Abstract
Daily efavirenz 400 mg (EFV400) was virologically noninferior to 600 mg (EFV600) at 48 weeks in treatment‐naïve patients. We evaluated EFV400 and EFV600 pharmacokinetics (NONMEM v. 7.2), assessing patient demographics and genetic polymorphisms (CYP2B6, CYP2A6, CYP3A4, NR1I3) as covariates and explored relationships with efficacy (plasma HIV‐RNA (pVL) <200 copies/mL) and safety outcomes at 48 weeks in 606 randomized ENCORE1 patients (female = 32%, African = 37%, Asian = 33%; EFV400 = 311, EFV600 = 295). CYP2B6 516G>T/983T>C/CYP2A6*9B/*17 and weight were associated with efavirenz CL/F. Exposure was significantly lower for EFV400 (geometric mean ratio, GMR; 90% confidence interval, CI: 0.73 (0.68–0.78)) but 97% (EFV400) and 98% (EFV600) of evaluable pVL was <200 copies/mL at 48 weeks (P = 0.802). Four of 20 patients with mid‐dose concentrations <1.0 mg/L had pVL ≥200 copies/mL (EFV400 = 1; EFV600 = 3). Efavirenz exposure was similar between those with and without efavirenz‐related side effects (GMR; 90% CI: 0.95 (0.88–1.02)). HIV suppression was comparable between doses despite significantly lower EFV400 exposure. Comprehensive evaluation of efavirenz pharmacokinetics/pharmacodynamics revealed important limitations in the accepted threshold concentration.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
-
1
ENCORE1 demonstrated noninferior HIV suppression with EFV400 compared to EFV600 at 48 weeks in treatment‐naïve patients.
WHAT QUESTION DID THIS STUDY ADDRESS?
-
1
Pharmacokinetic parameters achieved with EFV400 and EFV600 once daily were estimated and patient characteristics and genetic polymorphisms assessed. Differences in pharmacokinetic parameters between doses and in patients experiencing adverse events were determined and associations at 48 weeks between pVL <200 copies/mL or adverse events with predicted pharmacokinetic parameters, dose, and polymorphisms were evaluated.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
-
1
Significantly lower EFV400 exposure (GMR (90% CI): 0.73 (0.68–0.78)) was not associated with pVL <200 copies/mL at 48 weeks (EFV400 = 97% vs. EFV600 = 98%; P = 0.802). Efavirenz‐related side effects were more common with EFV600 than EFV400 but not associated with plasma concentrations. Four of 20 patients with C12 <1.0 mg/L had pVL ≥200 copies/mL (EFV400 = 1; EFV600 = 3).
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY AND THERAPEUTICS
-
1
Efavirenz dose‐reduction retained good virological outcomes and reduced toxicities and has the potential to cut HIV therapy costs, allowing greater global coverage to patients. These data also challenge the validity of the widely accepted MEC (1.0 mg/L).
Approximately 10 million people receive antiretrovirals, although there is disproportionate global access to treatment.1 Current funds are unlikely to cover a planned scale‐up of treatment to 15 million by 2015.2 Exploration of strategies to lower drug costs may help attain this goal.
Daily therapy including efavirenz (600 mg) with tenofovir (300 mg) and emtricitabine (200 mg) or lamivudine (300 mg) is recommended as first‐line treatment for HIV‐infected individuals aged ≥3 years.3 Historical data suggest similar efficacy of lower efavirenz doses4 and ENCORE1, a multicenter, double‐bind, placebo‐controlled trial comparing reduced dose (400 mg once daily; EFV400) with standard dose efavirenz (600 mg once daily; EFV600) in treatment‐naïve, HIV‐infected adults showed that EFV400 was noninferior to EFV600 at 48 weeks (plasma HIV‐RNA (pVL) <200 copies/mL: 94% vs. 92%, respectively; modified intent‐to‐treat (ITT)).5 Based on 12 million receiving recommended first‐line treatment, a reduction to 400 mg efavirenz would save $16 per person in direct drug costs ($192 million worldwide per year).6
Various demographic and genetic factors impact efavirenz disposition.7, 8, 9, 10 Plasma efavirenz concentrations were associated with HIV suppression and toxicity when dosed 600 mg once daily.11, 12, 13 ENCORE1 provided an opportunity to examine such factors in a geographically and genetically diverse group of patients and to explore relationships between efficacy and safety outcomes with reduced dose efavirenz. We aimed to determine pharmacokinetic (PK) parameters of EFV400 and EFV600 and investigate the impact of patient characteristics and genetics by population PK modeling. Additionally, efavirenz PK/pharmacodynamic (PD) relationships in patients enrolled in ENCORE1 at 48 weeks were assessed.
RESULTS
Patients and sampling
Of 630 patients included in the ENCORE1 ITT,5 concentrations (n = 1,543) were available from 619 individuals (Figure 1). In total, 3% (n = 52) of samples were excluded (no recorded time postdose, time postdose >30 hours, concentration below assay lower limit of quantification (LLQ) or combination thereof). Overall, 1,491 samples (median (range) 2 (1–9) per patient; 1–3 occasions per patient) from 606 patients randomized to EFV400 (n = 311, 51%) or EFV600 (n = 295, 49%) were included. Forty‐six patients also participated in the intensive PK substudy (EFV400, n = 28; EFV600, n = 18) (Figure 1).
Figure 1.
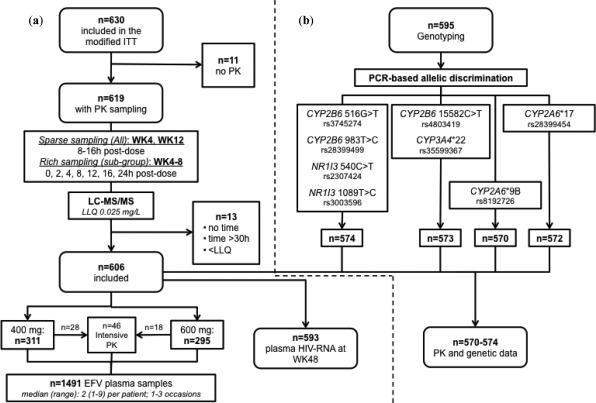
Flow diagram summarizing (a) the data included in the population PK model and (b) genetic data available for analysis. EFV: efavirenz; PK: pharmacokinetics; WK: week; LLQ: lower limit of qualification; ITT: intent to treat.
During the study, eight patients (n = 7 included in the model) commenced rifampicin‐containing tuberculosis (TB) therapy and switched to open‐label efavirenz throughout this phase. Four patients received rifampicin at weeks 4 or 12 (random sampling): two in EFV400 (one received 600 mg both weeks; one received 800 mg at week 12) and two in EFV600 (one received 800 mg both weeks; one remained at 600 mg). Sensitivity analyses compared model‐derived population parameters with and without patients on rifampicin; there were no differences in estimates and patients were retained in the analysis. Patient demographics and clinical characteristics are shown (Table 1).
Table 1.
