Significance
The tropical forests of South America are among the most diverse and unique habitats in the world in terms of plant and animal species. One of the most popular explanations for this diversity and endemism is the idea that forests retracted and fragmented during glacial periods, forming ecological refuges, surrounded by dry lands or savannas. These historically stable forest refuges would have been responsible for maintaining the pattern of diversity and endemism observed today. Here, we show that the Atlantic Forest of eastern South America probably expanded, rather than contracted, during the last glacial period. In addition, the emerged Brazilian continental shelf played a major, yet neglected, role on the evolution of this biodiversity hotspot during the last glacial period.
Keywords: last glacial maximum, sea level, Atlantic Forest, Quaternary, continental shelf
Abstract
The forest refuge hypothesis (FRH) has long been a paradigm for explaining the extreme biological diversity of tropical forests. According to this hypothesis, forest retraction and fragmentation during glacial periods would have promoted reproductive isolation and consequently speciation in forest patches (ecological refuges) surrounded by open habitats. The recent use of paleoclimatic models of species and habitat distributions revitalized the FRH, not by considering refuges as the main drivers of allopatric speciation, but instead by suggesting that high contemporary diversity is associated with historically stable forest areas. However, the role of the emerged continental shelf on the Atlantic Forest biodiversity hotspot of eastern South America during glacial periods has been ignored in the literature. Here, we combined results of species distribution models with coalescent simulations based on DNA sequences to explore the congruence between scenarios of forest dynamics through time and the genetic structure of mammal species cooccurring in the central region of the Atlantic Forest. Contrary to the FRH predictions, we found more fragmentation of suitable habitats during the last interglacial (LIG) and the present than in the last glacial maximum (LGM), probably due to topography. We also detected expansion of suitable climatic conditions onto the emerged continental shelf during the LGM, which would have allowed forests and forest-adapted species to expand. The interplay of sea level and land distribution must have been crucial in the biogeographic history of the Atlantic Forest, and forest refuges played only a minor role, if any, in this biodiversity hotspot during glacial periods.
The extreme biological diversity of tropical forests has inspired and puzzled naturalists and scientists for centuries, and the forest refuge hypothesis (FRH) has long been one of the major paradigms to explain it. According to the FRH, forest retraction and fragmentation during glacial periods would have promoted isolation and consequently allopatric speciation in forest patches, or ecological refuges, surrounded by open habitats in the Amazon (1). Although originally based on climate fluctuations in the Pleistocene, the FRH was subsequently invoked for climate changes irrespective of the time period (2). The FRH was also applied to South America’s Atlantic Forest (3), one of the top-five biodiversity hotspots on Earth (4). The FRH gained broad acceptance during the 1980s when empirical paleoecological data from neotropical rainforests were still lacking. Nevertheless, heavy criticism came upon the FRH because some paleobotanical data showed that forests had persisted throughout glacial cycles (5). As paleoclimatic models of species and habitats became widely used, recent studies revitalized the FRH, not by considering refuges as the main drivers of allopatric speciation, but instead by suggesting that high contemporary diversity and endemism are associated with historically stable Atlantic Forest areas (6). This hypothesis is based on the assumption that populations were restricted to refugia within the bounds of the current observed distribution of a species, thus disregarding paleogeography. Here, we used coalescent simulations to test alternative demographic models and found potentially larger past distributions. Contrary to the refuge models, here, we show that forest specialist small mammal populations actually expanded during the last glacial period, after the expansion of the Atlantic Forest onto the Brazilian continental shelf, a process that has been neglected in paleoenvironmental reconstructions. This new idea is what we call the Atlantis Forest hypothesis, in homage to Plato’s legendary continent, which was eventually swallowed up by the sea.
We combined the results of distribution models with coalescent simulations based on DNA sequences to explore the congruence between scenarios of forest dynamics through time and the genetic diversity of mammal species cooccurring in the central region of the Atlantic Forest. First, we used climatic models to estimate range shifts from the last interglacial (LIG) [∼120,000 y before present (120 kybp)], through the last glacial maximum (LGM) (∼21 kybp), to the present. For LGM simulations, we extended the baseline climate data onto the exposed continental shelf, in contrast to other studies (6). Then, we used coalescent simulations to test the genetic data for congruence with historic demographic models of population fragmentation, bottlenecks, and expansions during the LGM, the LIG, and the penultimate glacial maximum, which corresponds to marine isotope stage 6 (MIS6) (∼160 kybp) (7). To generate and test alternative biogeographical hypotheses, herein we used data from five forest specialist small mammal species, whose occurrence is restricted to forested habitats. Considering their habitat fidelity (Table S1), these species should provide appropriate proxy records of forest changes through time.
Table S1.
Number and percentage of sites where the five small mammal species occurred within two distinct habitats (forests and open areas) and the calculated index of forest fidelity
| Species | Continuously forested areas | Open areas | Forest fidelity index (FD) | ||
| No. of sites | % sites | No. of sites | % sites | ||
| Euryoryzomys russatus | 18 | 1.00 | 1 | 0.03 | 0.97 |
| Marmosops incanus | 17 | 0.94 | 0 | 0.00 | 0.94 |
| Monodelphis americana | 17 | 0.94 | 0 | 0.00 | 0.94 |
| Delomys sublineatus | 17 | 0.94 | 1 | 0.03 | 0.92 |
| Gracilinanus microtarsus | 10 | 0.56 | 3 | 0.08 | 0.47 |
| Total no. sampled sites | 18 | 36 | |||
Results and Discussion
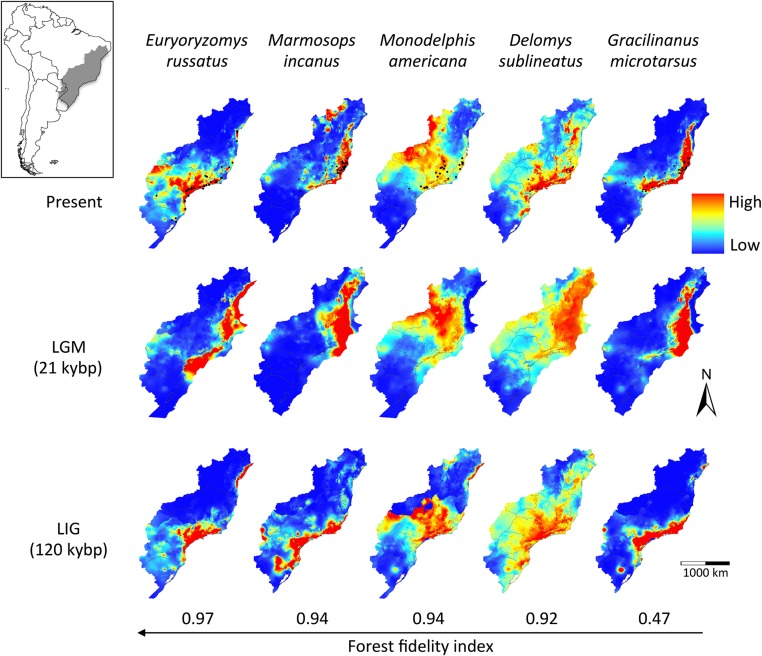
The results of species distribution models were in general congruent among species, and three major trends emerged (Fig. 1). First, species showed suitable habitat displacement toward northern and lower elevation (i.e., warmer) areas during the LGM. Second, we found more fragmentation of suitable habitats during the LIG and the present than in the LGM. Third, we detected expansion of suitable climatic conditions onto the emerged continental shelf during the LGM, which would have allowed forests and forest-adapted species to expand as well. The first trend is a well-known biogeographic response to climate change, but the other two were unexpected because they are in the opposite direction to that predicted by the FRH, prompting us to hypothesize alternative biogeographic scenarios that incorporate forest expansion during glacial periods.
