Abstract
Post-translational modification (PTM) is recognized as a major process accounting for protein structural variation, functional diversity, and the dynamics and complexity of the proteome. Since PTMs can change the structure and function of proteins, they are essential to coordinate signaling networks and to regulate important physiological processes in eukaryotes. Plants are constantly challenged by both biotic and abiotic stresses that reduce productivity, causing economic losses in crops. The plant responses involve complex physiological, cellular, and molecular processes, with stomatal movement as one of the earliest responses. In order to activate such a rapid response, stomatal guard cells employ cellular PTMs of key protein players in the signaling pathways to regulate the opening and closure of the stomatal pores. Here we discuss two major types of PTMs, protein phosphorylation and redox modification that play essential roles in stomatal movement under stress conditions. We present an overview of PTMs that occur in stomatal guard cells, especially the methods and technologies, and their applications in PTM identification and quantification. Our focus is on PTMs that modify molecular components in guard cell signaling at the stages of signal perception, second messenger production, as well as downstream signaling events and output. Improved understanding of guard cell signaling will enable generation of crops with enhanced stress tolerance, and increased yield and bioenergy through biotechnology and molecular breeding.
Keywords: phosphorylation, redox, guard cell, signaling, abiotic and biotic stresses
Introduction
Stomata are composed of a pair of specialized epidermal cells termed guard cells, which are responsible for regulating gas exchange and water loss through changing the size of the stomatal pores. The opening and closing of stomatal pores are affected by numerous factors, such as humidity, CO2, temperature, light, hormones, and pathogens. Changes in the turgor and volume of guard cells accordingly are required for stomatal movement, which are controlled by complex signaling networks (Azoulay-Shemer et al., 2015).
Abscisic acid (ABA) plays important roles in a broad range of plant physiological processes (e.g., seed germination and seedling growth) and plant responses to abiotic and biotic stresses (Lee and Luan, 2012). Under high salinity and drought conditions, the increased levels of ABA are perceived by the guard cells to promote stomatal closure and to inhibit of stomatal opening (Assmann, 2003). The mechanisms underlying ABA signaling in guard cells have been extensively studied (Pei et al., 1997; Schroeder et al., 2001; Assmann, 2003; Acharya et al., 2013; Zhang et al., 2015), which involve the binding of ABA to the receptors, activation of protein kinases, production of second messengers such as reactive oxygen species (ROS) and nitric oxide (NO), regulation of membrane ion channels, and eventually the decrease in turgor and stomatal closure (Schroeder et al., 2001; Zhang et al., 2015). In addition to abiotic stress, guard cells play an important role in limiting pathogen entrance to the plant body. The guard cell response to bacteria is triggered by the recognition of pathogen associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) on the plasma membrane. Upon PAMP recognition, one of the earliest responses is the change in ion fluxes across the membrane, leading to a rapid and transient extracellular alkalization and increase of Ca2+ in the cytosol (Boller and Felix, 2009). Ca2+ functions as a second messenger, activating downstream signaling players such as calcium-dependent protein kinases (CDPKs) to promote stomatal immunity responses. In addition, the apoplastic production of ROS by NADPH oxidase (Boller and Felix, 2009) is a hallmark of successful recognition of plant pathogens. Subsequent plant immune responses include transcriptional reprogramming, which involves the regulation of ROS homeostasis and activation of other protein kinases such as mitogen-activated protein kinases (MAPKs) (Boudsocq et al., 2010).
Stomatal studies are technically challenging because guard cells are small and of low abundance in leaves (Tallman, 2006). Methods for isolating guard cell protoplasts with relatively high purity have been reported over the past 30 years (Outlaw et al., 1981; Gotow et al., 1982, 1984; Zhu et al., 2010, 2014; Obulareddy et al., 2013). They have contributed considerably to the understanding of guard cell signaling. However, these methods are usually laborious and the yield is relatively low. The general principle of guard cell isolation is to release the guard cells from epidermal peels in a two-step process. In the first step the pavement and mesophyll cells are removed, and in the second step the guard cell wall is digested to facilitate the release of the guard cell protoplasts. It is important to note that there are important variations of the procedures according to different plant species (Zhu et al., 2016).
Stomatal movement in response to abiotic and biotic stresses is a fast process, which requires an efficient molecular regulation mechanism to relay the signals. Phosphorylation and redox control of the key players during both the signal perception and transduction in plant responses to abiotic and biotic stresses have demonstrated the high efficiency of protein PTMs in cell signaling (Grennan, 2007; Waszczak et al., 2015; Zhang et al., 2015). As the relevance of PTMs in plant stress responses has been demonstrated by independent studies over the years (Kodama et al., 2009; Lindermayr et al., 2010; Stecker et al., 2014; Kim et al., 2015; Yang et al., 2015), there is a growing interest to understand how specific PTMs control various aspects of stomatal guard cell functions. In this review, the frequently used approaches and methods in identification and quantification of PTMs are described. The main objective is to focus on the phosphorylation and redox events, and the recently identified proteins that undergo PTMs in guard cells in response to phytohormone and stress signals. We also discuss the different types of PTMs in the regulation of stomatal movement, and the challenges and perspectives of PTM proteomics.
Advances in protein PTM technologies
Significance of PTMs in biological processes
PTMs include chemical modifications of specific amino acid residues of a protein and/or cleavage of the translated sequence. They greatly increase the structural and functional diversity of proteins in a proteome. Currently, more than 300 different types of PTMs have been identified (Zhao and Jensen, 2009), including phosphorylation, glycosylation, acetylation, nitrosylation, ubiquitination, and proteolytic cleavage. These modifications affect the properties of the proteins (e.g., charge status and conformation), resulting in changes of activity, binding affinity, localization as well as stability. Most PTMs are highly controlled in the cells, and they often serve as rapid, specific, and reversible molecular switches to regulate biochemical and physiological processes. Different PTMs have also been shown to crosstalk in the modulation of molecular interactions between proteins or regulation within the same protein through multiple site modification, e.g., the histone code (Bannister and Kouzarides, 2011). Therefore, identification and functional characterization of PTMs are critical toward deciphering their roles in cellular processes in many different areas of biology and biomedical research.
Qualitative analysis of PTMs
In the past, PTMs were often studied at a specific amino acid residue of a particular protein level using molecular and biochemical approaches (Zhu et al., 2000; Reimer et al., 2002). Nowadays, the advances in biological mass spectrometry (MS) have allowed accurate identification and quantification of PTMs at the proteome scale. Two-dimensional gel electrophoresis (2-DE) was widely used in the early years of proteomics to identify PTMs, such as phosphorylation, nitrosylation, acetylation, and glycosylation (Llop et al., 2007; Roux et al., 2008; Scheving et al., 2012). Because PTMs can alter the isoelectric point and/or molecular weight, they may be detected when a change of spot location on the gel is observed between different samples. Different PTM protein stains have been developed to reveal specific PTMs, such as ProQ diamond and ProQ emerald to detect phosphoproteins and glycoproteins in the gels, respectively (Steinberg et al., 2001; Schulenberg et al., 2003; Ge et al., 2004). A big challenge has been to identify the PTM peptides and map the sites of modifications due to the low abundance nature of the modified protein species.
To overcome the challenge of capturing the relatively low abundance of PTM proteins compared with unmodified proteins, fractionation, and/or enrichment strategies have been employed during sample preparation (Lenman et al., 2008; Guo et al., 2014a; Aryal et al., 2015). The MS-based proteomics coupled with PTM enrichment typically has four steps. First, samples containing the total protein of interest are digested by a protease, such as trypsin. Second, the resulting peptides are subject to enrichment, in order to separate the PTM peptides of interest from the often abundant non-modified peptides. Third, the isolated PTM-peptide is analyzed by liquid chromatography (LC)-MS/MS for peptide identification and PTM site mapping. Finally, the MS spectra of the peptides are analyzed using different software algorithms and/or evaluated manually to ensure the accuracy and statistical significance of the data.
