ABSTRACT
Usutu virus (USUV), one of the most neglected Old World encephalitic flaviviruses, causes epizootics among wild and captive birds and sporadic infection in humans. The dynamics of USUV spread and evolution in its natural hosts are unknown. Here, we present the phylogeny and evolutionary history of all available USUV strains, including 77 newly sequenced complete genomes from a variety of host species at a temporal and spatial scaled resolution. The results showed that USUV can be classified into six distinct lineages and that the most recent common ancestor of the recent European epizootics emerged in Africa at least 500 years ago. We demonstrated that USUV was introduced regularly from Africa into Europe in the last 50 years, and the genetic diversity of European lineages is shaped primarily by in situ evolution, while the African lineages have been driven by extensive gene flow. Most of the amino acid changes are deleterious polymorphisms removed by purifying selection, with adaptive evolution restricted to the NS5 gene and several others evolving under episodic directional selection, indicating that the ecological or immunological factors were mostly the key determinants of USUV dispersal and outbreaks. Host-specific mutations have been detected, while the host transition analysis identified mosquitoes as the most likely origin of the common ancestor and birds as the source of the recent European USUV lineages. Our results suggest that the major migratory bird flyways could predict the continental and intercontinental dispersal patterns of USUV and that migratory birds might act as potential long-distance dispersal vehicles.
IMPORTANCE
Usutu virus (USUV), a mosquito-borne flavivirus of the Japanese encephalitis virus antigenic group, caused massive bird die-offs, mostly in Europe. There is increasing evidence that USUV appears to be pathogenic for humans, becoming a potential public health problem. The emergence of USUV in Europe allows us to understand how an arbovirus spreads, adapts, and evolves in a naive environment. Thus, understanding the epidemiological and evolutionary processes that contribute to the emergence, maintenance, and further spread of viral diseases is the sine qua non to develop and implement surveillance strategies for their control. In this work, we performed an expansive phylogeographic and evolutionary analysis of USUV using all published sequences and those generated during this study. Subsequently, we described the genetic traits, reconstructed the potential pattern of geographic spread between continents/countries of the identified viral lineages and the drivers of viral migration, and traced the origin of outbreaks and transition events between different hosts.
INTRODUCTION
Isolated for the first time from a Culex neavei mosquito in South Africa in 1959 (1, 2), Usutu virus (USUV) was subsequently detected in different species of mosquitoes and birds throughout sub-Saharan countries (1, 3). USUV is an Old World flavivirus included in the Japanese encephalitis virus (JEV) antigenic complex along with several human and animal pathogens (e.g., JEV, West Nile virus [WNV], Murray Valley encephalitis virus [MVEV], and Saint Louis encephalitis virus [SLEV]) (4). Its genome is a single-stranded RNA molecule of positive polarity coding one long open reading frame (ORF) that is flanked by a type 1 capped 5′-terminal noncoding region (NCR) and a 3′-terminal NCR required for genome replication and translation. The polyprotein includes four 5′ structural genes (coding for capsid [C], premembrane [prM], membrane [M], and envelope [E]) and seven nonstructural (NS) genes (coding for the NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 proteins) produced as a result of co- and posttranslationally proteolytic processing by viral and cellular proteases (4). The virus is transmitted and maintained in the natural cycle by mosquitoes (mostly of the Culex genus) as vectors with birds as the main amplifying hosts, while humans are considered incidental or dead-end hosts. A very recent study showed that bats could also be infected by USUV and might act as amplifying hosts (5). In 1996, USUV emerged outside Africa and caused deaths among Eurasian blackbird (Turdus merula) populations in the Tuscany region of Italy (6). In the following years, the virus could be detected in several Central European countries, including Austria, Hungary, Spain, Switzerland, Belgium, Czech Republic, and Germany (7–16). In addition, USUV-specific seroconversion among birds has been demonstrated in England, Poland, and Greece (17–19). Within the last years, there is increasing evidence of clinically apparent human USUV infections characterized by fever, rash, jaundice, headache, nuchal rigidity, hand tremor, and hyperreflexia (20–24). The detection of USUV-specific antibodies in blood donors in Germany and Italy highlights the fact that USUV can also be transmitted to humans without causing any symptoms (25, 26). However, nothing is known about the evolutionary dynamics of USUV and how the virus interacts with its various host and vector species. Although few studies related to USUV genetic diversity at the country level have been conducted, there have been no studies specifically addressing the evolutionary events responsible for adaptation to the hosts and spread of USUV, making it a successful pathogen responsible for neuroinvasive disease in multiple host species, including humans. To understand the evolutionary mechanisms of USUV, we obtained the complete genome sequences of 77 USUV strains sampled from a variety of host species (principally mosquitoes and birds). With these and all available USUV sequences with known geographic and temporal information data, we conducted an expansive analysis of USUV to assess the drivers and barriers of viral migration and the pattern of evolutionary dynamics and to trace the origin of the outbreaks, as well as movement patterns of USUV strains between continents/countries and transitions between host species.
RESULTS
Genome characterization.
The USUV genomes ranged in size from 11,032 to 11,066 nucleotides (nt). The predicted ORFs were 10,305 nt (3,434 amino acids [aa]) flanked by a 5′ NCR consisting of 95 to 96 nt and highly variable 3′ termini ranging from 631 to 664 nt in length. The genetic variation across the viral genome (see Fig. S1 in the supplemental material) was relatively heterogeneous and indicated that USUV has maintained a highly conserved genome since its first detection in 1959 in Africa. The identity matrices for the genome and individual genes were greater than 94%, except for Central African Republic strain ArB1803 (GenBank accession no. KC754958), which exhibited an overall 80% nucleotide and 94% amino acid similarity to all known USUV strains (see Fig. S1). Several structural and nonstructural genes (coding for prM, E, NS1, NS3, and NS5) exhibited higher nucleotide than amino acid divergence. Strikingly the greatest variation in both amino acids and nucleotides was apparently observed in the nonstructural genes coding for the NS1, NS2A, NS3, and NS5. Conversely, the C, NS2B, and NS4A genes were the most conserved genes.
Phylogenetic analysis.
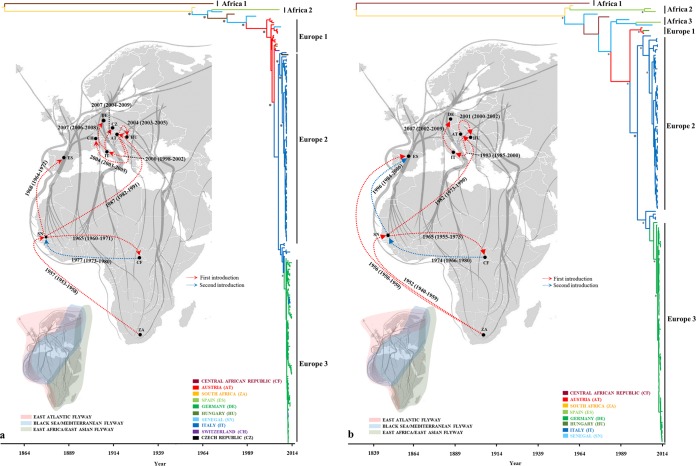
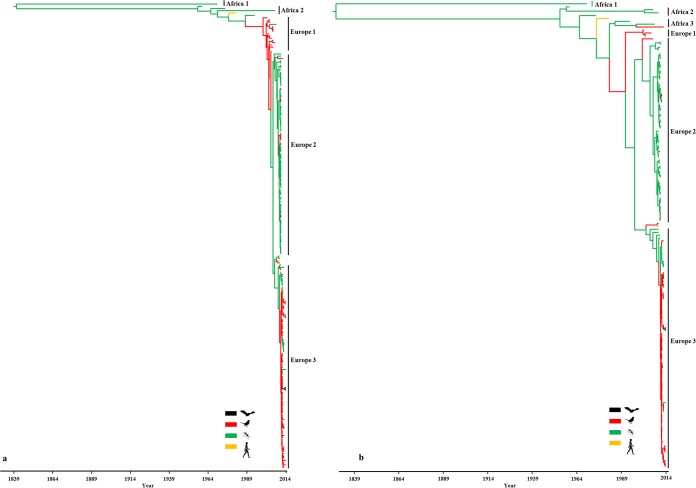
The similar topologies inferred by maximum likelihood (ML) and Bayesian maximum clade credibility (MCC) phylogenies of the E, NS5, and complete genome data sets revealed that all European strains (except the Spanish strains) fell into a monophyletic group, suggesting a single introduction event into Central Europe (Fig. 1; see Fig. S2 and S3 in the supplemental material). The detailed analysis of the European strains demonstrated that all USUV strains detected in Central Europe form 3 major lineages (Europe 1 to 3) defined by place of sampling. Europe 1 consists of strains from Austria, Hungary, Switzerland and one from Senegal (based on the E gene data set [Fig. 1A; see Fig. S3]), and Europe 2 includes Italian and Czech Republic strains, while Europe 3 consists of all German strains and some Italian strains (based on the E gene data set [Fig. 1A; see Fig. S3]). However, the latter two are not monophyletic, as the German lineage evolved from a subset of Italian viruses (Fig. 1; see Fig. S2 and S3). The European group is also notable for a star-like structure in which the three dominant lineages connect viruses sampled from multiple time points. The MCC and ML phylogenies also revealed that all strains from Africa and Spain fell within 2 (E gene) or 3 (NS5 and complete genome) distinct lineages, designated Africa 1, which includes the highly divergent singleton ArB1803 strain (KC754958), and poorly differentiated lineages Africa 2 and 3, which comprise sub-Saharan and Spanish strains (Fig. 1; see Fig. S2 and S3). The ML and MCC trees of NS5 and complete genome data sets reinforced the fact that USUV could be classified into 6 distinct lineages (5). Our phylogenetic analyses (Fig. 1B; see Fig. S2 and S3) further revealed that the USUV from Spain belong to the African monophyletic lineages 2 and 3, indicating multiple introduction of the virus in this European country. Although the host-to-host transmission analysis revealed possible phylogenetic clustering (relatively small clusters of sequences) of mosquitoes in Europe 2 and birds in Europe 3, it is likely the effect of an unbalanced sampling of different hosts (Fig. 2).