Fig. 1.
Habitat suitability of Atlantic Forest small mammals through time. Modeled ranges of small mammal species sorted according to their habitat fidelity (Table S1) were projected to the present, last glacial maximum (LGM) including the exposed continental shelf, and last interglacial (LIG). Habitat fidelity reflects the proportion of forest sites where each species occurs compared with open areas, ranging from 0 (lack of forest fidelity) to 1 (complete forest fidelity).
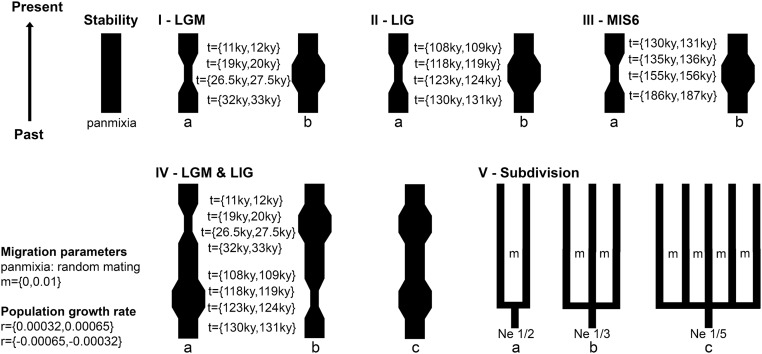
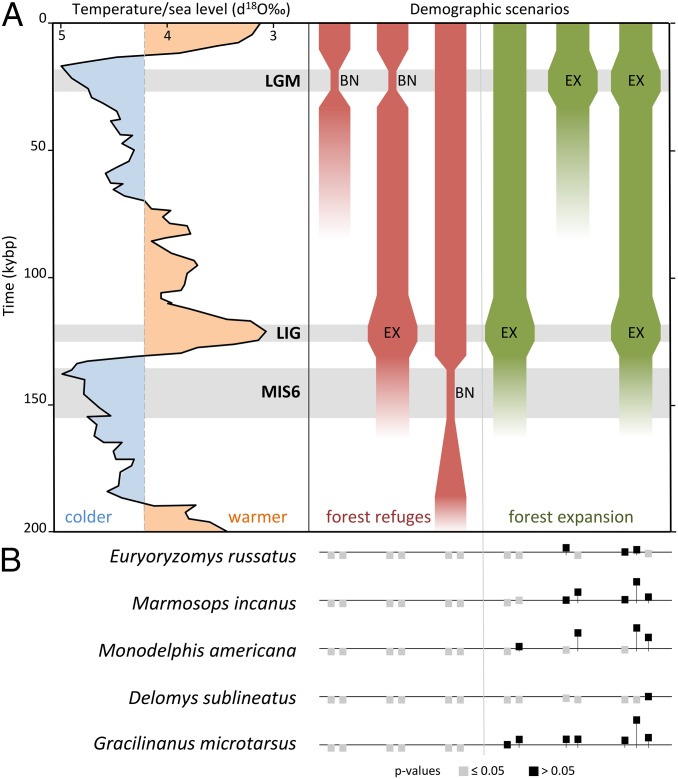
We used intraspecific Bayesian coalescent simulations to evaluate the fit of the empirical genetic data to distinct historical scenarios (Fig. S1 and Tables S2–S6). We first tested a single panmictic population against subdivision into two to five populations, either in a stable island system or including bottleneck or expansion during the LGM. Because all scenarios of population subdivision were rejected, we explored demographic changes in a single panmictic population. The stability scenario was rejected for all species, as well as bottlenecks [1–20% of the effective population size (Ne)] in any of the three events (LGM, LIG, or MIS6) (Fig. 2). Population expansions (1–20% of the Ne) during either LGM (which indicate population expansion onto the emerged continental shelf) or LIG, or both, were not rejected for all forest specialist species, usually confirming population expansion during both LIG and LGM (Fig. 2).
Fig. S1.
Historical demographic models evaluated by coalescent simulations and corresponding parameters. For all models, we simulated populations that suffered three types of historical events: bottleneck, expansion, or population fragmentation. The prior distribution of time of events (t), migration (m), and of the demographic growth rate (r) are specified. Time is expressed in thousand of years (ky) before the present. LGM, last glacial maximum; LIG, last interglacial; MIS6, marine isotope stage 6 (penultimate glacial); Ne, effective population size.
Table S2.
Number of individual sequences (n), localities (loc), base pairs (bp), haplotypes (h), polymorphic sites (S), haplotype diversity (Hd) and nucleotide diversity (π), raggedness statistics (r), and deviation from neutrality tests (D, Fs, R2) for each small mammal species
| Taxon | n | loc | bp | h | S | Hd | π | r | Mismatch | D | Fs | R2 |
| Euryoryzomys russatus | 86 | 30 | 801 | 47 | 66 | 0.964 | 0.01125 | 0.0082** | Bimodal | −1.1727 | −23.547*** | 0.0622 |
| Marmosops incanus | 59 | 28 | 801 | 34 | 161 | 0.968 | 0.04109 | 0.0099 | Multimodal | −0.5688 | −0.111 | 0.0928 |
| Monodelphis americana | 45 | 23 | 773 | 23 | 102 | 0.966 | 0.0326 | 0.0122 | Multimodal | −0.0783 | 1.809 | 0.1155 |
| Delomys sublineatus | 38 | 13 | 801 | 26 | 43 | 0.972 | 0.00969 | 0.0132 | Bimodal | −0.8873 | −10.749** | 0.0817 |
| Gracilinanus microtarsus | 70 | 19 | 801 | 38 | 103 | 0.973 | 0.04389 | 0.0068 | Multimodal | 0.4598 | −0.864 | 0.1329 |
**0.001 < P < 0.05; ***<0.001.
Table S6.
Results of the coalescent simulations in BayeSSC of the five small mammal species
| Taxon | Stability | LGM | LIG | MIS6 | LGM & LIG | Subdivision | |||||||
| Ia | Ib | IIa | IIb | IIIa | IIIb | IVa | IVb | IVc | Va | Vb | Vc | ||
| E. russatus | 0 | 0/0 | 0.12/0 | 0/0 | 0.002/0 | 0/0 | 0/0 | 0/0 | 0.006/0.038 | 0.052/0.088/0.028 | 0 | 0 | 0 |
| M. incanus | 0 | 0/0 | 0.054/0.182 | 0/0 | 0.012/0.05 | 0/0 | 0/0 | 0/0 | 0/0.018 | 0.058/0.349/0.102 | 0 | 0 | 0 |
| M. americana | 0 | 0/0 | 0.024/0.302 | 0/0 | 0.004/0.08 | 0/0 | 0/0 | 0/0 | 0/0.004 | 0.03/0.383/0.23 | 0 | 0 | 0 |
| D. sublineatus | 0 | 0/0 | 0.024/0.002 | 0/0 | 0/0.004 | 0/0 | 0/0 | 0/0 | 0/0 | 0.004/0.004/0.052 | 0 | 0 | 0 |
| G. microtarsus | 0 | 0/0 | 0.14/0.142 | 0/0 | 0.054/0.136 | 0/0 | 0/0 | 0/0 | 0/0.026 | 0.124/0.452/0.166 | 0 | 0 | 0 |
The demographic scenarios are depicted in Fig. S1. Slash separates results of historical demographic events with Ne = 1–10% of the population and Ne = 5–20% of the population, respectively. Simulations of combined expansions (IVc) were also conducted with an increase of Ne of 30–50% of the population during LIG followed by an increase of 5–20% during LGM, respectively. The scenario was rejected when P ≤ 0.05; the scenario was not rejected when P > 0.05 (numbers in bold).
