Abstract
CD1c is abundantly expressed on human dendritic cells (DC) and B cells, where it binds and displays lipid antigens to T cells. Here we report that CD1c tetramers carrying M. tuberculosis phosphomycoketide bind γδ T cell receptors (TCRs). An unbiased method of ligand-based TCR selection detects interactions only with Vδ1+ TCRs, and mutational analyses demonstrate a role of the Vδ1 domain during recognition. These results strengthen evidence for a role of CD1c in the γδ T cell response, providing biophysical evidence for CD1c-γδ TCR interactions and a named foreign antigen. Surprisingly, TCRs also bind CD1c complexes formed with diverse lipids such as lysophosphatidylcholine, sulfatide or mannosyl-phosophomycoketide, but not lipopeptide ligands. Dissection of TCR interactions with CD1c carrying foreign antigens, permissive ligands and non-permissive lipid ligands clarifies the molecular basis of the frequently observed but poorly understood phenomenon of mixed self and foreign antigen reactivity in the CD1 system.
Keywords: CD1c, Mycobacterium tuberculosis, T cells, T cell receptor, phosphomycoketide
Introduction
T cells are classified into αβ and γδ T cell lineages based on the T cell receptor (TCR) gene usage. Many γδ T cells are tissue resident and thus are thought to serve as a first line of defense in cancer surveillance, stress responses, microbial infection and tissue homeostasis (1). In broad terms, the rapid response to a few general stimuli byγδ T cells is contrasted with the delayed and more complex patterns of acquired responses to MHC-peptide (2). The evidence for γδ T cell involvement in immunity comes from work showing correlations with different disease states in the presence or absence of these cells, expansion of certain γδ T cell populations during infection and findings that γδ T cells are primary producers of IL-17, a potent cytokine for inducing early immune responses in infection and auto-immune disorders (1).
αβ T cells recognize cell-surface expressed, Major Histocompatibility Complex (MHC) proteins that have clefts that bind small (< 2 kDa) antigenic molecules. αβ T cells show one general mechanism for antigen recognition: their TCR α and β chains bind to the hybrid surface formed by antigenic peptides, lipids or vitamin metabolites, which are presented by classical MHC, CD1 or MR1 molecules, respectively (3–6). In contrast, the mechanisms of antigen recognition by γδ T cells are much less well understood, but appear to involve both soluble and cell surface bound targets using diverse molecular mechanisms. The known proteins involved in human γδ T cell stimulation include phycoerythrin as well as cellular proteins such as butryophilin3A1, soluble RNA synthetases and several MHC-class I like molecules, including MHC class I-related sequence A (MICA), UL16 binding protein (ULBP), endothelial protein C receptor (EPCR) and CD1c (7). Recently, Vδ1+ γδ T cells were shown to recognize a composite surface of CD1d in complex with lipid antigens (8–10). Thus, in contrast to early ideas that γδ T cells use their immunoglobulin-like TCR to recognize soluble antigens (11), evidence is rapidly mounting that at least one of the major types of human γδ T cells can recognize ligands that are produced, processed and displayed in antigen presenting molecules on antigen presenting cells (APCs).
The two most abundant and biologically important subtypes of human γδ T cells are Vδ1 and Vδ2, which are defined by the TCR δ variable region genes encoding the TCRδ chain, TRDV1 and TRDV2, respectively. These two subsets are considered to be largely non-overlapping in function as they differ in their tissue localization and antigen recognition. Vδ2 expressing Vγ9Vδ2 T cells, while broadly found in most human tissues, predominate in the blood, whereas Vδ1 T cells are mostly found in peripheral, mucosal tissues.
This study focuses on human CD1c proteins as candidate ligands for γδ TCR recognition. Most studies of CD1c have emphasized its role in theαβ T cell response, including display of unknown self antigens (12), tumor antigens (13), a synthetic lipopeptide known as acyl-12 (14) and mycobacterial lipid antigens (15). Recent studies have identified the mycobacterial cell wall lipid antigens known as mannosyl-β1-phosphomycoketide (MPM) and phosphomycoketide (PM) (15–17). Structural studies show that MPM and PM are presented by CD1c predominantly through accommodation of the methylated mycoketide tails in the A’ pocket of the CD1c molecule, with the phosphate or phosphomannose head groups extending out of the F’ portal to mediate interactions with αβ TCRs (17, 18). Focus on CD1c as a target of γδ TCR recognition derives from prior findings showing that individual T cell clones (IDP2, J2B7, JR.2.1 and XV.1.14) are activated when CD1c proteins are expressed on cells and on inhibition studies with anti-CD1c antibodies (19–22).
Although the response of γδ T cell clones to CD1c has been known for more than 25 years (21), characterized antigens and direct biophysical evidence for γδ TCR interactions with CD1c have been elusive. Human γδ T cells are highly abundant in tissues, where the full complement of activating receptors is presumably present, but a major and general hindrance to studying their antigen specificity has been that individual clones proliferate poorly in vitro. Recently CD1c tetramers were validated as reagents that bind CD1c-reactive TCRs (16). We reasoned that if CD1c is a physiological target and functions by binding γδ TCRs, then this new reagent could be used to enrich T cells from whole blood in the ex vivo state. Here we show that CD1c tetramers allow reproducible capture of γδ T cell clones from peripheral blood mononuclear cells (PBMC) of human donors. In all cases, the γδ T cells express the Vδ1 domain, and our binding studies show that the δ chain dominates the specificity of the response to CD1c-lipid complexes. After cloning four TCRs, we studied their patterns of response to CD1c-lipid complexes in detail and demonstrate that mycobacterial PM increases TCR affinity for CD1c, providing a named antigen for this system. For most models of TCR activation, the antigen presenting molecule and antigen are both necessary for activation. In contrast, we found that CD1c can also carry diverse self-ligands that permit TCR binding, or non-permissive ligands that block TCR binding. These data suggest a distinct mode of TCR discrimination in which TCR recognition is heavily biased towards CD1c itself with the lipid ligands playing a secondary role in modulating binding.
Materials and Methods
Source of lipids
MPM and PM were synthesized using two separate methods (23, 24), yielding identical molecules with the same biological properties. Lipo12 was synthesized as previously described (14, 18). LPA was purchased from Cayman Chemicals, Michigan, USA (catalog number 62215), LPC (catalog number 845875C) from Avanti Polar Lipids, Alabama, USA and mixed sulfatide from Matreya, Pennsylvania, USA (catalog number 1049).
Tetramer staining and flow cytometry
Phosphomycoketide-loaded CD1c tetramers were generated as previously described (16). For cell sorting and ex vivo tetramer staining, T cell enriched PBMC were blocked using human AB serum (Gemini) for 10 min at RT and washed. T cell lines were stained with APC-labelled or PE-labelled tetramer at 10 μg/ml in PBS containing 0.5% bovine serum albumin (FACS buffer) for 45 min at room temperature in the dark, and subsequently stained with phycoerythrin-labelled (PE) anti-TCR γδ (clone B1) or anti-TCRVδ1 (clone TS8.2, ThermoScientific) or Vδ2 (clone B6) for an additional 20 min at 4 °C. Additionally, γδ T cell lines were stained with PE-labelled anti-CD8 (clone HIT8a), FITC-labelled anti-CD4 (RPA-T4) anti-NKG2D (clone 1D11), anti-NKp44 (clone P44-8), anti-NKp46 (clone 9E2), or APC-labelled anti-NKp30 (clone P30-15) in FACs buffer for 25 min at 4 °C in the dark. Cells were washed in FACs buffer and collected on BD FACS Canto II (BD Biosciences). For blocking experiments, γδ T cell lines were incubated in the presence of increasing concentrations of anti-TCRδ1 (clone TCS-1, ThermoScientific) for 30 min at 37 °C, before addition of phosphomycoketide-loaded CD1c tetramers and staining as described above. Collected samples were analyzed using FlowJo software (Treestar Inc). All antibodies were purchased from BD Bioscience, eBiosciences or Biolegend unless indicated otherwise.
