Summary
Cell recognition molecules are key regulators of neural circuit assembly. The Dscam family of recognition molecules in Drosophila has been shown to regulate interactions between neurons through homophilic repulsion. This is exemplified by Dscam1 and Dscam2, which together repel dendrites of lamina neurons, L1 and L2, in the visual system. By contrast, here we show that Dscam2 directs dendritic targeting of another lamina neuron, L4, through homophilic adhesion. Through live imaging and genetic mosaics to dissect interactions between specific cells, we show that Dscam2 is required in L4 and its target cells for correct dendritic targeting. In a genetic screen we identified Dscam4 as another regulator of L4 targeting which acts with Dscam2 in the same pathway to regulate this process. This ensures tiling of the lamina neuropil through heterotypic interactions. Thus, different combinations of Dscam proteins act through distinct mechanisms in closely related neurons to pattern neural circuits.
Introduction
Precise patterns of synaptic connections between neurons are a striking feature of nervous system organization. Studies over the past several decades have shown that homophilic recognition molecules contribute to patterning neural circuits by mediating interactions between neuronal processes, axons and dendrites (Edelman, 1983; Hatta and Takeichi, 1986; Hirano and Takeichi, 2012; Maness and Schachner, 2007). Several strategies have evolved to expand the repertoire of mechanisms by which these molecules contribute to circuit formation. First, variation in expression of the same adhesion protein on processes of different cells leads to selective association of those expressing similar levels (Foty and Steinberg, 2013; Schwabe et al., 2014). Second, massive exon expansion and divergence has given rise to large families of related homophilic recognition molecules with discrete specificities and related functions (Chen and Maniatis, 2013; Zipursky and Grueber, 2013). Finally, in some systems modest expansion via gene duplication followed by divergence gives rise to small families of paralog-specific homophilic recognition molecules (Millard et al., 2007; Yamagata and Sanes, 2008).
Several features of the Drosophila visual system are particularly well suited to exploring the cellular recognition strategies underlying the assembly of neural circuits. First, the visual system, containing the retina that detects light and the underlying optic ganglia that process visual information, is modular. Each module is repeated some 750 times in each brain hemisphere. Within each module there is a discrete set of neurons of different types and well-defined patterns of synapses between them (Meinertzhagen and O’Neil, 1991; Meinertzhagen and Sorra, 2001; Rivera-Alba et al., 2011; Takemura et al., 2013). Thus, within a single animal one can assess the development of many different neurons of the same or different types. Second, neuron-specific markers allow one to characterize the patterns of axons and dendrites of each cell type in detail (Jenett et al., 2012). Finally, this system is particularly amenable to single cell genetic analysis, in which single mutant neurons can be generated surrounded by wild-type neighbors (Lee and Luo, 1999). This level of analysis is essential, as most cellular recognition molecules are expressed by multiple cells in close contact with one another (Tan et al., submitted). Thus, single cell analysis allows one to tease apart the genetic contributions to phenotypes one cell at a time.
The lamina region of the optic lobe (Figure 1A) is particularly well suited to studying circuit assembly. Here, each module or cartridge contains axons and dendrites from 19 different neurons. These include the axons of the outer photoreceptors (R cells; R1-R6) that form synapses onto the dendrites of a subset of L1-L5 lamina neurons. These synapses are called tetrads and comprise a single presynaptic active zone juxtaposing the postsynaptic elements of an invariant pair of dendrites from L1 and L2 neurons and variable contributions from two other neurons. Circuit assembly in the lamina has been studied using light and electron microscopy, genetics and, more recently, through live imaging (Langen et al., 2015; Meinertzhagen and O’Neil, 1991; Meinertzhagen and Sorra, 2001; Rivera-Alba et al., 2011; see also references below).
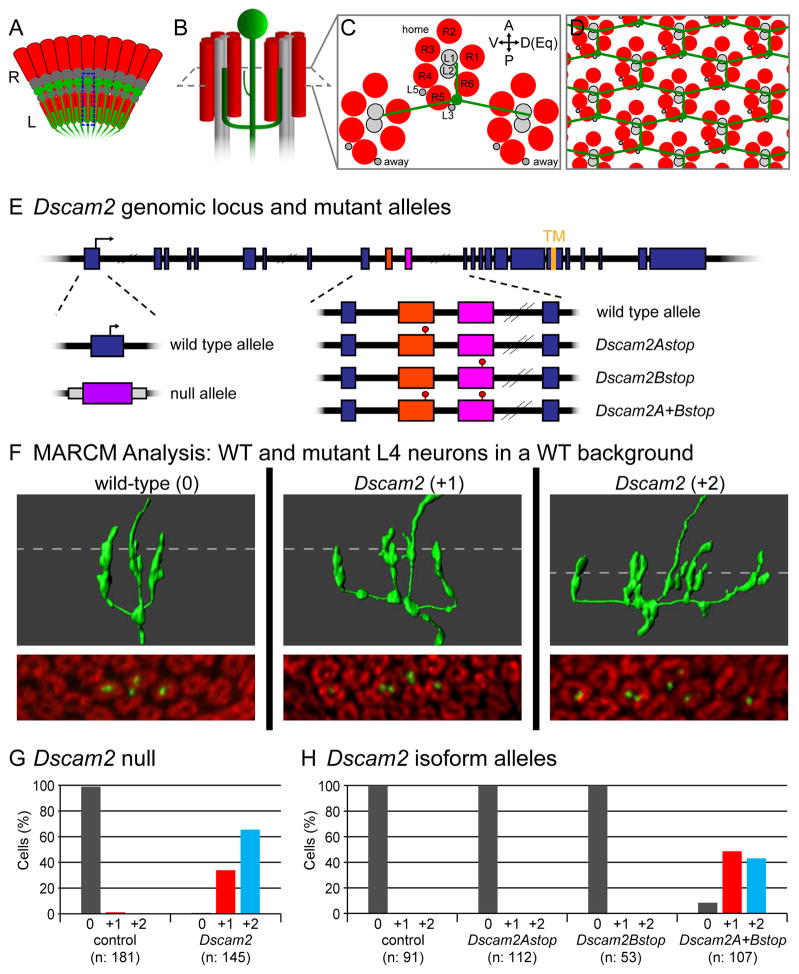
Figure 1. Dscam2 is required for dendritic patterning of L4 neurons.
(A) Schematic of the retina (R) and lamina (L) of the Drosophila visual system showing the location and arrangement of L4 neurons (green). A single L4, highlighted by the dotted box is detailed in (B).
(B) Schematic of L4 neuron (green) innervating two posterior (away) cartridges containing R cells (red) and lamina neurons (grey). Home cartridge (see C) omitted for clarity.
(C) Cross-sectional view of B (including home and away cartridges). Positions of R1-R6 and lamina neurons (L1-L5) indicated. Anterior (A), posterior (P), dorsal (D), ventral (V) and equatorial (Eq) orientations as indicated. The equator is the midline of the D-V axis.
(D) L4 dendrites tile the lamina neuropil.
(E) Schematic of Dscam2 genomic locus and mutant alleles. Location of translational start site (arrow), exons (blue boxes) and sequences encoding the transmembrane domain (TM) are indicated. Variable exons 10A and 10B in Dscam2 are demarcated in orange and pink, respectively (see also Figure S2A) Dscam2 null allele is a replacement of exon 1 with exogenous DNA bearing the white (w) gene (purple box; Millard et al., 2007). Isoform specific stop mutants, Dscam2Astop, Dscam2Bstop and Dscam2A+Bstop contain stop codons in exons 10A, 10B or both, respectively.
(F) MARCM analysis of Dscam2 in adult L4 neurons. 3D renderings (top 3 panels) taken from serial confocal slices (bottom 3 panels) of wild-type and Dscam2 mutant L4 neurons. L4 neurons were labeled with GFP (green) and cartridges with anti-Highwire (Hiw; red). Dotted line indicates position of corresponding confocal slice. Axons (Ax; arrowheads) and additional dendritic branches (*) are indicated. Scale bar, 5 μm.
(G) Quantification of dendritic phenotypes seen in MARCM-generated L4 neurons with Dscam2 null mutation.
(H) Quantification of dendritic phenotypes seen in MARCM-generated L4 neurons with Dscam2 isoform-specific alleles.