Fig. 2.
Species responses to demographic scenarios of forest refuges (red) and forest expansion (green) during climatic oscillations from 200 kybp. (A) Benthic d18O records reflecting changes in seawater temperature and ice volume (7), and corresponding demographic scenarios of population bottleneck (BN) and/or expansion (EX) in different periods: last glacial maximum (LGM), last interglacial (LIG), and penultimate glacial maximum (MIS6). (B) Results of the coalescent simulations for each species, confirming population expansions during both LGM and LIG. The horizontal line represents the threshold P value (0.05) for each species. Scenarios resulting in P values at or below this level were rejected (gray), and those above this level were not rejected (black). See Fig. S1 for additional scenarios tested and rejected.
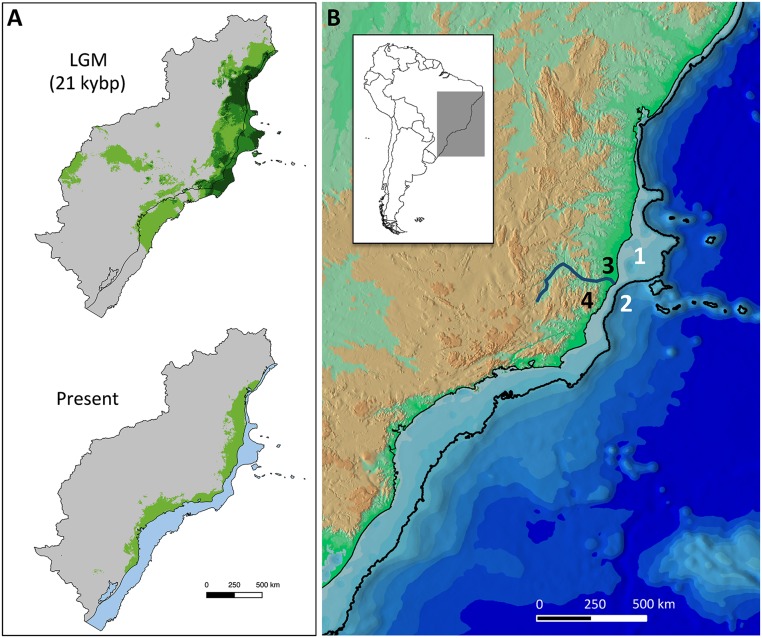
The FRH posits that forest specialist species went through cyclical population subdivision and bottlenecks during past glacial periods and population stability or expansion during interglacials. Our results, however, support the alternative hypothesis that populations of forest specialist small mammals actually expanded during both LIG and LGM. We therefore suggest that the Atlantic Forest also expanded during both periods, especially onto the exposed continental shelf during the LGM. Global sea levels dropped to a maximum of −150 m during Quaternary Glacial Maxima, and the average Quaternary sea level was around −62 m (8). The southeastern South American coastline thus would have been displaced hundreds of kilometers to the east during glacial periods, exposing one of the widest shelves in the world, the Brazilian continental shelf (9) (Fig. 3). This expanded available surface area has not been taken into account in any regional paleoecological reconstruction to date. Rather, the area covered by Atlantic rainforest during the LGM has been estimated at only 150,000 km2 (excluding the continental shelf), which is nearly half of its current size (320,000 km2) (10). But the shelf represents an additional 270,000 km2 (9) of land available for potential expansion. A key question, therefore, is what kind of vegetation covered this shelf during the LGM. To address this question, we projected modeled climatic conditions for the Atlantic Forest and confirmed that extensive suitable areas of broadleaf evergreen rainforest were available on the shelf during this period (Fig. 3A), corroborating simulated LGM biome distributions based on global vegetation models (11). Analyses of a marine sediment core from the southeastern Brazilian continental margin revealed a wide spectrum of pollen types, including those representative of dense and semideciduous forests, coastal scrublands, and a low frequency of grasses (12). In contrast, another marine core from the same region suggested maximal grassland expansion and rainforest reduction during the LGM (13), with the two datasets thus reflecting regional variation in forest cover on the shelf. Other proxy data, such as changes in the oxygen isotopic composition of planktonic foraminifera from the same continental margin imply that maximum precipitation actually occurred around the LGM (14), when the shelf was exposed to its maximum extent. Paleochannels are also evidence of active continental sedimentary processes along the exposed shelf during low sea level periods (15), which point to the presence of riparian forests on the shelf, further supporting our results.
Fig. 3.
Connections between the Brazilian continental shelf and the Atlantic Forest. (A) Projected extent of suitable broadleaf evergreen rainforest areas (green) during the last glacial maximum (LGM) and the present. The LGM map shows the overlap of three models (dark green), two models (medium green), and one model (light green). Suitable areas for rainforest on the continental shelf during the LGM (green) are submersed in the present (blue). (B) Topographic map of the eastern Brazilian coast emphasizing the continental shelf (light blue, bounded by the −150 m isobath) and four key features mentioned in the text: the Abrolhos Bank (site 1), Tubarão Bight (site 2), Doce River (site 3), and Espírito Santo Mountains (site 4). The lowlands (<500 m) are represented in green, the highlands (>500 m) in orange, and ocean depth increases progressively toward darker blue areas.
Interglacial forest expansion on the continent is expected and confirmed by a long-term (140 kybp) pollen record from the Atlantic Forest (16). Forest extent on the continent during the LGM is, however, a controversial topic. Pollen records suggest that grasslands dominated south and southeastern Brazil during this period, indicating a markedly drier and cooler climate (17), and giving support to the FRH. In contrast, other pollen and diatom records combined with carbon and nitrogen isotopes suggest instead that cool, but humid, forests persisted, with no evidence of forest retraction throughout the LGM (18). Oxygen isotopes from speleothems, which are secondary carbonate cave formations, also indicate wetter climate from 70 to 17 kybp, especially during the LGM (19), thus supporting the Atlantis Forest hypothesis and refuting the FRH. We argue that, even if large forested areas were replaced by grassland and the tree line shifted 800–1,000 m downward on the continent during glacial periods (20), the Atlantic rainforest would have not been restricted to small patches as predicted by the FRH because forest cover would have extended onto the continental shelf (Fig. 3A).
