Abstract
CD46 is a cell surface complement inhibitor widely expressed in human tissues, in contrast to mice, where expression is limited to the testes. In humans, it has been identified as an important T cell costimulatory receptor, and patients deficient in CD46 or its endogenous ligands are unable to mount effective Th1 T cell responses. Stimulation of human CD4+ T cells with CD3 and CD46 also leads to the differentiation of a ‘switched’ Th1 population, which shuts down IFN-γ secretion and upregulates IL-10, and is believed to be important for negative feedback regulation of the Th1 response. We show here that CD46 costimulation leads to amplified microRNA expression changes in human CD4+ T cells, with associated increases in activation more potent than that mediated by the ‘classic’ costimulator CD28. Blockade of cell-surface CD46 inhibited CD28-mediated costimulation, identifying autocrine CD46 signaling as downstream of CD28. We also identify a downregulation of microRNA-150 in CD46-costimulated T cells, and identify the glucose transporter-1 (GLUT1) encoding transcript SLC2A1 as a target of microRNA-150 regulation, connecting microRNA-150 with modulation of glucose uptake. We also investigated microRNA expression profiles of CD46-induced ‘switched’ IL-10-secreting Th1 T cells and found increased expression of microRNA-150, compared to IFN-γ-secreting Th1 cells. Knockdown of microRNA-150 led to a reduction in IL-10 but not IFN-γ. CD46 therefore controls both Th1 activation and regulation via a miRNA-150-dependent mechanism.
Keywords: CD46, IL-10, Th1, microRNA, miR-150, GLUT1, FLT3, costimulation, complement
Introduction
T helper type 1 (Th1) cells are pro-inflammatory effector cells characterised by their production of IFN-γ and expression of transcription factor Tbet (T-box expressed in T cells (1)), and function to protect the host against intracellular pathogens. However, excessive Th1 responses can lead to pathological tissue damage, and inappropriately induced or unregulated Th1 responses can contribute to autoimmunity. The role of IL-10 in negative regulation of Th1 responses has been highlighted by lethal outcomes of IL-10 knockout or neutralization in toxoplasma or trypanosome mouse infection models, associated with hyper-production of type I cytokines (2, 3). In these infections, IL-10 is predominantly produced by Th1 cells, and blocking this negative feedback leads to increased pathogen clearance, but also immune-mediated damage to the host (4-6). In mice, IL-10 secretion from Th1 cells can be induced following repeated antigen stimulation (7), and a similar Th1-IL-10 negative feedback also develops in humans after repeated antigen exposure (8). IL-12-induced STAT4 activation (9) and subsequent IFN-γ and IL-2 (10) production are a prerequisite for optimal development of these regulatory Th1 cells, ensuring that Th1-derived IL-10 negative feedback only develops during or after the peak of the response, therefore contributing to contraction.
A mechanism for induction of IL-10 secretion from human Th1 cells has been found with the discovery that CD46 (membrane cofactor protein, MCP) acts as a T cell costimulatory receptor (11), and is crucial for Th1 induction and production of the pro-inflammatory cytokine IFN-γ (12). Although essential for this initial induction of pro-inflammatory Th1 responses, CD46 stimulation in the presence of high IL-2 also mediates the subsequent co-induction of IL-10 in Th1 cells and their ‘switch’ into a (self)regulatory phenotype (13). It has been hypothesised that the CD46-driven induction of IL-10 may be one strategy behind the targeting of CD46 by pathogens such as Neisseria meningitides and measles virus (14). CD46 is expressed on all human nucleated cells, but absent in murine somatic tissues (15), meaning that murine-based immunological studies do not contribute to the understanding of the role of CD46 in human T cell responses. As a complement inhibitor, CD46 binds C3b and acts as co-factor for its inactivation by the serine protease factor I. C3b can be generated in an autocrine fashion by activated T-cells, and has also been identified as a ligand for the T cell costimulatory function of CD46 (13, 16). This activation-associated T cell-derived generation of C3b, and dependence of human CD28-costimulated Th1 responses on CD46/C3, leads to a model of autocrine CD46 signaling functioning downstream of CD28 during human Th1 cell activation.
The clinical importance of CD46-mediated regulation of Th1 responses is supported by the altered expression of CD46 isoforms in T cells from multiple sclerosis patients (17), and by the failure of T cells from patients with rheumatoid arthritis to develop the full IL-10-secreting regulatory phenotype upon sustained CD46 costimulation, compared to healthy controls (10). It is therefore of interest to investigate and potentially therapeutically harness the mechanisms by which this mode of immunoregulation functions. The downstream molecular effector pathways are still incompletely mapped, and we have focused on CD46-mediated alterations in microRNA (miRNA) expression. MiRNAs have important roles as regulators of immune cell differentiation and function (18), and more specifically, have been shown to affect T cell regulation, development, signaling, and metabolism (19-21). Several miRNAs have high specificity of expression in lymphocytes, and their expression is required for normal lymphocyte function. We found that CD46 signaling in CD4+ T cells leads to a strong reduction in miRNA-150 (miR-150) levels, and we then identified miR-150 targets, which include regulators of T cell metabolism and cytokine secretion. Furthermore, miR-150 is required for IL-10 secretion from CD46-stimulated Th1 cells. We therefore highlight the role of miR-150 in CD46-induced Th1 activation and regulation.
Materials and methods
Purification and activation of T cells
All primary cells were purified from fresh peripheral blood collected from healthy volunteers according to the permission of the local ethics committee in Lund and with informed, written consent. Blood was taken using EDTA-coated vacuum tubes, diluted in PBS EDTA at room temperature, and PBMCs purified using Lymphoprep (Axis Shield) according to manufacturer’s instructions. CD4+ T cells were then purified using positive selection magnetic cell sorting (Miltenyi biotech) and purity (above 95%) verified by staining with Allophycocyanin-labeled anti-CD4 (Immunotools). Cells were washed and resuspended in RPMI (Invitrogen) with 10% FCS and 50 U/ml IL-2 (Immunotools), and 3.5×105 plated out per well in 48 well plates, coated overnight with 2 μg/ml anti-CD3 (OKT3, BD biosciences) and 2 μg/ml of either anti-CD28 (CD28.1, BD Biosciences) or anti-CD46 (Tra2-10, Sheffield university hybridoma biobank, UK).
Antibodies and Proteins
Anti-CD25-FITC, anti-CD69-Allophycocyanin, and anti-CD46-Phycoerthyrin (PE) (Immunotools) were used to assess CD antigen expression. Fc-CD46 and Fc-CD55 were expressed in CHO cells and purified on protein A columns as described in (22). For flow cytometry of purified T cells, viable cells were gated by exclusion of cells stained with fluorophore-labeled AnnexinV (Immunotools).
Cytokine detection
Cytokine secretion was measured using Miltenyi biotech flow cytometry cytokine capture kits for IL-10 and IFN-γ, according to manufacturer’s instructions. Alternatively, cytokines in supernatant were measured by ELISA (Peprotech/Mabtech).
RNA purification
RNA was purified using microRNeasy RNA purification kit (Qiagen). RNA integrity was verified by automated electrophoresis (Experion RNA analyser, Biorad), and purity assessed by 260:230 nm and 260:280 nm absorbance ratios. Cut-offs of 1.95 for 260:280 nm and 1.4 for the 260:230 nm ratios were used, as recommended by the microarray supplier.