Clinical characteristics and demographics of patients included in the model (n = 606)
| Parameter | Median (range)a |
|---|---|
| Female (n (%)) | 191 (32) |
| Age (years) | 35 (18–69) |
| Weight (kg) | 65 (39–148) |
| Height (m) | 1.68 (1.44–1.90) |
| BMI (kg/m2) | 23 (15–50) |
| FFM (kg) | 87 (41–206) |
| CrCL (ml/min) | 113 (54–264) |
| CD4+ T cell count (cells/mm3) | 270 (40–679) |
| HIV‐RNA at week 0 (copies/mL) | 56803 (162–10000000) |
| HIV‐RNA <200 copies/mL at 48 weeksb (n (%)) | 577 (97) |
| Randomised dose (n (%)) | |
| 400 mg efavirenz once daily | 311 (51) |
| 600 mg efavirenz once daily | 295 (49) |
| Ethnicity (n (%)) | |
| Aboriginal and Torres Strait Islander (ATSI) | 1 (0.2) |
| African | 226 (37) |
| Asian | 201 (33) |
| Caucasian | 76 (13) |
| Hispanic | 102 (17) |
| CYP2B6 516G>Tc (n (%)) | |
| GG | 253 (44) |
| GT | 262 (46) |
| TT | 59 (10) |
| CYP2B6 983T>Cc (n (%)) | |
| TT | 535 (93) |
| TC | 36 (6.0) |
| CC | 3 (0.5) |
| CYP2B6 15582C>Td (n (%)) | |
| CC | 320 (56) |
| CT | 222 (39) |
| TT | 31 (5.0) |
| CYP2A6 *9Be (n (%)) | |
| CC | 466 (82) |
| CA | 90 (16) |
| AA | 14 (3.0) |
| CYP2A6 *17f (n (%)) | |
| CC | 514 (90) |
| CT | 55 (10) |
| TT | 3 (0.5) |
| CYP3A4 *22d (n (%)) | |
| GG | 545 (95) |
| GA | 28 (5.0) |
| AA | 0 (0.0) |
| NR1I3 540C>Tc (n (%)) | |
| CC | 285 (50) |
| CT | 205 (36) |
| TT | 84 (15) |
| NR1I3 1089T>Cc (n (%)) | |
| TT | 153 (27) |
| TC | 277 (48) |
| CC | 144 (25) |
BMI: body mass index; FFM: fat free mass; CrCL: creatinine clearance.
Unless stated otherwise.
n = 593 available viral load measurements at week 48 (13/606 missing).
n = 574;
n = 573;
n = 570;
n = 572.
Genotyping
Blood samples for genotyping were available for 595 patients, 21 of whom were not included in the model (see Methods). Of 606 patients with PK data, 32 did not have a genotyping sample (Figure 1). Amplification failed in one, two, and four patients for CYP2B6 15582C>T and CYP3A4*22, CYP2A6*17 and CYP2A6*9B, respectively. PK and genetics were available for n = 574 for CYP2B6 516G>T, CYP2B6 983T>C, NR1I3 540C>T and NR1I3 1089T>C, n = 573 for CYP2B6 15582C>T and CYP3A4*22, and n = 572 and n = 570 for CYP2A6*17 and CYP2A6*9B, respectively (Figure 1). Genotype frequencies are summarized (Table 1); all were in Hardy–Weinberg equilibrium and this was also confirmed when stratified by ethnicity.
PK modeling
Efavirenz plasma concentrations over time are shown (Figure 2). A one‐compartment model with first‐order absorption best described the data, parameterized by apparent oral clearance (CL/F), apparent volume of distribution (V/F), and absorption rate constant (ka). Given the lack of absorption phase samples, ka was fixed to a literature value (0.6h‐1).14 Interindividual (IIV) and interoccasion variability (IOV) were estimated on CL/F and residual error described by a proportional model.
Figure 2.
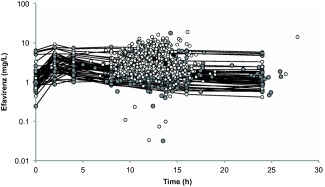
Efavirenz concentrations on a log‐linear scale following 400 mg once daily (gray), 600 mg once daily (white), and 800 mg once daily (black) dosing (n = 1,491 concentrations; n = 606 patients). Black lines connect the points of the full pharmacokinetic profiles of those patients with intensive sampling included in the pharmacokinetic substudy (n = 46 patients).
Following univariate analysis, CYP2B6 516G>T, CYP2B6 983T>C, and CYP2B6 15582C>T genotypes were significantly associated with CL/F (Supplementary Table 1). Weight and fat‐free mass (FFM) on CL/F and V/F produced a significant drop in objective function value (OFV); however, the change was greater for weight (ΔOFV –67.7 vs. –44.0). Ethnicity (Asian and African separately vs. Caucasian and Hispanic combined; CL/F decreased by ∼5% in Hispanics compared to Caucasians and combining in the model did not affect OFV) and sex on CL/F were also significant. Neither NR1I3 nor CYP3A4*22 single nucleotide polymorphisms (SNPs) improved model fit (Supplementary Table 1).
For multivariate analysis, inclusion of weight and CYP2B6 983T>C with CYP2B6 516G>T on CL/F was significant, but addition of ethnicity was not. Addition of CYP2A6*9B/CYP2A6*17 composite genotype consisting of wildtype (CC/CC) and carriers for either SNP or both (CC/CT or TT, CA or AA/CC, CA or AA/CT or TT; combined as such due to lower numbers for the variants) to CYP2B6 516G>T/983C>T composite genotype (GG/TT, GG/TC or CC, GT/TT, GT/TC or CC, TT/TT) produced a greater change in OFV than addition of CYP2B6 15582C>T, so was added into the model first. Addition of CYP2B6 15582C>T and sex did not improve the fit. Following backwards elimination, CYP2B6 516G>T/983T>C/CYP2A6*9B/*17 on CL/F and weight on CL/F and V/F were retained. For the composite genotype, those wildtype for both CYP2B6 SNPs with combinations of CYP2A6*9B/*17 were used as the reference genotype due to no change in CL/F between groups and given that the impact of CYP2A6 is more prominent in those without fully functional CYP2B614 (GG/TT/CC/CC, GG/TT/CC/CT or TT, GG/TT/CA or AA/CC, GG/TT/CA or AA/CT or TT); this did not significantly affect estimates. Overall, there were 16 genotype groups (Supplementary Table 1). IIV CL/F was decreased 15% by the addition of covariates, with the reduction largely from inclusion of CYP2B6 516G>T (↓9.5%) and CYP2B6 983T>C (↓3.5%). Grouping patients as extensive, intermediate, or slow metabolizers (see Methods) after the final model was obtained or as a final step of the modeling process did not influence the individual parameter estimates.
Final model parameters and diagnostic plots are shown (Supplementary Table 2 and Supplementary Figure 1, respectively). Compared to reference genotypes (11.9 L/h), efavirenz CL/F decreased by 4.5%–82%, depending on genotype group with typical population CL/F between 2.2 and 11.4 L/h for an individual weighing 70 kg (Supplementary Table 2).
Based on 1,000 simulated patients (51% EFV400) with the same distribution of bodyweights and CYP2B6/CYP2A6 genotypes as the original dataset and three sampling occasions per patient, 90% of observed concentrations were within the prediction interval. Stratified for dose, 90% and 91% of concentrations were within the prediction interval for EFV400 and EFV600, respectively, indicative of an adequate model (Supplementary Figure 2).
