Abstract
Purpose
Genetic risk prediction models such as BRCAPRO are used routinely in genetic counseling for identification of potential BRCA1 and BRCA2 mutation carriers. They require extensive information on the counselee and her family history, and thus are not practical for primary care.
Methods
To address this gap, we develop and test a two-stage approach to genetic risk assessment by balancing the tradeoff between the amount of information used and accuracy achieved. The first stage is intended for primary care wherein limited information is collected and analyzed using a simplified version of BRCAPRO. If the assessed risk is sufficiently high, more extensive information is collected and the full BRCAPRO is used (stage two; intended for genetic counseling). We consider three first-stage tools: BRCAPROLYTE, BRCAPROLYTE-Plus, and BRCAPROLYTE-Simple. We evaluate the two-stage approach on independent clinical data on probands with family history of breast and ovarian cancers, and BRCA genetic test results. These include population-based data on 1344 probands from Newton-Wellesley Hospital and mostly high risk family data on 2713 probands from Cancer Genetics Network and MD Anderson Cancer Center. We use discrimination and calibration measures, appropriately modified to evaluate the overall performance of a two-stage approach.
Results
We find that the proposed two-stage approach has very limited loss of discrimination and comparable calibration as BRCAPRO. It identifies a similar number of carriers without requiring a full family history evaluation on all probands.
Conclusions
We conclude that the two-stage approach allows for practical large-scale genetic risk assessment in primary care.
Keywords: BRCA1, BRCA2, BRCAPRO, BayesMendel, CancerGene
Introduction
The risk of developing breast and ovarian cancers is high for the carriers of deleterious mutations of the BRCA1 and BRCA2 genes, and thus it is critical to identify carriers as early as possible [1,2]. Yet there is a lack of streamlined procedures for identifying mutation carriers from a general population. As a result, many carriers remain unaware of their status [3]. The identification of potential carriers can be ideally initiated by primary care providers; however, they need tools that can help them in efficiently identifying high-risk patients within their constraints of limited time and resources. Genetic risk prediction models that are used currently in genetic counseling are effective but too complex for the primary care setting, unless they can be simplified and incorporated into the Electronic Medical Records (EMR) or other Health Information Technology (HIT) solutions [4]. With a simplified adaptation, potential carriers can be identified in primary care and referred for further risk assessment and genetic testing.
The BRCAPRO genetic risk prediction model [5] is used extensively in genetic counseling and is available in the BayesMendel R package [6], the CancerGene genetic counseling package [7], the web-based risk service [8], the HughesRiskApps (HRA) package [9] and other computing environments. Based on the family history information provided by a counselee, BRCAPRO estimates the probability that she/he carries a BRCA1/2 mutation as well as her/his prospective risk of developing cancer. Several improvements to BRCAPRO in recent years allow it to use a variety of information [10–15]. Primary care settings and breast imaging centers are ideal for fully reaping the preventative benefits of such risk prediction models at a large population level. However, in such settings, collecting and assembling the exhaustive family history used by BRCAPRO is not practical.
We have recently proposed three simplified versions of BRCAPRO: BRCAPROLYTE, BRCAPROLYTE-Plus, and BRCAPROLYTE-Simple [16]. We evaluated these tools on datasets collected in genetic counseling settings, and found that they entail only a modest loss of accuracy compared to BRCAPRO, especially BRCAPROLYTE-Plus and BRCAPROLYTE-Simple. This suggests that we can use these tools to achieve a balance between simplicity (an issue of utmost importance in primary care) and accuracy by carrying out genetic risk prediction in two stages. In the first stage, intended for primary care, risk will be assessed using a simplified version of BRCAPRO. Those found to be at sufficiently high risk will be referred to the second stage (counseling), where the full BRCAPRO will be used. A software implementation of such a two-stage approach is already available in HRA [17] which includes two sequentially administered surveys — “Short breast” and “Risk Clinic”, using limited and more exhaustive family information. The two-stage procedure has not been evaluated in primary care settings thus far.
Our aim is to formally develop and investigate the two-stage approach. We evaluate three versions of this approach, each with a different first stage tool – BRCAPROLYTE, BRCAPROLYTE-Plus, and BRCAPROLYTE-Simple. In each case, the second stage uses BRCAPRO on all available information.
Methods
Cohorts
We use retrospective data from three sources: Cancer Genetics Network (CGN), MD Anderson Cancer Center (MDA), and Newton-Wellesley Hospital (NWH). The first two have been described and analyzed earlier [14,16,18]; here we analyze them for the first time using two-stage approaches. In particular, the pedigree characteristics for each of seven sites in CGN and that of MDA can be found in Table 1 of Biswas et al (2013) [16]. For completeness, in Table 1, we list the characteristics of the two datasets combined (referred as CGN+MDA and analyzed as a whole throughout). MDA as well as all CGN sites except one consist of high risk families, i.e., the probands entered the study at least in part because of their personal and/or family history. The third cohort captures all probands referred for genetic counseling at the NWH. The vast majority of these individuals enter the sample through a primary care encounter, typically a breast imaging visit. They are subsequently referred to counseling if they have a prior history of ovarian cancer, meet National Comprehensive Cancer Network (NCCN) guidelines [19], their BRCAPROLYTE probability exceeds 10%, or are referred by a clinician based on his/her interpretation of risk. A fraction of this cohort sought genetic counseling after a relative had already been tested; they will be analyzed separately also. Pedigree characteristics of NWH data are listed in Table 1. Less than 10% of NWH probands are BRCA mutation carriers compared to 21% carriers in CGN+MDA data. The latter has also higher proportions of probands with breast and/or ovarian cancers along with younger affection ages. The median reported family size is about 20, highlighting the practical difficulty of collecting complete family information in primary care.
