Abstract
While performing several functions, adherent cells deform their surrounding substrate via stable adhesions that connect the intracellular cytoskeleton to the extracellular matrix. The traction forces that deform the substrate are studied in mechanotrasduction because they are affected by the mechanics of the extracellular milieu. We review the development and application of two methods widely used to measure traction forces generated by cells on 2D substrates: i) traction force microscopy with polyacrylamide hydrogels and ii) calculation of traction forces with arrays of deformable microposts. Measuring forces with these methods relies on measuring substrate displacements and converting them into force. We describe approaches to determine force from displacements and elaborate on the necessary experimental conditions for this type of analysis. We emphasize device fabrication, mechanical calibration of substrates and covalent attachment of extracellular matrix proteins to substrates as key features in the design of experiments to measure cell traction forces with polyacrylamide hydrogels or microposts. We also report the challenges and achievements in integrating these methods with platforms for the mechanical stimulation of adherent cells. The approaches described here will enable new studies to understand cell mechanical outputs as a function of mechanical inputs and the understanding of mechanotransduction mechanisms.
Keywords: Traction Force Microscopy, Polyacrylamide Hydrogels, PDMS Microposts, Displacements, Mechanical Calibration, Surface Functionalization
1. Introduction
Mechanical forces between cells and the extracellular matrix (ECM) evolve during growth and development [1], tissue homeostasis [2, 3], and wound healing [4]. Changes in tissue mechanical properties have been linked to cancer metastasis [5, 6] and disease progression [7–9]. Understanding how mechanical cues affect cellular responses in these processes requires methods to measure cell-generated forces while modulating their mechanical environment. Forces between cells and their environment are transmitted across cell-ECM and cell-cell adhesions (Figure 1, inset). These forces are either generated externally and applied to cells or generated by cells and applied to the ECM. Cells convert externally applied forces to biological signals by mechanotransduction mechanisms, leading to changes in cell phenotypes (reviewed concisely in Vogel & Sheetz [10]). Some of the same structures involved in mechanotransduction also transmit forces generated by the contractile machinery of cells to the ECM. By generating forces, cells deform their local environment [9, 11], which can lead to remodeling of the extracellular environment [12].
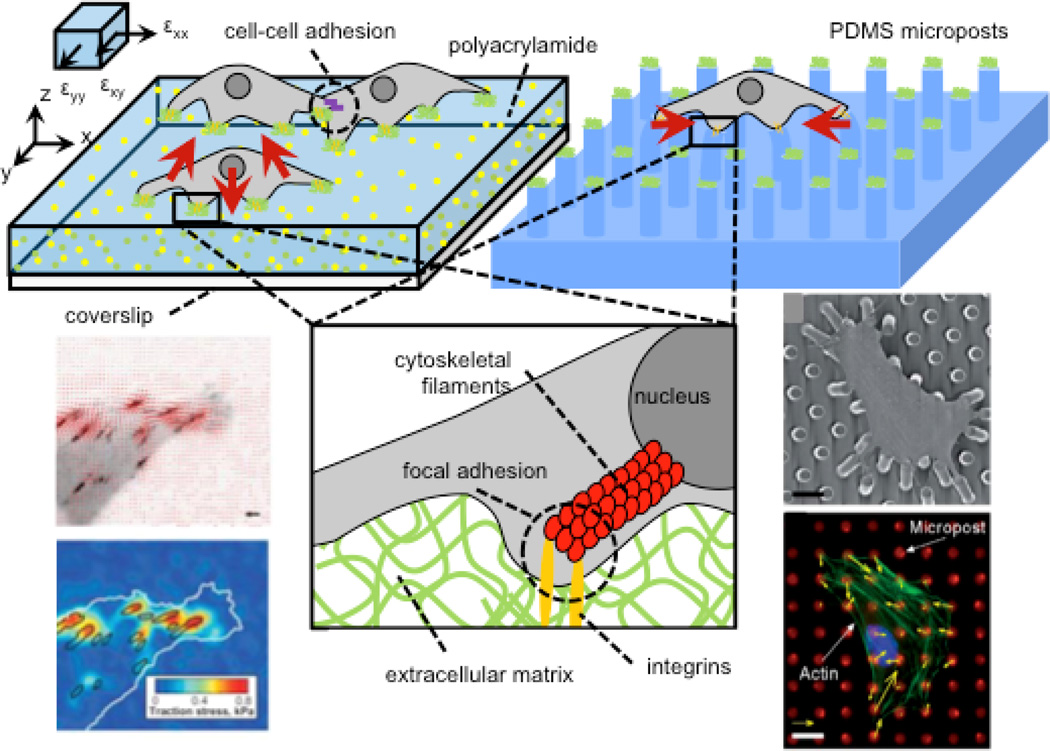
Figure 1. Measurement of cell traction forces with hydrogel traction force microscopy (TFM) and arrays of elastomeric microposts.
Cell contractile machinery produces traction forces (shown as red vectors), which are transmitted to the extracellular environment via focal adhesions, which transmit forces to the nucleus via cytoskeletal (figure inset) to enable mechanotransduction. Hydrogel platforms offer continuous substrates for cell adhesion whereas microposts provide discrete binding “islands.” In both systems, cell tractions are calculated from substrate displacements. 3D tractions can be computed on hydrogels if x-, y-, and z-axis deflections of fiducial markers are visualized by confocal microscopy. Figure adapted with permissions from [14, 19, 199]
In this review, we describe methods to measure traction forces exerted by cells through substrate interactions in 2D using hydrogel substrates and elastomeric micropost arrays. We discuss the importance of protein functionalization methods for both platforms and highlight their applications in mechanobiology studies. To guide those interested in measuring cell traction forces with these methods, we note key considerations and assumptions in: 1) calculation of force from acquired data, 2) fabrication of devices, 3) design of experiments, 4) engineering cell attachment to the surfaces of devices and 5) integration of these methods with platforms for mechanical stimulation. For detailed step-by-step fabrication and data analysis discussions, we refer readers to more targeted methods reviews. Style and Plotnikov provide methods and sample analysis code for traction force microscopy (TFM) [13, 14]. Yang, Fu and colleagues offer in-depth discussions of micropost arrays: [15, 16].
2. Traction Force Measurements using Hydrogel TFM and Elastomeric Micropost Arrays
The first qualitative reports of cell-ECM forces began in the 1980s when Harris and colleagues observed wrinkling of a silicone membrane due to fibroblast traction forces [17]. These observations inspired the design and fabrication of systems that could quantitatively measure traction forces. In 1999, Dembo and Wang introduced “traction force microscopy” as a method to quantify forces exerted by adherent cells on a hydrogel substrate with fiducial markers [18] (Figure 1). Fiducial markers are observed with a microscope, and their movement is computationally determined from images acquired before and after changes in cell contractile activity. Adherent cells deform the substrate, and the resulting surface deformation field is determined from the displacement of fiducials and used to calculate cell traction forces using continuum mechanics models. In 2003, Tan and colleagues developed another commonly used system for measuring cell-ECM forces [19]. This system relies on an array of compliant microposts of constant controllable dimensions (diameter and height). Cells attach and contract to displace the microposts. Researchers observe the microposts using microscopy and the traction forces exerted by the cell are estimated from micropost displacements using beam bending theory.
TFM on hydrogel substrates and micropost deflection measurements allow for cell traction force analysis with spatial and temporal resolution set by microscopy acquisition parameters. These methods have similar aims, but they are fundamentally different systems, which may lead to differences in force calculations between both methods. Table 1 shows the range of forces that have been calculated in a set of studies with the two systems and highlights that the force values vary considerably between studies, even when using similar cell types. Comparisons across these methods and studies are difficult because experiments for both TFM on hydrogels and analysis of micropost deflection must be designed with self-consistent sets of control experiments using the same methods. We provide detail on TFM on hydrogel substrates and micropost deflection measurements in the next sections and discuss the factors in manufacturing, experimentation and data analysis that can lead to discrepancies in estimated force values and affect the accuracy of traction force measurements.
Table 1.
Cell tractions reported using hydrogel and micropost platforms vary widely with cell type, ECM, and substrate conditions.
| Substrate stiffness (if reported, kPa) |
Range of Traction Stress (kPa) |
Cell Type | Reference |
|---|---|---|---|
| Polyacrylamide hydrogels analyzed by traction force microscopy | |||
| 2.8 – 30 | 0.25 – 0.5 | T3T fibroblasts | [188] |
| 15.6 | 0.5 – 10 | Mouse embryo fibroblast | [43] |
| 10 | 0.15 – 0.80 | Invasive epithelial bladder cancer cells (T24) | [189] |
| 1.95 – 9.9 | 0.2 | Invasive epithelial bladder cancer cells (T24) | [50] |
| 6.2 | 1.32 – 2.48 | 3T3 fibroblasts | [47] |
| PDMS micropost arrays | |||
| Micropost stiffness (nN/µ m) | Force per post (nN) | Cell Type | Reference |
| 32 | 2 – 78 | Bovine pulmonary artery smooth muscle cells, 3T3 fibroblasts | [19] |
| 31 | 1 – 32 | Human pulmonary artery endothelial cells | [190] |
| 1.9 | 0.14 – 1.2 | Murine dendritic cells | [191] |
Comparison between these systems is complicated due to assumptions required about the substrate and cell-ECM adhesions.
2.1 Hydrogels for Traction Force Microscopy
One can estimate the traction forces exerted by cells cultured on compliant substrates by processing the displacement of fiducial markers in these materials (Figure 2). Polyacrylamide hydrogels are commonly used as TFM cell culture substrates [13] due to the deformability and widely tunable stiffness of these materials (in the range of 0.2–35 kPa [20]). Fluorescent microbeads are added into the polyacrylamide to act as fiducial markers and are imaged with fluorescence microscopy. Conventional TFM with epifluorescent microscopy assumes dominant 2D stresses and necessarily neglects the z-axis as it tracks only in-plane lateral microbead displacements (x- and y- axes in Figure 1). 3D TFM bypasses the assumption that normal forces are negligible [21–23]. Therefore, 3D TFM requires confocal microscopy to scan for z-axis substrate displacements and image processing to determine bead position with high resolution [23, 24]. This approach has revealed that substrate displacements, the direction of cell tractions and cell adhesions of cells on hydrogel surfaces have 3D complexity [23, 25], which are not detected with 2D TFM methodologies. As it occurs in 2D TFM, successful assessment of substrate force fields with 3D TFM also requires knowledge of substrate mechanical properties. Other deformable substrate materials, such as collagen [26–28] and fibrin [29], are used to encapsulate cells in 3D environments and track the forces they generate during cell-substrate interactions [30]. These materials are mechanically more complex than polyacrylamide gels [31, 32]. Collagen fibers remodel under applied force and imaging this remodeling with confocal reflectance microscopy provides a semi-quantitative measurement of cell-generated forces [33]. However, compared with 2D TFM, 3D TFM is more time consuming, more computationally expensive and involves longer imaging acquisition times. Such characteristics limit the use of 3D TFM for analyzing fast traction force measurements of contractile activities involved in cell motility [34, 35] and beating of cardiomyocytes [36, 37]. These methods can also be extended to estimate traction stresses for cells embedded in 3D environments using digital volume correlation algorithms [25, 29, 38, 39].
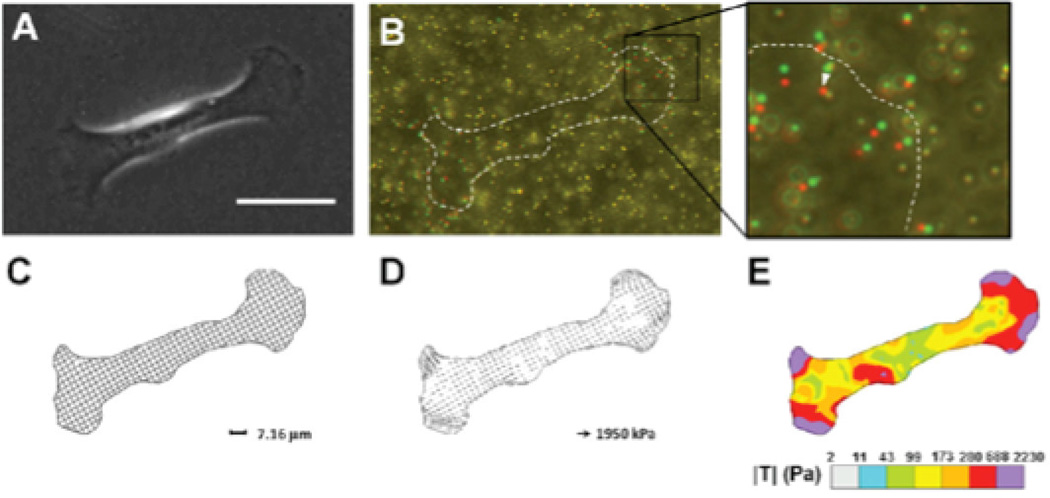
Figure 2. Traction Force Microscopy.