Secondary PK parameters
Efavirenz area under the concentration–time curve over 24 hours (AUC0‐24), maximum concentration (Cmax), trough concentration 24 hours postdose (C24), and concentration 12 hours postdose representing the mid‐dose interval concentration (C12) were significantly lower for EFV400 (Table 2). PK parameters are summarized, stratified by dose and metabolizer status (extensive, intermediate, and slow; Table 3). The proportion of patients with C12 below the recommended minimum effective concentration (MEC) of 1·0 mg/L12 for each group was similar between doses (P = 0.09, 1.00, 1.00, respectively; Fisher's Exact test); moreover, the number of patients with C12<MEC was low (n = 20). There was overlap in concentrations between groups but generally followed the trend of slow > intermediate > extensive (Figure 3).
Table 2.
Geometric mean (90% CI) of mean individual predicted PK parameters for 400 mg and 600 mg efavirenz
| Parameter | 400 mg EFV | 600 mg EFV | GMR (90% CI)a |
|---|---|---|---|
| AUC0‐24 (mg·h/L) | 49.2 (47.0–51.5) | 67.2 (63.8–70.9) | 0.73 (0.68–0.78) |
| Cmax (mg/L) | 2.52 (2.42–2.62) | 3.66 (3.51–3.81) | 0.69 (0.65–0.73) |
| C24 (mg/L) | 1.40 (1.32–1.49) | 1.82 (1.68–1.97) | 0.77 (0.70–0.85) |
| C12 (mg/L) | 2.10 (2.01–2.20) | 2.85 (2.70–3.0) | 0.74 (0.69–0.79) |
Differences in parameters were assessed by geometric mean ratios (GMR) and 90% CI (n = 605).
Efavirenz 400 mg/efavirenz 600 mg.
EFV, efavirenz; CI, confidence interval; AUC0‐24, area under the concentration‐time curve over 24 hour interval; Cmax, maximum concentration; C24, trough concentration, 24 hours postdose; C12, concentration 12 hours postdose representing the mid‐dose interval.
Table 3.
Mean individual predicted PK parameters stratified by dose and composite CYP2B6 516G>T/983T>C/CYP2A6*9B/*17 genotype (defined as extensive, intermediate, slow) predicted by the final model (n = 568 patients)a
| Efavirenz 400 mg once daily | Efavirenz 600 mg once daily | |||||
|---|---|---|---|---|---|---|
| Extensive (n = 119) | Intermediate (n = 128) | Slow (n = 48) | Extensive (n = 107) | Intermediate (n = 127) | Slow (n = 39) | |
|
CL/F (L/h) CV (%) |
10.7 (4.69–25.6) 33 |
8.05 (2.14–27.3) 37 |
4.12 (1.45–45.9) 124 |
12.4 (5.11–69.7) 55 |
8.93 (2.98–65.4) 62 |
3.55 (1.69–58.2) 155 |
|
AUC0‐24 (mg·h/L) CV (%) |
37.6 (17.6–85.4) 32 |
49.9 (14.8–187) 37 |
97.5 (20.7–285) 49 |
49.2 (8.61–117) 34 |
67.6 (12.4–202) 38 |
171 (19.2–359) 47 |
|
Tmax (h) CV (%) |
3.98 (3.35–4.22) 3.5 |
4.08 (3.28–4.32) 3.4 |
4.27 (3.11–4.39) 5.1 |
3.93 (2.23–4.23) 6.3 |
4.04 (2.64–4.32) 4.4 |
4.27 (2.74–4.38) 7.1 |
|
Cmax (mg/L) CV (%) |
1.97 (0.95–4.14) 28 |
2.51 (1.10–8.34) 32 |
4.38 (1.38–12.2) 45 |
2.80 (1.53–5.79) 27 |
3.71 (1.69–9.03) 32 |
7.80 (1.75–15.6) 42 |
|
C24 (mg/L) CV (%) |
0.970 (0.305–2.79) 44 |
1.46 (0.169–7.00) 48 |
3.54 (0.341–11.3) 56 |
1.28 (0.00247–3.94) 47 |
1.94 (0.0789–7.55) 51 |
6.24 (0.243–14.1) 52 |
|
C12 (mg/L) CV (%) |
1.60 (0.734–3.65) 32 |
2.14 (0.579–7.89) 37 |
4.13 (0.845–12.0) 49 |
2.11 (0.119–5.02) 35 |
2.90 (0.377–8.52) 38 |
7.25 (0.684–15.09) 47 |
|
Half‐life (h) CV (%) |
17.5 (7.71–37.0) 31 |
22.8 (6.09–69.1) 36 |
48.0 (8.78–152) 49 |
15.9 (2.13–40.4) 34 |
20.5 (3.70–68.6) 39 |
49.4 (4.92–125) 46 |
| C12<1.0 mg/L (n (%)) | 10 (8.4) | 2 (1.6) | 2 (4.2) | 3 (2.8) | 1 (0.79) | 2 (5.1) |
| VL≥200 copies/mL (n (%))b | 1 (0.85) | 6 (4.8) | 2 (4.3) | 2 (1.9) | 2 (1.6) | 2 (5.6) |
Data presented as median (range).
37/605 with missing composite genotype (n = 15 400 mg EFV; n = 22 600 mg EFV).
555/568 patients with a viral load at 48 weeks (n = 13 missing).
CL/F, apparent oral clearance; AUC0‐24, area under the curve over the 24‐hour dosing interval; Tmax, time of maximum concentration; Cmax, maximum concentration; C24, trough concentration, 24 hours postdose; C12, concentration 12 hours postdose representing the mid‐dose interval; CV, coefficient of variation; VL, viral load.
Figure 3.
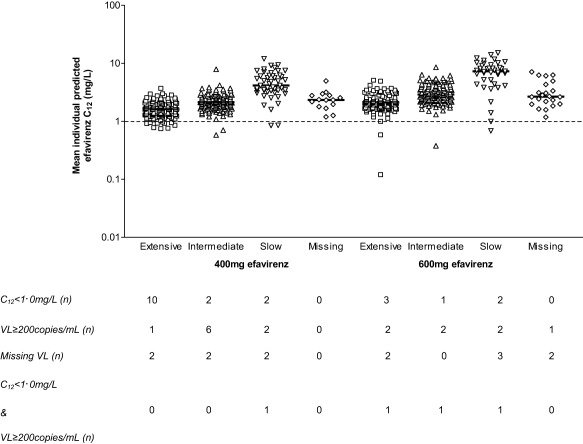
Mean individual predicted efavirenz concentrations at 12 hours postdose (C12) on a log scale stratified for metabolizer status (extensive, intermediate, slow) and dose (400 and 600 mg once daily; n = 295 and n = 273, respectively). The black dashed line illustrates the recommended minimum effective concentration for efavirenz (MEC) of 1.0 mg/L. Each point represents an individual patient and the solid black line the median concentrations. Numbers of patients with C12 below the MEC or with detectable or missing viral load at 48 weeks are shown. C12: concentration 12 hours post‐dose representing the mid‐dose interval concentration; VL: viral load.
PK‐PD analysis
At screening, 231 patients had pVL >100,000 copies/mL and 375 had pVL ≤100,000 copies/mL. At 48 weeks, 2% (n = 13) were missing and 577/593 patients (97%; 97% EFV400, 98% EFV600) had pVL <200 copies/mL and 3% (n = 16) had pVL ≥200 copies/mL.