FIG 1 .
Bayesian maximum clade credibility (MCC) trees representing the time scale phylogeny, reconstruction of spatiotemporal spread pattern of USUV in Africa and Europe and the three major flyway (East Atlantic, Black Sea/Mediterranean, and East Africa/West Asian flyways; see color codes on the small map) networks (marked with gray lines onto the map) of migratory birds (66). The colored branches of MCC trees represent the most probable geographic location of their descendant nodes (see color codes). Bayesian posterior probabilities (≥90%) and 1,000 parallel maximum likelihood bootstrap replicates (≥70%) are indicated at the nodes (asterisks). The main lineages are indicated to the right of the tree. Time is reported in the axis below the tree and represents the year before the last sampling time (2014). Dotted lines on the maps between locations represent the temporal dynamics of USUV spatial diffusion with the most probable sources and target localities of branches in MCC trees. The distinct introductions within or between Africa and Europe are represented by different colored lines, while the estimated TMRCA (oldest possible year of introduction) of USUV strains from different countries are shown with 95% posterior time intervals in parentheses.
FIG 2 .
Maximum clade credibility trees of E (a) and NS5 (b) gene sequences colored by reconstructed host species (Markov jump process). Branches are colored by host and represent the transitions between different host species for the virus and the host of the common ancestor of all USUV strains.
Phylogeography and spatiotemporal dynamics of USUV.
In order to assess the drivers of viral migration and explore the origin of the outbreaks, a discrete-trait phylogeography analysis (27) using the Markov jumps method (28) was used to reconstruct the USUV movements between continents/countries and to compare them with the three major African-European flyways of migratory birds. The data sets exhibit a strong temporal signal and the coefficient of rate variation supports the use of a relaxed clock model (Table 1; see Fig. S4 in the supplemental material). Phylogenetic comparisons and the phylogeny-trait association tests indicated a very strong geographical clustering of European lineages (P < 0.001), reflecting a significant population subdivision, while for African lineages, there was evidence of significant gene flow between distinct regions (P < 0.05) (Fig. 1; see Table S1 in the supplemental material). This further strengthens the assumption that the genetic diversity of USUV in Central Europe is shaped primarily by in situ evolution rather than by extensive migration. Phylogenetic analysis also revealed that the long-distance movement pattern of USUV between countries and continents occurred and our estimate of 4 intercontinental and 8 continental viral migration events (Fig. 1 and 3) based on available data sets surely underestimates the real number. The time scale phylogenies indicate that all predicted viral migration events occurred in the last 60 years (Fig. 1). Although the USUV diversity in Europe appears to have emerged in the last decade, the phylogenies suggest relative long-term circulation of USUV in Europe (Fig. 1; see Fig. S3 in the supplemental material). The limited number of available sequences and the lack of USUV data from several sub-Saharan and North-African countries make it difficult to infer with confidence the spatiotemporal pattern of African lineages. However, in order to explore how the network of the flyways of migratory birds may drive movements of USUV, we performed a Bayesian phylogeny-trait association test to reveal the extent of clustering by migratory flyway and superimposed the three major flyways (East Atlantic, Black Sea/Mediterranean, and East Africa/West Asian flyways) onto the map with simulated patterns of viral dissemination. The results suggest that these flyway networks are strongly consistent with the spatial movements observed in the genetic data (significant clustering for the East Atlantic and/or Black Sea/Mediterranean flyways [Fig. 1; see Table S1]), suggesting possible viral export via migratory birds from Africa to Western Europe (Spain) using the east Atlantic flyway and/or Black Sea/Mediterranean flyway and to Central Europe through the Black Sea/Mediterranean corridor (Fig. 1). Whereas the USUV strains in Central Europe are monophyletic, resulting from a single introduction from Africa, our estimates of two introductions of African virus variants into Spain likely underestimate the real number. Furthermore, the singleton and ungrouped variants among African viruses complicate the estimation of the number of viral migration events into sub-Saharan countries.
Table 1 .
Comparison of nucleotide substitution rates and times to common ancestry of USUV from the E and NS5 genes and complete genome phylogenetic reconstructions using MCMC analysis
| Parameter | E gene (n = 205) | NS5 gene (n = 161) | Complete genome (n = 91) |
|---|---|---|---|
| Sampling time interval | 1959–2014 | 1959–2014 | 1959–2014 |
| Evolutionary rate (95% HPD) | 1.13 × 10−3 (8.20 × 10−4 to 1.46 × 10 −3) | 8.29 × 10−4 (6.09 × 10−4 to 1.27 × 10−3) | 6.49 × 10−4 (3.67 × 10−4 to 9.02 × 10−4) |
| Coefficient of variation | 0.61 | 0.90 | 0.78 |
| TMRCA (95% HPD) | |||
| Global | 1852 (1807–1891) | 1829 (1681–1956) | 1464 (1131–1713) |
| Africa 1 | 1852 (1807–1891) | 1829 (1681–1956) | 1464 (1131–1713) |
| Africa 2 | 1957 (1953–1958) | 1952 (1940–1959) | 1943 (1920–1961) |
| Africa 3 | N/Aa | 1974 (1967–1981) | 1962 (1950–1974) |
| Europe 1 | 1987 (1982–1991) | 1982 (1973–1990) | 1974 (1966–1980) |
| Europe 2 | 2004 (2003–2005) | 1993 (1985–2000) | 1995 (1993–2000) |
| Europe 3 | 2007 (2006–2008) | 2007 (2002–2009) | 2007 (2004–2009) |
N/A, not applicable.
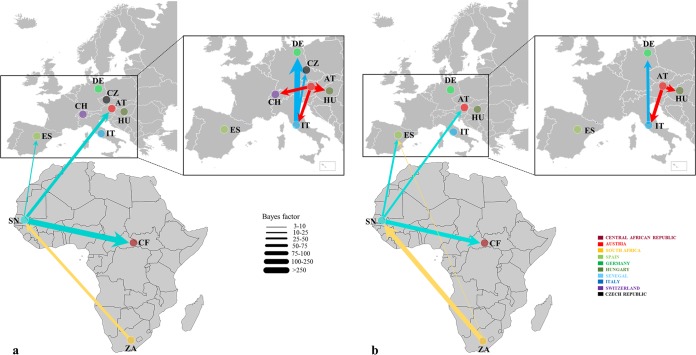
FIG 3 .
Migration pattern of USUV between or within Africa and Europe. Viral migration patterns based on E (a) and NS5 (b) genes are indicated between the different regions and are proportional to the strength of the transmission rate (Bayes factor [BF]). The color of the connections indicates the origin and the direction of migration. Only connections with a BF of <3 are shown.