Microarray
T cells were collected from four healthy donors using heparin anti-coagulant, diluted with equal amounts of PBS EDTA, then layered over Lymphoprep and spun to collect PBMCs. CD4+ T cells were purified and plated out for stimulation as described above. Purity was assessed by flow cytometry and was in all cases ≥ 95%. RNA was purified from a set of unactivated cells before plating, to represent resting, 0 hr time point samples. For early activation samples, CD28 or CD46 costimulated cells were harvested 2 hrs after plating. After 36 hours, the remaining CD46-costimulated cells were harvested and labeled for IL-10 or IFN-γ secretion using the Miltenyi Cytokine Secretion Kits. Cells were sorted into separate cytokine-secreting populations (IFN-γ+/IL-10−, IFN-γ+/IL-10+, and IFN-γ−/IL-10+) using a BD Biosciences FACS machine.
RNA from separate populations was labeled using the Hy3 microRNA Power labeling kit (Exiqon) and hybridised to an LNA-based miRCURY miRNA microarray slides (Exiqon, version 11) using a Maui hybridisation chamber (BioMicrosystems). Spot signals were acquired with an Agilent array scanner and GenePix Pro 4.1 was used for annotation (mirBase 14.0), signal processing and quantification using default background subtraction. The arrays were normalized using the global Lowess (Locally Weighted Scatterplot Smoothing) regression algorithm. To determine expression changes, Significant Analysis of Microarrays (SAM) (23) was performed as implemented in MeV v4.3 for Multi Class without using the fold-change criterion (24). Briefly, each four treatment replicates were grouped together, forming six classes, and a delta value of 0.102 corresponding to a strict mean false discovery rate (FDR) of 0% was set. Validation of significant miRNA expression changes was performed as described below. Normalized and pre-processed array data were deposited to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-2658.
qPCR
Array validation and mRNA/miRNA quantification were carried out using specific assays from Applied Biosystems, according to manufacturer’s instructions. For mature miRNA quantification, cDNA was produced using miR-specific hairpin primers, then qPCR run using an Applied Biosystems 7900HT qPCR machine. Beta 2-microglobulin, cyclophilin A and TATA-box binding protein were used as mRNA endogenous controls, while RNU-44 and RNU-38B were used for miRNA endogenous controls. Normalization using geometric means of multiple endogenous controls were performed according to GeNorm method (25). Relative expression levels of genes were compared using one-way or two-way analysis of variance (ANOVA).
CFSE labeling
T cells were washed in PBS then resuspended at 2 × 106/ml in PBS with 0.2 μM CFSE (Sigma) at 37°C for 15 minutes, then washed twice in culture medium, before resuspension and plating for activation.
AntagomiR transfection of T cells
Purified CD4+ cells were transfected using a nucleofector 4D device (Lonza) using a primary cell kit with 100 nM of miR-150-specific LNA antagomiR (Exiqon), or control LNA. Cells were then rested overnight in 96-well plates in 10% RPMI before plating for activation as described above.
Construction of Luciferase reporter plasmids
The 3’ UTRs of c-Myb (residues 1 to 1163) and SLC2A1 (residues 11 to 854) were cloned into the pMIR-REPORT vector (a kind gift from Dr Yvonne Ceder), which produces a firefly luciferase mRNA transcript with the cloned 3’ UTR. Human CD4+ T cell cDNA was used as a template for amplification of the c-Myb UTR using forward and reverse primers GCCGGCGACATTTCCAGAAAAGCATTATG and AAGCTTCCCAAATTACCAAGAACC respectively, and for amplification of the SLC2A1 UTR using forward and reverse primers GAGCTCATCACCAGCCCGGCCTG and GTTTAAACGTAAGTAACAGGAGTGAGGTG respectively. The amplicons contained all described miR-150 binding sites within the c-Myb UTR and the conserved miR-150 site in SLC2A1. PCR products were gel purified and cloned using the StrataClone Blunt cloning system (Agilent), amplified in DH5a cells, then the c-Myb amplicon removed by NaeI and HindIII double digestion, purified from the gel, and ligated into pMIR-REPORT opened with PmeI and HindIII digestion, to create the pMIR c-Myb plasmid. The SLC2A1 insert was released by SacI and PmeI double digestion, purified from the gel and then ligated into pMIR-REPORT opened by digestion with SacI and PmeI to create the pMIR-GLUT1 plasmid. Mutation of the SLC2A1 miR-150 binding site was then carried out using the Quikchange lightning site directed mutagenesis kit (Agilent), using forward and reverse primers CTGAGACCAGTTCGGTGCACTGGAGTG and CACTCCAGTGCACCGAACTGGTCTCAG. Sequences of all UTR inserts were verified by Sanger sequencing (GATC biotech). To perform luciferase assays, HEK293 cells were plated in 24-well plates for 24 hrs before co-transfection with 20 ng of Renilla luciferase expression vector (a kind gift from Dr Yvonne Ceder), 100 ng of pMIR-REPORT plasmid, and either miR-150 mimic or negative control miRNA mimic (Dharmacon) to a final concentration of 50 nM, using Dharmafect Duo transfection reagent (Dharmacon). After 24 hrs, cells were lysed and Renilla/Firefly luciferase expression assessed using the Dual-Glo Luciferase Assay (Promega) and a Wallac Victor2 fluorometer (Perkin Elmer).
pre-miR-150 expression vector construction
Complementary DNA oligomers consisting of human pre-miR-150, flanked by 30 additional nucleotides from the pri-miRNA sequence and BamHI/XhoI sticky ends, were synthesised (Eurofins MWG Operon) and hybridised together in hybridisation buffer (10 mM TrisHCl, 100 mM NaCl, 1 mM EDTA). Phosphorylation at the 5′ end was carried out using polynucleotide kinase (Invitrogen) before the dsDNA was ligated into the pcDNA3 expression vector opened with BamHI/XhoI (Fermentas), and verified by sequencing. Jurkat cells were transfected using the nucleofector 4D machine and stable clones selected using G418 (Gibco).
Northern and Western blot analyses
For Western blot analyses, cell lysates were separated using 10% SDS-PAGE gels under reducing conditions, transferred to PVDF membranes with the TransBlot turbo system (BioRad) and membranes blocked using fish gelatin (Nordic). Anti-c-Myb (Millipore) and anti-β-actin (Abcam) were used in combination with goat anti-mouse-HRP secondary antibody (Dako), while anti-GLUT1 (Millipore) was used in combination with goat anti-rabbit-HRP (DAKO), and proteins visualised using ECL reagents (Santa Cruz) and a ChemiDoc CCD camera (BioRad). The small RNA Northern blot protocol was adapted from reference (26). Briefly: 100 - 500 ng total RNA was separated in denaturing urea 15% PAGE gels in TBE buffer before electrophoretic transfer to positively charged nylon membranes (Amersham). RNA was cross-linked to the membrane in a UV chamber (Stratagene), and then blocked using Ultrahyb (Ambion) for 1 hr at 50°C. Double-DIG labeled LNA probe (1 nM, Exiqon) diluted in Ultrahyb was incubated overnight at 50°C, before the membrane was washed in 0.2× SSC buffer, followed by probing using anti-DIG-HRP antibody (Roche) in combination with DIG wash & block buffer kit (Roche), and detection by ECL reagents.
Glucose uptake
Jurkat cells were pre-incubated for 1 hr in pre-warmed PBS supplemented with 0.9 mM MgCl2 and 0.5 mM CaCl2 and 25 mM glucose, then spun and resuspended at 2×106/ml in 500 μl of the same solution spiked with 30 μCi/ml 3H D-glucose (Perkin Elmer), and incubated rotating at 37°C for 10 min. The incubation was then stopped by addition of ice-cold buffer and immediate washing. After three washes, the cell pellet was lysed and 3H D-glucose uptake counted using a beta-counter (Wallac), and normalized to total protein content as assessed by BCA assay (Pierce). A control of uptake at a zero timepoint was used by addition of 3H-D-glucose to cells on ice followed by immediate washing.