Following univariate analysis, no categorical variables (dose, composite genotype, individual SNPs, screening pVL strata) were found to have significant relationships with pVL <200 copies/mL at 48 weeks (Supplementary Table 3). Following univariate logistic regression of PK parameters, mean individual predicted efavirenz logCL/F (odds ratio (OR) (95% CI): 0.037 (0.005–0.303), P = 0.002), logAUC0–24 (OR (95% CI): 17.56 (1.61–192.95), P = 0.019), logC24 (OR (95% CI): 7.53 (2.04–27.75), P = 0.002), and logC12 (OR (95% CI): 21.01 (2.94‐150.11), P = 0.002) were associated with pVL <200 copies/mL at 48 weeks, although confidence intervals (CIs) were generally wide.
Proportions of patients with pVL ≥200 copies/mL at 48 weeks stratified by metabolizer status was similar between doses (P = 0.604, 0.172, 1.00, respectively; Fisher's Exact test, Table 3; Figure 3).
Of 14 and six patients with C12<MEC for EFV400 and EFV600, one and three had pVL ≥200 copies/mL at 48 weeks, respectively (Figure 3). Composite CYP2B6 516G>T/983T>C/CYP2A6*9B/*17 genotype was TT/TT/CC/CC (slow), GG/TT/CC/CC (extensive), GT/TT/CC/CC (intermediate), and GG/TC or CC/CA or AA/CC (slow), respectively, for these four patients. The ranges of C12 stratified by metabolizer status of the nine (EFV400) and seven patients (EFV600) with pVL ≥200 copies/mL at 48 weeks (n = 16 total) were: EFV400: 1.31 mg/L (extensive, n = 1), 1.45–2.90 mg/L (intermediate, n = 6), 0.85 mg/L and 2.99 mg/L (slow, n = 2); EFV600: 0.12 mg/L and 1.60 mg/L (extensive, n = 2), 0.37 mg/L and 3.02 mg/L (intermediate, n = 2), 0.68 mg/L and 8.15 mg/L (slow, n = 2), and 6.24 mg/L (missing, n = 1).
Treatment discontinuation and adverse events
Overall, 42/606 (7%) discontinued therapy during the 48 weeks (median (range) 15 weeks (0.1–49)). Seven percent had severe adverse events and 65% and 48% experienced efavirenz‐related and CNS adverse events, respectively. Significantly more patients administered EFV600 had efavirenz‐related (70% vs. 61%; P = 0.017) and CNS events (52% vs. 44%; P = 0.042), compared to EFV400 (Pearson's chi‐square).
Predicted PK parameters were not significantly different between those who did and did not discontinue efavirenz or had adverse events (Table 4). After adjusting for age, sex, and dose and stratifying by country, those with CYP2B6 516TT and CYP2A6*9B heterozygote or homozygous variant allele (CA or AA) were at increased risk of discontinuation (HR (95% CI): 2.81 (1.12–7.06); P = 0.028 and 2.14 (1.05–4.35); P = 0.035, respectively). EFV600 was independently associated with 24% increased risk of efavirenz‐related side effects when adjusted for age and sex and stratified by country (HR (95% CI): 1.24 (1.01–1.53); P = 0.039).
Table 4.
Differences in mean individual predicted PK parameters for toxicity endpoints, assessed by calculation of geometric mean ratios (GMR) and 90% CI (n = 606).
| GMR (90% CI)a | ||||
|---|---|---|---|---|
| Parameter | Discontinuation | Efavirenz‐related adverse event | Serious adverse event | CNS adverse event |
| AUC0‐24 | 0.90 (0.78–1.04) | 0.95 (0.88–1.02) | 0.97 (0.84–1.12) | 0.97 (0.90–1.05) |
| Cmax | 0.88 (0.78–1.00) | 0.93 (0.87–1.00) | 0.99 (0.87–1.12) | 0.96 (0.90–1.02) |
| C24 | 0.94 (0.77–1.14) | 0.96 (0.87–1.07) | 0.93 (0.76–1.14) | 1.00 (0.90–1.10) |
| C12 | 0.90 (0.78–1.05) | 0.95 (0.88–1.03) | 0.96 (0.83–1.12) | 0.98 (0.91–1.05) |
Did not have the event/had the event.
CI, confidence interval; AUC0‐24, area under the concentration‐time curve over 24‐hour interval; Cmax, maximum concentration; C24, trough concentration, 24 hours postdose; C12, concentration 12 hours postdose representing the mid‐dose interval.
DISCUSSION
Efavirenz dose was not associated with virological response at 48 weeks despite significantly lower exposure with EFV400. Predicted C12 was <1.0 mg/L (suggested MEC12) in 5% (n = 14) and 2% (n = 6) for EFV400 and EFV600, respectively, following 4–12 weeks of therapy. Of these patients, only one EFV400 and three EFV600 patients had detectable pVL at 48 weeks. Moreover, patients randomized to EFV400 experienced significantly fewer efavirenz‐related adverse events.
The impact of CYP2B6 516G>T and 983T>C on efavirenz PK have been well documented in different populations8, 9, 10, 13, 15 and replicated in ENCORE1. CYP2A6 polymorphisms including CYP2A6*9B and/or *17 carriers8, 16 have been associated with higher efavirenz concentrations, particularly in those with impaired CYP2B6 function. Composite CYP2B6 516G>T/983T>C/CYP2A6*9B/*17 genotype and bodyweight were significantly associated with efavirenz CL/F in ENCORE1. The model described the data well and parameters were within reported ranges.9, 14, 15 Addition of covariates reduced CL/F variability by 15%. The patient population was diverse and although 58% of variability remained unexplained (37% and 21%; IIV and IOV, respectively), it could be attributed to covariates not captured in the study, e.g., unidentified host genetic factors and potentially variable adherence.
Lower efavirenz concentrations have been associated with NR1I3 540C>T and NR1I3 1089T>C carriers (TT, CC, respectively).7, 17, 18 Relationships between NRII3 540C>T or 1089T>C and CL/F was not confirmed in ENCORE1 patients.
The impact of CYP2B6 15582C>T SNP on efavirenz C24 in combination with CYP2B6 516G>T and 983T>C was recently described.19 Patients wildtype for all three SNPs (CYP2B6 15582CC/516GG/983TT) had the lowest median efavirenz C24, potentially placing them at risk of virological failure. Despite CYP2B6 15582C>T not meeting statistical criteria to remain in the ENCORE1 population PK model, post‐hoc analysis was performed to estimate each patient's C24 based on CYP2B6 15582C>T/516G>T/983T>C composite genotype as previously described.19 Patients wildtype for all three SNPs had the lowest model‐predicted median C24 and concentrations were lower for EFV400. No patients with this genotype had pVL≥200 copies/mL at 48 weeks (Supplementary Figure 3). Given the small number of patients with pVL ≥200 copies/mL, interpretation of this observation is limited.
Dose was not associated with pVL <200 copies/mL at 48 weeks, despite lower concentrations with EFV400. No relationships between genetic polymorphisms and viral suppression were observed. Patients were at higher risk of suppressing virus in association with increasing AUC0‐24, C24, and C12, although the CIs were generally wide. Due to low numbers of detectable viral loads at 48 weeks, the models should be interpreted with caution.