The fragmentation of suitable habitats we found during both current and previous (LIG) interglacial periods is unexpected, given that a warmer and wetter climate is more suitable to forest expansion. This putative fragmentation might have been the result of topographic heterogeneity across eastern Brazil. Our species distribution models show that, during glacial periods, ranges are displaced to the north, where the coastal plain is very wide and the topography is monotonously flat, allowing for continuous species ranges. During interglacials, on the other hand, ranges moved south, where the steep coastal mountains (Serra do Mar) drastically replace a narrow coastal plain, leading to a very complex inland topography (Fig. 3B). The seaward side of the Serra do Mar has the highest mean annual rainfall (up to 3,600 mm) of the entire Atlantic Forest range whereas the inland side has typical seasonal climates (1,300–1,600 mm) (21). This heterogeneous topography is associated with distinct rainfall patterns and a mosaic of vegetation types, potentially leading to habitat and range fragmentation.
The Atlantic Forest shows strong latitudinal differentiation, and the geomorphological history of the continental shelf must have played an important, yet ignored, role in this process. A north–south floristic differentiation is caused by distinct temperature and rainfall regimes as mountain ranges are progressively farther from the coast and lower in altitude to the north of the Doce River (21) (Fig. 3B), which is a major biogeographic contact zone for many terrestrial vertebrate groups (22). Furthermore, an analysis of phylogeographic endemism of 25 vertebrate species indicated that this region was recently a location for a subdivision of the Atlantic Forest in two bioclimatic domains (23). Rivers and river valleys are frequently associated with biogeographic breaks in the Atlantic Forest (24), but contrasting results raised some questions about their roles as primary barriers whereas currently hidden geological or climatic barriers have been invoked (25). We argue that the Brazilian continental shelf is a putative barrier because it changes drastically at the Abrolhos Bank, from very narrow to the north to very wide to the south, where significantly more land was exposed during glacial periods (Fig. 3). Also, the steep slopes of the Tubarão Bight and the Espírito Santo Mountains form one of the narrowest lowland areas on the Brazilian coast (Fig. 3B). This region therefore marks major geomorphological and climatic transitions and must have been crucial in shaping the evolutionary history of the adjacent biota (26). Statistical phylogeographic studies on other Atlantic Forest organisms showed mixed results regarding demographic changes during the LGM, but moderate population growth and demographic stability were found for a warbler (27), toads (25), and one spider (28).
Eustatic changes play a major role on the evolution of islands and coastal habitats (29, 30), but such changes have been overlooked in the FRH debate. In addition, the vast continental shelf has been completely ignored in paleomodeling studies of the Atlantic Forest, despite its evolutionary significance in other tropical areas, such as Southeast Asia, where widespread rainforest covered the exposed Sunda shelf during the LGM (31, 32). These facts show that the predominantly glacial Quaternary environment is the norm, characterized by cooler climate, significantly lower sea levels, and slightly reduced precipitation, which was still enough to keep large stretches of rainforest instead of small refugia. This greatly expanded land area, resulting from the exposure of the continental shelf, supported widespread forests (31, 33). The current high sea level condition is therefore the exception, reflecting in reduced forest area in Southeast Asia and the Brazilian Atlantic coast. Our results show that the interplay of sea level and land distribution has been crucial in the biogeographic history of the Atlantic Forest and that forest refuges may have played only a minor role, if any, in this biodiversity hotspot during glacial periods.
Materials and Methods
Selection of Taxa.
Five species of small mammals, three didelphid marsupials [Marmosops incanus (Lund), Monodelphis americana (Müller), and Gracilinanus microtarsus (Wagner)] and two cricetid sigmodontine rodents [Euryoryzomys russatus (Wagner) and Delomys sublineatus (Thomas)], were included in this study. These small mammals are all forest specialist species, for which ecological data indicate a clear preference for forests compared with open habitats, and should respond to both current and past alterations in the distribution of the Atlantic Forest. All five species are widespread and locally abundant in most of the Atlantic Forest (34). To verify and quantify forest fidelity, we used occurrence data from previous studies (35, 36), obtained through standardized samplings (i.e., using the same type, number, and disposition of traps, and the same number of sampling days) in 54 sites in the Atlantic Plateau of the state of São Paulo, southeastern Brazil. The information on species occurrence is therefore comparable among the 54 sites. One third of the sites were located in continuously forested areas within one of the largest tracts of remaining Atlantic Forest. The other 36 sites were located in open, deforested areas used for agriculture (plantations of annual crops) within two 10,000-ha fragmented landscapes located adjacent to the sampled continuously forested areas (18 sites within a fragmented landscape containing only 10% of remaining forest, and 18 sites within a fragmented landscapes containing 50% of remaining forest). We computed the proportion of forest and open area sites where each of the five species occurred (PF and PO, respectively). We then used these proportions to calculate a simple index of forest fidelity (FD = PF − PO) that varies from −1 (preference for open habitats) to +1 (preference for forests), and for which a value close to zero indicates species equally well-distributed in both habitats. All five species were well distributed in continuously forested areas and were absent from most of the open area sites, and fidelity values ranges from 0.47 to 0.97 (Table S1). As expected from their high forest fidelity, these selected forest specialist species negatively respond to current deforestation and forest fragmentation whereas habitat generalist species do not respond or benefit from these processes (35, 37). The obtained values of forest fidelity corroborate several other ecological studies on habitat selection or preference for all five small mammal species (34).
Species and Habitat Distribution Modeling.
Georeferenced occurrence localities of the five target species were gathered from data from museums, scientific articles, theses, and dissertations (38). The museum data were obtained from the online platforms SpeciesLink (splink.cria.org.br/), Global Biodiversity Information Facility (GBIF) (www.gbif.org), and Mammal Networked Information System (MaNIS) (manisnet.org/). The locations of the points of occurrence were verified using Google Maps (www.google.com.br/maps), Google Earth 7.0 (www.google.com/earth/), and SpeciesLink tools (geoLoc and infoXY), and possible mislabeled coordinates were excluded. Occurrence points for each species were then compared with distribution maps from the most recent taxonomic reviews, and discrepancies were further scrutinized to remove suspicious records. In total, we used 44 points of occurrence for E. russatus, 114 for M. incanus, 29 for M. americana, 17 for D. sublineatus, and 73 for G. microtarsus, scattered throughout the study area to create the suitability maps (Dataset S1). To estimate historical broadleaf evergreen rainforest occurrence, we used 395 points (Dataset S1) randomly extracted from the extent of this vegetation (39). To prevent spatial autocorrelation, only points distant at least 10 km of each other were included. To estimate habitat suitability for each species and habitat, we used 19 climate variables from WorldClim (40) in the 2.5-min resolution (www.worldclim.org/). For each species and habitat, correlated environment variables were rejected, as indicated by principal component analysis (PCA) and Mantel tests conducted in R 3.0.1 (41). Climatic layers were cropped to the Atlantic Forest river basins that encompass the distribution of the target species: Paraná, South Atlantic, Southeastern Atlantic, East Atlantic, and São Francisco. The algorithm chosen for modeling was that of maximum entropy, as implemented in MaxEnt 3.3.3 (42). Models were run with default regularization and 10 replicates subsampled, using 20% of the points for test and 80% for training each replicate. To determine whether the model's discrimination capacity is better than random chance, models were validated by accessing area under the curve (AUC), sensitivity, specificity, and accuracy values averaged across the replicates. The adequate environment conditions for each species and habitat in the present were projected into past layers by creating suitability maps for the last glacial maximum (LGM) period 21,000 y ago (including the continental shelf) and for the last interglacial (LIG) period 120,000 y ago. We used General Circulation Models (GCMs) for LIG and LGM estimated by the Community Climate System Model (CCSM). To estimate broadleaf evergreen rainforest occurrence during the LGM, we also used GCMs estimated by the Model for Interdisciplinary Research on Climate-Earth System Model (MIROC-ESM) and the Max-Planck-Institute Earth System Model running in low resolution grid and paleo mode (MPI-ESM-P). All layers of bioclimatic variables are available at the WorldClim website (www.worldclim.org/). We created binary maps of broadleaf evergreen rainforest by estimating the presence/absence of suitable areas using a 10th percentile training presence logistic threshold.