A discrepancy in the accuracy of the estimated global times to the most recent common ancestor (TMRCA) for the E, NS5, and complete genome data sets has been observed (Table 1). However, the estimated TMRCAs for each lineage (except Africa 1) were very similar in the E, NS5, and complete genome MCC trees (Table 1). Based on the available data, we estimated that the virus likely emerged in South Africa from an ancestor that existed at least from the beginning of the 16th century (Table 1; see Fig. S3 in the supplemental material). The TMRCA of the European lineages indicates a very recent emergence (Table 1). We estimate that USUV was likely first introduced in Western Europe (Spain) during the period from 1950 through the 1960’s, followed by a second introduction in Central Europe (Austria) between 1970 and the 1980’s and a recent third introduction in Western Europe (Spain) around 1996 (Fig. 1; see Fig. S3). Europe 1 seems to be a descendant of an ancestor that probably existed in Senegal and is likely the origin of the first epizootics in Central Europe. Europe 2 was estimated to have arisen from an ancestor that existed in Austria around 1993 (95% high-probability density [HPD] for 1985 to 2000; posterior probability [pp] = 0.98), whereas Europe 3 emerged from an Italian variant around 2007 (95% HPD for 2004 to 2009; pp = 0.97) (Fig. 1). Figure S5 gives the pp distribution for the country of origin of each European epizootic. The Europe 1 lineage (mainly of Austrian origin) shares a common ancestor with basal Austrian strains. Similar relationships of the members within Europe 2 and 3 have been observed. These results further provide strong support for in situ evolution of the European lineages.
The scenario of the USUV spatial diffusion patterns between/within Africa and Europe has been reconstructed using a Bayes factor (BF) test under Bayesian stochastic search variable (BSSVS) analysis (i.e., non-zero rates supported by a BF of >3). The earliest migration event was detected from South Africa to Senegal between 1943 and 1957 (Fig. 3), after which the virus dispersed to Central African Republic around 1965. The virus then spread northward from Senegal into Europe in two distinct directions, to Spain and Austria, respectively (Fig. 3). From Austria, the virus probably spread into Hungary and Italy. From Italy, the virus was introduced into Germany and probably in Czech Republic. Nevertheless, the strongest epidemiological links based on the BF estimates have been detected between South Africa and Senegal, Senegal and the Central African Republic, Senegal and Austria, Austria and Italy, Austria and Hungary, and Italy and Germany (Fig. 3). In addition, the identified links between Africa and Europe were across countries from which samples were not available and tended to exhibit lower BF support in comparison with short-distance linkages (Fig. 3).
Population dynamics and host species analysis of USUV strains.
Results of the evolutionary time-resolved analyses for each data set are summarized in Table 1. Thus, the E gene exhibited a mean rate of 1.13 × 10−3 (95% HPD, 8.20 × 10−4 to 1.46 × 10−3) substitutions site−1 year−1, about twice that for NS5, 9.29 × 10−4 (95% HPD, 6.09 × 10−4 to 1.27 × 10−3) substitutions site−1 year−1, with rates for individual lineages in a narrow range of 6 × 10−4 to 2 × 10−3 substitutions site−1 year−1. Furthermore, the rate estimates for complete genome were lower than that for E and NS5, 6.49 × 10−4 (3.67 × 10−4 to 9.02 × 10−4) substitutions site−1 year−1 (Table 1). These rates are comparable to that previously estimated for other flaviviruses (29).
In order to investigate the rate of transition events (Markov jumps) of USUV between different host species, we performed a discrete-trait analysis using strains derived from mosquitoes, birds, humans, and bats. The median number of jumps was non-zero for mosquito-avian [15 (E) and 19 (NS5)] avian-mosquito [9 (E) and 12 (NS5)] mosquito-human [2 (E) and 1 (NS5)] and avian-bat [2 (E) and 3 (NS5)] transitions, but only the avian-mosquito and mosquito-avian transitions were supported by a pp of 95%. The most likely host species of the common ancestor of all USUV strains was the mosquito (pp = 0.42) (Fig. 2). Our host-based transition phylogenies revealed that European lineages possibly have arisen from a bird-derived USUV strain, further supporting potential introduction into Europe via migratory birds.
Potential cleavage sites, cysteine residues, and glycosylation sites in USUV polyprotein.
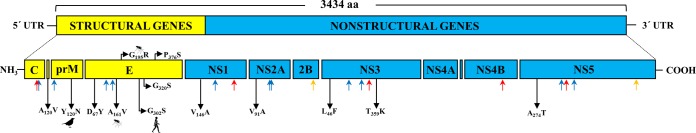
Highly conserved potential cleavage sites for each gene junction have been identified in all studied USUV ORFs (see Table S2 in the supplemental material). Only slight differences in the first or second protein residues directly flanking the protein cleavage sites at the C/AnchC and NS1/NS2a junctions in Africa 1 and Europe 1 strains have been observed (see Table S2). Except for the NS4A gene, each viral protein presented glycosylation sites being conserved among all USUV strains, except NS2A-1267 (absent in the Africa 1 and 2) and NS3-1968 (present only in Africa 1 and 2) (Fig. 4; see Table S3 in the supplemental material). If these changes affect the virus assembly or RNA replication still remains to be elucidated. The number of cysteine residues was also conserved among all USUV strains.
FIG 4 .
Schematic representation of the genome organization of USUV. Red arrows below indicate O-glycosylation sites, blue arrows N-glycosylation sites, and orange arrows C-glycosylation sites. A number indicating the position of amino acid mutations and the single-letter amino acid codes are used to denote the geography- and/or host-specific mutations along the polyprotein of the African and European USUV strains.
Secondary structure patterns in 5′ and 3′ NCRs.
Both 5′ and 3′ NCRs exhibited variability in USUV strains. In the 5′ NCR, specific nucleotide mutations between the African and European strains (A3T, T4C, C10T, and T14C) were observed. One deletion and 5 unique mutations in the highly divergent Africa 1 singleton were also detected. In all USUV strains, the 5′ secondary structures represented by a long stem loop (SL) with an internal loop (absent in the Africa 1) and lateral loop were conserved (see Fig. S6 in the supplemental material). Highly variable size heterogeneity in the 3′ NCR was detected. Two long deletions were found in Africa 1 (ArB1803), denominated deleted motif 1 (DM1) and DM2. These deletions were observed among the region corresponding to the stem loops SL-I and SL-V (see Fig. S6). Furthermore, a unique motif (UM1) located in SL-I was also found. A long deletion in a German bird-derived USUV strain (V10) located in SL-V (DM3) and a short deletion motif, DM4, in African strain HB81P08 were observed. The 3′ secondary structure was not conserved in these strains and was characterized by the lack of SL-V. Thus, the highly divergent ArB1803 strain 3′ secondary structure was affected by a lack of SL-I and SL-V and prediction of a long SL-II (see Fig. S6).
Protein changes and analysis of selection pressure.
The analysis of the E, NS5, and complete genome sequences revealed geography- and/or host-specific point mutations (Fig. 4 and 5). The nonsynonymous geographic-specific mutations C-A120V, E-V126M, NS3-S603A, NS4B-M16I, NS5-A274T, and NS5-S898N are fixed in all members of African lineages, while C-N11S, C-K85R, M-N246Y, NS2A-I74V, NS2A-V91A, NS3-L46F, NS3-T359K, NS4B-R174K are in Europe 3, and E-D67Y, E-A168T are in Europe 2. Host- and country-specific mutations have been found at prM-Y120N (birds, Germany), E-R195G (mosquitoes, Italy), and E-G302S (humans, Italy) (Fig. 4 and 5).
FIG 5 .
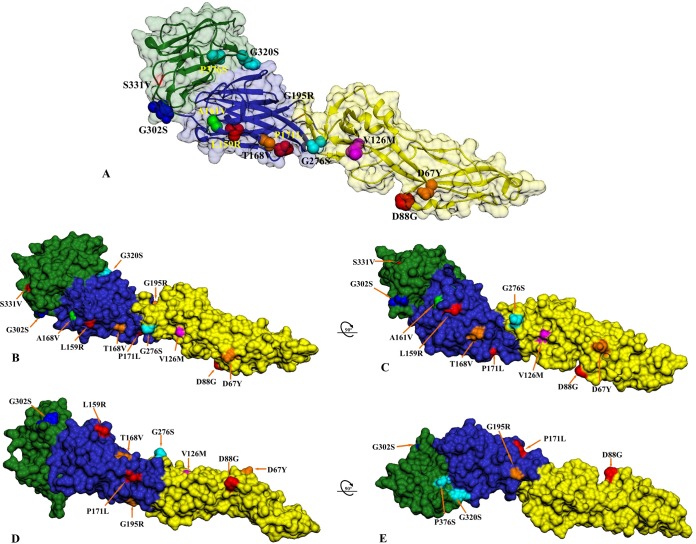
Structural location of the USUV mutations in different hosts from Africa and Europe depicted on the predicted USUV envelope glycoprotein structure. (A) The three-dimensional ribbon structure of a single monomer of the USUV envelope glycoprotein is shown with the corresponding three viral domains (domain I in blue, domain II in yellow, and domain III in green) and surface-exposed variable residues. (B and C) Structural mapping of the variable sites in the mosquito-, bird-, and human-derived USUV E protein on the surface rendition is shown from the side view and in a counterclockwise ~90° rotation. (D and E) The same model as in panel C in a counterclockwise ~90° plus ~90° rotation showing the external surface of the predicted USUV E monomer. All unique mutations in the singleton Africa 1 strain are shown in red and Europe 2-specific mutations in orange. Specific amino acid mutations found only in the African strains are indicated in cyan, and the Spanish-specific mutation is in green, while the unique mutation detected in the human USUV case from Italy is shown in blue. Amino acid V126M (magenta) is also indicated.
