Abstract
Mutational hotspots indicate selective pressure across a population of tumor samples, but their prevalence within and across cancer types is incompletely characterized. An approach to detect significantly mutated residues, rather than methods that identify recurrently mutated genes, may uncover new biologically and therapeutically relevant driver mutations. Here we developed a statistical algorithm to identify recurrently mutated residues in tumour samples. We applied the algorithm to 11,119 human tumors, spanning 41 cancer types, and identified 470 hotspot somatic substitutions in 275 genes. We find that half of all human tumors possess one or more mutational hotspots with widespread lineage-, position-, and mutant allele-specific differences, many of which are likely functional. In total, 243 hotspots were novel and appeared to affect a broad spectrum of molecular function, including hotspots at paralogous residues of Ras-related small GTPases RAC1 and RRAS2. Redefining hotspots at mutant amino acid resolution will help elucidate the allele-specific differences in their function and could have important therapeutic implications.
Among the best-studied therapeutic targets in human cancers are proteins encoded by genes with tumor-specific mutational hotspots, such as KRAS, NRAS, BRAF, KIT, and EGFR. The acquisition of somatic mutations is one of the major mechanisms responsible for the dysregulation of proliferation, invasion, and apoptosis that is required for oncogenesis. Comprehensive genomic characterization of tumors has produced significant insights into the somatic aberrations that define individual cancer types1,2, broadening our understanding of the dysfunctional molecular pathways that govern tumor initiation, progression, and maintenance. These data have spurred the development of computational algorithms to identify cancer driver genes, defined as those in which molecular abnormalities lead to a fitness advantage for the affected cancer cells (Methods).
These computational approaches develop either gene-level statistical models that exploit different mutational patterns3–6 to identify significantly mutated genes or use weight-of-evidence-based methods1,7 that are heuristic and ratiometric in approach. Together, these methods focus on identifying cancer genes from a multitude of diverse molecular abnormalities affecting the gene. However, not all genomic alterations in cancer genes are driver alterations. Furthermore, not all driver alterations in a cancer gene have the same functional impact, and are therefore likely to have varying clinical significance. The potentially diverse functional effects of different lesions in the same gene are not captured and reported by gene-level models, but are rather assumed to be equivalent. However, emerging data indicate that different hotspot mutations in the same cancer gene can be functionally distinct in vitro and in vivo and display different clinical phenotypes and drug sensitivity8–11. Moreover, it is unknown how widespread such hotspot-specific functional differences may be.
To date, studies of hotspot mutations in cancer have been limited to within individual tumor types12–14 or have focused on individual cancer genes across tumor types15. A systematic population-scale, cross-cancer, genome-wide analysis of mutational hotspots has not been performed and the extent to which mutant allele and lineage-specific effects exist remains unknown. As broad-based clinical sequencing has begun to inform the care of individual cancer patients, this would begin to address one of the greatest challenges in the practice of genomically driven cancer medicine: interpreting the biological and clinical significance of mutations in even presumed actionable cancer genes as they arise in oncology clinics.
To address this challenge, we develop a computational algorithm to identify driver mutations, rather than driver genes. We assembled and rigorously curated a large repository of cancer genome data consisting of the sequenced tumor exomes and whole genomes of 11,119 human tumors representing 41 tumor types. We developed a biologically aware, statistically principled computational model by combining observed biological phenomena such as nucleotide mutability and varying gene-specific mutation rates into coefficients that we incorporate into binomial statistics. From this, we systematically identify individual recurrent mutations and associate these with related temporal and transcriptional data to investigate lineage-specific variation in mutations, and identify novel hotspots with likely clinical implications.
Results
Landscape of hotspots mutations in primary human cancer
We collected the mutational data from the sequenced exomes and genomes of 11,119 human tumors in 41 tumor types (Supplementary Table 1). These originate from diverse sources including large international consortia and various published studies (Methods). This cohort represents a broad range of primary human malignancies with three or more tumor types in each of nine major organ systems (Fig. 1a). The repository consists of 2,007,694 somatic substitutions in protein-coding regions with a median of 57 mutations (25 and 125 mutations; 25th and 75th percentile respectively) per tumor-normal pair with significant variability in mutation rates among and between tumors and types4,16. In total, 19,223 human genes harbor at least one somatic mutation in this dataset.
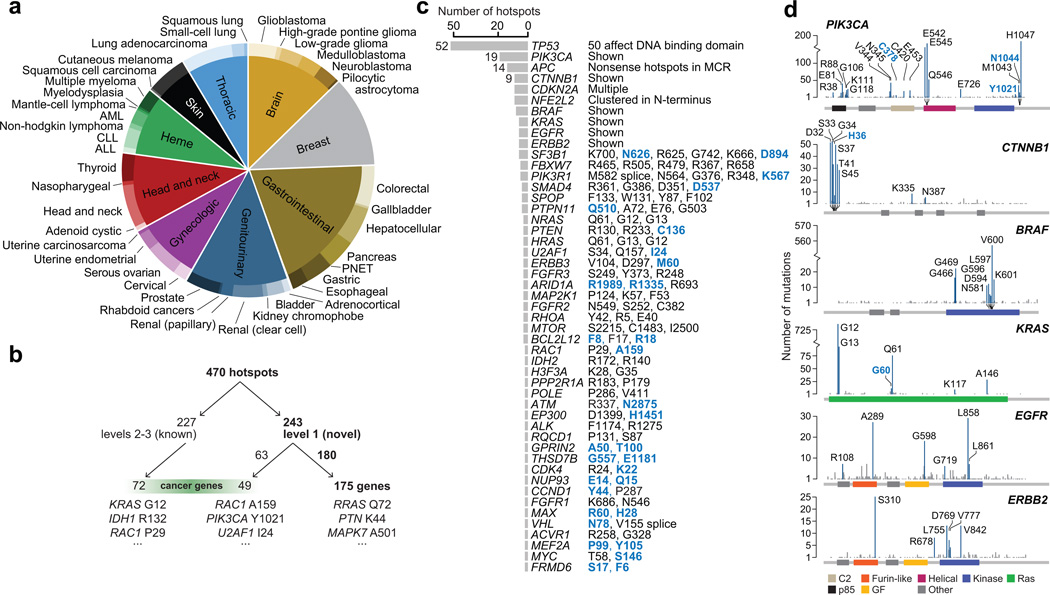
Figure 1. Mutational data and hotspot detection.
a) The distribution of tumor types included in this analysis. b) Breakdown of known and classified novel hotspots and genes (see Methods). c) The number of hotspots in each of 49 genes with two more hotspots detected across the cohort. At right, a summary of hotspots identified. Novel hotspots are bolded blue. d) The distribution of mutations and hotspots in six oncogenes refines known patterns and reveals new hotspots.
Here, we define a mutational hotspot as an amino acid position in a protein-coding gene that is mutated (by substitutions) more frequently than would be expected in the absence of selection (Methods). In this analysis, we focus exclusively on individual substitutions rather than other somatic abnormalities such as translocations, amplifications, deletions, or epigenetic modifications. To identify mutational hotspots, including low-incidence mutations, we developed a binomial statistical model that incorporates several aspects of underlying mutational processes including nucleotide context mutability, gene-specific mutation rates, and major expected patterns of hotspot mutation emergence (Supplementary Fig. 1a and 2, Supplementary Code; Methods). As considerable variability exists in the methods and standards for mutation calling used by individual studies and centers, we also developed several evidence-based criteria for eliminating probable false positive hotspots (Methods, Supplementary Fig. 1b). In total, we identified 470 statistically significant hotspots (q-value < 0.01) affecting 275 protein-coding genes (Supplementary Tables 2 and 3). Overall, more than half of all hotspots were determined to be novel (Fig. 1b, Table 1, Supplementary Table 2, and Methods) and 54.8% of all tumors assessed here possessed one or more hotspot mutations.