DNA Extraction, Amplification, and Sequencing.
Genomic DNA was isolated from tissue samples (muscle or liver samples) preserved in ethanol (Dataset S2) using the salt extraction method (43). For each species, we sampled between 13 and 30 localities and between 1 and 15 specimens per locality. Polymerase chain reactions (PCRs) were performed to amplify 801 base pairs (bp) of the Cytochrome b gene (Cyt b), using primers MVZ05 and MVZ16 (44). PCR (12.5 μL) contained 1.25 μL of 10× buffer, 0.5 μL of MgCl2 (50 mM), 0.25 μL of dNTP solution (10 mM per nucleotide), 0.15 μL of each primer (10 mM), 0.15 μL of Taq Platinum (Invitrogen Life Technologies), and 1 μL of DNA (20 ng/μL). The PCR profile was as follows: initial denaturation at 94 °C for 5 min, followed by 39 cycles at 94 °C for 30 s, 48 °C for 45 s, 72 °C for 45 s, and a final extension at 72 °C for 5 min. PCR products were purified using ExoSAP enzymes (GE Healthcare Life Sciences), and the cycle-sequencing reactions were performed with a Big Dye v3.1 kit (Applied Biosystems Inc.), following the manufacturer's protocol. Samples were sequenced in both directions using automated DNA sequencer ABI 3500 (Thermo Fisher Scientific, Applied Biosystems), with the same primers listed above. Electropherograms were inspected, and sequences were aligned using the CLUSTAL W method implemented in MEGA 6 (45). All alignments were inspected and corrected manually. All generated sequences have been deposited in GenBank (Dataset S2).
Summary Statistics.
Newly generated and GenBank sequences were used to estimate summary statistics for each target species. Numbers of haplotypes (h), polymorphic sites (S), haplotype diversity (Hd), and nucleotide diversity (Π) for each species were estimated with DnaSP v.5 (46). Deviation from neutrality was tested through different statistics: D (47), Fs (48), and R2 (49), mismatch distribution, and raggedness (50), which were calculated using DnaSP v.5. Coalescent simulations with 1,000 replicates were applied to determine the P value of each statistics. Significant P values (<0.05) were taken as evidence of demographic expansion (Table S2). These summary statistics were further used in coalescent simulations, to test the fit of each simulated scenario to observed data.
Substitution Rate Estimation.
To estimate substitution rates for five taxa to be used in subsequent analyses, we used a fossil-calibrated approach (51). Estimates were performed independently for marsupials and for rodents. Both estimates were implemented in BEAST 2.1.3 (52) assuming a Yule speciation model and using a relaxed molecular clock with lognormal distribution. For marsupials, we used a Cyt b matrix containing previously published sequences (Table S3) and a published phylogeny (53) to set monophyletic constraints. We used two calibration points: the diversification of Didelphidae and the diversification of Didelphimorphia (Table S4). These calibration points were set as minimum and maximum ages, being the minimum age of the oldest unequivocal fossil belonging to clade (54). The maximum age of Didelphidae was based on stratigraphic bounding, which encompasses two stratigraphic units without fossil records of this clade (54). The maximum age of Didelphimorphia was based on phylogenetic bounding, which encompasses the age of the stem of didelphimorph (Peradectes) (54, 55). For rodents, we also used a Cyt b matrix containing previously published sequences (Table S3) and followed a published phylogeny (56) to set monophyletic constraints. We used six calibration points that represent multiple shallow (e.g., genera) and deep (e.g., tribes) calibrations of sigmodontines (56, 57) (Table S4). These calibration points were set as lognormal prior distributions, where fossil ages were set as offsets, and mean and SD were set as 1 (57). We performed two independent runs of 100,000,000 generations each and sampled every 10,000 generations. Analyses were performed in the CIPRES Science Gateway (https://www.phylo.org). Convergence of the Markov Chain Monte Carlo (MCMC) chains was verified in Tracer v.1.6 (58), making sure that all effective sample size (ESS) values were higher than 200. The resulting trees were combined using LogCombiner, and the consensus tree was generated in TreeAnotator, after a burn-in of 10%. Consensus trees were visualized in FigTree 1.4. The substitution rates obtained for each species were further used to estimate time to the most recent common ancestor (tMRCA) and in the coalescent simulations.
Table S3.
GenBank accession numbers of the sequences used in estimating substitution rates of marsupials (Didelphimorphia) and rodents (Rodentia)
| Taxon | GenBank accession number |
| Didelphimorphia | |
| Caluromys lanatus | U34664 |
| Chironectes minimus | AJ628363 |
| Didelphis albiventris | JF280984 |
| Didelphis marsupialis | JF281001 |
| Gracilinanus agilis | JF281013 |
| Gracilinanus microtarsus | JF281020 |
| Lutreolina crassicaudata | AJ628364 |
| Marmosa murina | JF281060 |
| Marmosops incanus | KT952153 |
| Monodelphis americana | KM071582 |
| Philander opossum | JF281037 |
| Thylamys karimii | EF051700 |
| Rodentia | |
| Abrothrix olivaceus | AF297879 |
| Akodon montensis | AY273905 |
| Auliscomys pictus | U03545 |
| Calomys callosus | DQ447282 |
| Calomys lepidus | AF159294 |
| Chelemys macronyx | U03533 |
| Cricetus cricetus | AJ490302 |
| Delomys sublineatus | AF108687 |
| Deltamys kempi | AY195862 |
| Euryoryzomys russatus | EU579486 |
| Holochilus brasiliensis | GU126517 |
| Juscelinomys huanchacae | AY275119 |
| Kunsia tomentosus | AY275121 |
| Oligoryzomys fulvescens | GU126529 |
| Oligoryzomys nigripes | GU126530 |
| Oxymycterus nasutus | AF175286 |
| Phyllotis xanthopygus | U86833 |
| Pseudoryzomys simplex | GU126547 |
| Rheomys raptor | KJ921706 |
| Scapteromys tumidus | AY275133 |
| Sigmodon hispidus | AF425227 |
Table S4.
Fossil calibration points used in the substitution rate estimation of didelphid marsupials and sigmodontine rodents
| Taxon | Fossil | Stage | Age | Ref(s). |
| Didelphimorphia | Micoureus laventicus and Peradectes | Miocene to Puercan | 11,608–65,800* | (54, 55) |
| Didelphidae | Micoureus laventicus | Miocene to Chattian | 11,608–28,500* | (54, 72) |
| Sigmodontalia | Prosigmodon oroscoi | Late Hemphilian | 4,800 | (57, 73, 74) |
| Oxymycterys | Oxymycterus cf. rufus | Ensenadan | 1,300 | (57, 75) |
| Akodon s.l. | Akodon lorenzinii | Vorohuensan | 2,700 | (57, 75) |
| Phyllotini | Auliscomys sp. | Early Chapadmalalan | 4,000 | (57, 76, 77) |
| Oligoryzomys | Oligoryzomys cf. flavescens | Late Ensenadan | 800 | (57, 75, 78) |
| Holochilus | Holochilus brasiliensis | Late Ensenadan | 800 | (57, 75, 79) |
Age is given in thousands of years before the present.
