Abstract
ZMYND8 (zinc finger MYND (Myeloid, Nervy and DEAF-1)-type containing 8), a newly identified component of the transcriptional coregulator network, was found to interact with the Nucleosome Remodeling and Deacetylase (NuRD) complex. Previous reports have shown that ZMYND8 is instrumental in recruiting the NuRD complex to damaged chromatin for repressing transcription and promoting double strand break repair by homologous recombination. However, the mode of transcription regulation by ZMYND8 has remained elusive. Here, we report that through its specific key residues present in its conserved chromatin-binding modules, ZMYND8 interacts with the selective epigenetic marks H3.1K36Me2/H4K16Ac. Furthermore, ZMYND8 shows a clear preference for canonical histone H3.1 over variant H3.3. Interestingly, ZMYND8 was found to be recruited to several developmental genes, including the all-trans-retinoic acid (ATRA)-responsive ones, through its modified histone-binding ability. Being itself inducible by ATRA, this zinc finger transcription factor is involved in modulating other ATRA-inducible genes. We found that ZMYND8 interacts with transcription initiation-competent RNA polymerase II phosphorylated at Ser-5 in a DNA template-dependent manner and can alter the global gene transcription. Overall, our study identifies that ZMYND8 has CHD4-independent functions in regulating gene expression through its modified histone-binding ability.
Keywords: chromatin, chromatin modification, chromatin remodeling, gene expression, gene regulation, gene silencing, gene transcription
Introduction
ZMYND8 (zinc finger MYND (Myeloid, Nervy and DEAF-1)-type containing 8) is a putative chromatin reader/effector harboring a PWWP domain, a bromodomain, and a PHD type zinc finger. It associates with the CHD4 protein of NuRD chromatin remodeling complex implicated in gene transcription, cell cycle progression, and genome integrity (1, 2). Reader proteins specifically recognize histone post-translational modifications (PTMs)5 and translate such recognition(s) into meaningful biological outcomes by virtue of either their intrinsic activities or those of their interacting partners (3). These readers may interpret histone PTMs via “monovalent” or “multivalent” recognition (4). In monovalent recognition, a chromatin reader recognizes one histone PTM. In contrast, in multivalent recognition, readers recognize multiple histone modifications intra/inter-nucleosomally. An accessible surface is provided by these readers (such as a cavity or surface groove), which accommodates the modified histone residues, and determines the modification (e.g. acetylation versus methylation) or state specificity (such as mono- versus trimethylation of lysine) (3).
ZMYND8 is a well known component of the transcription coregulator complex and is associated with several demethylase machinery components, including KDM5A, KDM5C, or LSD1 (5, 6). Through its ability to interact with Xenopus RCoR2, ZMYND8 plays a significant role in embryonic neural differentiation (7). Apart from this, ZMYND8 is also involved in T-cell lymphoma and breast and cervical cancer (8–10). Another interesting feature is that ZMYND8 is significantly involved in transcription activation (5). ATRA, a vitamin A metabolite, is a well known inducer of transcription of several genes. It modulates RA receptor (retinoic acid receptor/retinoid X receptor) dimerization at the retinoic acid-response element (RARE) (11). We show for the first time that ZMYND8 is ATRA-responsive and a substantial number of genes regulated by ZMYND8 harbor the RARE sequence. Interestingly, we found that ZMYND8 is involved in regulating transcription initiation through its interaction with the RNA polymerase II complex in a DNA-mediated manner. Furthermore, ZMYND8 can regulate global gene expression in a CHD4-independent manner. In an attempt to understand its mode of recruitment to chromatin, we found that it has a selective interaction with H3.1K36Me2/H4K16Ac marks through its chromatin-binding module. Thus, this study establishes a new role of ZMYND8 in the context of its epigenetic recognition thereby regulating ATRA-induced gene transcription.
Experimental Procedures
Cell Culture and ATRA Treatment
HeLa and HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Gibco, Invitrogen), and SH-SY5Y cells were maintained in DMEM/F-12 (1:1) (Gibco, Invitrogen). Neuro2A cells were maintained in Eagle's minimum essential medium (Sigma). All cell lines were supplemented with 10% fetal bovine serum (Gibco, Invitrogen) and penicillin/streptomycin (10 μl/ml medium, Gibco, Invitrogen) at 37 °C in 5% (v/v) CO2. SH-SY5Y cells were treated with 10 μm ATRA (Sigma) or DMSO (Sigma, solvent control) for 2 or 4 days. Every other day, media change was given to the SH-SY5Y cells with ATRA.
Overexpression and siRNA Transfection
HEK293 cells were transiently transfected with FLAG-H3.1/H3.3 (for co-IP experiments) and FLAG-ZMYND8 and WT-/FLAG-ZMYND8 ΔPBP (for co-IP and ChIP), and HeLa cells were transfected with FLAG-ZMYND8 WT/ΔPBP (for coimmunofluorescence) using Lipofectamine2000 (Invitrogen) as per manufacturer's protocol.
HeLa and SH-SY5Y cells were transfected with ZMYND8 siRNA (Santa Cruz Biotechnology) or negative control siRNA (Invitrogen) using INTERFERin® transfection reagent (Polyplus) according to the manufacturer's protocol and incubated for 24 h. For SH-SY5Y cells, prior to siRNA transfection, cells were treated with 10 μm ATRA or DMSO for 24 h.
Cloning and Site-directed Mutagenesis
Individual domains PHD, Bromo, PWWP and a combination of all three (designated as PBP) of ZMYND8 were cloned in pDEST15 bacterial expression vector. PBP domain was also cloned in pDEST17 bacterial expression vector. ZMYND8 wild type and ΔPBP were also cloned in pCMV-FLAG mammalian expression vector. All the cloning vectors were from Gateway® cloning system (Invitrogen), and standard protocols were followed. All the clones were confirmed by sequencing. The point mutation was generated using QuikChange site-directed mutagenesis kit from Stratagene as per standard protocols (12).
Protein Purification
Transformed cells were grown until optical density reached 0.8 and was induced at 1 mm isopropyl 1-thio-β-d-galactopyranoside (Sigma) at 20 °C for 16 h. The cells were lysed in Lysis Buffer: 20 mm Tris-HCl (pH 8), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT, 2 mm PMSF, 1× protease inhibitor mixture (EDTA-free). After sonication, the lysate was spun down twice at 13,000 rpm, 4 °C, 30 min. The supernatant was subjected to glutathione-Sepharose beads binding for 2 h at 4 °C. The beads were spun down and washed with Wash Buffer: 20 mm Tris-HCl (pH 8), 500 mm NaCl, 1 mm DTT. The bead bound proteins were eluted with Elution Buffer: 20 mm Tris-HCl (pH 8), 150 mm NaCl, 100 mm reduced glutathione, 2 mm DTT. The eluted proteins were concentrated in Amicon Ultra Filter tubes (Millipore) and purified further by gel filtration chromatography using a Superdex75 column (GE Healthcare).
Chromatosome Isolation
Chromatosome was prepared from freshly harvested HeLa cells as described elsewhere (13). Briefly, nuclear pellet of HeLa cells was digested with micrococcal nuclease (Sigma 0.2 units/μl) and extracted with TE buffer for 1 h. Concentrated chromatin was separated by sucrose gradient (5–40%) ultracentrifugation using Sorvall WXUltra100 (Thermo Scientific) in an AH650 rotor for 16 h at 35,000 rpm. Chromatin fractions were analyzed in agarose gel; chromatosomal fractions were pooled and dialyzed in TE buffer for further analysis.
CelluSpots Peptide Array Analysis
MODified Histone Peptide Array (CelluSpots array) was procured from Active Motif (catalogue no. 13005). CelluSpots array analysis was done as per standard protocol (14, 15). Briefly, the array was blocked with the Blocking Buffer overnight at 4 °C, washed three times with Wash Buffer, and incubated with GST-fused protein at a concentration of 1 μm. The array was probed with anti-GST antibody.
Peptide Pulldown Assay
Peptides purchased from AnaSpec were as follows: H4K5Ac (64844-1); H4K8Ac (64843-1); H4K12Ac (64849-1); H4K16Ac (64847-025); H4K20Ac (65212-1); H3.1K36Me1 (64602-1); H3.1K36Me2 (64442-1); H3.1K36Me3 (64441-1); H3.1K9Ac (63680); H3.1K14Ac (63682), and H3.1K27Ac (64637-1). Peptides purchased from EpiCypher were as follows: H3.3(22–44) (12-0020); H3.3K36Me1 (12-0022); H3.3K36Me2 (12-0023), and H3.3K36Me3 (12-0024).
Peptide pulldown assays were done as per standard protocol (16). Briefly, biotinylated histone peptides were incubated with GST-tagged protein in IP Buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT) at 4 °C, overnight. The reactions were subjected to Streptavidin beads (GE Healthcare) binding for 2 h at 4 °C and washed with IP Buffer, and the complex was pulled down and analyzed by Western blotting.
Protein-Protein Interaction Studies
The core histone and chromatosome interactions with GST and GST fusion proteins were performed as per standard protocol (5, 14). Briefly, proteins were incubated in an equal molar ratio in binding buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT), pulled down by pre-blocked glutathione-Sepharose beads, and washed in binding buffer, and the protein complex was analyzed by Western blotting.
Fluorescence Spectroscopy and Analysis of the Binding Data
The fluorescence measurements were performed in an LS 55 luminescence spectrometer (PerkinElmer Life Sciences) at 25 °C. In the case of H3K36Me3, H3K36Me2, H3K36Me1, and H3K36 unmodified peptides, tryptophan fluorescence measurements were carried out on samples containing 0.45 μm ZMYND8 (His-PHD/Bromo/PWWP) and dissolved in 20 mm HEPES (pH 6.5), 200 mm NaCl, 300 mm ammonium sulfate, 1 mm β-mercaptoethanol, while progressively increasing the peptide concentration in each case until there was no further change in the emission intensity with an increase in the peptide concentration. Samples were excited at 295 nm, and emission was recorded at 320 nm with a scan speed of 200 nm/s, and averaged over three scans.
In case of FAM-conjugated H4K16-acetylated and H4K16-unmodified peptides, fluorescence measurements were carried out using 0.6 μm of the peptides dissolved in 20 mm Tris-HCl (pH 6.5), 150 mm NaCl, 1 mm β-mercaptoethanol at 25 °C with increasing addition of ZMYND8 (His-PBP). Samples were excited at 494 nm, and emission was recorded at 520 nm with scan speed 200 nm/s and averaged over three scans.
Following peptides were custom-synthesized from GL Biochem (Shanghai, China) Ltd. for the fluorescence spectroscopic studies 5-FAM-H4K16Ac, 5-FAM-H4K16 unmodified, H3.1K36Me3(29–40), H3.1K36Me2(29–40), H3.1K36Me1(29–40), H3.1K36 unmodified(29–40). Results from fluorometric titrations were analyzed with the following method (17). The apparent dissociation constant (Kd) was determined using non-linear curve fitting analysis (Equation 1) according to Reaction 1,
 |
Here, L is the ligand ZMYND8 (His-PBP), and P is the histone peptide used.
All experimental points for binding isotherms were fitted by least squares analysis as shown in Equation 1.
where ΔF is the change in fluorescence emission intensity at 340 nm (excitation of tryptophan residue(s) at 295 nm) for each point of the titration curve. ΔFmax is the same parameter when the ligand (ZMYND8 His-PBP) is totally bound to histone. Cp is the concentration of histone, and C0 is the initial concentration of the ligand. A double-reciprocal plot was used for determination of ΔFmax using Equation 2,
In the case of titration of ZMYND8 (His-PBP) with H4K16 unmodified and H4K16Ac histone peptides, the data analysis was done following the same theory as reported above, and the non-linear curve fitting was done following Equations 1 and 2. The only difference being that in this case, C0 is the initial concentration of the FAM-conjugated histone H4K16 unmodified and Cp is the concentration of the ligand ZMYND8 His-PBP.
Molecular Modeling Studies
Bromo-PWWP module of ZMYND8 was generated by MODELLER (18) package using chain A of 4N4G as a reference structure.
