Abstract
Better characterization of the preclinical phase of Alzheimer's disease (AD) is needed in order to develop effective interventions. Neuropathological changes in AD, including neuronal loss and the formation of proteinaceous deposits, begin up to 20 years before the onset of clinical symptoms. As such, the emergence of cognitive impairment should not be the sole basis used to diagnose AD nor to evaluate individuals for enrollment in clinical trials for preventative AD treatments. Instead, early preclinical biomarkers of disease and genetic risk should be used to determine most likely prognosis and enroll individuals in appropriate clinical trials. Neuroimaging-based biomarkers and genetic analysis together present a powerful system for classifying preclinical pathology in patients. Disease modifying interventions are more likely to produce positive outcomes when administered early in the course of AD. In this review, we examine the utility of the neuroimaging genetics field as it applies to AD and early detection during the preclinical phase. Neuroimaging studies focused on single genetic risk factors are summarized. However, we particularly focus on the recent increased interest in polygenic methods and discuss the benefits and disadvantages of these approaches. We discuss challenges in the neuroimaging genetics field, including limitations of statistical power arising from small effect sizes and the over-use of cross-sectional designs. Despite the limitations, neuroimaging genetics has already begun to influence clinical trial design and will play a major role in the prevention of AD.
Keywords: neuroimaging, genetics, Alzheimer's disease, polygenic risk score, preclinical, clinical trials
Introduction
A long prodrome precedes the emergence of the clinical symptoms of Alzheimer's disease (AD) (1–3). Increasingly, the time between the first silent pathological changes in the brain and the earliest stages of cognitive impairment is understood to be a critical window during which prevention and treatment strategies may be most effective (4). This preclinical phase of AD pathogenesis that occurs before clinical symptoms emerge is not well characterized. By definition, individuals with preclinical AD are not aware that they are affected by any neurological pathology, nor are their deficits detectable with cognitive testing. Preclinical AD is distinct from mild cognitive impairment (MCI), which is characterized by subtle cognitive decline and can sometimes progress to a clinical diagnosis of AD (5; 6). In the absence of detectable cognitive decline, we have access to a limited set of research tools to explore preclinical AD in humans. These include neuroimaging, genetic testing, and biochemical assays of the blood and cerebrospinal fluid (CSF). Thus, neuroimaging genetics research is poised to play a critical role in improving the characterization of the earliest phases of AD pathophysiology. In the following sections, we will discuss the important role of neuroimaging genetics in AD prevention and treatment with a particular focus on the preclinical phase of the disease. Specifically, we will review findings resulting from both candidate gene and polygenic approaches to neuroimaging genetics studies in AD. The goal of this review is to educate readers on the status of the field, including its many limitations, and to argue that neuroimaging genetics research utilizing polygenic approaches will lead to better characterization of preclinical AD, which is necessary to achieve effective AD prevention.
Neuroimaging Preclinical Alzheimer Disease
A common approach for studying preclinical AD is to use a group at increased risk for AD as a potential preclinical cohort and compare them to a cohort of controls without the risk factor. Increased risk can be defined by the presence of a particular genetic risk variant, such as the apolipoprotein E ε4 (APOEε4) allele, a positive family history of AD, subjective memory impairment as well as the presence of an early neuroimaging or cerebral spinal fluid (CSF) biomarker. Well validated neuroimaging-based biomarkers for AD in these types of cohorts include hippocampal volume loss or thinning, cortical thinning of key AD-related cortical regions, beta-amyloid positivity measured by positron emission tomography (PET) and default mode network (DMN) dysfunction measured by resting state functional MRI (rs-fMRI) (7–16). There is evidence from familial AD patients that these biomarkers precede the emergence of clinical symptoms by at least 3-5 and up to 20 years (1). A thorough description of the literature supporting these biomarker data is outside our focus and there are many excellent reviews available on these topics (17–21).
Clinical neuroimaging positive for biomarker changes, such as thinning of the hippocampus as measured with structural MRI, have been added to the updated AD diagnostic criteria (22). The acquisition of MRI-based biomarkers is minimally invasive, making these methods preferable to lumbar punctures. Both MRI and PET imaging can and have been used in longitudinal studies and provide a quantitative measure of change over time that is not influenced by cognitive performance, which can be affected by sleep patterns, illness, stress and other confounding factors. However, characteristics of imaging biomarkers are not yet sufficient for a preclinical AD diagnosis on the individual level. This is due to several factors, including the lack of extensive longitudinal data to map biomarker changes over time in an individual as well as the limitations in resolution and measurement of modern imaging techniques. Combining known biomarker trajectories with genetic risk stratification may increase prediction power, especially in clinical trial settings, giving greater relative importance to possible disease-related changes in individuals at the highest genetic risk for AD.
Neuroimaging and AD Candidate Genes
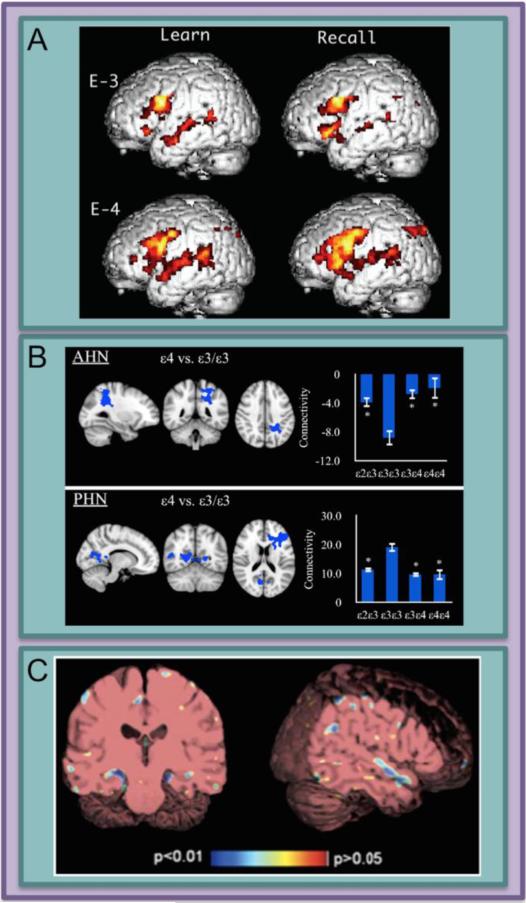
In 2000, the first study to combine neuroimaging and genetic risk for AD in healthy subjects found that carriers of the APOEε4 allele had higher activation across several cortical regions during a memory task compared to non-carriers (Figure 1A; (23)). This approach, examining a selected variant(s) within a single gene and the association of that variant with brain structure and function, is a type of candidate gene study. Candidate gene studies in neuroimaging are very common, but they are controversial due to difficulties in both interpretation and replication of results (24). The now common practice of restricting candidates to genes for which a disease association has already been demonstrated has helped to make findings more robust. Still, a gene with a relatively large effect on disease incidence in a genome wide association study (GWAS) is not necessarily related to neuroimaging phenotypes to the same degree. APOE is the most commonly studied candidate gene for AD. Because of the large proportion of the variance in AD heritability that is accounted for by APOE, investigators have been successful in identifying differences in many neuroimaging modalities based on APOE genotype (Figure 1; see (17–19; 21); for updated review including recent findings see Supplement).