A) Phase contrast image of a mammary epithelial cell on a polyacrylamide gel with fiducial microbeads. Scale bar 50 µm. B) Composite image of microbeads during cell force generation (red) and after removing the cell from the substrate with trypsin (green). C) Discretized mesh of cell area defines the region of interest for traction calculation. D) Traction stresses calculated across the mesh. E) Color contour plot visualizes the distribution of traction stresses under the cell. Reproduced with permission from [42]
For all TFM processes, at least two images are required to estimate traction [40, 41]: an image of the deformed beads (with the cell on the substrate) and the undeformed beads (after removal for the cell from the substrate). Measuring changes in traction due to an applied stimulus requires images before and during the stimulus application.
In the most basic approaches of 2D TFM analyses, several key assumptions are imposed in order to use linear elastic theory and estimate traction forces from a displacement field [42]:
Forces normal to the surface are negligible because cell-substrate interactions mainly develop along the substrate surface. Only the εxx, εxy, and εyy strain vectors are nonzero.
The hydrogel substrate is a 2D elastic plane extending laterally to infinity (a semi-infinite half space).
Strains are infinitesimally small and thus the substrate material deforms within a regime of linear elasticity. The substrate material is mechanically described by a Young’s modulus and Poisson’s ratio.
The substrate’s material properties remain homogenous.
Once these assumptions are experimentally satisfied, (Figure 2C, D), several methods can be used to estimate traction forces from the displacement data. Images of particles before and after movement can be converted to displacement data by either direct tracking of fiducial markers using particle tracking velocimetry (PTV) or employing cross-correlation functions to statistically interpolate bead movements using particle image velocimetry (PIV) [43]. Trepat and colleagues have used PIV to evaluate the migration of cell sheets, cell-cell rearrangements within the sheet, and traction forces produced by the moving groups of cells [44]. Using the displacement data, one can solve the ‘inverse problem’ by fitting equilibrium solutions to displacements resulting from applied forces using the Boussinesq formulation [45]. For this method, an equation for the displacement field in the semi-infinite half space is derived into a convolution integral using Green’s tensor [46]. This equation is then discretized and inverted to determine surface traction forces [47]. The inversion method is computationally expensive and requires regularization steps to account for noise in the displacement data. Fourier Transform Traction Cytometry (FTTC) addresses these issues by transforming the integrated displacements of microbeads to Fourier space and performing calculations using matrix multiplication [48]. Another method solves the inverse problem by estimating the force magnitude and minimizing the distance between measured and computed displacements by solving the direct and adjoint elastic problems using finite element discretization [49, 50]. del Álamo and colleagues developed an exact Boussinesq solution using Fourier expansions to account for the finite thickness of the gel when interpolating traction forces [51, 52]. The semi-infinite approximation holds as long as the displacements of beads and the cell length are much smaller than the substrate thickness. Thus, del Álamo and colleagues enabled analysis of many combinations of gel thicknesses and cell lengths. Further, the updated solution allows for non-zero net force for each cell, meaning that z-displacements and cell migration can be better quantified [51, 52]. An alternate approach to Boussinesq solutions is to assign boundary conditions to bead displacements, discretizing the gel, and then apply finite element methods (FEM) to solve for stress equilibrium [53]. FEM enables calculations of traction stress without the need for a Boussinesq formulation, thus relaxing the substrate constraints to satisfy assumptions of a semi-infinite elastic half space [54]. FEM methods can characterize 2D and 3D systems with nonlinear materials and arbitrary gel geometries [55]. Finally, experimental simulations of TFM can also optimize TFM algorithms, test optimal experimental setups, and compare predicted cell-generated forces to experimentally derived values [56].
The accessibility to analysis codes that convert images of moving beads to force measurements has been a bottleneck for the implementation of these methods in biology labs. Several research groups have publically shared computational tools for TFM. Specifically, Martiel and colleagues [57] have shared computational packages that prepare images for processing, that extract bead displacement using PIV, and that finally determine traction stresses using an FTTC method. The authors also provide discussion on experimental and numerical approaches for robust determination of cell-generated forces. Style and colleagues have also provided TFM analysis tools for particle tracking and for calculating traction stresses using inverse Fourier transforms and elasticity theory [13].
2.2 Micropost Arrays
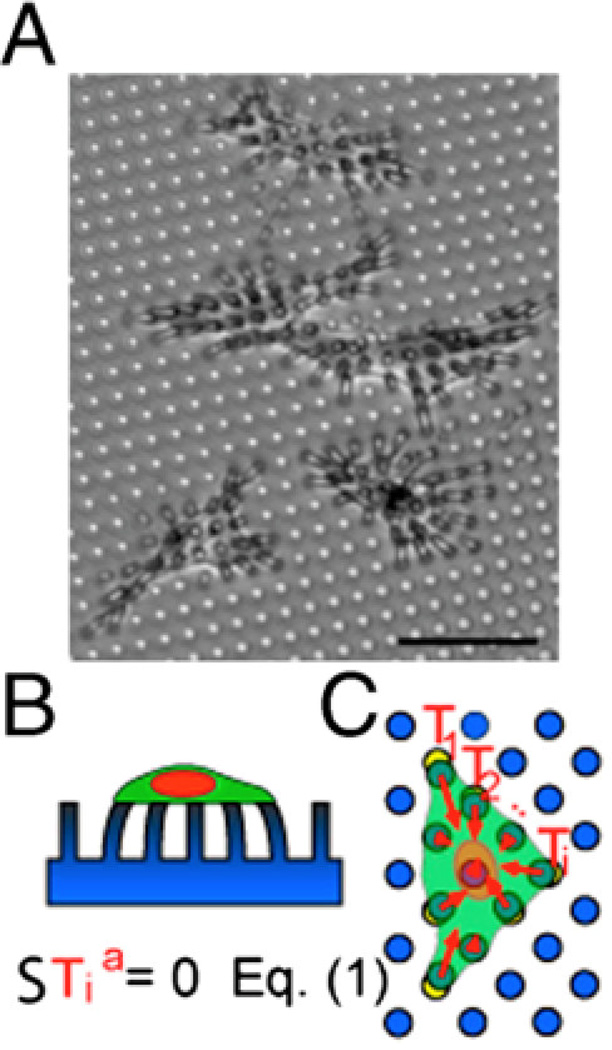
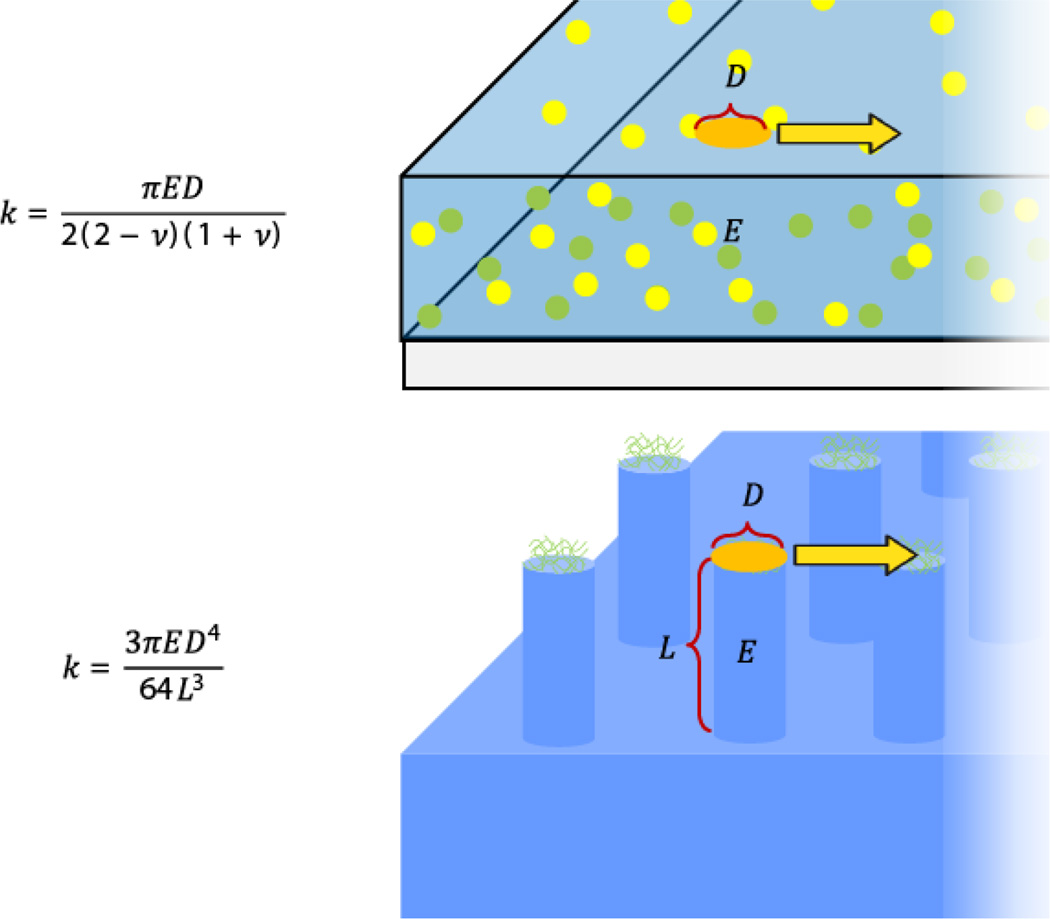
Upon attachment to arrays of elastomeric microposts, cells exert traction forces that displace the tops of the microposts away from their unloaded position (Figure 3). Displacement of the top portions of the microposts is directly linked to the traction forces generated by the cell through the Euler-Bernoulli beam theory:
| (1) |
where k is the effective post stiffness and δ is the displacement of the micropost. Effective spring constant is determined by the material properties of the post (E is the Young’s modulus) and the structure geometry (D is the post diameter and L is the post length). A number of assumptions are also used to calculate forces in micropost traction measurements:
Microposts do not undergo elongation or compression because their resistance to out of plane deformation is orders of magnitude larger than their resistance to lateral displacement [58].
Micropost displacements are small.
The substrate material is linearly elastic [15, 19, 59, 60].
Figure 3. Micropost arrays.
A) Endothelial cells tug on microposts (dia. = 3 µm). The microposts were coated with fibronectin by microcontact printing to restrict cell adhesion to a specific area. Scale bar is 50 µm. B) Side view cartoon of the cell on the micropost array. C) The individual traction force vectors exerted by the cell sum to zero. Reproduced with permission from [200].
Analyzing the displacement of micropost forces is a distinct method from calculating cell traction forces through TFM with hydrogel substrates. Few studies have employed both methods for a direct comparison [61]. Figure 4 compares the relative stiffness resisting traction forces and deformation modes on hydrogels and microposts. Importantly, the execution of these methods does not rely solely on the optimization of data analysis, but also in additional factors involved in device fabrication and experiments with cells.
Figure 4. Comparison of how cells pull and induce traction forces on hydrogels and microposts.
Cells bind to substrates via coupling to extracellular matrix proteins in focal adhesions. On both continuous hydrogel substrates and microposts, these focal adhesions occupy distinct areas of the material. Cell-binding area can be defined on hydrogels and micropost arrays by patterning of extracellular matrix proteins (and in addition by micropost geometry). The effective spring constant of hydrogels and microposts is based on contract area between the substrate and the cell exerting traction forces. Figure adapted with permission from [201].
In summary, four main steps are needed to obtain cell traction data with these methods: 1) device fabrication, 2) cell attachment to device surfaces, 3) imaging of displacements in the device and 4) derivation of traction forces from displacements. We have detailed different options to derive forces from displacements and commented on the effects of noise in TFM analysis of hydrogel deformation and deflection of microposts. Computational challenges in force calculation decrease when using microscopy capabilities that can register with high resolution the movement of microbeads in hydrogels or the displacement of the tops of microposts. Now we elaborate on how the fabrication and calibration of devices affect the outcome of these force-measuring methods.