Homophilic recognition molecules belonging to two evolutionary conserved families, the cadherin and immunoglobulin (Ig) superfamilies, play key roles in regulating circuit assembly in the developing lamina. N-Cadherin (CadN) and the protocadherin Flamingo (Fmi) are proposed to integrate adhesive forces between R cell growth cones allowing them to orient and target to the correct lamina neuron targets (Chen and Clandinin, 2008; Lee et al., 2003; Schwabe et al., 2013). In addition, CadN is required to promote interactions between R cell growth cones and lamina neuron targets through homophilic interactions (Lee et al., 2001; Prakash et al., 2005). Dscams act at a later step in lamina circuit formation, the assembly of the tetrad synapse (Millard et al., 2010). Meinertzhagen et al. (2000) reported that dendrites of developing L1 and L2 neurons randomly associate at nascent tetrads and that, as these structures mature, the L1-L1 and L2-L2 pairs are lost. Dscam1 and Dscam2 act together in a redundant fashion to ensure that only one L1 and one L2 dendrite are present in the tetrad (Millard et al., 2010). Genetic analyses support a model in which these proteins act in parallel through homophilic recognition followed by repulsion between L1-L1 and L2-L2 dendritic pairs. Thus, two families of homophilic recognition proteins contribute to patterning circuits in the lamina, one through adhesive, and the other through repulsive interactions.
These studies raised the question of whether other lamina neurons also use Dscam proteins to promote cell recognition during circuit assembly. To address this issue we focused on L4 lamina neurons. By contrast to L1 and L2 neurons, each of which elaborates many short primary dendrites, L4 neurons produce three primary dendrites each innervating a separate cartridge with an invariant geometric relationship (Figure 1B,C). One dendrite projects anteriorly within its own cartridge (referred to as the “home” cartridge) while the other two project to cartridges located immediately posteriodorsal and posterioventral to the home cartridge (i.e. “away” cartridges). Thus, L4 dendrites tile the entire lamina (Figure 1D) with each cartridge receiving dendrites from three spatially segregated L4s. It has been proposed that this wiring pattern contributes to directional selectivity in optomotor behavior (Tuthill et al., 2013). Here, we show that Dscam2, but not Dscam1, is essential to establish this pattern. Dscam2 mutant L4 dendrites innervate additional cartridges at unique dorsal and ventral locations thereby disrupting tiling. We demonstrate that this phenotype emerges from a primary defect early in development due to loss of selective adhesion between L4 dendrites and their targets. In an unbiased genetic screen for other similar dendritic targeting phenotypes, we discovered that loss of Dscam4 from L4 neurons exhibits the same phenotype as Dscam2 mutants. Thus, Dscam proteins act in different combinations to promote both adhesive and repulsive interactions between processes to regulate the assembly of neural circuits.
Results
Dscam2 is required for patterning of L4 dendrites
As Dscam1 and Dscam2 are required for L1 and L2 dendritic patterning, we assessed whether they are also required in L4 dendrites. We used Mosaic Analysis of a Repressible Cell Marker (MARCM; Lee and Luo, 1999) to generate clones of individual L4 neurons homozygous for null alleles of Dscam1 (see below) and Dscam2 (Figure 1E). Whereas Dscam1 mutant L4 neurons were wild-type (see below), Dscam2 mutants exhibited a stereotyped dendritic phenotype (Figure 1F,G). Here, an additional branch often extended from one or both of the posterior dendrites and innervated an adjacent cartridge. We refer to these phenotypes as “+1” or “+2”, respectively (with the wild-type dendritic pattern as “0”). The phenotypes were highly penetrant with 33.8% and 65.5% of Dscam2 L4 neurons displaying +1 and +2 phenotypes, respectively. As a consequence of this patterning phenotype, the dendritic fields of neighboring L4 neurons overlap thereby disrupting tiling. Thus, Dscam2 is necessary for patterning L4 dendrites.
Dscam2 has two ectodomain isoforms, Dscam2A and Dscam2B, each displaying isoform-specific homophilic binding (Millard et al., 2007). We, therefore, sought to determine which isoforms were required for dendritic patterning. To do this, we generated mutants lacking Dscam2A, Dscam2B or both by introducing stop codons within the alternatively spliced exons using ends-out homologous recombination and recombination-mediated cassette exchange (Figure 1E; we refer to these as Dscam2Astop, Dscam2Bstop and Dscam2A+Bstop). The morphology of L4 neurons lacking either Dscam2A or Dscam2B was indistinguishable from wild type, while the L4 dendritic phenotype in flies with stop codons in both alternative exons was fully penetrant (Figure 1H). In summary, loss of Dscam2 resulted in a highly specific dendritic phenotype and the activity of either Dscam2A or 2B was sufficient for wild-type dendritic patterning.
Dscam2 is required at an early step in dendritic targeting
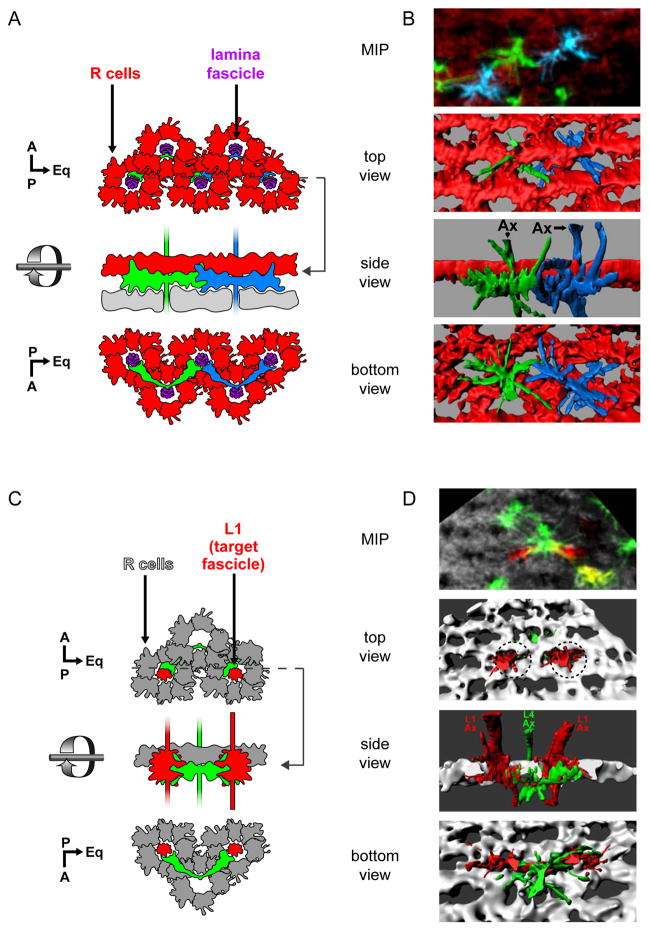
To understand the role of Dscam2 in patterning L4 dendrites, we first examined the development of wild-type L4 neurons beginning in early pupa in fixed whole mount preparations using confocal microscopy. At 24 hours after puparium formation (h APF), the axons of lamina neurons from a single cartridge form a fascicle encircled by six R cell growth cones (R1-R6; Meinertzhagen and Hansen, 1993; Schwabe et al., 2014). The lamina axon fascicle continues to extend into the medulla neuropil. This pattern is repeated across the entire developing lamina to form a planar lattice of R cell growth cones (lamina plexus) pierced at regular intervals by lamina axon fascicles (Figure 2A). A layer of glial cells lies beneath the R cell growth cones (Edwards et al., 2012). Each L4 neuron initially produces two incipient dendrites from a region of the axon just below the lamina plexus and above the glial cells (Figure 2A, B). These early dendrites form elaborate fan-shaped processes from the posterior aspect of the L4 axon making extensive contacts with the two immediately posterior lamina fascicles (i.e. target fascicles; Figure 2C, D). At the same time, extensive contacts are also seen between dendrites from neighboring L4s which converge onto the same target fascicle (Figure 2A, B). At 30 h APF the L4 dendrites remain largely unchanged (Figure S1A). By 36 h APF these dendrites take on a U shaped morphology where the position from which the dendrites emerge from the L4 axon is displaced from the lamina plexus (Figure S1B). Between 42 and 48 h APF, as the lamina plexus expands, L4 dendrites extend distally to achieve their final adult morphology (Figure S1C–E). The anterior dendrite begins to emerge at 48 h APF innervating the developing home cartridge (Figure S1D).
Figure 2. L4 dendrites project to their target fascicles at an early stage in their development.
(A,B) Dendrites from neighboring L4 neurons contact each other as they converge on a common target fascicle at 24 h APF.
(A) Schematic representation of two adjacent L4 neurons (green and blue) imaged in (B). A lattice of R cell growth cones (red) surrounds each lamina fascicle (purple). Glia (grey) lie immediately beneath L4 dendrites. Dotted line indicates the cross-section at which the side view is taken.
(B) Multicolor Flip Out (MCFO; Nern et al., 2015) was used to simultaneously label two adjacent L4s. Confocal sections (top panel; MIP: maximum intensity projection) were used to generate 3D renderings of L4 neurons and R cell growth cones displayed from different perspectives (bottom 3 panels).
(C,D) L4 dendrites make contact with neurons in their target fascicles at 24 h APF.