Coimmunofluorescence and Confocal Microscopy
Coimmunofluorescence staining was performed as per standard protocol (19). Briefly, the cells were fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100, and blocked with 3% BSA. Cells were incubated with anti-FLAG antibody (Sigma) and either anti-H4K16Ac (Active Motif) or H3K36Me2 (Abcam) antibody for 1 h. Following washes with PBST, the cells were incubated with Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies for 1 h at room temperature. The coverslips were washed with PBST and mounted after DAPI staining. OLYMPUS FLUOVIEW FV1000 confocal scanning microscope was used for imaging.
Co-IP
Cross-linked cells were subjected to co-IP as delineated elsewhere (20). In brief, after cross-linking the cells were lysed with 50 mm HEPES (pH 7.5), 150 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 0.5% sodium deoxycholate, 5% glycerol, 1 mm DTT, 2 mm PMSF, 1 mm trichostatin A, 5 mm sodium butyrate, and complete protease inhibitor mixture and incubated on ice for 1 h followed by centrifugation at 13,000 rpm for 10 min at 4 °C. After pre-clearing the lysates, immunoprecipitation was done with the antibodies, followed by washes with the same buffer, and the complexes were analyzed by Western blotting.
Chromatin Fractionation
Chromatin fractionation was done as per standard protocol (21) with some modifications. Briefly, cells were resuspended in TB Buffer (20 mm HEPES (pH 7.3), 110 mm KOAc, 5 mm NaOAc, 2 mm Mg(OAc)2, 1 mm EGTA, 2 mm DTT, 2 mm PMSF, 1× protease inhibitor cocktail) containing increasing concentrations of Nonidet P-40 (0.1–0.5%). The pellet fractions were separated from soluble fractions by centrifugation at 13,000 rpm for 10 min at 4 °C. The pellet was treated with DNase I in TB Buffer containing 0.1% Nonidet P-40, 10 mm MnCl2 at 37 °C for 30 min.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as per standard protocol (22). Briefly, cross-linking was done with 1% formaldehyde and then the reaction was stopped by 0.125 m glycine, followed by cell lysis in Cell Lysis buffer (5 mm PIPES (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40 (with fresh protease inhibitor)). The isolated nuclei were resuspended in nuclei lysis buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 1% SDS (with fresh protease inhibitor)). After sonication of the chromatin, pre-clearing was done. Subsequently, chromatin was immunoprecipitated with anti-ZMYND8 (Sigma Prestige Group), anti-H4K16Ac (Active Motif), anti-H3K36Me2 (Abcam), anti-H4 (Abcam), and anti-H3 (Abcam) antibodies overnight at 4 °C. Dynabeads were blocked with 0.3 mg/ml salmon sperm DNA and set for interaction. Beads were washed with RIPA buffer, high salt buffer, LiCl buffer, and TE buffer, consecutively. Following RNase A and proteinase K treatment, the beads were kept for de-cross-linking at 65 °C. Phenol/chloroform extraction followed by ethanol precipitation was performed. The DNA pellet was dissolved in H2O and then qPCR analysis was done using gene-specific primers.
ChIP assay in SH-SY5Y cells was performed after treating with 10 μm ATRA or DMSO for 4 days. The primers used in ChIP assays were designed inside the gene body or promoter elements using the UCSC Genome Browser. The detailed primer information is available on request.
Western Blot Analysis
Whole cell extracts were prepared with Laemmli Buffer (4% SDS, 20% glycerol, and 120 mm Tris-HCl (pH 6.8)) and sonicated followed by boiling at 100 °C. The samples were electrophoresed on 7.5, 11, or 15% SDS-PAGE and transferred on nitrocellulose membrane followed by blocking with nonfat skimmed milk and probed for specific antibodies. Primary antibodies used are listed in Table 1.
TABLE 1.
The list of antibodies used in Western blotting, co-IP, ChIP, and immunofluorescence
| Antibody | Company | Catalog No. |
|---|---|---|
| Anti-GST-HRP | GE HealthCare | RPN1236 |
| Anti-FLAG-HRP | Sigma | A8592 |
| Anti-FLAG monoclonal | Sigma | F1804 |
| Anti-ZMYND8 | Sigma | HPA020949 |
| Anti-Tau | Santa Cruz Biotechnology | sc20172 |
| Anti-GAPDH | Abcam | ab9485 |
| Anti-tubulin α | AbD-Serotec | MCA78G |
| Anti-H3 | Abcam | ab10799 |
| Anti-H4 | Abcam | ab10158 |
| Anti-H3K36Me1 | Abcam | ab9048 |
| Anti-H3K36Me2 | Abcam | ab9049 |
| Anti-H3K36Me3 | Abcam | ab9050 |
| Anti-H3K9Ac | Millipore | 07–352 |
| Anti-H4K16Ac | Active Motif | 39167 |
| Anti-RNA pol II (N-20) | Santa Cruz Biotechnology | sc899 |
| Anti-RNA polymerase II phospho-Ser-5 | Abcam | ab5131 |
| Anti-RNA polymerase II phospho-Ser-2 | Abcam | ab24758 |
| Anti-ZMYND11 | Sigma | AV34704 |
| Anti-rabbit IgG-HRP | Sigma | A1949 |
| Anti-rat IgG-HRP | Sigma | A5795 |
| Anti-mouse IgG-HRP | Promega | W402B |
| Anti-rabbit Alexa Fluor 488 | Invitrogen | A11034 |
| Anti-rabbit Alexa Fluor 594 | Invitrogen | A11037 |
| Anti-mouse Alexa Fluor 488 | Invitrogen | A11029 |
| Anti-mouse Alexa Fluor 594 | Invitrogen | A11032 |
Reporter Construct and Luciferase Assay
The 527-bp upstream promoter region of mouse Zmynd8 gene was amplified by PCR using mouse genomic DNA as template and then cloned into pGL3 basic vector (Promega) at KpnI/HindIII site. Similarly, the 873-bp upstream region of human ZMYND8 gene was cloned into pGL3-basic vector at KpnI/SacI site. The primer sequences used to clone the promoter regions are mentioned in Table 2 where the restriction enzyme sites are underlined. All constructs were sequenced to confirm their identity. The mutation within the RARE in both human and mouse promoters was done by using QuikChange II XL site-directed mutagenesis kit (Agilent).
TABLE 2.
Primer list
Fw is forward, and Rv is reverse.
| Gene name | Sequence |
|---|---|
| For qRT-PCR | |
| ZMYND8 | Fw: 5′-CAG AAA ATG AAA CAG CCA GGG-3′ |
| Rv: 5′-ACT TTG CAT CAG CCA GGA AG-3′ | |
| TAU | Fw: 5′-GCG GCA GTG TGC AAA TAG TCT ACA A-3′ |
| Rv: 5′-GGA AGG TCA GCT TGT GGG TTT CAA T-3′ | |
| TUBB3 | Fw: 5′-GCC TCA AGA TGT CCT CCA CC-3′ |
| Rv: 5′-CGT ACA TCT CGC CCT CTT CC-3′ | |
| DRD2 | Fw: 5′-TGC AGA CCA CCACCA ACT ACC TGA T-3′ |
| Rv: 5′-GAG CTG TAG CGC GTA TTG TAC AGC AT-3′ | |
| REST | Fw: 5′-CCT TTC GCT GTA AGC CAT GC-3′ |
| Rv: 5′-TGG TGT TTC AGG TGT GCT GT-3′ | |
| GAPDH | Fw: 5′-AAT CCC ATC ACC ATC TTC CAG-3′ |
| Rv: 5-ATG ACC CTT TTG GCT CCC-3′ | |
| E2F3 | Fw: 5′-AATATCCCTAAACCCGCTTCC-3′ |
| Rv: 5′-AGTTCACAAACGGTCCTTCTAG-3′ | |
| TFEB | Fw: 5′-AACAGTGCTCCCAATAGCC-3′ |
| Rv: 5′-TGCTGTACACATTCAGGTGG-3′ | |
| BMP8A | Fw: 5′-TGGTCATGAGCTTCGTCAAC-3′ |
| Rv: 5′-TGGGCACCTTGTAAATCCG-3′ | |
| HIPK3 | Fw: 5′-ACACAGATTCTATATGCCAGCC-3′ |
| Rv: 5′-ATTCCACTCTTGATGCCCAG-3′ | |
| BARD1 | Fw: 5′-TGAAATTCCTGAAGGTCCACG-3′ |
| Rv: 5′-GGCTTTCTACTGAGGATCTGG-3′ | |
| ZNF687 | Fw: 5′-ATCCCAGCTTCCAAACTCAG-3′ |
| Rv: 5′-AAAGAGCAGAGGACAAGACG-3′ | |
| WDR13 | Fw: 5′-TGGAAGGGACGGAACTAGATC-3′ |
| Rv: 5′-CTGTGGAAACGTTGGTGTG-3′ | |
| PCGF6 | Fw: 5′-CTCTAAGGGAAATCCGACGTG-3′ |
| Rv: 5′-TTTGGTGAGATGCAGTCCTG-3′ | |
| PBX1 | Fw: 5′-AAAGGAGATTGAGCGGATGG-3′ |
| Rv: 5′-GGGTTGCTGAGATGGGAATAG-3′ | |
| WSB1 | Fw: 5′-TCACAAGGACATCGCACAG-3′ |
| Rv: 5′-ACCCAAAAGCAAGACTCCAG-3′ | |
| WFS1 | Fw: 5′-GGAAGCTCAACCCCAAGAA-3′ |
| Rv: 5′-TCTCCACAAAGTCATCCAGC-3′ | |
| HTR3A | Fw: 5′-GACATCTACAACTTCCCCTTCG-3′ |
| Rv: 5′-CGGATTTCACCTTTTCTGGC-3′ | |
| CD47 | Fw: 5′-AGCCATTCTTTTCGTCCCAG-3′ |
| Rv: 5′-CCAACCACAGCGAGGATATAG-3′ | |
| BTG1 | Fw: 5′-TCATCTCCAAGTTTCTCCGC-3′ |
| Rv: 5′-TGCGAATACAACGGTAACCC-3′ | |
| KIF2B | Fw: 5′-TGGTTATGGTGCATGAGTCC-3′ |
| Rv: 5′-TCGTTGGAGGCTTTGTCATC-3′ | |
| ZFP3 | Fw: 5′-GATCTGACTGCCATCTACGAG-3′ |
| Rv: 5′-CCCTCCACTAGGCTGGTG-3′ | |
| For cloning | |
| Zmynd8 (mouse promoter) | Fw: 5′-GGGGTACCGGAGCCCCAACAAATCAG-3′ |
| Rv: 5′-GGAAGCTTCTCTCTCTCTCTCTGCATCT-3′ | |
| ZMYND8 (human promoter) | Fw: 5′-GGGGGTACCGGAAGTTTGTAGATCCAGGC-3′ |
| Rv: 5′-GGGGAGCTCGGCTCCTAGTTCTCACAGAA-3′ | |
| Zmynd8 (mouse promoter with RARE mut) | Fw: 5′-GCCACCCCTAAGCCGATCCTAAAAGA-3′ |
| Rv: 5′-TCTTTTAGGATCGGCTTAGGGGTGGC-3′ | |
| ZMYND8 (human promoter with RARE mut) | Fw: 5′-GCTCTTGCTAACCCGAGCCTAACAAAG-3′ |
| Rv: 5′-CTTTGTTAGGCTCGGGTTAGCAAGAGC-3′ | |
For reporter assay, 5 × 104 cells were seeded on 12-well culture plates. After 24 h, the pGL3-ZMYND8 construct (0.4 μg) was transiently transfected with Lipofectamine 2000 (Invitrogen). After 4 h of transfection, the medium was replaced with a fresh incomplete one supplemented with either RA or DMSO for the next 16 h. Each transfection was normalized with pRL-CMV vector (0.04 μg). The following day, cells were harvested, and firefly/Renilla luciferase activity was determined using the manufacturer's protocol. Each transfection was performed in triplicate, and the experiments were repeated three times.
