). The principal components of fecal gut microbiota were significantly different in the three groups. The FA group was located between the FN and FC both on the X-axis in the PC1 direction and on the Y-axis in the PC2 direction (Figure 1A and 1B), which indicated that structural transformation of fecal gut microbiota might occur from normal colorectal tissues to CRA and then to CRC (P < 0.05, ANOVA).
Figure 1. Intestinal microbiota structures among the FN group (n = 52), FA group (n = 47) and FC group (n = 42).
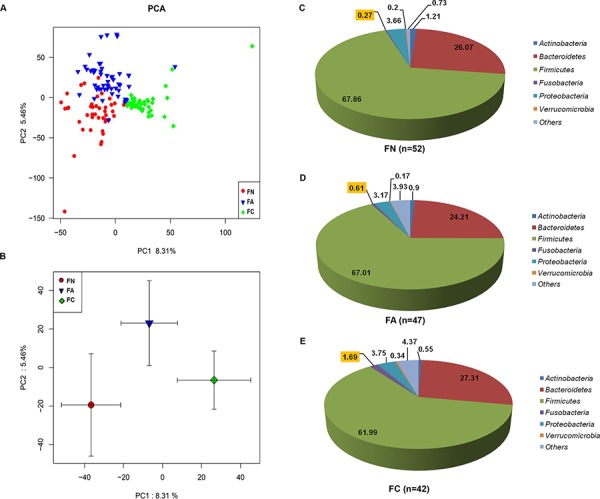
A. Principal component analysis (PCA) scores plot based on unweighted UniFrac metrics. Each symbol represents a sample. B. PCA scores plot based on unweighted UniFrac metrics. Each point represents the mean principal component scores of all populations in a different group, and the error bar represents the standard deviation. C. Relative abundance of lumen microbiota distribution in the negative control (FN) group (n = 52) at the phylum level. D. Relative abundance of lumen microbiota distribution in the CRA (FA) group (n = 47) at the phylum level. E. Relative abundance of lumen microbiota distribution in the CRC (FC) group (n = 42) at the phylum level.
More than half of the microbial community comprised the Firmicutes phylum: 67.86% in the FN group (n = 52), 67.01% in the FA group (n = 47), and 61.99% in the FC group (n = 42). The next most abundant phyla were Bacteroidetes, Proteobacteria, Fusobacteria, Actinobacteria, and Verrucomicrobia. These six bacterial phyla accounted for more than 90% of the microbial community in the three groups. Furthermore, the proportions of the Fusobacterial phylum showed a significant, gradually increase during the colorectal adenoma-carcinoma sequence: from normal colorectal tissue (0.27% in the FN group) to adenoma tissues (0.61% in the FA group) and to CRC tissues (1.69% in the FC group) (P = 0.016; 2.26 fold change (FA/FN) and 6.26 fold change (FC/FN), respectively, Figure 1C–1E).
We analyzed the fecal microbial community among the three groups at the genera level. A heatmap was generated with 55 genera whose relative abundance was more than 0.1% of the total bacteria in the stool samples (Supplementary Figure 2A). The data revealed that the levels of opportunistic pathogens, such as Fusobacterium, Streptococcus, and Enterococcus spp. increased during the colorectal adenoma-carcinoma sequence in the lumen (all P < 0.05, Table 1 and Supplementary Figure 2B).
Table 1. Significant differences in some genera identified among the fecal microbiota of the FN group (n = 52), FA group (n = 47) and FC group (n = 42).