Fossil constraints established with minimum and maximum ages.
Divergence Time Estimation.
The tMRCA was estimated independently for each of five taxa, using all available sequences of each species (Dataset S2) and two close outgroups. Before this estimation, we tested the molecular clock for each dataset by performing maximum likelihood (ML) analyses in MEGA 6. The ML values of the obtained topologies were compared with and without molecular clock constraints also using MEGA 6. We rejected the presence of a strict molecular clock for the five small mammal species and used relaxed molecular clock approaches in the all of the subsequent analyses. The time to the most recent common ancestor (tMRCA) for each species was estimated assuming a Yule speciation model. We used a relaxed molecular clock with lognormal distribution as indicated by ML tests, our estimation of the mean substitution rates as the clock rate, and evolution models as selected using MrModeltest (59) for each species (Table S5). To obtain absolute dates in this analysis, we used our estimation of the mean substitution rates for each species (Table S5). Estimates were implemented in BEAST 2.1.3 and ran in the CIPRES Science Gateway. Runs and MCMC chain convergence, as well as resulting consensus trees, were defined as described above. Mean divergence dates and 95% highest posterior densities (HPDs) were obtained from the deepest node of the species in the tree.
Table S5.
Models of nucleotide substitution for each species as selected using MrModeltest
| Taxon | Model | Mutation rate | 95% HPD | T/T ratio | Θ | 95% HPD | Ne | 95% HPD | tMRCA | 95% HPD |
| E. russatus | GTR+I | 0.0239 | 0.0152–0.0326 | 7.45 | 0.013 | 0.010–0.017 | 275,314 | 205,021–345,816 | 593 | 365–877 |
| M. incanus | GTR+I+G | 0.0373 | 0.0123–0.0667 | 6.82 | 0.018 | 0.013–0.023 | 237,131 | 173,324–305,630 | 1,153 | 557–1,931 |
| M. americana | GTR+G | 0.0347 | 0.0120–0.0623 | 7.39 | 0.009 | 0.006–0.012 | 128,242 | 80,692–176,801 | 1,124 | 419–2,123 |
| D. sublineatus | HKY+I | 0.0284 | 0.0195–0.0382 | 8.38 | 0.007 | 0.004–0.010 | 118,134 | 66,901–169,014 | 678 | 275–1,218 |
| G. microtarsus | GTR+I | 0.0489 | 0.0150–0.0934 | 3.40 | 0.023 | 0.020–0.062 | 235,685 | 199,693–267,894 | 1,111 | 616–1,748 |
Mean mutation rates, generation time, transition/transversion (T/T) ratios, theta (Θ), population effective size (Ne), and mean time to most recent common ancestor (tMRCA) for each small mammal species, followed by 95% highest posterior density intervals (95% HPD). Mean time and 95% highest posterior density intervals are given in thousands of years before the present (kybp).
Hypotheses Tests of Historical Scenarios.
To test putative scenarios of population diversification, we implemented coalescent simulations in the Bayesian Serial SimCoal (BayeSSC) (60, 61), and we then evaluated the fit of the empirical genetic data to distinct historical models. To evaluate fitting of each simulated scenario to the empirical data (summary statistics: h, S, Π, Tajima’s D, and pairwise differences), we applied the statistical framework described in the literature (27, 62–65). We started with simple models as a baseline against which more complex ones were compared (66). First, we simulated a scenario of a single panmictic population without any demographic event throughout time, which was then compared with three population subdivision models, where the ancestral population was separated into two, three, and five subpopulations (Fig. S1). The subdivision date was based on the estimates of tMRCA of each species (Table S5). We then simulated bottlenecks and expansions during the last glacial maximum (LGM) [∼11,000–33,000 y before present (11–33 kybp)], the last interglacial (LIG) (∼109–130 kybp), and the penultimate glacial period, which corresponds to marine isotope stage 6 (MIS6) (∼130–185 kybp) (67, 68). Bottlenecks and expansions were first simulated for each period separately and were then conjugated in both LGM and LIG (Fig. S1). These demographic events were simulated to reduce or increase the population effective size (Ne) to 1–10% and 5–20% of the current size (under a uniform prior). To further test a scenario of broader expansion, we also simulated an increase of 30–50% (under a uniform prior) during LIG followed by an increase of 5–20% (under a uniform prior) during the LGM. Ne was estimated from theta (Θ), according to the following formula: Θ = 2μNe, where μ is the mutation rate previously obtained for each species (Table S5). Θ was estimated in MIGRATE-N 3.6 (69) using transition/transversion ratios for each species estimated in MEGA 6 (Table S5), a static heating scheme, and the Bayesian Inference option with the default search parameters. For population subdivision models, we assumed that Ne from each present population was 1/2, 1/3, and 1/5 of the species Ne. In these scenarios, we assumed a linear stepping-stone model, with migration (m) equal to 0.01. BayeSSC input files for each species included the estimated range of population effective size Ne (under a uniform prior), mutation rates, generation time of 1 y (70), and transition/transversion (T/T) ratios (Table S5). For each model, we ran 1,000 simulations, in which five simulated summary statistics were obtained (h, S, Π, Tajima’s D, and pairwise differences). These simulations were then tested against observed data for each species (71). For assessing the significance of our data, we followed the procedure described in the literature (62, 63), where each summary statistic was compared with values representing the empirical distribution of the statistic from simulation. We then compared the combined empirical likelihood of each summary statistic (Cobs) against a null distribution of C (62, 63) and rejected simulated scenarios when P ≤ 0.05 (Table S6).
Supplementary Material
Acknowledgments
We thank J. Justino, L. Zanchetta, J. Dalapicolla, J. Agrizzi, R. Guimarães, R. Duda, and the many undergraduate students for laboratory work. We thank J. L. Patton and A. B. Rylands for helpful discussions and advice; M. Nascimento, J. Guedes, V. Colombi, V. Vale, E. Guerra, R. Duda, and K. Alves for field assistance; and J. Hoppe and F. Barreto for assistance with distribution models. This study was funded by Fundação de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES, Brazil) and Conselho Nacional de Desenvolvimento Científico e Tenológico (CNPq, Brazil).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Dataset S2.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513062113/-/DCSupplemental.
References
- 1.Haffer J. Speciation in amazonian forest birds. Science. 1969;165(3889):131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- 2.Haffer J. Alternative models of vertebrate speciation in Amazonia: An overview. Biodivers Conserv. 1997;6:451–476. [Google Scholar]
- 3.Prance GT. Biological Diversification in the Tropics. Columbia Univ Press; New York: 1982. [Google Scholar]
- 4.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 5.Colinvaux P, de Oliveira P, Moreno J, Miller M, Bush M. A long pollen record from lowland Amazonia: Forest and cooling in glacial times. Science. 1996;274:85–88. [Google Scholar]
- 6.Carnaval AC, Moritz C. Historical climate modeling predicts patterns of current biodiversity in the Brazilian Atlantic forest. J Biogeogr. 2008;35:1187–1201. [Google Scholar]
- 7.Cohen KM, Gibbard P. 2011 Global Chronostratigraphical Correlation Table for the Last 2.7 Million Years (Subcommission on Quaternary Stratigraphy, International Commission on Stratigraphy, Cambridge). Available at quaternary.stratigraphy.org/charts. Accessed June 30, 2015.