Figure 1. Differences between carriers and non-carriers of the APOE ε4 (APOEε4) allele have been identified using both structural and functional neuroimaging.
The association between APOEε4 and AD risk has a moderate effect size. This may increase the likelihood of observing differences in neuroimaging phenotypes, which are relatively gross measures of neural structure and function. A) Carriers of the APOEε4 risk allele show potentially compensatory cortical activity in language areas during the learning and recall phases of a word-based paired-associates task. B) The anterior hippocampal network (AHN) and posterior hippocampal network (PHN) connectivity is modulated by APOE genotype. Bar graphs represent the network as a region of interest and denotes average connectivity in each genotype group. C) Structural MRI shows that healthy older carriers of APOEε4 have a greater atrophy rate over time in hippocampus and superior temporal gyrus when compared to non-carriers. Panels reprinted: A (23), B (82) and C (7).
In addition to APOE, other GWAS-identified AD risk genes have been studied using a candidate gene approach. These include CLU, PICALM, and CR1 as well as BIN1, ABCA7 and EPHA1. Of these genes, the one that has received the most attention in the neuroimaging literature is CLU. First linked to AD by May and colleagues in 1990, the coincident discovery of CLU in two independent GWASs in 2009 renewed the interest in CLU and its role in AD (25–27). The association of rs11136000 to AD has been replicated several times (28–30).
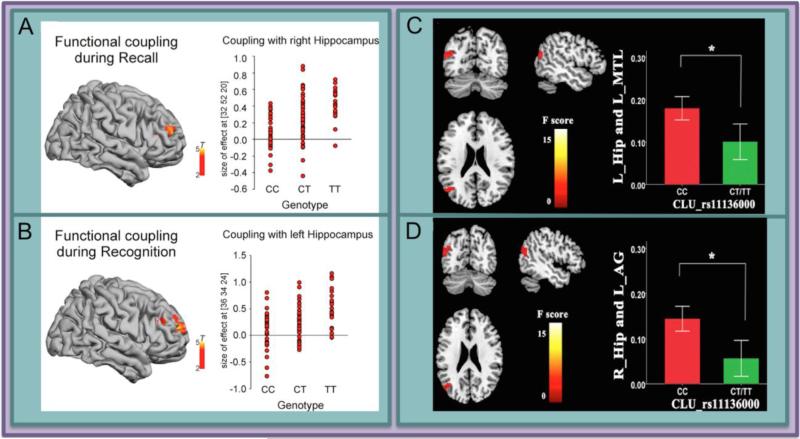
Several functional imaging studies have reported an effect of CLU genotype in both task-based and resting functional MRI (fMRI) paradigms. One fMRI experiment that tested for additive effects of CLU and APOE on blood-oxygen-level dependent (BOLD) signal during an executive attention task found a negative correlation between genetic risk and the BOLD signal associated with executive attention in the medial temporal lobe, as well as other regions (31). In another study, healthy older carriers of the CLU risk variant showed decreased coupling of the hippocampus and prefrontal cortex during memory retrieval tasks (recall and recognition) (Figure 2A,B) (32). In a resting-state fMRI experiment, subjects who were homozygous for the CLU risk allele had the same general pattern of positive and negative functional connectivity compared to carriers of the protective allele, but the magnitude of the connectivity was stronger in both the positive and negative directions (Figure 2C,D) (33). Taken together, these studies indicate a modulatory relationship between BOLD signal and CLU genotype.
Figure 2. A single nucleotide polymorphism within the gene CLU that is associated with higher risk for AD has been associated with decreased functional connectivity of the hippocampus in two distinct studies.
Functional connectivity between the hippocampus and frontal regions during both recall (A) and recognition (B) is modulated by CLU genotype such that individuals who carry the risk allele show lower connectivity in a dose-dependent manner. In another study, individuals who are homozygous for the CLU risk allele show greater connectivity between left hippocampus and left medial temporal lobe, as well as higher connectivity between right hippocampus and angular gyrus (D). Panels A and B reprinted (32). Panels C and D reprinted (33).
PICALM, a gene whose protein product is involved in synaptic transmission, has also been linked to imaging phenotypes in both structural and functional imaging (34; 35; 33; 36). An epistatic effect of PICALM and BIN1, another gene involved in synaptic transmission, on amyloid deposition has been reported (36). BIN1 was also linked to smaller entorhinal cortex and temporal pole volume in a structural imaging study (34). CR1 has been shown in several studies to be associated with smaller entorhinal cortex volume in both young and older healthy adult subjects (34; 37). Finally, a positron emission tomography (PET) study found that there was a relationship between amyloid deposition and polymorphisms in ABCA7 and EPHA1 such that carrying the risk variant of ABCA7 increases likelihood of amyloid positivity while the low-risk polymorphism of EPHA1 decreases likelihood of amyloid positivity (38). A more complete description of imaging studies focused on these GWAS-identified risk genes can be found in Table 1. See Supplement for more details.