Table 1.
Pedigree Characteristics
| CGN+MDA | NWH | |
|---|---|---|
| Total pedigrees | 2713 | 1344 |
| Probands of AJ descent: n (%) | 744 (27.4) | 366 (27.2) |
| Probands tested BRCA1+: n (%) | 377 (13.9) | 49 (3.6) |
| Probands tested BRCA2+: n (%) | 202 (7.4) | 76 (5.7) |
| No. of members per pedigree: median (IQR) | 20 (15) | 19 (20) |
| Age of proband: median (IQR) | 49 (17) | 53 (16) |
| Males tested: n (%) | 87 (3.2) | 14 (1) |
| Males tested with BC: n (%) | 48 (1.8) | 6 (0.4) |
| Probands with unilateral BC: n (%) | 1628 (60) | 698 (51.9) |
| Probands with bilateral BC: n (%) | 244 (9) | 72 (5.4) |
| Probands with OC: n (%) | 245 (9) | 55 (4.1) |
| Probands with BC & OC: n (%) | 88 (3.2) | 17 (1.3) |
| BC age for proband: median (IQR) | 43 (14) | 47 (13.75) |
| OC age for proband: median (IQR) | 51 (14) | 53 (16.5) |
IQR: Inter-Quartile Range; BC: Breast Cancer; OC: Ovarian Cancer
Two-Stage Approach
In this approach, the first stage tool is either BRCAPROLYTE, BRCAPROLYTE-Plus, or BRCAPROLYTE-Simple. We had earlier also considered the Family History Assessment Tool (FHAT) [20] and another simpler version of BRCAPRO based on first-degree relatives only. Their performance was inferior compared to the three selected and thus we do not pursue them here. In the second stage, the complete BRCAPRO is used on those probands whose first stage probability exceeds a chosen cutoff.
BRCAPRO
BRCAPRO utilizes information on all available relatives including family structure, ages of diagnosis, current age/age at death for unaffected members, ethnicity, and additional information such as breast tumor markers and BRCA genetic test results, if available. We used the version implemented in BayesMendel 2.0–9.
BRCAPROLYTE
BRCAPROLYTE applies the BRCAPRO model using only information on the numbers and types of first- and second- degree relatives, which relatives are affected with breast and ovarian cancer, and their affection ages. We performed BRCAPROLYTE calculations using BRCAPRO after setting the current age/age at death of all relatives as missing to mimic the scenario when ages are not collected/known to the proband.
BRCAPROLYTE-Plus
BRCAPROLYTE does not collect data on ages of unaffected relatives leading to inflated carrier probabilities in general [16]. As the numbers of first- and second-degree relatives are collected, we can impute the ages of unaffected relatives to compensate for the inflation of probabilities, and thereby offset false positives. This idea is implemented in BRCAPROLYTE-Plus. The imputation of ages is based on an external large dataset as described elsewhere [16].
BRCAPROLYTE-Simple
BRCAPROLYTE-Plus needs knowledge of the numbers of each type of relative. A further simplification can be achieved by imputing this information when it is unknown. BRCAPROLYTE-Simple does this through two levels of imputation: number of relatives of each type and ages of unaffected relatives, based on an external dataset [16]. The burden of data collection is therefore the least with BRCAPROLYTE-Simple.
Evaluation Strategy
To establish a baseline, we apply each simplified tool and BRCAPRO separately to all probands and compare the results for NWH data. We earlier reported results for each of the simplified tools on the CGN and MDA data [16]. This is the first analysis addressing the two-stage approach.
We then evaluate the clinical impact of two-stage approaches by quantifying the reduction in genetic counseling burden as compared to applying BRCAPRO to all probands. We consider various combinations of cutoffs for the two stages (denoted as c1 and c2 respectively) and note the number of counselees whose carrier probabilities exceed c1 and/or c2. We compare scenarios constructed so that the numbers of carriers captured by the two-stage approaches and BRCAPRO are about the same.
The statistical evaluation of a two-stage approach is more involved than that of a single stage tool as results from both stages as well as dependence of the second stage on the first stage results must be considered. In Supplementary Methods, we show that the overall sensitivity (Se.O), specificity (Sp.O), Area Under ROC Curve (AUC.O), and predictive value positive and negative (PVP.O and PVN.O) can be written in terms of sensitivity and specificity of the first stage and of the second stage given the results of the first stage.
Next we calculate the ratio of the observed (O) number of carriers to the expected (E) number (O/E). To compute E, we need one carrier probability per proband. We use the first stage probability for counselees who undergo the first stage only (first stage probability < c1) while for the rest we use their second stage probability. We plot O/E, along with their 95% confidence intervals (CI), for a set of c1 values ranging from 0.01 to 0.1, the cutoffs typically used in practice.