3. Device Fabrication and Calibration as Potential Sources of Error in Force Estimates
For both polyacrylamide and PDMS-based devices, the measured displacement of the material and calculated traction forces can substantially change with variable mechanical properties. A wide range of Young’s moduli (Table 2) have been reported for specific formulations of both polyacrylamide hydrogels and PDMS substrates. The variability in these reports can originate from: 1) polymerization methods [62] and from 2) differences in assays for mechanical testing used in device calibration [63, 64].
Table 2.
Elastic moduli of PDMS and polyacrylamide materials
The mechanical properties of polyacrylamide and PDMS depend on the protocols used to polymerize these substrates, as well as on the batch, lot, storage or age of used polymerization materials [65–67]. Variations in material properties contribute to difficulties in quantitatively comparing calculated traction forces after using devices fabricated with different polymerization materials or between labs with different fabrication methods (Figure 4) [13, 38, 59, 68]. For example, we found variations in the effective micropost stiffness ranged from 0.01 to 0.065 N/m batch-to-batch and 2.3 – 28.6% measurement standard deviations within each tested batch due to variability in fabrication and PDMS batch [69]. The mechanical parameters of substrates and devices are determined by mechanical tests, which yield different results depending on the type of testing apparatus, the geometry of the material sample [63] and the length scale of testing [64]. The dimensional reproducibility of devices also affects the device performance. Specifically, the thickness of hydrogels [63, 70] and the geometry of microposts (diameter and aspect ratio) should be considered when comparing results across batches of fabricated devices. These differences introduce offset errors in cell traction measurements, which require careful experimental design to determine traction forces.
Direct calibration of mechanical properties for materials and devices under experimental conditions reduces the uncertainty of measurements when compared to relying on estimates performed in other studies. For example, some formulations of polyacrylamide exhibit time-dependent viscoelasticity [71] or poroelastic properties [72, 73] that should be considered when measurements address cell-generated deformations of the substrate exerted at different rates: fast or slow cell contraction. Micropost effective stiffness differently varies with distinct types of post-fabrication treatments of the PDMS surface [74]. In general, the effective post stiffness of a set of representative microposts in an array can be directly calibrated using a force sensor to observe top displacement under an applied transverse load. With this approach, we previously found that direct calibration of k reduced measurement uncertainty from about 100%, based on material property and geometry assumptions alone, to as low as 10% [68, 69]. Calibration also addresses nonlinear mechanical effects due to substrate warping or large deformations [15, 59].
The differences in material properties between hydrogels and PDMS may also affect how cells interact with these surfaces and thus cell traction force measurements. Recent studies have tried to understand how nanoscale substrate surface properties such as hydrogel porosity, PDMS network density and surface roughness impact the surface density and presentation of cell-binding protein ligands and how these influence cell adhesion [75, 76]. For instance, Wen et al. determined that stem cell differentiation was influenced by substrate stiffness but not by substrate porosity or protein tethering [77]. Another study verified that the same cell type exhibited similar cell spreading area on both TFM polyacrylamide substrates and PDMS micropost substrates of similar stiffness [61]. However, this study did not compare cell-generated forces across the two platforms. Such quantitative comparisons may require a deeper understanding and control of cell-ECM adhesion to provide controllable cell attachment to polyacrylamide and PDMS surfaces.
4. Enabling cell adhesion by engineering surface-ECM protein interactions
Cell membranes do not directly bind to the surfaces of hydrogels, PDMS devices or any other material surface. Instead, varied types of ECM proteins stably bind to specific classes of cell transmembrane proteins and link them to surfaces, triggering cell-surface interactions [78]. The strength and robustness of cellular adhesion is crucial for the stable attachment of cells to surfaces [79, 80]. Only then, can contractile tension propagate between the cell and the substrate [79, 81, 82]. Under physiological conditions, the ECM ligands bind readily to transmembrane proteins while cells are selective for specific ECM types that match in vivo conditions [78, 79]. Interactions between ECM proteins and substrate surfaces are the main regulators of cell adhesion to artificial materials [83] and are thus commonly engineered by changing the physical-chemical properties of surfaces to induce cell adhesion [84]. Here, we detail specific strategies to promote stable adhesion of ECM proteins to surfaces of polyacrylamide hydrogels and PDMS devices. First, we review the governing factors of protein-surface interactions to note the benefits of covalently attaching ECM proteins to the surfaces of traction force devices (Figure 5). In general, protein-surface interactions depend on the properties of both the protein and the surface [83].
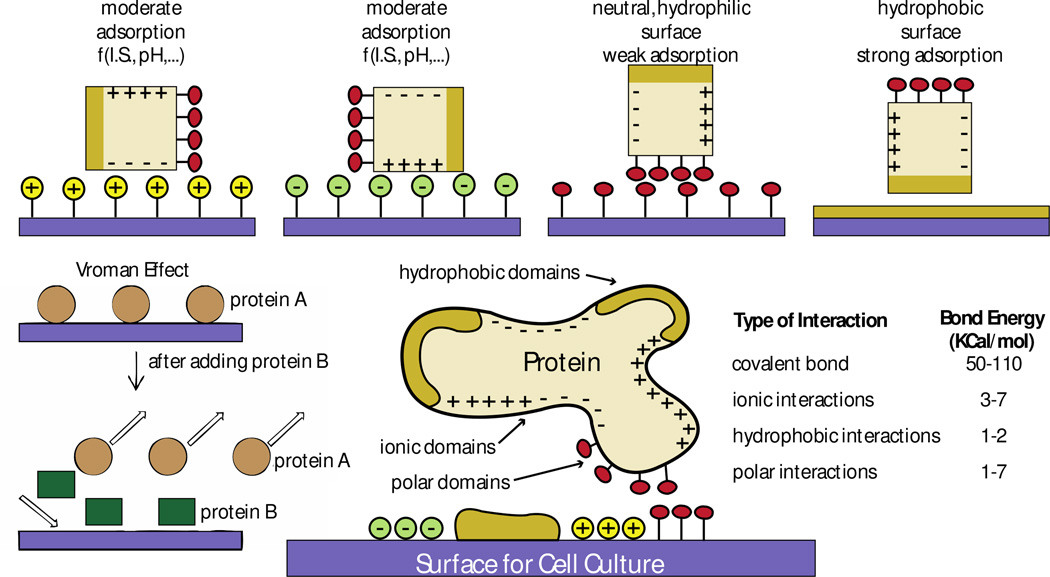
Figure 5. General mechanisms that govern protein-surface interactions based on surface chemistry.
Proteins interact with material surfaces and these interactions are regulated by both protein and surface properties. Interactions between ionic domains result in moderate protein adsorption, which is unstable because it relies on the ionic state (I.S) and the pH of the culture media. Interactions between hydrophilic polar domains are weak and also unstable. Interactions between hydrophobic domains lead to strong adsorption. Based on the mechanisms that regulate protein interaction (the Vroman effect), a protein B with higher affinity to a surface can replace a protein A of lower surface affinity previously adsorbed to the surface. Ideally, to maximize surface-protein interactions, surfaces should have different engineered regions with different chemical properties to match the properties of the protein of interest. Covalent interactions are the strongest among all types of interactions between proteins and surfaces. Figure adapted with permission from [86, 87]
Protein size, net charge, stability and unfolding rate regulate the affinity of proteins to surfaces. These properties also play strong roles when using mixtures of proteins, such as serum in common cell culture media formulations. Smaller proteins adsorb faster to surfaces because of faster diffusion. Larger proteins present a higher surface for interactions with material surfaces, but diffuse slower and take a longer time for surface adsorption. Diffusion and binding kinetics underlie the “Vroman effect” [85], which describes the exchange of proteins adsorbed on surfaces based on protein affinity properties (Figure 5): over time, proteins with stronger and more stable interactions replace proteins with low surface affinity. The overall combination of protein properties is what dictates the stability of the protein’s adhesion to and stability of interaction with the surface [83].
Surface properties of materials such as topography, chemical composition, hydrophobicity, heterogeneity and surface potential affect protein adsorption and adhesion [86, 87] (Figure 5). Proteins expose hydrophilic amino acids, which are readily available to interact with material surfaces because physiological conditions involve aqueous environments. However, the interactions of hydrophobic surfaces with the hydrophobic domains of proteins lead to the strongest protein adsorption states [88]. Protein affinity to charged surfaces is stronger when the pH of the environment is near the protein’s isoelectric point due to decreased protein-protein interactions in solution, as proteins assume a neutral net charge. The distribution of charged residues on proteins and protein-protein interactions in solution therefore affect these adsorption events and their stability easily varies with the pH and ionic state of the culture media (Figure 5) [89]. The rate of protein unfolding accelerates protein-material interactions. Yet unfolding can also lead to protein denaturing and loss of protein stability. For example, hydrophobic surfaces (e.g., Teflon) with low surface energy denature proteins [90].
In summary, the effects of surface charges, surface chemical groups, other proteins in solution and the surface potential of the material determine the amount and strength of protein interactions with material surfaces. Details on how material surface modifications affect protein adhesion can be found elsewhere: [91, 92]. The surface of polyacrylamide hydrogels is very hydrophilic and thus sterically hinders protein adsorption (Figure 5). PDMS surfaces are hydrophobic, but present low surface energy, which promotes weak interactions with proteins [93]. Protocols to attach proteins to these materials first modify the undesirable material surface properties and then functionalize the material surface to promote stable and strong protein attachment using polar interactions, hydrophobic interactions, ionic bonds, and/or covalent bonds (Figure 5). Of these, covalent bonds are the strongest and least dependent on the electrochemistry of the extracellular environment [94–96].
Covalent attachment of ECM proteins to material surfaces increases the strength of cell adhesion [97], which affects mechanotransduction [98]. For example, we have shown that the nature of protein attachment for beating heart muscle cells cultured on micropost arrays also influences their force generation and contractile properties [99]. The covalent attachment of collagen or fibronectin to PDMS surfaces was also reported to enable mesenchymal stem cells to maintain their undifferentiated state [100]. Cells cultured on surfaces with covalently attached proteins also exhibited higher adhesion area and better-developed actin fibers. Taken together, these studies demonstrate the importance of covalently attaching ECM proteins to the surface of devices to induce strong and stable cell adhesion that better transduces intracellular forces to a deformable extracellular milieu.
4.1 Approaches to covalently attach proteins to the surface of polyacrylamide hydrogels and PDMS microposts
4.1.1 Functionalizing Polyacrylamide Hydrogels
Polyacrylamide is hydrophilic and repels protein adsorption by steric hindrance effects. Thus, the surface of polyacrylamide must be functionalized with coupling chemical species to bind proteins that promote cell adhesion [65]. Polyacrylamide hydrogel functionalization approaches alter the hydrogel fabrication protocol and thus fall into two categories (see Figure 6): i) adding functional groups to the prepolymer solution to form a link between components of the hydrogel and proteins and ii) modifying the hydrogel surface after polymerization.
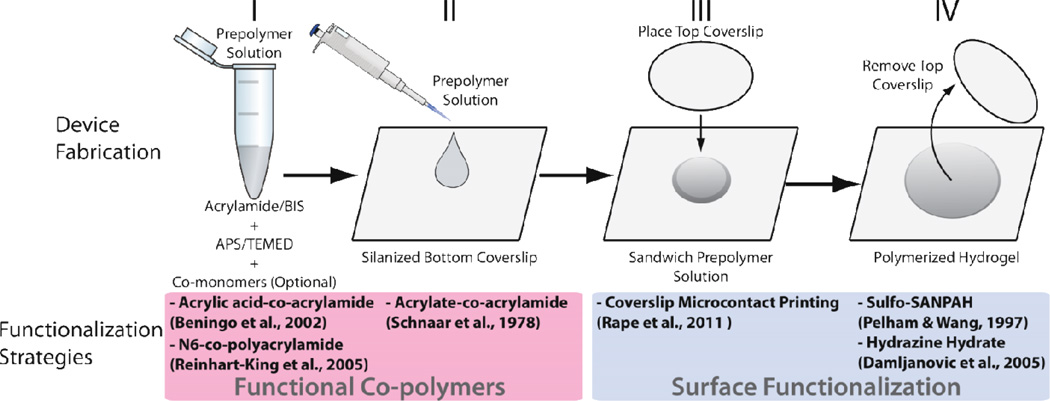
Figure 6. Approach to fabricate hydrogel substrates while considering different methods for functionalization of surfaces with proteins.