- 8.Rabineau M, et al. Paleo sea levels reconsidered from direct observation of paleoshoreline position during Glacial Maxima (for the last 500,000 yr) Earth Planet Sci Lett. 2006;252:119–137. [Google Scholar]
- 9.Nagai RH, Sousa SHM, Mahiques MM. The southern Brazilian shelf. In: Chiocci FL, Chivas AR, editors. Continental Shelves of the World: Their Evolution During the Last Glacio-Eustatic Cycle. Geological Society Memoirs; London: 2014. pp. 47–54. [Google Scholar]
- 10.Behling H. Carbon storage increases by major forest ecosystems in tropical South America since the Last Glacial Maximum and the early Holocene. Global Planet Change. 2002;33:107–116. [Google Scholar]
- 11.Prentice IC, Harrison SP, Bartlein PJ. Global vegetation and terrestrial carbon cycle changes after the last ice age. New Phytol. 2011;189(4):988–998. doi: 10.1111/j.1469-8137.2010.03620.x. [DOI] [PubMed] [Google Scholar]
- 12.Freitas AG, Carvalho MA, Mendonça CBF, Gonçalves-Esteves V. Pollen grains in Quaternary sediments from the Campos Basin, state of Rio de Janeiro, Brazil: Core BU-91-GL-05. Acta Bot Brasilica. 2013;27:761–772. [Google Scholar]
- 13.Behling H, Arzb HW, Pätzold J, Wefer G. Late Quaternary vegetational and climate dynamics in southeastern Brazil, inferences from marine cores GeoB 3229-2 and GeoB 3202-1. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;179:227–243. [Google Scholar]
- 14.Pivel MAG, Toledo FAL, Costa KB. Foraminiferal record of changes in summer monsoon precipitation at the southeastern Brazilian continental margin since the Last Glacial Maximum. Revista Brasileira de Paleontologia. 2010;13:79–88. [Google Scholar]
- 15.Dominguez JML, da Silva RP, Nunes AS, Freire AFM. The narrow, shallow, low-accommodation shelf of central Brazil: Sedimentology, evolution, and human uses. Geomorphology. 2013;203:46–59. [Google Scholar]
- 16.Ledru MP, Mourguiart P, Riccomini C. Related changes in biodiversity, insolation and climate in the Atlantic rainforest since the last interglacial. Palaeogeogr Palaeoclimatol Palaeoecol. 2009;271:140–152. [Google Scholar]
- 17.Behling H. South and southeast Brazilian grasslands during Late Quaternary times: A synthesis. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;177:19–27. [Google Scholar]
- 18.Pessenda LCR, et al. The evolution of a tropical rainforest/grassland mosaic in southeastern Brazil since 28,000 14C yr BP based on carbon isotopes and pollen records. Quat Res. 2009;71:437–452. [Google Scholar]
- 19.Cruz FW, et al. Orbital and millennial-scale precipitation changes in Brazil from speleothem records. In: Vimeux F, Sylvestre F, Khodri M, editors. Past Climate Variability in South America and Surrounding 29 Regions: From the Last Glacial Maximum to the Holocene. Springer; Dordrecht, The Netherlands: 2009. pp. 29–60. [Google Scholar]
- 20.Behling H. Tropical mountain forest dynamics in Mata Atlantica and northern Andean biodiversity hotspots during the late Quaternary. In: Gradstein SR, Homeier J, Gansert D, editors. The Tropical Mountain Forest: Patterns and Processes in a Biodiversity Hotspot. Universitätsverlag Göttingen; Göttingen, Germany: 2008. pp. 25–33. [Google Scholar]
- 21.Oliveira-Filho AT, Fontes MA. Patterns of floristic differentiation among Atlantic forests in Southeastern Brazil and the influence of climate. Biotropica. 2000;32:793–810. [Google Scholar]
- 22.Costa LP, Leite YLR. Historical fragmentation shaping vertebrate diversification in the Atlantic Forest biodiversity hotspot. In: Patterson BD, Costa LP, editors. Bones, Clones, and Biomes: The History and Geography of Recent Neotropical Mammals. Chicago Univ Press; Chicago: 2013. pp. 283–306. [Google Scholar]
- 23.Carnaval AC, et al. Prediction of phylogeographic endemism in an environmentally complex biome. Proc Biol Sci. 2014;281(1792):20141461. doi: 10.1098/rspb.2014.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellegrino KCM, et al. Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): Genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc Lond. 2005;85:13–26. [Google Scholar]
- 25.Thomé MTC, Zamudio KR, Haddad CFB, Alexandrino J. Barriers, rather than refugia, underlie the origin of diversity in toads endemic to the Brazilian Atlantic Forest. Mol Ecol. 2014;23(24):6152–6164. doi: 10.1111/mec.12986. [DOI] [PubMed] [Google Scholar]
- 26.Pereira TL, et al. Dispersal and vicariance of Hoplias malabaricus (Bloch, 1794) (Teleostei, Erythrinidae) populations of the Brazilian continental margin. J Biogeogr. 2013;40:905–914. [Google Scholar]
- 27.Batalha-Filho H, Cabanne GS, Miyaki CY. Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol Phylogenet Evol. 2012;65(3):892–902. doi: 10.1016/j.ympev.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Peres EA, et al. Pleistocene niche stability and lineage diversification in the subtropical spider Araneus omnicolor (Araneidae) PLoS One. 2015;10(4):e0121543. doi: 10.1371/journal.pone.0121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali JR, Aitchison JC. Exploring the combined role of eustasy and oceanic island thermal subsidence in shaping biodiversity on the Galápagos. J Biogeogr. 2014;41:1227–1241. [Google Scholar]
- 30.Huntley B, Allen JRM, Barnard P, Collingham YC, Holliday PR. Species distribution models indicate contrasting late-Quaternary histories for Southern and Northern Hemisphere bird species. Glob Ecol Biogeogr. 2013;22:277–288. [Google Scholar]
- 31.Wang XM, Sun XJ, Wang PX, Stattegger K. Vegetation on the Sunda Shelf, South China Sea, during the Last Glacial Maximum. Palaeogeogr Palaeoclimatol Palaeoecol. 2009;278:88–97. [Google Scholar]
- 32.Raes N, et al. Historical distribution of Sundaland’s Dipterocarp rainforests at Quaternary glacial maxima. Proc Natl Acad Sci USA. 2014;111(47):16790–16795. doi: 10.1073/pnas.1403053111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannon CH, Morley RJ, Bush ABG. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA. 2009;106(27):11188–11193. doi: 10.1073/pnas.0809865106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi NF. 2011 Non-volant small mammals of the Atlantic Plateau of São Paulo: Identification, natural history and threats. Master’s thesis (Universidade de São Paulo, São Paulo, Brazil). Available at www.teses.usp.br/teses/disponiveis/41/41133/tde-21092011-091459/pt-br.php. Accessed June 30, 2015.