We also consider scenarios where the percentage of counselees to be followed-up in the second stage is fixed in advance in consideration of resource constraints. This requires setting a specific cutoff c1 for the first stage. Depending on whether a proband was evaluated at the second stage, we use either the first or second stage probability and calculate AUC referred as AUC.p, where “p” is the fixed percentage follow-up and is set to 25%, 50%, and 75% in turn.
For all analyses, we also report the results when all probands are evaluated using BRCAPRO. We refer to each two-stage approach by the name of the corresponding first stage tool, as BRCAPRO is always used in the second stage. We used the statistical software R 3.1.0 for all computations.
Results
In NWH, BRCAPROLYTE (applied to all probands) overestimates carrier probabilities compared to BRCAPRO as reflected in O/E values smaller than 1 in the Supplementary Table 1. We also observe higher sensitivity and lower specificity of BRCAPROLYTE for a fixed cutoff. BRCAPROLYTE-Plus and BRCAPROLYTE-Simple are better calibrated with O/E values closer to 1, and show a trend towards better calibration than BRCAPRO. The AUCs of all three first stage tools are close to 0.65, the AUC of BRCAPRO in this dataset. We reported similar results for CGN and MDA [16].
Next, we quantify the consequences of using the two-stage approach on the clinical workflow. There are 576/2713 BRCA mutation carriers in CGN+MDA and 125/1344 in NWH. As shown in Figure 1, when BRCAPRO is applied to all probands, the carrier probabilities of 1036 and 363 probands exceed 10%, a cutoff traditionally used in clinical practice, though not necessarily optimum [21]. Of these, only 414 and 57 are carriers. The genetic counseling burden with this single-stage approach is the totality of probands (2713 and 1344) and genetic testing is done for 1036 and 363 probands. The sensitivity of BRCAPRO at this cutoff is 0.72 and 0.46 in CGN+MDA and NWH, respectively (Supplementary Table 1). The corresponding specificities are 0.71 and 0.75. Now let us compare these numbers with those for the two-stage approaches.
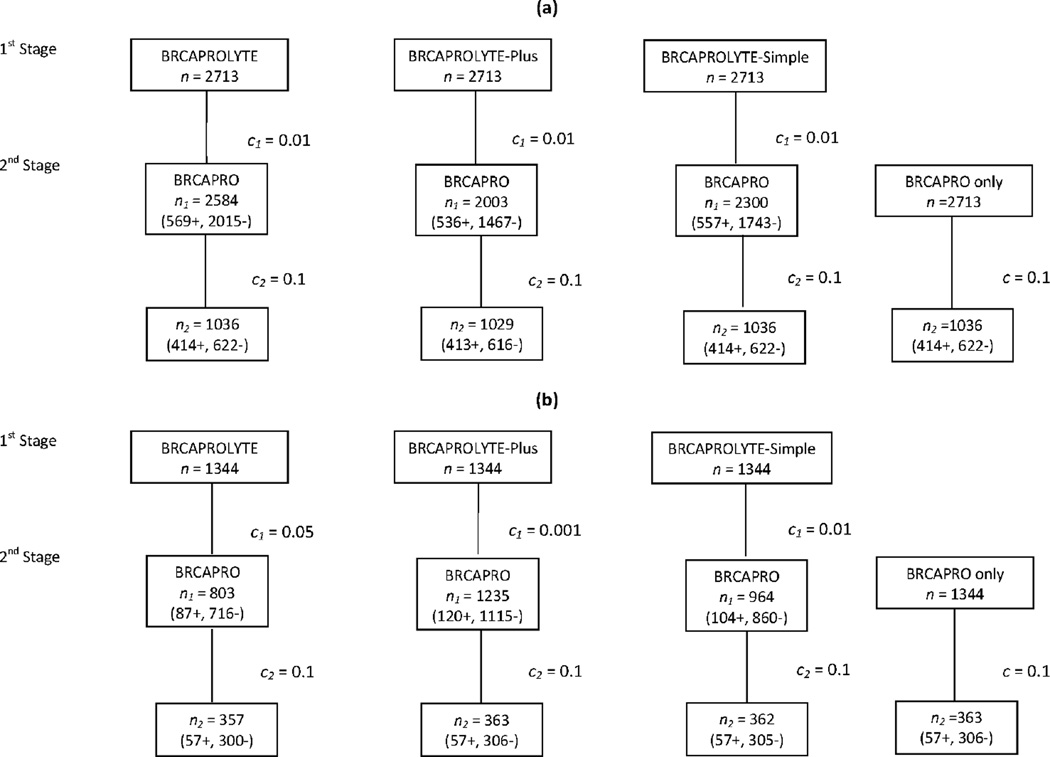
Figure 1.
Numbers of referrals made at each stage using a two-stage approach, as compared to using BRCAPRO only on all probands for (a) CGN+MDA and (b) NWH data. We denote by c1 and c2 the cutoffs used at the first and second stages, respectively. The number of probands whose carrier probability at the first stage exceed c1 is denoted by n1, and out of n1, the number of probands with second stage carrier probability exceeding c2 is denoted by n2. When we evaluate BRCAPRO alone, the cutoff is labeled as c and the number of probands with carrier probability exceeding c is n2. These numbers are further stratified by the carrier status in parentheses.