Top: the acrylamide-based prepolymer solution is mixed before being added to the top of a coverslip pre-treated to bind polyacrylamide during its polymerization. Another coverslip that will define the topography of the substrate is placed on top of the prepolymer droplet and the hydrogel polymerizes. After hydration, this coverslip is removed. Bottom: different strategies to functionalize these substrates are integrated at different stages of this fabrication method. Strategies grouped at the left involve co-polymerization of acrylamide with species added to the pre-polymer solution that bind acrylamide and proteins. Strategies grouped at the right consist of surface modifications and do not affect the bulk properties of hydrogels. Figure adapted with permission from [65].
Polyacrylamide gels result from the free radical polymerization of vinyl groups among acrylamide and the crosslinker bis-acrylamide [101]. The reaction is initiated by tetramethylethylenediamine (TEMED) and catalyzed by ammonium persulfate (Figure 6). The final material consists of a hydrogel with acrylamide chains of variable length stochastically crosslinked with bis-acrylamide [102]. To obtain a thin polyacrylamide layer, the prepolymer solution is sandwiched between two coverslips (Figure 6), which dictates the gel surface area and thickness. The bottom coverslip is functionalized with silanes that generate a monolayer of free chemical groups that covalently bind the polyacrylamide during its polymerization [103, 104]. A coverslip is placed on a solution droplet to define the hydrogel surface topography while applied pressure sets the hydrogel thickness.
Adding reactive monomers to the prepolymer solution alters the bulk chemical and mechanical properties [105]. Acrylic acid or other carboxyl-containing molecules that co-polymerize with acrylamide result in hydrogels with carboxyl groups on the surface [106]. Then, proteins can be bound to the carboxyl groups using N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDAC or EDC), a reagent which couples amine groups on protein backbone to the carboxylic groups on the hydrogel surface. An alternative approach is to add N-hydroxysuccinimidyl acrylate (NHS-acrylate) to the prepolymer solution [107, 108]. The acrylate group of NHS-acrylate co-polymerizes with acrylamide and the NHS moiety randomly distributes throughout the hydrogel, including the gel surface where it binds amines found in protein backbones. A third co-polymerization technique is to use the monomer 6-Acrylaminohexylaminohexanoic acid N-succinimidyl ester (N6) [109]. The succinimidyl ester group in one end of N6 also binds amine groups in the protein backbone and the acrylate group on the opposing end co-polymerizes with acrylamide. N6 has a higher degree of freedom and longer length, increasing its efficiency due to higher accessibility for protein binding when compared to the acrylic acid or NHS-acrylate approaches.
Another method to conjugate proteins on hydrogel surfaces during polymerization involves patterning proteins on the top coverslip that sandwiches the hydrogel prepolymer (Figure 6) [110]. Proteins patterned on the glass coverslip transfer to the hydrogel surface during polymerization. Deep UV functionalization of hydrogel surfaces also leads to stable protein attachment and cell culture [111]. With these methods, cells stably attach to hydrogel surfaces where ECM proteins are immobilized. However, the mechanism of protein immobilization, entanglement versus chemical binding, remains unknown. Protein micropatterns on glass can also be developed with methodologies such as microcontact printing by coating a PDMS block with features to stamp proteins onto the glass [112], lift-off of a photoresist layer and glass passivation with PLL-g-PEG [113], and UV micropatterning of glass homogeneously coated with PLL-g-PEG [114]. These micropatterns can be transferred to hydrogel surfaces for TFM measurements of cells confined to specific sizes or shapes [74].
Methods that involve surface functionalization after hydrogel polymerization (Figure 6) rely on changing the reactivity of the material surface. A simple method involves surface activation with ultra-violet (UV) light in the presence of the photoactivatable heterobifunctional reagent sulfosuccinimidyl-6-(4’-azido-2’-nitrophenylamino) hexanoate (sulfo-SANPAH) [107]. The photoactivatable region of sulfo-SANPAH binds polyacrylamide and the sulfosuccinimidyl group at the other end reacts with protein primary amine groups. Sulfo-SANPAH is an expensive reagent and the efficiency of protein crosslinking varies among batches due to chemical instability in aqueous media. Functionalizing polyacrylamide with hydrazine hydrate is a less expensive but more laborious alternative to sulfo-SANPAH [115]. The hydrogel surface is activated by hydrazine hydrate to bind aldehyde groups on oxidized proteins. After the hydrogel is activated, ECM proteins oxidized with sodium periodate are introduced onto the surface for stable coupling.
4.1.2 Functionalizing PDMS Microposts
PDMS surfaces have low surface energy that inhibits protein adsorption. Surface oxidation of PDMS is the most common method to induce protein adsorption to PDMS surfaces [93] or to prepare PDMS for microcontact printing (Figure 7). The most common methods to oxidize PDMS surfaces include oxygen plasma treatment [74], UV-ozone deposition [16], incubation in piranha solution [116], and chemical deposition [117]. Oxidation alters micropost stiffness so this must be taken into consideration when calculating cell-generated forces from micropost bending after this treatment [74]. Adsorption of ECM to oxidized PDMS alone does not induce strong adhesions of cells. However, additional treatments will promote covalent ECM protein attachment to PDMS: i) silanization of oxidized PDMS, ii) surface photografting, or iii) adding functionalized PDMS to the inert PDMS prepolymer mixture.
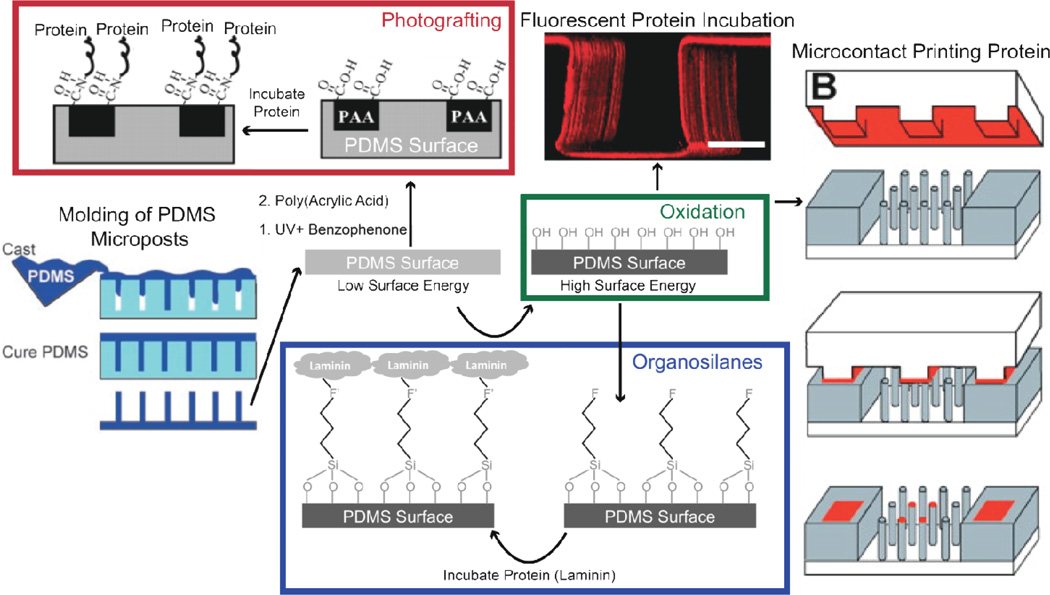
Figure 7. Different methods to functionalized PDMS surfaces.
A degassed viscous prepolymer mixture is cast on molds and cured to form arrays of PDMS microposts. PDMS surfaces are functionalized by increasing their surface energy with oxidation using plasma, UV-ozone, piranha, and chemical deposition methods. Once oxidized, proteins can be adsorbed or stamped on surfaces for stable cell adhesion or incubated in organosilanes that contain chemical groups to covalently bind proteins and oxide to bind to the PDMS surface. Laminin is a potential ECM protein that binds to PDMS with this strategy. An alternative approach not involving oxidation consists of incubating PDMS in benzophenone and exposing it to UV to polymerize the acrylic acid on the surface and bind proteins. Figure adapted with permission from [19, 99, 120, 202].
i) Silanization
Organosilanes can covalently link the oxidized PDMS surface to ECM proteins [99, 100]. However, care must be taken with protocols as the stability of organosilanes varies with environmental storage and reaction conditions [118, 119]. A silane-based approach that induces stable, covalent protein binding to PDMS consists of functionalizing oxidized surfaces with 3-aminopropyltriethoxysilane (APTES) [119]. APTES contains a free amine group that readily binds one of the aldehyde groups in gluteraldehyde. The exposed primary amines on proteins will then bind the other free aldehyde group of gluteraldehyde upon incubation.
ii) Photografting
PDMS can be photografted for covalent attachment of ECM proteins. Photografting involves surface UV activation, incubation with non-aqueous benzophenone solution, and incubation with acrylic acid in water to add carboxyl groups to the material surface [120]. EDC chemistry, as described for polyacrylamide functionalization, can then bind carboxyl groups exposed on the surfaces of proteins [120]. A particular challenge during PDMS micropost functionalization using photografting is avoiding stiction of microposts to one another, as observed with many different solvents [121, 122].
iii) Functionalized PDMS prepolymer
Functional molecules can also be added to the PDMS prepolymer mixture. PDMS devices are fabricated by mixing a viscous prepolymer with a curing agent to induce polymerization [121] and the mixture is cast and cured on microfabricated molds. Mixing PDMS with functional molecules enables the material surface to be crosslinked to other chemical species via the added reactive surface functional groups [123–126]. This method has high potential for binding proteins to PDMS surfaces, but it has not yet been developed to fabricate devices for robust cell culture.
4.2 ECM protein type, structure, and function regulate the biology of cells in culture
When functionalizing the surface of polyacrylamide and PDMS, the chosen ECM protein must also be suitable for the cell type under study. Different cell types secrete different sets of ECM proteins and will typically exhibit more physiological phenotypes when cultured on proteins that mimic attributes of in vivo ECM composition [78, 127, 128]. In addition, ECM composition can mediate certain phenotypes, such as malignancy of mammary epithelium, acting together with stiffness as determinants for invasiveness [129]. This study shows that high ligand density of the ECM can resemble high substrate stiffness in enhancing cell malignancy. Transmembrane interactions with ECM proteins are complex, however the use of recombinant or isolated ECM proteins to culture cells provides a reductionist approach that mimics cell adhesion pathways in vitro [130].
Synthetic polypeptides consisting of conserved ECM cell-binding motifs have been used to promote adhesion of cells [131], the most popular being Arginine-Glycine-Aspartic Acid (Arg– Gly–Asp; RGD). RGD is present in fibronectin and fibrinogen and binds transmembrane integrins [83]. RGD binding to specific cell surface receptors and cell adhesion depend on the synergy of this tripeptide with adjacent peptide sequences [132]. Therefore, the efficiency of RGD-functionalized surfaces for cell adhesion depends on the physical chemistry of the macromolecules that harbor RGD and connect this tripeptide to surfaces. In general, the conformation of ECM proteins plays a strong role in cell-ECM binding by making certain areas accessible to cell binding regions [133].
Collagen, fibronectin, laminin, and vitronectin are among the most used families of ECM proteins for in vitro cell culture [134]. ECM proteins can also alter cell function and gene expression [128, 135] and changes in ECM composition can induce malignant phenotypes [136, 137]. Thus, improper ECM matching to the cell type for in vitro culture may induce uncontrollable cell phenotypes. For this reason, ECM formulations isolated from in vivo organisms, like Matrigel [138], fail as systems with controllable protein composition, even if they promote robust cell adhesion. Culturing endothelial cells on collagen or laminin triggers different signaling cascades that affect cell shape, intercellular interactions, and eventually tissue morphogenesis [139]. Fibronectin-functionalized culture substrates lead to early malignant phenotypes for epithelial cells, while laminin functionalization does not [140]. In addition, smooth muscle cells are sensitive to substrate mechanics when cultured on fibronectin, but not on laminin [141]. Altogether, these studies exemplify the need for proper selection of ECM in the design of traction force experiments to avoid non-controllable effects of changes in cell biology from the type of used ECM.