Table 1. Select new hotspots in cancer genes.
A subset of newly identified hotspots are shown, a complete listing is available in Supplementary Table 2.
| Pathway/Symbol | Codon | Q-value | No. of affected tumors | No. of tumor types |
|---|---|---|---|---|
| Signaling effectors | ||||
| KRAS | GQ60 | 2.28E-06 | 11 | 7 |
| PIK3CA | Y1021 | 3.18E-06 | 9 | 6 |
| C378 | 0.0018 | 6 | 5 | |
| N1044 | 0.0008 | 6 | 3 | |
| PIK3CB | D1067 | 0.0068 | 5 | 5 |
| PIK3R1 | K567 | 0.0002 | 5 | 4 |
| PTEN[a] | C136 | 2.27E-05 | 9 | 5 |
| RAC1 | A159 | 2.27E-06 | 10 | 5 |
| RRAS2 | Q72 | 8.00E-15 | 9 | 6 |
| GNAQ | T96 | 7.04E-08 | 7 | 5 |
| ERBB3 | M60 | 0.0083 | 4 | 4 |
| MAPK7 | A501 | 9.50E-06 | 6 | 4 |
| PTPN11[a] | Q510 | 1.84E-06 | 7 | 4 |
| PTN | K44 | 1.46E-05 | 7 | 4 |
| ARHGAP28 | L259 | 0.0061 | 5 | 3 |
| Cell cycle | ||||
| CDK4[a] | K22 | 0.0008 | 4 | 2 |
| CCND1 | Y44 | 3.48E-07 | 7 | 2 |
| CDKN2A | E88 | 4.24E-05 | 15 | 5 |
| L130 | 0.007 | 6 | 3 | |
| Transcription Factors | ||||
| NFE2L2 | E82 | 1.60E-13 | 11 | 7 |
| T80 | 1.96E-10 | 9 | 7 | |
| Q26 | 9.26E-08 | 7 | 5 | |
| G81 | 1.34E-09 | 10 | 7 | |
| L30 | 4.52E-06 | 8 | 5 | |
| G31 | 0.0001 | 8 | 5 | |
| R34 | 0.0001 | 13 | 6 | |
| MEF2A | P99 | 2.91E-05 | 7 | 6 |
| Y105 | 0.0061 | 4 | 4 | |
| MYC | S146 | 0.0046 | 6 | 4 |
| MAX | R60 | 0.0006 | 9 | 6 |
| H28 | 0.004 | 4 | 1 | |
| FOXA1 | I176 | 0.0001 | 7 | 2 |
| Epigenetic modifiers | ||||
| ARID1A | R1989 | 2.45E-08 | 17 | 5 |
| R1335 | 0.0062 | 9 | 6 | |
| ING1 | R196 | 1.06E-06 | 11 | 5 |
| EP300 | H1451 | 0.008 | 4 | 4 |
| HIST1H3C | K37 | 0.0008 | 5 | 2 |
| SMARCA4 | G1232 | 0.0006 | 9 | 6 |
| DNA Damage | ||||
| ATM | N2875 | 4.66E-05 | 6 | 4 |
| RNA splicing | ||||
| SF3B1 | N626 | 2.06E-06 | 6 | 4 |
| D894 | 0.009 | 5 | 4 | |
| U2AF1 | I24 | 0.0002 | 4 | 4 |
| Wnt pathway | ||||
| CTNNB1 | H36 | 0.0001 | 6 | 2 |
| Nuclear transport | ||||
| NUP93 | E14 | 1.59E-10 | 11 | 6 |
| Q15 | 0.0082 | 4 | 2 | |
| TGF beta signaling | ||||
| SMAD2 | S464 | 1.19E-07 | 11 | 5 |
| SMAD4 | D351 | 0.0003 | 8 | 6 |
| SMAD4 | D537 | 0.0033 | 9 | 3 |
| TGFBR2 | R528 | 0.0013 | 10 | 5 |
While previously identified in the germline of patients with associated syndromes and familial cancers, this is the first documentation of somatic mutations.
Most affected genes possessed only a single hotspot (Supplementary Fig. 3a). A subset of genes, however, possessed many hotspots of varying frequency. In total, 49 genes possessed two or more hotspots (Fig. 1c), with many of these also arising in the greatest number of tumor types (Supplementary Fig. 3b). TP53 R248 was the most disseminated hotspot, observed in 25 tumor types. Among a subset of even well characterized oncogenes, a pattern of both known and novel hotspots emerge (Fig. 1d). Moreover, the number of observed mutant amino acids at a given hotspot generally increases with its mutational frequency across tumors types (Supplementary Fig. 3c), though 35% (n=164) of hotspots mutate to only a single variant amino acid. In most genes, hotspots bear only a fraction of the total mutational burden across the gene, whereas in a subset of cancer genes, the dominant mutational hotspot constitutes the vast majority of mutations independent of total mutational burden (Fig. 1d and Supplementary Fig. 3d). Overall, we identified considerable variability in the patterns of mRNA expression of individual hotspots in even canonical oncogenes (Supplementary Fig. 4), indicating that levels of expression are often not correlated with the biologic significance of known activating mutations.
The patterns by which some hotspots emerge support new clinical paradigms for testing targeted agents. Some hotspots that dominate the mutational landscape in one or a few cancer types also arise as uncommon subsets of many others. For instance, IDH1 R132 is most common in low-grade gliomas, glioblastomas, acute myeloid leukemias (AMLs), and cutaneous melanomas; but it is also present in 1 to 6 tumors in each of 11 additional cancer types. AKT1 E17K arises in greatest numbers in breast cancer, but also in 1 to 3 tumors of 10 additional cancer types. The distribution of CREBBP R1446 mutations is qualitatively different. They were originally identified in relapsed acute lymphoblastic leukemias17, but in this cohort of mostly primary disease, we find that they arise in only a small minority (1–3; 0.17–1.7%) of many (11) cancer types. Such patterns reaffirm the value of basket study designs that test mutant-specific inhibitors in early phase clinical trials, where enrollment is based on specific mutations in patients instead of tissue of origin.
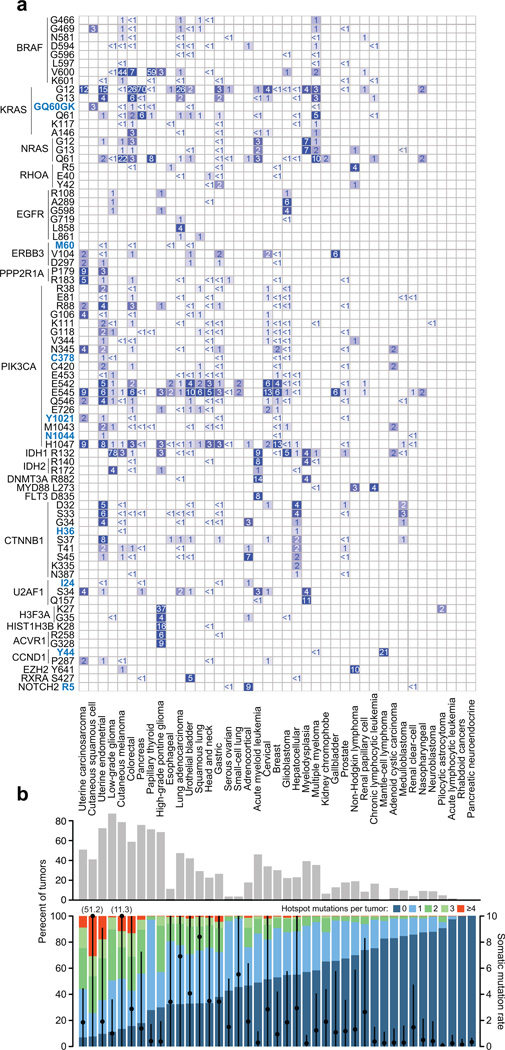
A lineage map of all hotspots in genes with at least one common hotspot (Fig. 2a and Supplementary Fig. 5) indicates most hotspots are defined more by the tissue types rather than the organ systems in which they arise. Of all hotspots, 81% arise in two or more tumor types, suggesting that many hotspot mutations may confer a growth advantage across diverse lineages. Indeed, of hotspots present in multiple tumor types, only 7.6% (n = 36) are confined to a single organ system (Table 2). Thus, hotspot mutations that arise in a single tumor type may reflect organ-specific growth advantages but they represent only a small minority of all hotspot mutations in cancer. Likewise, a subset of hotspots arises in a cell-type specific manner. Twenty-seven hotspots (5.7%) were more frequently mutated in tumors of a squamous cell lineage (Supplementary Fig. 6), the most significant of which were MAPK1 E322 and EP300 D1399 (q-value = 6×10−13 and 1×10−11 respectively, χ2) and may potentially confer a squamous cell-type specific growth advantage.