Generation of CD1c-restricted γδ T cell lines
PBMC were collected as discarded buffy collars during patient plateletpheresis at the Kraft Family Blood Donor Center at Dana-Farber Cancer Institute. PBMCs were separated by Ficoll density gradient centrifugation and enriched for T cells using a Dynabeads Untouched Human T Cells Primary negative selection kit (Invitrogen). After resting overnight, cells were stained with PM-loaded-CD1c tetramer and anti-CD3 (clone SK7). PM-loaded-CD1c tetramer+ CD3+ cells were sorted with a FACSAria flow cytometer. C32-PM CD1c tetramer binding cells were expanded polyclonally or by limiting dilution in the presence of an expansion mixture, consisting of irradiated PBMC, EBV-transformed B cells and 50 ng/ml anti-CD3 (clone OKT3) for two weeks. After an initial round of expansion, expanding wells were screened for C32-PM CD1c tetramer binding, and positive wells were screened for T cell receptor usage by staining anti-TCR αβ (clone WT31) or anti-TCR γδ (clone B1). PM-loaded-CD1c tetramer+ TCR γδ+ T cell lines were further enriched with anti-TCR γδ and sorted on FACSAria flow cytometer. All expanded lines were restimulated every other week with either expansion mixture or with Human T-activator CD3/CD28 Expansion and Activation beads (Invitrogen).
Expression and purification of TCRs in insect cell expression system
Standard RT-PCR with IMGT degenerate primer sets was used to determine the sequence of TCRs from the T cell clones. The variable ectodomains of γδ TCRs of CD1c-reactive T cell clones 12.9–2, 12.9–10, 12.16–3 and 22.4 were amplified from cDNA and fused with αβ TCR constant ectodomains using an overlapping PCR method. Reconstructed γδ TCR chains were modified to favor proper heterodimer formation by inserting T48C and S57C mutations in the α and β constant domains, respectively, and elimination of the wild type inter-chain disulfide cysteines (25). TCR chains were cloned in modified pACGP67 vectors with acid and basic zipper sequences for dimerization and recombinant baculoviruses were prepared in Sf9 cells. Baculoviruses with γ and δ chains were co-infected to express TCRs in Hi5 insect cells. TCRs were purified from insect cell supernatant as previously described (17).
Expression of CD1c and loading with lipid molecules
CD1c constructs have been previously described (17, 18). In brief, hybrid CD1c proteins were engineered for increased stability by swapping of the α3 domain for that of CD1b and fusing, using a glycine-serine linker, the β2m light chain to the N-terminus of the heavy chain. All constructs were expressed in Hi5 insect cells with the baculovirus expression system and purified as previously described. Lipids were loaded in CD1c through overnight incubation with lipids at 37 °C. CD1c was incubated with PM, MPM, LPC (Avanti polar lipids), LPA (Cayman chemicals) and mixed sulfatide (Matreya) at least 20–40 fold higher molar excess and purified with chromatography to remove excess lipids.
CD1c and TCR biophysical interaction analysis
All interaction measurements for TCRs with CD1c-ligands were performed using Biolayer Interferometry (BLI) with either Streptavidin or Ni-NTA sensors (Blitz, Forte bioscience). Hepes buffered saline (10 mM HEPES, pH 7.4, 150 mM NaCl) was used in all BLI measurements. In several cases BSA was added to the buffer to block non-specific interactions. Equilibrium analysis and dissociation constant (KD) calculation was done using GraphPad Prism. Dissociation constants (KD) were calculated with shared Bmax and identical immobilization of CD1c on the sensor in case of mycobacterial lipids and endogenous lipids. Binding analyses with MPM, endogenous lipids and 12.16-3-DP10.7 TCR are representative of one experiment; all other interactions were measured at least twice.
Jurkat transductions and activation assays
The full-length γ and δ chains of the 12.9-2, 12.9-10 and 12.16-3 TCR were cloned into the pMSCV-P2 and pMSCV-Z4 vectors with puromycin and zeocin resistant genes (a gift of M. Kuhns) to make retroviruses for transduction into TCRβ deficient Jurkat J.RT3-T3.5 cells. Since Jurkat J.RT3-T3.5 cells express CD1c on their cell surface, to measure CD1c-phosphomycoketide specific stimulation, PM was added directly to 5x103 stable TCR transduced cells in 96-well U-bottomed plates and incubated overnight. The JR.2 TCR J.RT3-T3.5 transductants were included in the assay as a control TCR (9), and Lipo12 antigen was also added to the TCR transductant as a control antigen. Activation was measured by CD69 expression (FN50, Biolegend). Cellular activation was measured twice for 12.16–3 TCR.
Results
Phosphomycoketide-loaded CD1c-tetramers identify antigen specific γδ T cells
Using CD1c tetramers loaded with phosphomycoketide antigens (CD1c-PM tetramers) (16, 17), we detected staining on the γδ TCR+ subpopulation of peripheral blood mononuclear cells (PBMC) (Figure 1A). Although detected at low frequencies in the peripheral blood, CD1c-PM tetramer/TCR γδ double positive cells could be sorted by flow cytometry, diluted and then expanded as oligoclonal lines. Using PBMC from two random donors (12, 22) we isolated four γδ T cell lines that were named X.Y, where X is the donor designation and Y denotes the well number(s) from which the line was derived (lines 12.9–2, 12.9–10, 12.16–3, and 22.4) (Figure 1B). At early stages of culture, the number of γδ T cells varied (1–65%), but were clearly detectable using a monoclonal antibody that recognizes all γδ TCRs, suggesting that CD1c tetramers allowed γδ T cell enrichment ex vivo.
Figure 1. Phosphomycoketide-loaded CD1c tetramer stains γδ T cells.
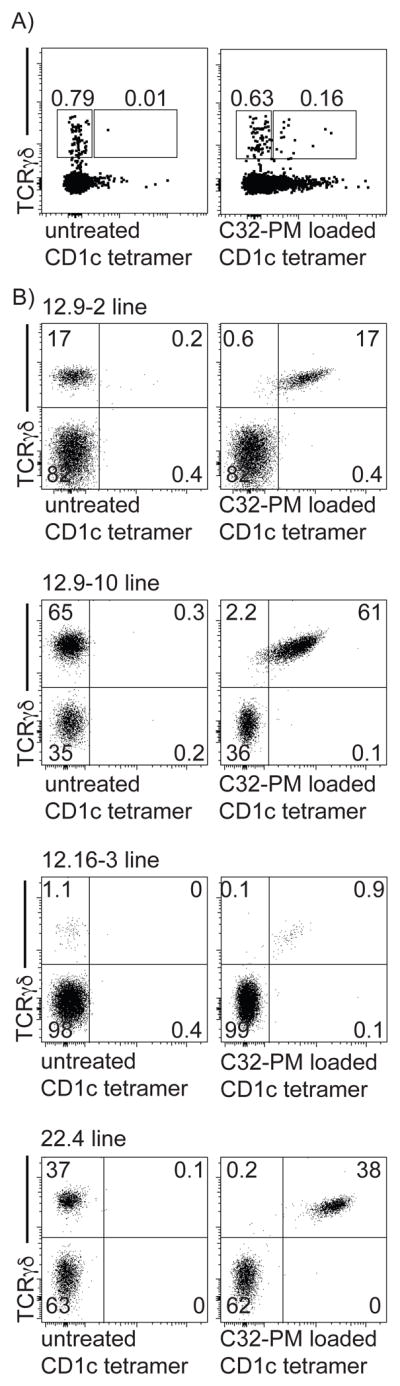
(A) CD1c tetramer staining of human PBMC. (B) After cloning at limiting dilution, oligoclonal T cell lines were expanded and stained using a pan-γδ TCR mAb and CD1c tetramers that were untreated or loaded with PM. Numbers indicate frequency of gated population.