A therapeutic range (1.0–4.0 mg/L) proposed in 2001 by Marzolini et al. (n = 130 HIV‐infected patients of unreported ethnicity) was adopted for efavirenz threshold concentrations. Seventy‐six percent suppressed virus below 400 copies/mL; of those who failed therapy, 50% had a mid‐dose interval concentration <1.0 mg/L.12 Efavirenz cutoffs have not been reevaluated in a large, randomized, controlled trial, particularly in combination with more active therapies such as tenofovir and emtricitabine. ENCORE1 data strongly indicate that this putative MEC does not represent the efficacy cutoff for current efavirenz‐based regimens. Indeed, of 16 patients with pVL ≥200 copies/mL at 48 weeks (nine vs. seven for EFV400 and EFV600, respectively), only one and three individuals had predicted C12 <1.0 mg/L. Furthermore, Ugandan patients (n = 99) receiving efavirenz with zidovudine/lamivudine also experienced a low number of failures (n = 6; pVL ≥40 copies/mL) that were not associated with plasma exposure.20
Limited failures in ENCORE1 meant robust reassessment of the 1.0 mg/L cutoff was not feasible; however, these data question the validity of this MEC. Self‐reported adherence was recorded at weeks 4 and 48, and although the impact of adherence on clinical outcome cannot be assessed, patients with detectable viral load at 48 weeks (n = 16) reported taking all or most of their medications (except therapy discontinuations) including four patients with C12 <1.0 mg/L.
Efavirenz therapy is associated with early neuropsychiatric adverse events in over half of patients21, 22 that generally subside with time. Although relationships between CNS toxicity and efavirenz plasma concentrations have been observed,11, 12, 23, 24 no association was seen in ENCORE1. Despite this, the incidence of efavirenz‐related and CNS adverse events was lower with EFV400. Furthermore, EFV600 was associated with higher risk of efavirenz‐related adverse effects. Efavirenz and its 8‐hydroxy (8OH) metabolite produced damage in rat neuronal cultures in vitro, with 8OH‐efavirenz exhibiting toxicity 10‐fold higher than the parent drug.25 Although translating in vitro observations to clinical settings is difficult, these data suggest that patients with higher 8OH‐efavirenz CNS concentrations (e.g., extensive metabolizers) could be more susceptible to CNS toxicities.21 Alternatively, those without fully functional CYP2B6 could be at lower risk. In ENCORE1, CYP2B6 983T>C carriers (TC or CC) had 33% lower risk of CNS side effects compared to wildtype (TT), suggesting lower 8OH‐efavirenz formation and CYP2B6 15582CT/TT had a higher risk (53%) (Supplementary Table 4).
Another ENCORE1 substudy (n = 28) found concentrations of efavirenz and its metabolites were slightly lower in plasma and CSF for EFV400 and exposure within the two compartments were correlated and associated with CYP2B6 516G>T genotype.26 Conversely, no relationships were observed between dose, plasma efavirenz, or 8OH‐efavirenz or genotype with CSF 8OH‐efavirenz. Although the CSF substudy was small and exploratory, a potential connection was noted between CSF 8OH‐efavirenz and patient‐reported CNS symptoms by questionnaire26 and most individuals achieved CSF 8OH‐efavirenz concentrations greater than those responsible for neuronal damage in rat cultures.25, 26 ENCORE1 contributes significantly to our understanding of efavirenz‐induced toxicity, although the mechanisms remain to be fully elucidated.
Efavirenz discontinuation in ENCORE1 occurred at a rate consistent with previous reports.27, 28, 29 Possession of CYP2B6 516G>T homozygous variant (TT) and CYP2A6*9B heterozygous or homozygous variant alleles (CA or AA) but not dose were associated with higher risk of discontinuation. The relationship observed with CYP2B6 516TT agrees with Wyen et al., who evaluated risk factors for early discontinuation (<3 months).29 Increased risk was also reported in patients with NR1I3 540CC; this was not seen in ENCORE1. EFV400 discontinuation was not associated with genotypes when analyzed separately (Supplementary Table 5).
Pharmacogenetic testing has been suggested to aid efavirenz dose optimization.30, 31 Genetic risk scores were used to predict early efavirenz discontinuation; individuals scoring 6 (CYP2B6 homozygous loss of function and loss of function in an accessory metabolic pathway of CYP2A6 or CYP3A4) had a higher risk of discontinuation compared to those with lower scores.32 Although statistically significant and partially confirmed in ENCORE1, the number of patients scoring 6 was small (13/272),32 thus questioning the feasibility of translating this approach into a cost‐effective, population‐wide clinical tool, particularly in resource‐limited settings. The data presented here indicate that pharmacogenetic analysis of known alleles is no more useful at predicting outcome, such as efficacy or discontinuation, for the efavirenz doses assessed (400 mg and 600 mg).
The antitubercular agent rifampicin is a cytochrome P450 enzyme inducer, markedly lowering concentrations of some antiretrovirals, potentially jeopardizing virological success.33 Coadministration with efavirenz has generated conflicting data and a systematic review reported a time‐dependent interaction between rifampicin and efavirenz,34 with efavirenz concentrations decreased in individuals receiving a single dose or ≤8 days of therapy, while PK parameters increased in those established on efavirenz.34 Furthermore, the STRIDE study observed good virological control in coinfected patients on and off TB therapy including rifampicin.35 Further PK‐PD studies of efavirenz and rifampicin in coinfected populations are warranted.
Current WHO guidelines recommend efavirenz as first‐line treatment during pregnancy3, 36; PK‐PD data of efavirenz during pregnancy and postpartum are sparse. A recent review concluded that pregnancy had little clinical impact on efavirenz PK, with good rates of viral suppression achieved in mothers at delivery.37 However, others observed higher efavirenz CL/F and lower AUC0‐24 and Cmin during pregnancy compared to postpartum.38 When stratified by CYP2B6 516G>T, PK changes were markedly greater in wildtype patients (516GG). Impact on clinical outcome requires investigation.38 Speculatively, these findings together with ENCORE1 outcomes suggest scope for dose reduction to 400 mg in HIV/TB coinfected and potentially pregnant patients. Transition to new recommendations would require careful prospective monitoring.
Although plasma efavirenz concentrations were reduced with EFV400 compared to EFV600, virus suppression was similar. Fewer adverse events with the reduced dose may improve quality of life. Genetic polymorphisms were associated with treatment discontinuation but the biological importance is uncertain. Furthermore, genetic testing is unlikely to be widely implemented, particularly in resource‐limited settings and would be no more useful for EFV400 than EFV600. Antiretroviral costs pose a barrier to rollout of treatment to meet WHO targets. Reducing efavirenz dose to 400 mg could provide an economically viable solution, cutting costs and expanding access while maintaining good efficacy and reducing adverse events.
METHODS
Patients
ENCORE1 has been described.5 Briefly, HIV‐infected individuals ≥16 years with pVL ≥1,000 copies/mL and CD4 cell counts between 50–500 cells/mm3 without preexisting CDC AIDS‐defining illnesses or active opportunistic infections and no prior exposure to antiretrovirals were eligible to participate in this randomized, double‐blind, placebo‐controlled trial. Patients were recruited from 13 countries (38 sites) across Africa, Asia, South America, Europe, and Oceania; ethical and regulatory approval and written informed consent were obtained.5 Patients were randomized to receive daily EFV400 or EFV600 in combination with tenofovir/emtricitabine (Truvada, 300/200 mg once daily). After randomization, patients requiring treatment with rifampicin for TB were switched to open‐label efavirenz 600 mg (or 800 mg) once daily, then returned to randomized efavirenz on completion of rifampicin.