| Phylum | Genus | FN (n = 52) | FA (n = 47) | FC (n = 42) | P value | Tendency from FN-FA-FC |
|---|---|---|---|---|---|---|
| Actinobacteria | Actinomyces | 0.09 ± 0.02 | 0.07 ± 0.02 | 0.04 ± 0.02 | <0.001 | ↓ |
| Actinobacteria | Bifidobacterium | 0.05 ± 0.01 | 0.03 ± 0.01 | 0.01 ± 0.00 | 0.008 | ↓ |
| Firmicutes | Blautia | 14.18 ± 1.94 | 12.28 ± 2.02 | 3.01 ± 0.55 | <0.001 | ↓ |
| Firmicutes | Clostridium | 2.08 ± 0.41 | 0.88 ± 0.20 | 0.31 ± 0.09 | <0.001 | ↓ |
| Firmicutes | Dorea | 1.70 ± 0.27 | 1.45 ± 0.35 | 0.29 ± 0.06 | <0.001 | ↓ |
| Firmicutes | Lactobacillus | 1.79 ± 1.16 | 0.70 ± 0.37 | 0.14 ± 0.09 | 0.011 | ↓ |
| Firmicutes | Roseburia | 2.67 ± 0.50 | 1.22 ± 0.27 | 0.75 ± 0.15 | 0.003 | ↓ |
| Firmicutes | Eubacterium | 0.08 ± 0.03 | 0.04 ± 0.02 | 0.02 ± 0.01 | 0.013 | ↓ |
| Fusobacteria | Fusobacterium | 0.26 ± 0.18 | 0.61 ± 0.29 | 1.22 ± 0.50 | 0.031 | ↑ |
| Proreobacteria | Escherichia-Shigella | 1.32 ± 0.32 | 1.82 ± 0.41 | 2.74 ± 0.66 | 0.025 | ↑ |
| Firmicutes | Coprococcus | 0.44 ± 0.07 | 0.73 ± 0.12 | 1.24 ± 0.27 | 0.034 | ↑ |
| Firmicutes | Streptococcus | 2.33 ± 0.50 | 3.75 ± 0.65 | 5.55 ± 1.22 | 0.016 | ↑ |
| Firmicutes | Enterococcus | 0.26 ± 0.15 | 1.42 ± 0.51 | 2.75 ± 0.92 | 0.004 | ↑ |
Note: 1. Relative contribution of a genus was calculated as percentage of the sequences of this genus to all sequences in this population. 2. Mean ± SEM (standard error of mean) was calculated according to percentage of the sequences of this genus to all sequences in each individual. 3. The differences of relative abundances were calculated using ANOVA assay. Two-sided P-values < 0.05 were considered statistically significant. The increasing or decreasing tendency during FN-FA-FC sequence was represented as ↓ or ↓, respectively.
Structural transformation of colonic mucosa microbiota during the colorectal adenoma-carcinoma sequence
The conclusions from our fecal pyrosequencing data agreed with previous reports [2, 3, 24]. However, there were few reports concerning the mucosa microbial structures transformed during the colorectal adenoma-carcinoma sequence [25, 26]. To identify the changed structures in the mucosa microbiota, pyrosequencing was also performed with DNA samples extracted from 98 colonic tissues, including 37 cases with normal colonic mucosa from negative control subjects (MN), 30 cases of adenoma tissues (MA), and 31 cases of carcinoma tissues (MC). From the pyrosequencing analysis, similar trends in the variation of mucosal microbiota were observed to those in the lumenal microbiota (Supplementary Figure 2B and 2C).
Enrichment of Fusobacterium nucleatum during the colorectal adenoma-carcinoma sequence
To further clarify the enrichment of F. nucleatum during the adenoma-carcinoma sequence pathway, we used a targeted qPCR assay to determine its abundance in feces or colonic tissues from various populations. QPCR measured the abundance of Fusobacterium, and the data was consistent with the results from the pyrosequencing data (Spearman correlation r = 0.827, P < 0.05). The qPCR data showed that Fusobacterium nucleatum gradually increased with the sequence from healthy controls to CRA and eventually to CRC, both in fecal and mucosal samples (Supplementary Figure 3A and 3B). Our results demonstrated that F. nucleatum accumulated in the human intestine at the early stage of colon cancer development, and might play a role during the colorectal adenoma-carcinoma sequence.
Fusobacterium nucleatum promoted colon tumorigenesis in mice
Colon tumorigenesis was analyzed by calculating the number of ACF and colon tumors in wild-type C57BL/6 mice and C57BL/6-APCMin/+ mice. The mice with DMH injected all generated obvious ACF in their colons (Figure 3A), and mice administered with both DMH and F. nucleatum (Fn+DMH) generated the largest numbers of ACF (Figure 2A) and colon tumors (Figure 2B). Interestingly, all the APCMin/+ mice administered with F. nucleatum for 8 weeks developed more colon tumors than the APCMin/+ control (Ctr) and the Escherichia coli (Ec) bacteria control mice (P < 0.01, Figure 2C). These data suggested that F. nucleatum might accelerate tumorigenesis in vivo.
Figure 3. Lumen microbiota distribution at the phylum level in mice treated with A. bacteria or B. BBR.