Quantitative Real Time PCR (qRT-PCR)
Total RNA was extracted using the RNeasy mini kit (Qiagen). Complementary DNA was synthesized from 2 μg of total RNA using RevertAid fast strand cDNA synthesis kit (Thermo Scientific) according to the manufacturer's protocol followed by qRT-PCR using ABI-SYBR GREEN mix (ABI). qRT-PCR was performed using 7500 real time PCR machine. Primers used are listed in Table 2.
Microarray Analysis
Total cellular RNA was extracted, and RNA quality was determined using a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples with RNA Integrity number of greater than 7.0 were taken for Affymetrix Human Whole Genome Gene Chip (Human HT Prime View Gene Chip)-based microarray hybridization as per the manufacturer's recommended protocol (Affymetrix, Santa Clara, CA). Raw data were RMA (Robust Multiarray Average)-normalized, and baseline transformation was done to median of all samples using Gene Spring GX 12.5 software (Agilent Technologies).
Statistical Analysis of Differentially Expressed Genes from Microarray
Differentially expressed probe sets (genes) upon ZMYND8 siRNA treatment in comparison with control siRNA-treated cells were identified by applying a fold change threshold of absolute fold change greater than or equal to 1.1, and a statistically significant t test p value threshold was adjusted for the false discovery rate of less than or equal to 0.05. The same was visualized using a volcano plot to understand the differential expression pattern. Unsupervised hierarchical clustering of differentially expressed genes upon treatment in comparison with untreated were done using Euclidian algorithm with Centroid linkage rule to identify gene clusters whose expression levels are significantly reproduced across the replicates.
Biological Pathways and Gene Ontology Enrichment Analysis of Microarray Data
Differentially expressed gene list was subjected to biological significance analysis by GOElite tool. 21,887 protein coding genes were used as the background, and differentially expressed gene lists were used as query. The database of Gene Ontology categories, Wikipathways, KEGG Pathways, Pathway Commons, Pheno Ontology, Diseases, Protein Domains, transcription factor targets, and tissue expression was configured for significance analysis. Each query list was subjected to over-representation analysis against each of the above databases. A Z score and permutation or Fisher's Exact Test p value were calculated to assess over-representation of enriched biological categories.
Results
ZMYND8 Is a Histone H3/H4-interacting Protein
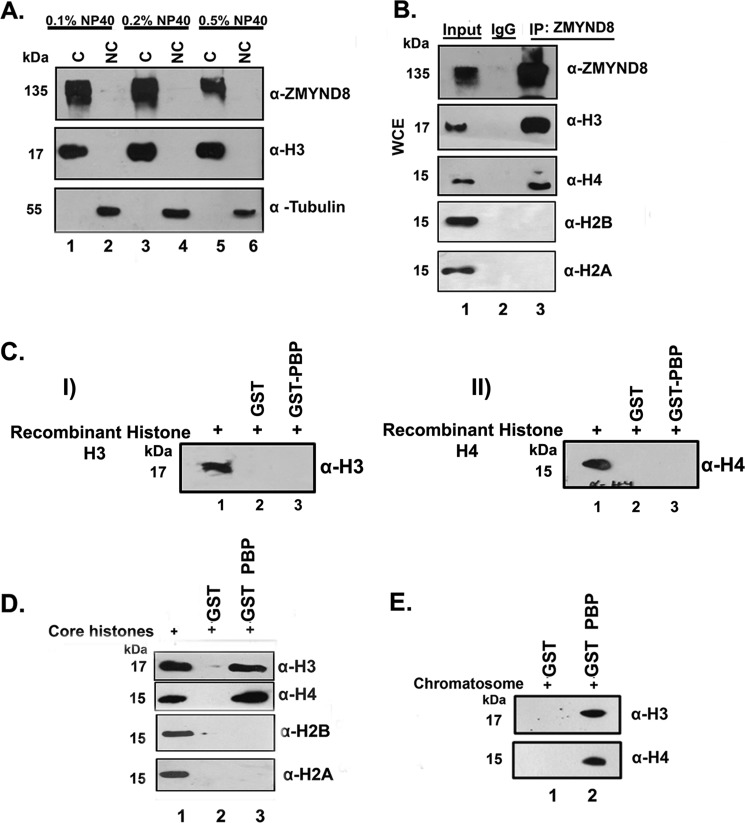
The human transcription factor ZMYND8 is a component of the transcription coregulator network having a wide range of interacting partners, including chromatin remodeling complexes and histone demethylase/deacetylase enzymes (5). It has been reported as a novel DNA damage response factor that recruits NuRD complex to damaged chromatin (23). Its involvement in chromatin template-dependent processes, viz. transcription and repair, prompted us to speculate that ZMYND8 could be a chromatin-associated protein. Indeed, we found that ZMYND8 had a tight association to chromatin by fractionation experiment, where even 0.5% Nonidet P-40 treatment could not release the protein to the non-chromatin fraction in HEK293 cells (Fig. 1A). Histone H3 and tubulin were used as nuclear and cytosolic markers, respectively (Fig. 1A). Next, we investigated the ability of ZMYND8 to interact with core histones in HEK293 cells. By co-IP with anti-ZMYND8 antibody, we observed a strong interaction with histone H3 and H4 (Fig. 1B) but not with histone H2A and H2B. However, the ability of ZMYND8 to interact with core histones could also be an indirect effect of preferential binding of ZMYND8 to other chromatin protein(s). To elucidate that, we performed an in vitro interaction of the PBP module of ZMYND8 with recombinant histones and core histones. Although ZMYND8 did not interact with individual histone H3 and H4 (Fig. 1C, panel I and panel II), it showed a strong interaction with histone H3 and H4 of core histone (Fig. 1D). These results indicate that the interaction with core histones is direct, although there is a requirement of a histone post-translational modification. Similar interaction of the PBP module of ZMYND8 with purified chromatosome from HeLa cells also showed its association to histone H3 and H4 (Fig. 1E). These results clearly indicate that ZMYND8 is a core histone H3/H4-interacting protein having a tight association with chromatin.
FIGURE 1.
Human transcription factor ZMYND8 is a chromatin-associated protein. A, chromatin fractionation experiment with increased detergent concentration (0.1%-0.5%) in HEK293 cells was performed. ZMYND8 is associated with chromatin (C) fraction even upon 0.5% Nonidet P-40 treatment. Histone H3 and tubulin were used as markers for chromatin (C) and non-chromatin (NC) fractions. B, co-IP with α-ZMYND8 antibody from HEK293 cells followed by immunoblotting with α-H3, α-H4, α-H2B, and α-H2A antibodies. IgG pull down served as a negative control. IP, immunoprecipitation; WCE, whole cell extract. C, GST-PBP (of ZMYND8) or GST protein interaction with recombinant histone H3 (panel I) and H4 (panel II). GST pulldown assay was performed and probed with α-H3 (panel I) and α-H4 (panel II) antibodies. D, core histone (from chicken erythrocytes) interaction with GST-PBP or GST proteins. GST pulldown assay followed by Western blot analysis with α-H3, α-H4 α-H2B, and α-H2A antibodies was performed. E, interaction of chromatosome (from HeLa cells) with GST-PBP or GST proteins. GST pull down was performed followed by and immunoblotting with α-H3 and α-H4 antibodies.
ZMYND8 Interacts with Modified Histone H3.1 and H4 through Its Comprehensive Chromatin-binding Module in Vitro
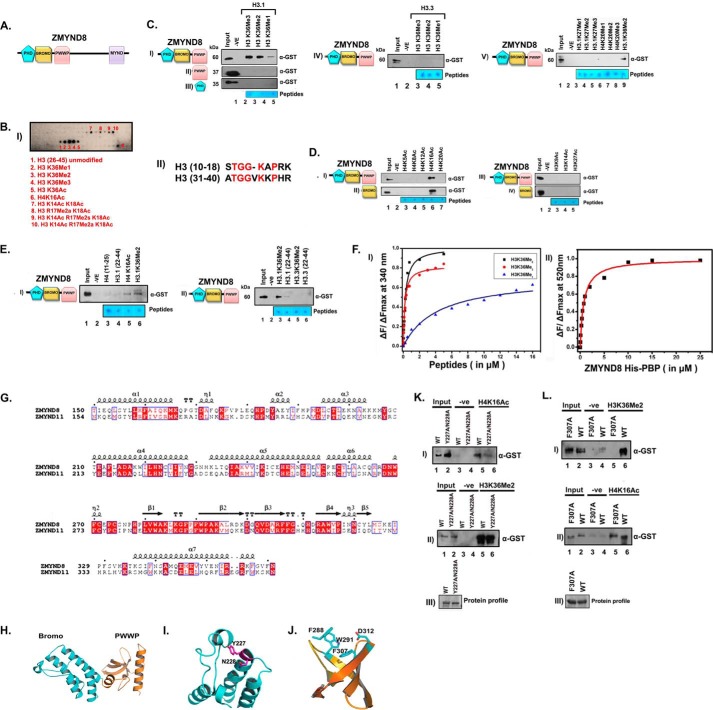
The N terminus of ZMYND8 includes the chromatin-interacting domains, viz. PHD, bromodomain, and PWWP motif (PBP module in combination), followed by the MYND finger at the C terminus (Fig. 2A). We speculated that the histone interaction ability of ZMYND8 is through its PBP module. For that we expressed and purified GST-PBP chromatin-binding modules of ZMYND8. We also purified GST-tagged PWWP, PHD, and bromodomain individually. To characterize the modified histone-binding ability of ZMYND8, MODified Histone Peptide Array (CelluSpots array) analysis was performed with purified PBP protein. H3K36Me2 (spot 3), H3K36Me3 (spot 4), and H4K16Ac (spot 6) were the most prominent interactors (Fig. 2B, panel I). H3K18Ac in combination with H3K14Ac (spot 7) or with R17Me2s/2a (spot 8/9/10) also showed positive interaction, which can be attributed to the high sequence homology between the H3K36- and H3K14/18-encompassing sequence (Fig. 2B, panel II). Furthermore, peptide pulldown assays were performed to validate the interaction biochemically. H3.1K36Me2 and H3.1K36Me3 showed a better interaction in comparison with H3.1K36Me1 peptide (Fig. 2C, panel I, compare lanes 3–5) with PBP module. The PHD finger and PWWP motifs are also well characterized methyl binders (24–26). Similar experiments were done with isolated PWWP motif or PHD finger and histone peptides H3.1K36Me1, H3.1K36Me2, and H3.1K36Me3. Neither of the domains in isolation could bind to methylated histone H3.1K36 peptides (Fig. 2C, panels II and III, compare lanes 3–5). Remarkably, GST-PBP showed no preferential interaction for methylated histones on H3.3 backbone (Fig. 2C, panel IV). We also investigated its ability to interact with other H3.1- or H4-methylated peptides (Fig. 2C, panel V) and found no other significant interactions. This clearly indicates that integrated PBP module is critical for recognition of H3.1K36Me2/Me3 only. We subsequently investigated the binding ability of the PBP module for acetylated histones and found that H4K16Ac has a better preference compared with any of the other site-specific acetylated histone H4 (at Lys-5, Lys-8, Lys-12, and Lys-20) peptides (Fig. 2D, panel I, compare lanes 3–7). However, histone H3 peptides acetylated at Lys-9, Lys-14, or Lys-27 did not show any interaction with ZMYND8 (Fig. 2D, panel III, lanes 3–5). Similar experiments done with isolated bromodomain, the acetylation binding module, also showed a good interaction with H4K16Ac but not with other acetylated H4 (Fig. 2D, panel II, compare lanes 3–7) or acetylated histone H3 (Fig. 2D, panel IV, compare lanes 3–5) peptides. We observed minimal binding of unmodified histone peptides, H3 (22–44 residues) and H4 (11–25 residues) to the PBP module of ZMYND8 confirming its preference for site-specifically modified histones at H3.1K36Me2/Me3 and H4K16Ac (Fig. 2E, panel I, compare lanes 3–6). Similarly, no significant interaction was observed for H3.3 (22–44 residues) or H3.3K36Me2 peptides with PBP module indicating preferential interaction with H3.1K36Me2/Me3 peptides (Fig. 2E, panel II, compare lanes 3–6).
FIGURE 2.