Also, the study of oligoclonal lines at an early stage provided internal staining controls, which allowed the specific assessment of CD1c-PM tetramer versus γδ TCR staining (Figure 1B). In all cases γδ TCR+ but not γδ TCR− cells stained with the CD1c-PM tetramer. Few cells stained with untreated-CD1c tetramer, which presumably carries diverse lipids derived from the CD1c-expression system. CD1c tetramer staining for γδ TCR− cells was low or absent. Combined, these three observations validated CD1c tetramers as a reliable and reproducible means for isolating γδ T cells from PBMC and provided evidence for PM as a named lipid antigen for the CD1c-restricted γδ T cells. In all cases the CD1c-tetramer+ cells also stained with an anti-Vδ1 antibody, which, in agreement with previous anecdotal studies of individual clones derived by other methods (21, 22), suggested selective enrichment for CD1c recognition among the Vδ1 compartment of the human γδ T cell repertoire. The strong correlation of anti-γδ TCR and CD1c-PM tetramer staining was most consistent with binding of CD1c-PM to the TCR, a tentative conclusion that was more formally addressed in later experiments.
γδ TCR sequences
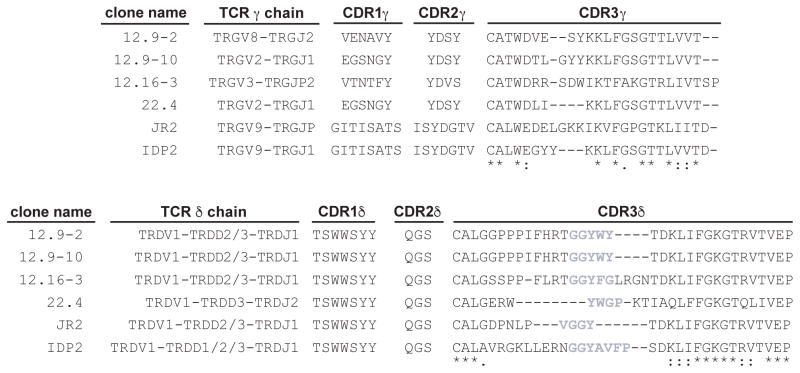
Using primer sets specific for γδ T cell receptor genes, we found only one γ and δ chain in each of the four lines, which were sequenced and compared to the previously published CD1c-specific lines JR2 and IDP2 (21, 22) (Figure 2). In all cases the Vδ domain was encoded by the TRDV1 variable region gene, which is consistent with staining results with an anti-Vδ1 antibody. In contrast to the conserved TRDV1 usage among these TCRs, different joining (TRDJ) genes were used to yield distinct CDR3δ sequences with the exception of the 12.9–2 and 12.9–10 TCRs, which have the same δ chain sequence. This finding was unlikely to have resulted from contamination, since no other δ chain sequence was found, and all lines expressed diverse γchains that were the product of different Vγ-Jγ rearrangements. The 12.9-2, 12.9–10 and 12.16–3 TCRs have long CDR3δ loops that are rich in proline and hydrophobic amino acid residues. These loops derive from Dδ segments TRDD2, TRDD3 and joining segment TRDJ1. In contrast, clone 22.4 has a comparably shorter CDR3δ loop and uses one Dδ segment, TRDD3, joined with the Jδ segment, TRDJ2. Thus, similar to CD1d-restricted γδ TCRs, the panel of CD1c-specific TCRs all express TRDV1, but otherwise have distinct clonotypic TCRs without observable sequence motifs.
Figure 2. TRDV1+ γδ TCR clones specific for CD1c.
Amino acid sequences of the CDR1, CDR2, and CDR3 loops and gene segments used in the γδ TCRs of the four clones examined in this study and previously identified clones JR2 and IDP2 (19, 21). An asterisk indicates a position of fully conserved amino acid residues; a colon indicates a position of amino acids with strongly similar properties. Grey highlights hydrophobic amino acids within the CDR3δ loop.
The γδ TCR directly binds CD1c-lipid
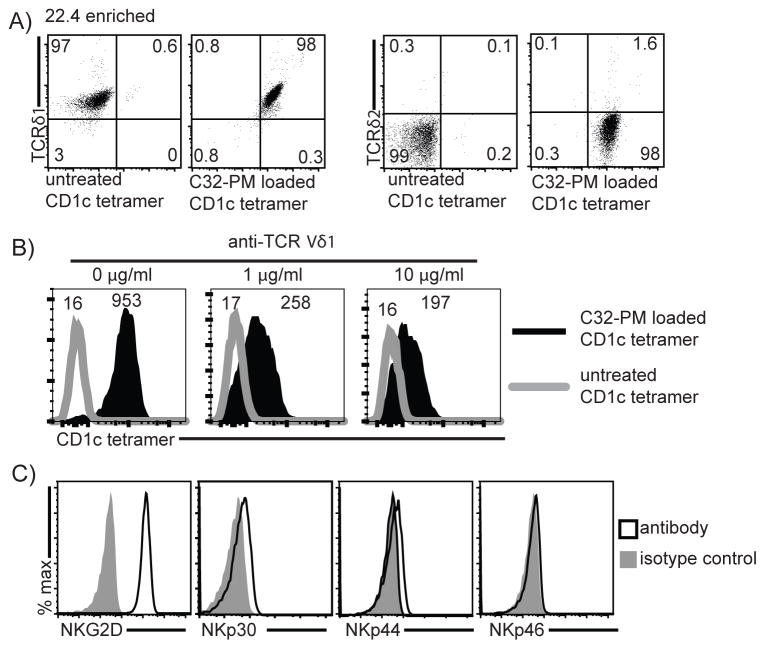
The structural basis of antigen recognition by γδ TCRs remains obscure and controversial. The simplest hypothesis to explain the γδ T cell response to CD1c+ APCs is direct binding of the TCR to CD1c-lipid complexes. CD1d recognition occurs by such a mechanism (9, 10), and antigen-loaded CD1 tetramers usually stain cells by binding to TCRs (Figure 1). However, CD1c-γδ TCR interactions have not been directly demonstrated or measured. Additionally, analysis of PBMC staining by CD1c-PM tetramers detects staining in the CD3− populations in some cases, raising the question of non-specific staining or the existence of CD1c ligands other than the TCR (16). In fact, CD1c has been shown to bind the Ig-like transcript 4 (ILT4) receptor (26) and thus could potentially be binding other cell surface receptors. To further assess the role of the TCR in CD1c recognition, we resorted and expanded the line 22.4 until it expressed nearly homogenous TRDV1+ TCRs (line 22.4 enriched) (Figure 3A). After treating this line with an anti-Vδ1 antibody, we observed a dose-dependent decrease in PM-loaded CD1c tetramer staining (Figure 3B). This preliminary finding suggested that the TCR binds to CD1c-lipid complexes, a finding that prompted further study of the TCRs role in binding CD1c through subsequent TCR cloning experiments described below. Line 22.4 expressed TCR co-receptors that are typical of most γδ T cells. The line was mostly double negative for CD4 and CD8 expression, with low expression of CD8 (data not shown). Further, 22.4 expressed a high density of the co-activating receptor NKG2D, but not NKp30, NKp44, or NKp46 (Figure 3C).
Figure 3. T cell receptor engagement by phosphomycoketide-loaded CD1c tetramer.
(A) After sorting the oligoclonal line 22.4 until anti-TRDV1+ expression was nearly homogenous (22.4 enriched), T cells could be reliably analyzed for known co-receptor expression. Numbers indicate frequency of gated population. (B) Pre-incubation with anti-TRDV1 inhibits binding of C32-PM-loaded CD1c tetramer, numbers indicate MFI of histogram. (C) Co-receptor expression of 22.4 enriched T cells.