Among the different fractionation and enrichment strategies, affinity-based approaches are commonly used to enrich PTM proteins/peptides (Blagoev et al., 2004; Rush et al., 2005; Zhang et al., 2005; Fíla and Honys, 2012; Wang et al., 2015b). The affinity-based enrichment has the advantage of relatively high specificity and significant reduction of sample complexity for downstream LC-MS/MS analyses. For example, antiphosphotyrosine antibodies were successfully used to enrich for peptides with phosphotyrosines residues (Blagoev et al., 2004; Rush et al., 2005; Zhang et al., 2005). However, the antibody-based method is often limited by the availability and quality of the antibodies for the specific PTM of interest. Thus, in order to overcome this limitation, several non-antibody based strategies have been developed. For instance, immobilized metal affinity chromatography (IMAC) utilizes a metal chelating agent to bind trivalent metal cation, such as Fe3+ or Ga3+ (Thingholm and Jensen, 2009). The charged resin is used to bind phosphoproteins or phosphopeptides. Although this strategy is widely used, it has the following shortcomings: (1) If multiply phosphorylated peptides are present in high abundance, they may saturate the IMAC resin, resulting in retention of few singly and doubly phosphorylated species (Thingholm et al., 2008). (2) Acidic peptides will be enriched along with the phosphopeptides (Thingholm et al., 2008). In order to overcome this issue, the incubation buffer needs to be acidified to pH 2–2.5. At this pH, most acidic amino acids will be protonated, which will mask the negative charge of the carboxyl groups, preventing acidic peptides from binding onto the column. In contrast, at this pH most of the phosphate moieties are deprotonated and will bind to the column (Fíla and Honys, 2012). Another approach is to use titanium dioxide (TiO2) as a substitute for the metal chelating resin. The use of TiO2 resin under acidic conditions also prevents the retention of acidic peptides (Fíla and Honys, 2012). Interestingly, these two approaches are complementary in that IMAC has higher affinity for multiply phosphorylated peptides, while TiO2 preferentially binds singly phosphopeptides (Silva-Sanchez et al., 2015). Therefore, application of both approaches in a single experiment leads to a high coverage of the phosphoproteome.
For cysteine redox modifications, such as S-nitrosylation, a classic biotin-switch method developed by Jaffrey et al. (2001) was often used. Free cysteines of proteins are firstly blocked by a thiol-reactive reagent through alkylation. The S-nitrosylated cysteines are then reduced using ascorbate, which is not a strong reducing reagent allowing specific reduction of the S-NO bonds. After chemical substitution with a biotin-containing affinity molecule, N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP), the biotinylated proteins/peptides can be enriched by avidin chromatography. Although the classic biotin-switch method has been widely used, and over 300 proteins have been reported to be S-nitrosylated using this method (Lefièvre et al., 2007; Forrester et al., 2009), there are some technical issues inherent to this approach. The disulfide bonds in the proteins may decrease the efficiency of trypsin digestion and further peptide identification (Imai and Yau, 2013). Furthermore, the decomposition of biotin-HPDP may lead to a side reaction with free thiols, which can introduce false-positive signals through disulfide interchange (Forrester et al., 2007). Alternatively, a Thiopropyl Sepharose 6B (TPS6b) enrichment method was developed. The free thiols are alkylated during protein extraction. The proteins are then digested and further reduced prior to enrichment. TPS6b captures reduced thiols via disulfide exchange. TPS6b was initially used to increase the depth of proteome coverage for discovery experiments (Tambor et al., 2012). To date, it has been applied in several redox proteomics studies using cyanobacteria (Guo et al., 2014b), rat myocardium (Paulech et al., 2013), at enrichment efficiencies >95%. Recently, a six-plex iodoTMT technology has been developed to identify and quantify redox cysteines, including S-nitrosylation. Similar to the biotin-switch, free thiols are labeled with iodoTMT, and the TMT-labeled proteins or peptides can be enriched using an anti-TMT resin. This technology allows analysis of up to six samples simultaneously, thus increases throughput and reproducibility.
Quantitative analysis of PTMs
Multiple proteomics tools are available to quantify the absolute or relative abundances of proteins and their specific PTMs. The quantification of PTMs is crucial, since simple identification of a modification may not provide adequate information for determining its functional importance. In vivo and in vitro labeling methods have been developed to couple with MS in order to identify, map, and quantify PTMs (Gygi et al., 1999; Goodlett et al., 2001; Ong et al., 2002; Ross et al., 2004; Balmant et al., 2015; Glibert et al., 2015; Parker et al., 2015). Stable isotopes can be used to label proteins in vivo via metabolic incorporation. In this approach, one set of sample is grown in a natural nitrogen source (N14) and the other set is grown in a substituted isotopic nitrogen source (N15) as either an amino acid (stable isotopic labeling of amino acids in cell culture, SILAC) or an inorganic nitrogen source (K15NO3) (Thelen and Peck, 2007; Stecker et al., 2014; Minkoff et al., 2015). In SILAC, since the isotopes are introduced as a specific amino acid, the mass differences between the heavy and light peptides in the MS scan can be predicted, making the quantification easy. However, this approach is challenging in plant studies, since plants can synthesize amino acids from inorganic nitrogen. For example, the labeling efficiency achieved using exogenous amino acid in Arabidopsis cell cultures has been reported to only 70–80% (Gruhler et al., 2005). In contrast, metabolic labeling with 15N as a inorganic source has been shown to achieve 98% incorporation in both intact plants (Ippel et al., 2004) and cell cultures (Engelsberger et al., 2006). However, the mass difference between differentially labeled samples cannot be easily predicted. Sophisticated software is needed to perform quantitative analysis, which can be challenging when working with highly complex samples (Thelen and Peck, 2007).
Alternatively, isotope labeling can be done to extracted proteins/peptides in vitro through several different approaches, e.g., isotope-coded affinity tag (ICAT), isobaric tag for relative and absolute quantification (iTRAQ), tandem mass tag (TMT), and iodoTMT. Except for ICAT, the relative quantification of peptides between samples is obtained by comparing the ion intensities of the different tags in the MS/MS spectra. The use of stable isotope labeling for absolute quantification requires internal standards, which are pre-selected synthetic peptides with isotope amino acids from a protein of interest. An absolute quantification of a PTM can be achieved by measuring the abundances of the modified and unmodified peptides and comparing them with the known amount of the isotope standard used (Xie et al., 2011). Recently, the use of label-free approaches to quantify PTMs has shown promise. Label-free analysis allows direct comparison of MS signals between any numbers of samples, which makes it applicable to any types of samples, avoiding isotope reagent costs. One label-free approach is spectral counting, where the levels of a modified form of a protein can be estimated by counting the number of the MS/MS spectra of the modified peptide from the protein. It has been noted that the number of assigned MS/MS spectra directly correlates with protein amount (Cooper et al., 2010; Olinares et al., 2011). Although spectral counting is fairly reliable in the measurement of large changes, its accuracy decreases considerably when measuring small changes of proteins (Jurisica et al., 2007) This is why peptide precursor peak alignment and peak area based label-free approach has been more popular in accuracy and robustness (Zhu et al., 2009; Zhang et al., 2010; Lin et al., 2014b).
It is important to note that although all the approaches mentioned above have found utility in the identification and quantification of PTMs, they do not often address the issue of protein turnover in the course of the experiment. Overlooking this important issue may lead to misleading results (Muthuramalingam et al., 2013; Go et al., 2014). In order to account for differences in global protein level change, which could lead to a false positive or false negative result, researches have started to acquire PTM proteomics results and total protein proteomics results from parallel or different studies (Rose et al., 2012; Zhu et al., 2014). However, the success of this strategy is often low because some proteins identified in the PTM proteomics experiments are either absent or not quantified with confidence in the total proteomics experiments (vice versa) due to experimental variation and MS2 stochastical sampling (Chong et al., 2006; Lee and Koh, 2011). To overcome this problem, Parker et al. (2015) developed a double-labeling strategy, called cysTMTRAQ, where the isobaric tags iTRAQ and cysTMT are employed in a single experiment for the simultaneous determination of quantifiable cysteine redox changes and protein level changes. This notion of normalizing against total protein turnover can certainly be applied in the studies of other PTMs. PTMs exist in many different forms, are highly dynamic and important in rapid adjustment of protein functions as molecular switches (Lothrop et al., 2013). The aforementioned approaches and the development of new tools are expected to advance the PTMs studies in many areas of biology.