Non-synonymous mutations mapped on the three-dimensional (3D) model of predicted E glycoprotein structure are present across the three E protein domains (DI, DII, and DIII) (Fig. 5). In DI, geographic-specific amino acid changes were observed at positions L159R and P171L (Africa 1), T168V and G195R (Europe 2), and A161V (Africa 2). For DII, specific amino acid substitutions were found at D67Y (Europe 2), D88G (Africa 1), V126M (Europe 1 to 3), and G276S (Africa 2). DIII of the USUV strains revealed mutations at positions G302S (present only in human USUV from Italy), G320S and P376S (Africa 2), and S331V (Africa 1).
We also determined the nature of the selection pressure acting on the USUV polyprotein gene. The results showed that the overall ratio of non-synonymous to synonymous substitutions (dN/dS ratio) was 0.048, indicating that most amino acid changes are deleterious polymorphisms removed by purifying selection (Table 2). Significant evidence of adaptive evolution was found only at amino acid site 898 in the NS5 gene, which was detected as positively selected by at least 4 methods implemented (Table 2). In addition, there was also evidence for some form of adaptive evolution, which suggest that a larger number of sites in the USUV polyprotein identified by MEME method may be subjected to positive pressure evolving under episodic directional selection. When selection pressure was analyzed gene by gene, one or more sites for the NS1, NS3, and NS5 genes were found to be subjected to positive selection by at least two of the methods employed (Table 2). Using the branch site random-effects likelihood (BS-REL) method, we were not able to detect lineage-specific evolutionary patterns in our data sets. Furthermore, to determine whether a viral population is characterized by different selection pressures, we conducted an additional analysis by estimating the ω values (i.e., dN/dS ratios) of the African and European lineages, separately. Thus, the overall ω values for the European lineages were significantly higher (E, ω = 0.143; NS5, ω = 0.070; polyprotein, ω = 0.164; P < 0.05) than the African lineages (E, ω = 0.051; NS5, ω = 0.005; polyprotein, ω = 0.033). These results suggest that the USUV genes contain transient deleterious mutations evolving under purifying selection, an effect that seems to be strongest in the European lineages.
Table 2 .
Results of selection pressure analysis showing the positions of positively selected codons of USUV mature peptides
| Gene product | Position by: |
dN/dS | |||||
|---|---|---|---|---|---|---|---|
| SLAC | FEL | REL | IFEL | MEME | FUBAR | ||
| C | 11 | 0.11 | |||||
| prM | 32 | 0.066 | |||||
| E | 64, 126, 159, 320, 490 | 0.068 | |||||
| NS1 | 274 | 91, 96, 175, 182, 272, 274 | 0.052 | ||||
| NS2A | 25, 34, 43 | 0.087 | |||||
| NS2B | 0.024 | ||||||
| NS3 | 61 | 115 | 9, 53, 61, 115, 274, 344 | 0.051 | |||
| NS4A | 0.019 | ||||||
| 2K | 0.005 | ||||||
| NS4B | 217 | 0.042 | |||||
| NS5 | 731, 898 | 898 | 84, 898 | 84, 291, 437, 697, 725, 731, 896 | 898 | 0.039 | |
DISCUSSION
The rate of globalization accelerates the migration of exotic pathogens and their hosts to new environments facilitating contacts with vulnerable new hosts (e.g., introduction, spread, and establishment of West Nile virus in North America; introduction and autochthonous transmission of chikungunya virus and Zika virus in the Americas). Thus, understanding evolutionary processes that contribute to the emergence, maintenance, and spread of viral diseases is the sine qua non to develop and implement surveillance strategies for their control. In this study, we sought to elucidate the possible origin, pattern of spatiotemporal dynamics, and eco-epidemiological factors that shape the evolution of USUV becoming a very successful pathogen responsible for neuroinvasive disease in multiple vertebrate species, including humans.
Our phylogenies showed an important spatial differentiation between Central European and African USUV that resulted in phylogeographic clustering of 6 distinct lineages. We found evidence that the phylogenetic structure is shaped by the geographic location and pattern of migratory flyways, which likely facilitates rapid long-distance virus dispersal. This demonstrates that the major migratory bird flyways (East Atlantic and Black Sea/Mediterranean [Fig. 1; see Table S1 in the supplemental material]) could predict the continental and intercontinental dispersal patterns of USUV in our data sets (long-distance linkages between African countries, and Africa-Europe supported by high BF values) and that the birds might act as potential long-distance dispersal vehicles. These results are comparable with the predicted dispersal pattern of West Nile virus across the United States via avian flyways (30–32). A recent study revealed that the migratory status did not appear to influence WNV viremia titers (birds remain viremic for several days), as might be expected if individuals were immunosuppressed during migration. Furthermore, the infection does not inhibit migratory behavior, demonstrating that long-distance migratory birds are able to carry the virus over long distances (33, 34). Likewise, there are many evidences of USUV-seroconverted migratory bird species detected throughout Europe, which use East Atlantic and/or Black Sea/Mediterranean flyways (18, 35–37). However, the long-distance spread of the virus through ship- or aircraft-borne transportation of USUV-infected mosquitoes cannot be excluded.
The phylogeography results show a westward spread of USUV in Europe that matched the chronological and geographical incidence of USUV epizootics in Central Europe. The BSSVS model confirmed the multiple introduction of the virus into Europe from Africa, with Senegal as a possible origin for the progenitor of Central European epizootics (Fig. 1 and 2). The inferred spread of USUV indicates that in Africa, Senegal was probably the major source population, whereas in Central Europe, Austria represented the primary source and Italy the source of USUV diffusion (Fig. 1 and 3). The limited number of sequences from Africa reduces our ability to predict viral migrations within African countries. Given that many intervening countries are unsampled and long branches may obscure additional spatial movements of the USUV between Africa and Europe, all inferences of spatial connections between African countries should be interpreted with caution.
The existence of geographically distinct lineages in Europe (likely due to adaptation to the local ecological conditions and overwintering in enzootic foci) reflects the fact that USUV circulates in multiple areas that are separated from each other by geographic barriers such as climate, vegetation, different host species, and other unknown ecological conditions. Thus, the adaptation of USUV to naïve vector and host populations can lead to the emergence of local virus variants. The most likely scenario for European lineages might be enzootic maintenance (in situ evolution) similar to that observed for West Nile virus and St. Louis encephalitis virus in United States (30, 31, 38–41). This hypothesis is supported by the observation that European lineages form a star-like structure (population expansion after a single viral introduction) in which the variant viral strains accumulate changes during the rapid adaptation to the local ecological conditions (e.g., USUV epizootics in Germany and Austria). Furthermore, the phylogeny-trait association tests indicated a very strong geographical clustering of European lineages (P < 0.001), supporting the in situ evolution scenario. It has also been observed that the African lineages are driven mostly by extensive migration and introduction of viral variants from different geographic origins (e.g., emergence of 2 lineages in Spain).
The estimated substitution rate of 1.13 × 10−3 substitutions site−1 year−1 is very similar to the rate 1.37 × 10−3 given by Nikolay et al. (42) for their analysis of the E gene using a much smaller data set. However, the mutation rates observed for E, NS5, and the complete genome are within the confidence interval estimated for other flaviviruses (29), and the differences per gene are expected given their particular biological role.
Although USUV has received more attention only recently after the first epizootics and human cases in Europe, our estimates suggest that the virus emerged in Africa at least 500 years ago. This estimation is compatible with those observed for other members of the Japanese encephalitis virus group (500 to 1,500 years) (39, 43, 44). It should be pointed out that the accuracy of the TMRCA of USUV could be influenced by several factors, including the limited amount of complete sequence data, the short and unbalanced time span, and the presence of many intervening unsampled African countries that can lead to underestimation of the lengths of long branches. However, the estimated TMRCAs of European lineages (~40 years) were similar in E, NS5, and complete genome MCC trees (Fig. 1; see Fig. S3 in the supplemental material). The apparent absence of virus activity for such a long period can be explained by several hypotheses: introduction of less virulent viral strains; cross-reactive flavivirus antibodies (heterotypic flavivirus antibodies as a result of frequent exposure to numerous flaviviruses [45]) among birds and other vertebrate hosts, which could have modulated or downregulated clinical illness and viremia, reducing the transmission (45); the absence of long periods of hot and dry weather, which influence the abundance of competent vectors and transmission to susceptible hosts (46); reduced population of susceptible hosts; herd immunity of resident bird populations supporting a silent spread of the virus (47); and other extrinsic factors that could have influenced selection of less virulent virus strains.