Random, single blood samples were drawn from all patients at weeks 4 and 12 (between 8–16 hours postdose). Intensive sampling was also undertaken in a subgroup (from Argentina, South Africa, Thailand, United Kingdom) between weeks 4 and 8 at predose (0 hour), 2, 4, 8, 12, 16, and 24 hours postdose. Plasma efavirenz concentrations were determined by a fully validated high‐performance liquid chromatography tandem mass spectrometry (HPLC‐MS/MS) method with LLQ and upper limit of quantification (ULQ) of 0.025 and l0.0 mg/L, respectively.39
Genotyping
Total genomic DNA was extracted from patient blood using the QI Amp DNA mini kit (Qiagen, West Sussex, UK) according to the manufacturer's instructions. Samples were quantified and normalized to 20 ng/μL. Genotyping for SNPs previously associated with efavirenz PK (CYP2B6 516 G>T (rs3745274),8, 9, 13, 15 CYP2B6 983 T>C (rs28399499),10 CYP2B6 15582C>T (rs4803419),19 CYP2A6*9B (rs8192726),8 CYP2A6*17 (rs28399454),8 CYP3A4*22 (rs35599367),40 NR1I3 (CAR) 540C>T (rs2307424)7 and NR1I3 1089T>C (rs3003596)18) was performed using real‐time polymerase chain reaction (PCR) allelic discrimination assays (C_7817765_60, C_60732328_20, C_7817764_10, C_29560333_20, C_34816076_20, C_59013445_10, C_25746794_20, and C_16194070_10, respectively; Applied Biosystems, Foster City, CA) as previously described.17, 40
PK modeling
Nonlinear mixed effects modeling (NONMEM v. 7.2, ICON Development Solutions, Ellicott City, MD) was applied to the data41 with initial parameter estimates taken from the literature.9, 14, 42
Covariates including age, weight, FFM, body mass index (BMI), sex, ethnicity, concomitant rifampicin, and CYP2B6 516G>T, CYP2B6 983T>C, CYP2B6 15582C>T, CYP2A6*9B, CYP2A6*17, CYP3A4*22, NR1I3 540C>T, and NR1I3 1089T>C genotypes on efavirenz CL/F were explored. Covariates were initially assessed (univariate), with the exception of CYP2A6*9B and CYP2A6*17, which were tested in combination with CYP2B6 SNPs due to more prominent effects observed in those without fully functional CYP2B6.14
Model fit was assessed by statistical and graphical methods. Decreased minimal OFV by at least 3.84 units was required to accept a model with one extra parameter (P = 0.05, χ2 distribution, 1 d.f.). Once significant covariates were included a backwards elimination process was performed and biologically plausible covariates producing an increase in OFV of at least 10.83 units (P = 0.001, χ2 distribution, 1 d.f.) were retained.
Model evaluation was performed by simulation and visual predictive check. Using model estimates a 90% prediction interval (P5‐P95) was generated from 1,000 simulated patients with the same distribution of doses and significant covariates as the original dataset; observed data were superimposed. Inclusion of ≥90% of data points within the prediction interval indicated an adequate model.
Secondary PK parameters
Along with model‐defined parameters, secondary PK parameters were estimated for each patient at each sampling occasion using the final model: AUC0‐24, Cmax, C24, and C12. If patients had more than one sampling occasion, the mean of individual predicted PK parameters were determined and carried forward into the analyses outlined below.
Differences between doses for efavirenz AUC0‐24, Cmax, C24, and C12, were evaluated by geometric mean ratios (GMR) and 90% CI using log‐transformed data then expressed as linear values. Differences were considered significant if the 90% CI did not include 1.
Genotypes were distributed into three groups based on change in efavirenz CL/F and CYP2B6/CYP2A6 alleles:
Extensive metabolizers: homozygous wildtype for CYP2B6 SNPs with combinations of CYP2A6 alleles (GG/TT/CC/CC, GG/TT/CC/CT or TT, GG/TT/CA or AA/CC, GG/TT/CA or AA/CT or TT).
Intermediate metabolizers: homozygous wildtype CYP2B6 516G>T and combinations of CYP2A6 alleles with heterozygous or homozygous variant CYP2B6 983T>C or heterozygous variant CYP2B6 516G>T with CYP2B6 983T>C homozygous wildtype and combinations of CYP2A6 alleles (GG/TC or CC/CC/CC, GT/TT/CC/CC, GT/TT/CC/CT or TT, GT/TT/CA or AA/CC, GT/TT/CA or AA/CT).
Slow metabolizers: heterozygous variant CYP2B6 516G>T with heterozygous or homozygous variant CYP2B6 983T>C, or homozygous variant CYP2B6 516G>T with homozygous wildtype CYP2B6 983T>C with combinations of CYP2A6 alleles (GG/TC or CC/CC/CT or TT, GG/TC or CC/CA or AA/CC, GG/TC or CC/CA or AA/CT, GT/TC or CC/CC/CC, GT/TC or CC/CC/CT or TT, GT/TC or CC/CA or AA/CC, TT/TT/CC/CC, TT/TT/CC/CT or TT, TT/TT/CA or AA/CC, TT/TT/CA or AA/CT).
Proportions of patients with C12 below the recommended MEC of 1.0 mg/L12 were determined for each group.
PK‐PD analysis
The primary PD endpoint was proportion of patients with pVL <200 copies/mL at 48 weeks by randomized dose. Patients without a recorded viral load were excluded. Logistic regression was performed to assess associations between pVL <200 copies/mL at 48 weeks and mean individual predicted logCL/F, logAUC0‐24, logCmax, logC24, and logC12. Univariate analyses (Fisher's Exact test or Pearson's chi‐square, as appropriate) were carried out for dose, SNPs, and screening pVL ≤/>100,000 copies/mL.
Treatment discontinuation and adverse events
Discontinuation was defined as interruption in efavirenz >30 days. Efavirenz‐related adverse events and CNS‐related side effects (including abnormal dreams, anxiety, cerebellar disorder and ataxia, dizziness, headache and migraine, impaired concentration, insomnia, seizure, and somnolence) were categorized as those defined in the Sustiva Prescribing Information.27
Comparison of PK parameters between those who did or did not stop therapy and experience adverse events was performed by GMR (90% CI). Differences in proportions of each endpoint by efavirenz dose were assessed by Fisher's Exact test or Pearson's chi‐square. Evaluation of relationships between efavirenz discontinuation and adverse events with dose and SNPs was performed by Cox regression adjusted a priori for potential confounders (age, sex). Post‐hoc exploratory analysis of the crude association of PK parameters and CNS‐related side effects (as a subset of adverse events) was undertaken using logistic regression.
Statistical analyses were performed using SPSS (v. 21, IBM, Armonk, NY).
AUTHOR CONTRIBUTIONS
L.D. wrote the article; M.B., D.C., D.A.C., S.E., and R.P. designed the research; L.D., J.A., L.E., M.B., D.E., A.O., S.K., D.B., C.O., A.C., M.L., P.P., D.C. and D.A.C. performed the research; L.D. analyzed the data.