Several experimental variables can further influence the bioactivation of device surfaces for stable cell adhesion, including other proteins present in the cell culture media [83] and the production of ECM proteins by certain cell types [79]. These proteins may interact with device surfaces and affect expected protein-material interactions. In summary, cell type-specific biological properties, media composition, the type of ECM and its attachment to the surface must be carefully considered when designing experiments to measure cell traction forces.
5. Measuring Cell Traction Forces in Dynamic Cell Culture Environments
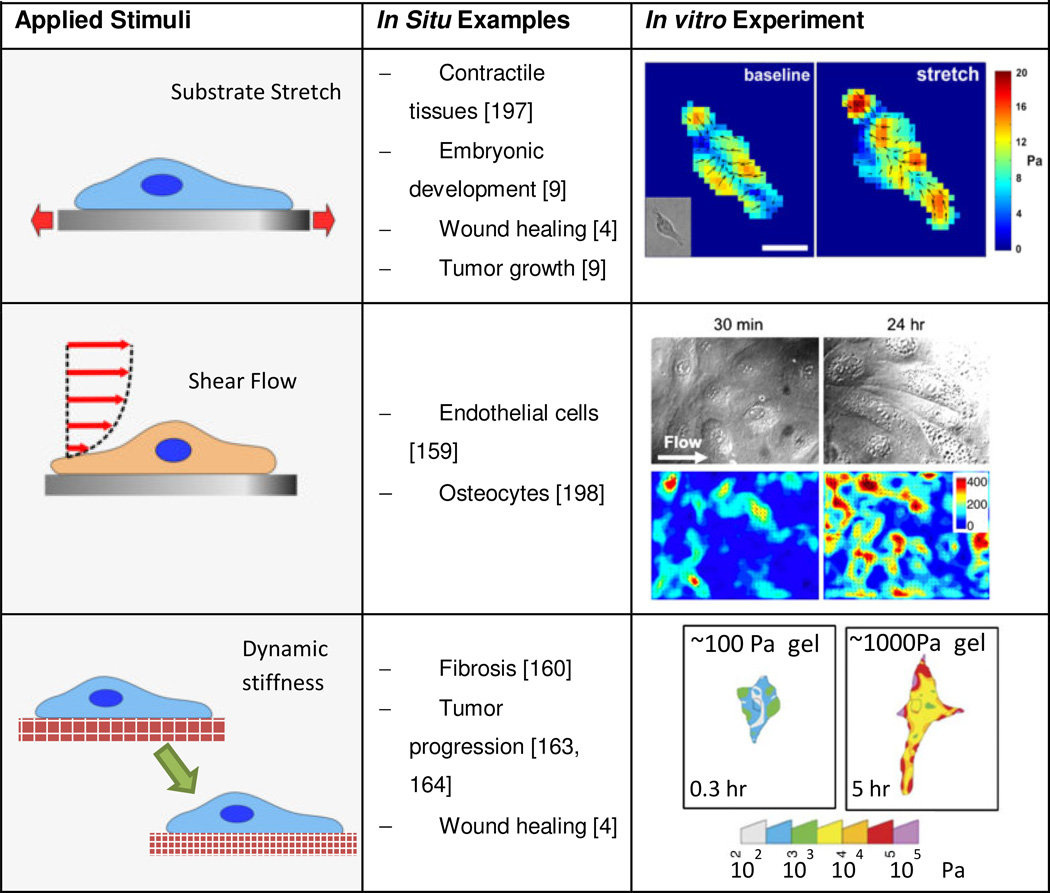
Cell mechanics and mechanotransduction studies often aim to replicate key extracellular mechanical cues in in vitro cell culture systems including stretch, shear flow and stiffness. Studying the effects of these mechanical cues on the ability of cells to generate forces will unravel the mechanisms underlying the interplay between force generation and mechanical sensitivity in cells [142]. For these studies, devices to calculate traction forces must be integrated with platforms that mechanically stimulate cells. Extensions of hydrogel TFM and micropost force measurement assays include investigating cell responses to dynamically varied stimuli such as stiffness, shear flow and stretch (Table 3). Implementations vary in complexity based on how the stimulus is applied and how it affects force measurements; particular concerns include maintaining optical access for imaging live cell features and substrate deformations. Applying mechanical stimuli in vitro is of interest to emulate how cells sense and respond to mechanical cues in vivo in mechanical tissues like the heart, lung, muscles, etc. [143, 144]. In this section, we discuss recent progress on integrating cellular force measuring devices in platforms for mechanical stimulation of cells, including: i) applied stretch, ii) applied shear flow, and iii) dynamically varied substrate stiffness (Table 3).
Table 3.
i) Stretch
Stretch is generally involved in active or passive movement of tissues, such as during embryonic development, wound healing, tissue remodeling, tumor growth and muscle contractility [9]. To understand mechanotransduction mechanisms related to stretch, both hydrogel and compliant micropost arrays can be stretched. The most common method to apply stretch is co-fabricating the hydrogel or compliant microposts on a deformable membrane and stretching the membrane with different approaches including in-plane stretch via mechanical actuation [145], deformation over a post using vacuum pressure [146], or out of-plane post actuation [147]. These strategies to stretch substrates may be able to control the stretch magnitude, profile (uniaxial or biaxial), and type (static or cyclic) [146, 148, 149]. To ensure a consistent strain field, the stretching actuators on the hydrogel or micropost substrates must be characterized and calibrated prior to cell experiments.
Another method to stretch cells on hydrogels is by direct indentation of the gel, which creates local substrate deformations [150–153]. This local strain field surrounding the indenter depends on the indentation depth, indenter geometry, and substrate properties [150–153]. In either approach for combining applied strain with hydrogel TFM, it is crucial to image the fiducial markers in both the unstretched and stretched states. To apply TFM analysis to hydrogels on stretched membranes, the hydrogel must be securely bonded to the deformable membrane to ensure transmission of the applied strain to the cells [145, 146, 154]. In addition, the gel must be thin enough to transmit the strain but thick enough to maintain the apparent stiffness on the hydrogel surface.
Compliant microposts can also be fabricated to include a PDMS membrane for inducing stretch by controlling the amount of excess PDMS on the mold. The membrane with microposts can potentially be used with any stretching apparatus. However, one difficulty is maintaining optical properties, which are necessary to obtain accurate traction data. Because the PDMS microposts impede imaging, the microposts should not be in the optical path between the microscope objective and cells [155].
The transfer of stretch from the substrate to cells depends on how the ECM is attached to these materials. When silicone is globally stretched, Wipf et al. [156] showed that covalent attachment of ECM proteins to the silicone increases the transfer of strain up to 15% from the substrate to cell monolayers. When silicone substrates were functionalized by surface oxidation and passive adsorption of collagen or layer-by-layer deposition of collagen, fibroblast monolayers failed to stretch as much as the underlying substrate. Covalent attachment of collagen on these surfaces increased cell adhesion area and proliferation and corresponded with increased cell adhesion force and structural stability [100].
ii) Shear flow
Shear flow is most often associated with blood flow through the vasculature. Application of shear flow to TFM usually involves bonding the hydrogel on a glass coverslip inside a flow chamber. TFM measurements are then obtained at multiple time points during the application of shear flow [157, 158]. Shear flow can also be applied with compliant microposts, demonstrated by integration of PDMS micropost arrays into microfluidic flow chambers [159]. Flow applied with an external pump submits cell to shear flow in the microfluidic chamber [159]. Small microfluidic dimensions enable high flow velocities at low flow rates.
iii) Tunable Stiffness
Changes in substrate stiffness are relevant in tissue fibrosis by fibroblast collagen deposition [160, 161], wound healing [162] and tumor progression [163, 164]. The mechanisms that govern the effects of stiffness in cell behavior are not understood [165]. The cross-linking density of polyacrylamide hydrogels governs their stiffness [76]. The effective stiffness of microposts varies with post height and diameter. Although PDMS microposts have not been modified to change stiffness over time, stiffness gradients can be fabricated on micropost arrays by changing micropost dimensions along a specific direction of the array. This is useful in understanding how migrating cells sense and respond to stiffness [166].
Ideally, the stiffness of force-sensing substrates should be tunable at any time point during experiments to test the effects of dynamic stiffness variation in cell biology. Currently, only dynamic hydrogels have been modified to allow for time-dependent stiffness variations in conjunction with cellular force measurements [167] and the most used method has been time-dependent degradation. Strategies for time-dependent gel degradation include use of copolymers with biodegradable blocks [168], modification of alginate oxidation [169] and hydrolytically degradable hyaluronic acid [170]. An alternative to time-dependent degradation is to use UV- activatable chemistries to mediate changes of gel crosslinking. This method enables the control of the initiation of stiffness changes and thus facilitates comparisons of cell traction before and after stimuli.
More recently, a number of techniques to dynamically modify hydrogel stiffness have been combined with TFM. For example, hyaluronic acid hydrogels have been modified to methacrylated hyaluronic acid (MeHA) gels to form additional crosslinks upon UV treatment [171]. The increased extracellular stiffness of these substrates has effects on the fate of mesenchymal stem cell differentiation [172]. Obtaining accurate TFM estimates from these dynamically tunable hydrogels also requires knowledge of both of the dynamic substrate stiffness and how cells remodel the surface as function of time.
Along with MeHA hydrogels [171], other types of artificial hydrogels for cell culture have tunable mechanical properties with spatial resolution, are biocompatible and bioactivie. Hyaluronic acid-based hydrogels can harbor cell adhesive ligands [173–175], be mechanically modified [171, 173, 175], release soluble factors [174] and be patterned [173, 175]. Polyethylene glycol-based hydrogels have been developed for mechanical tunability [176], microenvironmental modifications [177], biocompatibility and harboring of cell ligands and soluble factors [178]. Hydrogels composed of synthetically produced polypeptides can also provide substrate properties of interest for assessing the ability of cells to generate forces [179] [180, 181].
The forces produced by cells on both gel substrates and micropost arrays are transduced with the involvement of proteins that play mechanical roles, generating intracellular tension. Recent advances in combining fluorophores capable of Forster Resonance Energy Transfer (FRET) with molecular tension sensors enable the measurement of intracellular forces generated under tension [182–184]. Using a FRET tension biosensor linked to E-cadherin, Borghi and colleagues showed that forces at the picoNewton scale are exerted between the actin cytoskeleton and E-cadherin [185]. Sim and colleagues studied patterned pairs of cells on gels expressing an E-cadherin FRET biosensor to simultaneously estimate cell-generated extracellular and intracellular forces, while controlling cell shape [186] Those studies revealed that epithelial cell pairs maintain a relatively constant molecular tension in E-cadherin even while cell-cell forces doubled. To study how changing substrate stiffness impacts tension within the ECM due to cell generated traction forces, Kong and colleagues used a FRET pair conjugated to a synthetic ECM adhesion ligand [187]. Future advances that combine FRET force biosensors with hydrogels for TFM or with PDMS surfaces of micropost arrays will enable new studies leveraging simultaneous analysis of molecular- and cell-level forces under a variety of environmental, pharmacological or genetic manipulations.
6. Conclusion
In this review, we discussed the use of hydrogels and elastomeric micropost arrays as systems for measuring cell traction forces in mechanobiology experiments. We detailed methods and challenges from the initial fabrication steps, device implementation including functionalization and integration of mechanical stimulation, data acquisition and data analysis. Key considerations include the mechanical properties of the substrate, force transmission in the different geometries and the engineering of cell attachment. We also presented strategies for dealing with uncertainty in both systems. Such approaches will enable new studies to relate mechanical inputs to outputs and to examine mechanisms of mechanotransduction related to cell-ECM interaction.
Highlights.
We review two methods to measure forces generated by cells: TFM and microposts.
Traction force microscopy (TFM) relies on measuring substrate displacements.
Force is calculated from the bending of posts upon cell attachment to microposts.
We detail strategies to functionalize these devices for stable cell attachment.
These methods are also combined with platforms for cell mechanical stimulation.