Results
CD46-stimulated T cells show distinct miRNA expression profiles
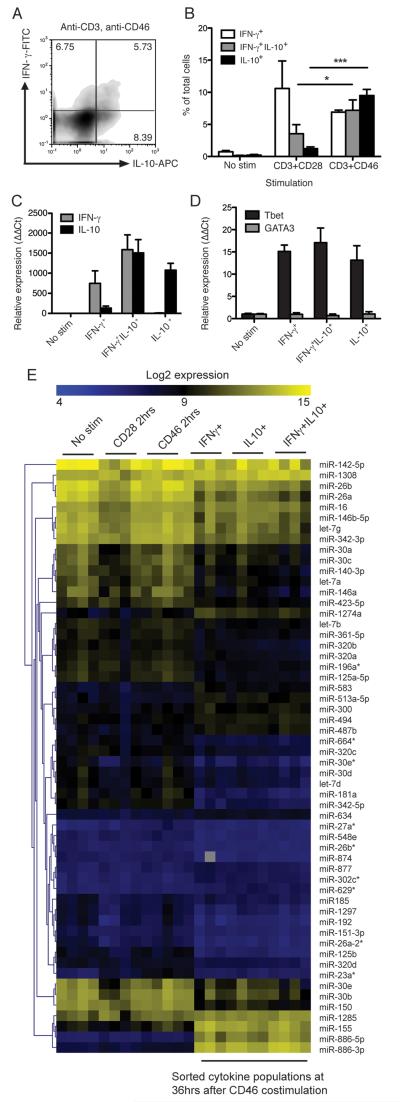
Studies of CD46 signaling have identified interactions of CD46’s intracellular domain with SPAK kinase, α-E-catenin and modulation of LAT activation and ICER/CREM nuclear translocation (10, 12, 27). However, CD46 signaling’s effect on miRNA expression, which is critical to normal T cell differentiation (28), is completely unexplored. Therefore, we investigated miRNA markers of T cell activation following CD46 activation. To study the molecular signatures engaged by CD46 activation of human CD4+ T cells, we first established the CD46-driven generation of pro-inflammatory IFN-γ+ and ‘switched’ IL-10-secreting (self)regulatory Th1 cells. Peripheral blood CD4+ T cells were purified and stimulated in 48-well plates with plate-bound antibodies against CD3 in combination with either CD28 or CD46. After 40 hrs of activation in the presence of IL-2, flow cytometry was used with cytokine capture staining reagents to quantify proportions of cells secreting IFN-γ, IL-10, or both cytokines (figure 1A). As previously described, CD46 costimulation leads to an increased proportion of cells which secrete IL-10, while CD28 costimulated cells lead to a measureable but substantially smaller population of IL-10+ cells (figure 1B).
Fig. 1. miRNA microarrays for miRNA profiles in cytokine-secreting Th1 sub-populations.
A) Purified human CD4+ T cells were stimulated for 36 hrs using immobilised plate-bound antibodies as indicated then stained for secreted IFN-γ and IL-10. Numbers denote percent of cells in each quadrant. B) The amounts of cells secreting IL-10 and IFN-γ from three independent experiments is shown, each using cells from an independent donor. C) Cells from 4 healthy donors were stimulated for 36 hrs stimulation, and IFN-γ+, IL-10+, or IL-10+IFN-γ+ cytokine-stained sub-populations were then separated from each by automated cell sorting, and their total RNA purified. To verify the integrity of these cytokine-secreting sub-populations, cytokine transcript levels were assessed by qPCR, results show mean expression data from three of the four donors, n=3. D) qPCR was also used to verify the Th1 identity of the sorted cell populations in three of the four donors, n=3. E) RNA samples from the same cytokine-secreting sub-populations from the 4 biological replicates, as well as RNA from unstimulated cells, and cells stimulated for only 2 hrs with either CD3/CD28 or CD3/CD46, were analysed using hybridisation microarrays with LNA probes covering 840 human miRs. Significant analysis of microarrays (median FDR=0%) identified 56 differentially-regulated miRNAs in different human T cell populations before/after activation. qPCR results show mean ± S.D. of at least 3 biological replicates. *, p < 0.05 **, p < 0.01, ***, p < 0.001 by one-way ANOVA. mRNA expression is relative to unstimulated cells (ΔΔCt method).
To investigate the influence of miRNAs on CD46-induced cytokine secretion, we isolated the three cytokine-secreting populations which develop after CD46 stimulation (IFN-γ+, IL-10+, and an intermediate IFN-γ+IL-10+ population (13)) at 36 hrs, and compared their miRNA profiles. To verify the integrity of the cell-sorted cytokine-secreting populations, mRNA coding for IFNG and IL10 were quantified by qPCR (figure 1C). In addition, the Th1 profile of the cells was verified by Tbet/GATA3 qPCR, which demonstrated that these cells were Tbet-positive but GATA3-negative (figure 1D). RNA was also purified from resting CD4+ T cells and either CD3+CD28 or CD3+CD46-stimulated T cells (2 hrs activation time). The miRNA expression profiles of these populations were then assessed using miRNA arrays covering over 840 human miRNAs. Expression data of miRNAs which were differentially expressed between any of the populations are shown in figure 1E, and mostly identified expression differences between the resting or early-activated T cells, and cytokine-secreting cells purified after 36 hrs. Interestingly, the most strongly upregulated miRNAs in the activated cytokine-secreting populations were mature products of pre-miR-886, which clustered together with activation marker miR-155. Pre-miR-886 has since been classified as a small non-coding RNA regulating activation of protein kinase R (PKR) (29). We have since determined that this RNA is upregulated during both CD28 and CD46 costimulation (although much more strongly in response to CD46) and regulates PKR activity in human T cells (manuscript in preparation).
CD46 costimulation drives enhanced CD4+ activation and functional changes in miRNA expression
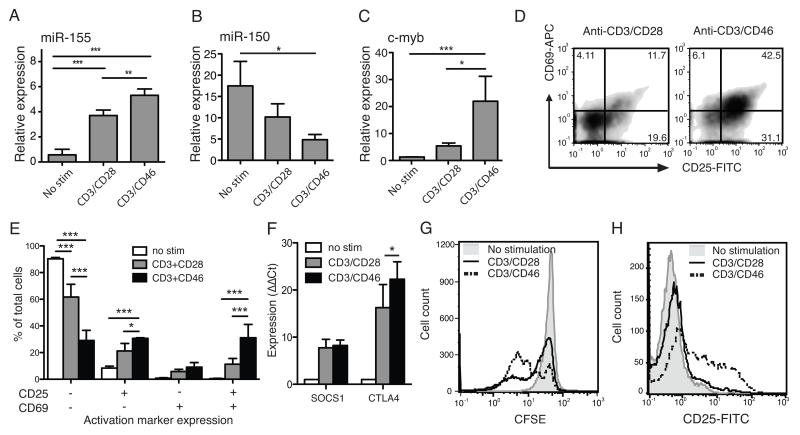
Array data showed strong upregulation of miR-155 and downregulation of miR-150 after CD46-mediated costimulation. To compare this effect with CD28, purified CD4+ T cells were stimulated with either CD3/CD28 or CD3/CD46 and subsequent miRNA expression evaluated by qPCR. miR-155, which is upregulated in activated lymphocytes and is necessary for their normal function (30), was more highly upregulated in cells costimulated with CD46 in comparison to CD28 (figure 2A), whereas miR-150 was more strongly downregulated in CD46-costimulated cells (figure 2B). A known target of miR-150 is the transcription factor c-Myb (31), and in negative correlation to miR-150 downregulation, c-Myb was most strongly upregulated in CD46-stimulated T cells (figure 2C), showing a functional outcome of stronger miR-150 downregulation after CD46 costimulation.