CONFLICT OF INTEREST
All authors had full access to study data and agreed to submit for publication. The corresponding author had final responsibility for the decision to submit for publication. No medical writers were used and no agency made any payments for writing. Neither the funding agency nor pharmaceutical companies supporting the trial played any role in collection, analysis, interpretation, or reporting of these data.
L.D. is supported by Pre‐DiCT‐TB. D.B., S.K., A.O., and L.E. have received research grants and/or travel bursaries from Merck, Bristol Myers and Squibb, GlaxoSmithKline, Pfizer, Abbott, ViiV, Boehringer Ingelheim and Janssen Pharmaceuticals. MB has received travel and research grants from and has been an adviser for Janssen, Roche, Pfizer, ViiV, Bristol‐Myers Squibb, Merck Sharp & Dohme, and Gilead. R.P., J.A., D.E., A.C., D.C., and P.P. report no conflict of interest. C.O. has received a travel bursary from Tibotec (2013) and an honorarium from Abbott (2011). M.L. has received research grant support from Abbott, Merck Research Laboratories, and Pfizer. D.A.C. has received honoraria and research grant support from Gilead Sciences, Merck Research Laboratories and Bristol‐Myers Squibb. S.E. has received research grant support from Abbvie, Gilead Sciences, Merck Research Laboratories, Pfizer, and ViiV Healthcare.
PROTOCOL STEERING COMMITTEE
Janaki Amin, Stephen Becker, Waldo Belloso, Marta Boffito, David Cooper, Brenda Crabtree‐Ramirez, Chris Duncombe, Sean Emery, Sharne Foulkes, Andrew Hill, Heiko Jessen, Suresh Kumar, Man Po Lee, Marcelo Losso, Chidi Nwizu, Praphan Phanuphak, David Ripin, Tim Read, Jim Rooney, Kim Schaffer, Eduardo Shahar, Alan Winston, Marcelo Wolff, Barnaby Young.
PROJECT TEAM
Cecilia Abela, Maria Arriaga, Janaki Amin, Waldo Belloso, Mark Boyd, Dianne Carey, Amanda Clarke, David Cooper, Kymme Courtney‐Vega, Carlo Dazo, Marina Delfino, Anna Donaldson, Sean Emery, Natalie Espinosa, Tanya Johannesen, Peeraporn Kaew‐on, Enmoore Lin, Marcelo Losso, Alejandra Moricz, Jessica Taylor, Praphan Phanupak, Rebekah Puls, Kanitta Pussadee, Parinya Sutheerasak, Louise Tomkins, Sasiwimol Ubolyam.
SITE INVESTIGATORS: MAIN STUDY
Waldo Belloso, Raja Iskandar Shah bin Raja Azwa, Emiliano Bissio, Liliana Calanni, Arnaldo Casiro, Ploenchan Chetchotisakd, Jorge Contarelli, David Cooper, Brenda Crabtree‐Ramirez, Nicholas Doong, Julian Elliott, Sharne Foulkes, Brian Gazzard, Mark Kelly, Suresh Kumar, Man Po Lee, Marcelo Losso, Norma del Carmen Luna, Sergio Lupo, Oscar Garcia Messina, Lerato Mohapi, Richard Moore, David Nolan, Chidi Nwizu, Catherine Orrell, Carlos Perez, Sarah Pett, Praphan Phanuphak, Jürgen Rockstroh, Eduardo Shahar, Khuanchai Supparatpinyo, Don Smith, Jaime Andrade Villanueva, Emanuel Vlahakis, Alan Winston, Marcelo Wolff, Barnaby Young.
SITE INVESTIGATORS AND STAFF: INTENSIVE PK STUDY
Chelsea & Westminster Hospital, London: Akil Jackson, Marta Boffito, Serge Federle, Brian Gazzard, Sophie Scott. Hospital Ramos Mejia, Buenos Aires: Patricia Burgoa, Juan Ebenrstejin, Mariana Kundro, Marcelo Losso. Desmond Tutu HIV Centre, Cape Town: Christie Heiberg, Richard Kaplan, Catherine Orrell, Maureen Rattley. HIV‐NAT AIDS Research Centre, Bangkok: Amanda Clarke, Kanitta Pussadee, Praphan Phanuphak. Central Laboratory: Alex Carrera, Philip Cunningham, Bertha Fsadni, Tony Kelleher, Melanie Lograsso, Kate Merlin, Ansari Shaik, Julie Yeung. Liverpool University Laboratory: Alieu Amara, Deirdre Egan, Laura Else.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
We thank all those who volunteered to participate in this study. Funding: ENCORE1 substudies were funded through a project grant from the Australian Government National Health and Medical Research Council (NHMRC), APP1048402. The Kirby Institute is funded in part by the Australian Government Department of Health and Ageing. Gilead Sciences Inc. donated Truvada and Mylan Inc. provided Efamat and donated matched placebo.
REFERENCES
- 1. World Health Organisation . Global update on HIV treatment 2013: results, impact and opportunities. <http://wwwwhoint/hiv/data/global_treatment_report_presentation_2013pdf> (2013). Accessed 10 September 2014.
- 2. World Health Organisation . Key facts on global HIV epidemic and progress in 2010. <http://wwwwhoint/hiv/pub/progress_report2011/global_facts/en/indexhtml> (2011). Accessed 9 September 2014.
- 3. World Health Organisation . Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach. <http://appswhoint/iris/bitstream/10665/85321/1/9789241505727_engpdf?ua=1> (2013). Accessed 9 September 2014. [PubMed]
- 4. Haas, D.W. et al A phase II, double‐blind, placebo‐controlled, dose ranging study to assess the antiretroviral activity and safety of efavirenz in combination with open‐label zidovudine with lamivudine at 36 weeks [DMP 266‐005]. The XII International AIDS Conference, Geneva, Switzerland. Abstract 22334 (1998).