(C) Schematic representation of (D).
(D) MCFO was used to label an L4 neuron projecting dendrites that contact two L1 neurons, each within its target fascicles (dotted circles). Confocal sections (top panel) were used to create 3D renderings (bottom 3 panels).
Immunofluorescent signals from the MCFO epitopes were false colored in green and blue for L4s in panels B and D. L1 neurons in D are shown in red. R cell growth cones highlighted by anti-Chaoptin (Chp; red in B and grey in D). Other labeled cells are indicated (*), but were not reconstructed. Axons, Ax. Scale bar, 5 μm.
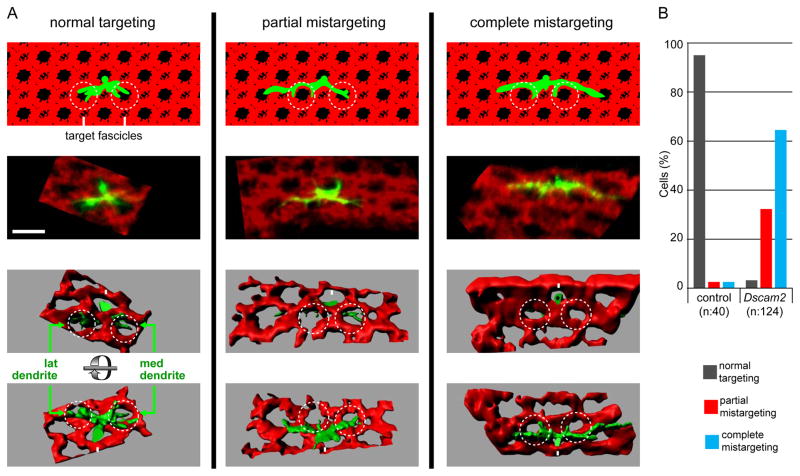
Dscam2 mutant dendrites analyzed by MARCM displayed dramatic morphological differences from wild type at 24 h APF (Figure 3A). Nascent mutant L4 dendrites made little (partial mistargeting) or no (complete mistargeting) contact with the target fascicles (Figure 3B). Instead, Dscam2 mutant L4 neurons extended their dendrites along the proximal surface of R cell growth cones reaching beyond the target fascicles along the dorsoventral axis. This over-extension placed these dendrites in close proximity to an additional lamina fascicle on either side of their normal targets. Thus, the adult morphology of mutant dendrites results from an early defect in targeting. This is followed by the formation of a secondary branch within the normal fascicle and its extension distally. The dendritic terminal also extends distally within the additional fascicle.
Figure 3. Dscam2 acts early in dendritic targeting of L4.
MARCM analysis of L4 neurons at 24 h APF
(A) Representative L4 neurons are shown for each phenotypic category. For each neuron schematic representations (top row of panels), confocal image (MIPs) and top and bottom view of 3D renderings as indicated. At this stage normal targeting is defined as L4 dendrites making contact with their target fascicles (dark hole within dotted circles. MARCM clones were labeled with GFP (green) and R cells with anti-Chp (red). L4 axons (Ax), lateral (lat) and medial (med) dendrites are indicated. Scale bar, 5 μm.
(B) Quantification of control and Dscam2 mutants.
In summary, L4 dendrites project to their target fascicles early in their development whereas Dscam2 mutant L4 dendrites extend along the dorsal-ventral axis in close contact with the lattice of R cell growth cones. To address how Dscam2 regulates the behavior of L4 dendrites, we determined both its expression pattern and its requirement in different cell types using genetic mosaic analyses.
Dscam2 is expressed in the developing lamina
To understand how Dscam2 mediates interactions between L4 dendrites and their environment, we first set out to assess its expression during the development of the visual system (Figure S2). We previously showed that Dscam2 immunoreactivity was associated with the developing lamina and medulla neuropils (Millard et al., 2007). Due to the density of processes of many different cell types within the neuropil, however, it was not possible to determine which cells express Dscam2. To identify Dscam2-expressing cells, and which isoforms they express, we modified the endogenous Dscam2 locus to generate splice traps as previously described (Figure S2A; Lah et al., 2014; Miura et al., 2013). When either isoform reporter is combined with nuclear envelope-targeted GFP (nuc-GFP) under the control of a LexA specific promoter (i.e. LexAop), the cells expressing the particular isoform were labeled. Using these reporters, expression in the retinal and lamina neurons was assessed from 24–48 h APF. In the lamina from 24 – 36 h APF, L1 and L4 neurons express Dscam2B, whereas L2, L3 and L5 exclusively express Dscam2A (Figure S2B). Between 36 and 42 h APF, however, L4 also begins to express Dscam2A. Dscam2 also displayed isoform-specific expression in R cells. Dscam2B is present in all R cell sub-types except for R4 (Figure S2C) while Dscam2A is not expressed in R cells at any stage (data not shown; Lah et al., 2014).
According to the isoform reporters, L4 neurons exclusively express Dscam2B at the time (24 h APF) the dendritic targeting phenotype is first observed. This is surprising as either Dscam2A or Dscam2B is sufficient for normal L4 dendritic patterning (see above). Several possible explanations could account for this inconsistency. First, the isoform reporters may not be accurately reflect Dscam2 protein expression. This is unlikely as knock-in protein tags recapitulate the patterns seen with the reporters (Figure S2D). Another possibility is that Dscam2Bstop mutant L4s may have an early dendritic targeting defect that is repaired at a later stage in development as Dscam2A levels rise. This is not the case, as dendritic development at 24 h APF in this mutant is indistinguishable from wild type (Figure S2E). Alternatively, the Dscam2Bstop allele may promote upregulation of splicing of the 10A exon, thereby compensating for the absence of Dscam2B. As the Dscam2A reporter shows no premature expression in the background of the Dscam2Bstop allele this is also unlikely (Figure S2F). Thus, we conclude that Dscam2A is expressed at 24 h APF at a level sufficient to support normal development, though below that required for detection using the splice trap.
In summary, developing L4 dendrites express both Dscam2 isoforms at 24 h APF. At this stage of development they interact with processes expressing Dscam2A (L2, L3, L4 and L5) and Dscam2B (L1, L4, and all R cell growth cones except R4). Neither Dscam2 isoform is expressed in glial cells. Thus Dscam2 isoforms exhibit a complex expression pattern in the lamina. To determine which cells guide L4 dendritic targeting, we turned to genetic mosaic analysis.
L4 dendrites use Dscam2 to adhere to lamina neurons in the target fascicle
We considered three mechanisms by which Dscam2 may mediate dendritic targeting. First, interactions between L4 dendrites converging onto the same target fascicle could repel each other and prevent further growth along the dorsoventral axis. This would be analogous to the role proposed for Dscam2 in L1 axonal tiling (Millard et al., 2007). Second, Dscam2 could mediate adhesion between L4 dendrites and one or more lamina neurons in the target fascicle. And third, L4 dendrites could be directed towards their target fascicles via repulsion from R cell growth cones.
Testing a Dscam2 requirement in surrounding lamina neurons
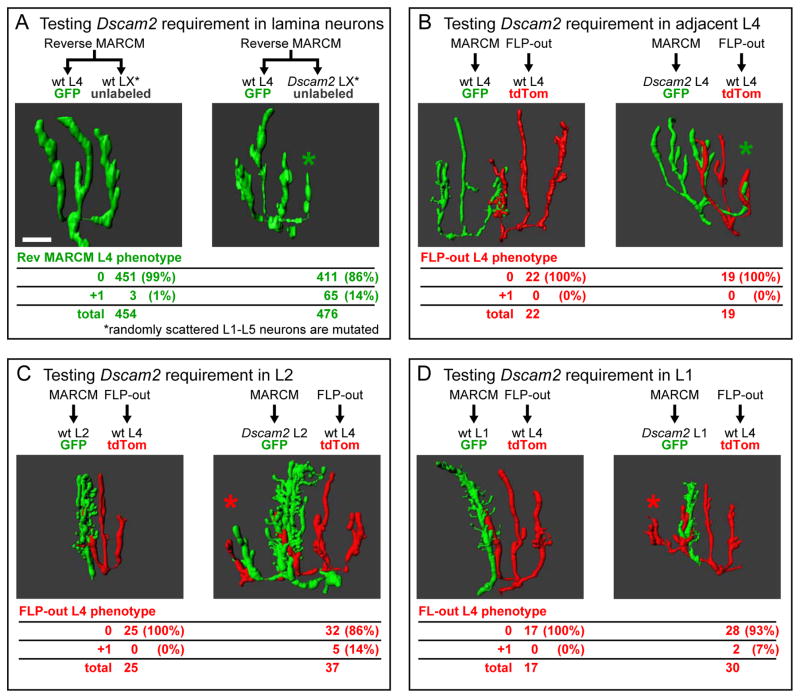
To test the first two possibilities we removed Dscam2 from surrounding lamina neurons using reverse MARCM (Lee et al., 2000). Here, mitotic recombination induced selectively in the lamina produces GFP-labeled wild-type L4 neurons in the background of unmarked homozygous Dscam2 lamina neurons (Figure S3A). To restrict the production of mutant neurons to the lamina, we used an enhancer to drive FLP recombinase specifically in lamina neuron precursor cells (Figure S3B). In reverse MARCM only a small fraction of labeled wild-type L4 neurons will have mutant neighboring lamina neurons due to the low activity of the FLP recombinase used. As a consequence, if interactions with these neurons were key to dendritic targeting, we would expect few of these wild type L4 neurons to exhibit an ectopic branch. In addition, most, if not all, phenotypes would be unilateral (i.e. +1 rather than +2). This is because each of L4’s two posterior dendrites interacts with a distinct groups of lamina neurons. The chance that both groups of lamina neurons would be mutated for Dscam2, and thereby result in +2 phenotype, is extremely low given the low frequency of clone production achieved using this protocol. Using this technique, 14% (65/476) of wild-type L4 neurons displayed an additional branch (Figure 4A) when generated in the background of Dscam2 mutant lamina neurons, compared to < 1% (3/454) of L4s generated in a wild-type control background. Furthermore, the L4s displayed dendritic phenotypes that were exclusively +1. Thus, L4 dendritic targeting relies on homophilic interactions between L4 dendrites and one or more lamina neurons.