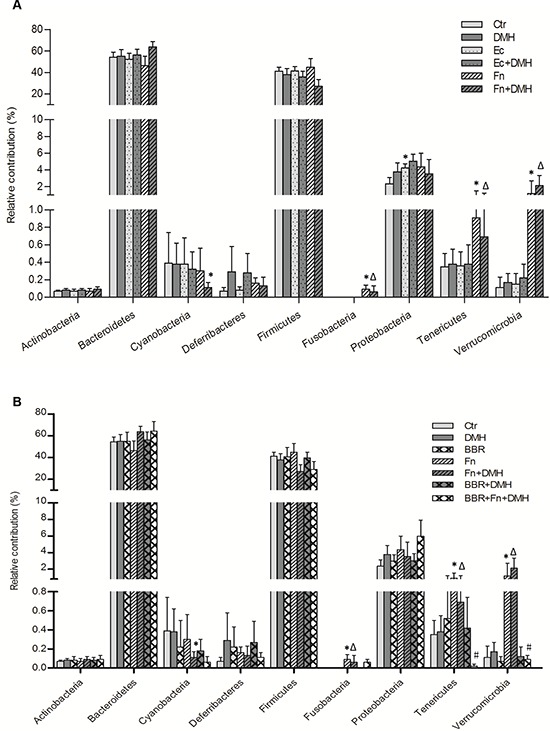
The relative contribution of a phylum was calculated as the percentage of the sequences of this phylum to all sequences in this population. Mean ± SEM (standard error of mean) was calculated according to percentage of the sequences of this phylum to all sequences in each individual. The differences in relative abundances were calculated using Mann-Whitney U test. Two-sided P-values < 0.05 were considered statistically significant. *P < 0.05, compared with Ctr group; ΔP < 0.05, compared with DMH group; #P < 0.05, compared with Fn+DMH group.
Figure 2. Intestine tumorigenesis and lumenal microbiota structures in mice with treated with Fusobacterium nucleatum or BBR.
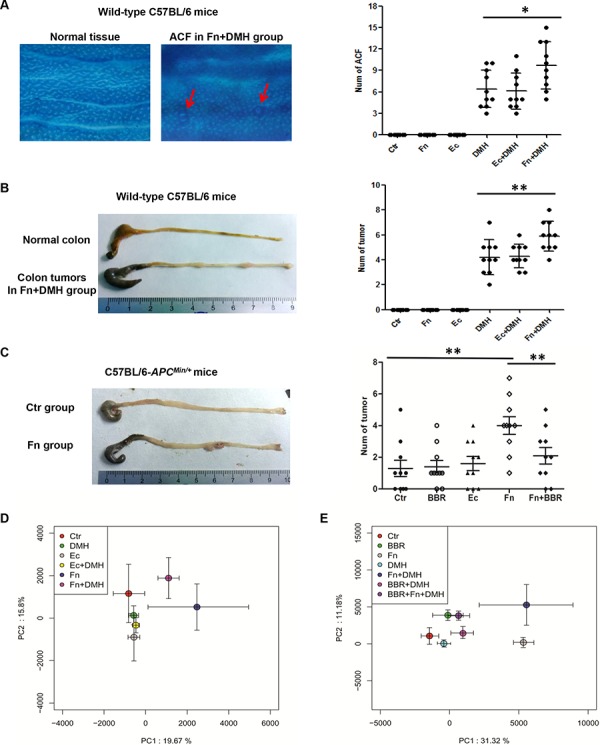
A. Colon ACF in wild-type C57BL/6 mice observed by staining with 0.2% methylene blue under a light microscope at 8 weeks (n = 10 per group). ACF are highlighted with red arrows. B. Intestine tumorigenesis in wild-type C57BL/6 mice at 20 weeks (n = 10 per group). C. Intestine tumorigenesis in C57BL/6-APCMin/+ mice at 8 weeks (n = 10 per group). Two-sided P-values < 0.05 were considered statistically significant. *P < 0.05 **P < 0.01. D. F. nucleatum colonization altered intestinal microbiota structures in mice. E. BBR modulated the microbiota disturbance induced by F. nucleatum. Each point represents the mean principal component scores of all population in the different groups, and the error bar represents the standard deviation.
Fusobacterium nucleatum altered lumen microbial structures in mice
We next pyrosequenced the lumenal microbial components in the different treatment groups of wild-type C57BL/6 mice. PCA of lumenal microbiota was performed among six groups of mice. As shown in Figure 2D, the microbial structures of the lumen in the mice after F. nucleatum treatment were significantly different from mice without bacterial administration or those receiving E. coli administration (P < 0.05, ANOVA), indicating that in addition to promoting colon tumorigenesis, introduction of F. nucleatum altered the lumenal microbial structures in mice.