A very recent study showed that USUV is able to emerge in other hosts (bats) (5). This observation raised questions regarding the USUV host range and the ability to adapt to the new hosts. Due to limitations of the available sequences, mostly from African countries, the host species analysis should be interpreted with caution. While the coloring of branches in Fig. 3 indicates the mosquito as the most probable host for the common ancestor of all USUV strains, this is unlikely to be reliable, due to the unbalanced sampling of different hosts. Nevertheless, that USUV originated in the mosquito is comparable with the fact that the mosquito is the main host for USUV. Transition from mosquitoes to birds and vice versa has been reconstructed (95% pp), being consistent with the enzootic cycle of the USUV. Although humans are considered incidental or dead-end hosts for USUV, adequate molecular surveillance is essential for public health (diagnostic and blood/organ donors) due to the increasing number of human USUV infections. Several genetic signatures were found in both African and European strains, which could be useful for development of molecular methods (e.g., real-time PCR) capable of differentiating USUV lineages. The virulence of the specific USUV lineages is currently unknown, and further studies are necessary to determine the biological characteristics of each lineage.
The overall low dN/dS ratio in the polyprotein gene indicates that most amino acid residues are subjected to purifying selection as results of genetic drift characteristic for arboviruses (48). We found strong evidence of adaptive evolution only at codon 898 in the NS5 gene. Although the function of this residue remains to be determined, it is located adjacent to codon D896E observed in the human USUV strain, which was associated with viral replication efficiency and neuroinvasive capacity in certain strains of JEV and WNV (25). In addition, a single amino-acid change in NS5 has been found to influence WNV replication in different hosts (49). To determine whether NS5-898 may be involved in the replication of USUV in different hosts, further studies are required. We also observed that a larger number of sites in the USUV polyprotein might be subjected to positive pressure evolving under episodic directional selection, indicating past occurrence of positive selection. The purifying selection observed is expected given the transmission and infection modes of arboviruses allowing accumulation of synonymous mutations and negatively selected sites as the effect of alternation between the arthropod vector and avian or mammal hosts (48). It is interesting to note that the pattern of increased positive selection in nonstructural genes compared to that in structural genes is similar to those observed in WNV, indicating that the host immune selection pressure does not caused increases in viral fitness (31). Mutations observed at amino acid positions V91A (NS2A), L46F and T359K (NS3), and D67Y (E) have been found to be involved in the formation of Europe 1 to 3, while A120V (C), G320S and P376S (E), and A274T (NS5) are specific for African lineages (convergent evolution). Similar patterns of parallel or convergent evolution have been observed for WNV. This suggests that a limited number of residue changes are permitted due to functional constraints (41). It is interesting to note that the V91A (NS2A) and L46F (NS3) mutations are specific for German strains and likely occurred due to introduction of USUV in this country. Although the impact of these mutations is unclear, similar changes in the related WNV modulated the host antiviral response by inhibition of interferon signaling (50). The residue exchange E-G302S observed in human USUV cases is considered important because it might have played a role in the human-specific neuroinvasive capacity of this virus (22). Our homology model suggested that each envelope protein domain is an important site for USUV evolution. In particular, specific amino acid substitutions within the DIII domain have been observed only in African lineages and in human cases from Italy (Fig. 5). It is known that DIII of flaviviruses represents a receptor binding domain and a major determinant of virus cellular tropism (51). Such mutations in WNV DIII have been involved in virus infectivity, virulence, antigenicity, and escape from neutralizing antibodies (52). It is unknown if these mutations in DIII observed are aftereffects of the mechanisms of antigenic escape or further adaptation to the host, but they might confer an evolutionary advantage to the virus. Potential cleavage sites for generation of USUV proteins and cysteines were conserved in all USUV strains, suggesting that their biological roles are preserved (see Table S2 and S3 in the supplemental material).
In all USUV strains, the 5′ secondary structures were conserved, whereas highly variable size heterogeneity in the 3′ NCR was detected. Five distinct 3′ NCR patterns were detected (see Fig. S6 in the supplemental material): three of them revealed long deletions in stem loops SL-I and SL-V, representing the hypervariable region of 3′ NCR. It has been shown that the variation in this region may have evolved as a function of dengue virus (DENV) transmission and replication in different mosquito and non-human primate/human host cycles (53).
Viral adaptation in mosquitoes and vertebrate hosts by local overwintering or reintroduction of the virus and migratory bird flyways could be considered key determinants in the spatial dispersal and establishment of USUV. Thus, further studies preferentially based on complete genomes, including those from previously unsampled intervening countries, are clearly necessary to fully understand the impact of ecological/immunological/virological factors on USUV epidemiology and evolution in different ecological habitats. This should be more feasible in the era of next-generation sequencing.
MATERIALS AND METHODS
Data sets.
A total of 77 complete USUV genomes were newly acquired as part of this study. These strains were sampled as part of German Arbovirus Surveillance Program (54) from different mosquito, bird, and bat species and different time points and locations (2010 to 2014) from the area of USUV endemicity in Germany (see Fig. S7 in the supplemental material). RNA extraction, reverse transcription-PCR (RT-PCR) of the complete genome, and sequencing were performed as previously described (5, 54). All complete genomes and E and NS5 sequences of USUV with known time (year) and geographical origin (country) of detection were retrieved from GenBank (15 complete genomes, 128 partial E gene sequences, and 84 NS5 gene sequences, representing data up to July 2015) and combined with those sequenced here. The initial USUV data sets were pruned of sequences representing duplicates. This resulted in a final data set of 92 complete or near complete genomes and 205 partial E and 161 NS5 gene sequences. This pruning process resulted in data sets of 55 year-dated sequences sampled between 1959 and 2014. A gene approach (E and NS5) was preferred over a genomic one because many more sequences are available and provided the greatest coverage of geographical regions and time of collection than the complete genomes, resulting in less biased data sets. Nevertheless, for comparison with the partial data sets, the complete genome data set has been included in the analyses. Although one USUV isolate (SAAR-1776, South Africa) exhibits passage history, we decided to include it in our analyses because the phylogenetic clustering and the TMRCAs were not influenced (Fig. 1; see Fig. S3 in the supplemental material). Sequences were aligned using the MAFFT algorithm and then visually inspected in Geneious v7.1.8. All sequences were confirmed as non-recombinant by the various methods for recombination detection implemented in RDP4 (55).
Genome characterization, prediction of signal peptidase cleavage, and N/O/C-glycosylation sites.
Genomes obtained for the German USUV strains were compared with all complete or near complete genomes and partial E and NS5 gene sequences publicly available. Potential cleavage sites for polyprotein were predicted based on neural networks (NN) and hidden Markov models (HMM) using SignalP 3.0 (http://www.cbs.dtu.dk/services/) and Geneious v7.1.8. Predictions of potential glycosylation sites and cysteine residues were investigated on complete viral polyprotein sequences using NetCGlyc v1.0, NetOGlyc v3.1, and NetNGlyc v1.0 (http://www.cbs.dtu.dk/services/) and Geneious v7.1.8.
Evolutionary dynamics, spatial and host species analysis.
Phylogenetic trees were inferred using the maximum likelihood (ML) method implemented in PhyML (56) and Bayesian Markov chain Monte Carlo (MCMC) approach available in BEAST v1.8 (57). Analyses were performed under the best fit nucleotide substitution model identified as the TN93+Γ for partial data sets and GTR+Γ for complete genome using jModelTest 2 (58). To search among ML trees, we employed both nearest neighbor interchange (NNI) and subtree pruning and regrafting (SPR) branch swapping. To assess the robustness of each node, a bootstrap-resampling process was performed (1,000 replicates) again using the NNI branch-swapping method available in PhyML. The clock-likeness and temporal signal of each data set were visualized using regression of the root-to-tip divergence inferred from the ML trees against the sampling time in Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/). Given that the time of sampling of all sequences included in the study was available, we were able to assess the spatial temporal dynamics of USUV in Africa and Europe. Thus, the rates of nucleotide substitution per site per year, the time to most recent common ancestor (TMRCA), and the effective population dynamics of USUV were estimated employing a relaxed uncorrelated log normal (UCLN) molecular clock, a flexible Bayesian skyline plot demographic model as the best demographic scenario detected, and the model of nucleotide substitution described above. In all cases, each of the MCMC chain lengths was run for 108 generations (with 10% burn-in) with subsampling every 104 iterations to achieve convergence as assessed using Tracer v1.5 (59). The MCC trees visualized using FigTree v1.4.1 (http://tree.bio.ed.ac.uk/software/figtree/) were topologically similar to that obtained in the ML analysis (see Fig. S2 and S3 in the supplemental material). To test the hypothesis that USUV is periodically imported from Africa into Europe, a phylogeographic analysis was conducted using a discrete model attributing state characters representing the detection locality of each of the strains and the Bayesian stochastic search variable (BSSVS) algorithm implemented in BEAST (27) and BEAGLE to enhance the computational speed. The spatiotemporal dispersal of USUV within Europe using the continuous probabilistic model of viral diffusion was also employed. The Bayesian MCC phylogenies were annotated at each node with posterior probability (pp) values, TMRCA, and state pp values for each plausible geographic location of origin (country). In order to detect which epidemiological factor (country and/or migratory bird flyway) best fit the data, we performed phylogeny traits tests using the Bayesian tip-significance testing (BaTS) program (60), calculating the parsimony score (PS), the association index (AI) (for overall association), and the monophyletic clade (MC) (for each state) statistics. Host species transition analysis was performed using the same tree settings described above implementing the Markov jump process (28), which summarizes the expected number of jumps (host-to-host transmission) from the MCMC phylogenies.