Protein phosphorylation in stomatal functions
Protein phosphorylation provides plants with a rapid and versatile mechanism to allow guard cells to respond rapidly to different environmental changes and adjust stomatal aperture accordingly (Zhang et al., 2015; Zou et al., 2015). Although the involvement of protein kinases and phosphorylation in stomatal movement has been known for decades, detailed molecular mechanisms connecting the key components have just emerged during the past 5 years. For instance, blue-light triggered stomatal opening is featured with phosphorylation and activation of the plasma membrane H+-ATPase by Blue Light Signaling 1 (BLUS1) (Takemiya et al., 2013). Here we focus on recent progress on the functions of protein phosphorylation in stomatal movement under abiotic and biotic stresses.
Protein phosphorylation in guard cells under abiotic stresses
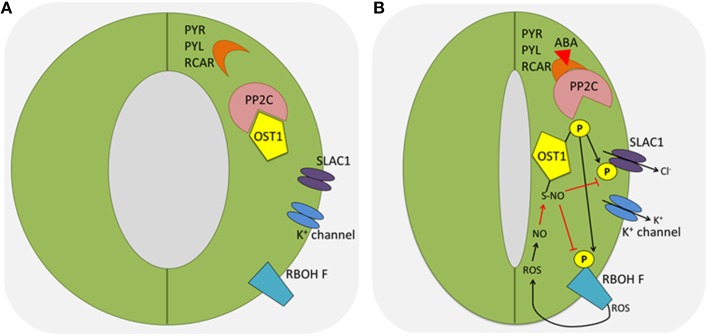
Guard cells are responsive to a plethora of environmental factors. The drought stress induced ABA signaling pathway has been well studied. In guard cells, the central node in the core ABA network is the Sucrose non-fermenting Receptor Kinase 2.6 (SnRK2.6), also known as Open Stomata 1 (OST1). In the absence of ABA, type A Protein Phosphatase 2C (PP2C) inhibits the kinase activity of OST1. In the presence of ABA, ABA binds to its receptor PYRabactin resistance/ PYrabactin-Like/Regulatory Components of ABA Receptor (PYR/PYL/RCAR). This hormone-receptor complex further binds and inhibits PP2C, thus releasing OST1 (Geiger et al., 2011; Lee et al., 2013; Zhang et al., 2015). Activated OST1 phosphorylates an array of substrates (Figure 1), including Respiratory Burst Oxidase Homolog (RBOH F) (Sirichandra et al., 2009; Acharya et al., 2013), SLow Anion Channel-associated 1 (SLAC1) (Vahisalu et al., 2008), QUickly-activating Anion Channel 1 (QUAC1) (Imes et al., 2013), K+ inward rectifying channel (KAT1) (Sato et al., 2009; Takahashi et al., 2013), and membrane water channel Plasma membrane Intrinsic Protein 2;1 (PIP2;1) (Grondin et al., 2015, Table 1). Phosphorylation of the substrates leads to the ROS burst, the promotion of anion and water efflux, and the inhibition of K+ influx. ROS can activate Ca2+ spikes in the cytosol, which can be further transduced by CDPK and CIPKs via phosphorylation of downstream target proteins (Drerup et al., 2013; Ye et al., 2013). Genetic and biochemical data indicated that MAPKs and some CDPKs such as CPK8 are activated by ABA downstream of ROS production in guard cells (Jammes et al., 2009; Wang et al., 2011; Marais et al., 2014; Zhang et al., 2015; Zou et al., 2015).
Figure 1.
Protein phosphorylation and redox modification in stomatal closure triggered by ABA. (A) In the absence of ABA, type A PP2C inhibits the kinase activity of OST1. (B) In the presence of ABA, ABA binds to its receptor PYR/PYL/RCAR, which further binds and inhibits PP2C, releasing and activating OST1. Activated OST1 phosphorylates an array of substrates, including RBOH F and SLAC1. Phosphorylated and active RBOH F promotes ROS burst. Later, ROS can activate Ca2+ spikes in the cytosol, which can be further transduced by CDPK and CIPKs via phosphorylation of downstream target proteins. In addition, ROS can modify OST1 and RBOH to inhibit their activities as a feedback mechanism to tune down ABA signaling (red arrows). ABA, abscisic acid; PP2C, protein phosphatase 2C; OST1, Open Stomata 1; PYR, pyrabactin resistance; PYL, PYR like; RCAR, regulatory components of ABA receptors; SLAC1, slow anion channel-associated 1; ROS, reactive oxygen species; NO, nitric oxide; RBOH F, respiratory burst oxidase protein F; CDPK, calcium dependent protein kinase; CIPK, CBL (Calcineurin B-like)-interacting protein kinase.
Table 1.
List of proteins undergoing PTMs in plant response to abiotic and biotic stresses.
| Organism | PTM | Modifier | Target | Evidence in GC | References |
|---|---|---|---|---|---|
| A. thaliana | Phosphorylation | OST1 protein kinase | AtRBOH F | Yes | Sirichandra et al., 2009 |
| A. thaliana | Phosphorylation | OST1 protein kinase | SLAC1 | Yes | Maierhofer et al., 2014 |
| A. thaliana | Phosphorylation | CPK6 | SLAC1 | Yes | Brandt et al., 2012 |
| A. thaliana | Phosphorylation | CPK21/23 | SLAC1 | Yes | Geiger et al., 2010 |
| A. thaliana | Phosphorylation | OST1 protein kinase | QUAC1 channel | Yes | Imes et al., 2013 |
| A. thaliana | Phosphorylation | OST1 protein kinase | K+ inward channel | Yes | Sato et al., 2009 |
| A. thaliana | Phosphorylation | OST1 protein kinase | PIP 2;1 aquaporin | Yes | Grondin et al., 2015 |
| A. thaliana | Phosphorylation | FLS2 | BAK1 | No | Schulze et al., 2010 |
| A. thaliana | Phosphorylation | BAK1 | FLS2 | No | Schulze et al., 2010 |
| A. thaliana | Phosphorylation | BAK1 | BIK1 | No | Lin et al., 2014a |
| A. thaliana | Phosphorylation | BIK1 | RBOH D | No | Li et al., 2014 |
| A. thaliana | Phosphorylation | CPK5 | RBOH D | Yes | Dubiella et al., 2013 |
| A. thaliana | Phosphorylation | BIK1 | MPK3 | Yes | Montillet et al., 2013 |
| A. thaliana | Phosphorylation | BIK1 | MPK6 | Yes | Montillet et al., 2013 |
| A. thaliana | Phosphorylation | RPM1-Induced kinase | RIN4 | Yes | Lee et al., 2015 |
| A. thaliana | Nitrosylation | NO | OST1 | Yes | Wang et al., 2015a |
| B. napus | Redox | ROS | BnSnRK2.4 | Yes | Zhu et al., 2014 |
| B. napus | Redox | ROS | IPMDH1 | Yes | Zhu et al., 2014 |
| A. thaliana | Nitrosylation | NO | NPR1 | No | Mou et al., 2003; Waszczak et al., 2015 |
| A. thaliana | Nitrosylation | NO | TGA transcriptional factor | No | Lindermayr et al., 2010 |
| A. thaliana | Nitrosylation | NO | RBOH D | No | Yun et al., 2011 |
| A. thaliana | Nitrosylation | NO | SABP 3 | No | Wang et al., 2009 |
Please refer to the text for abbreviations.
Evidence also indicates there are protein kinases that function in parallel to OST1. For example, CPK6 (Brandt et al., 2012b), CPK21/23 (Geiger et al., 2010), and Guard cell Hydrogen peroxide-Resistant 1 (GHR1) phosphorylate SLAC1 to activate the anion channels upon ABA treatment, forming a redundant signaling pathway (Table 1). However, detailed characterizations using a loss- and gain-of-function approach imply that OST1 is still the central node and limiting factor in ABA guard cell signaling (Acharya et al., 2013). In addition to drought stress, protein phosphorylation may also play a role in stomatal movement in response to other abiotic stresses. For example, mutants of MPK9 and MPK12 are partially impaired in cold-induced stomatal closure, suggesting that the two kinases may function in a cold signaling pathway (Jammes et al., 2009). Further studies are needed to identify the MPK9 and MPK12 targets, and their roles in guard cell cold stress signaling cascade.