Table 1. GWAS-Identified Risk Genes for AD: Neuroimaging Modalities in the Literature and Representative References.
OR = odds ratio, from (78) ; sMRI = structural magnetic resonance imaging; DWI = diffusion weighted imaging; t-fMRI = task-based functional MRI; rs-fMRI = resting state functional MRI; PET = positron emission tomography
| Gene | OR | sMRI | DWI | t-fMRI | rs-fMRI | PET | Comment |
|---|---|---|---|---|---|---|---|
| CLU | 0.86 (0.84 – 0.89) | Bralten et al., 2011a; Stevens et al., 2014 (37; 79) | Braskie et al., 2011 (80) | Erk et al., 2011; Green et al., 2014 (31; 32) | Zhang et al., 2014 (33) | Protein co-chaperone | |
| PICALM | 0.87 (0.85 – 0.89) | Biffi et al., 2010; Bralten et al., 2011a; Furney et al., 2011 (34; 35; 37) | Zhang et al., 2014 (33) | Hohman et al., 2013 (36) | Synaptic transmission | ||
| CR1 | 1.18 (1.14 – 1.22) | Biffi et al., 2010; Bralten et al., 2011b (34; 81) | Innate immunity | ||||
| BIN1 | 1.22 (1.18 – 1.25) | Biffi et al., 2010 (34) | Hohman et al., 2013 (36) | Synaptic transmission | |||
| ABCA7 | 1.15 (1.11 – 1.19) | Hughes et al., 2014 (38) | Lipid homeostasis | ||||
| EPHA1 | 0.90 (0.88 – 0.93) | Hughes et al., 2014 (38) | Adhesion and contact mediated signaling |
Relatively little genetic variance is accounted for by differentiating experimental groups based on carrier status of a single risk variant. In the next sections, we will cover polygenic scores and regression-based polygenic modeling approaches. These efforts aim to measure genetic risk as a continuous metric or as a set of predictors capable of revealing important relationships between genetic risk, brain structure and function and preclinical AD.
Polygenic Risk Scores
Combining multiple genetic risk loci into a single metric or score is an attractive way to modernize the candidate gene approach by using the metric or score as your “candidate” rather than a single gene. Associations between a risk score and, for example, an imaging endophenotype cannot be attributed to a single gene, but these associations may be clinically useful in the effort to better characterize preclinical AD (39). Such metrics are designed on one of two main theoretical bases: first, that multiple risk polymorphisms in the same disease-related biological pathway will be more likely to disrupt normal functioning of that pathway or second, that multiple risk polymorphisms affecting various neuronal functions will together predispose or lead to disease. A polygenic risk score (PRS) can be calculated in several ways. Unweighted approaches simply tally the number of known risk alleles carried by a given individual. Weighted risk scores apply a statistic that captures the strength of the relationship between the genetic variant and disease to differentially weight each risk allele. When GWAS data is available, odds ratios are often used to weight risk alleles in a polygenic risk score but other effect size measures can be used (39). Another method of quantifying polygenic risk is assessing genotype patterns and binning subjects by their genotypes at multiple loci. A limitation of this approach is that a large sample is needed in order to have large enough sub-groups for meaningful statistical analysis. Finally, testing for interaction effects, or epistatic effects, between two or more genes is also technically a polygenic approach, although it differs in that risk effects are not additive but rather emerge from specific interactions between loci.
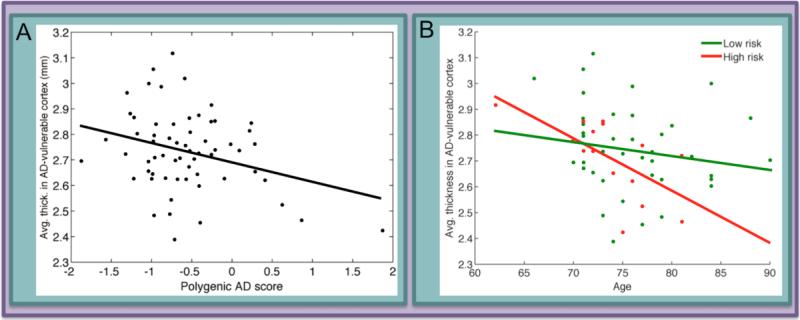
Using a PRS weighted by GWAS-reported odds ratios, Sabuncu and colleagues showed that increased genetic risk for AD was associated with decreased cortical thickness in AD-vulnerable regions, including entorhinal, lateral temporal, inferior parietal and posterior cingulate cortices (Figure 3; (40)). In another structural imaging study a large cohort of over 8,000 cognitively healthy older individuals was used to assess the relationship between a GWAS-loci based weighted PRS and several measures including intracranial volume, total brain volume, and hippocampal volume (41). The authors reported that higher PRS was associated with smaller hippocampal volume, a result that remained significant even after removing APOE from the PRS. Decreases in fractional anisotropy (FA) have emerged in the APOE literature as a possible early indicator of disease-susceptibility (42; 43). More work is needed to ascertain whether or not there is an additive effect of AD risk genes on FA, but preliminary efforts in polygenic approaches to account for white matter integrity are promising (44).
Figure 3. Polygenic risk scores have been used to show relationships between aggregate genetic risk for AD and morphological differences in AD-vulnerable cortical regions.
A) A polygenic score for AD risk based on 26 common variants was negatively correlated with average thickness in a set of AD-vulnerable cortical regions in healthy older adults. The 26 variants, based on closest gene, were within or near DAB1, CR1, BIN1, SSB, C6orf155, ARID18, CLU (two SNPs), KCNU1, MS4A6A, C11orf30, PICALM, CNTN5, BCL3 (two SNPs), PVRL2 (5 SNPs), TOMM40 (3 SNPs) and APOE (see Supplementary Table 2 in 40). B) The relationship between risk score and cortical thickness was driven by a strong age-associated decline in cortical thickness amongst individuals at highest genetic risk for AD. Panels reprinted (40).