Figure 2. Lineage landscape of hotspot mutations.
a) Both common and rare hotspots are largely disseminated across a broad range of malignancies. All hotspots detected in genes with at least one hotspot affecting >5% of tumors of one or more tumor types are shown. Novel hotspots are bolded blue. Genes are grouped broadly by functional similarity, hotspots are ordered by amino acid position, and tumor types (columns, labeled at bottom) are sorted according to the fraction of tumors affected by 1 or more hotspots overall (panel B). The percent of samples altered is represented by colored squares and indicated text. Hotspots in tumor suppressors TP53, PTEN, APC, and FBXW7 were excluded here (see Supplementary Fig. 5). b) The fraction of tumors of a given type (as indicated) affected by one or more hotspots. Black circles represent the median mutation rate (right axis) in the indicated tumor type (bar is the median absolute deviation). Shown at top is the number of tumors of each type with a hotspot mutation affecting a known or candidate oncogene1.
Table 2. Organ system-specific hotspots.
Shown are the 10 most significant hotspots that arise in multiple tumor types of a single organ system.
| Symbol | Position | Tumor types[a] | Organ system |
No. affected tumors |
Q-value[b] |
|---|---|---|---|---|---|
| H3F3A | K27M | High-grade pontine glioma (26) Pilocytic astrocytoma (1) |
Brain | 27 | 2.2×10−50 |
| MYD88 | L273P | Chronic lymphocytic leukemia (12) Non-hodgkin lymphoma (2) Multiple myeloma (1) |
Hematologic | 15 | 1.9×10−26 |
| STK19 | D89N | Cutaneous melanoma (13) Squamous cell carcinoma (5) |
Skin | 18 | 3.5×10−21 |
| EGFR | G598V/A | Glioblastoma (15) Low-grade glioma (3) High-grade pontine glioma (1) |
Brain | 19 | 5×10−16 |
| PPP2R1A | P179R/L | Endometrial (8) Uterine carcinosarcoma (5) |
Gynecologic | 13 | 1.6×10−12 |
| FGFR3 | Y375C | Urothelial bladder (7) Renal papillary cell carcinoma (1) |
Genitourinary | 8 | 2.8×10−10 |
| KNSTRN | S24F | Cutaneous melanoma (11) Squamous cell carcinoma (2) |
Skin | 13 | 7.7×10−10 |
| CCND1 | Y44D/S/H/F/C/* | Mantel cell lymphoma (6) Multiple myeloma (1) |
Hematologic | 7 | 3.5×10−7 |
| CRNKL1 | S128F | Cutaneous melanoma (8) Squamous cell carcinoma (2) |
Skin | 10 | 4×10−7 |
| EGFR | L861Q | Lung adenocarcinoma (5) Lung squamous cell carcinoma (2) |
Thoracic | 7 | 5.4×10−7 |
Number in parentheses is the count of affected tumors of the indicated cancer type.
Q-value is estimated from p-values produced by the binomial model described in Methods.
Overall, the presence, type, and frequency of hotspots by tumor type vary widely (Fig. 2b). In some tumor types, a large proportion of tumors possess one or more hotspot mutations including a significant fraction of tumors with a hotspot in a candidate oncogene (Fig. 2b, top). Conversely, other tumor types never or rarely possess a tumor defined by a hotspot identified here. Some of these differences are certainly attributable to the fact that hotspots are only one of many possible driver genomic aberrations, including specific gene fusions or focal amplifications and deletions. These other aberrations may define tumors of a given type, but they are not mutually exclusive with hotspots in many cancers. Other differences could not, alone, be explained by the overall mutational burden in these tumor types. For instance, uterine carcinosarcomas and prostate cancers have a similar mutation rate while there is 3-fold greater frequency of hotspot-bearing tumors among the former. Likewise, while papillary thyroid and high-grade pontine gliomas have mutations rates similar to nasopharyngeal tumors and neuroblastomas, the former far more commonly bear hotspot mutations (Fig. 2b).
Unconventional hotspots
In addition to missense mutations, we identified a variety of unconventional hotspot mutations with varied impact. Among these were 13 splice site hotspots. For each of these hotspots, an associated transcript abnormality was identified from RNA sequencing of affected tumors (exon skipping, intron retention, in-frame deletions; Supplementary Fig. 7a), including two previously characterized in-frame activating mutations (MET D1010_splice and PIK3R1 M582_splice, both exon 14 skipping events). We also identified 70 hotspots in 34 genes for which a nonsense mutation was among a diversity of changes at the affected residue, including 28 hotspots in which only a nonsense mutation was present (Supplementary Fig. 7b). While nonsense mutations scattered throughout a gene may reflect a pattern of loss-of-function consistent with tumor suppressor activity, a nonsense hotspot would appear to indicate the selection for the selective truncation of specific functional domains. Such events are consistent with the loss of some functions and the retention of others, as has been observed previously in genes such as PIK3R1, NOTCH1, and MET18,19. These hotspots aside, there was a depletion of nonsense mutations in hotspots in constitutively essential genes (p-value<10−16, those genes predicted or experimentally verified to be essential across all cell and tissue types and developmental states20). Otherwise, the specific impact of nonsense hotspots is generally unknown and belies the disseminated pattern of truncating mutations in likely or proven tumor suppressors (Supplementary Fig. 7c).