Fig. 2. CD46 delivers a stronger costimulatory signal than CD28.
A) Purified human CD4+ T cells were stimulated for 36 hrs using immobilised plate-bound antibodies as indicated, then RNA purified and expression of miR-155 (A) and miR-150 (B) assessed by qPCR. C) The expression of miR-150 target c-Myb was also assessed by qPCR in activated cells. D) Cells activated overnight as indicated were also stained for activation markers CD25 and CD69. Numbers denote cell percentage in each quadrant. E) The proportions of cells expressing CD25 and/or CD69 from three independent repeats are shown. F) SOCS1 and CTLA4 expression in cells stimulated for 36 hrs was quantified by qPCR. G) Cell proliferation after either CD28 or CD46 costimulation was assessed by CFSE dilution. H) Purified naïve CD4+ T cells were stimulated with anti-CD3 and either anti-CD28 or CD46 and upregulation of CD25 assessed after overnight incubation. Flow cytometry plots and histograms are from one representative experiment of at least three repeats. Bar graphs show mean ± standard deviation (S.D.) of at least three independent repeats. *, p < 0.05, **, p < 0.01, ***, p < 0.005 by one-way (A-C) two-way (E, F) ANOVA.
miR-155 upregulation is strongly associated with T cell activation, and we therefore investigated whether CD46 costimulation led to increased activation compared to CD28. Cell activation was assessed by measurement of CD25 (the high affinity IL-2 receptor α-chain) and CD69 expression after overnight stimulation, demonstrating that CD46 costimulation resulted in higher proportions of activated cells (figure 2D, E). However, despite increased levels of miR-155, we did not detect measurable decreases in the negative regulators of T-cell activation, CTLA4 or suppressor of cytokine signaling 1 (SOCS1), which have been described as targets of miR-155 (32, 33) (figure 2F). Both these genes are upregulated during activation, and CTLA4 upregulation was significantly higher after CD46 compared to CD28 costimulation, also reflecting the greater costimulatory potential of CD46. In addition, a significantly lower number of CD4+ T cells remained undivided 90 hrs after CD28 costimulation as compared to CD46 costimulation (figure 2G). In sum, these data support a significantly stronger costimulatory potential of CD46 when compared to CD28. This was not due to the preferential CD46-mediated activation and expansion of a specific T cell subset, because CD46 costimulation also resulted in increased CD25 upregulation in purified naïve CD4+ T cells, compared to CD28 (figure 2H). Titration experiments show that this is not due to a difference in affinity of the anti-CD28 and CD46 antibodies (data not shown).
miR-150 controls GLUT1 expression in human T cells
T cell activation results in rapid proliferation, and costimulation is a requirement for the associated upregulation of glucose metabolism that is needed for increased energy expenditure (34). CD28 activation in mice, and particularly CD46 costimulation in human CD4+ T cells, have been shown to be critical in the upregulation of the high-affinity cell surface glucose transporter 1 (GLUT1), encoded by the SLC2A1 gene, leading to a subsequent increase in glycolytic glucose consumption (35, 36). We have already identified that CD46 gives a stronger costimulatory signal in terms of the outcomes of activation and proliferation, and as such it is expected that there would also be a greater energy requirement. Positions 573-580 of the 3’ UTR of SLC2A1 mRNA contains an 8-mer match to the seed sequence of miR-150, which is conserved amongst several mammalian species, including chimpanzee and rhesus macaque (but not rat or mouse), and we therefore investigated whether the greater miR-150 downregulation associated with CD46 stimulation may result in enhanced upregulation of GLUT1 expression.
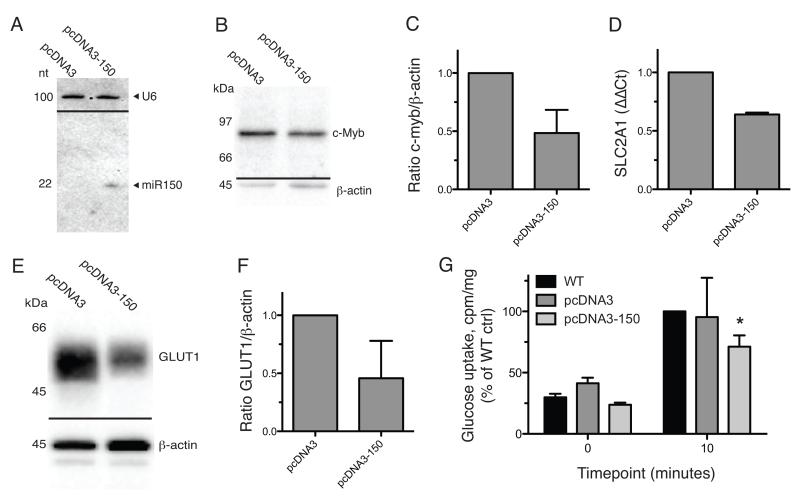
We generated stably transfected Jurkat clones which overexpress mature miR-150, as confirmed by Northern blot (figure 3A). miR-150 expression was within physiological levels as assessed by qPCR (not shown), and was functional as confirmed by 52±19% down-regulation of c-Myb protein in these cells (figure 3B, quantified in 3C). Compared to cells carrying the empty vector alone, Jurkat cells expressing miR-150 had significantly lower levels of both the SLC2A1 transcript (figure 3D) and the encoded GLUT1 protein (figure 3E, quantified in 3F, 56±27% downregulation). These same cells were also less efficient at glucose uptake in comparison to controls (figure 3G), verifying the functional significance of the measured GLUT1 downregulation.
Fig. 3. SLC2A1 (GLUT1) is a target of miR-150 in T cells.
In order to test miR-150 targeting of the SLC2A1 transcript, Jurkat cells stably transfected with a pre-miR-150 expressing vector were established. A) Processing of pre-miR-150 to the mature 22nt miR-150, in cells transfected with pcDNA3 carrying pre-miR-150, was assessed by northern blot. Panel shows overlayed images of the same membrane probed for miR-150 probe and U6 loading control. Representative of three repeats. B) To assess function of the expressed miR-150, Jurkat lysates were blotted for known miR-150 target c-Myb. Results from 3 repeats were assessed by densitometry as shown in panel C. D) the relative levels of SLC2A1 transcript in Jurkat and Jurkat-150 cells were assessed by qPCR. E) Protein levels of GLUT1 were assessed in Jurkat lysates by western blot. Results from 3 repeats were assessed by densitometry (F). G) Uptake of radio-labeled glucose in Jurkat cell lysates was assessed and normalized to total protein, showing lower uptake rates in miR-150 over-expressing cells. *, p < 0.05 compared to WT, by 2-way ANOVA.