There is also evidence from the functional imaging literature that epistatic effects are detectable. One study tested interactions between single nucleotide polymorphisms (SNPs) from 9 AD risk genes identified in GWASs and found that carrying BIN1 risk variants and the PICALM protective variant was associated with increased amyloid deposition as measured by PET imaging (36). In young adults, it was reported that the effect of APOE and CLU risk on BOLD signal during an executive attention task was decreased activation of medial temporal structures with increasing genetic risk load (31). Another study of young adults using resting state fMRI found that an interaction effect between PICALM and CLU risk modulated hippocampal connectivity (33).
Regression Approaches to Polygenic Risk
The use of predictive regression models in clinical biostatistics is extremely common (45). Neuroimaging genetics presents a unique problem with millions of genetic markers (in whole genome data) that can be used as predictors and many outcome phenotypes of interest. Furthermore, linkage disequilibrium, or the tendency of certain genetic loci to be inherited together, must be considered when using any regression method since many of these models assume that predictors are independent (46). The numerous data reduction or selection methods used in regression analyses can be categorized as follows: stepwise regression, regularized regression, mixed linear modeling, projection and prior biological knowledge (47–51). While the methods are too numerous to review in detail, we highlight a few important perspectives with respect to AD.
Stepwise regression optimizes a linear model by successively removing, adding or alternating between adding and removing predictors. One study specifically demonstrated there is an advantage to using machine-learning based, cross-validated genetic algorithms over stepwise regression to predict conversion from MCI to AD (47). Regularized regression is similar to stepwise in that it assumes that a small number of the predictors will be the most informative. These approaches, like Lasso or sparse regression (e.g., ridge, elastic net), penalize larger models in favor of more parsimonious models. Silver and colleagues used sparse reduced-rank (Lasso) regression to model groups of SNPs that are all within a single biological pathway and calculate the strength of the relationship of that pathway to AD-related neuroimaging phenotypes (48). The authors reported that SNPs belonging to insulin signaling, vascular smooth muscle contraction and focal adhesion pathways were the strongest predictors of structural change over 24 months of follow-up. Another study used an elastic net regularization method to explore genetic risk factors for AD affecting the hippocampal surface and found that APOE and TOMM40 were associated with hippocampal surface differences in anterior and middle regions (52).
Genome-wide complex trait analysis (GCTA; http://cnsgenomics.com/software/gcta/) is an example of an optimized linear modeling approach to polygenic risk for phenotypes. Developed to determine the portion of variability of a given trait that can be explained by all available SNPs rather than those that survive genome-wide significance, GCTA takes advantage of linear mixed effect modeling to combine fixed effects like age and sex with SNPs as random effects (53). A recent update to the approach ensures that this procedure can be completed in reasonable time despite the high computational demand of considering millions of SNPs and many phenotypes (54). The authors of the updated GCTA approach used a cohort of 1,320 subjects to compute heritability estimates for several structural neuroimaging measures including whole-brain cortical thickness (54). Ridge and colleagues used the GCTA approach to examine the proportion of the variance in AD status explained by 11 known, common genetic risk loci for AD and found that only 8% (standard error 0.03) of phenotypic variance was accounted for by these markers, while 33% (standard error 0.0072) of the variance was due to common SNPs, known and unknown (49). These results suggest that there are many more common AD-associated SNPs that have not been identified yet and that genetic variants that explain a large proportion of phenotypic variance are rare.
To test across many millions of SNP-SNP interactions it is necessary to apply a method that is capable of performing the computationally intensive task of high-dimensional predictor selection. Hibar and colleagues used a machine learning approach that was designed to perform well when the number of predictors is greater than the number of observations, as is the case when examining human SNP data, by ranking the normalized predictors by their correlation to the dependent variable (55). The authors discovered that the volume of a region of the temporal lobe was associated with the interaction between two SNPs across the clinical categories in the ADNI sample. Another study, also using ADNI, reduced the number of SNP-SNP interactions they tested using a linear regression approach by only testing for interactions between SNPs that were members of a common biological pathway, such as calcium signaling or axon guidance, which were both associated with entorhinal cortex and hippocampal atrophy in their cohort (51). This approach based on prior biological knowledge has been shown to be an effective method of predictor selection (56). Similarly, SNP data reduction using projection techniques like independent component analysis has been used to identify independent groups of genes affecting a given trait (50). Post-hoc pathway analysis of the components then can reveal whether they are enriched for genes related to, for example, as Meda and colleagues found in their ADNI sample, inflammation, diabetes, obesity and cardiovascular disease (50).
In addition to more traditional regression approaches, advanced association models, such as canonical correlation, can be used to efficiently analyze large neuroimaging genetics datasets. These methods are outside the scope of this review but please see the Supplement for a summary.
Limitations
Power: Effect Sizes and Variant Frequency
A major challenge in neuroimaging genetics is sufficiently powering studies to detect hypothesized effects. One problem is the low effect size of common genetic associations to disease in human polygenic disorders (57; 58). An exception to this pattern is the APOE locus where a commonly occurring variant is strongly associated to increased AD risk. In fact, APOE accounts for a larger amount of the variance in AD heritability than any single known genetic locus in another human neurobehavioral, polygenic disorder. Theoretically, because APOE accounts for a relatively large proportion of the heritability variance in AD, it is possible that accurately modeling polygenic risk for AD will be simpler than in other common polygenic neurobehavioral diseases. Thus, AD is an attractive neurological disorder to neuroimaging genetics investigators who are anxious to demonstrate that their field is uniquely positioned to identify early, preclinical predictors of disease.