Figure 4. L4 dendrites use Dscam2 to adhere to multiple lamina neurons in the target fascicle.
(A) Reverse MARCM analysis establishes a non-autonomous Dscam2 requirement in lamina neurons (see Figure S3). Representative 3D renderings of L4 neurons displaying wild-type (left panel) and +1 (right panel) dendritic patterning phenotypes. Quantification of these phenotypes, scored in the presence of unlabeled wild-type or unlabeled Dscam2 mutant lamina neurons, is shown in the table.
(B–D) DL-MARCM analyses reveals Dscam2 requirement in specific lamina neurons (see Figure S4). 3D renderings showing MARCM generated control (left panel) or Dscam2 mutant (right panel) lamina neurons (green) in contact with FLP-out labeled L4 neuron (red). Phenotypes of FLP-out L4s are quantified in tables. (B) FLP-out generated L4 neurons do not produce an ectopic branch when adjacent to a Dscam2 MARCM generated L4. (C,D) FLP-out L4s produce an ectopic +1 branch when targeting to a fascicle containing a MARCM generated Dscam2 L2 (C) or L1 (D). As previously described (Lah et al., 2014), we also observed L1 and L2 dendrites lacking Dscam2 occasionally project into adjacent cartridges (see example in C). In most cases, however, this ectopic branch is not correlated with a defect in L4 targeting (see Figure S5). Additional dendritic branches indicated (*). Scale bar, 5 μm.
Testing a Dscam2 requirement in neighboring L4 neurons
To identify which lamina neurons provide the Dscam2 signal to L4, we modified MARCM to independently label both the mutant and wild-type cells. We refer to this modification as dual labeled MARCM (DL-MARCM; Figure S4). First, we tested whether Dscam2 mediates interactions between adjacent L4 neurons. We labeled Dscam2 mutant L4 neurons in green (GFP) using MARCM, while labeling wild-type L4 neurons in red (tandem tomato; tdTom) in the same lamina using FLP-out mediated excision. These two classes of L4 neurons were labeled using orthogonal expression systems (i.e. GAL4/UAS and LexA/LexAop). As both labeling methods are stochastic in nature, pairs of wild type and mutant L4 neurons are infrequent (i.e. ~1 pair/10 laminas). In every pair (n=19), the targeting of wild-type L4 dendrites was normal (Figure 4B). Thus, L4 targeting defects do not result from a loss of repulsive interactions between L4 dendrites.
Testing a Dscam2 requirement in L1 or L2 neurons in the target fascicle
We next sought to address whether Dscam2 is required in L1 or L2 axons within the target fascicle using DL-MARCM with an L1-L2 marker. Abnormal targeting was observed in L4 dendrites encountering Dscam2 mutant L2 neurons (5/37; Figure 4C), or Dscam2 mutant L1 neurons (2/30; Figure 4D) in the target fascicle. By contrast, no targeting defects were seen in control experiments in which dendrites of wild type L4 neurons encountered labeled wild type L1(0/17) or L2 (0/25) neurons. The low penetrance of the effects on L4 dendritic targeting suggests that L1 and L2 function in a partially redundant manner in their interactions with L4. As L1 and L2 mutant cells were generated at only low frequency, we were not able to test this by assessing targeting of L4 dendrites into cartridges in which both L1 and L2 were mutant.
A possible caveat with the DL-MARCM experiments described above is that although mutant clones are generated in all five lamina neuron sub-types, we only label one (L4) or two (L1 and L2) subtypes at a time. Thus, it is possible that the non-cell autonomous effects we see on L4 dendrites are the result of multiple lamina neurons (e.g. any combination of L1-L5 neurons) being mutant for Dscam2 in their vicinity. Given, as indicated above, we never saw mutant L1 and L2 neurons within the same fascicle, this seems unlikely.
We conclude L4 targeting relies upon Dscam2-mediated interactions with multiple neurons in the target fascicle.
Testing a Dscam2 requirement in R cells
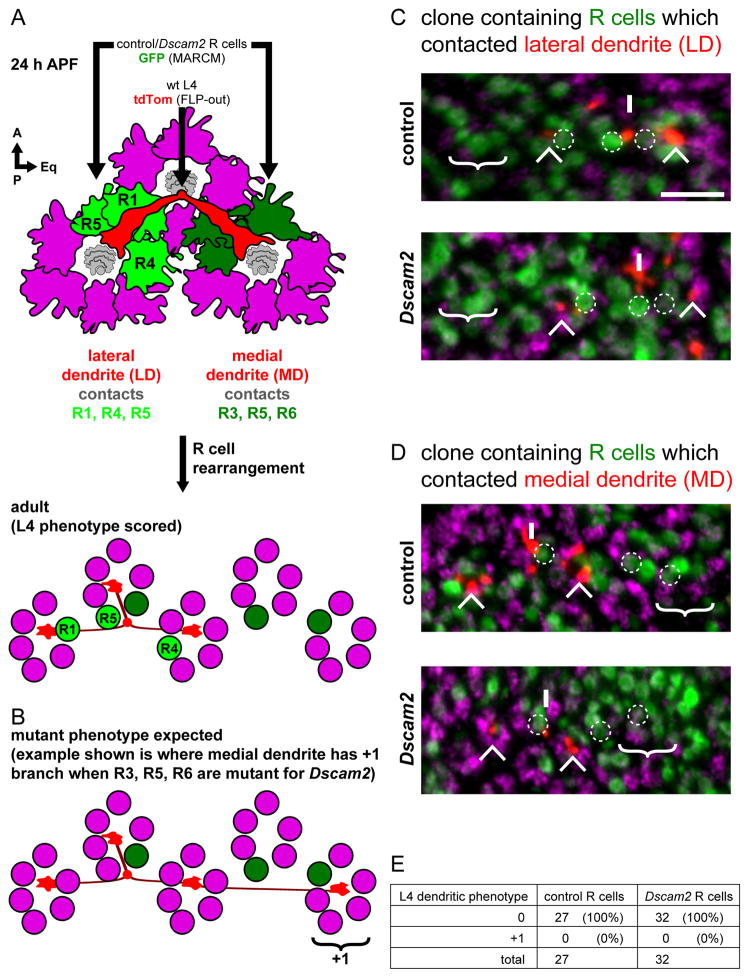
To test whether Dscam2 mediates interactions between L4 dendrites and R cell growth cones, we removed Dscam2 from R1-R6 while simultaneously assessing the effect on L4 targeting using DL-MARCM (Figure S6). As R1-R6 growth cones rearrange in a stereotyped fashion between 24 h APF and the adult, it was necessary to identify the position of different R1-R6 axon terminals in the adult and, from this, infer their positions at 24 h APF (Figure 5A, B). As the lateral L4 dendrite extends, it contacts the growth cones of R1, R4 and R5. The medial L4 dendrite contacts R3, R5 and R6 growth cones. Both dendrites target normally even in mosaics in which the three R cells they contact lacked Dscam2 (n=32; Figure 5C-E). Thus, L4 dendritic targeting does not rely upon Dscam2-mediated interactions with R cell growth cones.
Figure 5. Dscam2 directs L4 dendritic targeting independent of R cells.