Selection pressure analysis.
To estimate the nature of the selection pressures acting on USUV envelope, NS5 and complete genome, the ratio of non-synonymous substitutions per non-synonymous site to synonymous substitutions per synonymous site (dN/dS) was estimated using three codon-based maximum likelihood tools for the identification of sites subjected to positive or negative selection: single likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), and internal fixed-effects likelihood (IFEL) implemented in the HyPhy package from the Datamonkey web interface (http://www.datamonkey.org). We also implemented the fast, unconstrained Bayesian approximation (FUBAR) and mixed-effects model of evolution (MEME) methods (61), which can consider transient (episodic) selective pressures. The branch site random-effects likelihood (BS-REL) (62) tool for evidence of adaptive evolution in individual lineages was also used. The positive selection results detected with FEL, SLAC, IFEL, and MEME analyses were considered significant at P = 0.05 and pp ≥ 0.95 for the FUBAR.
Secondary structure prediction and structural modeling.
Secondary structures involving the 5′ and 3′ NCRs of the USUV strains were assessed using Mfold (63) and Geneious v7.1.8 based on alignment of bird-, mosquito-, bat-, and human-derived USUV strains. Secondary structure patterns were detected according to the lowest fold energy and the presence of conserved secondary structures (conserved sequences [CS], repeated conserved sequence [RCS], long stable hairpin [LSH], cycling sequence within CS1 of the 3′ NCR [3′CYC], dumbbell [DB], and stem loop [SL]). The three-dimensional protein models of the USUV E were generated using the initial homology search and template selection method Phyre2 v2.0 (64). The template sequence used to create the USUV E protein models was the crystal structure of the West Nile virus envelope glycoprotein (PDB 2I69) with 403/500 residues (81%) modeled at 100% accuracy. The final 3D structures were prepared and visualized with Chimera v1.8.1 (65).
Nucleotide sequence accession numbers.
The nucleotide sequences generated in this study have been deposited in GenBank under accession no. KJ438705 to KJ438782.
SUPPLEMENTAL MATERIAL
Nucleotide (a) and amino acid (b) sequence divergence scan of African and European USUV (uncorrected nucleotide and amino acid p-distances) sequences. Sequences were assigned into six different tag groups based on USUV lineages (Africa 1 to 3; Europe 1 to 3). Download
Maximum likelihood phylogeny based on nucleotide sequence information derived from the E (a) and NS5 (b) genes and complete genomes (c) of USUV strains. Numbers at the nodes represent bootstrap support values (≥70%) based on 1,000 replicates. The scale at the bottom of the trees represents the genetic distance in nucleotide substitutions per site. Download
Maximum likelihood and time-resolved phylogenies based on reduced E gene (panels A1 and A2, ML and Bayesian trees, including the SAAR-1776 strain; panels A3 and A4, ML and Bayesian trees without the SAAR-1776 strain), NS5 gene (panels B1 and B2, ML and Bayesian trees, including the SAAR-1776 strain; panels B3 and B4, ML and Bayesian trees without the SAAR-1776 strain), and the complete genome data set (panels C1 and C2, ML and Bayesian trees, including the SAAR-1776 strain; panels C3 and C4, ML and Bayesian trees without the SAAR-1776 strain), as well as Bayesian trees of the complete E gene (panel D1, without the SAAR-1776 strain), NS5 gene (panel D2, without the SAAR-1776 strain), and complete genome data set (panels E1 and E2, including/without the SAAR-1776 strain). Download
Regression of root-to-tip genetic distances against year of sampling for the E and NS5 genes and complete genome sequence alignment inferred from the ML phylogenetic trees. Download
Histograms indicate the location state probabilities for the estimated introductions of Usutu virus in African and European countries based on E and NS5 genes’ MCMC phylogenies. N/A, not applicable. Download
Schematic representation of the patterns of RNA structures that occurred in the 5′ and 3′ NCRs of 92 USUV strains. Predicted secondary structures generated by the Mfold program for the 5′ and 3′ NCR long stable hairpin (LSH) are shown. Five distinct patterns found for the 3′ NCR are described for USUV strains from Africa and Europe: 3′ NCR deleted motifs DM1 to -4 and unique motif UM1. Patterns I (DM1), II (UM1), and III (DM4) correspond to African strain ArB1803/KC754958 lacking SL-I, while patterns IV (DM2; V10/KJ438760) and V (DM5; ArB1803/KC754958) lack SL-V. Conserved structures are indicated by boxes. Abbreviations: CS, conserved sequences; RCS, repeated conserved sequence; 3′CYC, cycling sequence within CS1 of the 3′ NCR; DB, dumbbell; SL, stem loop. Download
(A) Map of Germany indicating the states from which the mosquitoes (blue) and dead birds (orange) were collected. In the tables are presented the collection period (2010 to 2013) and the numbers of mosquitoes and birds collected. (B) Map of Germany with states (blue, mosquitoes; orange, dead birds) indicating regions from which the USUV-positive mosquito pools and dead birds were collected. The tables show the collection period (2010 to 2013) and the number of USUV-positive mosquito pools and birds. The red rectangles represent the regions of USUV endemicity. Download
Phylogeny-trait association tests of the phylogeographic and migratory flyway structure of USUV strains based on E and NS5 genes using BaTS. The significance level is the P value for the statistical hypothesis test for equality between the index observed and that expected under no association. Each state was defined following the two-letter ISO code of the country where the sample was collected. The three major migratory flyways are east Atlantic, Black Sea/Mediterranean, and East Africa/west Asian. Values of <0.05 indicate statistical significance.
Predicted cleavage sites in the polyproteins of USUV strains from Africa and Europe. Boldface areas represent amino acid changes.
Number of cysteine residues and positions of glycosylation sites in USUV polyproteins.
ACKNOWLEDGMENTS
We thank Alexandra Bialonski, Mathis Petersen, and Claudia Poggensee for excellent technical assistance.
Funding Statement
This work was financially supported by the German Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE) under grant no. 2819104315.
Footnotes
Citation Engel D, Jöst H, Wink M, Börstler J, Bosch S, Garigliany M, Jöst A, Czajka C, Lühken R, Ziegler U, Groschup MH, Pfeffer M, Becker N, Cadar D, Schmidt-Chanasit J. 2016. Reconstruction of the evolutionary history and dispersal of Usutu virus, a neglected emerging arbovirus in Europe and Africa. mBio 7(1):e01938-15. doi:10.1128/mBio.01938-15.