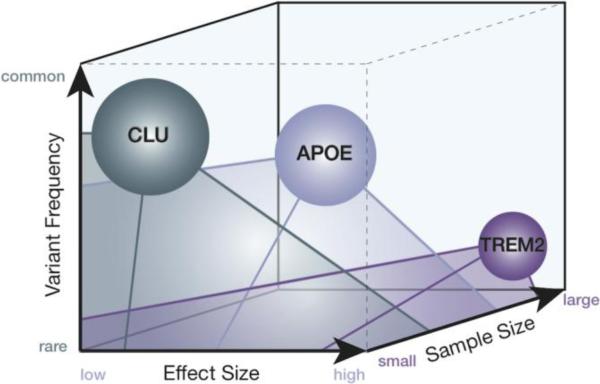
Today, it is not clear if the underlying genetics of AD are best described as many high-effect rare variants (e.g., TREM2 or MAPT) that, in different individuals, each lead to clinical AD or many low-effect common variants that together in a single individual can lead to clinical AD. To the neuroimaging genetics investigator, there are advantages and disadvantages to a common-variant or rare-variant theory of AD genetics. Of course, rare variants occur in so few individuals that it is difficult to amass a large cohort of carriers. However, increased emphasis on data sharing and access to continuously expanding reservoirs of pooled data means that reasonably sized samples of individuals with specific rare variants may be plausible (given a minor allele frequency of 0.002, a sample of 20,000 subjects would be needed to identify 40 carriers of the TREM2 risk variant) (59). Often, rare variants associated with a particular disease have a relatively high effect size, which may make differences between carrier groups easier to detect, even at smaller sample sizes. In contrast, carriers of common variants are more easily amassed in large numbers, but investigators need extremely large cohorts to detect the low-effect size association that usually accompanies a disease-related common variant (Figure 4). As discussed in previous sections, methods for modeling multiple genetic risk factors in a single experiment are actively being developed and may help to exploit the synergistic predictive power of many low-effect-size common variants. In a thorough analysis of the PRS literature, Dudbridge used heritability estimate, sample size, locus significance threshold and a PRS weighting method to generate formulae that allow investigators to estimate the likelihood that future studies will be sufficiently powered (39). The findings indicated that perhaps hundreds of thousands of subjects would be required to make PRS useful at the individual level. Sample sizes are generally not of this magnitude, but they are increasing quickly. Another simulation-based study based on 10,000 cases and controls reported that subjects in the top 5% of genetic risk for hypothetical disease are three to seven times more likely to be affected (60). A three to seven fold increase in risk is certainly clinically useful if not conclusive, as it suggests some individuals may be better candidates for clinical trials and that more frequent follow-up assessments are indicated.
Figure 4. Practical and theoretical parameters of genetic risk factors in AD.
The relationships between variant/allele frequency, effect size and sample size are such that designing adequately powered studies is challenging. CLU, APOE and TREM2 are plotted as representative genes for the following three scenarios: first, a commonly occurring risk variant with a small effect size (CLU, risk allele is major allele with frequency at 60% and effect size of 0.86 (78)), second, a moderately common risk variant with a moderate effect size (APOE ε4 risk allele frequency is 12-14% with an effect size of 2.5 (26; 78)), and third, a rare variant with a relatively large effect size (TREM2 risk variant is minor allele with a frequency of 0.2% and effect size of 3 or more (59; 83)). Note that there are no examples of genes in two extremes in this three dimensional space: high frequency variants that have large effect sizes and low frequency variants that have very small effect sizes. The lack of risk variants of the latter description could be due to the practical difficulties of measuring very small risk effects mediated by very uncommon variants.
Cross-sectional Versus Longitudinal Designs
Another major challenge in the field of neuroimaging genetics of AD is the predominant use of cross-sectional experimental designs to uncover the pathophysiological trajectory of AD. In the literature, inferences about the trajectory of AD are overwhelmingly made from cross-sectional studies in which data is collected from each subject only once and all the subjects are randomly distributed across the age range under investigation with equal number of males and females. This approach is makes it particularly difficult to make conclusions on the subject level because cross-sectional studies confound between-subject and within-subject variation (61). Given this limitation, drawing longitudinal conclusions based on cross-sectional evidence, even from many studies, is precarious and should be done cautiously (62).
The importance of early detection in neurodegenerative diseases like AD is illustrated by the extensive neuronal loss already present in mildly symptomatic AD patients (65). In addition, recent work has established that AD risk genes are associated with differences in brain structure and function even in young people, including children and infants (66; 67). In light of these associations in young people, how can investigators optimize experimental design for the study of AD risk and preclinical AD? Following subjects in longitudinal designs better allows for making inferences about disease trajectory but these studies are difficult in practice. In the modern pro-collaboration atmosphere though, multi-cohort longitudinal designs are feasible because many sites can each collect longitudinal data on a reasonably small number of subjects and then, assuming proper standardization and oversight is in place, these subjects can be combined to create a much larger cohort. The Alzheimer's Disease Neuroimaging Initiative (ADNI) is a good example of a multi-center effort in neuroimaging genetics of AD (63; 64). Optimized longitudinal mapping of AD progression will help identify individuals who are in the preclinical phase of AD. These individuals are likely to benefit the most from intervention, especially a progression-slowing or halting drug. Such a drug is not available today, but the accurate and precise definition of preclinical AD will be an essential component to the success of any candidate.
Discussion: Implications for Clinical Trials
Despite major challenges related to statistical power, polygenic risk modeling and generalizability, the field of neuroimaging genetics is poised to play a major role in the development of effective treatments for AD. Phase 3 AD treatment trials in humans have all had negative outcomes, not meeting endpoints despite promising data in model organisms and in preceding trial phases (68; 69). This high failure rate may be the result, in part, of heterogeneity across the study participants enrolled in these clinical trials. One source of heterogeneity is neuropathological variation. The clinical-neuropathological correspondence of AD (both pure and AD-vascular mixed pathology) occurs in about 87% of clinical AD cases that come to autopsy (70). Thus, more than 10% of clinically diagnosed AD patients actually suffer from some other neurodegenerative disorder, such as frontotemporal lobar degeneration (FTLD) or corticobasal degeneration (CBD). It is reasonable to assume that subjects with each of these diseases, from pure AD and mixed AD pathology to FTLC and CBD, will respond differently, if at all, to potential treatments that target a single molecular species, like Aβ oligomers or plaques. One way to help minimize neuropathological heterogeneity is through the use of PET imaging. The use PET imaging of Aβ and tau as a pre-screening technique in clinical trials, while costly, will allow investigators to amass a more neuropathologically homogeneous cohort. Indeed, neuropathological pre-screening using PET imaging is currently being implemented for the first time as part of the Anti-Amyloid Treatment in Asymptomatic AD (A4) trial, the protocol of which requires a positive Aβ florbetapir-PET scan for enrollment into the treatment arm (71). Another imaging-based method for neuropathological prescreening is diffusion-weighted MRI which can be used to estimate the severity of vascular pathology (72).