(A) Schematic of L4 neuron dendrites (red) and surrounding R cell growth cones (magenta) at 24 h APF. Each dendrite projects beneath two groups of R cell growth cones; the lateral dendrite (LD) contacts R1, R4 and R5 (light green) while the medial dendrite (MD) contacts R3, R5 and R6 (dark green). After R cell rearrangement (see text), the R cells end up in different cartridges relative to the L4 neuron in the adult. This is the stage at which we could score for L4 phenotypes (see C and D).
(B) Schematic showing the L4 dendritic phenotype expected should Dscam2 in R cells be required for targeting. Here the medial dendrite produces a +1 branch after having contacted R3, R5 and R6 which are mutant from Dscam2.
(C, D) DL-MARCM was used to label L4 neurons (red; myr-tdTom) in the background of MARCM generated clones of R cells (green; GFP) (see Figure S6). (C) Example of R cells contacted by the lateral dendrite (i.e. R1, R4 and R5; dotted circles) were in a control (top panel) or Dscam2 (bottom panel) clone. In both control and Dscam2 experiments, no ectopic branching was observed; the cartridge where an ectopic branch would be expected is devoid of any L4 dendrites (‘no +1’). (D) Clones of R cells that contacted the medial dendrite (i.e. R3, R5 and R6; dotted circles) are identified in using the same experimental approach as in (C). Again, in no case was an ectopic branch observed in the expected cartridge (‘no +1’). L4 axons (Ax), lateral (LD) and medial dendrites (MD) are indicated. Processes from non-relevant L4s are also indicated (*). Anti-Hiw (magenta) labels all cartridges. Scale bar, 5 μm. (E) Quantification of L4 dendrites having a 0 or +1 phenotype when in the background of control or Dscam2 mutant R cells.
Summary
These genetic mosaic experiments establish that Dscam2 mediates interactions between L4 dendrites and multiple neurons (including L1 and L2) in the target fascicle. Furthermore, there is no evidence that Dscam2 mediates interactions between neighboring L4s or between L4 dendrites and R cell growth cones. Based on these findings and our developmental analysis (see above), we propose that Dscam2 mediates homophilic adhesive interactions between L4 dendrites and multiple axons in the target fascicle acting in a partially redundant fashion. We consider this redundancy and the pattern of Dscam2 isoform expression in the model presented in Figure S7G.
Dscam4 is required for L4 dendritic patterning
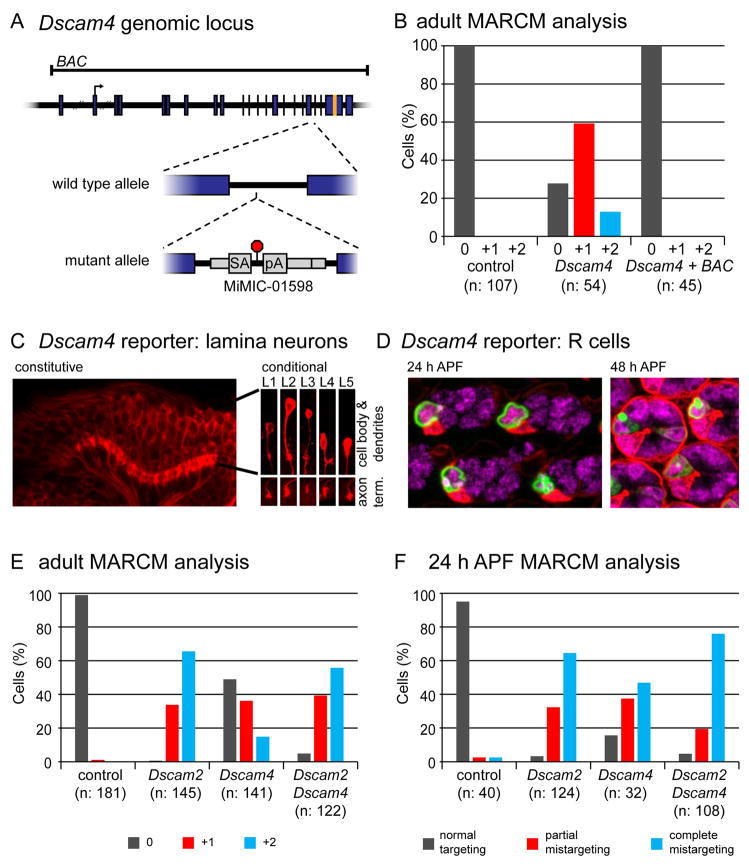
To identify other genes involved in L4 dendritic patterning, we carried out an unbiased genetic mosaic screen using small chromosomal deletions, or deficiencies. We identified one deficiency with a dendritic branch phenotype similar to Dscam2 mutants. This deficiency uncovers 54 genes, including Dscam4. To test if loss of Dscam4 is responsible for this phenotype, we identified a Minos transposon, Mi01598 (Venken et al., 2011) inserted between two exons encoding part of the ectodomain of Dscam4, which is predicted to arrest both transcription and translation (Figure 6A). Using primers downstream of the insertion site, Dscam4 mRNA was not detected by qPCR in homozygous animals (Figure S7A). An antibody produced against the cytoplasmic domain detected a signal in the neuropils of the developing visual system in wild type (Figure S7B) that was lost in animals homozygous for Mi01598 and restored in the presence of a BAC containing the Dscam4 locus (Figure S7C). Thus, Mi01598 is a strong loss-of-function, if not a null, mutation in Dscam4.
Figure 6. Dscam4 acts in the same pathway as Dscam2 to direct L4 dendritic targeting.
(A) Diagram of Dscam4 locus and mutant allele. Dscam4 allele is a MiMIC insertion bearing a splice acceptor (SA), followed by translational (red octagon) and transcriptional stops (pA). Translation start site and sequences encoding transmembrane domain are denoted with an arrow and orange band, respectively. Genomic sequence contained within a BAC is also indicated.
(B) MARCM analysis of Dscam4 mutant L4 neurons showing that the BAC rescues the dendritic patterning phenotype.
(C) Constitutive Dscam4 reporter (left panel) driving myr-tdTom (red) at 24 h APF. Scale bar, 15 μm. Dscam4 expression in each lamina neuron subtype was confirmed with a conditional reporter (right panels) with L1–L5 identified by their unique cell body positions, dendritic and axonal morphologies. This conditional reporter is only expressed in lamina neurons where the presence of a lamina-specific recombinase induces excision of an FRT-cassette, which otherwise prevents its expression. See Figure S7F for reporters. Scale bar, 5 μm.
(D) Constitutive Dscam4 reporter driving nuc-GFP reveals expression in R3 cells in the retina at 24 h APF (left panel). At 48 h APF, sporadic expression in R7 cells is seen in addition to R3 (right panel). mδ-GAL4 driving myr-tdTom and anti-ELAV staining are in red and magenta, respectively. Scale bar, 5 μm.
(E,F) MARCM analysis of Dscam4 in adults (E) and at 24 h APF (F). Data for control and Dscam2 samples are the same as in Figure 1G (for adults) and Figure 3B (for 24 h APF) as these were collected at the same time.
L4 neurons homozygous for this mutation generated by MARCM display the same dendritic patterning phenotype as seen in Dscam2 mutants (see below; Figure 6B). That these defects reflect loss of Dscam4 rather than other mutations on this chromosome was established by rescuing the phenotype with the Dscam4 BAC. By contrast, mutations in neither Dscam1 (Figure S7D; as described above) nor Dscam3 alter L4 dendritic patterning (Figure S7E). Thus, a subset of Dscam paralogs, Dscam2 and Dscam4, are required for L4 dendritic targeting.
Dscam4 is expressed in all lamina neurons
Dscam4 protein is highly localized to the neuropil (Figure S7B). The density of processes within the neuropil, however, precluded identifying which cells express Dscam4, as was also the case for Dscam2. To circumvent this problem, we modified the endogenous locus to generate, a “constitutive” and a “conditional” version of a transcriptional reporter (Figure S7F). The constitutive version reveals broad expression in the visual system consistent with the antibody staining (Figure 6C, left panel). The conditional version is only active in lamina neurons as its expression is conditional on the presence of a lamina specific recombinase (see Figure S7F). This enabled us to analyze expression in scattered lamina neurons, thereby allowing us to identify each cell type based on its morphology. In this way, we demonstrated that all five lamina neurons, L1–L5, express Dscam4 at 24 h APF (Figure 6C, right panel). By contrast, in the developing eye, Dscam4 is expressed only in R3 and sporadically in R7 (Figure 6D).