Protein phosphorylation in guard cells under biotic stress
Stomatal pores, as the major gate of pathogen entry, constitute the first line of defense to prevent infection of the plant body by efficient stomatal closure. This process is initiated with the detection of the conserved PAMPs by various immune receptors. One of the best characterized interactions is the flagellin N-terminal 22 amino acid peptide (flg22) and the PRR Flagellin-Sensitive 2 (FLS2) and co-receptor Brassinosteroid insensitive 1-Associated Kinase 1 (BAK1) (Chinchilla et al., 2007; Sun et al., 2013). Using genetic and biochemical approaches, Schulze et al. (2010) showed that phosphorylation of FLS2 and BAK1 were detected within 15 s after flg22 treatment of Arabidopsis plants, and the kinase activity of BAK1 was required for flg22 perception (Table 1). Although there is no study specific for guard cells showing that FLS2 and BAK1 are phosphorylated after flg22 perception, the same events are likely to occur in the guard cells. It is known that FLS2 plays an important role in flg22-induced stomatal closure, since stomata in Arabidopsis fls2 mutant are completely impaired by flg22 carrying pathogen Pst. DC3000 (Zeng and He, 2010). Genetics and biochemical approaches showed that activation of FLS2 and BAK1 in Arabidopsis plants promote formation of the receptor complex with the botrytis-induced kinase 1 (BIK1, Table 1). BIK1 phosphorylates RBOH D (Table 1), which directly modulates stomatal closure in response to flg22, as rboh D mutant and Arabidopsis carrying RBOH DS39A, S343A, S347A exhibited completely impaired stomatal closure under flg22 treatment (Li et al., 2014). Interestingly, Arabidopsis RBOH D was also shown to be phosphorylated by CPK5 upon flg22 treatment (Dubiella et al., 2013, Table 1). In addition to RBOH D activation, flg22-induced FLS2 receptor complex also activates MPK3 and MPK6 to induce stomatal closure (Montillet et al., 2013). Thus, phosphorylation is an essential and common mechanism in pattern triggered immunity (PTI) responses. Downstream of PTI signaling includes regulation of K+ channels, turgor decrease in guard cells, and closure of the stomatal pores to prevent pathogen entry (Zhang et al., 2008; Zeng et al., 2010).
Successful pathogens deliver effector proteins into the plant cells to overcome PTI, and the effectors trigger the second layer of plant immunity called effector triggered immunity (ETI). For example, the bacterial effectors AvrB can be recognized by the plant immune receptor Resistance to Pseudomonas syringae pv Maculicola 1 (RPM1). Recognition of AvrB by RPM1 causes phosphorylation of RPM1-Interacting Protein4 (RIN4) by RPM1-Induced Protein Kinase (RIPK, Table 1). Recently, Lee et al. (2015) showed that RIN4T21D∕S160D∕T166D, a mutant with three phosphorylation sites changed to phosphorylation mimic aspartate residues, rendered Arabidopsis plants to exhibit large stomatal apertures and decreased resistance to P. syringae. This exemplifies how an effector protein facilitates pathogen infection by modulating host cell protein phosphorylation events.
Current questions in guard cell protein phosphorylation research
As more aspects of phosphorylation in stomatal movement have been revealed, more questions have also been raised. The findings of OST1, as a central player in the core ABA pathway, open doors for questions such as how the activity of this key modulator is controlled. Is it activated by autophosphorylation or by an upstream kinase? How is OST1 dephosphorylated? Recently, Casein Kinase 2 (CK2) has been shown as a negative regulator of OST1 by increasing the binding of CK2-phosphorylated OST1 to PP2C (Vilela et al., 2015). With many key kinases identified in guard cells including SnRKs, CPKs, and MPKs, how are these kinase pathways crosstalk to minimize redundancy, and how is the signal specificity determined? What are the target proteins involved in stomatal movement? With the development of kinase substrates screening (Umezawa et al., 2013; Wang et al., 2013) and techniques in live-cell phosphorylation detection (Hayashi et al., 2011), more studies are forthcoming toward better understanding of the phosphorylation-mediated stomatal movement at high spatial and temporal resolution. In addition, since phosphorylation is essential in both ABA and flg22 triggered stomatal closure, what are the convergent nodes and edges? This question is still under debate. One study showed that the flg22 response was independent of ABA signaling (Montillet et al., 2013), while another study indicated that flg22 induced stomatal closure was impaired in the ost1 mutant (Guzel Deger et al., 2015). It should be noted that in the first study 10 times more flg22 was used to cause stomatal movement in the ost1 mutant. Therefore, it is likely that both ABA-dependent and independent pathways are functional. In addition, different protein kinases may be involved in different pathways. For example, MPK3 and MPK6 were shown to be important players in flg22 triggered stomatal closure (Montillet et al., 2013), while MPK9 and MPK12 play critical roles in the guard cell ABA and cold stress signaling (Jammes et al., 2009), as well as yeast elicitor signaling (Salam et al., 2013). Moreover, both protein kinases and phosphatases control the dynamics of protein phosphorylation in guard cell signaling. However, only a few phosphatases have been identified in guard cells (Tseng and Briggs, 2010; Sun et al., 2012; Takemiya et al., 2013), and their interactions with key signaling proteins remaining largely elusive.
Redox-dependent PTMs in stomatal functions
As with protein phosphorylation and other PTMs, redox-dependent PTMs may function as molecular switches to turn on or off signaling processes in plant response to abiotic and biotic stresses. Thiol is a nucleophile that when exposed to oxidative stress, undergoes reversible inter- and intra-molecular disulfide bond formation, nitrosylation, glutathionylation, sulfenic acid and sulfinic acid modification, and irreversible sulfonic acid modification. Additionally, the high pKa values of protein cysteines make these residues highly responsive to small redox perturbation (Spoel and Loake, 2011). The production of ROS and NO is a common event during stomatal closure (Xie et al., 2014). The ROS and NO can serve as signaling molecules by modifying the reactive protein thiol groups. Here we focus on recent progress on the roles of redox-dependent cysteine PTMs in stomatal movement under abiotic and biotic stresses.
Redox PTMs in guard cells under abiotic stress
As described in the previous section, under drought stress ABA-induced stomatal closure is associated with an increase in NO and ROS production in guard cells (Zhang et al., 2001; Neill et al., 2008). The ROS production is catalyzed mainly by two types of enzymes, the plasma membrane NADPH oxidases, and the cell wall peroxidases (Sharma et al., 2012). Other ROS-generating enzymes, such as apoplastic amine oxidases and oxalate oxidases, may also be involved in ROS production leading to stomatal closure (Tripathy and Oelmüller, 2012). The NADPH oxidases are regulated by direct binding of Ca2+ (Kadota et al., 2015), phosphatidic acid (Zhang et al., 2009), Rac GTPases (Wong et al., 2007), and via phosphorylation by OST1 (Sirichandra et al., 2009), CDPKs (Kadota et al., 2015), and BIK1 (Kadota et al., 2014). Consequently, NADPH oxidase may integrate multiple upstream signaling events to promote stomatal closure. NO is produced by the nitrite-dependent nitrate reductase pathway (Desikan et al., 2002) and a nitric oxide associated 1 (NOA1) protein-dependent pathway (Lozano-Juste and León, 2010). It is important to note that the NOA1 is not a NO synthase (Moreau et al., 2008).
Although the essential function of ROS and NO in stomatal closure has been widely accepted, little is known about the underlying molecular mechanisms, by which they achieve the PTM regulation in guard cells. Thus, direct evidence for thiol-based redox regulation under stress conditions and a link between protein redox regulation and stomatal movement need to be established. A recent study showed that NO resulting from the ABA signaling caused S-nitrosylation of OST1 at the cysteine residue (Cys137) close to the kinase catalytic site (Table 1), and the PTM abolished the kinase activity (Figure 1B). This represents an interesting negative feedback mechanism by which ABA-induced NO helps to desensitize ABA signaling. Additionally, the authors showed that the Cys137 is evolutionarily conserved in some AMPK/SNF1-related kinases and glycogen synthase kinase 3/SHAGGY-like kinases (SKs) in plants, yeast and mammals, and the S-nitrosylation-mediated inhibition may be a general regulatory mechanism (Wang et al., 2015a). This example also highlighted how redox changes regulate protein kinase phosphorylation and signaling cascade in stomatal movement.