Neuropathological differences are not the only source of heterogeneity in clinical trial subjects. It is also important to consider the heterogeneity of the underlying genetics in each individual subject. Depending on the mechanism of the candidate drug, it is possible there will be some variation of response in trial participants with different genetic risk factors for AD. (73). Also, it is likely that by examining genetic risk, the ability to identify asymptomatic individuals who will progress to show cognitive decline is improved. Thus, investigators should consider implementing genetic prescreening measures that select for clinical trial participants that have certain genetic risk factors for AD (74). Clinical trials in AD have already started to use carriage of one or two risk variants (APOE, TOMM40) as a prescreening measure (75). Kohannim and colleagues published a study in which they tested the hypothesis that a polygenic screening protocol would decrease the sample size necessary to detect an effect in a hypothetical trial (76). The authors ranked 394 cognitively healthy and MCI ADNI subjects in order of decreasing polygenic risk score, calculated based on multiplying risk alleles for APOE, CLU, CR1 and PICALM by the logarithm of the odds ratios reported for each gene in GWASs. They found that by selecting only the top 15% of subjects with highest genetic risk, the required sample size to show differences in temporal lobe atrophy decreased from 142 to 69 (76). This is excellent evidence that genetic pre-screening would increase statistical power in trials. Binning participants by genetic risk may well be the next frontier in AD clinical trial design.
Another important role for neuroimaging genetics in clinical trials is the development of hard, non-cognitive endpoints to assess treatment efficacy (77). Most AD trials to date have used soft endpoints such as paper-and-pencil memory measures or a composite dementia severity scores (68; 69). However, as trials shift their focus to preclinical individuals who are asymptomatic cognitive endpoints will no longer appropriate. Thus, neuroimaging-based biomarkers as well as others, such as CSF analyte levels, which capture pathological changes that precede cognitive decline must be refined for use as clinical endpoints (77).
A neuroimaging genetics approach uses minimally invasive technologies to characterize the earliest pathophysiological changes in preclinical AD. In the effort to prevent and treat AD, the proximal goal of combining multiple genetic factors, neuroimaging biomarkers and other measures to estimate AD-risk is to pre-select clinical trial and research participants. The distal goal is to provide more detailed prognoses in the clinic during the preclinical phase that can be used to create optimized treatment plans and enroll ideal candidates in specific clinical trials.
Supplementary Material
Acknowledgements
The authors would like to thank Ms. Therese Vasagas for her help preparing the figures included in this manuscript. This work was supported by the National Institute of Aging (grant number 5R01AG013308 to SYB; 1F31AG047041 to TMH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe V, et al. Short-term clinical outcomes for stages of NIA-AA preclinical Alzheimer disease. Neurology. 2012;78:1576–82. doi: 10.1212/WNL.0b013e3182563bbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mormino EC, Betensky RA, Hedden T, Schultz AP, Ward A, Huijbers W, et al. Amyloid and APOE ε4 interact to influence short-term decline in preclinical Alzheimer disease. Neurology. 2014;82:1760–7. doi: 10.1212/WNL.0000000000000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganguli M, Lee C-W, Snitz BE, Hughes TF, McDade EM. Chang C-CHHow well do MCI criteria predict progression to severe cognitive impairment and dementia? Alzheimer Dis Assoc Disord. 28:113–21. doi: 10.1097/WAD.0000000000000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–9. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu PH, Thompson PM, Leow A, Lee GJ, Lee A, Yanovsky I, et al. Apolipoprotein E genotype is associated with temporal and hippocampal atrophy rates in healthy elderly adults: a tensor-based morphometry study. J Alzheimers Dis. 2011;23:433–42. doi: 10.3233/JAD-2010-101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor JL, Scanlon BK, Farrell M, Hernandez B, Adamson MM, Ashford JW, et al. APOE-epsilon4 and aging of medial temporal lobe gray matter in healthy adults older than 50 years. Neurobiol Aging. 2014;35:2479–85. doi: 10.1016/j.neurobiolaging.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honea RA, Swerdlow RH, Vidoni ED, Goodwin J, Burns JM. Reduced gray matter volume in normal adults with a maternal family history of Alzheimer disease. Neurology. 2010;74:113–20. doi: 10.1212/WNL.0b013e3181c918cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donix M, Burggren AC, Suthana NA, Siddarth P, Ekstrom AD, Krupa AK, et al. Longitudinal changes in medial temporal cortical thickness in normal subjects with the APOE-4 polymorphism. Neuroimage. 2010;53:37–43. doi: 10.1016/j.neuroimage.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleisher AS, Sherzai A, Taylor C, Langbaum JBS, Chen K, Buxton RB. Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer's disease risk groups. Neuroimage. 2009;47:1678–90. doi: 10.1016/j.neuroimage.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Roe CM, Snyder AZ, Brier MR, Thomas JB, Xiong C, et al. Alzheimer disease family history impacts resting state functional connectivity. Ann Neurol. 2012;72:571–7. doi: 10.1002/ana.23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heise V, Filippini N, Trachtenberg AJ, Suri S, Ebmeier KP, Mackay CE. Apolipoprotein E genotype, gender and age modulate connectivity of the hippocampus in healthy adults. Neuroimage. 2014;98:23–30. doi: 10.1016/j.neuroimage.2014.04.081. [DOI] [PubMed] [Google Scholar]