In Figure 1b, for NWH, we see that out of a total of 1344 probands that go through the first stage, 803, 1235, and 964 are referred to the second stage by BRCAPROLYTE, BRCAPROLYTE-Plus, and BRCAPROLYTE-Simple, respectively. Correspondingly, the reduction in genetic counseling burden as compared to direct counseling of all 1344 probands is 541 (40%), 109 (8%), and 380 (28%) families. In the second stage, the numbers of probands referred for genetic testing are close to 363 as obtained using BRCAPRO only. The sensitivity and specificity for all two-stage approaches are about the same as those of BRCAPRO (0.46 and 0.75; Figure 2 discussed below). The comparison on CGN+MDA is similar, as shown in Figure 1a. In particular, the reduction in genetic counseling is 129 (5%), 710 (26%), and 413 (15%) by using BRCAPROLYTE, BRCAPROLYTE-Plus, and BRCAPROLYTE-Simple in the first stage, respectively. Thus, two-stage approaches are able to capture the same numbers of carriers and achieve same sensitivity and specificity as BRCAPRO with a reduction of genetic counseling burden. Additional scenarios are considered in Supplementary Tables 1 and 2. We see that similar numbers of carriers can be captured using other cutoff combinations as well. The genetic counseling burden can be further reduced through a higher first stage cutoff, albeit with larger number of genetic tests.
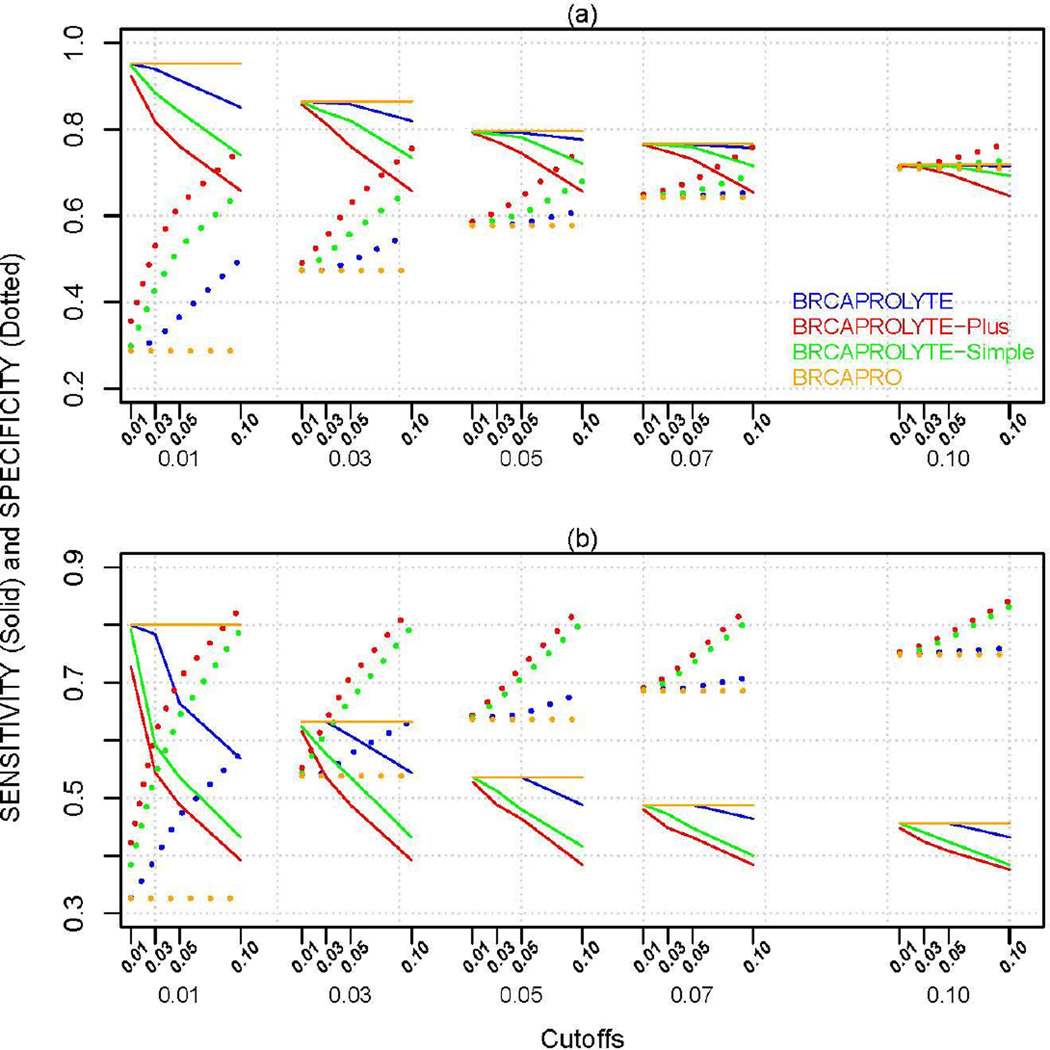
Figure 2.
Sensitivity (Se.O) and Specificity (Sp.O) of the two-stage approach and BRCAPRO for (a) CGN+MDA and (b) NWH data. The x-axis has two sets of cutoffs, c1 (first stage) followed by c2 (second stage) below it. For BRCAPRO, only one cutoff (indicated by the second level c2 values) is applicable.