A total of 952 OTUs were found in the wild-type C57BL/6 mice. Almost all of the OTUs were represented by the following phyla: Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia, Cyanobacteria, Tenericutes, and Unclassified bacteria, which were similar to humans. There was no Fusobacteria colonization in the lumen of wild-type C57BL/6 mice. However, in addition to promoting colon tumorigenesis, colonization by F. nucleatum dramatically altered the lumen microbial structures by increasing the Tenericutes and Verrucomicrobia in the mice (Figure 3A).
BBR could reverse the F. nucleatum-induced lumenal microbiota imbalance and colon tumorigenesis in mice
Previous studies have reported that some chemicals, such as dietary fiber [2, 27], folate [28, 29] or BBR [30–32] had a preventative effect on colonic tumorigenesis. Recently, BBR has been reported to improve obesity and insulin resistance in mice by changing microbiota structures [33]. Therefore, we next introduced BBR into mice to identify whether it could rescue the F. nucleatum-induced microbiota disturbance. The microbial structures alteration, which was characterized by the increase in Tenericutes and Verrucomicrobia, was dramatically reversed in F. nucleatum infected mice after BBR intervention (Figure 2E and Figure 3B). These findings suggested that BBR intervention significantly blocked the Fusobacteria-induced lumen microbiota imbalance. In addition, C57BL/6-APCMin/+ mice treated with BBR developed significantly fewer colon tumors compared with mice fed without BBR after F. nucleatum treatment (Figure 2C). The same phenomena were observed in wild-type C57BL/6 mice (Figure 4A and 4B).
Figure 4. BBR inhibited colon tumorigenesis induced by F. nucleatum in mice by modulating the expression of mucosa tumor-immune cytokines.
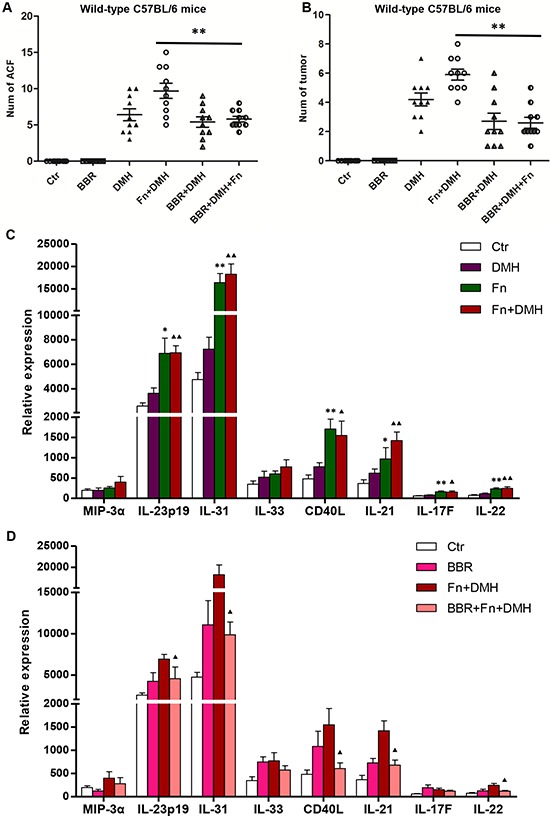
A. Colon ACF and B. tumors counted in wild-type C57BL/6 mice with BBR intervention at different times. Two-sided P-values < 0.05 were considered statistically significant. **P < 0.01. C. F. nucleatum colonization altered the expression of mucosa tumor-immune cytokines in mice. *P < 0.05 and **P < 0.01, compared with the Ctr group; ▲P < 0.05 and ▲▲P < 0.01, compared with the DMH group. D. BBR modulated the mucosa tumor-immune cytokines secretion induced by F. nucleatum. ▲P < 0.05 and ▲▲P < 0.01, compared with the Fn+DMH group.
BBR might modulate the F. nucleatum-mediated increase of tumor-immune cytokine secretion in mice
It has been reported that F. nucleatum potentiates intestinal tumorigenesis by modulating the tumor-immune microenvironment in mouse models [24]. In this study, we detected changes in the mucosal cytokines by performing Bio-Plex Pro™ Assays to explore the colon microenvironment alteration in mice after F. nucleatum or BBR treatment. As shown in Figure 4C, the levels of IL-17F/21/22/23/31 and CD40L increased after the introduction of F. nucleatum in mice, indicating that colonization by F. nucleatum stimulated the secretion of immune cytokines. Furthermore, the increased secretion of IL-21/22/31 and CD40L were reversed after the addition of BBR in F. nucleatum-treated mice (Figure 4D). The data indicated that BBR might modulate the increased secretion of mucosa immune cytokines in F. nucleatum colonized mice.