Lineage diversity and mutant allele-specificity
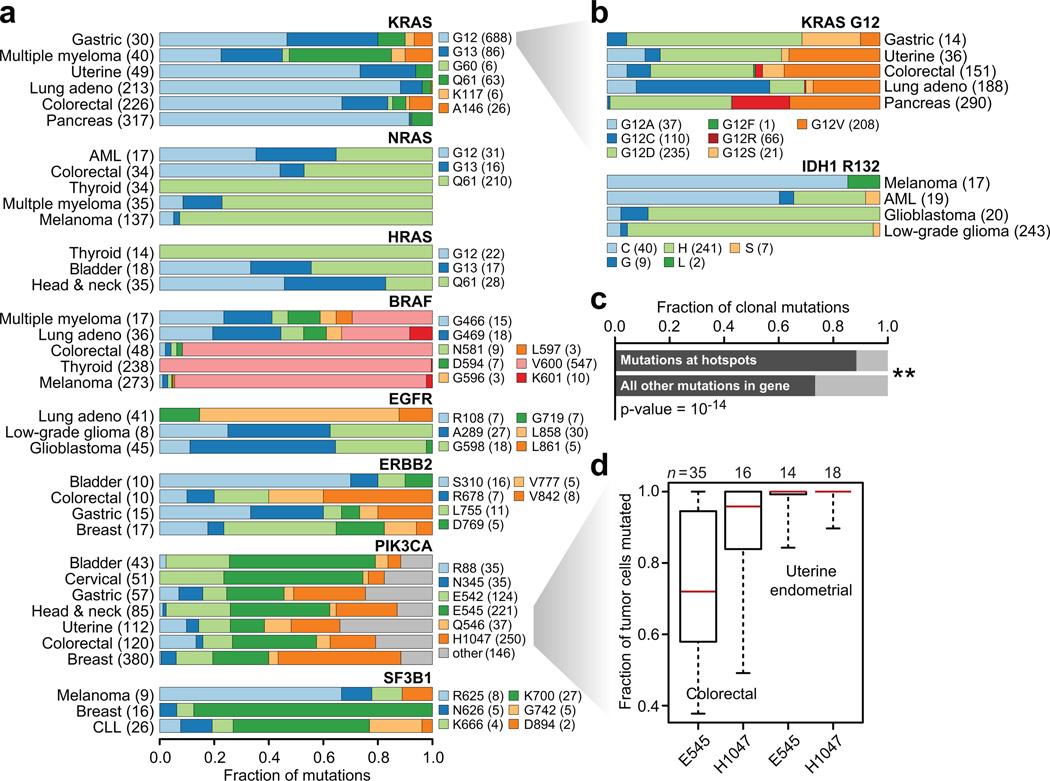
The majority of hotspot mutations arose in diverse tumor types and organ systems, yet widespread differences exist among individual residues and mutant amino acids in hotspots, genes, and tumor types (Fig. 3a). Examining the spectrum of KRAS mutations, which includes the most frequently mutated hotspot overall in our study (KRAS G12; n=736 mutant tumors, Fig. 1d and 2a), clarified patterns only incidentally observed in the past. We found that gastric cancers were more similar to multiple myeloma in the preponderance of non-G12 mutations compared to endometrial, lung, colorectal, and pancreas tumors (p-value = 5.3×10−18, Supplementary Table 4). Only colorectal tumors had KRAS A146 mutations whereas pancreas tumors lacked G13 mutations (p-values = 4×10−7 and 2.8×10−15 respectively). Many of these lineage-specific patterns were present at finer resolution as well. Among KRAS G12 mutations, the abundance of G12C mutations are highest in lung adenocarcinomas (p-value = 4×10−42), an event that may be associated with prognostic differences compared with non-G12C KRAS mutations21–23. Such mutant amino acid specificity was also apparent in pancreas tumors, where KRAS G12R was more common than in any other tumor type (21% versus between 0 and 2.6%; χ2 p-value = 4.8×10−19). Gastric cancers, on the other hand, had the fewest G12V mutations among all KRAS G12-mutant tumor types, but the highest proportion of G12S (p-value = 0.007, Fig. 3b). There is a different balance among hotspots in the other Ras genes. While papillary thyroid cancers nearly exclusively possessed codon Q61 mutations in HRAS and NRAS (p-value = 4×10−7), there was a higher prevalence of G12 and 13 codon mutations in these genes in AMLs, colorectal, bladder, and head and neck cancers, which together share few mutational processes in common (p-value = 4×10−10, Fig. 3a).
Figure 3. Lineage diversity and mutant allele specificity.
a) The fraction of cases mutated for each of the most common hotspots in 8 frequently mutated genes in the most commonly mutated lineages indicate substantial lineage diversity and hotspot specificity. b) Same as in panel (a), but for KRAS G12 and IDH1 R132 mutations, showing that mutant amino acid specificity exists within individual hotspots across affected tumor types. c) The fraction of clonal mutations, those present in 80% or more of the tumor cells of affected samples, was higher among mutations in hotspots versus all other non-recurrent mutations in the same genes (χ2 p-value = 1×10−14). d) The fraction of tumor cells mutated for PIK3CA E545 and H1047 hotspots in affected colorectal and uterine endometrial cancers indicates a pattern of allele-specific subclonality for E545 mutations in colorectal cancer.
Similar differences emerged in other driver cancer genes with multiple hotspots. V600E mutations describe nearly all BRAF hotspot mutations in melanoma, papillary thyroid, and colorectal carcinomas, whereas multiple myelomas are similar to lung adenocarcinoma in which non-V600E hotspots predominate (p-value = 1.9×-10−32). The balance between extracellular and kinase domain mutations in EGFR between brain tumors and lung adenocarcinoma (p-value = 3.3×10−12) respectively have been documented previously and affect their biological impact and the efficacy of genotype-directed therapy10. ERBB2 followed a similar pattern, where extracellular domain mutations typified by S310F are far more common than are kinase domain mutations in bladder cancers compared to breast cancers (p-value = 0.006, Fig. 3a). Another notable gene was PIK3CA. While bladder and cervical cancers are similar in their distribution of PIK3CA hotspot mutations, they vary significantly from breast cancers in the overall balance of helical to kinase domain mutations, possessing far fewer H1047R mutations among PIK3CA-mutated cases (p-value = 4.8×10−19). Endometrial and colorectal cancers also have a similar pattern of PIK3CA hotspots, but both have a higher prevalence of R88Q mutations than any other tumor type (p-value = 1.3×10−11, Fig. 3a). Such patterns extend beyond essential MAPK or PI3K signaling components, such as with SF3B1 K700 mutations that predominate in breast cancers and chronic lymphocytic leukemias whereas melanomas more frequently possess R625 mutations (p-value = 0.0001). Finally, mutant amino acid specificity was not limited to hotspots in Ras genes. The IDH1 R132H hotspot mutation predominated in multiple brain tumor types, but cysteine was the most common IDH1 R132 mutant amino acid in melanoma, which is unlikely to be exclusively related to UV light exposure, as this is also true in AMLs that lack a UV-driven etiology (p-value = 3.9×10−21). Together, these results indicate that substantial mutant amino acid specificity exists among hotspot mutations across highly diverse tumor lineages. Two related conclusions may be drawn from these data. First, different hotspots in the same gene may possess in many cases different function, much of which may be lineage-dependent, while not excluding the possibility that some may still arise as a function of differing underlying mutational mechanisms. Second, that perhaps different mutant amino acids within the same hotspot can be functionally different, support for which is growing8,11.
Timing of individual hotspots
We next sought to determine if hotspot mutations, many of which are likely driver mutations and in some cases may serve as the initiating lesion, typically arise earlier than do non-recurrent mutations in the same genes and are therefore more often clonal. Overall, mutations at hotspot residues more often resided in a greater fraction of tumor cells (see Methods) and were therefore earlier arising (presumptive clonal), than were non-hotspot mutations in the same genes (Fig. 3c). So, while prior work has shown that driver genes in lung adenocarcinomas were enriched for clonal mutations24, we found that this was true of hotspot mutations across a broad class of cancer genes and tumor types. However, there was considerable variability among hotspots. While colorectal and endometrial cancers have a similar pattern of PIK3CA hotspot mutations (Fig. 3a) and share hypermutated subtypes of tumors driven by MSI and POLE exonuclease domain mutations25,26, colorectal tumors were unique in the clonality of the E545 and H1047 mutations. The majority of PIK3CA E545 helical domain mutations in colorectal cancers were subclonal, whereas H1047 kinase domain mutations were clonal, a difference that was not apparent in endometrial tumors, in which both are early clonal mutations (Fig. 3d). This may be a function of the pattern of oncogenic co-mutation in these tumors as PIK3CA E545, but not H1047, mutations were significantly associated with KRAS mutations in these colorectal cancers (χ2 p-value = 0.0004) and in previous cohorts27. Overall, these differences in the molecular timing of specific hotspots augurs potentially important differences in their function in tumor initiation versus progression that requires further study.