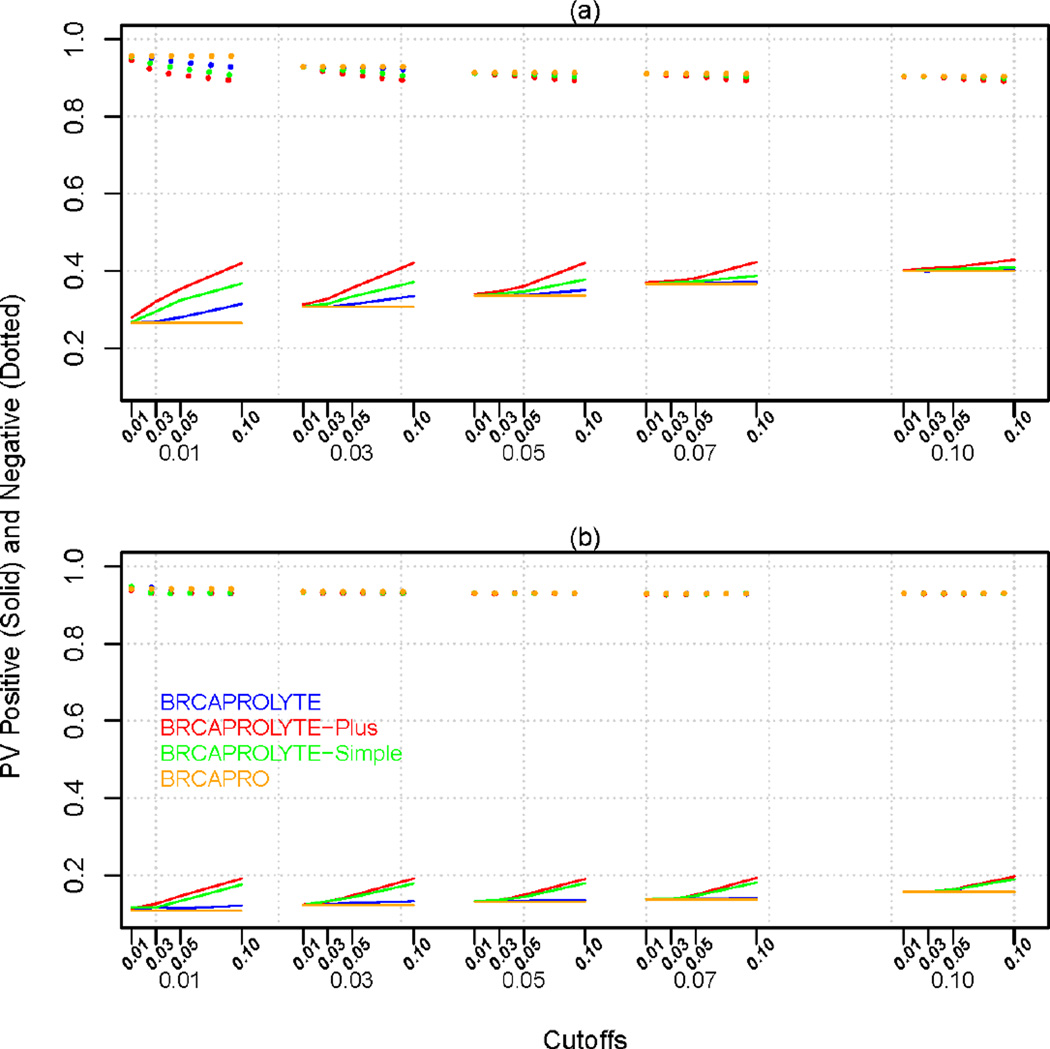
Figure 2 plots Se.O and Sp.O for specific cutoff combinations for the two stages. We see that if we want a two-stage approach to achieve same Se.O value (e.g., 80%) with a similar (or higher) Sp.O value as that of BRCAPRO, it is usually possible at different cutoff combinations for different models. In general, BRCAPROLYTE has high values of sensitivity while BRCAPROLYTE-Plus and BRCAPROLYTE-Simple give slightly higher values of specificity for the same sensitivity. If we choose the same cutoff for BRCAPRO when applied by itself and when applied as the second-stage (i.e., compare the curves within each column panel), then the former seems to have highest sensitivity and lowest specificity. However, by allowing the cutoffs to vary, the two-stage approaches can achieve similar values of sensitivity and specificity as BRCAPRO. Similar plots for PVP.O and PVN.O is in Figure 3, and the same trend is seen. Similar considerations apply to PVP.O and PVN.O.
Figure 3.
Predictive Value Positive (PVP.O) and Negative (PVN.O) of the two-stage approach and BRCAPRO for (a) CGN+MDA and (b) NWH data. The x-axis has two sets of cutoffs, c1 (first stage) followed by c2 (second stage) below it. For BRCAPRO, only one cutoff (c2) is applicable.
Table 2 lists AUC.O, AUC.25, AUC.50, and AUC.75. AUC.O values for all two-stage approaches are practically same as that of BRCAPRO. In fact, AUCs remain comparable even when the percentage of follow-up in the second stage is restricted to 25, 50, or 75%.
Table 2.