Quantitative assessment of γδ TCR binding to CD1c-PM
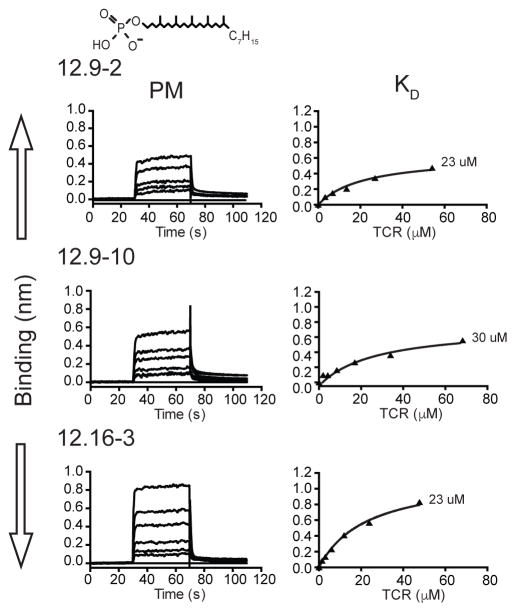
To formally and quantitatively establish a role of the γδ TCR in recognizing CD1c and PM, we expressed the extracellular domains of the 12.9–2, 12.9–10 and 12.16–3 TCRs in a baculovirus insect system and measured binding to CD1c-PM using Biolayer-interferometry (BLI) (Figure 4). We were unable to express the recombinant 22.4 TCR to adequate levels, so it was not included in this study. The CD1c used in this study was engineered for increased stability (18), and also expressed in the baculovirus insect system. The protein construct was biotinylated through a C-terminal Avitag and immobilized on a streptavidin sensor. All three TCRs bound to CD1c-PM complexes with similar measured affinity constants, measuring 23–30 μM. These values are within the physiological range for other TCRs binding to cellular antigen presenting molecules (27). More generally, these data provide direct evidence for human γδ TCRs binding to CD1c.
Figure 4. Binding analyses of human Vδ1+ γδ-TCRs to CD1c loaded with the phosphomycoketide, PM, using bio-layer interferometry (BLI).
BLI sensorgrams showing reference-subtracted binding (binding in nm) of CD1c-PM with increasing concentrations of the 12.9-2, 12.9-10 and 12.16-3 TCRs (top, middle and bottom panel, respectively). The 12.9-2 (concentrations used: 3.3–54.1 μM), 12.9-10 (2.0–68.25 μM) and 12.16-3 (1.5–47.75 μM) TCRs were flowed over sensor immobilized CD1c-PM. Associated equilibrium analysis fits and calculated dissociation constants (KD) are shown for each of the TCRs with CD1c loaded with PM (filled triangle) at right. The chemical structure of PM is shown above the sensorgrams.
The CDR3δ loop plays a key role in CD1c binding
Next we designed experiments to map the portions of the TCR involved in binding to CD1c. These studies were guided by the observation that all T cells reported here expressed the Vδ1 domain, and recent studies showed a bias towards the δ chain in governing γδ T cell recognition of CD1d (9, 10). Therefore, we focused on a likely role for the δ chain, which guided and simplified the design of loop-swapping experiments. Our previous studies showed that the DP10.7 and δ1A/B3 TRDV1+ TCRs bind to CD1d (9), so they were compared to the CD1c-reactive TCR 12.16–3. As shown in the aligned CDR3 sequences in Figure 5A, these TCR δ chains provide the equivalent small (DP10.7) or large (δ1A/B3) deletions in the longer native CDR3 loop sequence of 12.16-3. Specifically, the DP10.7 CDR3δ loop retains four hydrophobic amino acids that are like the 12.16-3 loop sequence, but this loop lacks eight residues. The δ1A/B3 loop has a larger, twelve-residue gap, including the hydrophobic tetrad sequence. The underlying logic is that these natural TCRs, unlike mutants that we might design, are known to fold properly and pair with TCR γ chains to generate intact, folded heterodimers. Therefore, any observed loss of target binding is less likely related to known artifacts that occur in other types of protein construct design.
Figure 5. CDR3δ loop and γ chain swapped γδ-TCR mutants probe the role of these domains on CD1c binding.
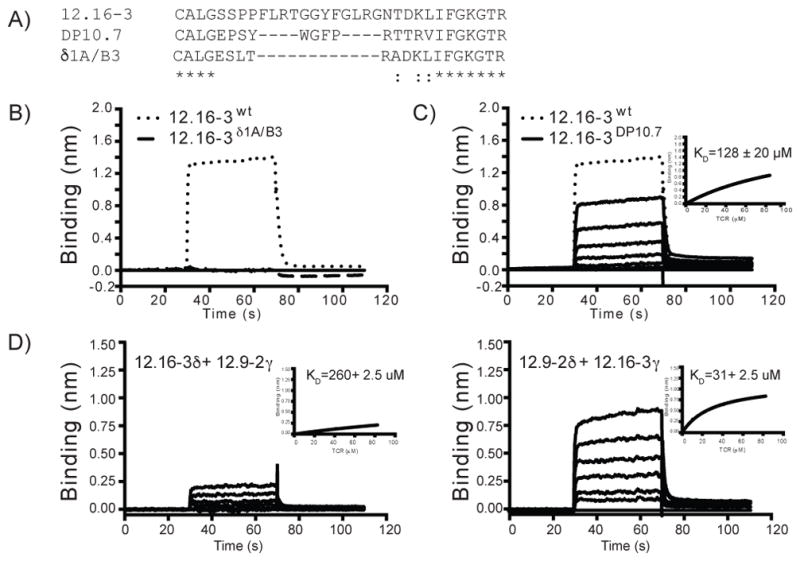
(A) Amino acid alignment of the CDR3δ sequences of the 12.16-3, DP10.7 and δ1A/B3 TCRs. An asterisk indicates position of fully conserved amino acid residues; a colon indicates position of amino acids with strongly similar properties. (B) BLI sensorgrams showing reference-subtracted binding (binding in nm) of CD1c-PM with 12.16-3 wild type (dotted line) and the CDR3δ swapped 12.16-3-δ1A/B3 mutant TCRs (dashed line). The 12.16-3 wild type and 12.16-3-δ1A/B3 TCRs are flowed at 84.0 μM and 120.0 μM concentrations, respectively. (C) BLI sensorgrams showing reference-subtracted binding of CD1c-PM with increasing concentrations of 12.16-3 wild type (dotted line) and the 12.16-3-DP10.7 mutant TCR (solid line). The 12.16-3 wild type and 12.16-3-DP10.7 TCRs are added at 84.0 μM and 2.6–84.0 μM, respectively. Equilibrium analysis fits and calculated dissociation constants (KD) are shown in inset panel. (D) BLI sensorgrams showing reference-subtracted binding of CD1c-PM with the chain swapped 12.16-3 and 12.9-2 TCRs as labeled. Concentrations for both the TCRs ranged from 2.57 to 82.5 μM. Equilibrium analysis fits and calculated dissociation constants (KD) for the chain swapped TCRs are shown in inset panels.
In agreement with this prediction, the 12.16-3 TCR framework with the CDR3δ loop sequences of the δ1A/B3 or DP10.7 TCRs expressed with high yield and were readily purified by size exclusion chromatography with a peak similar in size to the wild type 12.16-3 TCR, indicating stable protein (Supplementary Figure 1). These hybrid TCRs were then tested for binding to CD1c-PM, which showed complete loss of binding to the 12.16-3-δ1A/B3 fusion TCR (Figure 5B). Thus, the CDR3δ loop plays a key role in CD1c-PM engagement. In contrast, the 12.16-3-DP10.7 fusion TCR bound CD1c-PM with a low but detectable KD of 128 ± 20 μM (Figure 5C). This weaker affinity indicates a perturbation of TCR binding; however, the four hydrophobic residues (WGFP) present in the DP10.7 CDR3δ loop likely provide compensatory contacts, suggesting that the hydrophobic tetrad sequence present in the original 12.16-3 loop sequence may play an important role in CD1c-PM recognition. Overall, the strong bias for TRDV1+ TCRs in the TCR panel (Figure 2), as well as chain swaps independently point to a dominant role of the TCR δ chain in CD1c contact.
