Abstract
The structural features and dynamics of biological macromolecules underlie the molecular biology and correct functioning of cells. However, heterogeneity and other complexity of these molecules and their interactions often lead to loss of important information in traditional biophysical experiments. Single-molecule methods have dramatically altered the conceptual thinking and experimental tests available for such studies, leveraging their ability to avoid ensemble averaging. Here, I discuss briefly the rise of fluorescence single-molecule methods over the past two decades, a few key applications, and end with a view to challenges and future prospects.
Graphical Abstract
Introduction
Just two decades ago, most molecular biophysics studies used methods that interrogated an ensemble, i.e., millions or billions, of molecules. Results from these experiments typically pool information over the ensemble, providing mainly an “averaged” view of molecular properties. However, it has become increasingly evident that the molecular biophysics and biology underlying cellular function is not static or simple. Rather, proteins, DNA, RNA and other cellular macromolecules are in constant motion, resulting in complex spatial and temporal distributions in conformational states, dynamics, interactions and chemistry, many of which play key roles in the correct functioning of the cell. Many of these important details are lost to ensemble averaging.
More recently, single-molecule methods [1–3] have increasingly been included in the molecular biophysics toolkit, providing a powerful complement to ensemble methods of molecular biophysics such as NMR, crystallography, electron microscopy, spectroscopy and many others. Single-molecule methods leverage improvements in technology to provide the ultimate in molecular resolution, views of individual molecules. Therefore, they avoid averaging over an ensemble, and can provide unprecedented views of biomolecular complexity, including structural distributions and stochastic dynamics. Early single-molecule experiments using manipulation or force began in the 1980’s [1], while detection of a single chromophore was reported at the end of that decade [4, 5], enabled by advances in high-sensitivity detection instrumentation and other methodologies. Since then, the single-molecule field has grown enormously, impacting a number of areas of science, engineering, genomics [6] and medicine. One of the largest impacts has been in molecular biophysics, with novel mechanistic insights being gained into protein folding and disorder, functioning of cellular machinery such as involved in gene regulation and motor proteins, and enzyme and virus function. In the remainder of this review section, I discuss a few examples to highlight some of these developments focused on use of single-molecule fluorescence (SMF) methods in molecular biophysics (Figure 1), beginning with a brief introduction to some of the common methods, and end with a discussion of challenges and look to the future. I note that this short review can only provide very limited coverage of this exciting and rapidly expanding field. In addition, a number of interesting studies have utilized single-molecule force methods, such as with atomic force microscopy (AFM), and optical or magnetic tweezers. Additional information and avenues for further exploration of these and various other aspects of single-molecule biophysics can be found in these review articles and references therein [1–3, 7–9].
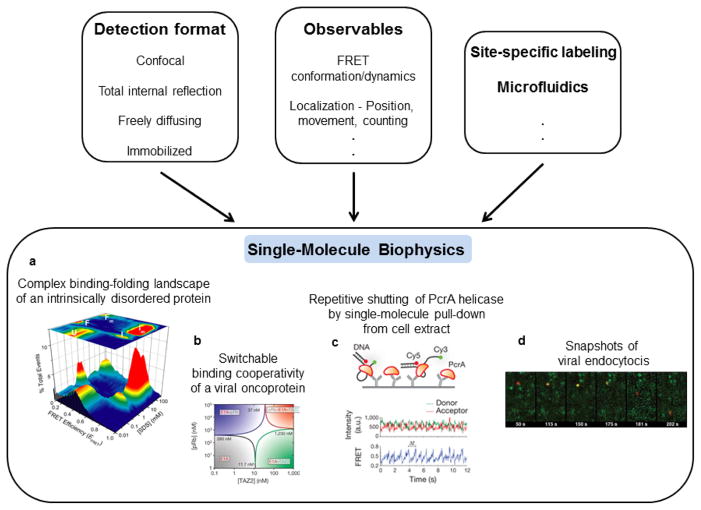
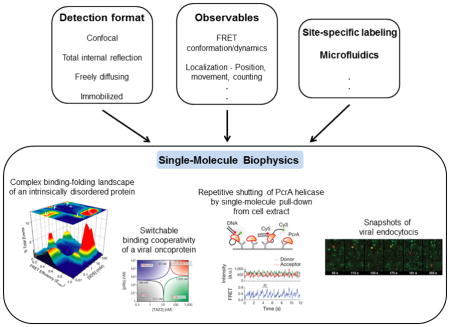
Figure 1. Methods and applications of single-molecule fluorescence.