Acknowledgements
We acknowledge support from the American Heart - postdoctoral fellowships 14POST18360018 (A.J.S.R.), National Science Foundation - grant MIKS-1136790 (B.L.P), National Institutes of Health - grant R01 EB006745 (B.L.P.) and National Science Foundation – Graduate Research Fellowship Program (A.D. and R.E.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wozniak Ma, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 4.Wong VW, Longaker MT, Gurtner GC. Soft tissue mechanotransduction in wound healing and fibrosis. Semin. Cell Dev. Biol. 2012;23:981–986. doi: 10.1016/j.semcdb.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo F, Ege N, Grande-Garcia a, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, Sahai E. Mechanotransduction and yap-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worman HJ, Courvalin JC. How do mutations in lamins a and c cause disease? J. Clin. Invest. 2004;113:349–351. doi: 10.1172/JCI20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: Architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 11.Eyckmans J, Boudou T, Yu X, Chen CS. A hitchhiker's guide to mechanobiology. Dev. Cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaman MH, Trapani LM, Siemeski A, MacKellar D, Gong HY, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3d matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl Acad Sci. U. S. A. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. Traction force microscopy in physics and biology. Soft Matter. 2014 doi: 10.1039/c4sm00264d. [DOI] [PubMed] [Google Scholar]
- 14.Plotnikov SV, Sabass B, Schwarz US, Waterman CM. High-resolution traction force microscopy. Methods Cell Biol. 2014;123:367–394. doi: 10.1016/B978-0-12-420138-5.00020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang MT, Fu J, Wang Y-K, Desai RA, Chen CS. Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat. Protocols. 2011;6:187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris a.K, Wild P, Stopak D. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 18.Dembo M, Wang Y-L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl Acad Sci. U. S. A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. 10.16.11-10.16.16. [DOI] [PubMed] [Google Scholar]
- 21.Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. Quantifying cellular traction forces in three dimensions. Proc. Natl Acad Sci. U. S. A. 2009;106:22108–22113. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delanoe-Ayari H, Rieu JP, Sano M. 4d traction force microscopy reveals asymmetric cortical forces in migrating dictyostelium cells. Phys. Rev. Lett. 2010;105:248103. doi: 10.1103/PhysRevLett.105.248103. [DOI] [PubMed] [Google Scholar]
- 23.Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl Acad Sci. U. S. A. 2013;110:881–886. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delanoë-Ayari H, Rieu JP, Sano M. 4D traction force microscopy reveals asymmetric cortical forces in migrating Dictyostelium cells. Phys. Rev. Lett. 2010;105:248103. doi: 10.1103/PhysRevLett.105.248103. [DOI] [PubMed] [Google Scholar]
- 25.Franck C, Maskarinec SA, Tirrell DA, Ravichandran G. Three-dimensional traction force microscopy: a new tool for quantifying cell-matrix interactions. PLoS ONE. 2011;6:e17833. doi: 10.1371/journal.pone.0017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjorevski N, Nelson Celeste M. Mapping of mechanical strains and stresses around quiescent engineered three-dimensional epithelial tissues. Biophys. J. 2012;103:152–162. doi: 10.1016/j.bpj.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall MS, Long R, Feng X, Huang Y, Hui C-Y, Wu M. Toward single cell traction microscopy within 3D collagen matrices. Exp. Cell Res. 2013;319:2396–2408. doi: 10.1016/j.yexcr.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piotrowski A, Varner V, Gjorevski N, Nelson C. In: Tissue morphogenesis. Nelson CM, editor. New York: Springer; 2015. pp. 191–206. [Google Scholar]
- 29.Toyjanova J, Hannen E, Bar-Kochba E, Darling EM, Henann DL, Franck C. 3d viscoelastic traction force microscopy. Soft matter. 2014;10:8095–8106. doi: 10.1039/c4sm01271b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grinnell F. Fibroblast-collagen-matrix contraction: growth-factor signalling and mechanical loading. Trends in Cell Biol. 2000;10:362–365. doi: 10.1016/s0962-8924(00)01802-x. [DOI] [PubMed] [Google Scholar]
- 31.Achilli M, Mantovani D. Tailoring mechanical properties of collagen-based scaffolds for vascular tissue engineering: the effects of pH, temperature and ionic strength on gelation. Polymers. 2010;2:664. [Google Scholar]
- 32.Lai VK, Lake SP, Frey CR, Tranquillo RT, Barocas VH. Mechanical behavior of collagen-fibrin co-gels reflects transition from series to parallel interactions with increasing collagen content. J. Biomech. Eng. 2012;134:011004–011004. doi: 10.1115/1.4005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey SP, Kraning-Rush CM, Reinhart-King CA. Single cell-mediated collagen reorganization in 3D matrices. Conf. Proc. IEEE ENg. Med. Biol. Soc. 2011:4333–4335. doi: 10.1109/IEMBS.2011.6091075. [DOI] [PubMed] [Google Scholar]
- 34.Lombardi ML, Knecht DA, Dembo M, Lee J. Traction force microscopy in dictyostelium reveals distinct roles for myosin ii motor and actin-crosslinking activity in polarized cell movement. J. Cell Sci. 2007;120:1624–1634. doi: 10.1242/jcs.002527. [DOI] [PubMed] [Google Scholar]
- 35.Borghi N, Lowndes M, Maruthamuthu V, Gardel ML, Nelson WJ. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13324–13329. doi: 10.1073/pnas.1002662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo PL, Lee H, Bray MA, Geisse NA, Huang YT, Adams WJ, Sheehy SP, Parker KK. Myocyte shape regulates lateral registry of sarcomeres and contractility. Am. J. Pathol. 2012;181:2030–2037. doi: 10.1016/j.ajpath.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCain ML, Lee H, Aratyn-Schaus Y, Kleber AG, Parker KK. Cooperative coupling of cell-matrix and cell-cell adhesions in cardiac muscle. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9881–9886. doi: 10.1073/pnas.1203007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyjanova J, Bar-Kochba E, López-Fagundo C, Reichner J, Hoffman-Kim D, Franck C. High resolution, large deformation 3d traction force microscopy. PloS One. 2014;9:e90976. doi: 10.1371/journal.pone.0090976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraning-Rush CM, Carey SP, Califano JP, Reinhart-King CA. In: Methods in cell biology. Anand RA, Adam PA, editors. Academic Press; 2012. pp. 139–178. [DOI] [PubMed] [Google Scholar]
- 41.Wang JHC, Li B. Cytoskeleton methods and protocols. Springer; 2010. pp. 301–313. [Google Scholar]
- 42.Kraning-Rush CM, Carey SP, Califano JP, Reinhart-King CA. Quantifying traction stresses in adherent cells. Methods Cell Biol. 2012;110:139–178. doi: 10.1016/B978-0-12-388403-9.00006-0. [DOI] [PubMed] [Google Scholar]
- 43.Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vedula SRK, Hirata H, Nai MH, Toyama Y, Trepat X, Lim CT, Ladoux B. Epithelial bridges maintain tissue integrity during collective cell migration. Nat. Mat. 2014;13:87–96. doi: 10.1038/nmat3814. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA. Calculation of forces at focal adhesions from elastic substrate data: The effect of localized force and the need for regularization. Biophys. J. 2002;83:1380–1394. doi: 10.1016/S0006-3495(02)73909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landau LD, Lifshits EM. Theory of elasticity. 2d English ed. Oxford, New York: Pergamon Press; 1970. [Google Scholar]
- 47.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 49.Ambrosi D. Cellular traction as an inverse problem. SIAM J. Appl. Mathematics. 2006;66:2049–2060. [Google Scholar]
- 50.Ambrosi D, Duperray A, Peschetola V, Verdier C. Traction patterns of tumor cells. J. Math. Biol. 2009;58:163–181. doi: 10.1007/s00285-008-0167-1. [DOI] [PubMed] [Google Scholar]
- 51.del Álamo JC, Meili R, Alonso-Latorre B, Rodríguez-Rodríguez J, Aliseda A, Firtel RA, Lasheras JC. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13343–13348. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meili R, Alonso-Latorre B, del Alamo JC, Firtel RA, Lasheras JC. Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol. Biol. Cell. 2010;21:405–417. doi: 10.1091/mbc.E09-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legant WR. In: Handbook of imaging in biological mechanics. Neu C, Genin G, editors. CRC Press; 2015. [Google Scholar]
- 54.Tang X, Tofangchi A, Anand SV, Saif TA. A novel cell traction force microscopy to study multi-cellular system. PLoS Comput. Biol. 2014;10:e1003631. doi: 10.1371/journal.pcbi.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wesley RL. Handbook of imaging in biological mechanics. CRC Press; 2014. pp. 291–298. [Google Scholar]
- 56.Jorge-Peñas A, Muñoz-Barrutia A, de-Juan-Pardo EM, Ortiz-de-Solorzano C. Validation tool for traction force microscopy. Comput. Methods Biomech. Biomed. Eng. 2014:1–9. doi: 10.1080/10255842.2014.903934. [DOI] [PubMed] [Google Scholar]
- 57.Martiel J-L, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Théry M. In: Methods in cell biology. Ewa KP, editor. Academic Press; 2015. pp. 269–287. [DOI] [PubMed] [Google Scholar]
- 58.Evans ND, Gentleman E. The role of material structure and mechanical properties in cell- matrix interactions. J. Mater. Chemistry B. 2014;2:2345–2356. doi: 10.1039/c3tb21604g. [DOI] [PubMed] [Google Scholar]
- 59.Schoen I, Hu W, Klotzsch E, Vogel V. Probing cellular traction forces by micropillar arrays: Contribution of substrate warping to pillar deflection. Nano Lett. 2010;10:1823–1830. doi: 10.1021/nl100533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gere JM, Timoshenko SP. Mechanics of materials. Boston: PWS Pub. Co.; 1997. [Google Scholar]
- 61.Ladoux B, Anon E, Lambert M, Rabodzey A, Hersen P, Buguin A, Silberzan P, Mege R-M. Strength dependence of cadherin-mediated adhesions. Biophys. J. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuard D, Tzvetkova-Chevolleau T, Decossas S, Tracqui P, Schiavone P. Optimization of poly-di-methyl-siloxane (PDMS) substrates for studying cellular adhesion and motility. Microelectron. Eng. 2008;85:1289–1293. [Google Scholar]
- 63.Kuo C-HR, Xian J, Brenton JD, Franze K, Sivaniah E. Complex stiffness gradient substrates for studying mechanotactic cell migration. Adv. Mater. 2012;24:6059–6064. doi: 10.1002/adma.201202520. [DOI] [PubMed] [Google Scholar]
- 64.Oyen ML. Mechanical characterisation of hydrogel materials. Int. Mater. Rev. 2013:1–16. [Google Scholar]
- 65.Kandow CE, Georges PC, Janmey PA, Beningo KA. In: Methods in cell biology. Yu-Li W, Dennis ED, editors. Academic Press; 2007. pp. 29–46. [DOI] [PubMed] [Google Scholar]
- 66.Gutierrez E, Groisman A. Measurements of elastic moduli of silicone gel substrates with a microfluidic device. PLoS One. 2011;6:e25534. doi: 10.1371/journal.pone.0025534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnston ID, McCluskey DK, Tan CKL, Tracey MC. Mechanical characterization of bulk sylgard 184 for microfluidics and microengineering. J. Micromech. Microeng. 2014;24 [Google Scholar]
- 68.Kim K, Taylor RE, Sim JY, Park SJ, Norman J, Fajardo G, Bernstein D, Pruitt BL. Calibrated micropost arrays for biomechanical characterisation of cardiomyocytes. Micro Nano Lett. 2011;6:317–322. [Google Scholar]
- 69.Taylor R, Kim K, Sun N, Park S-J, Sim J, Fajardo G, Bernstein D, Wu J, Pruitt B. Sacrificial layer technique for axial force post assay of immature cardiomyocytes. Biomed. Microdevices. 2013;15:171–181. doi: 10.1007/s10544-012-9710-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Long R, Hall MS, Wu M, Hui C-Y. Effects of gel thickness on microscopic indentation measurements of gel modulus. Biophys. J. 2011;101:643–650. doi: 10.1016/j.bpj.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Constantinides G, Kalcioglu ZI, McFarland M, Smith JF, Van Vliet KJ. Probing mechanical properties of fully hydrated gels and biological tissues. J. Biomech. 2008;41:3285–3289. doi: 10.1016/j.jbiomech.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 72.Kalcioglu ZI, Mahmoodian R, Hu Y, Suo Z, Van Vliet KJ. From macro-to microscale poroelastic characterization of polymeric hydrogels via indentation. Soft Matter. 2012;8:3393–3398. [Google Scholar]