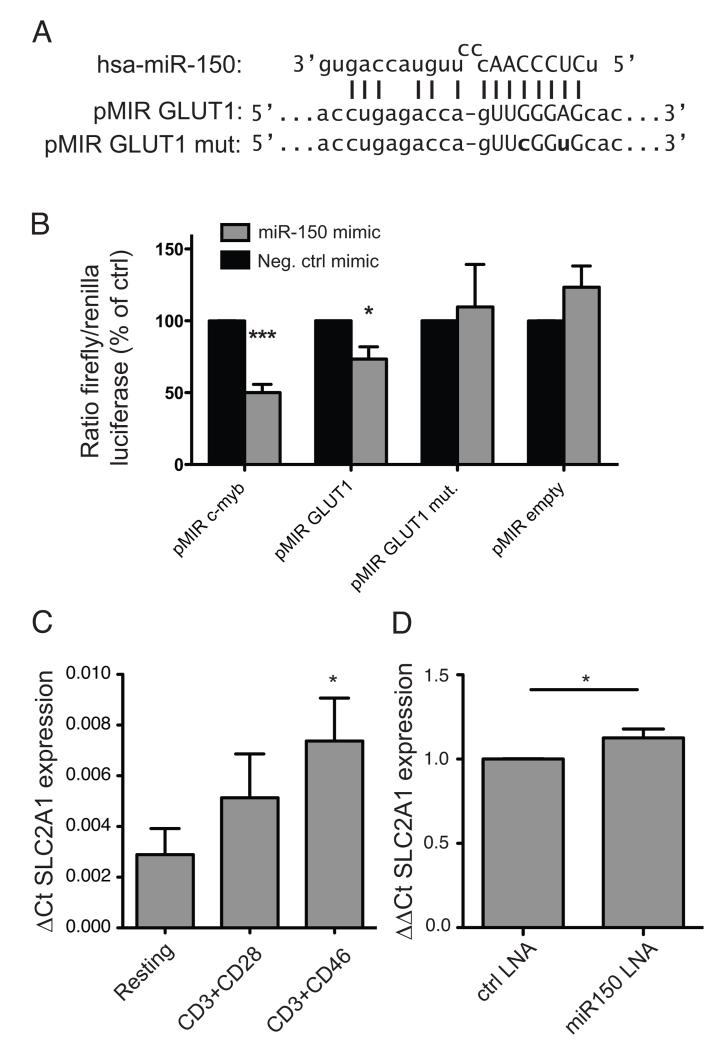
To confirm the active target site of miR-150, the 3’ UTR of SLC2A1 was cloned into a luciferase reporter vector, which was then subjected to targeted mutagenesis to alter the seed site and disrupt miR-150 binding (figure 4A). We found that although miR-150 caused a trend for increased luciferase signal in the control reporter vector, it significantly reduced the amount of luciferase signal for the transcript carrying the SLC2A1 3′ UTR (figure 4B). The positive control miR-150 target, carrying the c-Myb 3′UTR, was more strongly downregulated, possibly due to this sequence carrying multiple miR-150 seed sites (31). Mutating residues within the SLC2A1 miR-150 seed binding site prevented regulation by miR-150, returning luciferase signal to that of the control, confirming the identified miR-150 binding site. Consistent with miR-150 regulation of SLC2A1 translation, CD46 costimulation leads to a greater upregulation of SLC2A1 transcript (figure. 4C), compared to CD28 costimulation. Finally, nucleofection of primary human CD4+ T cells with miR-150 locked nucleic acid (LNA) antisense oligomers also resulted in a significant increase in the SCL2A1 mRNA, as detected by qPCR (figure. 4D). Together, these results confirm miR-150 regulation of the SLC2A1 transcript in human T cells.
Fig. 4. Identification of the miR-150 seed site in SLC2A1.
A) The predicted miR-150 binding site in the SLC2A1 transcript UTR is shown with the seed site in capitals and predicted complimentary nucleotides indicated by vertical lines. The mutant UTR sequence is shown below, containing seed sequence substitutions indicated in bold. B) The SLC2A1 UTR was cloned into the pMIR-Report firefly luciferase reporter vector, along with mutant version and c-Myb UTR control, and transfected into HEK cells together with a Renilla luciferase control vector and miR-150 or control miRNA mimic. Luciferase production was read after 24 hrs. C) To explore miR-150 regulation of SLC2A1 in primary T cells, SLC2A1 transcript levels in miR-150-low CD3/CD46 costimulated cells was assessed by qPCR, compared to resting or CD3/CD28 costimulated CD4+ cells. D) Nucleofection of LNA miR-150 inhibitor into primary CD4+ T cells also led to a significant increase in SLC2A1 transcript. All results show mean and SD of at least 3 independent repeats. *, p < 0.05, ***, p < 0.005 by two-way ANOVA (B) or one-way ANOVA (C) or two-tailed T-test (D).
Autocrine CD46 signaling affects miR-150 target levels and cytokine secretion in CD28-costimulated CD4+ T cells
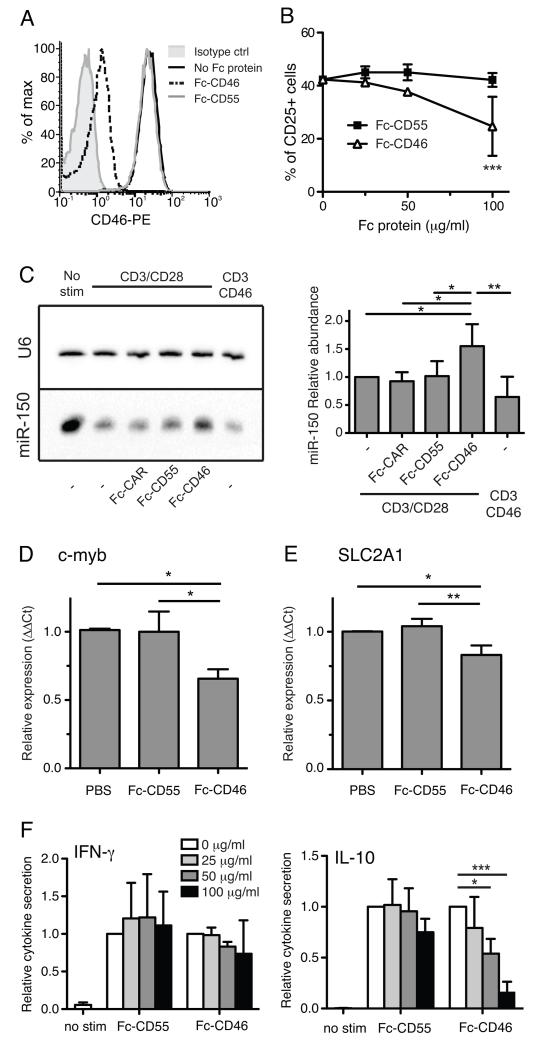
Previous experiments have shown that soluble forms of CD46 can compete with cell-surface CD46 for activating ligands, and impede CD46-mediated T cell signaling (10). In order to investigate physiological roles of autocrine CD46 signaling, we inhibited CD46 ligation during T cell activation (usually mediated by T cell-derived C3b or Jagged1 expression) by administration of a soluble Fc-CD46 fusion protein. Addition of Fc-CD46 to Jurkat cells blocked antibody binding of cell surface CD46 (figure 5A), demonstrating the ability of Fc-CD46 to competitively inhibit binding of ligands. Treatment of T cells during CD3 and CD28 stimulation with Fc-CD46, but not Fc-tagged CD55 (another complement regulator expressed on T cells), reduced the upregulation of activation marker CD25 (figure 5B), confirming that auto- or juxtacrine CD46 signaling is involved downstream of CD28 costimulation during T cell activation. Having already established that CD46 signaling leads to accelerated down-regulation of miR-150 expression, we also confirmed that addition of Fc-CD46 as a soluble competitor of autocrine CD46 signaling prevented miR-150 downregulation (figure 5C). We then assessed expression of the miR-150 target transcripts c-Myb and SLC2A1 in CD4+ T cells activated via CD3/CD28 in the presence of the blocking Fc-CD46. After overnight activation, cells treated with Fc-CD46 had lower expression of c-Myb (figure 5D) and SLC2A1 (figure 5E) transcripts, consistent with a higher miR-150 level caused by interruption of autocrine CD46 signaling. This data shows the importance of autocrine CD46 signaling during CD28-costimulated T-cell activation, and implies a role for CD46-induced regulation of miR-150 targets downstream of CD28. Blocking of CD46 signaling by addition of Fc-CD46 also inhibited secretion of IL-10, but not IFN-γ, from cells stimulated over 36 hrs by anti-CD3 and anti-CD28 (figure 5F), consistent with the known role of CD46 in induction of Th1 IL-10 secretion, downstream of CD28 (13). We therefore instigated further investigation of the function of CD46-mediated signaling, and in particular downstream miRNA expression, in the cytokine-secreting sub-populations of CD46-activated Th1 T cells.