Boxes at top show detection formats, observables and ancillary methods that are combined to carry out single-molecule biophysics experiments. Panels a-d illustrate some important types of information and questions that have been addressed using these methods. (a) The complex, multistate binding-folding landscape of α-synuclein in the presence of lipid/membrane mimics. Experiments were carried out using smFRET in a confocal and freely-diffusing detection format. Dual channel photon bursts were used to calculate FRET efficiencies, which were compiled into the depicted 3D histogram. Adapted from Ferreon et al. Proc. Natl. Acad. Sci. USA (2009) 106:5645. (b) Binding-folding phase diagrams of the viral oncoprotein E1A, found to be switchable as discussed in the text. Experiments were carried out as in (a), with subsequent analysis resulting in phase diagrams. Adapted from Ferreon et al. Nature (2013) 498:390. (c) smFRET time trajectory of PcrA helicase shows repetitive shuttling as discussed in text. Experiments were carried out with TIRF detection with immobilized DNA and pulling down PcrA from cell extracts. Adapted with permission from Jain et al. Nature (2011) 473:484. (d) Fluorescence time-trajectories of cell entry of individual virus particles, as discussed in text. Experiments were carried out in a wide-field imaging format, with virus particles red and clathrin green. Adapted with permission from Rust et al. Nat. Struct. Mol. Biol. (2004) 11:567.
Brief overview of common SMF techniques
Commonly used in SMF methods are either confocal or total internal reflection fluorescence (TIRF) microscopy detection formats. Lasers are used for excitation of dyes, and fluorescence is efficiently collected using high numerical aperture objectives, while minimizing background by use of the confocal or TIRF geometries. Detection in the confocal geometry is typically by avalanche photodiode detectors with time-resolution down to the ps timescale. Imaging can be carried out by scanning the sample. TIRF detection is typically imaging via CCD cameras, with time-resolution usually in the ms timescale, dependent on the fluorophores and imaging mode used. Experiments can be further classified into those on immobilized vs. freely diffusing or flowing molecules. The former permits longer trajectories to be measured for a given molecule but needs careful use of a non-perturbative immobilization technique, often a passivated quartz surface. Experiments on freely diffusing molecules are limited to shorter views of individual molecules, but avoid potential surface immobilization artifacts and permit higher throughput. Finally, several fluorescence observables can be used. A popular one is single-molecule FRET (smFRET), whose strong distance dependence on the 3–8 nm scale is useful for studies of biomolecular conformation and binding. Changes in fluorescence intensity and polarization can also report on structure, binding or enzyme activity. Histograms of these observables can provide information about distributions of states (Figure 1a,b) and time-trajectories can provide movies of molecular information (Figure 1c). The image of a single dye molecule is obtained as a diffraction-limited point-spread-function whose position can be localized to provide information on movement (Figure 1d) or superresolution imaging. Fluorescence correlation spectroscopy (FCS), a method that predates but is related to SMF, uses correlation analysis of fluorescence fluctuations from a small number of molecules to extract molecular information, such as about conformational fluctuations, with time-resolution down to ns or faster. Fluorescence lifetime analysis can also be used to study fast protein dynamics. Finally, as noted above, single-molecule manipulation methods have also been extensively used in studies of protein biophysics. For example, by using a sensitive cantilever whose deflection can be measured with high precision, AFM studies have measured forces and steps involved in protein folding or have imaged protein interactions. Additionally, by tethering molecules between a surface and a bead that can be manipulated using light or magnetic forces, numerous studies have measured protein folding pathways, protein motions on molecular tracks, or other protein functions. More detailed information about the different techniques and observables is available in these reviews [1, 9, 10].