- 5. Puls, R. et al Efficacy of 400 mg efavirenz versus standard 600 mg dose in HIV‐infected, antiretroviral‐naive adults (ENCORE1): a randomized, double‐blind, placebo‐controlled, non‐inferiority trial. Lancet 383, 1474–1482 (2014). [DOI] [PubMed] [Google Scholar]
- 6. Hill, A. Optimizing HIV treatment. Curr. Opin. HIV AIDS 8, 34–40 (2013). [DOI] [PubMed] [Google Scholar]
- 7. Cortes, C.P. , Siccardi, M. , Chaikan, A. , Owen, A. , Zhang, G. & la Porte, C.J. Correlates of efavirenz exposure in Chilean patients affected with human immunodeficiency virus reveals a novel association with a polymorphism in the constitutive androstane receptor. Ther. Drug Monit. 35, 78–83 (2013). [DOI] [PubMed] [Google Scholar]
- 8. Kwara, A. , Lartey, M. , Sagoe, K.W. , Rzek, N.L. & Court, M.H. CYP2B6 (c.516G‐‐>T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV‐infected patients. Br. J. Clin. Pharmacol. 67, 427–436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mukonzo, J.K. et al A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single‐dose efavirenz population pharmacokinetics in Ugandans. Br. J. Clin. Pharmacol. 68, 690–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wyen, C. et al Impact of CYP2B6 983T>C polymorphism on non‐nucleoside reverse transcriptase inhibitor plasma concentrations in HIV‐infected patients. J. Antimicrob. Chemother. 61, 914–918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gutierrez, F. et al Prediction of neuropsychiatric adverse events associated with long‐term efavirenz therapy, using plasma drug level monitoring. Clin. Infect. Dis. 41, 1648–1653 (2005). [DOI] [PubMed] [Google Scholar]
- 12. Marzolini, C. , Telenti, A. , Decosterd, L.A. , Greub, G. , Biollaz, J. & Buclin, T. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV‐1‐infected patients. AIDS 15, 71–75 (2001). [DOI] [PubMed] [Google Scholar]
- 13. Rotger, M. et al Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV‐infected patients. Pharmacogenet. Genomics 15, 1–5 (2005). [DOI] [PubMed] [Google Scholar]
- 14. Arab‐Alameddine, M. et al Pharmacogenetics‐based population pharmacokinetic analysis of efavirenz in HIV‐1‐infected individuals. Clin. Pharmacol. Ther. 85, 485–494 (2009). [DOI] [PubMed] [Google Scholar]
- 15. Nyakutira, C. et al High prevalence of the CYP2B6 516G—>T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur. J. Clin. Pharmacol. 64, 357–365 (2008). [DOI] [PubMed] [Google Scholar]
- 16. di Iulio, J. et al In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet. Genomics 19, 300–309 (2009). [DOI] [PubMed] [Google Scholar]
- 17. Olagunju, A. et al CYP2B6 516G>T (rs3745274) and smoking status are associated with efavirenz plasma concentration in a Serbian cohort of HIV patients. Ther. Drug Monit. 36, 734–738 (2014). [DOI] [PubMed] [Google Scholar]
- 18. Swart, M. , Whitehorn, H. , Ren, Y. , Smith, P. , Ramesar, R.S. & Dandara, C. PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC Med. Genet. 13, 112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holzinger, E.R. et al Genome‐wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet. Genomics 22, 858–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mukonzo, J.K. et al Pharmacogenetic‐based efavirenz dose modification: suggestions for an African population and the different CYP2B6 genotypes. PloS One 9, e86919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Decloedt, E.H. & Maartens, G. Neuronal toxicity of efavirenz: a systematic review. Expert Opin. Drug Saf. 12, 841–846 (2013). [DOI] [PubMed] [Google Scholar]
- 22. Kenedi, C.A. & Goforth, H.W. A systematic review of the psychiatric side‐effects of efavirenz. AIDS Behav. 15, 1803–1818 (2011). [DOI] [PubMed] [Google Scholar]
- 23. Clifford, D.B. et al Impact of efavirenz on neuropsychological performance and symptoms in HIV‐infected individuals. Ann. Intern. Med. 143, 714–721 (2005). [DOI] [PubMed] [Google Scholar]
- 24. Siccardi, M. et al Pharmacokinetic and pharmacodynamic analysis of efavirenz dose reduction using an in vitro‐in vivo extrapolation model. Clin. Pharmacol. Ther. 92, 494–502 (2012). [DOI] [PubMed] [Google Scholar]
- 25. Tovar‐y‐Romo, L.B. et al Dendritic spine injury induced by the 8‐hydroxy metabolite of efavirenz. J. Pharmacol. Exp. Ther. 343, 696–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winston, A. et al Cerebrospinal‐fluid exposure of efavirenz and its major metabolites when dosed at 400 and 600 mg once daily; a randomised controlled trial. Clin. Infect. Dis. 60, 1026–1032 (2015). [DOI] [PubMed] [Google Scholar]
- 27. Bristol‐Myers Squibb Company . Sustiva (efavirenz) capsules and tablets for oral use: highlights of prescribing information. <http://packageinsertsbmscom/pi/pi_sustivapdf> (2014). Accessed 19 August 2014.
- 28. Gallant, J.E. et al Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N. Engl. J. Med. 354, 251–260 (2006). [DOI] [PubMed] [Google Scholar]
- 29. Wyen, C. et al Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz‐containing regimens. J. Antimicrob. Chemother. 66, 2092–2098 (2011). [DOI] [PubMed] [Google Scholar]
- 30. Gatanaga, H. et al Successful efavirenz dose reduction in HIV type 1‐infected individuals with cytochrome P450 2B6 *6 and *26. Clin. Infect. Dis. 45, 1230–1237 (2007). [DOI] [PubMed] [Google Scholar]
- 31. Sukasem, C. & Sungkanuparph, S. Would a CYP2B6 test help HIV patients being treated with efavirenz? Pharmacogenomics 14, 999–1001 (2013). [DOI] [PubMed] [Google Scholar]
- 32. Lubomirov, R. et al Association of pharmacogenetic markers with premature discontinuation of first‐line anti‐HIV therapy: an observational cohort study. J. Infect. Dis. 203, 246–257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Regazzi, M. , Carvalho, A.C. , Villani, P. & Matteelli, A. Treatment optimization in patients co‐infected with HIV and Mycobacterium tuberculosis infections: focus on drug‐drug interactions with rifamycins. Clin. Pharmacokinet. 53, 489–507 (2014). [DOI] [PubMed] [Google Scholar]
- 34. Hill, A. , Khoo, S. & Back, D. The induction effect of rifampicin on efavirenz is time‐dependent: systematic review of 12 drug interaction studies. International Workshop on HIV Clinical Pharmacology, Washington DC. Abstract P29 (2014).
- 35. Luetkemeyer, A.F. et al Relationship between weight, efavirenz exposure, and virologic suppression in HIV‐infected patients on rifampin‐based tuberculosis treatment in the AIDS Clinical Trials Group A5221 STRIDE Study. Clin. Infect. Dis. 57, 586–593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. World Health Organisation . Technical update on treatment optimization. Use of efavirenz during pregnancy: a public health perspective. <http://appswhoint/iris/bitstream/10665/70920/1/9789241503792_engpdf?ua=1> (2012). Accessed 9 September 2014.
- 37. Hill, A. , Ford, N. , Boffito, M. , Pozniak, A. & Cressey, T.R. Does pregnancy affect the pharmacokinetics of efavirenz? AIDS 28, 1542–1543 (2014). [DOI] [PubMed] [Google Scholar]
- 38. Olagunju, A. et al Pharmacogenetics of pregnancy‐induced changes in efavirenz pharmacokinetics. Clin. Pharmacol. Ther. 97, 298–306 (2014). [DOI] [PubMed] [Google Scholar]
- 39. Amara, A.B. , Tjia, J. , Dutton, J. , Else, L.J. , Back, D.J. & Khoo, S. Development and validation of a HPLC‐MS/MS assay to quantify the antiretroviral (ARV) drug, efavirenz and its major metabolites in plasma. British Mass Spectrometry Society Meeting, Cardiff, UK. Abstract BMS S11–1240 (2011).
- 40. Olagunju, A. et al CYP3A4*22 (c.522‐191 C>T; rs35599367) is associated with lopinavir pharmacokinetics in HIV‐positive adults. Pharmacogenet. Genomics 24, 459–463 (2014). [DOI] [PubMed] [Google Scholar]
- 41. Beal, S. & Sheiner, L.B. NONMEM Users Guide. ICON Development Solutions, Ellicott City, Maryland (1989. –1998).
- 42. Kappelhoff, B.S. et al Population pharmacokinetics of efavirenz in an unselected cohort of HIV‐1‐infected individuals. Clin. Pharmacokinet. 44, 849–861 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