CD1c binding affinity is modulated by the TCR γ chain
In the ternary crystal structure of the DP10.7 TCR and CD1d-sulfatide, all TCR contacts with CD1d were mediated by the δ chain CDR loops, suggesting the γ chain played more of a stabilizing role for the TCR rather than providing important contacts for ligand recognition. To determine whether the γ chain is similarly dispensable in CD1c-PM recognition, we performed γ chain-swapping experiments with the 12.16-3 and 12.9-2 TCRs and compared them to the wild type TCRs. These two TCRs use different γ chains (12.9–2 uses TRGV8, or Vγ8, 12.16–3 uses TRGV3, or Vγ3) and also differ in their CDR3γ loop sequences. The chain swapped TCRs (12.9-2δ/12.16-3γ and 12.16-3δ/12.9-2γ) expressed with high yield and purified well, as evident by size-exclusion chromatography (Supplementary Figure 2). In binding experiments, the 12.9-2δ/12.16-3γ hybrid TCR bound CD1c-PM with an affinity similar to the wild type TCR (31 ± 2.5 μM) (Figure 5D). The lack of influence of the large γ-chain modification again pointed to a dominance of the δ chain in recognizing CD1c-PM. However, the reverse swapped, 12.16-3δ/12.9-2γ hybrid TCR, showed a dramatic reduction in binding (KD = ~260μM) (Figure 5D) demonstrating that for the 12.16-3 TCR, the γ chain contributes more to the binding affinity of CD1c-PM. This finding is consistent with the moderate differences in affinities observed between the 12.9-2 and 12.9-10 TCRs, which share the same Vδ1 domain sequence (including CDR3δ loop sequence), yet differ in Vγ chain usage (Vγ8 versus Vγ2, respectively). Curiously, the CDR3γ loop sequences are quite similar between these two TCRs, differing by only one insertion and three amino acid differences (WD*VESYK versus WDTLGYYK, Figure 2).
Cellular activation by phosphomycoketide antigen
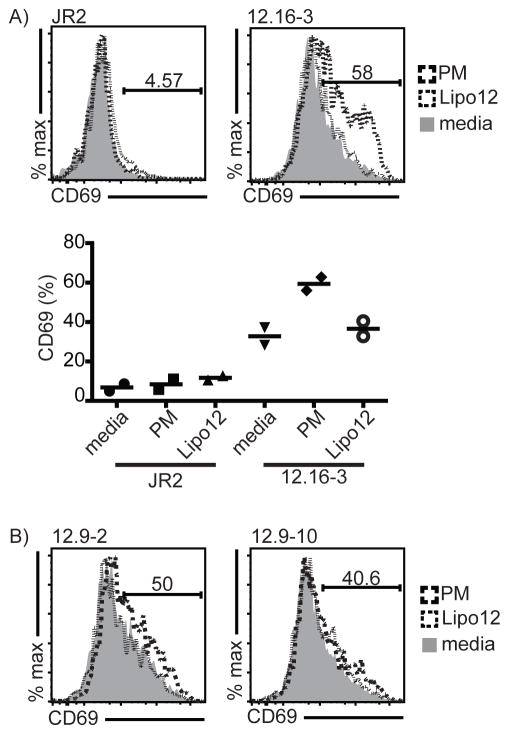
To determine if binding of CD1c-PM can lead to cellular activation, we transduced the 12.9-2, 12.9-10 and 12.16-3 TCRs into the β chain deficient Jurkat J.RT3-T3.5 T cell line. Clone JR.2, a CD1c-specific Vδ1 γδ T cell clone with unknown lipid specificity was included in the assay as a negative control TCR. Jurkat cells express both CD1c and CD1d at a basal level on the cell surface sufficient to mediate activation. Therefore, we directly added PM to the medium of transductants and incubated for 14 hours, similar to previous studies for NKT cells and δ/αβ T cells (28). Lipo12 was added in the medium as a negative control as γδ TCRs showed lower affinity for the lipopeptide antigen (see data below). The 12.16-3 transductants show a higher baseline for CD69 expression than JR2, indicating that these cells exhibit some autoreactivity towards endogenously presented Jurkat lipids, consistent with our binding studies to CD1c and unknown endogenous lipids (Figure 6A). Addition of PM caused upregulation of CD69 by 12.16-3, but not to JR2 (Figure 6A). Moreover addition of lipo12 did not up-regulate CD69 for either the JR2 or 12.16-3 transductants. Similar experiments performed with 12.9-2 and 12.9-10 TCR transductants yielded weak but detectable CD69 upregulation (Figure 6B) consistent with the much weaker binding affinity of these TCRs to CD1c-PM. These TCR transfer experiments confirm that direct recognition of CD1c-PM by the γδ TCR is sufficient to initiate T cell stimulation.
Figure 6. TCR transductants confirm TCR induced, lipid specific T cell activation specific to CD1c.
(A) Representative FACS plot of CD69 expression of the JR.2 (control) and 12.16-3 Jurkat transductants in presence of PM (dashed line), lipo12 (dotted line) and without any added antigen (shaded area). Cells are gated on CD3+ cells. Summary graph of two independent experiments showing percentage CD69 up-regulation shown at bottom. (B) Percentage of CD69 positive 12.9-2 and 12.9-10 Jurkat transductants in presence of PM, lipo12 and without any antigen.
γδ TCR binding to CD1c with altered ligands
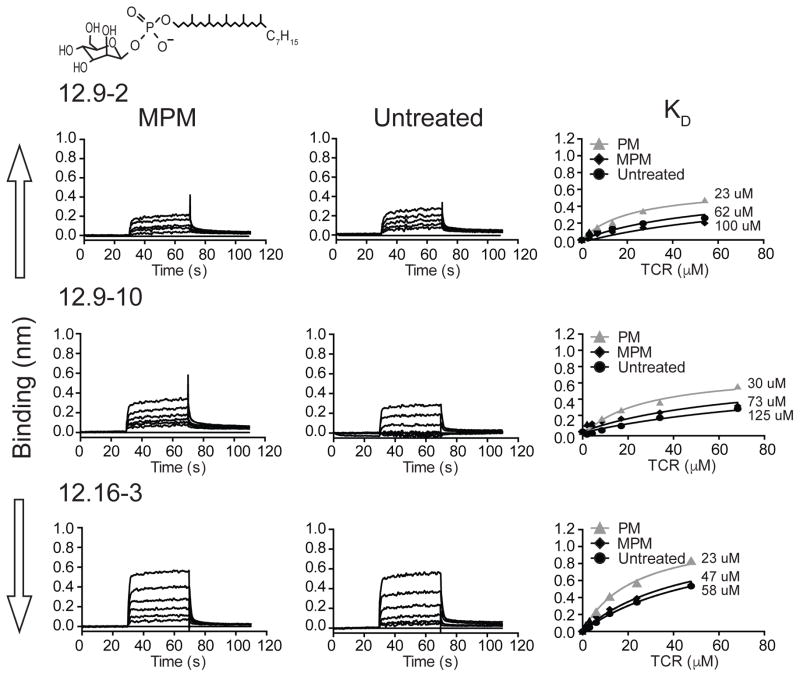
To further our understanding of TCR specificity, we loaded CD1c with MPM or used untreated CD1c proteins with endogenous lipids derived from the insect cell expression system. These were designed as negative controls based on the expectation that the TCRs would require MPM association with CD1c. However, we observed γδ TCR binding to both untreated CD1c and MPM-treated CD1c. Such results were highly surprising but considered reliable because similar results were seen with both types of altered ligands and among all three γδ TCRs tested, and binding was quantifiable and reproducible among experiments. The affinities of the three TCRs binding to CD1c-MPM, which ranged in KD from 47-100 μM, were approximately 2-fold weaker than that to CD1c-PM (Figure 7). TCRs binding to untreated CD1c proteins were also detectable but with weak affinity in the range of 58-125 μM (Figure 7). Untreated CD1c is not experimentally loaded with exogenous lipid, but carries lipids derived from the insect cell phospholipid membranes. Thus, while there exists a preference for PM, all three TCRs show detectable recognition of CD1c-lipid complexes composed of lipids that are different from the foreign phospholipid used to select the T cells. This conclusion suggests a broader recognition strategy in which CD1c is essential, and specific ligands influence but are not absolutely necessary for binding. The loading of phosphomycoketide antigens onto CD1c appears to be necessary for TCR binding measured in most tetramer assays (Figures 1, 3A) but not BLI assays (Figure 7). These apparently different results are likely due to the significantly higher concentration of monomer protein used in the BLI experiments and the higher sensitivity of BLI for detection of protein association over tetramer binding that reaches an avidity threshold. The potential for the heterogeneous loading of the tetrameric reagent, potentially with some lipid antigens that are non-permissive for TCR binding, could substantially reduce the avidity effect of the tetramer. Both methods suggest that TCR affinity or avidity is higher when phosphorylated mycoketides are bound to CD1c, but even weakly detectable binding to diversely liganded CD1c proteins suggests a large role of the CD1c protein in the binding interaction.