REFERENCES
- 1.Williams MC, Simpson DI, Haddow AJ, Knight EM. 1964. The isolation of West Nile virus from man and of Usutu virus from the bird-biting mosquito Mansonia aurites (Theobald) in the Entebbe area of Uganda. Ann Trop Med Parasitol 58:367–374. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh BM. 1985. Usutu (SAAr 1776), nouvel arbovirus du groupe B. Int Cat Arboviruses 3:1059–1060. [Google Scholar]
- 3.Nikolay B, Diallo M, Boye CS, Sall AA. 2011. Usutu virus in Africa. Vector Borne Zoonotic Dis 11:1417–1423. doi: 10.1089/vbz.2011.0631. [DOI] [PubMed] [Google Scholar]
- 4.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2011. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 5.Cadar D, Becker N, Campos RdM, Börstler J, Jöst H, Schmidt-Chanasit J. 2014. Usutu virus in bats, Germany, 2013. Emerg Infect Dis 20:1771–1773. doi: 10.3201/eid2010.140909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenböck H, Bakonyi T, Rossi G, Mani P, Nowotny N. 2013. Usutu virus, Italy, 1996. Emerg Infect Dis 19:274–277. doi: 10.3201/eid1902.121191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissenböck H, Kolodziejek J, Url A, Lussy H, Rebel-Bauder B, Nowotny N. 2002. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, Central Europe. Emerg Infect Dis 8:652–656. doi: 10.3201/eid0807.020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakonyi T, Erdélyi K, Ursu K, Ferenczi E, Csörgo T, Lussy H, Chvala S, Bukovsky C, Meister T, Weissenböck H, Nowotny N. 2007. Emergence of Usutu virus in Hungary. J Clin Microbiol 45:3870–3874. doi: 10.1128/JCM.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busquets N, Alba A, Allepuz A, Aranda C, Nuñez JI. 2008. Usutu virus sequences in Culex pipiens (Diptera: Culicidae), Spain. Emerg Infect Dis 14:861–863. doi: 10.3201/eid1405.071577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manarolla G, Bakonyi T, Gallazzi D, Crosta L, Weissenböck H, Dorrestein GM, Nowotny N. 2010. Usutu virus in wild birds in northern Italy. Vet Microbiol 141:159–163. doi: 10.1016/j.vetmic.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 11.Steinmetz HW, Bakonyi T, Weissenböck H, Hatt JM, Eulenberger U, Robert N, Hoop R, Nowotny N. 2011. Emergence and establishment of Usutu virus infection in wild and captive avian species in and around Zurich, Switzerland—genomic and pathologic comparison to other Central European outbreaks. Vet Microbiol 148:207–212. doi: 10.1016/j.vetmic.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Jöst H, Bialonski A, Maus D, Sambri V, Eiden M, Groschup MH, Günther S, Becker N, Schmidt-Chanasit J. 2011. Isolation of Usutu virus in Germany. Am J Trop Med Hyg 85:551–553. doi: 10.4269/ajtmh.2011.11-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vázquez A, Ruiz S, Herrero L, Moreno J, Molero F, Magallanes A, Sánchez-Seco MP, Figuerola J, Tenorio A. 2011. West Nile and Usutu viruses in mosquitoes in Spain, 2008–2009. Am J Trop Med Hyg 85:178–181. doi: 10.4269/ajtmh.2011.11-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker N, Jöst H, Ziegler U, Eiden M, Höper D, Emmerich P, Fichet-Calvet E, Ehichioya DU, Czajka C, Gabriel M, Hoffmann B, Beer M, Tenner-Racz K, Racz P, Günther S, Wink M, Bosch S, Konrad A, Pfeffer M, Groschup MH, Schmidt-Chanasit J. 2012. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS One 7:e32604. doi: 10.1371/journal.pone.0032604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garigliany MM, Marlier D, Tenner-Racz K, Eiden M, Cassart D, Gandar F, Beer M, Schmidt-Chanasit J, Desmecht D. 2014. Detection of Usutu virus in a bullfinch (Pyrrhula pyrrhula) and a great spotted woodpecker (Dendrocopos major) in north-west Europe. Vet J 199:191–193. doi: 10.1016/j.tvjl.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Hubálek Z, Rudolf I, Čapek M, Bakonyi T, Betášová L, Nowotny N. 2014. Usutu virus in blackbirds (Turdus merula), Czech Republic, 2011–2012. Transbound Emerg Dis 61:273–276. doi: 10.1111/tbed.12025. [DOI] [PubMed] [Google Scholar]
- 17.Buckley A, Dawson A, Gould EA. 2006. Detection of seroconversion to West Nile virus, Usutu virus and Sindbis virus in UK sentinel chickens. Virol J 3:71. doi: 10.1186/1743-422X-3-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubálek Z, Wegner E, Halouzka J, Tryjanowski P, Jerzak L, Sikutová S, Rudolf I, Kruszewicz AG, Jaworski Z, Wlodarczyk R. 2008. Serologic survey of potential vertebrate hosts for West Nile virus in Poland. Viral Immunol 21:247–253. doi: 10.1089/vim.2007.0111. [DOI] [PubMed] [Google Scholar]
- 19.Chaintoutis SC, Dovas CI, Papanastassopoulou M, Gewehr S, Danis K, Beck C, Lecollinet S, Antalis V, Kalaitzopoulou S, Panagiotopoulos T, Mourelatos S, Zientara S, Papadopoulos O. 2014. Evaluation of a West Nile virus surveillance and early warning system in Greece based on domestic pigeons. Comp Immunol Microbiol Infect Dis 37:131–141. doi: 10.1016/j.cimid.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Cavrini F, Gaibani P, Longo G, Pierro AM, Rossini G, Bonilauri P, Gerunda GE, Di Benedetto F, Pasetto A, Girardis M, Dottori M, Landini MP, Sambri V. 2009. Usutu virus infection in a patient who underwent orthotropic liver transplantation, Italy, August–September 2009. Euro Surveill 14:19448. [PubMed] [Google Scholar]
- 21.Pecorari M, Longo G, Gennari W, Grottola A, Sabbatini A, Tagliazucchi S, Savini G, Monaco F, Simone M, Lelli R, Rumpianesi F. 2009. First human case of Usutu virus neuroinvasive infection, Italy, August–September 2009. Euro Surveill 14:19446. [PubMed] [Google Scholar]
- 22.Gaibani P, Cavrini F, Gould EA, Rossini G, Pierro A, Landini MP, Sambri V. 2013. Comparative genomic and phylogenetic analysis of the first Usutu virus isolate from a human patient presenting with neurological symptoms. PLoS One 8:e64761. doi: 10.1371/journal.pone.0064761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilibic-Cavlek T, Kaic B, Barbic L, Pem-Novosel I, Slavic-Vrzic V, Lesnikar V, Kurecic-Filipovic S, Babic-Erceg A, Listes E, Stevanovic V, Gjenero-Margan I, Savini G. 2014. First evidence of simultaneous occurrence of West Nile virus and Usutu virus neuroinvasive disease in humans in Croatia during the 2013 outbreak. Infection 42:689–695. doi: 10.1007/s15010-014-0625-1. [DOI] [PubMed] [Google Scholar]
- 24.Santini M, Vilibic-Cavlek T, Barsic B, Barbic L, Savic V, Stevanovic V, Listes E, Di Gennaro A, Savini G. 2015. First cases of human Usutu virus neuroinvasive infection in Croatia, August–September 2013: clinical and laboratory features. J Neurovirol 21:92–97. doi: 10.1007/s13365-014-0300-4. [DOI] [PubMed] [Google Scholar]
- 25.Gaibani P, Pierro A, Alicino R, Rossini G, Cavrini F, Landini MP, Sambri V. 2012. Detection of Usutu-virus-specific IgG in blood donors from Northern Italy. Vector Borne Zoonotic Dis 12:431–433. doi: 10.1089/vbz.2011.0813. [DOI] [PubMed] [Google Scholar]
- 26.Allering L, Jost H, Emmerich P, Günther S, Lattwein E, Schmidt M, Seifried E, Sambri V, Hourfar K, Schmidt-Chanasit J. 2012. Detection of Usutu virus infection in a healthy blood donor from south-west Germany, 2012. Euro Surveill 17:20341. [PubMed] [Google Scholar]
- 27.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minin VN, Suchard MA. 2008. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 29.Twiddy SS, Pybus OG, Holmes EC. 2003. Comparative population dynamics of mosquito-borne flaviviruses. Infect Genet Evol 3:87–95. doi: 10.1016/S1567-1348(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 30.Añez G, Grinev A, Chancey C, Ball C, Akolkar N, Land KJ, Winkelman V, Stramer SL, Kramer LD, Rios M. 2013. Evolutionary dynamics of West Nile virus in the United States, 1999–2011: phylogeny, selection pressure and evolutionary time scale analysis. PLoS Negl Trop Dis 7:e2245. doi: 10.1371/journal.pntd.0002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Giallonardo F, Geoghegan JL, Docherty DE, McLean RG, Zody MC, Qu J, Yang X, Birren BW, Malboeuf CM, Newman RM, Ip HS, Holmes EC. 2015. Fluid spatial dynamics of West Nile virus in the USA: rapid spread in a permissive host environment. J Virol 90:862–872. doi: 10.1128/JVI.02305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann BR, McMullen AR, Swetnam DM, Salvato V, Reyna M, Guzman H, Bueno R Jr, Dennett JA, Tesh RB, Barrett AD. 2013. Continued evolution of West Nile virus, Houston, Texas, USA, 2002–2012. Emerg Infect Dis 19:1418–1427. doi: 10.3201/eid1909.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen J, Moore F, Panella N, Edwards E, Bru R, Hughes M, Komar N. 2006. Migrating birds as dispersal vehicles for West Nile virus. EcoHealth 3:79–85. doi: 10.1007/s10393-006-0025-9. [DOI] [Google Scholar]