BBR could block F. nucleatum-induced colon tumorigenesis in mice by inhibiting the activation of JAK/STAT and MAPK pathways
Numerous studies have indicated some tumor-immune cytokines play crucial roles in tumor development and progression by activating specific signaling pathways [34–36]. It has been reported that IL-21/22/31 and CD40L activate the JAK/STAT, MAPK/ERK and PI3K/AKT pathways [25, 26]. Therefore, we next examined whether F. nucleatum-induced increases in the secretion of these cytokines via activation of JAK/STAT, MAPK/ERK and PI3K/AKT pathways. As shown in Figure 5, the expressions of p-STAT3, p-STAT5 and p-ERK1/2 increased after introduction of F. nucleatum both in Wild-type C57BL/6 mice and C57BL/6-APCMin/+ mice, indicating that F. nucleatum colonization may activate JAK/STAT and MAPK/ERK pathways. Furthermore, the F. nucleatum-induced increases in p-STAT3, p-STAT5 and p-ERK1/2 expressions were significantly reduced by BBR treatment in mice. The data indicated that BBR treatment significantly blocked the Fusobacteria-induced activation of the JAK/STAT and MAPK/ERK pathways.
Figure 5. BBR inhibited colon tumorigenesis induced by F. nucleatum in mice by modulating JAK/STAT and MAPK pathways.
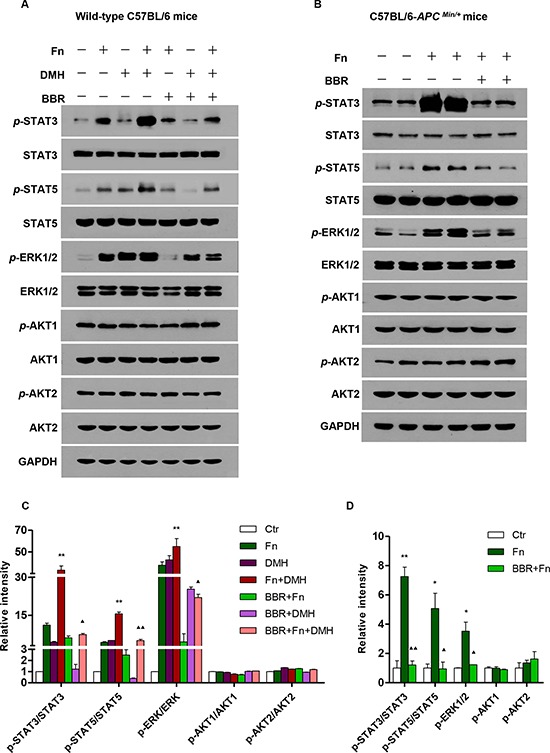
Western blot assays were performed to measure the expressions of STAT3, STAT5, ERK1/2, AKT1/2, p-STAT3, p-STAT5, p-ERK1/2 and p-AKT1/2 with different treatments in wild-type C57BL/6 mice A. and in C57BL/6-APCMin/+ mice B, C. The summarized analysis of the western blot is shown in wild-type C57BL/6 mice with different treatments. *P < 0.05 and **P < 0.01, compared with the Ctr group; ▲P < 0.05 and ▲▲P < 0.01, compared with the Fn+DMH group. D. The summarized analysis of western blot is shown in C57BL/6-APCMin/+ mice with different treatments. *P < 0.05 and **P < 0.01, compared with the Ctr group; ▲P < 0.05 and ▲▲P < 0.01, compared with the Fn group.
DISCUSSION
Numerous studies have shown that the intestinal microbial structures are different in patients with CRC compared with healthy subjects [3, 18, 37]. Our results demonstrated that the intestinal microbial structures were altered in the lumen and the mucosa during the progression of the colorectal adenoma-carcinoma sequence. These alterations were characterized by reductions in some beneficial bacteria, such as Actinomyces, Bifidobacterium, Lactobacillus, butyrate-producing bacteria (including Clostridium, Rosuburia, Eubacterium, Blautia, and Dorea spp.), and by enrichment of opportunistic pathogens, such as Fusobacterium, Escherichia-Shigella, Coprococcus, Streptococcus and Enterococcus spp. The data indicated that these bacteria might mediate the progression of CRC.