Fig. 5. Soluble CD46 affects cytokine secretion during CD3/CD28 stimulation of purified CD4+ T cells.
A) To test the ability of soluble Fc-CD46 to competitively inhibit binding of ligands to cell-surface CD46, Jurkat cells were stained with anti-CD46-PE in the presence of either Fc-CD46 or Fc-CD55 (each at 15 mg/ml). B) The presence of titrated Fc-CD46, but not Fc-CD55, inhibited overnight activation of CD4+ T cells by anti-CD3/anti-CD28 antibodies, as measured by upregulation of activation marker CD25. C) Addition of Fc-CD46, but not Fc-DAF or Fc-CAR, inhibited down-regulation of miR-150 during CD3/CD28 stimulation, as detected by northern blotting. Panels show representative blot (left) and quantification from three independent repeats (right). D) Addition of Fc-CD46 (50 μg/ml) but not controls during CD3/CD28 costimulation led to a decrease in miR-150 target transcripts c-Myb (D) and SLC2A1 (E) in T cells stimulated overnight, as measured by qPCR. F) Presence of Fc-CD46 but not Fc-CD55 also inhibited the secretion of IL-10 (right panel) but not IFN-γ (left panel) from CD3/CD28 stimulated CD4+ T cells, as measured by ELISA. Histogram is representative of 3 independent experiments. Panels B-E show mean ± S.D. from 3 independent experiments. *, p < 0.05, **, p < 0.01, ***, p < 0.005 by one-way ANOVA (C, D, E) or two-way ANOVA (B, F).
Hsa-miR-150 is required for IL-10-secretion in CD46-stimulated regulatory Th1 cells
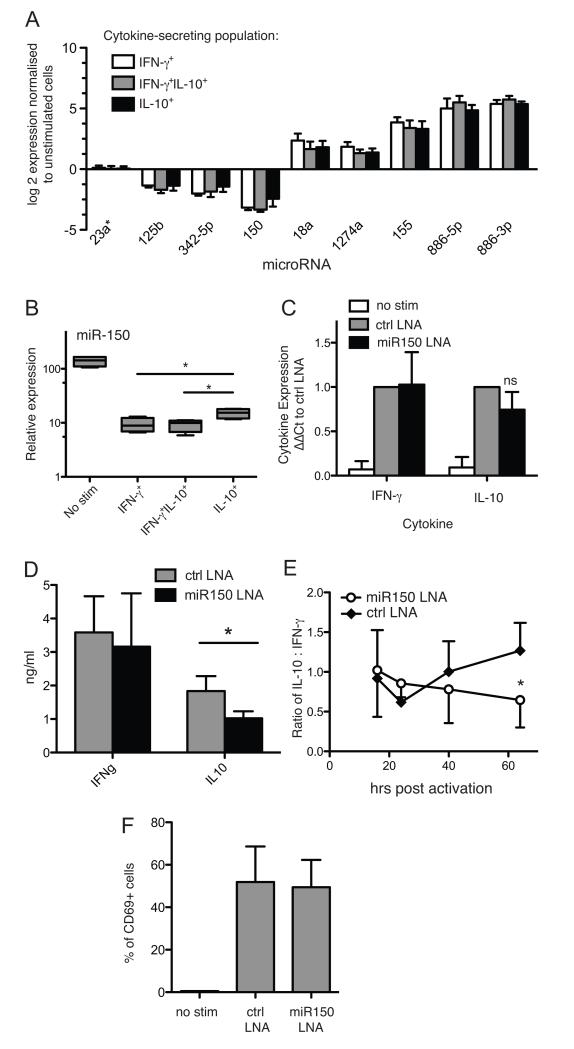
The array data was analysed to identify miRNAs, which were differentially expressed between the three CD46-induced cytokine-secreting populations (figure 6A). Differential expression of miR-150 was verified by qPCR (figure 6B), revealing a significantly increased expression in the regulatory IL-10+ cells compared to IFN-γ+ and IFN-γ+/ IL-10+ cells. To assess the function of elevated miR-150 in IL-10+ feedback, we used a specific LNA inhibitor. After transfection of primary T cells with miR-150 antisense LNA, there was a statistically non-significant trend for reduction of IL-10 mRNA in cells stimulated for 60 hrs via CD3 and CD46 (figure 6C). However, this translated into a significant reduction in IL-10 protein secretion as measured by ELISA, while IFN-γ secretion was unaffected (figure 6D). This was confirmed in separate time course experiments (figure 6E). Overall activation of cells, as measured by CD69 upregulation, were not affected (figure 6F), and neither was proliferation (not shown). These results identify miR-150 as a positive regulator of IL-10 cytokine secretion in Th1 cells.
Fig. 6. miR-150 is over expressed in IL-10 secreting CD46-induced CD4+ T cells, and controls IL-10 production.
A) Statistical analysis of the array data from the 3 cytokine-secreting CD46-activated populations from 4 donors revealed several miRNAs differentially expressed in the different populations. B) qPCR confirmed differential expression of miR-150 among the cytokine-secreting populations, using the same RNA from the four donors used for arrays in figure 1 (four biological replicates, 3 technical repeats). C, D) Purified human CD4+ T cells were transfected with miR-150 LNA antagonist or control LNA, and stimulated via CD3/CD46 for 60 hrs. IFN-γ and IL-10 expression and secretion were then assessed by qPCR (C) and ELISA (D). E) In separate experiments, CD4+ T cells were nucleofected with miR-150 LNA antagonist or control and stimulated via CD3/CD46 and supernatant samples taken over a time course. Values show ratio of measured IL-10 to IFN-γ. F) LNA nucleofected cells were stimulated via CD3/CD46 for 18 hrs and expression of activation marker CD69 assessed by flow cytometry. Graphs show means and S.D. from at least three biological replicates tested in independent experiments. *, p < 0.05, ***, p < 0.005 by two-way ANOVA, or one-way ANOVA for panel B.
Discussion
As miRNAs have now emerged as key players in the regulation of cellular functions, we have assessed if CD46 costimulation impacts on this important immune-regulative system in T cells. Indeed, CD46-mediated signals not only induce a specific miRNA landscape early upon T cell activation, but the levels of several specific miRNAs are then subsequently modulated during the CD46-induced Th1 switching phase. CD46 has dual roles in T cell activation, being required both for initiation of Th1 responses but also for inducing regulation in the form of Th1-derived IL-10 secretion. We have found evidence for CD46-induced miRNA expression patterns which affect both of these phases of the Th1 T cell response. miR-150 is expressed most abundantly in lymphocytes, and has an important role in regulating c-Myb expression in B cell development (31). c-Myb itself is a transcription factor with prolific binding sites across the genome (37), but has been shown to interact with GATA3 in induction and maintenance of Th2 responses (38). We found c-Myb to be upregulated during T cell activation, in concordance to a general downregulation in miR-150. Regulation of gene expression by miRNAs is an area of growing complexity, with modes of action determined at transcriptional and post-transcriptional levels (39), in manners which may or may not affect targets at the mRNA level. The action of a miRNA on any specific target can also depend on the relative levels of competing endogenous target mRNAs (40), as well as the expression of non-coding regulatory ‘sponge’ RNAs, which bind to and neutralise miRNAs in a sequence-specific manner (41). This therefore makes it hard to predict outcomes at the protein level from RNA data alone (42). This, in addition to the lack of a representative animal model for the immune functions of CD46, and ample evidence for its role in human disease (10, 12, 14, 43), makes the delineation of the molecular pathways of CD46 signaling in disease both an important and challenging task.