Key applications
This section is organized under subheadings that correspond to two of the most important capabilities and application areas of SMF. First, I discuss several examples that illustrate how the absence of ensemble averaging allows for more direct measurements of complex conformational distributions and stochastic dynamics in the folding, interactions and movement of proteins and other macromolecules. Second, I briefly discuss examples of progress towards directly making single-molecule measurements in cellular contexts.
Complex conformational distributions and stochastic dynamics
The structural features and folding landscapes of proteins are critically important for their cellular functions. smFRET and other SMF methods have been applied in several proteins to directly resolve mixtures of states and dynamics during folding [11, 12]. The first reported single-molecule test of the distance measurement capabilities smFRET [13] was followed soon after by an application of the method to directly probe and confirm the two-state model for a well-studied protein CI2 [14]. More complex folding mechanisms have been studied since. For example, Pirchi et al. used smFRET and found a folding landscape with multiple intermediates for adenylate kinase [15]. Also noteworthy is that the authors studied protein molecules trapped inside immobilized vesicles, allowing them to study longer folding time-trajectories, without direct immobilization on a surface and consequent perturbation. In this context, a recent microfluidic method can also enable recordings of longer single-molecule time-trajectories without surface immobilization [16]. Exciting recent work reports [17] on folding transition paths that proteins take while transitioning between folding states. The transitions contain important mechanistic information about the state basins. However, they are stochastic and hence are not directly visible in ensemble experiments, and are normally analyzed as “jumps” even in single molecule protein folding trajectories. By using improved analyses, the authors found quite similar transition path times for two proteins with dramatically different overall folding times, consistent with a simple model with different barrier heights but similar chain diffusion coefficients and well curvature. A number of single-molecule studies have also provided new insights into conformational heterogeneity and folding of RNA molecules [18, 19].
A related exciting and recent application of SMF is to intrinsically disordered proteins (IDPs) [20–22], which have long stretches of sequence that are relatively devoid of secondary structure, with some IDPs being almost entirely disordered. About 30% of the human proteome has been estimated to contain disorder, and the special biophysics of these systems is believed to be important in a number of cellular functions. However, their flexibility and dynamic complexity make them more difficult to study by traditional biophysics tools. Therefore, the capabilities of SMF have been particularly useful for these proteins [23]. In 2007, Mukhopadhyay et al. used smFRET along with other analyses to study structural features and dynamics of the amyloid determining region of the yeast prion protein Sup35 [24]. They found that it is disordered and compact, and yet is rapidly fluctuating on the sub-μs timescale. Another advantage leveraged in this study is that single-molecule experiments are typically carried out at low sub-nM concentrations, and hence can be used to study monomeric states of aggregation-prone IDPs and other biomolecules. Subsequent studies by the Schuler group have explored the polymer physics of a few different IDPs using smFRET and fluctuation analysis, finding varied degrees of compaction and effects of internal friction [25, 26]. Additional layers of conformational complexity play important roles in IDPs as many undergo binding-coupled folding, again a good venue for single-molecule studies. In the case of the Parkinson’s disease linked IDP α-synuclein, Ferreon et al. obtained direct views of a complex, multi-state binding-folding landscape (Figure 1a) upon interaction with the lipid/membrane mimic SDS [27]. Gambin et al. developed a rapid microfluidic mixing method, and used it to find that the reaction occurs via encounter complexes and that substantial folding occurs after initial binding of α-synuclein [28]. Finally, in a more complex case, smFRET was used to probe binding of the viral oncoprotein E1A to two important cellular partners by Ferreon et al., here again leveraging low-concentration experiments to avoid problems of aggregation [29]. The authors found that availability of different sequence regions in E1A can exert complex layers of control and switching of the binding cooperativity of the system (Figure 1b), important in the function of signaling hub IDPs. Many of these exciting measurements and insights into IDPs would have been very difficult or virtually impossible to obtain using conventional ensemble methods.