ZMYND8 interacts preferentially with histone H3.1K36Me2/Me3 and H4K16Ac through its putative chromatin-binding module in vitro. A, schematic domain organization of ZMYND8. B, interaction of GST-PBP module of ZMYND8 with MODified Histone Peptide Array (CelluSpots array). Preferential histone peptide interactors of GST-PBP were scored by probing the array with α-GST antibody. GST-PBP showed significant interaction with H3K36Me2 (spot 3), H3K36Me3 (spot 4), and H4K16Ac (spot 6). H3K14Ac K18Ac alone or in combination with R17Me2a/s or H3K18Ac R17Me2a also showed positive interaction (spots 7–10) (panel I). Sequence alignment of the canonical H3(10–18), H3(31–40) by ClustalW. The conserved amino acids are marked in red (panel II). C, interaction of GST-PBP (panel I), GST-PWWP (panel II), and GST-PHD (panel III) with biotinylated mono/di/tri-methylated H3.1K36. Similar interactions of GST-PBP were with H3.3K36-methylated peptides (panel IV). GST-PBP interaction were with biotinylated mono/di/tri-methylated H3K27 and H4K20 peptides (panel V). D, interaction of GST-PBP (panel I), GST-bromo (panel II), with biotinylated H4-K5/K8/K12/K16/K20 acetylated peptides. Interaction of GST-PBP (panel III) and GST-bromo (panel IV) module with other acetylated histone H3 peptides (H3K9Ac, H3K14Ac, and H3K27Ac). E, unmodified H4(11–25) and H3(22–44) (panel I, lanes 3 and 4) showed minimal interaction as compared with H4K16Ac and H3.1K36Me2 peptides (panel I, lanes 5 and 6) with GST-PBP. Furthermore, GST-PBP showed preferential interaction with H3.1K36Me2 compared with H3.3K36Me2 (panel II, compare lanes 3 and 5). F, binding isotherms for the interaction of His-PBP (of ZMYND8) with indicated histone peptides as obtained from steady state fluorescence spectroscopy. Data points for H3K36Me3, H3K36Me2, and H3K36Me1 are indicated by circle (red), square (black), and triangle (blue), respectively (panel I). Binding isotherm for the interaction of His-PBP and H4K16Ac (FAM-conjugated) peptide as obtained from steady state fluorescence spectroscopy (panel II). G, sequence alignment of ZMYND8 and ZMYND11 showing conserved critical residues in Bromo-PWWP (BP) module by using ESPript. H, molecular modeling of ZMYND8 with ZMYND11 BP module was done by using Modeler version 9.13. Modeled bromodomain (I) and PWWP domain (J) structure highlighting the predicted critical residues involved in interaction with acetylated H4K16 and methylated H3.1K36, respectively. K and L, comparative binding ability of GST-PBP wild type (WT) or Y227A/N228A mutant (K) or F307A mutant (L) with H4K16Ac and H3.1K36Me2 peptides (panels I and II).
The selective interaction of ZMYND8 with H3.1K36-methylated and H4K16-acetylated peptides was further quantified by fluorescence spectroscopy. The PBP module was titrated with H3.1K36Me0/Me1/Me2/Me3 peptides, and quenching of the fluorescence intensity of the tryptophan residue at the peptide binding pocket of the protein was measured. No appreciable binding was seen with H3.1K36Me0 peptide. H3.1K36Me2 peptide (Kd ∼0.23 μm) showed 2-fold higher affinity compared with H3.1K36Me3 (Kd ∼0.45 μm) (Fig. 2F, panel I, and Table 3). In comparison, binding affinity of H3.1K36Me1 (Kd ∼19.7 μm) was 85-fold weaker compared with H3.1K36Me2 peptide (Fig. 2F, panel I, and Table 3). Because the bromodomain, which is crucial for the interaction with H4K16Ac peptide, is devoid of tryptophan residues, we performed extrinsic fluorophore conjugation to H4 or H4K16Ac peptides with FAM. Subsequently, the FAM-conjugated peptides were titrated with the PBP module in increasing concentrations to score the interaction. Although no appreciable binding was seen for histone H4 peptide with the PBP module, H4K16Ac peptide showed significant interaction (Kd ∼0.75 μm) (Fig. 2F, panel II, and Table 3). Thus, the specificity in the interaction of ZMYND8 with modified histone H3.1K36Me2/H4K16Ac was reconfirmed.
TABLE 3.
The Kd values of fluorescence data are presented below
Apparent dissociation constant was obtained from steady state fluorescence spectroscopy. At 25 °C, the Kd values were averaged over three separate experiments, with error calculated as the standard deviation between runs.
| Peptide | Kd |
|---|---|
| μm | |
| H3.1K36Me3 | 0.45 ± 0.03 |
| H3.1K36Me2 | 0.23 ± 0.07 |
| H3.1K36Me1 | 19.7 ± 2.24 |
| H3.1K36 unmodified | Undetermined |
| H4K16Ac | 0.75 ± 0.1 |
| H4K16 unmodified | Undetermined |
As ZMYND8 shows similar domain organization (Bromo-PWWP) as well as the amino acid sequence identity to ZMYND11 (33% for the Bromo-PWWP module) (Fig. 2G), Bromo-PWWP domain of ZMYND8 was modeled with reference to ZMYND11 structure (Fig. 2H). The modeled bromodomain structure consists of a four-helix bundle interspersed by ZA and BC loops, which is a conserved canonical structural feature (Fig. 2I). The asparagine residue (Asn-228), which is involved in the acetylated lysine recognition, has also been highlighted in stick representation (Fig. 2I). The PWWP domain adopts a five-bladed β-barrel fold with an α-helix at the C terminus (Fig. 2J). The H3.1K36Me2-binding aromatic residues (Phe-288, Trp-291, Phe-307, and Asp-312) are in stick representation in the PWWP structure (Fig. 2J). We mutated the active site residues of the bromodomain (i.e. Tyr-227 and Asn-228) and the aromatic cage of PWWP (Phe-307) to alanine. The Y227A/N228A double mutation in PBP module of ZMYND8 showed compromised binding to H4K16Ac peptide (Fig. 2K, panel I, compare lanes 5 and 6). Similarly, PWWP F307A mutation in the PBP module showed minimal interaction to H3.1K36Me2 (Fig. 2L, panel I, compare lanes 5 and 6). Y227A/N228A double mutant, however, showed no alteration in H3.1K36Me2 binding as compared with the wild type protein (Fig. 2K, panel II, compare lanes 5 and 6). In parallel, wild type and F307A mutant proteins showed significant H4K16Ac binding (Fig. 2L, panel II, compare lanes 5 and 6). This indicates that Tyr-227 and Asn-228 residues are critical for H4K16Ac binding, and the aromatic cage in PWWP is critical for H3.1K36Me2 binding. These results confirm that the bromodomain and the PWWP motif are responsible for interacting with H4K16Ac and H3.1K36Me2 respectively, although the comprehensive organization of PBP, harboring certain critical residues in the modified histone binding pocket, is essential for mediating this interaction.
ZMYND8 Interacts with H3.1K36Me2 and H4K16Ac Histones ex Vivo
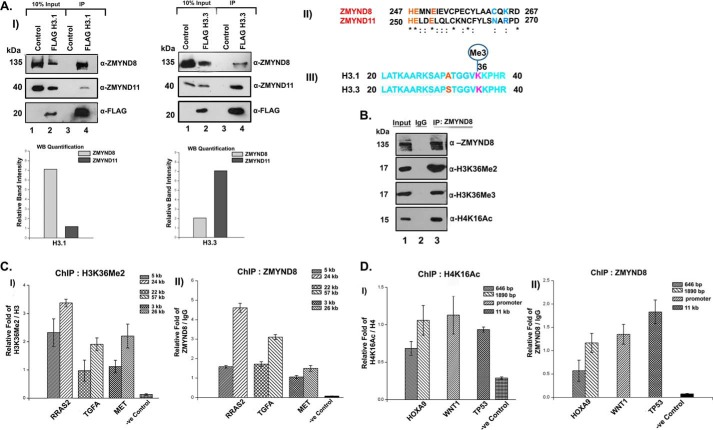
We subsequently wanted to score the binding preference of ZMYND8 for canonical or variant histone in an ex vivo context for which FLAG-H3.1- or FLAG-H3.3-transfected HEK293 cells were subjected to M2-agarose pull down and immunoblotted with anti-ZMYND8 antibody. We observed a preferential interaction of histone H3.1 with ZMYND8 and histone H3.3 with ZMYND11 (Fig. 3A, panel I) and subsequently quantified each of these interactions (Fig. 3A). A sequence alignment of ZMYND8 and ZMYND11 (Fig. 3A, panel II) and H3.1 and H3.3 (Fig. 3A, panel III) was performed to understand these binding preferences.
FIGURE 3.
Preferential interaction of ZMYND8 with H3.1K36Me2/H4K16Ac ex vivo. A, interaction of ZMYND8 with H3.1 or H3.3 histones. HEK293 cells were transiently transfected with FLAG-H3.1/FLAG-H3.3. M2-agarose pull down was done and subsequently probed with α-ZMYND8 antibody. ZMYND11 was used as a control. Western blot quantification showed the preferential interaction of ZMYND8 with H3.1 and ZMYND11 with H3.3 (panel I). Blots were quantified using ImageJ software from National Institutes of Health. Sequence alignment of histone H3 binding pocket of ZMYND11 and similar sequence stretch in ZMYND8 by ClustalW. Critical residues marked in blue mediating H3.3 binding in ZMYND11 (Asn and Arg) are substituted in ZMYND8 (by Cys and Lys) (panel II). Sequence alignment of histone H3.1 and H3.3 spanned amino acids 20–40. The variation at the N-terminal tail is at amino acid Ala-31 for H3.1 which is replaced by Ser-31 for H3.3 (marked in red) (panel III). B, interaction of ZMYND8 with modified histones. Endogenous ZMYND8 was co-immunoprecipitated from HEK293 cells and immunoblotted for ZMYND8, H3K36Me2, H3K36Me3, and H4K16Ac. IgG pull down served as a negative control. C, ChIP assays were performed in HEK293 cells with α-H3K36Me2 and α-ZMYND8 antibodies to check for enrichment of H3K36Me2 (panel I) and ZMYND8 (panel II) at RRAS2, TGFA, and MET gene loci and a negative control region (depleted of H3K36Me2 and H4K16Ac). Two separate regions of each of the genes (5 and 24 kb for RRAS2, 22 and 57 kb for TGFA, and 3 and 26 kb for MET, downstream from start site) were selected for designing primers. D, similarly, ChIP assays were performed in HEK293 cells with α-H4K16Ac and α-ZMYND8 antibodies to show enrichment of H4K16Ac (panel I) and ZMYND8 (panel II) at HOXA9, WNT1, and TP53 gene loci and a negative control region (depleted of H3K36Me2 and H4K16Ac). Two separate regions of 646 and 1890 bp downstream from the start site were selected for designing primers from HOXA9 gene. WNT1 primers were for promoter region, whereas the TP53 primer was designed 11 kb downstream. Quantitative PCR was done, and relative fold was plotted normalizing by IgG. For H3K36Me2 and H4K16Ac, ChIP relative fold was further normalized to endogenous H3 and H4, respectively. At least three separate experiments were performed. Error bars show standard deviation.
We subsequently performed co-IP assays to understand its modified histone binding preference ex vivo. Endogenous ZMYND8 showed robust interaction with H3K36Me2, compared with H3K36Me3 (Fig. 3B). Here, H4K16Ac was also found to be associated with endogenous ZMYND8 (Fig. 3B). Genome-wide distribution of H3K36Me2 and H4K16Ac epigenetic marks has been previously reported (27–29). The NSD2 (Nuclear receptor SET Domain-containing family of histone lysine methyltransferase) target genes, for example TGFA, MET, and RRAS2, were found to have high H3K36Me2 occupancy (Fig. 3C, panel I). However, several key developmental genes, including HOXA9, WNT1, and TP53, showed a high abundance of H4K16Ac (Fig. 3D, panel I). Interestingly, we observed a significant occupancy of ZMYND8 at these H3K36Me2- and H4K16Ac-enriched gene loci in HEK293 cells (Fig. 3, C, panel II, and D, panel II). In contrast, ZMYND8 had meager binding to the region of the genome with a low abundance of H3K36Me2 and H4K16Ac levels (marked as -ve Control in panels I and II of Fig. 3, C and D), implicating that the recruitment of the protein to the genome is through its ability to interact with these modified histones.