In a redox-proteomics study, Zhu et al. (2014) identified 65 and 118 potential redox responsive proteins in ABA and MeJA treated Brassica napus guard cells, respectively. The authors demonstrated that most of the proteins belong to functional groups such as energy, stress and defense, and metabolism. In addition, osmotic stress-activated protein kinase (BnSnRK2) and isopropylmalate dehydrogenase (IPMDH) were confirmed to be redox regulated and involved in stomatal movement (Table 1). These findings demonstrate the utility of redox-proteomics in discovering uncharacterized redox proteins and their roles in stomatal movement. Although some proteins have been identified to be redox regulated, their functions in regulating stomatal movement are still to be fully characterized.
Redox PTMs in guard cells under biotic stress
Pathogen perception initiates a signal transduction cascade including ROS and NO production, increase in Ca2+ influx, alkalization of the extracellular space, activation of MAPK, CDPK, salicylic acid (SA) pathway, and synthesis of ethylene (Arnaud and Hwang, 2015). The ROS and NO generated under biotic stresses are known to act as antimicrobial compounds. ROS are also known to be involved in cell wall cross-linking and blockage of pathogen infection (Torres et al., 2006). Furthermore, they play important signaling roles, e.g., in redox PTM of essential proteins in plant defense (Agurla et al., 2014). Methionine and cysteine residues of certain proteins are sensitive to H2O2 and NO (Hoshi and Heinemann, 2001). The sensitivity of the residues depends on the protein structure, neighboring residues, and solvent accessibility (Roos et al., 2013). H2O2 can react with a cysteine thiolate forming intra- or inter- disulfide bonds, sulfenic acid (-SOH), sulfinic acid (-SO2H), and sulfonic (-SO3H) acid (Dalle-Donne et al., 2006). NO can covalent bind to a cysteine thiol through S-nitrosylation.
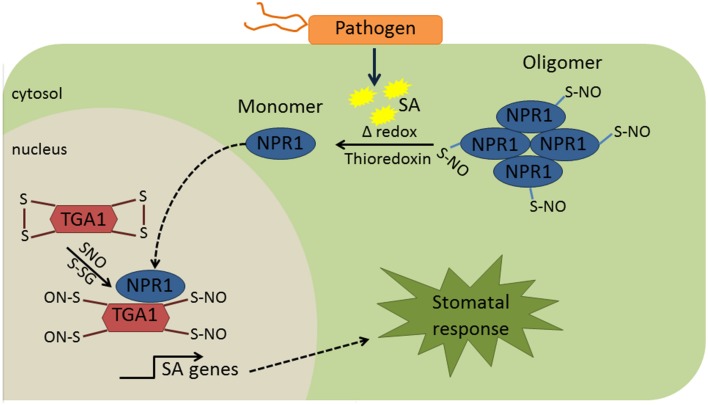
Although redox-dependent PTMs in biotic stresses is an emerging field, there are some examples showing the redox regulation of proteins in guard cells. In plant defense, Nonexpresser of PR gene 1 (NPR1) is one of a limited number of examples of protein redox regulation. NPR1 was detected primarily in the cytoplasm and nuclei of guard cells (Kinkema et al., 2000). Under normal conditions, NPR1 is retained in the cytoplasm as inactive disulfide-bonded oligomers, which is promoted by the S-nitrosylation at cysteine 156 (Table 1). In the presence of pathogen, an increase in SA mediates cellular redox changes, leading to thioredoxin-mediated reduction of the NPR1 oligomer to monomeric forms, which are then transported into the nucleus to activate plant immune processes (Mou et al., 2003; Waszczak et al., 2015). In the nucleus, SA mediated redox change causes de-nitrosylation and reduction of disulfide bonds in TGA transcriptional factors (Table 1) so that they can form an active transcriptional complex with NPR1 to turn on pathogenesis related (PR) genes (Lindermayr et al., 2010, Figure 2), and NPR1 is then phosphorylated and ubiquitinylated for degradation (Waszczak et al., 2015). Although protein redox regulation is not well studied in plant innate immunity, it is clear from the above example that modification of cysteine thiols can alter protein activity, function, and redox crosstalk with other modifications.
Figure 2.
Redox regulation of NPR1 and TGA1. Under normal conditions, NPR1 is retained in the cytosol as an oligomer. S-nitrosylation of NPR1 is known to promote NPR1 oligomerization. In the presence of pathogen, production of SA promotes cellular redox changes, which will contribute to reduction of the NPR1 oligomer to monomeric form. Monomeric form of NPR1 moves to the nucleus and binds to TGA1 that was nitrosylated due to cellular redox changes mediated by SA. The complex NPR1-TGA1 turns on the transcription of PR genes. Although this mechanism was not directly elucidated in the guard cells, it is likely to be the case since NPR1 was primarily in the cytosol and nucleus of guard cells (Kinkema et al., 2000). SA, salicylic acid; NPR1, nonexpresser of PR gene 1; TGA1, teosine glume architecture 1; SNO, S-nitrosylation; S-GS, S-glutathionylation.
Yun et al. (2011) demonstrated a NO biphasic control in pathogen triggered cell death. At the initial stage of pathogen infection, S-nitrosothiol (SNO) accumulation leads to accelerated cell death. Conversely, constitutively high SNO levels promote decreased cell death through S-nitrosylation of RBOH D (Table 1), leading to reduction in its activity and oxidative stress. This differential regulation seems important in fine tuning the extent of cell death under conditions of abiotic and biotic stresses, since both cause increases of NO levels. At a certain level of NO concentration, the signaling components of stomatal movement and plant response may be unresponsive or irreversibly regulated with detrimental effects on stress acclimation. During the NO burst, NO also promotes S-nitrosylation of an Arabidopsis SA-binding protein 3 (AtSABP3) at Cys280 (Table 1). The S-nitrosylation suppresses both SA binding, and its chloroplast carbonic anhydrase activity (Wang et al., 2009). Interestingly, in tobacco SABP3 showed antioxidant activity and plays a role in the hypersensitive defense response (Slaymaker et al., 2002). Although the role of SABP3 nitrosylation in stomatal closure in response to biotic stresses has not been studied, it may play a role in stomatal movement signaling as SA is known to promote stomatal closure (Khokon et al., 2011). The examples here demonstrated the great potential of redox regulation in stomatal movement in response to biotic stresses. The development of redox proteomics technologies such as the cysTMTRAQ (Parker et al., 2015) and application of genetics, biochemistry, metabolism, and bioinformatics tools would accelerate the discovery and characterization of redox-dependent PTMs of proteins and their roles in stomatal signaling and plant immunity.
Concluding remarks
Regulation of the size of the stomatal aperture is an essential mechanism in plants for optimizing the efficiency of water usage and photosynthesis. Stomatal movement through dynamic changes of the turgor of guard cells represents the output of integration of environmental signals with cellular signal transduction networks. Perception of abiotic and/or biotic stress signals triggers activation of signal transduction cascade, leading to rapid guard cell responses, which are known to be regulated by PTMs (e.g., protein phosphorylation and redox modification) of key players in the complex guard cell signaling networks. Over the past years, improvement and development of new tools in proteomics and MS have enabled the identification of PTMs of proteins involved in stomatal movement. In fact, LC-MS/MS based PTMomics technologies have become indispensable in identification and mapping of novel protein phosphorylation and redox modification sites. Additional sample preparation techniques, such as PTM enrichment and specific isotope labeling have greatly helped the detection and quantification of protein phosphorylation and redox changes, and thereby the understanding of PTM-controlled signaling pathways. The past decade has seen exciting discoveries in ABA and bacterial pathogen-triggered PTMs, especially phosphorylation and redox modification. Despite of current progress, guard cell PTMomics is still in its infancy and many aspects of protein level regulations remain elusive. For example, the crosstalk among different PTMs, and PTMs involved in regulating stomatal movement in response to other environmental factors are largely unknown. The fast advancement of proteomics technologies, together with genetics, molecular biology, biochemistry, and bioinformatics tools will accelerate the discovery and characterization of novel PTMs, and provide new insights into the complex protein phosphorylation and redox regulatory networks in guard cell signal transduction.
Author contributions
KB drafted the manuscript with assistance from TZ. KB drew the figures. TZ focused on phosphorylation sections. SC provided guidance, edited and finalized the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Jacqueline D. Schneider in the Chen lab for critical reading of the manuscript. Research on stomatal guard cell functions in the Chen lab has been supported by National Science Foundation (Projects 0818051, 1412547, and 1158000) and University of Florida.