One of the important areas of SMF application has been to cellular machines involved in gene regulation and transport. The three examples below illustrate the types of new insight available by using SMF to track movements of molecules. Myong et al. reported [30] on use of smFRET to track the motion of Rep helicase on DNA. Intriguingly, they observed that the helicase moved along single-stranded DNA till it hit a road block of duplex DNA, upon which it jumped back to the initial position on ssDNA and repeated the cycle. Yildiz et al. used the powerful localization capabilities of SMF to follow the stochastic movements of the motor proteins myosin V and kinesin on their molecular tracks, and found evidence for a hand-overhand walking mechanism for these proteins [31, 32]. Lee et al. used SMF in conjunction with DNA curtains (a large number of DNA molecules aligned side-by-side) to study how FtsK translocates along DNA, testing the particularly interesting (and biologically relevant) question of what happens where it runs into obstacles (other bound proteins) on the DNA [33]. Interestingly, they found that FtsK can bypass, push, or even dislodge other DNA-bound proteins.
SMF has also shed new light on enzyme chemistry. In the 1990s the groups of Yeung [34] and Dovichi [35] studied fluorescent product formation from single enzyme molecules, and discovered molecule-to-molecule variations in catalytic activity. A report in 1998 by Lu et al. [36] took a big step further by recording individual turnovers of single cholesterol oxidase molecules, imaging switching of fluorescence of a cofactor during the reaction. The authors found that in addition to differences between molecules, even individual molecules showed variations in turnover rates over time, which they ascribed to result from proteins switching between somewhat different folded substates. In another study, Noji et al. in 1997 reported measurements that directly visualized and confirmed the rotary motion of the protein F1-ATPase. Subsequent work by Itoh et al. strikingly demonstrated that reversing the rotation of the protein using optical tweezers could reverse the direction of this enzyme’s chemistry.
Overall, the above examples illustrate how SMF methods can resolve more directly several aspects of biomolecular complexity, including in structure, dynamics, interactions and movement.
Cellular studies
While most single-molecule studies have been performed in vitro with purified components, some studies have worked with cell extracts, moving the work closer to a cellular context, but maintaining advantages of in vitro studies. In one case, a single-molecule pull-down method was used by Jain et al. to study the helicase PcrA [37]. Labeled DNA was immobilized and used to pull down PcrA directly from unpurified cell lysate, and the authors were able to record repetitive shuttling activity of the protein on the DNA (Figure 1c). Another recent study [38] used SMF to study ubiquitilation by the anaphase-promoting complex/cyclosome (APC) from cell extracts, finding efficient polyubiquitination of substrates upon encounter with the APC and an interesting increase in affinity following initial ubiquitilation.
An increasing number of studies are utilizing SMF methods to study molecular processes directly in cells. A recent study reported on smFRET measurements of folding and conformational dynamics of proteins directly in live eukaryotic cells [39]. The proteins in this initial study showed characteristics similar to those in vitro, though future studies may show variations in other systems, for example due to interactions with cellular species. Other studies have used single-molecule tracking to study viral infection in cells, in one case showing that influenza virus particles may induce endocytosis by altering membrane curvature [40] (Figure 1d), and then follow a complex trajectory in the cell [41]. Another set of studies by multiple groups has used single-molecule imaging, tracking and counting methods, and revealed the stochastic nature of gene expression and that transcription occurs in bursts that may be important in regulation [42, 43]. More recent studies have even extended such studies to mammalian tissues [44], revealing bursty gene expression even in this context [45]. Another study developed a single-molecule method of fluorescence in-situ hybridization, revealing temporal and spatial patterns of 100s of RNAs in parallel in multiple cells [46]. Finally, a brief mention is made here of single-molecule based superresolution cellular microscopy methods which are discussed in more detail elsewhere in this issue. These methods are based on some of the same localization capabilities of single-molecule imaging that were mentioned above and previously used in the field, but applied here in a novel manner to enable new cellular applications. By compiling information from a number of images, each with a small number of stochastically activated fluorescent markers (of single molecules), and because these single-molecule images can be localized with high precision, these methods provide higher resolution biological imaging capabilities that overcome the diffraction limit.