Minimal PBP Module of ZMYND8 Is Critical for Its Recruitment to the Modified Histones of Chromatin
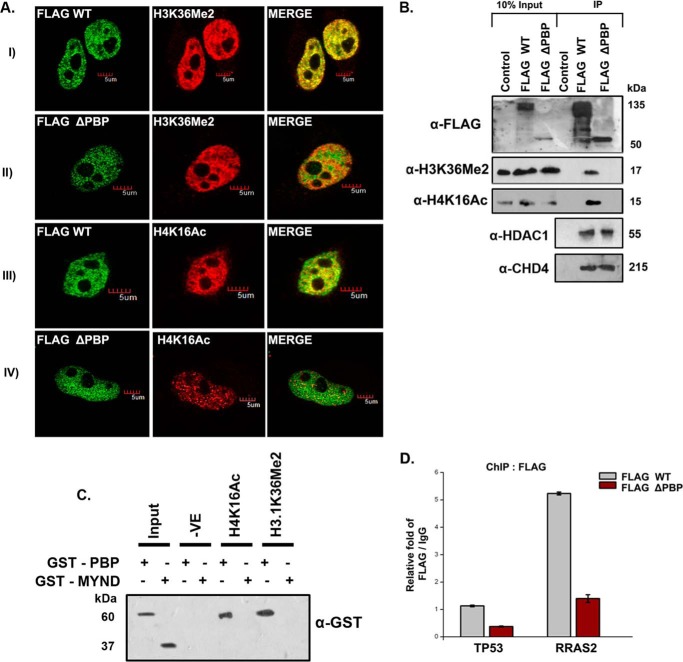
We hypothesized that the PBP module of ZMYND8 is the minimal requirement for its chromatin-binding ability. To confirm this, FLAG-conjugated constructs of wild type and PBP-deleted ZMYND8 were transfected into HeLa cells, and immunofluorescence staining with anti-FLAG and anti-H3K36Me2/-H4K16Ac was performed. The extent of co-localization of H3K36Me2 with wild type (Pearson's coefficient 0.78) was more significant than with PBP-deleted (Pearson's coefficient 0.47) (Fig. 4A, compare panels I and II). Similarly, H4K16Ac and wild type showed better co-localization (Pearson's coefficient 0.68) than with the mutant (Pearson's coefficient 0.29) (Fig. 4A, compare panels III and IV). Co-IP assays with anti-FLAG antibody also established that unlike wild type, PBP deleted-ZMYND8 showed a poor interaction with H3K36Me2 and H4K16Ac marks (Fig. 4B). However, both wild type and PBP-deleted ZMYND8 showed comparable interaction with HDAC1 and CHD4 (Fig. 4B). To further confirm whether the MYND domain is also involved in mediating the interaction with histones, we expressed and purified the GST-MYND domain of ZMYND8. In a pulldown assay with H4K16Ac and H3.1K36Me2 peptides, specific interaction could be detected with GST-PBP but not GST-MYND (Fig. 4C). Finally, the requirement of the PBP module of the protein for being targeted to its binding sites was assessed. Here, we selected TP53 and RRAS2 (24 kb) as they showed significant enrichment of H4K16Ac and H3K36Me2, respectively (Fig. 4D). The genes harboring H3K36Me2 and H4K16Ac had high occupancy of wild type but not the PBP-deleted construct as seen by ChIP assays (Fig. 4D). These results clearly indicate that the minimum PBP module of ZMYND8 is critical for its recruitment to chromatin through its modified histone interacting partners.
FIGURE 4.
Chromatin binding module of ZMYND8 is essential for its histone interaction. A, HeLa cells were transiently transfected with FLAG-WT ZMYND8 or FLAG-ΔPBP ZMYND8, co-stained with α-FLAG and either α-H3K36Me2 or α-H4K16Ac antibodies. The Pearson's coefficient for FLAG-WT ZMYND8 and H3K36Me2/H4K16Ac was >0.5, whereas that for FLAG-ΔPBP ZMYND8 and H3K36Me2/H4K16Ac was <0.5. B, HEK293 cells were transiently transfected with FLAG-WT or FLAG-ΔPBP ZMYND8, and whole cell extracts were subjected to M2-agarose FLAG pull down and immunoblotted with α-FLAG, α-H3K36Me2, α-H4K16Ac, α-HDAC1, and α-CHD4 antibodies. C, interaction of GST-PBP or GST-MYND with biotinylated H4K16Ac and H3.1K36Me2 peptides. Western blot analysis was done with α-GST antibody. D, ChIP assay was performed after transiently transfecting FLAG-WT or FLAG-ΔPBP ZMYND8 in HEK293 cells with α-FLAG antibody. Relative fold enrichment at TP53 (11 kb) and RRAS2 (24 kb) gene loci was scored by quantitative PCR normalized to IgG. At least three separate experiments were performed. Error bars show standard deviation.
ZMYND8 Is Itself an ATRA-responsive Gene and Regulates Other RA-inducible Genes
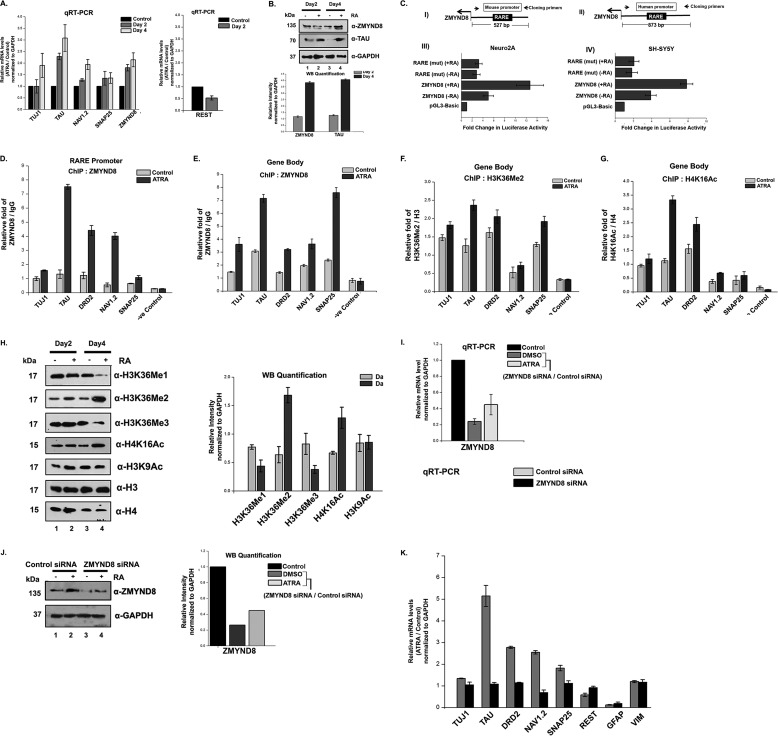
To understand the role of ZMYND8 in regulating transcription in chromatin context, we used ATRA, which is a well studied transcription inducer (11). At first, we attempted to check whether there was an alteration in expression of ZMYND8 upon ATRA treatment. For this, we selected SH-SY5Y cells that showed elevated expression of TAU, TUJ1, NAV1.2, and SNAP25 (30–32) and down-regulation of REST, a pluripotency marker leading to neurite formation (33) in response to RA. A similar alteration of gene expression upon RA stimulation was observed (Fig. 5A). ZMYND8 gene showed a progressive hyper-expression after 2 or 4 days of ATRA treatment (Fig. 5A). Similarly, ZMYND8 increased in protein abundance with progressive days of RA treatment (Fig. 5B) where TAU was used as a positive control.
FIGURE 5.
ZMYND8 is ATRA-responsive and regulates other RA-inducible genes. A, ATRA treatment of SH-SY5Y cells for 2 and 4 days, respectively, leads to an increase in ZMYND8 RNA level. TAU, TUJ1, NAV1.2, SNAP25, and REST were used as controls. Relative mRNA level was plotted for ATRA-treated differentiated cells over DMSO-treated undifferentiated cells and normalized to GAPDH. At least three individual experiments were performed. Error bars show standard deviation. B, alteration in the expression of ZMYND8 protein abundance in SH-SY5Y cells upon ATRA treatment for 2 and 4 days. Western blotting was done by probing with α-ZMYND8, α-TAU, and α-GAPDH antibodies. GAPDH was used as loading control. At least three individual experiments were performed. Error bars show standard deviation. Blots were quantified using ImageJ software from National Institutes of Health. C, schematic diagram shows the upstream regulatory region of mouse (panel I) and human (panel II) ZMYND8 genes with the arbitrary position of RARE and the primer pairs used to clone those regions into pGL3-basic vector. The respective reporter constructs were transiently transfected into Neuro2A (panel III) and SH-SY5Y (panel IV) cells, respectively, with/without ATRA treatment followed by luciferase assay. The activities are shown as mean fold enhancement compared with the empty vector after normalization with Renilla luciferase activity. Here, the constructs containing mutated RARE sequence are named as RARE-mut. Each transfection was performed in triplicate, and the experiments were repeated at least three times. Error bar shows standard deviation. D–G, relative enrichment of ZMYND8 on the RARE harboring sequence of promoter of RA-responsive genes (TUJ1, TAU, DRD2, NAV1.2, and SNAP25) was observed (D). Similar enrichment of ZMYND8 (E), H3K36Me2 (F), and H4K16Ac (G) onto gene body region of TUJ1, TAU, DRD2, NAV1.2, and SNAP25 genes was monitored. ChIP assays were performed after 4 days of ATRA treatment in SH-SY5Y cells with α-ZMYND8 antibody. Relative fold was calculated by normalizing ZMYND8 with IgG for both DMSO (control) and ATRA-treated cells. At least three separate experiments were performed. Error bars show standard deviation. H, alteration in the expression of different epigenetic marks upon ATRA treatment for 2 and 4 days. Western blotting was done by probing with α-H3K36Me1, α-H3K36Me2, α-H3K36Me3, α-H4K16Ac, and α-H3K9Ac antibodies from whole cell extracts and quantified by ImageJ software from National Institutes of Health. α-H3 and α-H4 are used as loading controls for respective modifications. At least three separate experiments were done. Error bars show standard deviation. I, ZMYND8 silencing was done in 2 days of ATRA- or DMSO- treated SH-SY5Y cells. Similar experiments were performed with non-targeting siRNA, as negative control. Total mRNA level was analyzed by quantitative PCR, and GAPDH was used for normalization. Reference level was considered as 1. J, ZMYND8 knockdown in SH-SY5Y cells was scored by Western blotting after ATRA treatment for 2 days, with α-ZMYND8 antibody. GAPDH was used for normalization. Blots were quantified using ImageJ software from National Institutes of Health. K, alterations in the expression of neuronal markers (TUJ1, TAU, DRD2, NAV1.2, and SNAP25), glial markers (GFAP and VIM), and pluripotency marker REST were measured after ZMYND8 siRNA (or a non-targeting siRNA as negative control) transfection during 2 days of ATRA or DMSO treatment in SH-SY5Y cells. Total mRNA level was scored by quantitative PCR normalizing to GAPDH. Relative mRNA level was plotted as ATRA-treated differentiated cells over DMSO-treated undifferentiated cells. Reference level considered as 1. At least three separate experiments were done. Error bars show standard deviation.
The increase in ZMYND8 in both RNA and protein levels led us to perform its promoter analysis. We observed the presence of RARE sequence at the upstream region of ZMYND8 in both human and mouse genome and subsequently cloned them in pGL3-basic vector to check their promoter activity. Indeed, we found an ATRA-induced firing of the luciferase fusion gene constructs in both mouse (Neuro2A) and human (SH-SY5Y) cells (Fig. 5C, panels III and IV). Mutating the RARE sequence led to a significant reduction in ZMYND8 promoter activity in both the cells (Fig. 5C, panels III and IV). These results clearly indicate that ZMYND8 is itself an ATRA-responsive gene.