References
- Acharya B. R., Jeon B. W., Zhang W., Assmann S. M. (2013). Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 200, 1049–1063. 10.1111/nph.12469 [DOI] [PubMed] [Google Scholar]
- Agurla S., Gayatri G., Raghavendra A. S. (2014). Nitric oxide as a secondary messenger during stomatal closure as a part of plant immunity response against pathogens. Nitric Oxide 43, 89–96. 10.1016/j.niox.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Arnaud D., Hwang I. (2015). A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol. Plant 8, 566–581. 10.1016/j.molp.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Aryal U. K., Ross A. R. S., Krochko J. E. (2015). Enrichment and analysis of intact phosphoproteins in Arabidopsis seedlings. PLoS ONE 10:e0130763. 10.1371/journal.pone.0130763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S. M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8, 151–153. 10.1016/S1360-1385(03)00052-9 [DOI] [PubMed] [Google Scholar]
- Azoulay-Shemer T., Palomares A., Bagheri A., Israelsson-Nordstrom M., Engineer C. B., Bargmann B. O. R., et al. (2015). Guard cell photosynthesis is critical for stomatal turgor production, yet does not directly mediate CO2- and ABA-induced stomatal closing. Plant J. 83, 567–581. 10.1111/tpj.12916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmant K., Parker J., Yoo M.-J., Zhu N., Dufresne C., Chen S. (2015). Redox proteomics of tomato in response to Pseudomonas syringae infection. Horticult. Res. 2:15043. 10.1038/hortres.2015.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev B., Ong S.-E., Kratchmarova I., Mann M. (2004). Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145. 10.1038/nbt1005 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M. R., McCormack M., Lee H., Shan L., He P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Brodsky D. E., Xue S., Negi J., Iba K., Kangasjärvi J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598. 10.1073/pnas.1116590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Chong P. K., Gan C. S., Pham T. K., Wright P. C. (2006). Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) reproducibility: implication of multiple injections. J. Proteome Res. 5, 1232–1240. 10.1021/pr060018u [DOI] [PubMed] [Google Scholar]
- Cooper B., Feng J., Garrett W. M. (2010). Relative, label-free protein quantitation: spectral counting error statistics from nine replicate MudPIT samples. J. Am. Soc. Mass Spectrom. 21, 1534–1546. 10.1016/j.jasms.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I., Scaloni A., Butterfield D. A. (2006). Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases. New Jersey, NJ: John Wiley & Sons. [DOI] [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J., Neill S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 16314–16318. 10.1073/pnas.252461999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup M. M., Schlücking K., Hashimoto K., Manishankar P., Steinhorst L., Kuchitsu K., et al. (2013). The Calcineurin B-Like Calcium Sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 6, 559–569. 10.1093/mp/sst009 [DOI] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.-S., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. 10.1073/pnas.1221294110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger W. R., Erban A., Kopka J., Schulze W. X. (2006). Metabolic labeling of plant cell cultures with K15NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2:14 10.1186/1746-4811-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fíla J., Honys D. (2012). Enrichment techniques employed in phosphoproteomics. Amino Acids 43, 1025–1047. 10.1007/s00726-011-1111-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester M. T., Foster M. W., Stamler J. S. (2007). Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J. Biol. Chem. 282, 13977–13983. 10.1074/jbc.M609684200 [DOI] [PubMed] [Google Scholar]
- Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009). Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559. 10.1038/nbt.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Rajkumar L., Guzman R. C., Nandi S., Patton W. F., Agnew B. J. (2004). Multiplexed fluorescence detection of phosphorylation, glycosylation, and total protein in the proteomic analysis of breast cancer refractoriness. Proteomics 4, 3464–3467. 10.1002/pmic.200400957 [DOI] [PubMed] [Google Scholar]
- Geiger D., Maierhofer T., Al-Rasheid K. A., Scherzer S., Mumm P., Ache P., et al. (2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4:ra32. 10.1126/scisignal.2001346 [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 107, 8023–8028. 10.1073/pnas.0912030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glibert P., Meert P., van Steendam K., van Nieuwerburgh F., de Coninck D., Martens L., et al. (2015). Phospho-iTRAQ: assessing isobaric labels for the large-scale study of phosphopeptide stoichiometry. J. Proteome Res. 14, 839–849. 10.1021/pr500889v [DOI] [PubMed] [Google Scholar]
- Go Y.-M., Roede J. R., Orr M., Liang Y., Jones D. P. (2014). Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of Cd toxicity. Toxicol. Sci. 139, 59–73. 10.1093/toxsci/kfu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett D. R., Keller A., Watts J. D., Newitt R., Yi E. C., Purvine S., et al. (2001). Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun Mass Spectr. 15, 1214–1221. 10.1002/rcm.362 [DOI] [PubMed] [Google Scholar]
- Gotow K., Kondo N., Syôno K. (1982). Effect of CO2 on volume change of guard cell protoplast from Vicia Faba L. Plant Cell Physiol. 23, 1063–1070. [Google Scholar]
- Gotow K., Shimazaki K.-I., Kondo N., Syôno K. (1984). Photosynthesis-dependent volume regulation in guard cell protoplasts from Vicia faba L. Plant Cell Physiol. 25, 671–675. [Google Scholar]
- Grennan A. K. (2007). Protein S-nitrosylation: potential targets and roles in signal transduction. Plant Physiol. 144, 1237–1239. 10.1104/pp.104.900228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A., Rodrigues O., Verdoucq L., Merlot S., Leonhardt N., Maurel C. (2015). Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27, 1945–1954. 10.1105/tpc.15.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhler A., Schulze W. X, Matthiesen R., Mann M., Jensen O. N. (2005). Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol. Cell. Proteomics 4, 1697–1709. 10.1074/mcp.M500190-MCP200 [DOI] [PubMed] [Google Scholar]
- Guo J., Gaffrey M. J., Su D., Liu T., Camp D. G. II. C., Smith R. D., et al. (2014a). Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 9, 64–75. 10.1038/nprot.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Nguyen A. Y., Dai Z., Su D., Gaffrey M. J., Moore R. J., et al. (2014b). Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics 13, 3270–3285. 10.1074/mcp.M114.041160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzel Deger A., Scherzer S., Nuhkat M., Kedzierska J., Kollist H., Brosché M., et al. (2015). Guard cell SLAC1-type anion channels mediate flagellin-induced stomatal closure. New Phytol. 208, 162–173. 10.1111/nph.13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999). Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999. 10.1038/13690 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Inoue S.-I., Takahashi K., Kinoshita T. (2011). Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 52, 1238–1248. 10.1093/pcp/pcr072 [DOI] [PubMed] [Google Scholar]
- Hoshi T., Heinemann S. T. (2001). Regulation of cell function by methionine oxidation and reduction. J. Physiol. 531(Pt 1), 1–11. 10.1111/j.1469-7793.2001.0001j.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Yau S. L. F. (2013). Quantitative Proteome Analysis: Methods and Applications. Boca Raton, FL: CRC Press. [Google Scholar]
- Imes D., Mumm P., Böhm J., Al-Rasheid K. A. S., Marten I., Geiger D., et al. (2013). Open Stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382. 10.1111/tpj.12133 [DOI] [PubMed] [Google Scholar]
- Ippel J. H., Pouvreau L., Kroef T., Gruppen H., Versteeg G., van den Putten P., et al. (2004). In vivo uniform (15)N-isotope labelling of plants: using the greenhouse for structural proteomics. Proteomics 4, 226–234. 10.1002/pmic.200300506 [DOI] [PubMed] [Google Scholar]
- Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001). Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197. 10.1038/35055104 [DOI] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20520–20525. 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurisica I., Wigle D. A., Wong B. (2007). Cancer Informatics in the Post Genomic Era: Toward Information-Based Medicine. New York, NY: Springer Science & Business Media. [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Khokon A. R., Okuma E., Hossain M. A., Munemasa S., Uraji M., Nakamura Y., et al. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34, 434–443. 10.1111/j.1365-3040.2010.02253.x [DOI] [PubMed] [Google Scholar]