During activation, we found that CD46 is a potent costimulatory receptor, leading to more rapid upregulation of miR-155 compared to CD28-costimulation. miR-155 is necessary for normal immune function (30), and becomes upregulated during activation. T cell activation leads to rapid growth and proliferation, and therefore places a great metabolic demand on the cell. Glycolysis upregulation is a hallmark of T cell activation, and it is already known that costimulation is required for GLUT1 upregulation and an increase in glucose uptake, which is a limiting step in T cell activation (34, 35). GLUT1 is the most important T cell glucose transporter, having been found essential for both activation and effector function (44). It is also now known that CD46 signaling acts downstream of CD28 costimulation during T cell activation, and is strictly required for initiation of Th1 responses (12). When we and others block signaling using soluble forms of CD46 which competitively inhibit interaction of cell surface CD46 with autocrine stimulatory ligands, T cell activation is diminished (45).
We have recently shown that increased glycolysis upon T cell activation, which is specifically required for Th1 induction (46), is dependent on CD46 signaling (36), and that CD28 activation alone is not sufficient to induce the high levels of GLUT1 (and LAT1) expression, which mediate increased glucose and amino acid influx. Our finding that CD46 signaling leads to a rapid downregulation of miR-150 expression is tied into this, by the finding that miR-150 targets the SLC2A1 transcript encoding the glucose transporter GLUT1. We have experimentally confirmed the conserved miR-150 seed site at position 573-580 of the SLC2A1 3’UTR, and shown that the SLC2A1 transcript is more abundant in T cell populations with lower miR-150 levels, and that CD46 costimulation results in lower miR-150 expression and higher SLC2A1/GLUT1. In addition, blocking of autocrine CD46 costimulation limits miR-150 downregulation and resulted in decreased SLC2A1 levels and limited activation. This reveals a novel level of control of cellular metabolism by CD46 during T cell activation, via regulation of miR-150 expression, and in turn, control of GLUT1-dependent glycolysis.
Cellular metabolism is thought to not only be important for T cell activation, but also subset differentiation (47). CD46 leads not only to potent T cell activation, but also to the development of an IL-10-secreting regulatory T-cell population (13). These cells develop from the IFN-γ+ pro-inflammatory population. We found that in this population of CD46-induced Tr1-like cells, there was an increased level of miR-150 compared to the pro-inflammatory cytokine-secreting populations, meaning that the initial reduction of miR-150 in activated Th1 cells is partially reversed as the cells switch to a regulatory phenotype. GLUT1 and glycolysis also have a role in defining cytokine secretion and different T cell subsets. Inhibition of glycolysis has been found to compromise the ability of T cells to produce IFN-γ, by freeing up GAPDH from the glycolysis pathway, when it is then able to play an alternative role by binding to the UTR of IFNG mRNA, limiting translation (46). This is of particular interest in the CD46-induced IL-10-secreting Th1 cells, which have higher levels of miR-150 compared to either IFN-γ or IFN-γ/IL-10 secreting Th1 cells, and have also undergone a switch to cease IFN-γ production. Lower rates of glycolysis are therefore linked to non-production of IFN-γ, consistent with the observation that regulatory T cells have a lower relative rate of glycolysis compared to effector T cells, relying instead on oxidative phosphorylation and lipid oxidation (48). It is therefore possible that increased miR-150 expression in the ‘switched’ IL-10 secreting Th1 cells contributes to the shutdown of IFN-γ secretion. miR-150 likely regulates several targets relevant for the establishment and maintenance of the regulatory Th1 phenotype, typical of the described function of miRNAs, which can achieve regulation of phenotypes by inhibiting multiple targets within the same processes or pathways, with a cumulative effect (49, 50).
miR-150 also has known roles in cancer. It has previously been described that miR-150 regulates FLT3 in leukaemia (51), but miR150 is also downregulated in mucosal associated lymphoid tissue lymphoma, Burkitt’s lymphoma, (52), Hodgkin lymphoma (53) and other B and T cell lymphoma cell lines (54) as well as primary human T and NK cell lymphomas (55, 56). Forced over-expression studies in lymphocytes leads to lower proliferation and increased apoptosis, mirroring the role for miR-150 downregulation as a requirement for initiation of T-cell proliferation after activation. Aerobic glycolysis is also highly upregulated in cancer, and the loss of miR-150 expression in various lymphomas may play a role in upregulation of GLUT1 in the development of this disease.
Currently we have not yet investigated the relationship between miR-150 and c-Myb in IL-10+ Th1 cells, although c-Myb has been previously identified as a marker of inducible Tregs (57). The binding of c-Myb to promoters is also regulated by the availability of those sequences, which alter with chromatin structure state and even cell cycle phase (58). C-Myb may therefore be regulating different gene sets in the IL-10+ IFN-γ− subset as compared to the IFN-γ+ cells. siRNA-mediated knockdown of CD4+ c-Myb led to a general suppression of T cell activation, limiting IFN-γ secretion and development of the subsequent IL-10+ populations (data not shown), a result which, unfortunately, does not help to shed light on any specific function for c-Myb within the specific IL-10+ IFN-γ− regulatory sub-population. The targeting of immune cell-expressed CD46 itself for therapy is complicated due to its differing roles at different phases of the response, as explained above, and its near-ubiquitous expression on other non-immune cell types. Better understanding of downstream effector systems may therefore offer improved opportunities to modulate this key immunological pathway. Recently, the targeting of T-cell metabolism has been demonstrated to be a potentially effective means of therapy for modulating T-cell mediated autoimmunity (59), and an improved understanding of the roles of CD46-dependent microRNAs involved in T-cell metabolism offer additional therapeutic opportunities. We therefore propose the highly lymphocyte-enriched miR-150 as a potential target for therapeutic intervention for modulation of IL-10 feedback and regulation of Th1 responses in infectious disease or autoimmunity.
Acknowledgements
The authors acknowledge the support of the MRC Centre for Transplantation. We are grateful to Dr Yvonne Ceder for reagents and advice, and to Liza Lind for technical assistance.