AUC (with CI) of the Two-Stage Approach and BRCAPRO. For AUC.p, the first stage cutoff corresponding to p (percentage followed-up in the second stage) is indicated as c1.
| Dataset | BRCAPROLYTE | BRCAPROLYTE- Plus |
BRCAPROLYTE- Simple |
BRCAPRO | |
|---|---|---|---|---|---|
| AUC.O | CGN+MDA | 0.79 (0.76, 0.81) | 0.78 (0.76, 0.81) | 0.79 (0.76, 0.81) | 0.78 (0.76, 0.81) |
| NWH | 0.66 (0.61 , 0.71) | 0.65 (0.6, 0.71) | 0.66 (0.61, 0.71) | 0.65 (0.59, 0.70) | |
|
AUC.75 (cutoff) |
CGN+MDA | 0.78 (0.76, 0.8), c1=0.049 |
0.78 (0.76, 0.8), c1=0.009 |
0.78 (0.76, 0.8), c1=0.018 |
|
| NWH | 0.65 (0.59, 0.7), c1=0.027 |
0.65 (0.6, 0.7), c1=0.005 |
0.65 (0.6, 0.7), c1=0.009 |
||
|
AUC.50 (cutoff) |
CGN+MDA | 0.78 (0.75, 0.8), c1=0.169 |
0.78 (0.76, 0.8), c1=0.039 |
0.78 (0.76, 0.8), c1=0.069 |
|
| NWH | 0.65 (0.6, 0.71), c1=0.08 |
0.64 (0.59, 0.7), c1=0.018 |
0.65 (0.59, 0.7), c1=0.028 |
||
|
AUC.25 (cutoff) |
CGN+MDA | 0.77 (0.74, 0.79), c1=0.479 |
0.77 (0.75, 0.8), c1=0.185 |
0.78 (0.75, 0.8), c1=0.274 |
|
| NWH | 0.63 (0.58, 0.69), c1=0.217 |
0.64 (0.58, 0.69), c1=0.068 |
0.64 (0.58, 0.7), c1=0.092 |
||
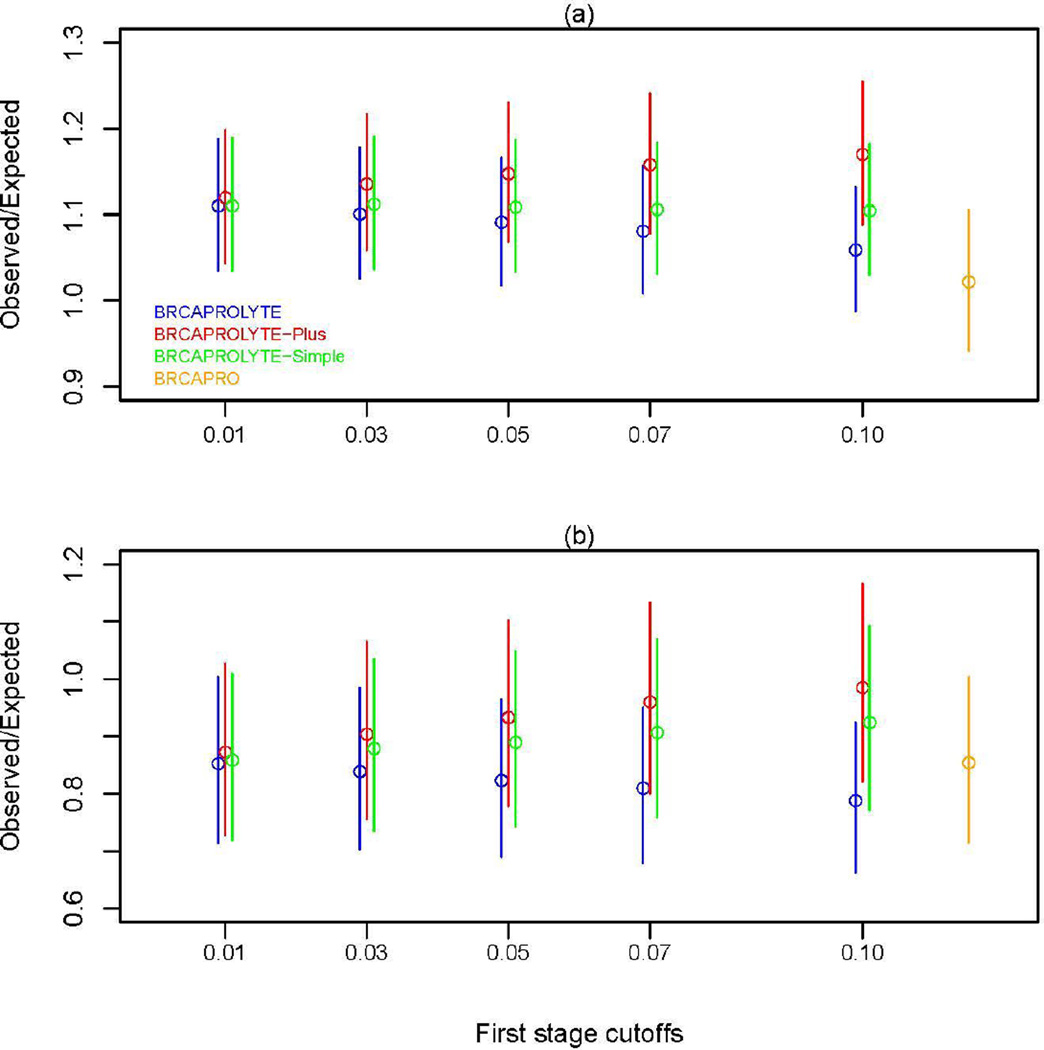
Next, we plot O/E values in Figure 4. In general, BRCAPROLYTE tends to overestimate the risk of mutation, BRCAPROLYTE-Plus tends to underestimate, and BRCAPROLYTE-Simple remains in between and is the most stable across varying cutoffs. When compared with BRCAPRO, the two datasets show somewhat different trends. In CGN+MDA, the two-stage approaches have somewhat worse calibration than BRCAPRO with BRCAPROLYTE performing the best (remains closest to 1) while in NWH, BRCAPROLYTE-Plus and BRCAPROLYTE-Simple have slightly better calibration than BRCAPRO. The CIs overlap substantially and so calibration of the two-stage approaches may not be very different from BRCAPRO although there seems to be some differences between high risk and community practice.