Dscam2 and Dscam4 act in the same pathway to regulate dendritic targeting
To understand the relationship between Dscam2 and Dscam4 in mediating L4 dendritic patterning, we compared the phenotypes of single and double mutant cells generated by MARCM. In both adults (Figure 6E) and at 24 h APF (Figure 6F) Dscam4 mutant L4 neurons displayed similar dendritic phenotypes to those of Dscam2, but with lower penetrance. Dscam2 Dscam4 double mutants, however, exhibited dendritic phenotypes with similar penetrance to those of Dscam2 single mutants. In adults, double mutant L4 neurons displayed the +1 and +2 ectopic branch phenotypes at frequencies of 39% and 56% (n=122), respectively. This was similar to the frequencies for these two phenotypes in Dscam2 single mutants (34% and 66% (n=145)). Likewise, the penetrance of the developmental phenotypes of the double and Dscam2 single mutants was similar; at 24 h APF, 95% (n=108) of Dscam2 Dscam4 double mutant L4 neurons either partially or completely mistargeted their dendrites compared with 97% (n=124) of Dscam2 single mutants. That the penetrance of single and double mutants was similar argues that these paralogs function in the same pathway mediating dendritic targeting.
Live imaging reveals that Dscam2 and Dscam4 anchor L4 dendrites to their target fascicles
We used MARCM and live imaging in intact pupae to gain further insight into how Dscam2 and Dscam4 regulate dendritic targeting. In the course of constructing different genotypes for these experiments, we found that cells homozygous for the UAS-myr-GFP insertion at the attP2 site on chromosome 3L generated via MARCM produced a fluorescence signal some 20X greater than cells carrying a single copy elsewhere (see Table S1). This allowed us to visualize L4 dendrites much earlier (i.e. from 16 h APF) than was possible in fixed preparations.
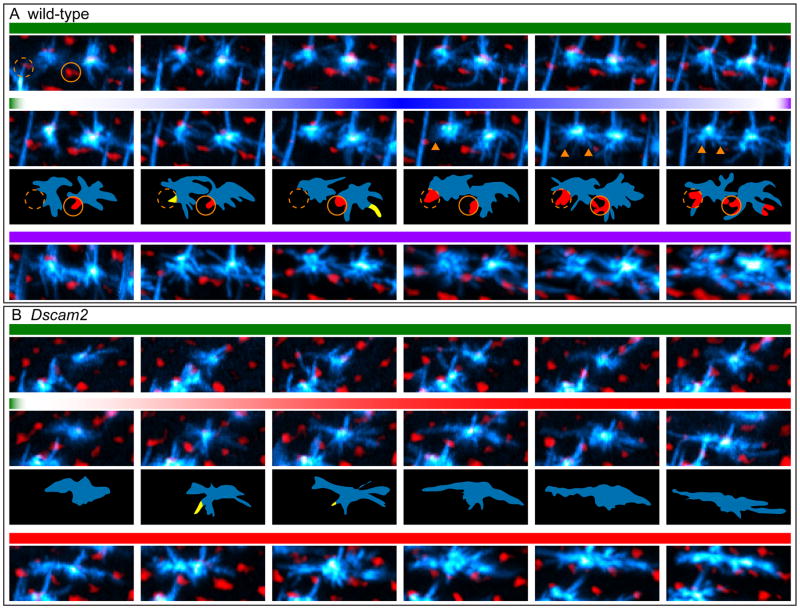
Dendritic filopodia of wild-type L4 neurons extend in an apparently random fashion prior to 17 h APF (Figure 7A; Movie S1). In this “Exploratory Phase” these dynamic projections occasionally make contact with the target fascicles only to quickly retract. Despite having no clear directional bias, the fascicles contacted by the dendritic filopodia during this exploratory phase are almost always the target fascicles. This is likely because the positioning of L4 axons at the posterior side of its home fascicle gives it greatest access to these two fascicles (Schwabe et al., 2014). Between 17.5 and 18.5 h APF, projections contacting the target fascicles become anchored to their anterior side. Between 18.5 and 26 h APF, these bound projections are progressively joined by more filopodia, which continue to contact a broader surface of the target fascicle (Consolidation Phase). These form the dendritic growth cones associated with the target fascicle seen in images from fixed samples at 24 h APF.
Figure 7. Live imaging of L4 neurons.
Frames (MIPs) from live imaging of MARCM-generated L4 neurons (cyan) at the indicated time points from 16–26 h APF (see Movies S1 and S2). In Movies S1 and S2 (left panels) myr-tdTom driven by a GMR promoter highlights the R cell lattice in red. Here, however, this red signal was inverted such that R cell lattice appears black and the holes in the lattice, i.e. positions of the lamina fascicles, are in red (see also Movies S1 and S2, right panels). This was done to allow for easier visualization of the thin and numerous filopodia in L4s. L4 axons (Ax) are also indicated. Scale bar, 5 μm.
(A) Two neighboring wild-type L4 neurons undergoing three phases in their dendritic targeting:1. Exploration: In this early phase (~16 hr to 17.3 h APF) dynamic filopodia branch out from the axon in all directions occasionally contacting their target fascicles. Target fascicles for left and right L4 neurons are indicated by orange and white dotted circles, respectively; 2. Anchoring: While some filopodia continue to make transient contacts with the target fascicles between 17.5 and 18.5 h APF, (yellow regions in schematics below), others become progressively anchored to the target fascicles (red regions in schematic; arrowheads in MIPs); 3. Consolidation: Filopodia continue to make stable contacts with the target fascicles.
(B) Dscam2 mutant L4 neurons have an ‘exploratory phase’ indistinguishable from wild type but fail to anchor their dendrites to their target fascicles between 17.5 and 18.5 h APF. Instead, their dendrites extend along the D-V axis in tight apposition to the R cell lattice (black).
In Dscam2 mutant neurons, the early exploratory phase of dendritic filopodia appears similar to wild type (Figure 7B; Movie S2). By contrast to wild type, however, anchoring is not observed. Instead, mutant dendrites exhibit striking polarization towards the dorsal and ventral poles. As seen in images from our fixed samples, these Dscam2 L4 dendrites extend in tight apposition to the R cell growth cones. Similar targeting defects were also observed in live imaging of Dscam4 and Dscam2 Dscam4 mutant L4 neurons (data not shown).
In summary, live imaging of wild-type and mutant L4 neurons indicate that Dscam2 and Dscam4 are required to anchor dendritic filopodia to their target fascicles. We conclude that these proteins act together to promote adhesion between L4 dendrites and their targets.
Discussion
In this paper we show that Dscam2 and Dscam4 act together to regulate dendritic targeting. The genetic data presented here, coupled with previous biochemical studies (Flanagan, 2007; Millard et al., 2007), are consistent with Dscam2 using homophilic interactions to adhere L4 dendrites to multiple lamina neurons. In all other contexts examined previously, Dscam2 promotes repulsion (Lah et al., 2014; Millard et al., 2007, 2010). To further understand the adhesive role of Dscam2 in circuit assembly, we carried out a genetic screen to identify other genes with a similar phenotype. This led to the identification of Dscam4 mutants with an L4 targeting phenotype indistinguishable from that of Dscam2. Dscam2 and Dscam4 promote adhesion between L4 dendritic filopodia and their target fascicles, as revealed through live imaging. In this way, these proteins regulate the tiling of L4 dendritic processes across the lamina neuropil. Thus, Dscam family members act in different combinations through either adhesion or repulsion to pattern neural circuits.
Dscam2 promotes tiling through heterotypic interactions
Previously, we reported that Dscam2 regulates tiling of the axon terminals of L1 neurons in the medulla (Millard et al., 2007). We proposed that this arrangement of processes is a consequence of homotypic interactions (i.e. interactions between cells of the same type) between L1 axon terminals in adjacent columns. This model was based on three observations. First, L1 axon terminals initially expand to contact those of their immediate neighbors and then retract. Second, the axon terminals of Dscam2 mutant L1 neurons remain expanded, invading the territory of their neighbors, in genetically mosaic animals. And third, wild-type L1s also exhibit the same defect when confronted with Dscam2 mutant lamina neurons in adjacent columns.
In this paper, we uncovered a role for Dscam2 in regulating L4 dendritic tiling that occurs in a mechanistically different fashion. Our developmental and reverse MARCM experiments suggested that Dscam2 mediates L4 tiling in a manner analogous to its role in L1 axons, that is via Dscam2-mediated homotypic interactions between L4 dendrites innervating the same cartridge. Additional genetic mosaic studies, however, ruled this out. By modifying MARCM (i.e. DL-MARCM) we were able to assess the targeting of wild type L4 dendrites encountering dendrites from neighboring L4 mutant neurons. In all cases, these wild type dendrites targeted normally. Thus, while L4 tiling relies upon Dscam2-mediated homophilic recognition, it does not result solely from homotypic interactions between neighboring L4 dendrites.