- 35.Pardini R, Bueno AdeA, Gardner TA, Prado PI, Metzger JP. Beyond the fragmentation threshold hypothesis: Regime shifts in biodiversity across fragmented landscapes. PLoS One. 2010;5(10):e13666. doi: 10.1371/journal.pone.0013666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umetsu F. 2010 Effect of landscape context at different scales on the distribution of small mammals in areas of agriculture and in forest remnants. PhD dissertation (Universidade de São Paulo, São Paulo, Brazil). Available at www.teses.usp.br/teses/disponiveis/41/41133/tde-17112010-190713/pt-br.php. Accessed June 30, 2015.
- 37.Banks-Leite C, et al. Using ecological thresholds to evaluate the costs and benefits of set-asides in a biodiversity hotspot. Science. 2014;345(6200):1041–1045. doi: 10.1126/science.1255768. [DOI] [PubMed] [Google Scholar]
- 38.Alves KS. 2013. Padrões de distribuição de pequenos mamíferos não-voadores na Mata Atlântica do Brasil e variáveis ambientais associadas. Master’s thesis (Universidade Federal do Espírito Santo, Vitória, Brazil)
- 39.IBGE . Mapa da Vegetação do Brasil. Fundação Instituto Brasileiro de Geografia Estatística; Rio de Janeiro: 1998. [Google Scholar]
- 40.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 41.R Development Core Team 2011 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org.
- 42.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–259. [Google Scholar]
- 43.Bruford MW, Hanotte O, Brookfield JFY, Burke T. Single-locus and DNA fingerprinting. In: Hoelzel AR, editor. Molecular Genetic Analyses of Populations: A Practical Approach. IRL Press; Oxford: 1992. pp. 225–269. [Google Scholar]
- 44.Smith MF, Patton JL. The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol J Linn Soc Lond. 1993;50:149–177. [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega 6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 47.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19(12):2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- 50.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66(4):591–600. [PubMed] [Google Scholar]
- 51.Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58(3):367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- 52.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voss RS, Jansa SA. Phylogenetic Relationships and Classification of Didelphid Marsupials, an Extant Radiation of New World Metatherian Mammals. 2009 (American Museum of Natural History, New York), Bulletin No. 322. [Google Scholar]
- 54.Meredith RW, et al. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 55.Horovitz I, et al. Cranial anatomy of the earliest marsupials and the origin of opossums. PLoS One. 2009;4(12):e8278. doi: 10.1371/journal.pone.0008278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leite RN, et al. In the wake of invasion: Tracing the historical biogeography of the South American cricetid radiation (Rodentia, Sigmodontinae) PLoS One. 2014;9(6):e100687. doi: 10.1371/journal.pone.0100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leite YLR, Kok PJR, Weksler M. Evolutionary affinities of the “Lost World” mouse suggest a late Pliocene connection between the Guiana and Brazilian shields. J Biogeogr. 2015;42:706–715. [Google Scholar]
- 58.Rambaut A, Drummond AJ. 2013 Tracer (University of Edinburgh, Edinburgh), Version 1.6. Available at beast.bio.ed.ac.uk/Tracer.
- 59.Nylander JAA. 2004 MrModeltest (Uppsala University, Uppsala), Version 2. Available at https://github.com/nylander/MrModeltest2.
- 60.Excoffier L, Novembre J, Schneider S. SIMCOAL: A general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J Hered. 2000;91(6):506–509. doi: 10.1093/jhered/91.6.506. [DOI] [PubMed] [Google Scholar]
- 61.Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA. Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics. 2005;21(8):1733–1734. doi: 10.1093/bioinformatics/bti154. [DOI] [PubMed] [Google Scholar]
- 62.Voight BF, et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci USA. 2005;102(51):18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabre V, Condemi S, Degioanni A. Genetic evidence of geographical groups among Neanderthals. PLoS One. 2009;4(4):e5151. doi: 10.1371/journal.pone.0005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maldonado-Coelho M, Blake JG, Silveira LF, Batalha-Filho H, Ricklefs RE. Rivers, refuges and population divergence of fire-eye antbirds (Pyriglena) in the Amazon Basin. J Evol Biol. 2013;26(5):1090–1107. doi: 10.1111/jeb.12123. [DOI] [PubMed] [Google Scholar]
- 65.Cabanne GS, Sari EHR, Meyer D, Santos FR, Miyaki CY. Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Greenish Schiffornis Schiffornis virescens (Aves: Tityridae) J Ornithol. 2013;154:371–384. [Google Scholar]
- 66.Nielsen R, Beaumont MA. Statistical inferences in phylogeography. Mol Ecol. 2009;18(6):1034–1047. doi: 10.1111/j.1365-294X.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- 67.Lisiecki LE, Raymo MEA. Pliocene-Pleistocene stack of 57 globally distributed benthic d18O records. Paleoceanography. 2005;20:PA1003. [Google Scholar]
- 68.Roucoux KH, Tzedakis PC, Lawson IT, Margari V. Vegetation history of the penultimate glacial period (Marine Isotope Stage 6) at Ioannina, north-west Greece. J Quat Sci. 2011;26:616–626. [Google Scholar]
- 69.Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185(1):313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacifici M, et al. Generation length for mammals. Nat Conserv. 2013;5:87–94. [Google Scholar]
- 71.Hickerson MJ, Dolman G, Moritz C. Comparative phylogeographic summary statistics for testing simultaneous vicariance. Mol Ecol. 2006;15(1):209–223. doi: 10.1111/j.1365-294X.2005.02718.x. [DOI] [PubMed] [Google Scholar]
- 72.Marshall LG. New didelphine marsupials from the La Venta fauna (Miocene) of Colombia, South America. J Paleontol. 1976;50:402–418. [Google Scholar]
- 73.Jacobs LL, Lindsay EH. Prosigmodon oroscoi, a new sigmodont rodent from the Late Tertiary of Mexico. J Paleontol. 1981;55:425–430. [Google Scholar]
- 74.Peláez-Campomanes P, Martin RA. The Pliocene and Pleistocene history of cotton rats in the Meade basin of southwestern Kansas. J Mammal. 2005;86:475–494. [Google Scholar]
- 75.Pardiñas UFJ, D’Elía G, Ortiz PE. Sigmodontinos fósiles (Rodentia, Muroidea, Sigmodontinae) de América del Sur: Estado actual de su conocimiento y prospectiva. Mastozool Neotrop. 2002;9:209–252. [Google Scholar]
- 76.Reig OA. Roedores cricétidos del Plioceno superior de la provincia de Buenos Aires (Argentina) Publ Mus Municip Cienc Natur Lorenzo Scaglia. 1978;2:164–190. [Google Scholar]
- 77.Pardiñas UFJ, Tonni EP. Procedencia estratigráfica y edad de los más antiguos muroideos (Mammalia, Rodentia) de América del Sur. Ameghiniana. 1998;35:473–475. [Google Scholar]
- 78.Voglino D, Pardiñas UFJ. Roedores sigmodontinos (Mammalia: Rodentia: Cricetidae) y otros micromamíferos pleistocénicos del norte de la provincia de Buenos Aires (Argentina): Reconstrucción paleoambiental para el Ensenadense cuspidal. Ameghiniana. 2005;42:143–158. [Google Scholar]
- 79.Pardiñas UFJ, Teta P, Voglino D, Fernandez FJ. Enlarging rodent diversity in westcentral Argentina: A new species of the genus Holochilus (Cricetidae, Sigmodontinae) J Mammal. 2013;94:231–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.