- 73.Galli M, Comley KSC, Shean TAV, Oyen ML. Viscoelastic and poroelastic mechanical characterization of hydrated gels. J. Mat. Res. 2009;24:973–979. [Google Scholar]
- 74.Sim JY, Taylor RE, Larsen T, Pruitt BL. Oxidation stiffening of PDMS microposts. Extreme Mech. Lett [Google Scholar]
- 75.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 76.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mat. 2014 doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J. Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayes JS, Czekanska EM, Richards RG. In: Tissue engineering iii: Cell - surface interactions for tissue culture. Kasper C, Witte F, Pörtner R, editors. Berlin Heidelberg: Springer; 2012. pp. 1–31. [Google Scholar]
- 80.Kirmse R, Otto H, Ludwig T, Interdependency of cell adhesion. force generation and extracellular proteolysis in matrix remodeling. J. Cell Sci. 2011;124:1857–1866. doi: 10.1242/jcs.079343. [DOI] [PubMed] [Google Scholar]
- 81.Gallant ND, Michael KE, Garcia AJ. Cell adhesion strengthening: Contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol. Biol. Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gallant ND, Garcia AJ. Quantitative analyses of cell adhesion strength. Methods Mol. Biol. 2007;370:83–96. doi: 10.1007/978-1-59745-353-0_7. [DOI] [PubMed] [Google Scholar]
- 83.Dee KC, Puleo DA, Bizios R. An introduction to tissue-biomaterial interactions. John Wiley & Sons, Inc.; 2003. pp. 37–52. [Google Scholar]
- 84.Gray JJ. The interaction of proteins with solid surfaces. Curr. Opin. Struct. Biol. 2004;14:110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Hirsh SL, McKenzie DR, Nosworthy NJ, Denman JA, Sezerman OU, Bilek MMM. The vroman effect: Competitive protein exchange with dynamic multilayer protein aggregates. Colloids Surf. B: Biointerfaces. 2013;103:395–404. doi: 10.1016/j.colsurfb.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 86.Palacio ML, Bhushan B. Bioadhesion: A review of concepts and applications. Philos. Trans. A. Math. Phys. Eng. Sci. 2012;370:2321–2347. doi: 10.1098/rsta.2011.0483. [DOI] [PubMed] [Google Scholar]
- 87.Fischer T, Hess H. Materials chemistry challenges in the design of hybrid bionanodevices: Supporting protein function within artificial environments. J. Mater. Chem. 2007;17:943–951. [Google Scholar]
- 88.Andrade JD, Hlady V. Biopolymers/non-exclusion hplc. Berlin Heidelberg: Springer; 1986. pp. 1–63. [Google Scholar]
- 89.Secundo F. Conformational changes of enzymes upon immobilisation. Chem. Soc. Rev. 2013;42:6250–6261. doi: 10.1039/c3cs35495d. [DOI] [PubMed] [Google Scholar]
- 90.Baier RE, Meyer AE, Natiella JR, Natiella RR, Carter JM. Surface properties determine bioadhesive outcomes: Methods and results. J. Biomed. Mater. Res. 1984;18:337–355. doi: 10.1002/jbm.820180404. [DOI] [PubMed] [Google Scholar]
- 91.Vasita R, Shanmugam IK, Katt DS. Improved biomaterials for tissue engineering applications: Surface modification of polymers. Curr. Top. Med. Chem. 2008;8:341–353. doi: 10.2174/156802608783790893. [DOI] [PubMed] [Google Scholar]
- 92.Roach P, Eglin D, Rohde K, Perry CC. Modern biomaterials: A review - bulk properties and implications of surface modifications. J. Mater. Sci. Mater. Med. 2007;18:1263–1277. doi: 10.1007/s10856-006-0064-3. [DOI] [PubMed] [Google Scholar]
- 93.Abbasi F, Mirzadeh H, Katbab A-A. Modification of polysiloxane polymers for biomedical applications: A review. Polym. Int. 2001;50:1279–1287. [Google Scholar]
- 94.Chen H, Yuan L, Song W, Wu Z, Li D. Biocompatible polymer materials: Role of protein- surface interactions. Prog. Polym. Sci. 2008;33:1059–1087. [Google Scholar]
- 95.Schwaderer P, Funk E, Achenbach F, Weis J, Bräuchle C, Michaelis J. Single-molecule measurement of the strength of a siloxane bond. Langmuir. 2008;24:1343–1349. doi: 10.1021/la702352x. [DOI] [PubMed] [Google Scholar]
- 96.Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. How strong is a covalent bond? Science. 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 97.Raynor J, Capadona J, Collard D, Petrie T, García A. Polymer brushes and self-assembled monolayers: Versatile platforms to control cell adhesion to biomaterials (review) Biointerphases. 2009;4:FA3–FA16. doi: 10.1116/1.3089252. [DOI] [PubMed] [Google Scholar]
- 98.Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ribeiro AJS, Zaleta-Rivera K, Ashley EA, Pruitt BL. Stable, covalent attachment of laminin to microposts improves the contractility of mouse neonatal cardiomyocytes. ACS Appl. Mater. Interfaces. 2014;6:15516–15526. doi: 10.1021/am5042324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuddannaya S, Chuah YJ, Lee MHA, Menon NV, Kang Y, Zhang Y. Surface chemical modification of poly(dimethylsiloxane) for the enhanced adhesion and proliferation of mesenchymal stem cells. ACS Appl. Mater. Interfaces. 2013;5:9777–9784. doi: 10.1021/am402903e. [DOI] [PubMed] [Google Scholar]
- 101.Menter P. Bio-Rad Laboratories. Hercules, CA: 2000. Acrylamide polymerization - a practical approach. [Google Scholar]
- 102.Chrambach A, Rodbard D. Polyacrylamide gel electrophoresis. Science. 1971;172:440–451. doi: 10.1126/science.172.3982.440. [DOI] [PubMed] [Google Scholar]
- 103.Wang YL, Pelham RJ., Jr Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- 104.Guo W-h, Wang Y-l. Micropatterning cell-substrate adhesions using linear polyacrylamide as the blocking agent. Cold Spring Harb. Protoc. 2011;2011 doi: 10.1101/pdb.prot5582. prot5582. [DOI] [PubMed] [Google Scholar]
- 105.Baker BA, Murff RL, Milam VT. Tailoring the mechanical properties of polyacrylamide-based hydrogels. Polymer. 2010;51:2207–2214. [Google Scholar]
- 106.Beningo KA, Wang YL. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J. Cell Sci. 2002;115:849–856. doi: 10.1242/jcs.115.4.849. [DOI] [PubMed] [Google Scholar]
- 107.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schnaar RL, Weigel PH, Kuhlenschmidt MS, Lee YC, Roseman S. Adhesion of chicken hepatocytes to polyacrylamide gels derivatized with n-acetylglucosamine. J. Biol. Chem. 1978;253:7940–7951. [PubMed] [Google Scholar]
- 109.Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys. J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rape AD, Guo WH, Wang YL. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32:2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, Filhol O, Piel M, Thery M, Balland M. A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip. 2011;11:2231–2240. doi: 10.1039/c0lc00641f. [DOI] [PubMed] [Google Scholar]
- 112.Ruiz SA, Chen CS. Microcontact printing: A tool to pattern. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 113.Moller J, Luhmann T, Chabria M, Hall H, Vogel V. Macrophages lift off surface-bound bacteria using a filopodium-lamellipodium hook-and-shovel mechanism. Sci. Rep. 2013;3 doi: 10.1038/srep02884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vignaud T, Ennomani H, Théry M. In: Methods in cellbiology. Matthieu P, Manuel T, editors. Academic Press; 2014. pp. 93–116. [Google Scholar]
- 115.Damljanovic V, Lagerholm BC, Jacobson K. Bulk and micropatterned conjugation ofextracellular matrix proteins to characterized polyacrylamide substrates for cellmechanotransduction assays. BioTechniques. 2005;39:847–851. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 116.Maji D, Lahiri SK, Das S. Study of hydrophilicity and stability of chemically modifiedPDMS surface using piranha and KOH solution. Surf. Interface Analysis. 2012;44:62–69. [Google Scholar]
- 117.Zhou J, Ellis AV, Voelcker NH. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis. 2010;31:2–16. doi: 10.1002/elps.200900475. [DOI] [PubMed] [Google Scholar]
- 118.Weetall H. Preparation of immobilized proteins covalently coupled through silane couplingagents to inorganic supports. Appl. Biochem. Biotechnol. 1993;41:157–188. doi: 10.1007/BF02916421. [DOI] [PubMed] [Google Scholar]
- 119.Lee MH, Boettiger D, Ducheyne P, Composto RJ. Silanes and other coupling agents. Volume 42007. [Google Scholar]
- 120.Wang Y, Lai H-H, Bachman M, Sims CE, Li GP, Allbritton NL. Covalentmicropatterning of poly(dimethylsiloxane) by photografting through a mask. Anal. Chem. 2005;77:7539–7546. doi: 10.1021/ac0509915. [DOI] [PubMed] [Google Scholar]
- 121.Xia Y, Whitesides GM. Soft lithography, Angewandte Chemie International Edition. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 122.del Campo A, Arzt E. Fabrication approaches for generating complex micro- andnanopatterns on polymeric surfaces. Chem. Rev. 2008;108:911–945. doi: 10.1021/cr050018y. [DOI] [PubMed] [Google Scholar]
- 123.van Poll ML, Khodabakhsh S, Brewer PJ, Shard AG, Ramstedt M, Huck WTS. Surface modification of PDMS via self-organization of vinyl-terminated small molecules. SoftMatter. 2009;5:2286–2293. [Google Scholar]
- 124.Matsuo Y, Konno R, Ishizone T, Goseki R, Hirao A. Precise synthesis of blockpolymers composed of three or more blocks by specially designed linking methodologies inconjunction with living anionic polymerization system. Polymers. 2013;5:1012–1040. [Google Scholar]
- 125.Kaplan S, Eddy CO, Badesha SS, Henry AW, Chow CC, Gervasi DJ, Klymachyov AN. Google Patents. 2004 [Google Scholar]
- 126.DeSimone JM, Rolland JP, Rothrock GMD, Resnick P. Google Patents. 2012 [Google Scholar]
- 127.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. New York: Garland Science; 2002. [Google Scholar]
- 128.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: Tissuearchitecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ. Extracellular matrix stiffness and composition jointly regulate the induction ofmalignant phenotypes in mammary epithelium. Nat Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 130.Nagaoka M, Jiang HL, Hoshiba T, Akaike T, Cho CS. Application of recombinantfusion proteins for tissue engineering. Ann. Biomed. Eng. 2010;38:683–693. doi: 10.1007/s10439-010-9935-3. [DOI] [PubMed] [Google Scholar]
- 131.Rahmany MB, Van Dyke M. Biomimetic approaches to modulate cellular adhesion inbiomaterials: a review. Acta Biomater. 2013;9:5431–5437. doi: 10.1016/j.actbio.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 132.Takagi J. Structural basis for ligand recognition by rgd (arg-gly-asp)-dependent integrins. Biochem. Soc. Trans. 2004;32:403–406. doi: 10.1042/BST0320403. [DOI] [PubMed] [Google Scholar]
- 133.Hytonen VP, Wehrle-Haller B. Protein conformation as a regulator of cell-matrixadhesion. Phys. Chem. Chem. Phys. 2014;16:6342–6357. doi: 10.1039/c3cp54884h. [DOI] [PubMed] [Google Scholar]
- 134.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions tothe third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 135.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J. Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spencer VA, Xu R, Bissell MJ. Extracellular matrix, nuclear and chromatin structure, and gene expression in normal tissues and malignant tumors: A work in progress. Adv. Cancer Res. 2007;97:275–294. doi: 10.1016/S0065-230X(06)97012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 2011;124:123–132. doi: 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hughes CS, Postovit LM, Lajoie GA. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 139.Liu Y, Senger DR. Matrix-specific activation of src and rho initiates capillary morphogenesis of endothelial cells. FASEB J. 2004;18:457–468. doi: 10.1096/fj.03-0948com. [DOI] [PubMed] [Google Scholar]