In line with other two research teams from America and Canada who reported the enrichment of Fusobacterium spp. in CRC tissue [16, 38, 39], our clinical data showed that Fusobacteria and Fusobacterium spp. accumulated in the human intestine during the colorectal adenoma-carcinoma sequence. More importantly, our data revealed that Fusobacterium expression in the mucosa was consistent with that in feces; therefore, fecal samples may replace tissue specimens as a simpler and more practical diagnostic method for the early detection of Fusobacterium enrichment [40].
In this study, we clarified that colonization of F. nucleatum in the intestine promoted the onset of colonic tumors in vivo. Kostic reported that F. nucleatum potentiated intestinal tumorigenesis by modulating the tumor-immune microenvironment [24]. However, the mechanism of F. nucleatum-mediated colorectal tumorigenesis is still poorly understood. We hypothesized that F. nucleatum might promote colorectal tumorigenesis because 1) F. nucleatum disturbs the intestinal microbiota balance via an increase in opportunistic pathogens and decreased probiotics; and 2) F. nucleatum induces tumor-related immune cytokine secretion, such as IL-21/22/31 and CD40L, and activates the JAK/STAT and MAPK/ERK pathways, which have been reported to promote tumorigenesis. Both IL-21 [41] and CD40L [42] have been linked to colorectal tumorigenesis. IL-22 may play a prominent role in the progression of CRC by activation of STAT3 (Signal Transducer and Activator of Transcription 3) [43]. IL-31 modulates cell proliferation and migration, suggesting a role in the regulation of the intestinal barrier function [36]. Both STAT3 and STAT5 increase tumor cell proliferation, survival and invasion, while suppressing anti-tumor immunity [44]. Activated MAPK/ERK plays an important part in progression of colorectal cancer [45].
The lumenal microbial community, combined with the mucosal immune system, determines the intestinal homeostasis balance, which represents the intestinal environment to some extent. There is growing evidence that the balance between symbiotic bacteria and the host defense response from the mucosa plays an essential role in the genesis, growth and metastasis of CRC [46–48]. Immune dysfunction might be an important mediating factor for the effects of intestinal symbiotic bacteria on inflammation and tumorigenesis. When intestinal microbial structures shift because of exterior factors, the link with the immune system could be broken, resulting in immune dysfunction, which ultimately leads to the occurrence and development of CRC [49, 50].
Some factors such as dietary fiber [2, 27], folate [28, 29] or BBR [30–32] prevent colonic tumorigenesis. BBR decreased the relative contribution of Firmicutes and Bacteroidetes in mice fed with a high-fat diet, and upregulated the expression of fasting-induced adipose factor genes in both intestinal and adipose tissues, which indicated that the antimicrobial activity of BBR contributed to its anti-obesity effects [12, 33]. In this study, we found that BBR not only reversed the microbiota imbalance induced by F. nucleatum, but also blocked the secretion of mucosal immune cytokines and the activation of the JAK/STAT and MAPK/ERK pathway, which was stimulated by F. nucleatum in vivo. Our data revealed a role for BBR in the prevention of colorectal carcinogenesis.
Recently, it has been reported that microbiota may promote carcinogenesis by activation of TLRs, NF-κB and STAT3 [51–53]. Han et al. demonstrated that F. nucleatum promotes colorectal carcinogenesis by modulating the Wnt pathway [54]. In addition, Chinese scientists later reported that BBR attenuates colorectal tumorigensis via inhibition of the Wnt pathway in vivo and in vitro [30, 31]. In the current study, we determined one of the mechanisms of F. nucleatum-mediated colorectal carcinogenesis. Firstly, we found that F. nucleatum treatment significantly increased the levels of p-STAT3, p-STAT5 and p-ERK1/2, both in wild-type C57BL/6 mice and C57BL/6-APCMin/+ mice. Furthermore, BBR may attenuate F. nucleatum-induced carcinogenesis by blocking the activation of JAK/STAT and MAPK/ERK pathways in mice. However, the specific molecule(s) that participates in the Fusobacterium-induced CRC carcinogenesis, and the mechanism of BBR-mediated blocking of Fusobacterium-induced CRC carcinogenesis, should be explored in the future.