- Kim J.-M., Sasaki T., Ueda M., Sako K., Seki M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. 10.3389/fpls.2015.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. 10.1105/tpc.12.12.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Tamura T., Hirasawa W., Nakamura K., Sano H. (2009). A novel protein phosphorylation pathway involved in osmotic-stress response in tobacco plants. Biochimie 91, 533–539. 10.1016/j.biochi.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Lee D., Bourdais G., Yu G., Robatzek S., Coaker G. (2015). Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 27, 2042–2056. 10.1105/tpc.114.132308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Koh H.-J. (2011). A label-free quantitative shotgun proteomics analysis of rice grain development. Proteome Sci. 9, 61. 10.1186/1477-5956-9-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C., Lim C. W., Lan W., He K., Luan S. (2013). ABA Signaling in guard cells entails a dynamic protein–protein interaction relay from the PYL-RCAR Family receptors to ion channels. Mol. Plant 6, 528–538. 10.1093/mp/sss078 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Luan S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35, 53–60. 10.1111/j.1365-3040.2011.02426.x [DOI] [PubMed] [Google Scholar]
- Lefièvre L., Chen Y., Conner S. J., Scott J. L., Publicover S. J., Ford W. C. L., et al. (2007). Human spermatozoa contain multiple targets for protein s-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics 7, 3066–3084. 10.1002/pmic.200700254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenman M., Sörensson C., Andreasson E. (2008). Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated Arabidopsis. Mol. Plant Microbe Interact. 21, 1275–1284. 10.1094/MPMI-21-10-1275 [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., et al. (2014). The FLS2-Associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Lin W., Li B., Lu D., Chen S., Zhu N., He P., et al. (2014a). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 3632–3637. 10.1073/pnas.1318817111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Yin H., Lo A., Ruffin M. T., Anderson M. A., Simeone D. M., et al. (2014b). Label-free relative quantification of alpha-2-macroglobulin site-specific core-fucosylation in pancreatic cancer by LC-MS/MS. Electrophoresis 35, 2108–2115. 10.1002/elps.201300376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Sell S., Müller B., Leister D., Durner J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. 10.1105/tpc.109.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llop E., Gallego R. G., Belalcazar V., Gerwig G. J., Kamerling J. P., Segura J., et al. (2007). Evaluation of protein N-glycosylation in 2-DE: erythropoietin as a study case. Proteomics 7, 4278–4291. 10.1002/pmic.200700572 [DOI] [PubMed] [Google Scholar]
- Lothrop A. P., Torres M. P., Fuchs S. M. (2013). Deciphering post-translational modification codes. FEBS Lett. 587, 1247–1257. 10.1016/j.febslet.2013.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J., León J. (2010). Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152, 891–903. 10.1104/pp.109.148023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer T., Diekmann M., Offenborn J. N., Lind C., Bauer H., Hashimoto K., et al. (2014). Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 7, ra86. 10.1126/scisignal.2005703 [DOI] [PubMed] [Google Scholar]
- Marais D. L. D., Auchincloss L. C., Sukamtoh E., McKay J. K., Logan T., Richards J. H., et al. (2014). Variation in MPK12 affects water use efficiency in Arabidopsis and reveals a pleiotropic link between guard cell size and ABA response. Proc. Natl. Acad. Sci. U.S.A. 111, 2836–2841. 10.1073/pnas.1321429111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff B. B., Stecker K. E., Sussman M. R. (2015). Rapid phosphoproteomic effects of abscisic acid (ABA) on wild-type and ABA receptor-deficient A. thaliana mutants. Mol Cell Proteomics 14, 1169–1182. 10.1074/mcp.M114.043307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet J.-L., Leonhardt N., Mondy S., Tranchimand S., Rumeau D., Boudsocq M., et al. (2013). An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11:e1001513. 10.1371/journal.pbio.1001513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Gyu I. L., Wang Y., Crane B. R., Klessig D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. 10.1074/jbc.M804838200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. 10.1016/S0092-8674(03)00429-X [DOI] [PubMed] [Google Scholar]
- Muthuramalingam M., Matros A., Scheibe R., Mock H.-P., Dietz K.-J. (2013). The hydrogen peroxide-sensitive proteome of the chloroplast in vitro and in vivo. Front. Plant Sci. 4:54. 10.3389/fpls.2013.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S., Barros R., Bright J., Desikan R., Hancock J., Harrison J., et al. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59, 165–176. 10.1093/jxb/erm293 [DOI] [PubMed] [Google Scholar]
- Obulareddy N., Panchal S., Melotto M. (2013). Guard cell purification and RNA isolation suitable for high throughput transcriptional analysis of cell-type responses to biotic stresses. Mol. Plant Microbe Interact. 26, 844–849. 10.1094/MPMI-03-13-0081-TA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares P. D., Kim J., Davis J. I., van Wijk K. J. (2011). Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23, 2348–2361. 10.1105/tpc.111.086454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., et al. (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 1, 376–386. 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Outlaw W. H., Mayne B. C., Zenger V. E., Manchester J. (1981). Presence of both photosystems in guard cells of Vicia faba L. Plant Physiol. 67, 12–16. 10.1104/pp.67.1.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Balmant K., Zhu F., Zhu N., Chen S. (2015). cysTMTRAQ—an integrative method for unbiased thiol-based redox proteomics. Mol. Cell. Proteomics 14, 237–242. 10.1074/mcp.O114.041772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulech J., Solis N., Edwards A. V. G., Puckeridge M., White M. Y., Cordwell S. J. (2013). Large-scale capture of peptides containing reversibly oxidized cysteines by thiol-disulfide exchange applied to the myocardial redox proteome. Anal. Chem. 85, 3774–3780. 10.1021/ac400166e [DOI] [PubMed] [Google Scholar]
- Pei Z. M., Kuchitsu K., Ward J. M., Schwarz M., Schroeder J. I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 Mutants. Plant Cell 9, 409–423. 10.1105/tpc.9.3.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer U., Reineke U., Schneider-Mergener J. (2002). peptide arrays: from macro to micro. Curr. Opin. Biotechnol. 13, 315–320. 10.1016/S0958-1669(02)00339-7 [DOI] [PubMed] [Google Scholar]
- Roos G., Foloppe N., Messens J. (2013). Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid. Redox Signal. 18, 94–127. 10.1089/ars.2012.4521 [DOI] [PubMed] [Google Scholar]
- Rose C. M., Venkateshwaran M., Volkening J. D., Grimsrud P. A., Maeda J., Bailey D. J., et al. (2012). Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744. 10.1074/mcp.M112.019208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., et al. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169. 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- Roux M. M., Radeke M. J., Goel M., Mushegian A., Foltz K. R. (2008). 2DE identification of proteins exhibiting turnover and phosphorylation dynamics during sea urchin egg activation. Dev. Biol. 313, 630–647. 10.1016/j.ydbio.2007.10.053 [DOI] [PubMed] [Google Scholar]
- Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., et al. (2005). Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101. 10.1038/nbt1046 [DOI] [PubMed] [Google Scholar]
- Salam M. A., Jammes F, Hossain M. A., Ye W., Nakamura Y., Mori I. C., et al. (2013). Two guard cell-preferential MAPKs, MPK9 and MPK12, regulate YEL signalling in Arabidopsis guard cells. Plant Biol. (Stuttg) 15, 436–442. 10.1111/j.1438-8677.2012.00671.x [DOI] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., et al. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424, 439–448. 10.1042/BJ20091221 [DOI] [PubMed] [Google Scholar]
- Scheving R., Wittig I., Heide H., Albuquerque B., Steger M., Brandt U., et al. (2012). Protein S-nitrosylation and denitrosylation in the mouse spinal cord upon injury of the sciatic nerve. J. Proteomics 75, 3987–4004. 10.1016/j.jprot.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Schroeder J. I., Allen G. J., Hugouvieux V., Kwak J. M., Waner D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 627–658. 10.1146/annurev.arplant.52.1.627 [DOI] [PubMed] [Google Scholar]
- Schulenberg B., Aggeler R., Beechem J. M., Capaldi R. A., Patton W. F. (2003). Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 278, 27251–27255. 10.1074/jbc.C300189200 [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., et al. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451. 10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Jha A. B., Dubey R. S., Pessarakli M., Sharma P., Jha A. B., et al. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions, reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:e217037 10.1155/2012/217037 [DOI] [Google Scholar]
- Silva-Sanchez C., Li H., Chen S. (2015). Recent advances and challenges in plant phosphoproteomics. Proteomics 15, 1127–1141. 10.1002/pmic.201400410 [DOI] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.-C., Davanture M., Lee S., Djaoui M., et al. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. 10.1016/j.febslet.2009.08.033 [DOI] [PubMed] [Google Scholar]