Our developmental analyses raised the alternative possibility that Dscam2 is used by L4 dendrites to promote interactions with the target fascicle. Indeed, we observed targeting defects in L4 dendrites innervating cartridges with mutant L1 or L2 neurons using DL-MARCM. Thus, in this context, tiling arises from heterotypic interactions. As the penetrance of this cell non-autonomous phenotype was weaker than removing Dscam2 from L4, we propose that L1 and L2 act redundantly, perhaps with other lamina neurons (e.g. L3 and L5 which were not tested), to promote Dscam2-dependent recognition between L4 dendrites and the target fascicle. This is consistent with the expression of Dscam2B and Dscam2A in L1 and L2 neurons, respectively, and the redundant role for these isoforms in L4 uncovered through the mosaic analysis of isoform-specific knock-in alleles (Figure S7G). Thus, L4 tiling arises through heterotypic interactions via Dscam2-mediated homophilic recognition. This is consistent with studies in both vertebrates and invertebrates in which cell ablation experiments indicate that, in some contexts, tiling may arise through heterotypic interactions (Gallegos and Bargmann, 2004; Lin et al., 2004).
As previously reported (Lah et al., 2014), removal of Dscam2 from either L1 or L2 results in ectopic branching of these neurons. This observation raised the possibility that wild type L4s follow these branches in a Dscam2-independent fashion and that this underlies the Dscam2 non-autonomous effect on wild type L4s. Although we cannot rule this out, we favor the interpretation that these data reflect a Dscam2-dependent effect independent of the ectopic L1 or L2 phenotype for two reasons. First, of the two instances in which a wild-type L4 produced an extra branch upon encountering a Dscam2 mutant L1, one L1 did not form an ectopic branch (see Figure S5). And second, in most cases where either L1 (2/3) or L2 (18/23) displayed an extra branch phenotype, the associated L4 did not (see Figure S5).
The functional significance of this tiling and its disruption in Dscam2 mutants remain open questions. A recent behavioral study (Tuthill et al., 2013) has suggested that L4 functions in responding to progressive, i.e. front-to-back, as opposed to regressive motion. This response would be consistent with L4’s asymmetric dendritic arrangement, having one dendrite projecting anteriorly and two posteriorly. While this is an attractive model, another study using slightly different methodology suggests no role for L4 in directional selectivity (Silies et al., 2013). Critically assessing the importance of the spatial arrangement of L4 dendrites on behavior will require selective removal of Dscam2 from all L4 neurons in an otherwise wild type background.
Dscam2 and Dscam4 act together to promote homophilic adhesive interactions with the target
Dscam paralogs promote cell adhesion in vitro (Flanagan, 2007; Matthews et al., 2007; Millard et al., 2007). However, in all cases examined previously, genetic and phenotypic analyses of Dscam1 and Dscam2 mutations are consistent with a repulsive function for these proteins upon homophilic binding in vivo (Hughes et al., 2007; Matthews et al., 2007; Millard et al., 2007, 2010; Soba et al., 2007; Wu et al., 2012; Zhan et al., 2004; Zhu et al., 2006; Similar studies for Dscam3 and Dscam4 have not been done.). In addition, gain-of-function studies in Drosophila support this view. Targeted expression of single Dscam1 isoforms promotes dendritic repulsion between class I and class III da sensory neurons in the developing fly embryo (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). This repulsive interaction is converted into adhesion by removing the cytoplasmic domain (Matthews et al., 2007) or selectively removing specific motifs (Kim and S.L.Z., unpublished observations) underscoring the role of cytoplasmic signaling in converting Dscam1-mediated homophilic binding to repulsion. And finally, as recently shown by Millard and colleagues (Lah et al., 2014), forced expression of either Dscam2A or Dscam2B isoforms in closely apposed neurons in the visual system promotes repulsion between them.
By contrast, several observations argue that Dscam2 and Dscam4 promote adhesion between L4 dendrites and their target fascicles. In fixed samples, wild-type L4 dendrites target to lamina axon fascicles while Dscam2, Dscam4 or Dscam2/4 double mutant dendrites extend laterally along the lattice of R cell growth cones surrounding the target fascicles. Indeed, using MCFO we observed extensive intermingling of L4 dendrites and L1 processes in the target fascicle. In live imaging, L4 dendritic processes become progressively anchored to their target fascicles. By contrast, in mutants the interactions between L4 dendrites and the target fascicles were transient, while interactions with R cell growth cones were sustained. This tight association in mutants could reflect a loss of repulsive interactions between L4 dendrites and R cells. Genetic mosaic analysis, however, indicate that this is highly unlikely. Together, these data support a model in which Dscam2 and Dscam4 act together to promote adhesion between L4 dendritic growth cones and their lamina targets (see Figure S7G). In the absence of this Dscam2/Dscam4-mediated adhesion, we suggest an adhesive interaction between L4 dendrites and the surrounding R cell growth cones is unmasked. Indeed, transcript profiling of R cells and L4 neurons during development suggest that they both express several recognition molecules that are candidates for mediating this interaction (Tan et al., in press, McEwen et al., unpublished results).
Although we favor a model in which Dscam2 and Dscam4 mediate selective adhesive interactions with the lamina fascicle, our data do not rule out a model in which Dscam2 and Dscam4 regulate L4 targeting in a permissive fashion. For instance, Dscam2/4 may restrict the exploratory phase to fascicles in the immediate vicinity through adhesive interactions with both lamina axons and R cell growth cones. Indeed, in our live imaging mutant L4 dendrites extend to additional fascicles on either side of their normal targets at a very early stage of filopodial exploration. In this model, another cell recognition molecule, acting in an instructive way, would promote association of L4 dendrites with the lamina axon fascicle. Characterization of other genes acting in this process and higher resolution live imaging may allow us to critically distinguish between an instructive and a permissive role for Dscam2 and Dscam4 in L4 dendritic targeting.
Dscam proteins use diverse molecular strategies to regulate circuit assembly
It is becoming increasingly clear that Drosophila Dscams regulate dendritic patterning in different ways. Dscam1 is alternatively spliced to give rise to greater than 19,000 ectodomain isoforms (Schmucker et al., 2000), with greater than 18,000 exhibiting homophilic binding specificity (Wojtowicz et al., 2004, 2007). This property, coupled with probabilistic expression endows each neuron with a unique identity (Miura et al., 2013) and allows for discrimination between self and non-self (i.e. self-avoidance). Dscam2, on the other hand, is alternatively spliced to give rise to only two isoforms. These act in combination with Dscam1 isoforms to regulate the appropriate association of L1 and L2 dendrites at multiple contact synapses (Millard et al., 2010). Here, L1 and L2 express different Dscam2 isoforms in a mutually exclusive fashion (this study; Lah et al., 2014). It is thought that the unique identity acquired by the dendrites of these cells, through alternative splicing of both Dscam1 and Dscam2, allows them to appropriately discriminate between self and non-self at multiple contact synapses. In these contexts, Dscam proteins promote repulsion. As we have shown here, Dscam2 in combination with Dscam4 promotes adhesive interactions during dendritic targeting. This may reflect a unique property of a Dscam2/Dscam4 protein complex, differences in output of different spliced variants of the cytoplasmic domains, or differences in levels of expression. Precedents for each of these potential mechanisms have been reported in other intercellular signaling proteins (Holmberg et al., 2000; Hong et al., 1999; Matsuoka et al., 2005). Additional experiments will be needed to distinguish between these different possibilities.
In summary, we have shown that two different combinations of Dscam proteins regulate the patterning of different dendritic elements in the developing lamina in diverse ways. As all four Dscam proteins are broadly expressed within the developing lamina, distinct paralogs, and isoforms of them, either alone or in combination may pattern dendrites in different ways. Furthermore, as Dscam proteins also interact with other proteins in both cis and trans (Andrews et al., 2008; Dascenco et al., 2015; Liu et al., 2009; Ly et al., 2008), they have the potential to pattern circuits in many different ways.
Experimental Procedures
Fly Stocks
Complete genotypes used in each experiment are detailed in Table S1. Flies were reared and staged at 25°C using standard protocols.
The following fly lines were used: Dscam2null-1, Dac-FLP (X), Dac-FLP20; Dscam123; Dscam4Mi01598; ap-GAL4(md544), tub-GAL80 (2R, 3L, 3R), UAS-CD8-GFP, Df(3L)Exel6112, Df(3R)BSC743, 10xUAS-myr-GFP (attP2; attP40), 10xUAS-myr-tdTom (attP2), GMR-GAL4, A5C-FSF-GAL4, L1-L2-GAL4, svp-GAL4, 27G05-FLP1 (attP8, attP18), 27G05-FLP2 (attP18), L4(31C06)-GAL4, L4(31C06)-LexA, 9B08-GAL4, A5C-GAL80, pJFRC201 pJFRC240, LexAop-nuc-GFP, mδ-GAL4, ey3.5FLPG5D, LexAop-myr-tdTom (attP2), LexAop-FSF-myr-tdTom, GMR-myr-tdTom, Dscam2Astop, Dscam2Bstop, Dscam2A+Bstop, Dscam2A-LexA, Dscam2B-LexA, Dscam2 Protein Tag, Dscam4-LexA (constitutive and conditional). Information on the source and construction (where applicable) of these lines is available in Supplemental Experimental Procedures.