This work was supported by Swedish Research Council (project K2012-66X-14928-09-5, to AB, and K2012-55X-13147-14-5, to LE), Cancerfonden (AB), and Foundations of Österlund (AB), Greta and Johan Kock (BK, AB), Gustav V 80-years anniversary (AB), Knut and Alice Wallenberg (AB) and Inga-Britt and Arne Lundberg (AB) as well as grants for clinical research (Skåne University Hospital and ALF, to AB), the Anna-Greta Crafoord foundation for rheumatological research (BK), The Wellcome Trust (grant 102932/Z/13/Z to CK), the Medical Research Council (grant G1002165 to CK) and the EU-funded Innovative Medicines Initiative BTCURE (CK). LE is a senior researcher at the Swedish Research Council. The research was also funded by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Abbreviations used in this article
- LNA
locked nucleic acid
- miRNA
microRNA
- miR-150
microRNA-150
- FLT3
fms-related tyrosine kinase 3
- GLUT1
Glucose transporter 1
- SLC2A1
Solute carrier family 2, facilitated glucose transporter membrane 1
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 2.Gazzinelli RT, Wysocka M, Hieny S, Scharton-Kersten T, Cheever A, Kuhn R, Muller W, Trinchieri G, Sher A. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J Immunol. 1996;157:798–805. [PubMed] [Google Scholar]
- 3.Hunter CA, Ellis-Neyes LA, Slifer T, Kanaly S, Grunig G, Fort M, Rennick D, Araujo FG. IL-10 is required to prevent immune hyperactivity during infection with Trypanosoma cruzi. J Immunol. 1997;158:3311–3316. [PubMed] [Google Scholar]
- 4.Trinchieri G. Interleukin-10 production by effector T cells: Th1 cells show self control. J Exp Med. 2007;204:239–243. doi: 10.1084/jem.20070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, Wynn TA, Kamanaka M, Flavell RA, Sher A. Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J Exp Med. 2007;204:273–283. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–297. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabrysova L, Nicolson KS, Streeter HB, Verhagen J, Sabatos-Peyton CA, Morgan DJ, Wraith DC. Negative feedback control of the autoimmune response through antigen-induced differentiation of IL-10-secreting Th1 cells. J Exp Med. 2009;206:1755–1767. doi: 10.1084/jem.20082118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meiler F, Zumkehr J, Klunker S, Ruckert B, Akdis CA, Akdis M. In vivo switch to IL-10-secreting T regulatory cells in high dose allergen exposure. J Exp Med. 2008;205:2887–2898. doi: 10.1084/jem.20080193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O’Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardone J, Le Friec G, Vantourout P, Roberts A, Fuchs A, Jackson I, Suddason T, Lord G, Atkinson JP, Cope A, Hayday A, Kemper C. Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat Immunol. 2010;11:862–871. doi: 10.1038/ni.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Astier A, Trescol-Biemont MC, Azocar O, Lamouille B, Rabourdin-Combe C. Cutting edge: CD46, a new costimulatory molecule for T cells, that induces p120CBL and LAT phosphorylation. J Immunol. 2000;164:6091–6095. doi: 10.4049/jimmunol.164.12.6091. [DOI] [PubMed] [Google Scholar]
- 12.Le Friec G, Sheppard D, Whiteman P, Karsten CM, Shamoun SA, Laing A, Bugeon L, Dallman MJ, Melchionna T, Chillakuri C, Smith RA, Drouet C, Couzi L, Fremeaux-Bacchi V, Kohl J, Waddington SN, McDonnell JM, Baker A, Handford PA, Lea SM, Kemper C. The CD46-Jagged1 interaction is critical for human TH1 immunity. Nat Immunol. 2012;13:1213–1221. doi: 10.1038/ni.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemper C, Chan AC, Green JM, Brett KA, Murphy KM, Atkinson JP. Activation of human CD4+ cells with CD3 and CD46 induces a T-regulatory cell 1 phenotype. Nature. 2003;421:388–392. doi: 10.1038/nature01315. [DOI] [PubMed] [Google Scholar]
- 14.Cattaneo R. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol. 2004;78:4385–4388. doi: 10.1128/JVI.78.9.4385-4388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujimura A, Shida K, Kitamura M, Nomura M, Takeda J, Tanaka H, Matsumoto M, Matsumiya K, Okuyama A, Nishimune Y, Okabe M, Seya T. Molecular cloning of a murine homologue of membrane cofactor protein (CD46): preferential expression in testicular germ cells. Biochem J. 1998;330(Pt 1):163–168. doi: 10.1042/bj3300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torok K, Kremlitzka M, Sandor N, Toth EA, Bajtay Z, Erdei A. Human T cell derived, cell-bound complement iC3b is integrally involved in T cell activation. Immunol Lett. 2012;143:131–136. doi: 10.1016/j.imlet.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J Clin Invest. 2006;116:3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limon P, Kaech SM, Nakayama M, Rinn JL, Flavell RA. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38:984–997. doi: 10.1016/j.immuni.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris CL, Spiller OB, Morgan BP. Human and rodent decay-accelerating factors (CD55) are not species restricted in their complement-inhibiting activities. Immunology. 2000;100:462–470. doi: 10.1046/j.1365-2567.2000.00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 25.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaffran Y, Destaing O, Roux A, Ory S, Nheu T, Jurdic P, Rabourdin-Combe C, Astier AL. CD46/CD3 costimulation induces morphological changes of human T cells and activation of Vav, Rac, and extracellular signal-regulated kinase mitogen-activated protein kinase. J Immunol. 2001;167:6780–6785. doi: 10.4049/jimmunol.167.12.6780. [DOI] [PubMed] [Google Scholar]
- 28.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY, Bang JY, Park HS, Leelayuwat C, Lee YS. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, Stenvang J, Kauppinen S, Alenius H, Lauerma A, Homey B, Winqvist O, Stahle M, Pivarcsi A. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581–589. e581–520. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 33.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM, Perret R, Muljo SA, Hebeisen M, Rufer N, Zehn D, Donda A, Restifo NP, Held W, Gattinoni L, Romero P. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 36.Kolev M, Dimeloe S, Le Friec G, Navarini A, Arbore G, Povoleri GA, Fischer M, Belle R, Loeliger J, Develioglu L, Bantug GR, Watson J, Couzi L, Afzali B, Lavender P, Hess C, Kemper C. Complement Regulates Nutrient Influx and Metabolic Reprogramming during Th1 Cell Responses. Immunity. 2015;42:1033–1047. doi: 10.1016/j.immuni.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintana AM, Liu F, O’Rourke JP, Ness SA. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer. 2011;11:30. doi: 10.1186/1471-2407-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakata Y, Brignier AC, Jin S, Shen Y, Rudnick SI, Sugita M, Gewirtz AM. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood. 2010;116:1280–1290. doi: 10.1182/blood-2009-05-223255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 42.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 43.Astier AL. T-cell regulation by CD46 and its relevance in multiple sclerosis. Immunology. 2008;124:149–154. doi: 10.1111/j.1365-2567.2008.02821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truscott SM, Abate G, Price JD, Kemper C, Atkinson JP, Hoft DF. CD46 engagement on human CD4+ T cells produces T regulatory type 1-like regulation of antimycobacterial T cell responses. Infect Immun. 2010;78:5295–5306. doi: 10.1128/IAI.00513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CH, Curtis JD, Maggi LB, Jr., Faubert B, Villarino AV, O Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM. Glucose metabolism regulates T cell activation, differentiation, and functions. Front Immunol. 2015;6:1. doi: 10.3389/fimmu.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 50.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, Wang J, Arnovitz S, Elkahloun A, Price C, Hong GM, Ren H, Kunjamma RB, Neilly MB, Matthews JM, Xu M, Larson RA, Le Beau MM, Slany RK, Liu PP, Lu J, Zhang J, Chen J. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai J, Liu X, Cheng J, Li Y, Huang X, Ma X, Yu H, Liu H, Wei R. MicroRNA-200 is commonly repressed in conjunctival MALT lymphoma, and targets cyclin E2. Graefes Arch Clin Exp Ophthalmol. 2012;250:523–531. doi: 10.1007/s00417-011-1885-4. [DOI] [PubMed] [Google Scholar]
- 53.Gibcus JH, Tan LP, Harms G, Schakel RN, de Jong D, Blokzijl T, Moller P, Poppema S, Kroesen BJ, van den Berg A. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, Wang Z, Dai X, Pan J, Ge J, Han X, Wu Z, Zhou X, Zhao T. Re-expression of microRNA-150 induces EBV-positive Burkitt lymphoma differentiation by modulating c-Myb in vitro. Cancer Sci. 2013;104:826–834. doi: 10.1111/cas.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, Shimizu N, Sawada K. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 56.Ito M, Teshima K, Ikeda S, Kitadate A, Watanabe A, Nara M, Yamashita J, Ohshima K, Sawada K, Tagawa H. MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma. Blood. 2014;123:1499–1511. doi: 10.1182/blood-2013-09-527739. [DOI] [PubMed] [Google Scholar]
- 57.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quintana AM, Zhou YE, Pena JJ, O’Rourke JP, Ness SA. Dramatic repositioning of c-Myb to different promoters during the cell cycle observed by combining cell sorting with chromatin immunoprecipitation. PLoS One. 2011;6:e17362. doi: 10.1371/journal.pone.0017362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med. 2015;7:274ra218. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]