Figure 7. Binding analyses of human Vδ1+ γδ-TCRs to CD1c loaded with the mycobacterial lipid antigen mannosylated phosphomycoketide (MPM) and CD1c loaded with endogenous lipids using bio-layer interferometry (BLI).
BLI sensorgrams showing reference-subtracted binding (binding in nm) of CD1c-MPM and CD1c-untreated (CD1c protein is expressed in Hi5 cells thereby loaded with endogenous insect cell lipids) with increasing concentrations of 12.9-2, 12.9-10 and 12.16-3 TCRs. The 12.9-2 (3.3–54.1 μM), 12.9-10 (2.0–68.25 μM) and 12.16-3 (1.5–47.75 μM) TCRs flow over sensor immobilized CD1c-lipid complexes. Associated equilibrium analysis fits and calculated dissociation constants (KD) are shown for each of the TCR with CD1c loaded with MPM and untreated CD1c; curves determined in Figure 4 for CD1c-PM are shown in grey and grey triangles for reference. The chemical structure of MPM is shown above the sensorgrams.
Roles of antigens, permissive ligands and non-permissive ligands
In most studied examples of ternary complexes involving TCRs, antigen presenting molecules and antigens, especially those involving MHC proteins, high TCR specificity for the antigen is observed (27). However, broad TCR cross-reactivities to lipid ligands are increasingly being observed in the CD1 system (29). For example, two recent studies show that γδ T cells bind to CD1d, when it carries diverse self-lipids, chemically defined sulfolipids or α-galactosyl ceramides (9, 10). Like the three CD1c-specific γδ TCRs observed here, the CD1a autoreactive αβ TCR known as BC2 can bind CD1a in complex with diverse endogenous lipids (CD1a-endog) (30). Similarly, other αβ T cells recognize CD1b bound to self phospholipids with defined patterns of cross-reactivities (31). Thus, the pattern of mixed CD1 autoreactivity and antigen dependent reactivity observed here for CD1c is being increasingly observed in the CD1 system. To explain this widely observed but poorly understood phenomenon, an emerging model emphasizes that certain TCRs have high intrinsic affinity for portions of the outer surface of CD1 proteins that are not covered by lipid ligands (29, 32). Such TCRs can directly bind the outer surface of CD1 as long as lipid ligands, known as permissive ligands, do not block binding. Whereas this ‘absence of interference’ model is based mainly on studies of CD1a and CD1d, the detection of binding of several γδ TCRs to CD1c-MPM (Figure 7) fulfilled a key prediction of this new model. Further, the binding of TCRs to untreated CD1c complexes carrying heterogeneous insect lipids suggests that other permissive ligands likely exist.
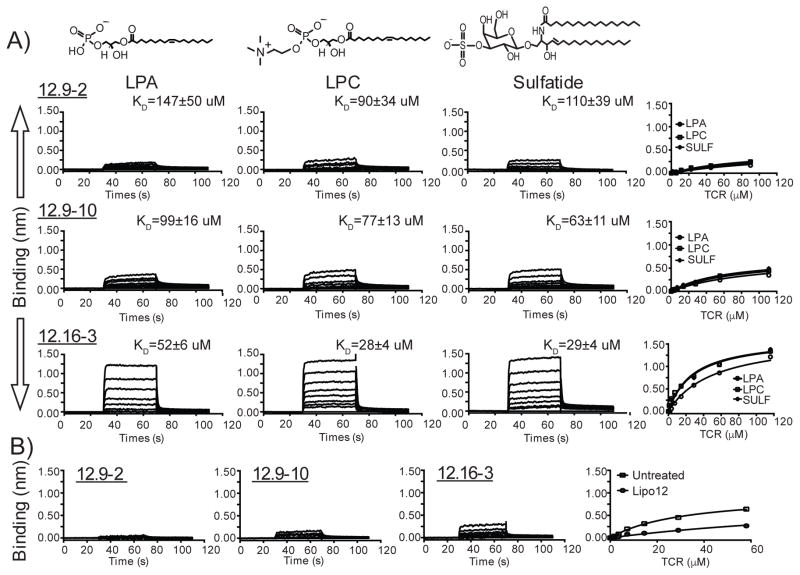
Therefore, we focused on known ligands of CD1c, measuring their effects on CD1c-TCR interactions in an attempt to define them as antigens that promote a response, permissive ligands that bind and do not affect the response, or non-permissive ligands that block a response. Lipo-12 is an acylated 12-mer peptide that is chemically unrelated to these phospholipids (14) but binds CD1c (18). Also, lysolipids in the phosphatidic acid family, including lysophosphatidylcholine (LPC) and lysophosphatic acid (LPA) are known CD1 ligands (33). Sulfatide is known to be bound and presented by all four human CD1 isoforms (34). Using the panel of recombinant TCRs, we measured binding to CD1c protein bound to LPA, LPC, sulfatide or acyl-12 (Figure 8). Loading protocols for LPA, LPC and sulfatide into CD1c were established by monitoring on Isoelectric Focusing (IEF) gels (Supplementary Figure 3), whereas protocols for loading of PM, MPM and lipo12 were previously determined (16, 18).
Figure 8. Binding analyses of human Vδ1+ γδ-TCRs to CD1c loaded with known endogenous lipids and lipo-peptide.
(A) BLI sensorgrams showing the reference-subtracted binding (nm) of CD1c loaded with the endogenous lipids lysophosphatidic acid (LPA), lysophosphatidylcholine (LPC) and sulfatide (left, middle and right panel, respectively) with increasing concentrations of the 12.9-2, 12.9-10 and 12.16-3 TCRs. The 12.9-2 (concentrations used: 1.4–90.0 μM), 12.9-10 (0.85–111.0 μM) and 12.16-3 (0.9–116.0 μM) TCRs are added as analytes over the CD1c molecule immobilized on the sensor. Associated equilibrium analysis fits and calculated dissociation constants (KD) are shown for each of the TCR with CD1c loaded with LPA, LPC or sulfatide. (B) BLI sensorgrams showing reference-subtracted binding of CD1c-lipo12 with increasing concentrations of the 12.9-2 (up to 45.0 μM), 12.9-10 (up to 55.5 μM) and 12.16-3 (0.9–58.0 μM) TCRs. In the far right panel, equilibrium analysis of 12.16-3 TCR binding to CD1c-lipo12 is shown in relation to that of untreated CD1c. Untreated CD1c is loaded with endogenous insect cell lipids. Chemical structures of LPA, LPC and sulfatide are shown above sensorgrams.