In conclusion, F. nucleatum might play an important role in the progression of CRC, especially in the colorectal adenoma-carcinoma sequence, where it disturbs the intestinal microbiota balance, alters tumor-related immune cytokine secretion and activates the JAK/STAT and MAPK/ERK pathways. Furthermore, BBR blocks F. nucleatum-mediated colorectal tumorigenesis in vivo by modulating the microbial balance, and regulating the mucosal immune system, the JAK/STAT pathway and the MAPK/ERK pathway. Further research is needed to reveal which specific cell signals are regulated by F. nucleatum at the molecular level.
SUPPLEMENTARY FIGURES
Acknowledgments
The authors thank Ya-Nan Yu, Ta-Chung Yu, Hui-Jun Zhao, Tian-Tian Sun and Yu-Rong Weng for animal experiment assistance; Hui-Min Chen and Hao-Yan Chen for statistical analysis; Min Li for bacteria culture; Hui-Fang An for analysis and interpretation of the data; Jun Yu, Wen-Xin Qin, Xiong Ma and Nan Shen for technical assistance; Ya-Nan Yu and Jie-Hong for drafting of the manuscript; and Jing-Yuan Fang for study design, supervision and critical review of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
GRANT SUPPORT
This work was supported by grants from the National Natural Science Foundation (Grant No. 81320108024 and 81421001) to FJY; The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. 201268) to HJ.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, Yang L, Liu ZJ, Yuan YZ, Liu F, Wu JX, Zhong L, Fang DC, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. The American journal of clinical nutrition. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 3.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME journal. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- 5.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial ecology. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 6.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Feng Y, Tsao S, Wang N, Curtain R, Wang Y. Berberine and Coptidis rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. Journal of ethnopharmacology. 2009;126:5–17. doi: 10.1016/j.jep.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Li X, Zou D, Liu W, Yang J, Zhu N, Huo L, Wang M, Hong J, Wu P, Ren G, Ning G. Treatment of type 2 diabetes and dyslipidemia with the natural plant alkaloid berberine. The Journal of clinical endocrinology and metabolism. 2008;93:2559–2565. doi: 10.1210/jc.2007-2404. [DOI] [PubMed] [Google Scholar]
- 11.Yin J, Xing H, Ye J. Efficacy of berberine in patients with type 2 diabetes mellitus. Metabolism: clinical and experimental. 2008;57:712–717. doi: 10.1016/j.metabol.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W, Gu D, Li J, Cui K, Zhang Y. Effects and action mechanisms of berberine and Rhizoma coptidis on gut microbes and obesity in high-fat diet-fed C57BL/6J mice. PloS one. 2011;6:e24520. doi: 10.1371/journal.pone.0024520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Lin H, Huang W. Modulating gut microbiota as an anti-diabetic mechanism of berberine. Medical science monitor : international medical journal of experimental and clinical research. 2011;17:RA164–167. doi: 10.12659/MSM.881842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macfarlane GT, Macfarlane S. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scandinavian journal of gastroenterology Supplement. 1997;222:3–9. doi: 10.1080/00365521.1997.11720708. [DOI] [PubMed] [Google Scholar]
- 15.Winawer SJ, Zauber AG, Fletcher RH, Stillman JS, O'Brien MJ, Levin B, Smith RA, Lieberman DA, Burt RW, Levin TR, Bond JH, Brooks D, Byers T, et al. Guidelines for colonoscopy surveillance after polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer and the American Cancer Society. CA: a cancer journal for clinicians. 2006;56:143–159. doi: 10.3322/canjclin.56.3.143. quiz 184-145. [DOI] [PubMed] [Google Scholar]
- 16.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Dechelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 18.Rhee KJ, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, Sartor RB, Golenbock D, Hamad AR, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infection and immunity. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JL, Lin YW, Chen HM, Kong X, Xiong H, Shen N, Hong J, Fang JY. Calcium prevents tumorigenesis in a mouse model of colorectal cancer. PloS one. 2011;6:e22566. doi: 10.1371/journal.pone.0022566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. e1841. [DOI] [PubMed] [Google Scholar]
- 21.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflammatory bowel diseases. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clinical and diagnostic laboratory immunology. 2003;10:133–139. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, Randall TA, Galanko J, Benson A, Sandler RS, Rawls JF, Abdo Z, Fodor AA, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. The ISME journal. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PloS one. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:799–803. doi: 10.1158/1078-0432.CCR-13-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Wang X, Sun DF, Tian XQ, Zhao SL, Chen YX, Fang JY. Folic acid and sodium butyrate prevent tumorigenesis in a mouse model of colorectal cancer. Epigenetics : official journal of the DNA Methylation Society. 2008;3:330–335. doi: 10.4161/epi.3.6.7125. [DOI] [PubMed] [Google Scholar]