- 14.Machulda MM, Jones DT, Vemuri P, McDade E, Avula R, Przybelski S, et al. Effect of APOE ε4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch Neurol. 2011;68:1131–6. doi: 10.1001/archneurol.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheline YI, Morris JC, Snyder AZ, Price JL, Yan Z, D'Angelo G, et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J Neurosci. 2010;30:17035–40. doi: 10.1523/JNEUROSCI.3987-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleisher AS, Chen K, Liu X, Ayutyanont N, Roontiva A, Thiyyagura P, et al. Apolipoprotein E ε4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol Aging. 2013;34:1–12. doi: 10.1016/j.neurobiolaging.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Cherbuin N, Leach LS, Christensen H, Anstey KJ. Dement Geriatr Cogn Disord. Vol. 24. Karger Publishers; 2007. Neuroimaging and APOE genotype: a systematic qualitative review. pp. 348–62. [DOI] [PubMed] [Google Scholar]
- 18.Bookheimer S, Burggren A. APOE-4 genotype and neurophysiological vulnerability to Alzheimer's and cognitive aging. Annu Rev Clin Psychol. 2009;5:343–62. doi: 10.1146/annurev.clinpsy.032408.153625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheline YI, Raichle ME. Resting state functional connectivity in preclinical Alzheimer's disease. Biol Psychiatry. 2013;74:340–7. doi: 10.1016/j.biopsych.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperling R, Mormino E, Johnson K. The Evolution of Preclinical Alzheimer's Disease: Implications for Prevention Trials. Neuron. 2014;84:608–622. doi: 10.1016/j.neuron.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trachtenberg AJ, Filippini N, Mackay CE. The effects of APOE-ε4 on the BOLD response. Neurobiol Aging. 2012;33:323–34. doi: 10.1016/j.neurobiolaging.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 22.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, Small GW. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–6. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krystal JH, State MW. Psychiatric disorders: diagnosis to therapy. Cell. 2014;157:201–14. doi: 10.1016/j.cell.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.May PC, Lampert-Etchells M, Johnson SA, Poirier J, Masters JN, Finch CE. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer's disease and in response to experimental lesions in rat. Neuron. 1990;5:831–9. doi: 10.1016/0896-6273(90)90342-d. [DOI] [PubMed] [Google Scholar]
- 26.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 28.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jun G, Naj AC, Beecham GW, Wang L-S, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–84. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green AE, Gray JR, Deyoung CG, Mhyre TR, Padilla R, Dibattista AM, William Rebeck G. A combined effect of two Alzheimer's risk genes on medial temporal activity during executive attention in young adults. Neuropsychologia. 2014;56:1–8. doi: 10.1016/j.neuropsychologia.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, et al. Hippocampal function in healthy carriers of the CLU Alzheimer's disease risk variant. J Neurosci. 2011;31:18180–4. doi: 10.1523/JNEUROSCI.4960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, Qin W, Wang D, Liu B, Zhang Y, Jiang T, Yu C. Impacts of PICALM and CLU variants associated with Alzheimer's disease on the functional connectivity of the hippocampus in healthy young adults. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0738-4. doi: 10.1007/s00429-014-0738-4. [DOI] [PubMed] [Google Scholar]
- 34.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–85. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol Psychiatry. 2011;16:1130–8. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hohman TJ, Koran ME, Thornton-Wells T. Epistatic genetic effects among Alzheimer's candidate genes. PLoS One. 2013;8:e80839. doi: 10.1371/journal.pone.0080839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bralten J, Franke B, Arias-Vásquez A, Heister A, Brunner HG, Fernández G, Rijpkema M. CR1 genotype is associated with entorhinal cortex volume in young healthy adults. Neurobiol Aging. 2011;32:2106.e7–11. doi: 10.1016/j.neurobiolaging.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 38.Hughes TM, Lopez OL, Evans RW, Kamboh MI, Williamson JD, Klunk WE, et al. Markers of cholesterol transport are associated with amyloid deposition in the brain. Neurobiol Aging. 2014;35:802–7. doi: 10.1016/j.neurobiolaging.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–61. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chauhan G, Adams HHH, Bis JC, Weinstein G, Yu L, Töglhofer AM, et al. Association of Alzheimer disease GWAS loci with MRI-markers of brain aging. Neurobiol Aging. 2015 doi: 10.1016/j.neurobiolaging.2014.12.028. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westlye ET, Hodneland E, Haász J, Espeseth T, Lundervold A, Lundervold AJ. Episodic memory of APOE ε4 carriers is correlated with fractional anisotropy, but not cortical thickness, in the medial temporal lobe. Neuroimage. 2012;63:507–16. doi: 10.1016/j.neuroimage.2012.06.072. [DOI] [PubMed] [Google Scholar]
- 43.Heise V, Filippini N, Ebmeier KP, Mackay CE. Mol Psychiatry. Vol. 16. Macmillan Publishers Limited; 2011. The APOE ε 4 allele modulates brain white matter integrity in healthy adults. pp. 908–16. [DOI] [PubMed] [Google Scholar]
- 44.Kohannim O, Jahanshad N, Braskie MN, Stein JL, Chiang M-C, Reese AH, et al. Predicting white matter integrity from multiple common genetic variants. Neuropsychopharmacology. 2012;37:2012–9. doi: 10.1038/npp.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrodi SJ, Mukherjee S, Shan Y, Tromp G, Sninsky JJ, Callear AP, et al. Genetic-based prediction of disease traits: prediction is very difficult, especially about the future. Front Genet. 2014;5:162. doi: 10.3389/fgene.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devlin B, Roeder K, Wasserman L. Analysis of multilocus models of association. Genet Epidemiol. 2003;25:36–47. doi: 10.1002/gepi.10237. [DOI] [PubMed] [Google Scholar]