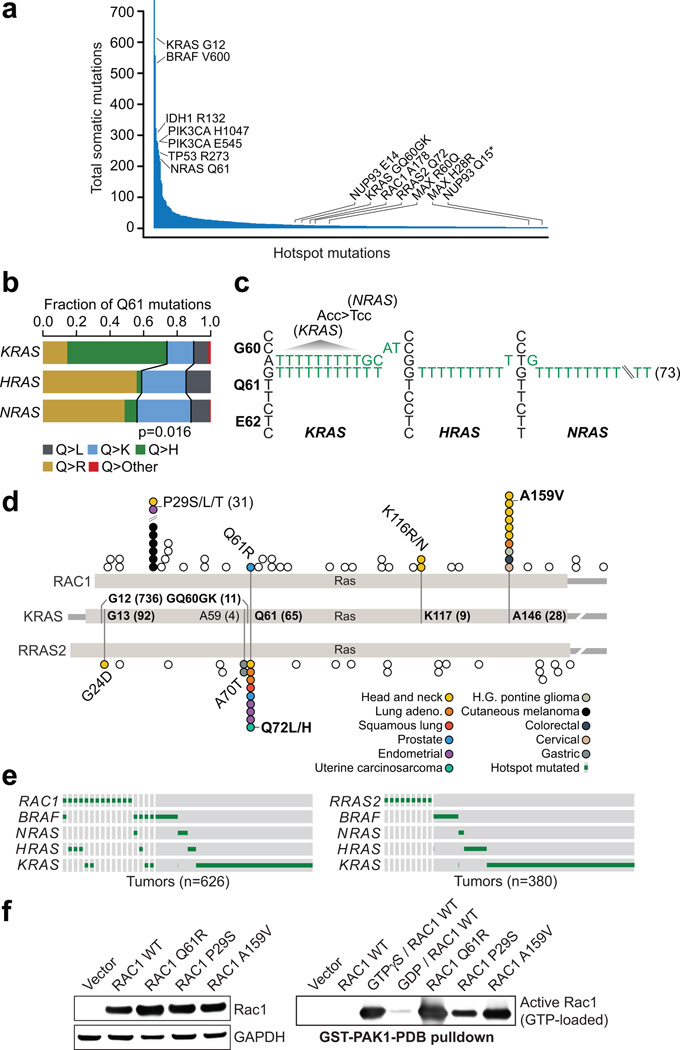
Population-level hotspots in the long tail
Consistent with the so-called long tail of the frequency distribution of somatically mutated genes across cancer2, we found that 85% of all hotspots identified here were mutated in less than 5% of tumors of all cancer types in which they were found (Fig. 4a). Such findings have led to calls for sequencing up to many thousands of additional specimens from every tumor type28. However, many hotspots present at low frequency across cancers are not mutated commonly or significantly in even a single cancer type. Indeed, 23% of all hotspots identified here were present in only one or two samples in the tumor types in which it was observed. This included 19 hotspots arising in only one sample of each affected cancer type such as U2AF1 I24, MYC T58, the hyperactivating MTOR I250029, PIK3CB D1067, EP300 H1451, and ERBB3 M60. Therefore, many driver mutations (rather than genes) may never be found mutated at even the minimal frequencies (2–3%) proposed by previous studies as a goal in each cancer type. Conversely, population-level analysis, rather than by individual cancer type or organ system, allows identification of hotspots that arise as even private mutations in rare malignancies, for which additional broad-scale sequencing is most challenging. While rare, such recurrent alleles are evidence of selection and may be associated with specific phenotypes, such as exceptional responses30,31, de novo resistance to cancer therapy, or reveal specific facets of pathway biology. Consequently, we found that notable long-tail hotspots affect a broad spectrum of abnormal molecular function including macromolecular transport and transcriptional regulation (Table 1, Supplementary Note, and Supplementary Fig. 8), as well as essential components of key signaling pathways.
Figure 4. Candidate Ras-related small GTPase driver mutations in the long tail.
a) The frequency distribution of hotspot mutations in cancer has a long right tail of mutated residues that while recurrent, are not common in any cancer type. b) There is a statistically significant difference in the pattern of Q61 codon mutations in KRAS, HRAS, and NRAS (χ2 p-value = 0.016). c) The sequence of Gly60-Glu62 of KRAS, HRAS, and NRAS are shown along with mutant alleles from affected cases indicating the GQ60GK dinucleotide mutation was the only source of KRAS Q61K mutation, whereas the far more common HRAS and NRAS Q61K mutations arose almost exclusively from single nucleotide events. The KRAS G60G synonymous mutation also creates a G60 codon in sequence (ACC>TCC) identical to wildtype sequence of NRAS G60, where Q61 mutations are the most common. d) RAC1, RRAS2, and KRAS are shown in schematic form indicating the position of novel hotspots RAC1 A159V and RRAS2 Q72L/H at paralogous residues in the Ras domain to known activating mutations in KRAS (A146 and Q61 respectively). e) The pattern of RAC1 (left) and RRAS2 (right) mutations along with those in BRAF and Ras genes in affected tumor types. f) RAC1 activation (GTP-bound RAC1) by PAK1 pull-down (right). RAC1 A159V was associated with significant RAC1 activation to levels equal to or exceeding the positive control GTPγS and greater than those of the known oncogenic RAC1 P29S.
Long-tail hotspots in Ras superfamily members
Mutations in the Ras family of small GTPases occur widely in human cancers. As expected, these were among the most significant hotspots detected here (Supplementary Table 2), affecting 1,335 tumors (12% of all cases). Whereas G12, G13, and Q61 codon hotspots predominate in KRAS, NRAS, and HRAS, albeit at varying frequencies in different tumor types (Fig. 2a and 3a), we also identified GQ60GK, K117, and A146 hotspots in KRAS. Both K117 and A146 are known activating hotspots in the long tail, but we also identified a previously occult GQ60GK dinucleotide substitution (q-value = 2.3×10−6) in 11 tumors. This dinucleotide substitution results in a Q61K mutation accompanied by a G60 synonymous mutation that are present in cis (in concomitant RNA sequencing, Supplementary Fig. 9). Although Q>K mutations at codon 61 can result from 3’G>T single-nucleotide mutations in KRAS, 100% of these tumors harbored the dinucleotide substitution, a rare spontaneous event in human genomes. Overall, the distribution of codon 61 mutations in KRAS, NRAS, and HRAS are very different, with Q>K mutations occurring significantly less frequently in KRAS (p-value=0.016; Fig. 4b). GA>TT mutations were the most common dinucleotide substitution producing GQ60GK (Fig. 4c) and converts the ACC codon at KRAS G60 to TCC, which is the sequence of the G60 codon in NRAS, in which Q61K mutations are far more common and arise nearly exclusively from single-nucleotide mutations. It remains to be determined whether KRAS GQ60GK is therefore driven by a pattern of codon usage at the −1 position. Notably, only one tumor had evidence of a non-KRAS GQ60GK mutation, an NRAS-mutant cutaneous melanoma (Fig. 4c and Supplementary Table 5).
We next explored whether KRAS GQ60GK may serve as a driver of Ras pathway activity as do conventional KRAS hotspots. GQ60GK is indeed present in diverse tumor types that all have well-established Ras-driven subsets (Supplementary Table 5). Reasoning that if GQ60GK were a passenger mutation in Ras-driven tumors, alternative MAPK activating mutations may be present in these tumors. Instead, we found that in every GQ60GK-mutant sample where another putative driver of MAPK signaling was present, that lesion was either 1) subclonal, defining a different clone than did GQ60GK; 2) low activity; or 3) a passenger mutation (Supplementary Table 5). Also, despite the frequency of GA>TT, there was no evidence that a common underlying mutational process or exogenous mutagen was the source of GQ60GK. There was no evidence of UV light exposure in the clinical histories or nucleotide contexts of most affected cases, only one of which was a cutaneous melanoma. Moreover, GQ60GK arose in both hypermutated (MSI-H colon lacking BRAF V600E) and non-hypermutated tumors. Finally, rare G60 missense mutations were evident in K- and HRAS in this dataset and in the literature (Supplementary Table 5)32. So, while we cannot exclude the possibility that the GQ60GK dinucleotide substitution is simply an alternative mechanism to achieve Q61K, the accompanying KRAS-specific G60 synonymous mutation may potentiate a different class of Q61-mutant tumors or cause signaling differences among Q61K-mutant tumors between K-N- or HRAS. Although further studies will need to explore the molecular properties of KRAS GQ60GK, this allele represents the most common dinucleotide substitution spanning two codons in human cancer and a mutation more common than other known hotspots in KRAS.