The 12.9-2 TCR binding to CD1c-LPA, CD1c-LPC and CD1c-sulfatide was weak but detectable with KDs of 147 ± 50, 90 ± 34 and 110 ± 39 μM, respectively. Similarly, the 12.9-10 TCRs bound all complexes with KDs ranging from 63 to 99 μM (Figure 8A). The 12.16-3 TCR also bound all three complexes but showed somewhat higher absolute affinity and some detectable discrimination of the bound lipid, recognizing CD1c-LPA, LPC and sulfatide with KDs of 52 ± 6, 28 ± 4 and 29 ± 4 μM, respectively (Figure 8A). Thus, LPA, LPC and sulfatide (Figure 8A) and MPM (Figure 7) are structurally diverse lipids with phosphate or sulfate head groups and one or two acyl chains, all of which allow TCR contact to CD1c. In contrast, the known CD1c-ligand, lipo12, can be considered a non-permissive ligand, because its loading into CD1c inhibits or blocks TCR binding in all cases.
The 12.16-3 TCR shows only trace binding to CD1c treated with lipo12 (KD = 143 ± 14 μM), which is reduced four-fold in comparison to untreated CD1c (KD = 32 ± 4 μM) (Figure 8B). For the 12.9-2 TCR and 12.9-10 TCRs, lipo12 suppressed TCR binding to trace or undetectable levels (Figure 8B). Thus, CD1c loaded with lipo-12 acts as a non-permissive ligand for γδ TCR interactions with CD1c.
Discussion
CD1c was the first ligand defined for γδ T cells (21), establishing that some γδ T cells require antigen presenting molecules for activation. Over the next 25 years, the phenomenon was confirmed with additional clones (19, 21, 22, 35), but more systematic interrogation of the CD1c-reactive repertoire has not been possible because tractable small animal models for CD1c are limited (36). Also, human γδ T cells, despite the large numbers present in vivo, often show anergic phenotypes (9, 10) and so are difficult to expand and study in vitro using conventional methods. Our results document that CD1c tetramers represent a new method for the rapid and reliable generation of human γδ T cell clones, which doubles the number of known TCRs responsive to CD1c. These resulting clones provide direct evidence for the involvement of theγδ TCR in binding CD1c as well as detailed molecular insights into the recognition mechanism, including the dominant role of the TCR δ chain and identification of PM as a characterized lipid antigen for the system.
Although the CD1c-reactive γδ TCR panel is not large, several clear patterns were observed, leading to several hypotheses that were formally ruled in using TCR gene transfer. First, using a method that does not require cytometric sorting on TCRs and therefore does not have any known mechanism for biasing TCR expression, all CD1c-reactive TCRs discovered here express Vδ1+ TCR δ chains. This agrees with Vδ1 expression of CD1c-reactive clones made by non-tetramer methods (19, 21, 22, 35), as well as Vδ1 expression by CD1d-reactiveγδ T cells (9, 10). Together these results provide strong correlative evidence that human CD1 proteins are a common, physiological target of the human Vδ1+ T cell repertoire. This correlative connection is further strengthened through controlled experiments in which genetic manipulation of native and altered TCRs demonstrate the essential role of γδ TCRs in CD1c recognition. These experiments also expose a bias towards CD1c binding to sequences in the TCRδ chain and specifically implicate a tetrad of hydrophobic residues in the CDR3δ sequence in binding CD1c. In contrast to earlier studies of individual clones, which used TRGV9 (Vγ9) in their TCR (19, 21, 22), these CD1c responsive γδ T cells conserve only the TRDV1 variable region gene. We demonstrated marked CDR3 diversity in the TCRs and found that TRDV1+ TCRs paired with members of divergent γ gene families (37): TRGV2 (Vγ2), TRGV3 (Vγ3) and TRGV8 (Vγ8). Swapping of the γ chains between TCRs demonstrated different patterns of bias of the δ chain in CD1c recognition.
A general question, which arises from and is partially answered by this TCR diversity study, is whether individual γδ clones, despite their sequence differences, have nearly identical antigen reactivities, or instead, differ and have even non-crossreactive patterns of antigen specificity. Because these clones were selected with one type of CD1c-lipid complex (CD1c- PM), non-crossreactive responses are not expected, but the in vitro TCR binding data document that sequence differences affect binding to CD1c-PM. The 12.9-2 TCR had a clear bias towards the δ chain in CD1c binding, as swapping of the γ chain had no substantial effect on affinity. In contrast, the binding affinity of the 12.16-3 TCR was drastically reduced when it was paired with an alternative γ chain, suggesting there is some contribution of γ loops to CD1c-PM binding in this TCR. This phenomenon is reminiscent of what was observed in the two structures of TRDV1+ TCRs in complex with CD1d; the DP10.7 TCR interacted with CD1d-sulfatide exclusively through the δ chain (9), whereas the 9C2 TCR had γ chain involvement in recognizing CD1d-αGalCer (10). Despite the shared use of the TRDV1 domain in these CD1-reactive TCRs, the apparently diverse pattern of γ chain usage in this small panel argues against one highly stereotyped recognition mode seen in T cell types that express more restricted repertoires of TCRs, such as in human invariant Natural Killer T cells (iNKT) (27, 38).
Finally, these data provide the first insights into ligand reactivity of CD1c-reactive γδ T cells. Whereas there is an increased affinity of binding to exogenous, microbial antigens such as PM, we observed weak binding to CD1c presenting endogenous lipid antigens from the insect expression system used to produce the recombinant protein, and also observed reactivity to known endogenous antigens presented by CD1c, including LPC, LPA and sulfatide. LPC and LPA both have one hydrocarbon tail and LPA shares the same headgroup structure as PM. All of these lipids showed weak to moderate binding to the γδ TCRs examined. In contrast, a lipid with a large, bulky headgroup, such as the lipopeptide, Lipo12, reduced binding significantly, suggesting that not all lipids are suitable for γδ TCR binding, either due to steric blocking or chemical differences in the headgroup. The measured autoreactivity may play a role in tissue surveillance, similar to what has been hypothesized for endogenous lipid-CD1d reactivity of Vδ1+ γδ T cells (7, 9). In this situation, “signal one” is mediated by CD1c-endogenous lipid/TCR interaction and it is complemented by a “signal two” provided by activating NK receptor/ligand interactions. NKG2D is such an activating NK receptor and is commonly found on Vδ1+ γδ T cells, as we have found here for the 22.4 T cell clone. The interaction of NKG2D with MICA expressed on tumor cells has been well documented for Vδ1+ γδ T cells (39).
Noting the similarities between the recently observed TCR binding to CD1a-endog (30) and the γδ TCR binding to CD1c-endog observed here (Figure 7), we tested some aspects of a new model for TCR recognition of CD1 (29, 30, 40, 41). This model derives from the unexpected finding that some CD1d- or CD1a-reactive TCRs, including BC2, have intrinsic affinity for CD1, as observed here for CD1c (Figures 4 and 7). In the BC2 TCR-CD1a complex, the crossreactivity to multiple lipids is explained by the fact that lipids emerge from the F’ portal on the right side of the protein, but the TCR binds CD1a closer to the A’ roof, on a region of the CD1a surface distal to where the lipid emerges. This structure provides proof of principle for a mechanism, which is absent in the MHC system, whereby the TCR contacts an ‘antigen-free’ region of the antigen presenting molecule, which plausibly provides a general mechanism by which many or most lipids, known as permissive ligands, permit TCR engagement of CD1. Certain rare ligands, known as non-permissive ligands, disrupt TCR binding by steric hindrance or alter the CD1 structure in ways that disrupt recognition of the CD1a binding epitope (29, 30, 40, 41). While the crystal structures of γδ TCR binding to CD1c remain to be elucidated, the structures of the CD1c-PM and CD1c-MPM complexes are known (17, 18). Like CD1a-lipid complexes, CD1c-lipid complexes form such that the phospholipid protrudes to the CD1c surface on the right side of the platform. Therefore a plausible basis by which the TCR could bind to the left of the protruding ligands is present and might represent a common theme in recognition of endogenous lipid ligands by CD1a- and CD1c-reactive T cells. Whereas classical models of antigen recognition emphasize all or nothing responses to rare foreign peptides, this model emphasizes graded responses of TCRs to the large cohort of CD1-lipid complexes on a cell.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grants R56 AI097386 and R01 AI073922 (to E.J.A.) and R01 AI049313 and R01 AR048632 (to D.B.M) and the Bill and Melinda Gates Vaccine Accelerator Grant.