- 47.Johnson P, Vandewater L, Wilson W, Maruff P, Savage G, Graham P, et al. Genetic algorithm with logistic regression for prediction of progression to Alzheimer's disease. BMC Bioinformatics. 2014;15(Suppl 1):S11. doi: 10.1186/1471-2105-15-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver M, Janousova E, Hua X, Thompson PM, Montana G. Identification of gene pathways implicated in Alzheimer's disease using longitudinal imaging phenotypes with sparse regression. Neuroimage. 2012;63:1681–94. doi: 10.1016/j.neuroimage.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridge PG, Mukherjee S, Crane PK, Kauwe JSK. Alzheimer's disease: analyzing the missing heritability. PLoS One. 2013;8:e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meda SA, Narayanan B, Liu J, Perrone-Bizzozero NI, Stevens MC, Calhoun VD, et al. A large scale multivariate parallel ICA method reveals novel imaging-genetic relationships for Alzheimer's disease in the ADNI cohort. Neuroimage. 2012;60:1608–21. doi: 10.1016/j.neuroimage.2011.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meda SA, Koran MEI, Pryweller JR, Vega JN, Thornton-Wells TA. Genetic interactions associated with 12-month atrophy in hippocampus and entorhinal cortex in Alzheimer's Disease Neuroimaging Initiative. Neurobiol Aging. 2013;34:1518.e9–18. doi: 10.1016/j.neurobiolaging.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan J, Kim S, Inlow M, Nho K, Swaminathan S, Risacheri SL, et al. Hippocampal surface mapping of genetic risk factors in AD via sparse learning models. Med Image Comput Comput Assist Interv. 2011;14:376–83. doi: 10.1007/978-3-642-23629-7_46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, et al. Massively expedited genome-wide heritability analysis (MEGHA). Proc Natl Acad Sci. 2015:201415603. doi: 10.1073/pnas.1415603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hibar DP, Stein JL, Jahanshad N, Kohannim O, Toga AW, McMahon KL, et al. Exhaustive search of the SNP-sNP interactome identifies epistatic effects on brain volume in two cohorts. Med Image Comput Comput Assist Interv. 2013;16:600–7. doi: 10.1007/978-3-642-40760-4_75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kerr WT, Douglas PK, Anderson A, Cohen MS. The utility of data-driven feature selection: re: Chu et al. 2012. Neuroimage. 2014;84:1107–10. doi: 10.1016/j.neuroimage.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Nat Rev Genet. Vol. 14. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved; 2013. Pitfalls of predicting complex traits from SNPs. pp. 507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson P V, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–8. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson WK, Hallmayer J, O'Hara R. Design considerations for characterizing psychiatric trajectories across the lifespan: application to effects of APOE-ε4 on cerebral cortical thickness in Alzheimer's disease. Am J Psychiatry. 2011;168:894–903. doi: 10.1176/appi.ajp.2011.10111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kraemer HC, Yesavage JA, Taylor JL, Kupfer D. How can we learn about developmental processes from cross-sectional studies, or can we? Am J Psychiatry. 2000;157:163–71. doi: 10.1176/appi.ajp.157.2.163. [DOI] [PubMed] [Google Scholar]
- 63.Manning EN, Barnes J, Cash DM, Bartlett JW, Leung KK, Ourselin S, Fox NC. APOE ε4 is associated with disproportionate progressive hippocampal atrophy in AD. PLoS One. 2014;9:e97608. doi: 10.1371/journal.pone.0097608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–7. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–28. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- 67.Dean DC, Jerskey BA, Chen K, Protas H, Thiyyagura P, Roontiva A, et al. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: a cross-sectional imaging study. JAMA Neurol. 2014;71:11–22. doi: 10.1001/jamaneurol.2013.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salloway S, Sperling R, Fox NC, Blennow K, Klunk W, Raskind M, et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N Engl J Med. 2014;370:322–33. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doody RS, Thomas RG, Farlow M, Iwatsubo T, Vellas B, Joffe S, et al. Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N Engl J Med. 2014;370:311–21. doi: 10.1056/NEJMoa1312889. [DOI] [PubMed] [Google Scholar]
- 70.Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann Neurol. 2009;66:200–208. doi: 10.1002/ana.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, Aisen P. The A4 study: stopping AD before symptoms begin? Sci Transl Med. 2014;6:228fs13. doi: 10.1126/scitranslmed.3007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayzel-Oreg O, Assaf Y, Gigi A, Ben-Bashat D, Verchovsky R, Mordohovitch M, et al. J Neurol Sci. Vol. 257. Elsevier; 2007. High b-value diffusion imaging of dementia: application to vascular dementia and alzheimer disease. pp. 105–13. [DOI] [PubMed] [Google Scholar]
- 73.Roden DM, George AL. The genetic basis of variability in drug responses. Nat Rev Drug Discov. 2002;1:37–44. doi: 10.1038/nrd705. [DOI] [PubMed] [Google Scholar]
- 74.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11:792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.TOMMORROW Study Alzheimer's Prev Initiat. n.d. Retrieved from http://tommorrowstudy.com.
- 76.Kohannim O, Hua X, Rajagopalan P, Hibar DP, Jahanshad N, Grill JD, et al. Multilocus genetic profiling to empower drug trials and predict brain atrophy. NeuroImage Clin. 2013;2:827–35. doi: 10.1016/j.nicl.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cash DM, Rohrer JD, Ryan NS, Ourselin S, Fox NC. Imaging endpoints for clinical trials in Alzheimer's disease. Alzheimers Res Ther. 2014;6:87. doi: 10.1186/s13195-014-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Nat Genet. Vol. 45. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved; 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. pp. 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stevens BW, DiBattista AM, William Rebeck G, Green AE. A gene-brain-cognition pathway for the effect of an Alzheimer's risk gene on working memory in young adults. Neuropsychologia. 2014;61:143–9. doi: 10.1016/j.neuropsychologia.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, et al. Common Alzheimer's disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–70. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bralten J, Franke B, Arias-Vásquez A, Heister A, Brunner HG, Fernández G, Rijpkema M. CR1 genotype is associated with entorhinal cortex volume in young healthy adults. Neurobiol Aging. 2011;32:2106.e7–11. doi: 10.1016/j.neurobiolaging.2011.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.