Novel long-tail hotspots were also identified in two other genes that encode members of the Ras superfamily of small GTPases. RAC1, in which we identified two hotspots, is a Rho subfamily member that plays a vital role in various cellular functions. RAC1 P29S is an oncogenic hotspot in melanomas12,33 that we also identified in head and neck and endometrial cancers (Fig. 4d). This mutation can confer resistance to RAF inhibitor treatment in vitro34, and may underlie early resistance in patients35. We also identified a novel RAC1 A159V hotspot present in 10 tumors (q-value = 2.27×10−6; Fig. 4d). Notably, RAC1 A159V is paralogous to KRAS A146, a known activating mutation36. Whereas activating KRAS A146T mutations arise predominantly in colorectal carcinomas (Supplementary Table 2), RAC1 A159V mutations are most common in head and neck cancers and were not present in any melanomas, despite the frequency of RAC1 P29S in this cancer type. Moreover, similar to P29S mutations, we observed RAC1 A159V mutations in tumors that are both Ras/Raf wildtype and mutant (Fig. 4e). To determine whether RAC1 A159V is an activating mutation, we assessed its effect in vitro. Active RAC1 is GTP-bound, interacting with PAK1 to activate downstream effectors. Therefore, to quantify RAC1 activation in vitro, we utilized a PAK1 pull-down assay. In HEK293T cells expressing RAC1 A159V, there was significant RAC1 activation to levels equal to or exceeding positive control RAC1 GTPγS cells and greater than even those levels induced by the known RAC1 P29S oncogenic mutation (Fig. 4f). Moreover, cells expressing RAC1 Q61R, a mutation we identified in a primary prostate cancer that is paralogous to KRAS Q61, also potently induced RAC1 activation (Fig. 4d,f).
RRAS2 is a Ras-related small GTPase37. RRAS2 is overexpressed or mutated in a small number of cancer cell lines of various origins38–40, and is oncogenic in vitro with transforming ability similar to established Ras oncoproteins41. However, it has not been documented as somatically mutated in human tumor specimens. Here, we identified a RRAS2 Q72 hotspot present in nine tumors (q-value = 8×10−15). Similar to RAC1 A159V, the RRAS2 Q72 hotspot is paralogous to KRAS Q61 (Fig. 4d). However, unlike RAC1, RRAS2 Q72 does not predominate in any individual tumor type. Also unlike RAC1, the RRAS2 Q72 mutation was present in Ras/Raf wildtype tumors among the affected types (Fig. 4e). This result suggests that RRAS2 activation may be an alternative avenue for tumors to acquire Ras-like activation as previous studies have shown that RRAS2 shares many Ras downstream signaling elements including phosphatidylinositol-3 kinase (PI3K)42,43, the Ral GDP dissociation pathway42, and Raf kinases44. Beyond these hotspots, several less common RAC1 and RRAS2 mutations affect paralogous residues of highly recurrent alleles in KRAS (Fig. 4d), some of which we validated were also activating in vitro (Fig. 4f), indicating that the landscape of potentially functional mutations in these genes extends beyond even these less common long-tail hotspots to private mutations as well.
Discussion
Although we focus only on recurrent substitutions, we did find that while a subset of hotspots were prevalent in individual cancer types, most hotspots are present infrequently across many cancer types. This indicates that studies of any individual cancer type may have limited power to identify novel alleles. We have also begun to detail best practices for the use of diverse public cancer sequencing data in the translational setting. Our approach for hotspot detection incorporates features such as the variable background mutational burden of individual codons and genes, thereby avoiding passenger mutations whose recurrence is due only to their presence in highly mutable amino acids. While the identification of private driver mutations remains challenging, our approach did uncover low-incidence hotspots in highly mutated genes. Though less common, these hotspots are under selection and may confer important clinical phenotypes in cancer patients, such as exceptional responses to cancer therapy30,31.
New mutant alleles in established genes are likely to emerge faster than new cancer genes are identified, extending the long tail of the frequency distribution of somatic mutations. This is especially true as clinical sequencing focuses on profiling advanced and metastatic disease for clinical trial enrollment. Such pre-treated, late-stage cases have been historically under-represented among such population-scale resources, including the one studied here. Moreover, at present there are fewer actionable mutations in cancer than there are cancer genes. Yet the near-term clinical utility of expanding the former is far greater. Our results suggest this will require an understanding of the function of different hotspot mutants in the same gene by lineage, as their function and response to therapy may be mutant amino acid specific. While positive selective pressure may produce the same hotspot mutation, or different variant amino acid changes within the same hotspot residue, it does not imply that they will confer similar selective advantages across lineages. Underlying functional distinctions may explain the differences observed here in the emergence and frequencies of hotspots across lineages. While this remains speculative or unknown for most hotspots, early evidence suggest that this will be true for even some of the most important alleles in human cancer8,9,11. Understanding this landscape of distinct molecular function is the necessary translational prerequisite for effective clinical implementation. This focus on mutations rather than genes will spur studies of the biochemical, biological, signaling impact, and drug sensitivity of candidate individual alleles. Collectively, the complementary study of both significantly mutated individual alleles as well as genes will prove indispensable in enabling precision oncology through clinical decision support for patients sequenced at the point of care.
Methods
Mutational dataset and pre-processing
Mutational data were obtained from three publically available sources: 1) The Cancer Genome Atlas (TCGA); 2) the data portal of the International Cancer Genome Consortium (ICGC); 3) various published studies in peer-reviewed journals in which mutational data was made available45,46. Mutation calling algorithms and mutation reporting practices varied from study to study in these curated data, so mutation data review and correction were undertaken where possible. Genomic coordinates of variants from alignments to human reference assembly NCBI36 (hg18) were converted to GRCh37 using LiftOver47 with an Ensembl chain file (see URLs). After standardization to GRCh37, the mutation calls were annotated to gene transcripts in Ensembl release 75 (Gencode release 19), and a single canonical effect per mutation was reported using Variant Effect Predictor (VEP) version 7748 and vcf2maf version 1.5. All possible pairs of any two samples with at least 10 somatic mutations were interrogated for sample duplication. For any pair of tumors that shared greater than 80% mutational identity and identical or near-identical clinic-pathological characteristics (upon review of data from the source site/publication), a single tumor in the pair was chosen at random and removed from further analysis as a presumptive duplicate specimen. Furthermore, we excluded small insertions and deletions (indels), despite their presence as true oncogenic hotspots in some genes, due to their greater variability in call quality across datasets. In total, the final dataset included 1,348,424 missense; 524,827 synonymous; 100,866 nonsense; 30,346 splice-site; and 3231 mutations affecting translational start or stop codons. There are also 21,130 oligo-nucleotide variants the majority of which are di-nucleotide mutations along with 71 tri-nucleotide mutations and 13 substitutions of 4bp or longer. Individual mutations and hotspots of interest (detected as described below) were inspected in individual BAM files from tumor and matched normal specimens of DNA and available RNA sequencing data downloaded from CGHub. When available, expression analyses were based on level-3 RNASeqV2 RSEM normalized gene expression counts from RNA sequencing available via the TCGA Data Coordinating Center. These values were log-transformed and scaled across all samples within each cancer type to facilitate comparisons between cancer types.