Next, we looked for the occupancy of ZMYND8 on the different ATRA-responsive genes, including TUJ1, TAU, DRD2, NAV1.2 and SNAP25 (34) by ChIP assays (Fig. 5, D and E). We have selected primers for two different regions of the gene, promoter (harboring the RARE element), and gene body. Upon ATRA treatment, the occupancy of ZMYND8 at the promoter as well as gene body of TUJ1, TAU, DRD2, NAV1.2, and SNAP25 showed significant increase (Fig. 5, D and E). The enrichment of ZMYND8 at the promoter and gene body of the ATRA-responsive genes could be through its ability to interact with transcription machinery components or modified histones. We at first investigated the cause of selective enrichment of ZMYND8 at the gene body. H3K36Me2 and H4K16Ac have been shown to be significantly enriched at the gene body by ChIP-Seq studies in different cell lines (27–29). To understand whether the preferential recruitment of ZMYND8 to the gene body of the ATRA-inducible genes in SH-SY5Y cells is through its modified histone-binding ability, we analyzed the histone PTM on these genes (Fig. 5, F and G). We observed an increased occupancy of H3K36Me2 and H4K16Ac by RA in SH-SY5Y cells in TUJ1, TAU, DRD2, NAV1.2 and SNAP25 (Fig. 5, F and G). The reason of recruitment of ZMYND8 at the promoter elements of these genes has been investigated later.
In addition, we monitored the altered level of different epigenetic marks in ATRA-treated SH-SY5Y cells. Among H3K36Me1, H3K36Me2, and H3K36Me3 signatures, H3K36Me2 mark showed a significant elevated level (Fig. 5H). However, RA treatment increased H4K16Ac appreciably, although the H3K9Ac signature did not show significant change (Fig. 5H). Thus, ATRA modulates specific histone modifications not only in gene-specific manner but also in a global context.
Our next attempt was to examine the involvement of ZMYND8 in the regulation of these ATRA-responsive genes. We knocked down ZMYND8 in DMSO or RA-treated SH-SY5Y cells. The reduction of ZMYND8 expression upon siRNA transfection was scored by qRT-PCR and Western blotting analysis (Fig. 5, I and J). Knocking down ZMYND8 compromised the expression of different ATRA-responsive genes, including TUJ1, TAU, DRD2, NAV1.2, and SNAP25 significantly (Fig. 5K). However, the expression of REST was marginally increased upon silencing of ZMYND8 (Fig. 5K). The expression of glial markers, including GFAP and VIM (36, 37), remained almost unaltered following ZMYND8 silencing (Fig. 5K). These results clearly show that ZMYND8 gets recruited to the ATRA-responsive genes and regulates their expression.
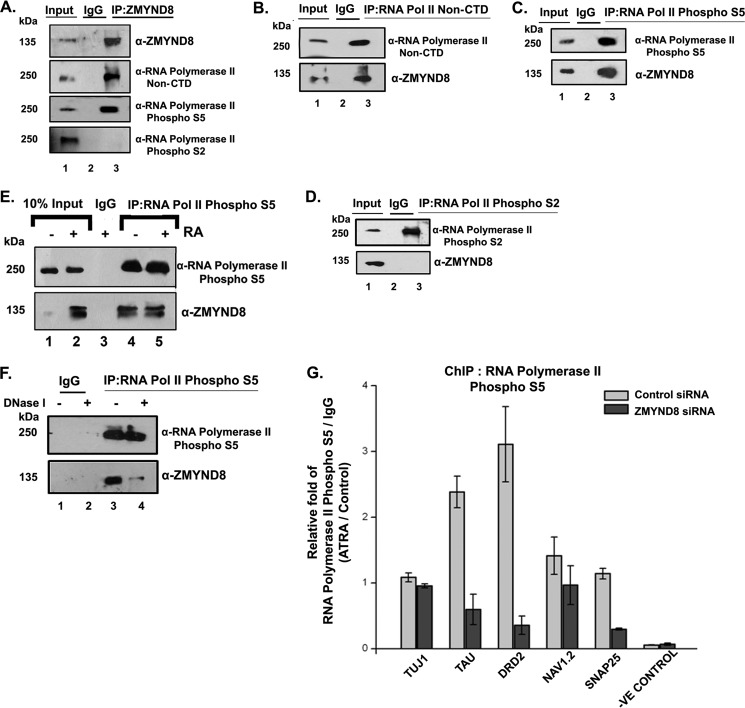
ZMYND8 Shows Selective Interaction with RNA Polymerase II Phosphorylated at Ser-5 in a DNA Template-dependent Manner
We have previously shown that upon ATRA treatment, ZMYND8 recruitment at the promoter element (harboring RARE sequence) was significantly high (Fig. 5D). Hence, we hypothesized its possible association with RNA polymerase II transcription machinery. Interestingly, by performing coimmunoprecipitation assays, we found that ZMYND8 interacts with transcription-initiation-competent RNA pol II S5P but not with transcription elongation/termination-competent RNA pol II S2P complex (Fig. 6A, compare lanes 1 and 3). Reciprocal interactions indicated that RNA pol II S5P but not S2P interacted with ZMYND8 (Fig. 6, C and D). Here, interaction with RNA pol II non-C-terminal domain was used as control (Fig. 6, A and B). We subsequently wanted to understand whether there is any alteration in its preferential interaction with RNA pol II S5P in presence or absence of ATRA. By immunoprecipitating RNA pol II S5P, the interaction with ZMYND8 did not alter in DMSO- or ATRA-treated SH-SY5Y cells (Fig. 6E). The interaction of ZMYND8 and RNA pol II machinery could be DNA template-dependent or -independent. Because the interaction between the two did not alter the presence or absence of ATRA, we predicted this complex formation onto the DNA template. We set immunoprecipitation with RNA pol II S5P after DNase I digestion of the lysates. In accordance with our speculation, we indeed see a sharp decrease in its association with ZMYND8 upon DNase I digestion of lysates (Fig. 6F). To further confirm the role of ZMYND8 in regulating transcription of ATRA-responsive genes, we performed ChIP assays with RNA pol II S5P following silencing of ZMYND8 gene expression. Indeed, we observed a significant reduction of RNA pol II S5P recruitment to chromatin in the absence of ZMYND8, even after ATRA induction (Fig. 6G). These results clearly indicate that ZMYND8 is a key component of transcription machinery complex having a significant role in mediating transcription initiation.
FIGURE 6.
ZMYND8 interacts with RNA polymerase II phospho-Ser5 in a DNA-dependent manner. A, ZMYND8 was co-immunoprecipitated from SH-SY5Y cells and immunoblotted with α-RNA polymerase II non-CTD (epitope at N terminus), α-RNA polymerase II phospho-Ser-5, and α-RNA polymerase II phospho-Ser-2 antibodies. IgG serves as negative control. B–D, similarly, RNA polymerase II non-CTD (B), RNA pol II phospho-Ser-5 (C), and RNA pol II phospho-Ser-2 (D) was co-immunoprecipitated from SH-SY5Y cells and immunoblotted with α-ZMYND8 antibody. E, co-IP of RNA pol II phospho-Ser-5 from 4 days ATRA- or DMSO-treated SH-SY5Y cells followed by immunoblotting with α-ZMYND8 antibody. IgG serves as negative control. F, RNA pol II phospho-Ser-5 was co-immunoprecipitated from SH-SY5Y cells after DNase I treatment of lysates and immunoblotted with α-ZMYND8 antibody. IgG serves as negative control. G, ChIP assays were performed after ZMYND8 siRNA or a non-targeting siRNA transfection during 2 days of ATRA or DMSO treatment in SH-SY5Y cells with α-RNA polymerase II phospho-Ser-5 antibody. Relative fold was calculated by normalizing RNA pol II phospho-Ser-5 with IgG and represented as ATRA-treated differentiated cells over DMSO-treated undifferentiated cells.
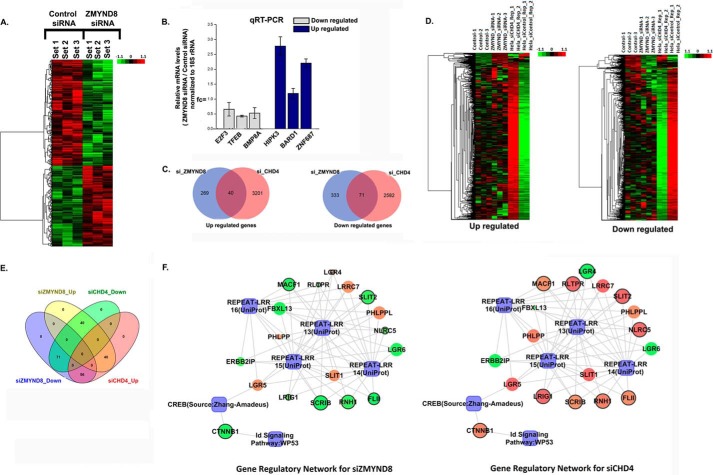
ZMYND8 Regulates Global Gene Transcription in CHD4-independent Manner
ZMYND8 is a component of the coregulator complex network having an extensive interaction with transcription machinery (5, 38). Recently ZMYND8 has been established to be a component of the NuRD chromatin remodeling complex (39) and can influence gene transcription (5). In an attempt to comprehend whether ZMYND8 can regulate global gene expression only when it remains associated with the NuRD complex, we performed microarray analysis in ZMYND8 knocked down HeLa cells. Overall, 331 genes were up-regulated and 438 genes were down-regulated upon ZMYND8 silencing. The cluster diagram is represented in Fig. 7A. The microarray data has been validated by qRT-PCR analysis where candidate up-regulated (E2F3, TFEB, and BMP8A) and down-regulated (HIPK3, BARD1, and ZNF687) genes showed differential expression upon ZMYND8 silencing (Fig. 7B). Previous studies have shown that silencing CHD4 (an important chromatin remodeling subunit of NuRD) can alter global gene expression in HeLa cells (40). To understand the NuRD-associated functions of ZMYND8, we analyzed the differentially expressed gene sets upon silencing ZMYND8 with that of CHD4 (40). On careful examination, we found that only 40 up- and 71 down-regulated genes were commonly regulated by these factors (Fig. 7C). A detailed comparison of the clusters (Fig. 7D) and overlapping Venn diagram (Fig. 7E) of up/down-regulated genes upon ZMYND8 or CHD4 silencing has been performed. Biological Analysis Network for the common gene regulatory pathways is represented in Fig. 7F. These results clearly indicate that ZMYND8 has a significant CHD4-independent role on a global platform.
FIGURE 7.
Regulation of global gene expression by ZMYND8 in a NuRD-independent mode. A, clustering and heat maps of expression values for differentially expressed genes. Down-regulated genes are marked in green, and up-regulated genes are marked in red. From left to right, first three samples are control siRNA-treated and latter three samples are ZMYND8 siRNA-treated HeLa cells. B, validation of microarray analysis. Bars show candidate up-regulated and down-regulated genes after knocking down ZMYND8 in HeLa cells. 18S rRNA was used for normalization. Reference level was considered as 1. At least three individual experiments were performed. Error bars show standard deviation. C, Venn diagram showing the overlap between up- and down-regulated genes in ZMYND8 or CHD4 knocked down HeLa cells. D, clustering and heat maps of up- and down-regulated genes in ZMYND8 knocked down and CHD4 knocked down HeLa cells. E, overlapping Venn diagram for common differentials between ZMYND8 and CHD4. The pink circle represents up-regulated genes in siCHD4, and the green circle is for down-regulated genes in siCHD4; the yellow circle is for up-regulated genes in siZMYND8, and the blue circle represents down-regulated genes in siZMYND8. As they have common differential genes, so the overlapping areas are in different colors. F, networks for ZMYND8 knocked down and CHD4 knocked down HeLa cells based on the common biology. The genes whose regulation are opposite in the two networks are bordered in black. The genes are colored according to their fold change and sized based on their p value which is <0.05. Green and red ellipses denote down-regulation and up-regulation, respectively, and the processes are denoted by a blue rectangle.