- Slaymaker D. H., Navarre D. A., Clark D., del Pozo O., Martin G. B., Klessig D. F. (2002). The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 11640–11645. 10.1073/pnas.182427699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S. H., Loake G. J. (2011). Redox-based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14, 358–364. 10.1016/j.pbi.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Stecker K. E., Minkoff B. B., Sussman M. R. (2014). Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 165, 1171–1187. 10.1104/pp.114.238816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg T. H., Pretty On Top K., Berggren K. N., Kemper C., Jones L., Diwu Z., et al. (2001). Rapid and simple single nanogram detection of glycoproteins in polyacrylamide gels and on electroblots. Proteomics 1, 841–855. [DOI] [PubMed] [Google Scholar]
- Sun X., Kang X., Ni M. (2012). Hypersensitive to red and blue 1 and its modification by protein phosphatase 7 are implicated in the control of Arabidopsis stomatal aperture. PLoS Genet. 8:e1002674. 10.1371/journal.pgen.1002674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Ebisu Y., Kinoshita T., Doi M., Okuma E., Murata Y., et al. (2013). bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 6, ra48. 10.1126/scisignal.2003760 [DOI] [PubMed] [Google Scholar]
- Takemiya A., Yamauchi S., Yano T., Ariyoshi C., Shimazaki K. (2013). Identification of a regulatory subunit of protein phosphatase 1 which mediates blue light signaling for stomatal opening. Plant Cell Physiol. 54, 24–35. 10.1093/pcp/pcs073 [DOI] [PubMed] [Google Scholar]
- Tallman G. (2006). Guard cell protoplasts: isolation, culture, and regeneration of plants. Methods Mol. Biol. 318, 233–252. 10.1385/1-59259-959-1:233 [DOI] [PubMed] [Google Scholar]
- Tambor V., Hunter C. L., Seymour S. L., Kacerovsky M., Stulik J., Lenco J. (2012). CysTRAQ - a combination of iTRAQ and enrichment of cysteinyl peptides for uncovering and quantifying hidden proteomes. J. Proteomics 75, 857–867. 10.1016/j.jprot.2011.09.027 [DOI] [PubMed] [Google Scholar]
- Thelen J. J., Peck S. C. (2007). Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346. 10.1105/tpc.107.053991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingholm T. E., Jensen O. N. (2009). Enrichment and characterization of phosphopeptides by immobilized metal affinity chromatography (IMAC) and mass spectrometry. Methods Mol. Biol. 527, 47–56. xi. 10.1007/978-1-60327-834-8_4 [DOI] [PubMed] [Google Scholar]
- Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008). SIMAC (Sequential Elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671. 10.1074/mcp.M700362-MCP200 [DOI] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D. G., Dangl J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. 10.1104/pp.106.079467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy B. C., Oelmüller R. (2012). Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 7, 1621–1633. 10.4161/psb.22455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.-S., Briggs W. R. (2010). The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses[C][W]. Plant Cell 22, 392–402. 10.1105/tpc.109.066423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8. 10.1126/scisignal.2003509 [DOI] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.-F., Nishimura N., Chan W.-Y., Valerio G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491. 10.1038/nature06608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela B., Nájar E., Lumbreras V., Leung J., Pagès M. (2015). Casein kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol. Plant 8, 709–721. 10.1016/j.molp.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Wang P., Du Y., Hou Y.-J., Zhao Y., Hsu C.-C., Yuan F., et al. (2015a). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. 10.1073/pnas.1423481112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.-J., Van Oosten M. J., et al. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110, 11205–11210. 10.1073/pnas.1308974110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.-S., Pandey S., Li S., Gookin T. E., Zhao Z., Albert R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216. 10.1186/1471-2164-12-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Q., Feechan A., Yun B.-W., Shafiei R., Hofmann A., Taylor P., et al. (2009). S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. 10.1074/jbc.M806782200 [DOI] [PubMed] [Google Scholar]
- Wang Z.-G., Lv N., Bi W.-Z., Zhang J.-L., Ni J.-Z. (2015b). Development of the affinity materials for phosphorylated proteins/peptides enrichment in phosphoproteomics analysis. ACS Appl. Mater. Interfaces 7, 8377–8392. 10.1021/acsami.5b01254 [DOI] [PubMed] [Google Scholar]
- Waszczak C., Akter S., Jacques S., Huang J., Messens J., Van Breusegem F. (2015). Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 66, 2923–2934. 10.1093/jxb/erv084 [DOI] [PubMed] [Google Scholar]
- Wong H. L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., et al. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034. 10.1105/tpc.107.055624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Liu T., Qian W.-J., Petyuk V. A., Smith R. D. (2011). Liquid chromatography-mass spectrometry-based quantitative proteomics. J. Biol. Chem. 286, 25443–25449. 10.1074/jbc.R110.199703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Mao Y., Zhang W., Lai D., Wang Q., Shen W. (2014). Reactive oxygen species-dependent nitric oxide production contributes to hydrogen-promoted stomatal closure in Arabidopsis. Plant Physiol. 165, 759–773. 10.1104/pp.114.237925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Mu J., Chen L., Feng J., Hu J., Li L., et al. (2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. 10.1104/pp.114.255216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Muroyama D., Munemasa S., Nakamura Y., Mori I. C., Murata Y. (2013). Calcium-dependent protein kinase CPK6 positively functions in induction by yeast elicitor of stomatal closure and inhibition by yeast elicitor of light-induced stomatal opening in Arabidopsis. Plant Physiol. 163, 591–599. 10.1104/pp.113.224055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.-W., Feechan A., Yin M., Saidi N. B. B., Le Bihan T., Yu M., et al. (2011). S-Nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. 10.1038/nature10427 [DOI] [PubMed] [Google Scholar]
- Zeng W., Melotto M., He S. Y. (2010). Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 21, 599–603. 10.1016/j.copbio.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., He S. Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae Pv tomato DC3000 in Arabidopsis. Plant Physiol. 153, 1188–1198. 10.1104/pp.110.157016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Ueberheide B. M., Waldemarson S., Myung S., Molloy K., Eriksson J., et al. (2010). Protein quantitation using mass spectrometry. Methods Mol. Biol. 673, 211–222. 10.1007/978-1-60761-842-3_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Chen S., Harmon A. C. (2015). Protein phosphorylation in stomatal movement. Plant Signal. Behav. 9:e972845. 10.4161/15592316.2014.972845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., He S. Y., Assmann S. M. (2008). The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 56, 984–996. 10.1111/j.1365-313X.2008.03657.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D. W., Song C.-P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126, 1438–1448. 10.1104/pp.126.4.1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., et al. (2005). Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250. 10.1074/mcp.M500089-MCP200 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., et al. (2009). Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377. 10.1105/tpc.108.062992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Jensen O. N. (2009). Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641. 10.1002/pmic.200900398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Klemic J. F., Chang S., Bertone P., Casamayor A., Klemic K. G., et al. (2000). Analysis of yeast protein kinases using protein chips. Nat. Genet. 26, 283–289. 10.1038/81576 [DOI] [PubMed] [Google Scholar]
- Zhu M., Jeon B. W., Geng S., Yu Y., Balmant K., Chen S., et al. (2016). Preparation of epidermal peels and guard cell protoplasts for cellular, electrophysiological, and -omics assays of guard cell function. Methods Mol. Biol. 1363, 89–121. 10.1007/978-1-4939-3115-6_9 [DOI] [PubMed] [Google Scholar]
- Zhu M., Simons B., Zhu N., Oppenheimer D. G., Chen S. (2010). Analysis of abscisic acid responsive proteins in Brassica napus guard cells by multiplexed isobaric tagging. J. Proteomics 73, 790–805. 10.1016/j.jprot.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Zhu M., Zhu N., Song W.-Y., Harmon A. C., Assmann S. M., Chen S. (2014). Thiol-based redox proteins in abscisic acid and methyl jasmonate signaling in Brassica Napus guard cells. Plant J. 78, 491–515. 10.1111/tpj.12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Smith J. W., Huang C.-M., Zhu W., Smith J. W., Huang C.-M. (2009). Mass spectrometry-based label-free quantitative proteomics, mass spectrometry-based label-free quantitative proteomics. Biomed Res. Int. 2010:e840518. 10.1155/2010/840518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.-J., Li X.-D., Ratnasekera D., Wang C., Liu W.-X., Song L.-F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. 10.1105/tpc.15.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]