Figure 4.
Ratio of observed number of carriers to the expected number of carriers as predicted by the two-stage approach and BRCAPRO for (a) CGN+MDA and (b) NWH data.
In summary, the two-stage approach has similar discrimination and calibration, and can achieve a similar clinical impact without requiring a full evaluation in primary care.
Discussion
We proposed and evaluated two-stage approaches for genetic risk assessment. The first stage can be implemented in a primary care setting using limited family history information, and can be efficiently integrated into an EMR or other HIT solutions. Patients with sufficiently high risk can then be assessed in further detail, typically, though not necessarily, within a genetic counseling clinic. We showed that the overall performance of the two-stage approach is comparable to using the more complete assessment on all patients. Even though it is more complex than performing a single full evaluation of all patients, the clinical importance of this result lies in the fact that the latter is not currently scalable to primary care delivery. Moreover, testing everyone is currently not a practical option in the U.S. due to financial and other considerations. The two-stage approach makes it possible to screen the general population for risk of carrying BRCA mutations. It entails an increase in the burden of data collection in primary care, and a duplicate assessment on a relatively large subset of families, but also results in a reduction in genetic counseling activities, the most challenging and less easily scalable stage.
A practical issue is the choice of cutoffs for use in clinical settings. We illustrated the clinical implications using specific combinations of first and second stage cutoffs to quantify the genetic counseling and testing burden associated with using different first stage tools in different populations. For practical applications, different clinical scenarios may require a different balance of specificity, sensitivity, and burden of data collection, and these considerations should guide the choice of appropriate combinations.
One of the strengths of our study is use of high-risk as well as population-based data. Although specific assessments of discrimination and calibration for the two datasets were different, both analyses gave the same conclusion about the viability of the two-stage approach. Thus, our overall conclusions are likely to be applicable to the more general population, at least qualitatively. However, due to limitations of the data (as discussed below), these results may not be fully representative of the use of the two-stage approach in an unselected population.
At present, the paucity of medical environments where the potential for genetic testing is routinely incorporated into primary care workflows makes it challenging to carry out a population-based evaluation of the two-stage approach. Our analysis of the NWH data comes close, but still has limitations. The main limitation is that the NWH cohort is enriched for patients with BRCAPROLYTE probability exceeding 10%, which makes it more difficult to generalize our conclusions on the operating characteristics of the two-stage approach involving BRCAPROLYTE as the first step. Generally our analyses are likely to overestimate the number of high-risk families found in the first stage, when compared to an application of a two-stage approach to a completely unselected population.
The discrimination of the BRCAPRO model in the NWH cohort, as measured by AUC, is lower than reported in other datasets [16, 18]. This indicates that the carrier probability distributions for BRCA carriers and non-carriers are not well-separated. For example, the median probabilities for carriers and non-carriers are 0.07 and 0.03, respectively, compared to 0.34 and 0.03 for CGN+MDA data. If we take each proband in CGN+MDA and find a matching proband (with the closest probability) from NWH, the BRCAPRO AUC for this subset of NWH probands is 0.72, compared to 0.65 observed in the whole NWH data. Also, 175 probands in NWH have genetic test results available for at least one relative. Including their test results in BRCAPRO calculations increases the AUC from 0.65 to 0.79 showing the strong impact of genetic test results. However, if information on relatives’ test results is not used (as it will be burdensome to collect this information in the primary care), the results of the two-stage approaches for this subset are similar to what we found for the whole NWH data.
Of the three two-stage approaches, BRCAPROLYTE tends to over-predict. BRCAPROLYTE-Plus gives less inflated estimates but appears to be affected by under-prediction of carrier probabilities. BRCAPROLYTE-Simple seems to provide a better balance between over- and under-prediction. This is somewhat unexpected as BRCAPROLYTE-Simple uses less information on family structure than BRCAPROLYTE-Plus. From a practical point of view, this is a useful result as the burden of data collection in the first stage is the least through use of BRCAPROLYTE-Simple. To provide a rough estimate of savings in time, consider that it takes about 10 to 30 minutes to collect the vital status and age of every relative. Compared to that, it may take only about 3 minutes to obtain the age and relationships of each person with cancer, which is sufficient for applying BRCAPROLYTE-Simple. Future work can focus on estimation of the burden of data collection as well as the acceptance rate of the simplified tools in clinical practice.