The authors would like to thank Dr. Anne Kasmar for discussions on tetramer development and analysis.
References
- 1.Chien YH, Meyer C, Bonneville M. gammadelta T cells: first line of defense and beyond. Annual review of immunology. 2014;32:121–155. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 2.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nature immunology. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annual review of immunology. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- 5.McWilliam HE, Birkinshaw RW, Villadangos JA, McCluskey J, Rossjohn J. MR1 presentation of vitamin B-based metabolite ligands. Current opinion in immunology. 2015;34C:28–34. doi: 10.1016/j.coi.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annual review of immunology. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 7.Luoma AM, Castro CD, Adams EJ. gammadelta T cell surveillance via CD1 molecules. Trends in immunology. 2014;35:613–621. doi: 10.1016/j.it.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai L, Picard D, Anderson B, Chaudhary V, Luoma A, Jabri B, Adams EJ, Savage PB, Bendelac A. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. European journal of immunology. 2012;42:2505–2510. doi: 10.1002/eji.201242531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luoma AM, Castro CD, Mayassi T, Bembinster LA, Bai L, Picard D, Anderson B, Scharf L, Kung JE, Sibener LV, Savage PB, Jabri B, Bendelac A, Adams EJ. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity. 2013;39:1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uldrich AP, Le Nours J, Pellicci DG, Gherardin NA, McPherson KG, Lim RT, Patel O, Beddoe T, Gras S, Rossjohn J, Godfrey DI. CD1d-lipid antigen recognition by the gammadelta TCR. Nature immunology. 2013;14:1137–1145. doi: 10.1038/ni.2713. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Lebedeva MI, Llera AS, Fields BA, Brenner MB, Mariuzza RA. Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor. Nature. 1998;391:502–506. doi: 10.1038/35172. [DOI] [PubMed] [Google Scholar]
- 12.de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, Mori L, De Libero G, Dellabona P, Casorati G. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. European journal of immunology. 2011;41:602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 13.Lepore M, de Lalla C, Gundimeda SR, Gsellinger H, Consonni M, Garavaglia C, Sansano S, Piccolo F, Scelfo A, Haussinger D, Montagna D, Locatelli F, Bonini C, Bondanza A, Forcina A, Li Z, Ni G, Ciceri F, Jeno P, Xia C, Mori L, Dellabona P, Casorati G, De Libero G. A novel self-lipid antigen targets human T cells against CD1c(+) leukemias. The Journal of experimental medicine. 2014;211:1363–1377. doi: 10.1084/jem.20140410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Rhijn I, Young DC, De Jong A, Vazquez J, Cheng TY, Talekar R, Barral DC, Leon L, Brenner MB, Katz JT, Riese R, Ruprecht RM, O'Connor PB, Costello CE, Porcelli SA, Briken V, Moody DB. CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. The Journal of experimental medicine. 2009;206:1409–1422. doi: 10.1084/jem.20082480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 16.Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR, Jr, Adams EJ, Minnaard AJ, Porcelli SA, Moody DB. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. The Journal of experimental medicine. 2013;210:729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB, Adams EJ. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4648–4657. doi: 10.1073/pnas.1408549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spada FM, Grant EP, Peters PJ, Sugita M, Melian A, Leslie DS, Lee HK, van Donselaar E, Hanson DA, Krensky AM, Majdic O, Porcelli SA, Morita CT, Brenner MB. Self-recognition of CD1 by gamma/delta T cells: implications for innate immunity. The Journal of experimental medicine. 2000;191:937–948. doi: 10.1084/jem.191.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, Porcelli SA, Spinozzi F. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. Journal of immunology. 2007;178:3620–3626. doi: 10.4049/jimmunol.178.6.3620. [DOI] [PubMed] [Google Scholar]
- 21.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 22.Faure F, Jitsukawa S, Miossec C, Hercend T. CD1c as a target recognition structure for human T lymphocytes: analysis with peripheral blood gamma/delta cells. European journal of immunology. 1990;20:703–706. doi: 10.1002/eji.1830200336. [DOI] [PubMed] [Google Scholar]
- 23.van Summeren RP, Moody DB, Feringa BL, Minnaard AJ. Total synthesis of enantiopure beta-D-mannosyl phosphomycoketides from Mycobacterium tuberculosis. J Am Chem Soc. 2006;128:4546–4547. doi: 10.1021/ja060499i. [DOI] [PubMed] [Google Scholar]
- 24.Li NS, Scharf L, Adams EJ, Piccirilli JA. Highly stereocontrolled total synthesis of beta-D-mannosyl phosphomycoketide: a natural product from Mycobacterium tuberculosis. The Journal of organic chemistry. 2013;78:5970–5986. doi: 10.1021/jo4006602. [DOI] [PubMed] [Google Scholar]
- 25.Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein engineering. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- 26.Li D, Hong A, Lu Q, Gao GF, Jin B, Screaton GR, Xu XN. A novel role of CD1c in regulating CD1d-mediated NKT cell recognition by competitive binding to Ig-like transcript 4. Int Immunol. 2012;24:729–737. doi: 10.1093/intimm/dxs082. [DOI] [PubMed] [Google Scholar]
- 27.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annual review of immunology. 2014 doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 28.Pellicci DG, Uldrich AP, Le Nours J, Ross F, Chabrol E, Eckle SB, de Boer R, Lim RT, McPherson K, Besra G, Howell AR, Moretta L, McCluskey J, Heemskerk MH, Gras S, Rossjohn J, Godfrey DI. The molecular bases of delta/alphabeta T cell-mediated antigen recognition. The Journal of experimental medicine. 2014;211:2599–2615. doi: 10.1084/jem.20141764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Rhijn I, Godfrey DI, Rossjohn J, Godfrey D, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birkinshaw RW, Pellicci DG, Cheng TY, Keller AN, Sandoval-Romero M, Gras S, de Jong A, Uldrich AP, Moody DB, Godfrey DI, Rossjohn J. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nature immunology. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Rhijn I, van Berlo T, Hilmenyuk T, Cheng T-Y, Wolf BJ, Tatituri RVV, Uldrich AP, Napolitani G, Cerundolo V, Altman JD, Willemsen P, Huang S, Rossjohn J, Besra GS, Brenner MB, Godfrey DI, Moody DB. Human autoreactive T cells recognize CD1b and phospholipids. PNAS. 2015 doi: 10.1073/pnas.1520947112. published ahead of print November 30, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Born WK, O'Brien RL. Antigen-restricted gammadelta T-cell receptors? Arch Immunol Ther Exp (Warsz) 2009;57:129–135. doi: 10.1007/s00005-009-0017-x. [DOI] [PubMed] [Google Scholar]
- 33.Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR, Cook ME, Adams EJ, Hildebrand WH, Gumperz JE. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie DS, Vincent MS, Spada FM, Das H, Sugita M, Morita CT, Brenner MB. CD1-mediated gamma/delta T cell maturation of dendritic cells. The Journal of experimental medicine. 2002;196:1575–1584. doi: 10.1084/jem.20021515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasmar A, Van Rhijn I, Moody DB. The evolved functions of CD1 during infection. Current opinion in immunology. 2009;21:397–403. doi: 10.1016/j.coi.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazen AR, Adams EJ. Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E332–340. doi: 10.1073/pnas.1105105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 39.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 40.de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, Van Rhijn I, Pena-Cruz V, Ruan DT, Altman JD, Rossjohn J, Moody DB. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nature immunology. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nature immunology. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.