Definition of a mutational hotspot
For the purposes of this analysis, we first define a driver cancer gene as one in which a molecular abnormality leads to a fitness advantage for the affected cancer cell. This is the broadest definition that encompasses both initiating lesions on which tumor growth depends as well as lesions arising later in tumor progression that perhaps confer a more modest fitness advantage. We then define a hotspot as an amino acid position in protein-coding gene mutated more frequently than would be expected in the absence of selection. Therefore, all of the following mutation types result in the same hotspot: 1) mutations in different nucleotide positions in the same codon of a gene, 2) different nucleotide substitutions at the same site in the same codon that result in different amino acid changes, and 3) mutations where the amino acid substitution is identical but the nucleotide change are different. At present, this analysis is limited to recurrent somatic substitutions, but can be expanded to other classes of somatic alterations such as small insertions and deletions, DNA copy number alterations, and structural rearrangements.
Determining significant mutational hotspots
To determine the statistical significance of individual mutational hotspots, we developed a truncated binomial probability model by incorporating not only underlying features of mutation rates in cancer but also anticipating the gene-specific pattern with which hotspots may arise in different classes of possible cancer genes. In its most general form, if X represents the count of mutations in n samples, the probability of observing k mutations is:
| (1) |
where p is the probability of a mutation in any sample. However, differences exist in the mutability of specific nucleotide contexts in cancer genomes. These vary as a function of the underlying mutational process, potential molecular abnormality in normal DNA maintenance pathways, and possible exposure to exogenous mutagens16. Moreover, individual genes have highly variably nucleotide composition and background mutation rates. To address these fundamental characteristics, we integrated a coefficient into a position-specific probability that incorporates both the mutability of the trinucleotide context in which the mutation arose and the trinucleotide composition of the affected gene. For each of the 32 possible trinucleotides, we estimate the mutability of a given trinucleotide t as:
| (2) |
where Ct is the number of mutations affecting the central position of trinucleotide t across all samples and Ft is the number of occurrences of the trinucleotide t in the coding genome. Too little data existed to compute tumor type- or underlying mutational process-specific mutability. Because a mutated codon in a given gene is comprised of mutations in any one of three trinucleotides that encode that codon, we estimate the mutability of a codon c in gene g as:
| (3) |
where nt,c is the number of mutations in the central position of trinucleotide t in codon c and nc is the number of mutations in codon c overall. We estimate the mutability of gene g as μg = Cg/(nLg), where Cg is the number of mutations affecting the gene across the n samples and Lg is the length of the gene in amino acids. We then estimated the expected mutability of a given gene g as:
| (4) |
where Nt,g is the number of occurrences of trinucleotide t in gene g. The relative mutability of a codon within a gene is then rc,g = mc,g/mg. This leads to a binomial parameter for hotspot detection of:
| (5) |
We sought to avoid overestimating the background mutation rate for a gene with several hotspots. This would limit the detection of lower frequency hotspots (warmspots) due to the rate of recurrence of one or a few dominant hotspots in the same gene. We therefore developed a truncated form by removing positions in gene g bearing greater than or equal to the 99th percentile of all mutations in the gene. The new background rate is therefore , calculated as before where the prime signifies the mutation counts and lengths modified using the above threshold. Then . Finally, in rarely mutated genes where the probability p is exceedingly small (relative to the size of the cohort N and the length of the protein L), we limited the number of false positive hotspots by allowing to get no smaller than the 20th percentile of all p′ dataset-wide. Therefore, the final binomial probability is:
| (6) |
Accordingly, we calculate one-sided p-values for all unique amino acids in every annotated gene per the binomial form given in eq. (1) with probability from eq. (6) and test whether more mutations are observed than would be expected by chance given the pattern of all mutations in the gene; its composition and length; the pattern of its mutability; and the number and type of samples assessed.
Multiple hypothesis correction
To correct for multiple hypotheses, we employed a method for false discovery rate correction that assumes dependence among tests. This correction was performed on the gene level in the following manner. P-values were aggregated per gene on the basis of their codon position. For codons that were not mutated in a given gene and therefore not formally assessed, we padded this with a vector of p-values equal to 1 such that the final set of p-values equaled the amino acid length of the given gene. For all resulting p-values in each gene, they were corrected with the Benjamini and Yekutieli49 method (implemented in p.adjust in the stats package in R) and significant hotspots were those sites with q-values < 0.01.
Mutation call quality filtering
Considerable variability exists in the processing and generation of mutational data in individual cohorts by originating centers. To address this variability, we developed several weight-of-evidence based criteria for eliminating presumptive false positives and sequencing artifacts from individual mutation calls as well as from hotspots across the dataset (Supplementary Fig. 1b). Initially, to exclude likely germline variants misattributed as somatic mutations we exclude any mutation identified by both 1000genomes and the NHLBI or those identified only by 1000genomes in two or more samples. We then reasoned that hotspots arising in genes not expressed in a given tumor type are less likely to exert biological impact. We therefore removed from consideration hotspot mutations in genes whose expression was <0.1 transcripts per million (TPM) in 90% or more of the tumors of that type, or for tumors that lacked RNA sequencing data, if more than 95% of all tumors independent of organ of origin had expression of TPM < 0.1. After determining statistically significant hotspots (described above), hotspots were removed from consideration based on a decision tree model as follows. First, a presumptive true positive (pTPs) list of hotspots was predetermined as coding positions harboring substitutions in five or more tumor samples (from the August 2013 release of the cBioPortal45,46) in one of 341 key cancer-associated genes sequenced as part of routine CLIA-certified sequencing of matched tumor and normal specimens at Memorial Sloan Kettering Cancer Center50. Initially, for all samples in which a hotspot was observed and for which the fraction of tumor cells mutated could be calculated from corresponding variant allele frequency and DNA copy number data, we calculated the fraction of tumors in which that site was mutated subclonally (in fewer than 90% of tumor cells). If the fraction of samples in which the hotspot arose subclonally exceeded the maximum such value among pTPs, it was excluded. For remaining sites, we excluded potential hotspots that arose from mutation calling bias from a single source center. We identified cohorts in which subsets of samples were called by different centers and excluded hotspots in which greater than 85% of contributing mutation calls originated from a single mutation-calling center. Next, as local sequence complexity can affect alignment accuracy in various ways based on the read lengths and chemistry of source studies in our dataset, we sought to exclude hotspots on the basis of sequence context. We excluded hotspots where the minimum of Shannon entropy calculated from both 12bp or 24bp of flanking sequence on either the 5’ or 3’ side of the mutated site was less than the minimum such value among pTPs. We then excluded hotspots that were positioned at either the 5’ or 3’ end of a mono-, di-, or tri-nucleotide homopolymer runs of 10bp or longer. Remaining hotspots were then excluded if either the sum of their ranked weighted 100 and 24bp alignability (determined by CRG Alignability; UCSC Genome Browser) was less than the minimum value of pTPs or their weighted 24bp alignability was lower than the 12.5 percentile of all sites. We also excluded any hotspot that while passing these criteria affected a gene that was 1) already rich in presumptive false positives by these criteria [the number of retained hotspots was less than two times the count of hotspots in the gene excluded by one or more of these criteria] or 2) one of 20 well-characterized presumptive “red-herring” cancer genes due to high mutation rates that co-vary with underlying features independent of selection4. Finally, we manually inspected the sequencing data contributing to the mutation call for select hotspots in a sampling of affected tumor and matched normal samples. The significant hotspots (q-value < 0.01) that were excluded from consideration on the basis of this model are available in Supplementary Table 3.