Challenges and future directions
Single-molecule biophysics experiments could be considered by some measures as fairly established, with some of its powerful capabilities available commercially and being used in an increasingly wide range of studies. However, there is still much room for improvement, with initial steps being taken in many directions.
Fluorescence observables such as FRET, while providing single-molecule sensitivity, can be somewhat limited in time and spatial resolution. For example, trajectories measured in smFRET experiments generally have time-resolution of a few hundred microseconds, primarily due to the limited photon flux detected from a single fluorophore. Limitations in photon flux also reduce signal-to-noise and therefore distance resolution. Improvements in dye photophysics and detection modalities have begun to improve temporal resolution, for example, use of additives or covalently coupled moieties [47] to reduce photobleaching and dark states, or combinations with microfluidic deoxygenation [48]. Such efforts coupled with creation of improved dyes by rational (based on mechanistic photophysics) or high-throughput means are expected to improve resolution in multiple ways. Improved methods and orthogonal means for site-specific dye labeling will continue to be made, building on efforts such as unnatural aminoacid, enzymatic, ligation and synthetic schemes, thus increasing the scope of SMF methods in more complex systems and for multicolor smFRET [49–52]. These newer multicolor experiments can provide additional layers of information on correlations between changes in different parts of molecules or complexes, but would suffer dramatically from a lack of site-specificity, for example by commonly used cysteine chemistry. Combined use of multiple fluorescence observables, including for multicolor studies, will also substantially assist with providing more global high-resolution views of cellular systems [53].
Improvement and use of combination techniques and closer integration with ensemble studies is also expected to be another venue of advancement. Techniques combining SMF methods with force methods such as optical tweezers/AFM [52] or electrical methods such as ion channel or nanotube transistors [54], or with interferometric scattering [55] or microfluidics [28, 56] could substantially increase the level of insight by not only combination but also time-correlation of multiple lines of molecular information. For example, a recent study combined optical tweezers and smFRET to simultaneously apply force and monitor conformation of nucleosomal DNA, finding asymmetric unwrapping of the DNA that depended on the relative flexibilities of the two ends [57]. More generally, closer integration of SMF methods with other biophysics and structural biology methods such as NMR, EM, and crystallography, as well as computational analysis, will help provide more accurate views of biomolecules across a range of temporal and spatial resolutions [58].
Finally, one of most important areas of advance and application will most certainly be to molecules in mesoscale and cellular contexts and beyond. SMF methods are especially well-suited for such studies because fluorescence techniques are relatively non-invasive, and can also be used to probe even low endogenous levels of molecules in individual cells. For example, one area will be the use of SMF in understanding the structure and dynamics of protein phases such as droplets, where IDPs are believed to play a major role [59]. The functional role of such phases in the context of membraneless cellular organelles as well as protein aggregation is an area of increasing study, and SMF studies can provide new views into mechanisms of formation and dissociation, and conformational and functional heterogeneity of molecules within such species. In vivo studies will build on existing work in cells, but are expected to also improve in resolution, techniques used and scope, and to increasingly move to tissues and organisms. One of the most exciting directions envisioned is leveraging single-molecule capabilities to provide us with an understanding of how the single-molecule properties for ensembles of molecules can contribute to the emergent characteristics of cellular systems and organisms.