Mosaic Analysis
Single cell mosaic analyses were done with genotypes listed in Table S1. Mitotic clones in lamina neurons were generated using either of two different genomic fragments, 27G05 (Pecot et al., 2013) or Dac (Millard et al., 2007), driving FLP1 or FLP2 recombinase. L4 neurons were labeled with either ap-GAL4 or L4(31C06)-GAL4. R cell mitotic clones were generated with ey3.5FLPG5D and labeled with GMR-GAL4. Sporadic FLP-out L4 clones were generated with Dac-FLP and LexAop-FSF-myr-tdTom. (Further details provided in Supplemental Experimental Procedures)
Multicolor Flip Out (MCFO) labeling
MCFO (Nern et al., 2015) was performed L1 and L4 specific markers and 27G05-FLP2 to stochastically label these neurons in seven different fluorescent color combinations. (Further details provided in Supplemental Experimental Procedures.)
Live Imaging
Image stacks were collected over ~10 hour of development using a custom built two photon microscope. Maximum intensity projections restricted to the lamina plexus were registered such that L4 neurons were aligned between stacks. (Further details provided in Supplemental Experimental Procedures.)
Immunostaining
Immunostaining was performed as described previously (Nern et al., 2008) with minor modification. For further details on this and the generation of the Dscam4 antibody see Supplemental Experimental Procedures.
Quantitative PCR (qPCR)
qPCR was performed on RNA extracted from adult flies using primers downstream of the Mi01598 insertion site. (Further details provided in Supplemental Experimental Procedures.)
Image Analysis
Images were acquired using Zeiss LSM 780 confocal microscope and a 63×/1.4 NA oil objective. Images were cropped, rotated and adjusted linearly for brightness and contrast using Photoshop (Adobe) or ImageJ (NIH).
3D Renderings of Confocal Data
Imaris (Bitplane) was used to transform stacks of 2D confocal images into 3D renderings. This was done using a combination of the FilamentTracer tool in AutoPath mode to trace the skeletal outline of neurites, followed the Surface creation tool to render the cellular outline in 3D.
Supplementary Material
Acknowledgments
We thank Dorian Gunning for producing protein used as antigen in generating the Dscam4 antibody. We thank Aljoscha Nern, Barret Pfeiffer, Gerald Rubin, Gilbert Henry, Tom Clandinin for sharing fly lines. We thank Michael Sawaya for advice in making the Dscam2 protein tag. We thank members of the S.L.Z. and S.S.M. labs for helpful comments on the manuscript. This work was supported by fellowships from the Human Frontiers Science Program to W.T., NINDS to S.X., Damon Runyon Cancer Research Foundation to O.A., Helen Hay Whitney Foundation to C.H.Y. S.S.M. was supported by a project grant from NHMRC (APP1021006) to S.S.M. S.L.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews GL, Tanglao S, Farmer WT, Morin S, Brotman S, Berberoglu MA, Price H, Fernandez GC, Mastick GS, Charron F, et al. Dscam guides embryonic axons by Netrin-dependent and -independent functions. Development. 2008;135:3839–3848. doi: 10.1242/dev.023739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron. 2008;58:26–33. doi: 10.1016/j.neuron.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140:3297–3302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascenco D, Erfurth ML, Izadifar A, Song M, Sachse S, Bortnick R, Urwyler O, Petrovic M, Ayaz D, He H, et al. Slit and Receptor Tyrosine Phosphatase 69D Confer Spatial Specificity to Axon Branching via Dscam1. Cell. 2015;162:1140–1154. doi: 10.1016/j.cell.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman GM. Cell adhesion molecules. Science. 1983;219:450–457. doi: 10.1126/science.6823544. [DOI] [PubMed] [Google Scholar]
- Edwards TN, Nuschke AC, Nern A, Meinertzhagen IA. Organization and metamorphosis of glia in the Drosophila visual system. J Comp Neurol. 2012;520:2067–2085. doi: 10.1002/cne.23071. [DOI] [PubMed] [Google Scholar]
- Flanagan JJ. Thesis. University of California; Los Angeles: 2007. Binding specificities of the Dscam family of axon guidance receptors. [Google Scholar]
- Foty RA, Steinberg MS. Differential adhesion in model systems: Differential adhesion in model systems. Wiley Interdiscip Rev Dev Biol. 2013;2:631–645. doi: 10.1002/wdev.104. [DOI] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Cadherins in Brain Morphogenesis and Wiring. Physiol Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisén J. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature. 2000;408:203–206. doi: 10.1038/35041577. [DOI] [PubMed] [Google Scholar]
- Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Bäumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam Interactions Control Complex Dendrite Morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah GJ, Li JSS, Millard SS. Cell-Specific Alternative Splicing of Drosophila Dscam2 Is Crucial for Proper Neuronal Wiring. Neuron. 2014;83:1376–1388. doi: 10.1016/j.neuron.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, Hiesinger PR. The Developmental Rules of Neural Superposition in Drosophila. Cell. 2015;162:120–133. doi: 10.1016/j.cell.2015.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM Is a Netrin Receptor that Collaborates with DCC in Mediating Turning Responses to Netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Matsuoka H, Obama H, Kelly ML, Matsui T, Nakamoto M. Biphasic functions of the kinase-defective Ephb6 receptor in cell adhesion and migration. J Biol Chem. 2005;280:29355–29363. doi: 10.1074/jbc.M500010200. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite Self-Avoidance Is Controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hansen TE. In The Development of Drosophila Melanogastor. CHCL Press; 1993. The development of the optic lobe; pp. 1363–1491. [Google Scholar]
- Meinertzhagen IA, O’Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Sorra KE. Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog Brain Res. 2001;131:53–69. doi: 10.1016/s0079-6123(01)31007-5. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Piper ST, Sun XJ, Fröhlich A. Neurite morphogenesis of identified visual interneurons and its relationship to photoreceptor synaptogenesis in the flies, Musca domestica and Drosophila melanogaster. Eur J Neurosci. 2000;12:1342–1356. doi: 10.1046/j.1460-9568.2000.00033.x. [DOI] [PubMed] [Google Scholar]
- Millard SS, Flanagan JJ, Pappu KS, Wu W, Zipursky SL. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature. 2007;447:720–724. doi: 10.1038/nature05855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA. Drosophila Dscam Proteins Regulate Postsynaptic Specificity at Multiple-Contact Synapses. Neuron. 2010;67:761–768. doi: 10.1016/j.neuron.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell. 2013;155:1166–1177. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL. Local N-Cadherin Interactions Mediate Distinct Steps in the Targeting of Lamina Neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci. 2015;112:E2967–E2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple Interactions Control Synaptic Layer Specificity in the Drosophila Visual System. Neuron. 2013;77:299–310. doi: 10.1016/j.neuron.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Alba M, Vitaladevuni SN, Mishchenko Y, Lu Z, Takemura S, Scheffer L, Meinertzhagen IA, Chklovskii DB, de Polavieja GG. Wiring Economy and Volume Exclusion Determine Neuronal Placement in the Drosophila Brain. Curr Biol. 2011;21:2000–2005. doi: 10.1016/j.cub.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam Is an Axon Guidance Receptor Exhibiting Extraordinary Molecular Diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Neuert H, Clandinin TR. A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell. 2013;154:351–364. doi: 10.1016/j.cell.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T, Borycz JA, Meinertzhagen IA, Clandinin TR. Differential adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr Biol CB. 2014;24:1304–1313. doi: 10.1016/j.cub.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. Modular Use of Peripheral Input Channels Tunes Motion-Detecting Circuitry. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila Sensory Neurons Require Dscam for Dendritic Self-Avoidance and Proper Dendritic Field Organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura S, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, Reiser MB. Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron. 2013;79:128–140. doi: 10.1016/j.neuron.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A Vast Repertoire of Dscam Binding Specificities Arises from Modular Interactions of Variable Ig Domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Ahlsen G, Baker D, Shapiro L, Zipursky SL. Complementary Chimeric Isoforms Reveal Dscam1 Binding Specificity In Vivo. Neuron. 2012;74:261–268. doi: 10.1016/j.neuron.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Grueber WB. The molecular basis of self-avoidance. Annu Rev Neurosci. 2013;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.