Hotspot mutation data and literature review
In addition to the mutation call filtering described above, we independently validated the accuracy of a subset of mutation calls contributing to the novel hotspots discussed in the text. We downloaded BAM files of the aligned and unaligned sequencing reads corresponding to the tumor and matched normal exomes from each patient harboring one of the novel hotspots we discuss in detail in the manuscript (RAC1 A159; RRAS2 Q72; NUP93 E14 and Q15; MAX R60; and MAX H28) and reprocessed these from raw FASTQ to mutation calls with an independent sequence analysis pipeline30,31. We re-identified the hotspot mutation of interest in 34 of 35 affected specimens. Only a single sample failed to reproduce the published mutation (NUP93 E14 in a hepatocellular carcinoma), but NUP93 E14K remains statistically significant even after excluding the tumor in which the mutation was not called by the independent pipeline. To determine novel hotspots among the 470 identified here, we classified each mutation into one of three levels of evidence from an extensive literature review (Supplementary Table 2). Level-1 mutations are those not previously identified in human tumors or have been identified in an individual sample, but never described as a hotspot of recurrent mutation. This also includes mutations previously documented in the germline of patients, but never identified previously as recurrently somatically mutated. Level-2 hotspots are those mutations that have been reported previously in one tumor type that we also identified in the same, but also find mutated in one or more additional tumor types not previously described. Level-3 hotspots are those mutations that have been previously identified in one or more tumor types and have been assessed functionally in vitro or in vivo. Using this categorization, we identified 243, 46, and 181 levels-1, -2, and -3 mutations from the 470 hotspots described here. These were classified as being present or not in established candidate cancer genes (n=880, Fig. 1b) compiled from the Sanger Cancer Gene Census and four additional published sources1,4,5,50.
Cell type specificity analysis
We determined the enrichment of individual hotspots in different cell types (squamous versus non-squamous). For this analysis, squamous cell cancers included head and neck squamous cell carcinoma, lung squamous cell carcinoma, cervical squamous cell carcinoma, and esophageal squamous cell carcinoma. Hotspots mutated in at least one squamous tumor were examined and their statistical significance was assed with Fisher’s exact test assuming the null hypothesis that squamous and non-squamous tumor samples are equally likely to possess a given hotspot. Resulting p-values were corrected for multiple hypothesis testing with Benjamini and Yekutieli49 method and squamous cell-type specific hotspots were those with q-values < 0.01.
Analysis of the fraction of cancer cells mutated
Level-3 segmentation of DNA copy number data was acquired when available without restriction for 15 cohorts (primarily TCGA). Utilizing this data, we estimated tumor purity using absCN-seq51, due to the ease of its automation. We calculated the fraction of tumor cells bearing each mutation (tumor/cancer cell fraction) in each tumor based on this purity estimate, local copy number, and mutant and reference allele sequencing coverage all as previously described52.
Functional validation
DNA coding sequences for wildtype RAC1 as well as RAC1P29S, RAC1Q61R, and RAC1A159V were generated via site-directed mutagenesis (Genewiz, NJ) to include an N-terminal 3xFLAG epitope tag and were subcloned into a pcDNA3 mammalian expression vector (Life Technologies, NY). The expression constructs were transfected into HEK293T cells using Lipofectamine 2000 (Life Technologies), and cells were harvested after 72 hours. GTP-bound Rac1 (active Rac1) was isolated via immunoprecipitation using recombinant p21-binding domain (PBD) of PAK1 (PAK1-PBD; Active Rac1 Detection Kit, Cat#8815, Cell Signaling, MA), according to the manufacturer's instructions. The Rac1 was detected using kit provided Rac1 primary antibody.
Supplementary Material
Acknowledgements
We would like to thank the members of the Taylor and Schultz labs and members of the Marie-Josée and Henry R. Kravis Center for Molecular Oncology for useful discussions throughout this work. M.T.C was supported by the NIH training grant T32 GM007175 and A.B.O was supported by NIH grant P30 CA82103. This work has been further supported by an NIH Core Grant (P30 CA008748, PI: Thompson); the Josie Robertson and Prostate Cancer Foundations (N.S. and B.S.T.), the Sontag Foundation and Cycle for Survival (B.S.T.).
Footnotes
Data Access and Availability
Publically available sequencing data, CGHub, https://cghub.ucsc.edu/; Broad Institute Genome Data Analysis Center, http://gdac.broadinstitute.org/; Mutation curation and visualization, cBioPortal for Cancer Genomics, http://cbioportal.org/; Reference genome remapping, Ensembl chain file, ftp://ftp.ensembl.org/pub/assembly_mapping/homo_sapiens/NCBI36_to_GRCh37.chain.gz; Mutational data analyzed in this study and source code, https://github.com/taylor-lab/hotspots.
Author Contributions
M.T.C., S.A., and B.S.T. conceived the study. N.S. and B.S.T supervised analyses. M.T.C., S.A., N.D.S., A.B.O., and B.S.T developed methods and algorithms. M.T.C., J.S.C., J.G., C.K., and N.S. acquired data. M.T.C., S.A., J.S.C., S.P.G., B.H.L., J.G., and D.B.S. performed experiments and analyses. M.T.C., N.S., and B.S.T. wrote the manuscript with input from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Tamborero D, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Perez A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 8.Burd CE, et al. Mutation-Specific RAS Oncogenicity Explains NRAS Codon 61 Selection in Melanoma. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menzies AM, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012;18:3242–3249. doi: 10.1158/1078-0432.CCR-12-0052. [DOI] [PubMed] [Google Scholar]
- 10.Vivanco I, et al. Differential sensitivity of glioma-versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westcott PM, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2014 doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CS, et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat Genet. 2014;46:1060–1062. doi: 10.1038/ng.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaiswal BS, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23:603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung LW, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26:479–494. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 20.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu HA, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol. 2014 doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ihle NT, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garassino MC, et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol. 2011;22:235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 24.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research, N et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitehall VL, et al. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131:813–820. doi: 10.1002/ijc.26440. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grabiner BC, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Ahmadie H, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 2014;4:1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forbes SA, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson IR, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrhardt A, Ehrhardt GR, Guo X, Schrader JW. Ras and relatives--job sharing and networking keep an old family together. Exp Hematol. 2002;30:1089–1106. doi: 10.1016/s0301-472x(02)00904-9. [DOI] [PubMed] [Google Scholar]
- 38.Barker KT, Crompton MR. Ras-related TC21 is activated by mutation in a breast cancer cell line, but infrequently in breast carcinomas in vivo. Br J Cancer. 1998;78:296–300. doi: 10.1038/bjc.1998.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark GJ, Kinch MS, Gilmer TM, Burridge K, Der CJ. Overexpression of the Ras-related TC21/R-Ras2 protein may contribute to the development of human breast cancers. Oncogene. 1996;12:169–176. [PubMed] [Google Scholar]
- 40.Huang Y, et al. A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene. 1995;11:1255–1260. [PubMed] [Google Scholar]
- 41.Erdogan M, Pozzi A, Bhowmick N, Moses HL, Zent R. Signaling pathways regulating TC21-induced tumorigenesis. J Biol Chem. 2007;282:27713–27720. doi: 10.1074/jbc.M703037200. [DOI] [PubMed] [Google Scholar]
- 42.Rosario M, Paterson HF, Marshall CJ. Activation of the Ral and phosphatidylinositol 3' kinase signaling pathways by the ras-related protein TC21. Mol Cell Biol. 2001;21:3750–3762. doi: 10.1128/MCB.21.11.3750-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong R, He Q, Liu Y, Sheikh MS, Huang Y. TC21 mediates transformation and cell survival via activation of phosphatidylinositol 3-kinase/Akt and NF-kappaB signaling pathway. Oncogene. 2002;21:1062–1070. doi: 10.1038/sj.onc.1205154. [DOI] [PubMed] [Google Scholar]
- 44.Rosario M, Paterson HF, Marshall CJ. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 1999;18:1270–1279. doi: 10.1093/emboj/18.5.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods-only References
- 45.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 50.Cheng DT, et al. MSK-IMPACT: A hybridization capture-based next generation sequencing clinical assay for solit tumor molecular oncology. J. Mol. Diagn. 2015 doi: 10.1016/j.jmoldx.2014.12.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao L, Pu M, Messer K. AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.