- 140.Chen QK, Lee K, Radisky DC, Nelson CM. Extracellular matrix proteins regulate epithelial-mesenchymal transition in mammary epithelial cells. Differentiation. 2013;86:126–132. doi: 10.1016/j.diff.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sazonova OV, Isenberg BC, Herrmann J, Lee KL, Purwada A, Valentine AD, Buczek-Thomas JA, Wong JY, Nugent MA. Extracellular matrix presentation modulates vascular smooth muscle cell mechanotransduction. Matrix Biol. 2015;41:36–43. doi: 10.1016/j.matbio.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 142.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim D-H, Wong PK, Park J, Levchenko A, Sun Y. Microengineered platforms for cell mechanobiology. Annu. Rev. Biomed. Eng. 2009;11:203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 144.Polacheck WJ, Li R, Uzel SGM, Kamm RD. Microfluidic platforms for mechanobiology. Lab Chip. 2013;13:2252–2267. doi: 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Quinlan AM, Sierad LN, Capulli AK, Firstenberg LE, Billiar KL. Combining dynamic stretch and tunable stiffness to probe cell mechanobiology in vitro. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Simmons CS, Ribeiro AJS, Pruitt BL. Formation of composite polyacrylamide and silicone substrates for independent control of stiffness and strain. Lab Chip. 2013;13:646–649. doi: 10.1039/c2lc41110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moraes C, Chen J-H, Sun Y, Simmons Ca. Microfabricated arrays for high-throughput screening of cellular response to cyclic substrate deformation. Lab Chip. 2010;10:227–234. doi: 10.1039/b914460a. [DOI] [PubMed] [Google Scholar]
- 148.Simmons CS, Sim JY, Baechtold P, Gonzalez A, Chung C, Borghi N, Pruitt BL. Integrated strain array for cellular mechanobiology studies. J. Micromech. Microeng. 2011;21:54016–54025. doi: 10.1088/0960-1317/21/5/054016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Higgs GC, Simmons CS, Gao Y, Fried A, Park SJ, Chung C, Pruitt BL. Mems-based shear characterization of soft hydrated samples. J. Micromech. Microeng. 2013;23 doi: 10.1088/0960-1317/23/8/085001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tang X, Bajaj P, Bashir R, Saif TA. How far cardiac cells can see each other mechanically. Soft Matter. 2011;7:6151–6158. [Google Scholar]
- 151.Hall MS, Long R, Hui CY, Wu M. Mapping three-dimensional stress and strain fields within a soft hydrogel using a fluorescence microscope. Biophys. J. 2012;102:2241–2250. doi: 10.1016/j.bpj.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Krishnan R, Park CY, Lin YC, Mead J, Jaspers RT, Trepat X, Lenormand G, Tambe D, Smolensky AV, Knoll AH, Butler JP, Fredberg JJ. Reinforcement versus fluidization in cytoskeletal mechanoresponsiveness. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen C, Krishnan R, Zhou E, Ramachandran A, Tambe D, Rajendran K, Adam RM, Deng L, Fredberg JJ. Fluidization and resolidification of the human bladder smooth muscle cell in response to transient stretch. PLoS ONE. 2010;5:16–21. doi: 10.1371/journal.pone.0012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gavara N, Roca-Cusachs P, Sunyer R, Farré R, Navajas D. Mapping cell-matrix stresses during stretch reveals inelastic reorganization of the cytoskeleton. Biophys. J. 2008;95:464–471. doi: 10.1529/biophysj.107.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lam RHW, Weng S, Lu W, Fu J. Live-cell subcellular measurement of cell stiffness using a microengineered stretchable micropost array membrane. Integr. Biol. 2012;4:1289. doi: 10.1039/c2ib20134h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wipff P-J, Majd H, Acharya C, Buscemi L, Meister J-J, Hinz B. The covalent attachment of adhesion molecules to silicone membranes for cell stretching applications. Biomaterials. 2009;30:1781–1789. doi: 10.1016/j.biomaterials.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 157.Shiu Y-T, Li S, Marganski Wa, Usami S, Schwartz Ma, Wang Y-L, Dembo M, Chien S. Rho mediates the shear-enhancement of endothelial cell migration and traction force generation. Biophys. J. 2004;86:2558–2565. doi: 10.1016/S0006-3495(04)74311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hur SS, del Alamo JC, Park JS, Li Y-S, Nguyen Ha, Teng D, Wang K-C, Flores L, Alonso-Latorre B, Lasheras JC, Chien S. From the cover: Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc. Natl Acad Sci. U. S. A. 2012;109:11110–11115. doi: 10.1073/pnas.1207326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lam RHW, Sun Y, Chen W, Fu J. Elastomeric microposts integrated into microfluidics for flow-mediated endothelial mechanotransduction analysis. Lab Chip. 2012;12:1865. doi: 10.1039/c2lc21146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires TGF-β. Journal of Clinical Investigation. 2010;120:3520–3529. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bercoff J, Chaffai S, Tanter M, Sandrin L, Catheline S, Fink M, Gennisson JL, Meunier M. In vivo breast tumor detection using transient elastography. Ultrasound Med. Biol. 2003;29:1387–1396. doi: 10.1016/s0301-5629(03)00978-5. [DOI] [PubMed] [Google Scholar]
- 162.Wang Y, Wang G, Luo X, Qiu J, Tang C. Substrate stiffness regulates the proliferation, migration, and differentiation of epidermal cells. Burns. 2012;38:414–420. doi: 10.1016/j.burns.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 163.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Butcher DT, Alliston T, Weaver VM. A tense situation: Forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 166.Sochol RD, Higa AT, Janairo RRR, Li S, Lin L. Unidirectional mechanical cellular stimuli via micropost array gradients. Soft Matter. 2011;7:4606. [Google Scholar]
- 167.Burdick Ja, Murphy WL. Moving from static to dynamic complexity in hydrogel design. Nat. Commun. 2012;3:1269. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- 168.Metters aT, Anseth KS, Bowman CN. Fundamental studies of a novel, biodegradable peg-b-pla hydrogel. Polymer. 2000;41:3993–4004. [Google Scholar]
- 169.Boontheekul T, Kong HJ, Mooney DJ. Controlling alginate gel degradation utilizing partial oxidation and bimodal molecular weight distribution. Biomaterials. 2005;26:2455–2465. doi: 10.1016/j.biomaterials.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 170.Sahoo S, Chung C, Khetan S, Burdick Ja. Hydrolytically degradable hyaluronic acid hydrogels with controlled temporal structures. Biomacromolecules. 2008;9:1088–1092. doi: 10.1021/bm800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guvendiren M, Burdick Ja. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 172.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gramlich WM, Kim IL, Burdick JA. Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials. 2013;34:9803–9811. doi: 10.1016/j.biomaterials.2013.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shen YI, Abaci HE, Krupsi Y, Weng LC, Burdick JA, Gerecht S. Hyaluronic acid hydrogel stiffness and oxygen tension affect cancer cell fate and endothelial sprouting. Biomater. Sci. 2014;2:655–665. doi: 10.1039/C3BM60274E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guvendiren M, Perepelyuk M, Wells RG, Burdick JA. Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J. Mech. Behav. Biomed. Mater. 2014;38:198–208. doi: 10.1016/j.jmbbm.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fairbanks BD, Singh SP, Bowman CN, Anseth KS. Photodegradable, photoadaptable hydrogels via radical-mediated disulfide fragmentation reaction. Macromolecules. 2011;44:2444–2450. doi: 10.1021/ma200202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kloxin AM, Lewis KJ, DeForest CA, Seedorf G, Tibbitt MW, Balasubramaniam V, Anseth KS. Responsive culture platform to examine the influence of microenvironmental geometry on cell function in 3d. Integr. Biol. 2012;4:1540–1549. doi: 10.1039/c2ib20212c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lewis KJ, Anseth KS. Hydrogel scaffolds to study cell biology in four dimensions. MRS Bull. 2013;38:260–268. doi: 10.1557/mrs.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chung C, Anderson E, Pera RR, Pruitt BL, Heilshorn SC. Hydrogel crosslinking density regulates temporal contractility of human embryonic stem cell-derived cardiomyocytes in 3d cultures. Soft Matter. 2012;8:10141–10148. doi: 10.1039/C2SM26082D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cai L, Dinh CB, Heilshorn SC. One-pot synthesis of elastin-like polypeptide hydrogels with grafted vegf-mimetic peptides. Biomater. Sci. 2014;2:757–765. doi: 10.1039/C3BM60293A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang H, Cai L, Paul A, Enejder A, Heilshorn SC. Hybrid elastin-like polypeptide-polyethylene glycol (ELP-PEG) hydrogels with improved transparency and independent control of matrix mechanics and cell ligand density. Biomacromolecules. 2014;15:3421–3428. doi: 10.1021/bm500969d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chang CW, Kumar S. Vinculin tension distributions of individual stress fibers within cell-matrix adhesions. J. Cell Sci. 2013;126:3021–3030. doi: 10.1242/jcs.119032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Letters. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sim JY, Moeller J, Hart KC, Ramallo D, Vogel V, Dunn AR, Nelson WJ, Pruitt BL. Spatial distribution of cell-cell and cell-ecm adhesions regulates force balance while maintaining e-cadherin molecular tension in cell pairs. Mol. Biol. Cell. 2015;26:2456–2465. doi: 10.1091/mbc.E14-12-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kong HJ, Polte TR, Alsberg E, Mooney DJ. Fret measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry regulates traction stresses in adherent cells. Biophys. J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peschetola V, Laurent VM, Duperray A, Michel R, Ambrosi D, Preziosi L, Verdier C. Time-dependent traction force microscopy for cancer cells as a measure of invasiveness. Cytoskeleton. 2013;70:201–214. doi: 10.1002/cm.21100. [DOI] [PubMed] [Google Scholar]
- 190.Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ. Decoupling substrate stiffness, spread area, and micropost density: A close spatial relationship between traction forces and focal adhesions. Biophys. J. 2012;103:640–648. doi: 10.1016/j.bpj.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ricart BG, Yang MT, Hunter CA, Chen CS, Hammer DA. Measuring traction forces of motile dendritic cells on micropost arrays. Biophys. J. 2011;101:2620–2628. doi: 10.1016/j.bpj.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Armani D, Liu C, Aluru N. Re-configurable fluid circuits by PDMS elastomer micromachining., Micro Electro Mechanical Systems. MEMS’99; Twelfth IEEE International Conference, IEEE; 1999. pp. 222–227. [Google Scholar]
- 193.Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, Delgado SM, Wakatsuki T, Crone WC, Pruitt BL. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int. J. Cell Biol. 2012;2012 doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, Smith ML. Micro-well arrays for 3d shape control and high resolution analysis of single cells. Lab Chip. 2007;7:1074–1077. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
- 195.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned young’s modulus. J. Biomed. Mater. Res. Part A. 2003;66:605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 196.Wang Z. Florida: University of South Florida; 2011. [Google Scholar]
- 197.Lammerding JAN, Kamm RD, Lee RT. Mechanotransduction in cardiac myocytes. Ann. N. Y. Acad. Sci. 2004;1015:53–70. doi: 10.1196/annals.1302.005. [DOI] [PubMed] [Google Scholar]
- 198.Bakker A, Klein-Nulend J, Burger E. Shear stress inhibits while disuse promotes osteocyte apoptosis. Biochem. Biophys. Res. Commun. 2004;320:1163–1168. doi: 10.1016/j.bbrc.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 199.Hersen P, Ladoux B. Biophysics: Push it, pull it. Nature. 2011;470:340–341. doi: 10.1038/470340a. [DOI] [PubMed] [Google Scholar]
- 200.Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc. Natl Acad Sci. U. S. A. 2010 doi: 10.1073/pnas.0914547107. 0914547107--0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schoen I, Pruitt BL, Vogel V. The yin-yang of rigidity sensing: How forces and mechanical properties regulate the cellular response to materials. Annual Rev. Mater. Res. 2013;43:589–618. [Google Scholar]
- 202.Moraes C, Kim BC, Zhu X, Mills KL, Dixon AR, Thouless MD, Takayama S. Defined topologically-complex protein matrices to manipulate cell shape via three-dimensional fiber-like patterns. Lab Chip. 2014;14:2191–2201. doi: 10.1039/c4lc00122b. [DOI] [PMC free article] [PubMed] [Google Scholar]