In summary, our work proposes a new paradigm of formally integrating genetic risk assessment in primary care. By implementing the process of risk assessment in two stages, the proposed approach strikes a balance between two competing issues — identifying potential carriers among large populations who are currently not receiving adequate risk counseling and ensuring that the burden of exhaustive genetic counseling (second stage) remains manageable. By adjusting the cutoffs for the two stages, this approach allows identification of as many carriers as is practically possible. Although we focused on breast and ovarian cancer risk here, the approach is general and can be used for risk prediction for other cancers, for which well-established genetic models exist such as pancreas, colon, and melanoma [22–24].
Supplementary Material
Acknowledgments
This work was supported in part by the following grants: 1R03CA173834-02 and 2P30CA006516-47 from the National Cancer Institute.
Footnotes
Conflict of Interest
KH is a founder of and has a financial interest in Hughes Risk Apps (HRA), LLC. Dr. Hughes’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. KH has received honoraria from Myriad Genetics Speaker’s Bureau and holds stock options in 5 AM solutions. GP is on the SAB of HRA and holds stock options in the company.
References
- 1.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 3.Drohan B, Roche CA, Cusack JC, Hughes KS. Hereditary breast and ovarian cancer and other hereditary syndromes: using technology to identify carriers. Ann Surg Oncol. 2012;19:1732–1737. doi: 10.1245/s10434-012-2257-y. 2012. [DOI] [PubMed] [Google Scholar]
- 4.Drohan B, Ozanne EM, Hughes KS. Electronic health records and the management of women at high risk of hereditary breast and ovarian cancer. Breast J. 2009;15(Suppl 1):46–55. doi: 10.1111/j.1524-4741.2009.00796.x. [DOI] [PubMed] [Google Scholar]
- 5.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer- susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. 2004; Article 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. [Accessed 31 December 2015];CancerGene: https://www4.utsouthwestern.edu/breasthealth/cagene/
- 8. [Accessed 31 December 2015];BayesMendel Web-based Risk Service: http://bayesmendel.dfci.harvard.edu/risk/
- 9. [Accessed 31 December 2015];HughesRiskApps: http://www.HughesRiskApps.com.
- 10.Tai YC, Chen S, Parmigiani G, Klein AP. Incorporating tumor immunohistochemical markers in BRCA1 and BRCA2 carrier prediction. Breast Cancer Res. 2008;10:401. doi: 10.1186/bcr1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katki HA, Blackford A, Chen S, Parmigiani G. Multiple diseases in carrier probability estimation: accounting for surviving all cancers other than breast and ovary in BRCAPRO. Stat Med. 2008;27:4532–4548. doi: 10.1002/sim.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katki HA. Incorporating medical interventions into Mendelian mutation prediction models. BMC Med Genet. 2007;8:13. doi: 10.1186/1471-2350-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Blackford A, Parmigiani G. Tailoring BRCAPRO to Asian-Americans. J Clin Oncol. 2009;27:642–643. doi: 10.1200/JCO.2008.20.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biswas S, Tankhiwale N, Blackford A, Barrera AM, Ready K, Lu K, et al. Assessing the added value of breast tumor markers in genetic risk prediction model BRCAPRO. Breast Cancer Res Treat. 2012;133:347–355. doi: 10.1007/s10549-012-1958-z. [DOI] [PubMed] [Google Scholar]
- 15.Mazzola E, Blackford A, Parmigiani G, Biswas S. Recent Enhancements to the genetic risk prediction model BRCAPRO. Cancer Inform. 2015;14(S2):147–157. doi: 10.4137/CIN.S17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas S, Atienza P, Chipman J, Hughes K, Gutierrez Barrera AM, Amos CI, et al. Simplifying Clinical Use of the Genetic Risk Prediction Model BRCAPRO. Breast Cancer Res Treat. 2013;139:571–579. doi: 10.1007/s10549-013-2564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozanne EM, Loberg A, Hughes S, Lawrence C, Drohan B, Semine A, et al. Identification and management of women at high risk for hereditary breast/ovarian cancer syndrome. Breast J. 2009;15:155–162. doi: 10.1111/j.1524-4741.2009.00690.x. [DOI] [PubMed] [Google Scholar]
- 18.Parmigiani G, Chen S, Iversen ES, Friebel TM, Finkelstein DM, Anton-Culver H, et al. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med. 2007;147:441–450. doi: 10.7326/0003-4819-147-7-200710020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. [Accessed on 31 December 2015];NCCN guidelines: http://www.nccn.org/professionals/physician_gls/f_guidelines.asp#detection.
- 20.Gilpin CA, Carson N, Hunter AG. A preliminary validation of a family history assessment form to select women at risk for breast or ovarian cancer for referral to a genetics center. Clin Genet. 2000;58:299–308. doi: 10.1034/j.1399-0004.2000.580408.x. [DOI] [PubMed] [Google Scholar]
- 21.American Society of Clinical Oncology. Statement of the American Society of Clinical Oncology: Genetic testing for cancer susceptibility. J Clin Oncol. 1996;14:1730–1736. doi: 10.1200/JCO.1996.14.5.1730. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. J Am Med Assoc. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani GP. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol. 2007;25:1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Niendorf KB, Patel D, Blackford A, Marroni F, Sober AJ, et al. Estimating CDKN2A carrier probability and personalizing cancer risk assessments in hereditary melanoma using MelaPRO. Cancer Res. 2010;70:552–559. doi: 10.1158/0008-5472.CAN-09-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.