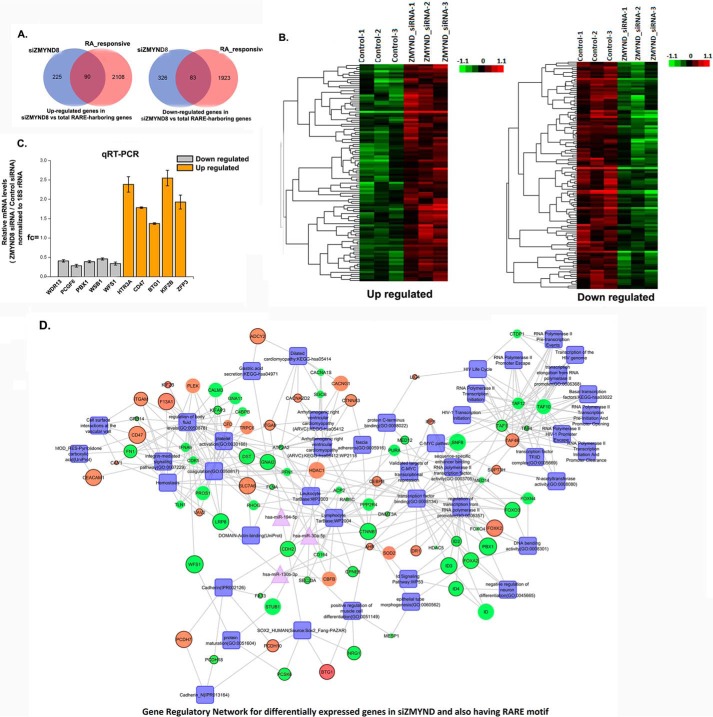
ZMYND8 Is a Key Regulator of a Subset of ATRA-inducible Genes
While analyzing the microarray results upon silencing ZMYND8, we found that a number of genes, which were regulated by ZMYND8, also harbored the RARE motif and hence can be considered as ATRA-responsive. A detailed list of differentially expressed genes as a sequel to ATRA treatment has been previously reported in HeLa cells (41). Upon ZMYND8 silencing, we found an overlap of at least 90 up- and 83 down-regulated candidates that are ATRA-inducible (Fig. 8A). Detailed cluster analyses of the up/down-regulated genes, which are simultaneously ATRA-responsive, have been represented in Fig. 8B. We further validated candidate up- and down-regulated genes upon silencing ZMYND8, which also harbors RARE motifs by qRT-PCR (Fig. 8C). Biological network for differentially expressed genes upon silencing of ZMYND8 as well as harboring the RARE motif are represented in Fig. 8D. These results clearly indicate that ZMYND8 is indeed a regulator of a fraction of ATRA-inducible genes in a global context.
FIGURE 8.
ZMYND8 modulates RARE-harboring genes. A, Venn diagram showing the overlap between up- and down-regulated genes in ZMYND8 knocked down and total RARE-harboring genes in HeLa cells. B, clustering and heat maps of up- and down-regulated genes in ZMYND8 knocked down and total RARE-harboring genes in HeLa cells. C, validation of microarray analysis. Bars show candidate RARE-harboring, up- and down-regulated genes after knocking down ZMYND8 in HeLa cells. 18S rRNA was used for normalization. Reference level was considered as 1. At least three separate experiments were done. Error bars show standard deviation. D, gene regulatory network for differentially expressed genes in ZMYND8 knocked down and total RARE-harboring genes in HeLa cells. Genes are in ellipses, and processes are in rectangles, and miRNAs are triangular in shape. The genes whose regulation are opposite in the two networks are bordered in black. Green denotes down-regulation, and red denotes up-regulation. The color shade of nodes differs due to their fold change. The size of the nodes is as per their p value which is <0.05.
Discussion
Multifunctional transcription factors harbor conserved chromatin-binding modules and show differential affinity for chromatin (42, 43). Essentially, these transcription factors are histone-interacting proteins (14), like ZMYND8, which has a distinct preference for modified histone H3 and H4 (Fig. 3B). Furthermore, ZMYND8 shows a preference for canonical histone H3.1, unlike ZMYND11 (Fig. 3A, panel I). A sequence alignment between these two proteins shows that certain key residues, including Asn-263 and Arg-265 (marked in blue), which were involved in mediating H3.3 binding through the H-bonding interaction in ZMYND11, is replaced by Cys-263 and Lys-265 in ZMYND8 (Fig. 3A, panel II). This possibly makes a difference in its choice for canonical histone H3.1 over its variant H3.3. However, among the five amino acids that are different between H3.1 and H3.3, Ala-31 of canonical H3.1 is substituted by Ser-31 in variant H3.3 and is probably an important determining factor for the preference of ZMYND8 (Fig. 3A, panel III).
The transcription factors with conserved chromatin-binding modules preferentially bind to modified histones (14). For example, the CREB-binding protein bromodomain binds to H3K36Ac/H3K56Ac/H4K20Ac (44, 45) in a monovalent manner, and bromodomain PHD finger transcription factor/PHD finger binds to H4K16Ac/H3K4Me3 (15) in a multivalent fashion. In our study, ZMYND8 preferentially binds to H3.1K36Me2/Me3 (Fig. 2, C, panel I, and F, panel I) and H4K16Ac (Fig. 2, D, panel I, and F, panel II). Although isolated bromodomain of ZMYND8 could bind to H4K16Ac (Fig. 2D, panel II), PWWP domain could interact with H3.1K36Me2/Me3 only when it was integrated along with bromodomain and PHD finger (Fig. 2C, panels I–III). For stabilization of conformation of the aromatic pocket in PWWP domain, the presence of other supporting domains, i.e. bromodomain and PHD finger, is essential. This comprehensive reader function can also be seen in case of ZMYND11 (14). To understand the binding preference of ZMYND8 for H4K16Ac in comparison with ZMYND11, we performed sequence alignment between the two proteins (Fig. 2G) and subsequently molecular modeling studies (Fig. 2, H–J). Several bromodomain proteins harbor YN residues in their active site (46). We found that ZMYND8 has Tyr-Y227 and Asn-228 amino acids in its bromodomain, which are absent in ZMYND11. The site-directed mutagenesis approach confirmed the Y227A/N228A mutant has reduced H4K16Ac binding ability (Fig. 2K). To find out the preference for modified histone recognition by ZMYND8 in cells, we performed the co-IP experiment that supported the better interaction between ZMYND8 and H3K36Me2 as compared with H3K36Me3 (Fig. 3B), in conformity with in vitro fluorescence titration assays (Fig. 2F). In parallel, the co-IP assay also confirmed the interaction of ZMYND8 with H4K16Ac ex vivo (Fig. 3B).
H3K36 methylation by Set2 recruits the Rpd3S histone deacetylase complex, which deacetylates the neighboring nucleosomes preventing cryptic transcription initiation within the gene body (47). Because ZMYND8 binds to H3K36Me2, it might have additional roles in modulating nucleosomal integrity over the coding regions preventing cryptic transcription initiation. Remarkably, the human orthologue of Drosophila melanogaster MOF (males-absent-on-the-first, a key member of the MYST family (hMOF)-mediated acetylation of H4K16) causes a recruitment of BRD4 and pTEFB to chromatin leading to a release of RNA polymerase II pausing (48, 49). In this context ZMYND8 through its chromatin reader ability might be instrumental in recruiting these factors onto chromatin thereby promoting processive transcription. The di- and tri-methylated H3K36 at the gene body shows a distinct distribution pattern, where H3K36Me2 is adjacent to promoters and H3K36Me3 accumulates at the 3′ end of the gene (47, 50). Although H3K36Me2 leads to elevated H4K16Ac, H3K36Me3 compensates it (50). The preferential interaction of ZMYND8 to H3K36Me2/H4K16Ac indicates its ability to modulate higher order chromatin structure and hence transcription. This could be possible through its ability to act as a chromatin reader, which further helps in recruiting a chromatin remodeling machinery complex steering transcription activation. Interestingly, as ZMYND8-enriched genes (RRAS2, TGFA, MET, HOXA9, TP53, and WNT1) (Fig. 3, E and F, panel II) are intricately involved in pathways regulating cell proliferation, differentiation, and development (51–56), our results indicate a possible involvement of ZMYND8 in these processes. We elucidated that a higher occupancy of ZMYND8 to these developmentally regulated genes is due to its chromatin reader function, as there is also a significant enrichment of H3K36Me2/H4K16Ac marks in these genes (Fig. 3, C and D, panel I). Furthermore, we observed an increased recruitment of ZMYND8 at the gene body of ATRA-inducible genes (TUJ1, TAU, DRD2, NAV1.2 and SNAP25) through its modified histone-binding ability (Fig. 5, E–G). However, we also observed robust enrichment of ZMYND8 at the promoter harboring the RARE sequence of these RA-responsive genes (Fig. 5D) and speculated its association with the core transcription machinery.
As RNA polymerase II transcribes a gene, there is a distinct alteration in its C-terminal domain phosphorylation status (57), which is instrumental in a dynamic association of factors that interact with the enzyme. We observed a strong interaction of ZMYND8 with RNA pol II S5P in a DNA-mediated manner (Fig. 6, A, C, and F). This interaction could be steered by some other factor(s) that promotes transcription initiation and brings ZMYND8 and RNA pol II complex to the promoter site. ZMYND8 might also be assisting in the recruitment of the RNA pol II complex through its selective binding to the H3K36Me2-enriched region of the genome located at the promoter proximal site. ZMYND11, however, modulates the activity of RNA pol II S2P at the 3′ end of the gene through its H3.3K36Me3 recognizing ability (14). Interestingly, the association of ZMYND8 with RNA pol II S5P is retained in absence of ATRA (Fig. 6E), further confirming that a stoichiometric complex formation between the two requires a DNA template. Indeed, we observed that the interaction is significantly reduced in the presence of DNase I (Fig. 6F). Furthermore, in the absence of ZMYND8, there was a significant reduction in RNA pol II S5P occupancy even upon ATRA induction, indicating that ZMYND8 is an indispensable component of initiating RNA pol II machinery (Fig. 6G).
Finally, we wanted to understand the role of ZMYND8, an integral component of transcription machinery, in regulating global gene expression. Previous studies have reported that ZMYND8 is a functional component of the transcription repression complex NuRD (58, 59) and targets it to the DNA damage response site (23). CHD3/4 is responsible for the ATP-dependent chromatin remodeling function of NuRD (35, 60). In this study, we tried to understand the CHD4-independent function of ZMYND8 in perspective of gene expression. Microarray analysis clearly indicated that ZMYND8 regulates a large class of genes that are unique from those regulated by CHD4 (Fig. 7). Interestingly, subsets of genes that are differentially expressed upon silencing of ZMYND8 also harbor the RARE motif and hence are ATRA-responsive (Fig. 8). ZMYND8 itself harbors the RARE sequence and is regulated by RA (Fig. 5, B and C). Because ZMYND8 has a preference for canonical histone H3.1 over variant H3.3, we also analyzed whether the gene targets of ZMYND8 have a depleted occupancy of H3.3. However, a significant number of ZMYND8 target genes is found to harbor H3.3 (data not shown). It would be interesting to understand the mechanism of transcription regulation by ZMYND8 in H3.1 versus H3.3-enriched genes.
In summary, we found that ZMYND8, a transcription regulator, is a putative chromatin reader. Through its specific motifs as well as amino acid residues, it exerts its chromatin-binding ability. Furthermore, it shows its preferential interaction with canonical histone H3.1K36Me2 and H4K16Ac. Interestingly, ZMYND8 is itself an RA-inducible gene, and through its modified histone-binding ability it regulates the expression of other ATRA-responsive genes. Furthermore, we show that ZMYND8 is indeed associated with transcription initiation-competent RNA pol II S5P in a DNA template-dependent manner. Although it is a transcription factor, the mechanism underlying the gene expression regulation by ZMYND8 has remained elusive. Hence, this study is an attempt to decipher the so far uncharacterized ability of ZMYND8 to modulate gene expression. Further study may uncover its possible role in lineage commitment during development.