Highlights.
Single-molecule methods can provide unparalleled insights into biomolecular complexity, including structural distributions and stochastic dynamics
Commonly used single-molecule fluorescence techniques are overviewed
Key applications including complex conformational distributions, stochastic dynamics and cellular studies are discussed
The challenges and further improvements of single-molecule methods can require combining them with force methods or other biophysics and structural biology methods
Acknowledgments
The author gratefully acknowledges funding from the NIH/NIGMS (Grant RO1 GM066833) and NSF (Grant MCB 1121959).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: at the interface of biology, physics and chemistry. J R Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orrit M, Ha T, Sandoghdar V. Single-molecule optical spectroscopy. Chem Soc Rev. 2014;43:973–6. doi: 10.1039/c4cs90001d. [DOI] [PubMed] [Google Scholar]
- 3.Walter NG, Bustamante C. Introduction to single molecule imaging and mechanics: seeing and touching molecules one at a time. Chem Rev. 2014;114:3069–71. doi: 10.1021/cr500059w. [DOI] [PubMed] [Google Scholar]
- 4.Moerner WE, Kador L. Optical detection and spectroscopy of single molecules in a solid. Phys Rev Lett. 1989;62:2535–8. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- 5.Orrit M, Bernard J. Single pentacene molecules detected by fluorescence excitation in a p-terphenyl crystal. Phys Rev Lett. 1990;65:2716–9. doi: 10.1103/PhysRevLett.65.2716. [DOI] [PubMed] [Google Scholar]
- 6.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–11. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratmann SA, van Oijen AM. DNA replication at the single-molecule level. Chem Soc Rev. 2014;43:1201–20. doi: 10.1039/c3cs60391a. [DOI] [PubMed] [Google Scholar]
- 8.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat Struct Biol. 2000;7:724–9. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 9.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee PR, Deniz AA. Shedding light on protein folding landscapes by single-molecule fluorescence. Chem Soc Rev. 2014;43:1172–88. doi: 10.1039/c3cs60311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deniz AA, Dahan M, Grunwell JR, Ha T, Faulhaber AE, Chemla DS, et al. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc Natl Acad Sci U S A. 1999;96:3670–5. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deniz AA, Laurence TA, Beligere GS, Dahan M, Martin AB, Chemla DS, et al. Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci U S A. 2000;97:5179–84. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirchi M, Ziv G, Riven I, Cohen SS, Zohar N, Barak Y, et al. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat Commun. 2011;2:493. doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi S, VanDelinder V, Banterle N, Fuertes G, Milles S, Agez M, et al. Continuous throughput and long-term observation of single-molecule FRET without immobilization. Nature Methods. 2014;11:297–U358. doi: 10.1038/nmeth.2809. [DOI] [PubMed] [Google Scholar]
- 17.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–4. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitchiaya S, Heinicke LA, Custer TC, Walter NG. Single molecule fluorescence approaches shed light on intracellular RNAs. Chem Rev. 2014;114:3224–65. doi: 10.1021/cr400496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forman-Kay JD, Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–9. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brucale M, Schuler B, Samori B. Single-molecule studies of intrinsically disordered proteins. Chem Rev. 2014;114:3281–317. doi: 10.1021/cr400297g. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–54. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci U S A. 2012;109:16155–60. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soranno A, Buchli B, Nettels D, Cheng RR, Muller-Spath S, Pfeil SH, et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci U S A. 2012;109:17800–6. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A. 2009;106:5645–50. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gambin Y, VanDelinder V, Ferreon AC, Lemke EA, Groisman A, Deniz AA. Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat Methods. 2011;8:239–41. doi: 10.1038/nmeth.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–4. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 31.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–5. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 32.