- 29.Witherspoon M, Chen Q, Kopelovich L, Gross SS, Lipkin SM. Unbiased metabolite profiling indicates that a diminished thymidine pool is the underlying mechanism of colon cancer chemoprevention by alpha-difluoromethylornithine. Cancer discovery. 2013;3:1072–1081. doi: 10.1158/2159-8290.CD-12-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu K, Yang Q, Mu Y, Zhou L, Liu Y, Zhou Q, He B. Berberine inhibits the proliferation of colon cancer cells by inactivating Wnt/beta-catenin signaling. International journal of oncology. 2012;41:292–298. doi: 10.3892/ijo.2012.1423. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Cao H, Zhang B, Xu X, Ruan H, Yi T, Tan L, Qu R, Song G, Wang B, Hu T. Berberine potently attenuates intestinal polyps growth in ApcMin mice and familial adenomatous polyposis patients through inhibition of Wnt signalling. Journal of cellular and molecular medicine. 2013;17:1484–1493. doi: 10.1111/jcmm.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iizuka N, Hazama S, Yoshimura K, Yoshino S, Tangoku A, Miyamoto K, Okita K, Oka M. Anticachectic effects of the natural herb Coptidis rhizoma and berberine on mice bearing colon 26/clone 20 adenocarcinoma. International journal of cancer Journal international du cancer. 2002;99:286–291. doi: 10.1002/ijc.10338. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, Li X, Ning G, Zhao L. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PloS one. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nature reviews Drug discovery. 2014;13:379–395. doi: 10.1038/nrd4296. [DOI] [PubMed] [Google Scholar]
- 35.Wolk K, Witte E, Witte K, Warszawska K, Sabat R. Biology of interleukin-22. Seminars in immunopathology. 2010;32:17–31. doi: 10.1007/s00281-009-0188-x. [DOI] [PubMed] [Google Scholar]
- 36.Dambacher J, Beigel F, Seiderer J, Haller D, Goke B, Auernhammer CJ, Brand S. Interleukin 31 mediates MAP kinase and STAT1/3 activation in intestinal epithelial cells and its expression is upregulated in inflammatory bowel disease. Gut. 2007;56:1257–1265. doi: 10.1136/gut.2006.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome research. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PloS one. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jauch D, Martin M, Schiechl G, Kesselring R, Schlitt HJ, Geissler EK, Fichtner-Feigl S. Interleukin 21 controls tumour growth and tumour immunosurveillance in colitis-associated tumorigenesis in mice. Gut. 2011;60:1678–1686. doi: 10.1136/gutjnl-2011-300612. [DOI] [PubMed] [Google Scholar]
- 42.Buning C, Kruger K, Sieber T, Schoeler D, Schriever F. Increased expression of CD40 ligand on activated T cells of patients with colon cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:1147–1151. [PubMed] [Google Scholar]
- 43.Ernst M, Thiem S, Nguyen PM, Eissmann M, Putoczki TL. Epithelial gp130/Stat3 functions: an intestinal signaling node in health and disease. Seminars in immunology. 2014;26:29–37. doi: 10.1016/j.smim.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. The Lancet Oncology. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 46.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS one. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen W, Durum SK. Synergy of IL-23 and Th17 cytokines: new light on inflammatory bowel disease. Neurochemical research. 2010;35:940–946. doi: 10.1007/s11064-009-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vipperla K, O'Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutrition in clinical practice : official publication of the American Society for Parenteral and Enteral Nutrition. 2012;27:624–635. doi: 10.1177/0884533612452012. [DOI] [PubMed] [Google Scholar]
- 49.Kryczek I, Wu K, Zhao E, Wei S, Vatan L, Szeliga W, Huang E, Greenson J, Chang A, Rolinski J, Radwan P, Fang J, Wang G, et al. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J Immunol. 2011;186:4388–4395. doi: 10.4049/jimmunol.1003251. [DOI] [PubMed] [Google Scholar]
- 50.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, Krishnareddy S, Hsu D, Xu R, Harpaz N, Dannenberg AJ, Subbaramaiah K, Cooper HS, Itzkowitz SH, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133:1869–1881. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tye H, Kennedy CL, Najdovska M, McLeod L, McCormack W, Hughes N, Dev A, Sievert W, Ooi CH, Ishikawa TO, Oshima H, Bhathal PS, Parker AE, et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer cell. 2012;22:466–478. doi: 10.1016/j.ccr.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell host & microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