Author Contributions
C. D. and S. R. conceived the study and designed the experiments. S. A., S. S., M. B., I. S., S. S., and D. K. S. performed the experiments and analyzed the data. C. D., S. R., M. B., S. A., and S. S. wrote the paper. All authors discussed the paper and commented on the manuscript.
Acknowledgments
We thank Prof. Hideaki Tagami for FLAG-H3.1/H3.3 plasmids. We thank Prof. Jessica K. Tyler for critical comments on the manuscript and Prof. Tapas K. Kundu and Prof. Dipak Dasgupta for helpful discussions. We thank Payal Chakraborty and Madavan Vasudevan from Bionivid Technology Pvt. Ltd. for their assistance with microarray experiments and data analysis. We also thank Rajarshi Chakrabarti for assistance with OLYMPUS FLUOVIEW FV1000 confocal imaging.
This work was supported in part by research grants from Biomolecular Assembly, Recognition and Dynamics Project, Grant 12-R&D-SIN-5.04-0103 from the Department of Atomic Energy, and a Ramalingaswami Fellowship, Department of Biotechnology, Government of India to C. D. The authors declare that they have no conflicts of interest with the contents of this article.
- PTM
- post-translational modification
- co-IP
- coimmunoprecipitation
- ATRA
- all-trans-retinoic acid
- RA
- retinoic acid
- RARE
- retinoic acid-response element
- FAM
- 6-carboxyfluorescein
- qRT
- quantitative RT
- PHD
- plant homeodomain
- pol II
- polymerase II.
References
- 1.Zhang Y., LeRoy G., Seelig H. P., Lane W. S., and Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 [DOI] [PubMed] [Google Scholar]
- 2.Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., and Schreiber S. L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature 395, 917–921 [DOI] [PubMed] [Google Scholar]
- 3.Yun M., Wu J., Workman J. L., and Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musselman C. A., Lalonde M. E., Côté J., and Kutateladze T. G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., and Qin J. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberl H. C., Spruijt C. G., Kelstrup C. D., Vermeulen M., and Mann M. (2013) A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 49, 368–378 [DOI] [PubMed] [Google Scholar]
- 7.Zeng W., Kong Q., Li C., and Mao B. (2010) Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem. Biophys. Res. Commun. 394, 1024–1029 [DOI] [PubMed] [Google Scholar]
- 8.Bierkens M., Krijgsman O., Wilting S. M., Bosch L., Jaspers A., Meijer G. A., Meijer C. J., Snijders P. J., Ylstra B., and Steenbergen R. D. (2013) Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer 52, 56–68 [DOI] [PubMed] [Google Scholar]
- 9.Edgren H., Murumagi A., Kangaspeska S., Nicorici D., Hongisto V., Kleivi K., Rye I. H., Nyberg S., Wolf M., Borresen-Dale A. L., and Kallioniemi O. (2011) Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 12, R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichmuller S., Usener D., Dummer R., Stein A., Thiel D., and Schadendorf D. (2001) Serological detection of cutaneous T-cell lymphoma-associated antigens. Proc. Natl. Acad. Sci. U.S.A. 98, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balmer J. E., and Blomhoff R. (2002) Gene expression regulation by retinoic acid. J. Lipid Res. 43, 1773–1808 [DOI] [PubMed] [Google Scholar]
- 12.Liu H., and Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das C., Hizume K., Batta K., Kumar B. R., Gadad S. S., Ganguly S., Lorain S., Verreault A., Sadhale P. P., Takeyasu K., and Kundu T. K. (2006) Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 26, 8303–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., Li B., Barton M. C., Li W., Li H., and Shi X. (2014) ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruthenburg A. J., Li H., Milne T. A., Dewell S., McGinty R. K., Yuen M., Ueberheide B., Dou Y., Muir T. W., Patel D. J., and Allis C. D. (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J. (2006) Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods 40, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti S., Bhattacharyya D., and Dasgupta D. (2000) Structural basis of DNA recognition by anticancer antibiotics, chromomycin A(3), and mithramycin: roles of minor groove width and ligand flexibility. Biopolymers 56, 85–95 [DOI] [PubMed] [Google Scholar]
- 18.Eswar N., Eramian D., Webb B., Shen M. Y., and Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 19.Das C., Gadad S. S., and Kundu T. K. (2010) Human positive coactivator 4 controls heterochromatinization and silencing of neural gene expression by interacting with REST/NRSF and CoREST. J. Mol. Biol. 397, 1–12 [DOI] [PubMed] [Google Scholar]
- 20.Zhan X., Shi X., Zhang Z., Chen Y., and Wu J. I. (2011) Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc. Natl. Acad. Sci. U.S.A. 108, 12758–12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J., and Stern D. F. (2005) DNA damage regulates Chk2 association with chromatin. J. Biol. Chem. 280, 37948–37956 [DOI] [PubMed] [Google Scholar]
- 22.Tsai W. W., Wang Z., Yiu T. T., Akdemir K. C., Xia W., Winter S., Tsai C. Y., Shi X., Schwarzer D., Plunkett W., Aronow B., Gozani O., Fischle W., Hung M. C., Patel D. J., and Barton M. C. (2010) TRIM24 links a non-canonical histone signature to breast cancer. Nature 468, 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong F., Chiu L. Y., Cox B., Aymard F., Clouaire T., Leung J. W., Cammarata M., Perez M., Agarwal P., Brodbelt J. S., Legube G., and Miller K. M. (2015) Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29, 197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., and Jeltsch A. (2010) The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansfield R. E., Musselman C. A., Kwan A. H., Oliver S. S., Garske A. L., Davrazou F., Denu J. M., Kutateladze T. G., and Mackay J. P. (2011) Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 286, 11779–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Zeng H., Lam R., Tempel W., Amaya M. F., Xu C., Dombrovski L., Qiu W., Wang Y., and Min J. (2011) Structural and histone-binding ability characterizations of human PWWP domains. PLoS ONE 6, e18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor G. C., Eskeland R., Hekimoglu-Balkan B., Pradeepa M. M., and Bickmore W. A. (2013) H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23, 2053–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo A. J., Cheung P., Chen K., Zee B. M., Kioi M., Lauring J., Xi Y., Park B. H., Shi X., Garcia B. A., Li W., and Gozani O. (2011) NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horikoshi N., Kumar P., Sharma G. G., Chen M., Hunt C. R., Westover K., Chowdhury S., and Pandita T. K. (2013) Genome-wide distribution of histone H4 lysine 16 acetylation sites and their relationship to gene expression. Genome Integr. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang H. B., Mi Y., Zhang Y., Wu N. H., and Shen Y. F. (2010) HDAC3 augments the autoregulation of neuroD gene in P19 cells. Neuroreport 21, 19–23 [DOI] [PubMed] [Google Scholar]
- 31.Musch T., Oz Y., Lyko F., and Breiling A. (2010) Nucleoside drugs induce cellular differentiation by caspase-dependent degradation of stem cell factors. PLoS ONE 5, e10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q., Zhou Z., Zhang L., Xu S., Chen C., and Yu Z. (2014) The cellular distribution and Ser262 phosphorylation of tau protein are regulated by BDNF in vitro. PLoS ONE 9, e91793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A., Rokes C., Gireud M., Fletcher S., Baumgartner J., Fuller G., Stewart J., Zage P., and Gopalakrishnan V. (2011) Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(β-TRCP) in neuroblastoma cells. Cancer 117, 5189–5202 [DOI] [PubMed] [Google Scholar]
- 34.Wernicke C., Hellmann J., Finckh U., and Rommelspacher H. (2010) Chronic ethanol exposure changes dopamine D2 receptor splicing during retinoic acid-induced differentiation of human SH-SY5Y cells. Pharmacol. Rep. 62, 649–663 [DOI] [PubMed] [Google Scholar]
- 35.Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16, 351–356 [DOI] [PubMed] [Google Scholar]
- 36.Widestrand A., Faijerson J., Wilhelmsson U., Smith P. L., Li L., Sihlbom C., Eriksson P. S., and Pekny M. (2007) Increased neurogenesis and astrogenesis from neural progenitor cells grafted in the hippocampus of GFAP−/− Vim−/− mice. Stem Cells 25, 2619–2627 [DOI] [PubMed] [Google Scholar]
- 37.Poloni A., Maurizi G., Foia F., Mondini E., Mattiucci D., Ambrogini P., Lattanzi D., Mancini S., Falconi M., Cinti S., Olivieri A., and Leoni P. (2015) Glial-like differentiation potential of human mature adipocytes. J. Mol. Neurosci. 55, 91–98 [DOI] [PubMed] [Google Scholar]
- 38.Poleshko A., Einarson M. B., Shalginskikh N., Zhang R., Adams P. D., Skalka A. M., and Katz R. A. (2010) Identification of a functional network of human epigenetic silencing factors. J. Biol. Chem. 285, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloet S. L., Baymaz H. I., Makowski M., Groenewold V., Jansen P. W., Berendsen M., Niazi H., Kops G. J., and Vermeulen M. (2015) Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 282, 1774–1785 [DOI] [PubMed] [Google Scholar]
- 40.Nishibuchi G., Shibata Y., Hayakawa T., Hayakawa N., Ohtani Y., Sinmyozu K., Tagami H., and Nakayama J. (2014) Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 289, 28956–28970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lalevée S., Anno Y. N., Chatagnon A., Samarut E., Poch O., Laudet V., Benoit G., Lecompte O., and Rochette-Egly C. (2011) Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp). J. Biol. Chem. 286, 33322–33334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., and Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falciola L., Spada F., Calogero S., Langst G., Voit R., Grummt I., and Bianchi M. E. (1997) High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 137, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das C., Roy S., Namjoshi S., Malarkey C. S., Jones D. N., Kutateladze T. G., Churchill M. E., and Tyler J. K. (2014) Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proc. Natl. Acad. Sci. U.S.A. 111, E1072–E1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng L., Zhang Q., Gerona-Navarro G., Moshkina N., and Zhou M. M. (2008) Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippakopoulos P., and Knapp S. (2012) The bromodomain interaction module. FEBS Lett. 586, 2692–2704 [DOI] [PubMed] [Google Scholar]
- 47.Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., and Mellor J. (2008) Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapoor-Vazirani P., Kagey J. D., and Vertino P. M. (2011) SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol. Cell. Biol. 31, 1594–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., and Oliviero S. (2009) Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 50.Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., and Schübeler D. (2007) Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 26, 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larive R. M., Abad A., Cardaba C. M., Hernández T., Cañamero M., de Álava E., Santos E., Alarcón B., and Bustelo X. R. (2012) The Ras-like protein R-Ras2/TC21 is important for proper mammary gland development. Mol. Biol. Cell 23, 2373–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamura K., Kawamura N., Kumagai J., Fukuda J., and Tanaka T. (2007) Tumor necrosis factor regulation of apoptosis in mouse preimplantation embryos and its antagonism by transforming growth factor α/phosphatidylinositol 3-kinase signaling system. Biol. Reprod. 76, 611–618 [DOI] [PubMed] [Google Scholar]
- 53.Li Y. Q., Ren X. Y., He Q. M., Xu Y. F., Tang X. R., Sun Y., Zeng M. S., Kang T. B., Liu N., and Ma J. (2015) MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. 6, e1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gersten M., Zhou D., Azad P., Haddad G. G., and Subramaniam S. (2014) Wnt pathway activation increases hypoxia tolerance during development. PLoS ONE 9, e103292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardiner D. M., Blumberg B., Komine Y., and Bryant S. V. (1995) Regulation of HoxA expression in developing and regenerating axolotl limbs. Development 121, 1731–1741 [DOI] [PubMed] [Google Scholar]
- 56.Sammons M. A., Zhu J., Drake A. M., and Berger S. L. (2015) TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 25, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phatnani H. P., and Greenleaf A. L. (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 58.Allen H. F., Wade P. A., and Kutateladze T. G. (2013) The NuRD architecture. Cell. Mol. Life Sci. 70, 3513–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Q., and Zhang Y. (2003) The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274, 269–290 [DOI] [PubMed] [Google Scholar]
- 60.O'Shaughnessy A., and Hendrich B. (2013) CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem. Soc. Trans. 41, 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]