- 34.Weaver SC, Barrett AD. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llopis IV, Rossi L, Di Gennaro A, Mosca A, Teodori L, Tomassone L, Grego E, Monaco F, Lorusso A, Savini G. 2015. Further circulation of West Nile and Usutu viruses in wild birds in Italy. Infect Genet Evol 32:292–297. doi: 10.1016/j.meegid.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Höfle U, Gamino V, de Mera IGF, Mangold AJ, Ortíz J, de la Fuente J. 2013. Usutu virus in migratory song thrushes, Spain. Emerg Infect Dis 19:1173–1175. doi: 10.3201/eid1907.130199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckley A, Dawson A, Moss SR, Hinsley SA, Bellamy PE, Gould EA. 2003. Serological evidence of West Nile virus, Usutu virus and Sindbis virus infection of birds in the UK. J Gen Virol 84:2807–2817. doi: 10.1099/vir.0.19341-0. [DOI] [PubMed] [Google Scholar]
- 38.May FJ, Davis CT, Tesh RB, Barrett AD. 2011. Phylogeography of West Nile virus: from the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J Virol 85:2964–2974. doi: 10.1128/JVI.01963-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMullen AR, Albayrak H, May FJ, Davis CT, Beasley DW, Barrett AD. 2013. Molecular evolution of lineage 2 West Nile virus. J Gen Virol 94:318–325. doi: 10.1099/vir.0.046888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis CT, Beasley DW, Guzman H, Siirin M, Parsons RE, Tesh RB, Barrett AD. 2004. Emergence of attenuated West Nile virus variants in Texas, 2003. Virology 330:342–345. doi: 10.1016/j.virol.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Armstrong PM, Vossbrinck CR, Andreadis TG, Anderson JF, Pesko KN, Newman RM, Lennon NJ, Birren BW, Ebel GD, Henn MR. 2011. Molecular evolution of West Nile virus in a northern temperate region: Connecticut, USA 1999–2008. Virology 417:203–210. doi: 10.1016/j.virol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolay B, Dupressoir A, Firth C, Faye O, Boye CS, Diallo M, Sall AA. 2013. Comparative full length genome sequence analysis of Usutu virus isolates from Africa. Virol J 10:217. doi: 10.1186/1743-422X-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuh AJ, Ward MJ, Brown AJ, Barrett AD. 2013. Phylogeography of Japanese encephalitis virus: genotype is associated with climate. PLoS Negl Trop Dis 7:e2411. doi: 10.1371/journal.pntd.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moureau G, Cook S, Lemey P, Nougairede A, Forrester NL, Khasnatinov M, Charrel RN, Firth AE, Gould EA, de Lamballerie X. 2015. New insights into flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences. PLoS One 10:e0117849. doi: 10.1371/journal.pone.0117849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gubler DJ. 2007. The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis 45:1039–1046. doi: 10.1086/521911. [DOI] [PubMed] [Google Scholar]
- 46.Rubel F, Brugger K, Hantel M, Chvala-Mannsberger S, Bakonyi T, Weissenböck H, Nowotny N. 2008. Explaining Usutu virus dynamics in Austria: model development and calibration. Prev Vet Med 85:166–186. doi: 10.1016/j.prevetmed.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Meister T, Lussy H, Bakonyi T, Šikutová S, Rudolf I, Vogl W, Winkler H, Frey H, Hubálek Z, Nowotny N, Weissenböck H. 2008. Serological evidence of continuing high Usutu virus (Flaviviridae) activity and establishment of herd immunity in wild birds in Austria. Vet Microbiol 127:237–248. doi: 10.1016/j.vetmic.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 48.Holmes EC. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol 77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Slyke GA, Ciota AT, Willsey GG, Jaeger J, Shi PY, Kramer LD. 2012. Point mutations in the West Nile virus (Flaviviridae; flavivirus) RNA-dependent RNA polymerase alter viral fitness in a host-dependent manner in vitro and in vivo. Virology 427:18–24. doi: 10.1016/j.virol.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol 78:12225–12235. doi: 10.1128/JVI.78.22.12225-12235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin JF, Chu JJ, Ng ML. 2007. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes Infect 9:1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. 2005. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol 86:405–412. doi: 10.1099/vir.0.80411-0. [DOI] [PubMed] [Google Scholar]
- 53.Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, Russell KL, Barrett AD. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- 54.Becker N, Krüger A, Kuhn C, Plenge-Bönig A, Thomas SM, Schmidt-Chanasit J, Tannich E. 2014. Mosquitoes as vectors for exotic pathogens in Germany. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 57:531–540 doi: 10.1007/s00103-013-1918-8. [DOI] [PubMed] [Google Scholar]
- 55.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 56.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 57.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darriba D, Taboada GL, Doallo R, Posada D. 2012. iModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Bairlein F, Dierschke J, Dierschke V, Salewski V, Geiter O, Hüppop K, Köppen U, Fiedler W. 2014. Atlas des Vogelzugs: Ringfunde Deutscher Brut- und Gastvögel. Aula-Verlag, Wiebelsheim, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide (a) and amino acid (b) sequence divergence scan of African and European USUV (uncorrected nucleotide and amino acid p-distances) sequences. Sequences were assigned into six different tag groups based on USUV lineages (Africa 1 to 3; Europe 1 to 3). Download
Maximum likelihood phylogeny based on nucleotide sequence information derived from the E (a) and NS5 (b) genes and complete genomes (c) of USUV strains. Numbers at the nodes represent bootstrap support values (≥70%) based on 1,000 replicates. The scale at the bottom of the trees represents the genetic distance in nucleotide substitutions per site. Download
Maximum likelihood and time-resolved phylogenies based on reduced E gene (panels A1 and A2, ML and Bayesian trees, including the SAAR-1776 strain; panels A3 and A4, ML and Bayesian trees without the SAAR-1776 strain), NS5 gene (panels B1 and B2, ML and Bayesian trees, including the SAAR-1776 strain; panels B3 and B4, ML and Bayesian trees without the SAAR-1776 strain), and the complete genome data set (panels C1 and C2, ML and Bayesian trees, including the SAAR-1776 strain; panels C3 and C4, ML and Bayesian trees without the SAAR-1776 strain), as well as Bayesian trees of the complete E gene (panel D1, without the SAAR-1776 strain), NS5 gene (panel D2, without the SAAR-1776 strain), and complete genome data set (panels E1 and E2, including/without the SAAR-1776 strain). Download
Regression of root-to-tip genetic distances against year of sampling for the E and NS5 genes and complete genome sequence alignment inferred from the ML phylogenetic trees. Download
Histograms indicate the location state probabilities for the estimated introductions of Usutu virus in African and European countries based on E and NS5 genes’ MCMC phylogenies. N/A, not applicable. Download
Schematic representation of the patterns of RNA structures that occurred in the 5′ and 3′ NCRs of 92 USUV strains. Predicted secondary structures generated by the Mfold program for the 5′ and 3′ NCR long stable hairpin (LSH) are shown. Five distinct patterns found for the 3′ NCR are described for USUV strains from Africa and Europe: 3′ NCR deleted motifs DM1 to -4 and unique motif UM1. Patterns I (DM1), II (UM1), and III (DM4) correspond to African strain ArB1803/KC754958 lacking SL-I, while patterns IV (DM2; V10/KJ438760) and V (DM5; ArB1803/KC754958) lack SL-V. Conserved structures are indicated by boxes. Abbreviations: CS, conserved sequences; RCS, repeated conserved sequence; 3′CYC, cycling sequence within CS1 of the 3′ NCR; DB, dumbbell; SL, stem loop. Download
(A) Map of Germany indicating the states from which the mosquitoes (blue) and dead birds (orange) were collected. In the tables are presented the collection period (2010 to 2013) and the numbers of mosquitoes and birds collected. (B) Map of Germany with states (blue, mosquitoes; orange, dead birds) indicating regions from which the USUV-positive mosquito pools and dead birds were collected. The tables show the collection period (2010 to 2013) and the number of USUV-positive mosquito pools and birds. The red rectangles represent the regions of USUV endemicity. Download
Phylogeny-trait association tests of the phylogeographic and migratory flyway structure of USUV strains based on E and NS5 genes using BaTS. The significance level is the P value for the statistical hypothesis test for equality between the index observed and that expected under no association. Each state was defined following the two-letter ISO code of the country where the sample was collected. The three major migratory flyways are east Atlantic, Black Sea/Mediterranean, and East Africa/west Asian. Values of <0.05 indicate statistical significance.
Predicted cleavage sites in the polyproteins of USUV strains from Africa and Europe. Boldface areas represent amino acid changes.
Number of cysteine residues and positions of glycosylation sites in USUV polyproteins.