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–8. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 33.Lee JY, Finkelstein IJ, Arciszewska LK, Sherratt DJ, Greene EC. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Mol Cell. 2014;54:832–43. doi: 10.1016/j.molcel.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Q, Yeung ES. Differences in the chemical reactivity of individual molecules of an enzyme. Nature. 1995;373:681–3. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- 35.Craig DB, Arriaga EA, Wong JCY, Lu H, Dovichi NJ. Studies on single alkaline phosphatase molecules: Reaction rate and activation energy of a reaction catalyzed by a single molecule and the effect of thermal denaturation - The death of an enzyme. J Am Chem Soc. 1996;118:5245–53. [Google Scholar]
- 36.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–82. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 37.Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–8. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Wang W, Kirschner MW. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015;348:1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konig I, Zarrine-Afsar A, Aznauryan M, Soranno A, Wunderlich B, Dingfelder F, et al. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat Methods. 2015;12:773–9. doi: 10.1038/nmeth.3475. [DOI] [PubMed] [Google Scholar]
- 40.Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567–73. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A. 2003;100:9280–5. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–8. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, van Oudenaarden A. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protoc. 2013;8:1743–58. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- 45.Bahar Halpern K, Tanami S, Landen S, Chapal M, Szlak L, Hutzler A, et al. Bursty gene expression in the intact mammalian liver. Mol Cell. 2015;58:147–56. doi: 10.1016/j.molcel.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Q, Juette MF, Jockusch S, Wasserman MR, Zhou Z, Altman RB, et al. Ultra-stable organic fluorophores for single-molecule research. Chem Soc Rev. 2014;43:1044–56. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemke EA, Gambin Y, Vandelinder V, Brustad EM, Liu HW, Schultz PG, et al. Microfluidic device for single-molecule experiments with enhanced photostability. J Am Chem Soc. 2009;131:13610–2. doi: 10.1021/ja9027023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi S, Lemke EA. Single-molecule FRET and crosslinking studies in structural biology enabled by noncanonical amino acids. Curr Opin Struct Biol. 2015;32:66–73. doi: 10.1016/j.sbi.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 51.Gambin Y, Deniz AA. Multicolor single-molecule FRET to explore protein folding and binding. Mol Biosyst. 2010;6:1540–7. doi: 10.1039/c003024d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hohng S, Lee S, Lee J, Jo MH. Maximizing information content of single-molecule FRET experiments: multi-color FRET and FRET combined with force or torque. Chem Soc Rev. 2014;43:1007–13. doi: 10.1039/c3cs60184f. [DOI] [PubMed] [Google Scholar]
- 53.Kalinin S, Peulen T, Sindbert S, Rothwell PJ, Berger S, Restle T, et al. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat Methods. 2012;9:1218–25. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 54.Choi YK, Moody IS, Sims PC, Hunt SR, Corso BL, Perez I, et al. Single-Molecule Lysozyme Dynamics Monitored by an Electronic Circuit. Science. 2012;335:319–24. doi: 10.1126/science.1214824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrecka J, Arroyo JO, Takagi Y, de Wit G, Fineberg A, MacKinnon L, et al. Structural dynamics of myosin 5 during processive motion revealed by interferometric scattering microscopy. Elife. 2015;4 doi: 10.7554/eLife.05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polinkovsky ME, Gambin Y, Banerjee PR, Erickstad MJ, Groisman A, Deniz AA. Ultrafast cooling reveals microsecond-scale biomolecular dynamics. Nat Commun. 2014;5:5737. doi: 10.1038/ncomms6737. [DOI] [PubMed] [Google Scholar]
- 57.Ngo TT, Zhang Q, Zhou R, Yodh JG, Ha T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015;160:1135–44. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339:913–5. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee T, Moran-Gutierrez CR, Deniz AA. Probing protein disorder and complexity at single-molecule resolution. Semin Cell Dev Biol. 2015;37:26–34. doi: 10.1016/j.semcdb.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]