Abstract
In order to evaluate whether rare regulatory variants in the vicinity of promoters are likely to impact gene expression, we conducted a novel burden test for enrichment of rare variants at the extremes of expression. After sequencing 2-kb promoter regions of 472 genes in 410 healthy adults, we performed a quadratic regression of rare variant count on bins of peripheral blood transcript abundance from microarrays, summing over ranks of all genes. After adjusting for common eQTLs and the major axes of gene expression covariance, a highly significant excess of variants with minor allele frequency less than 0.05 at both high and low extremes across individuals was observed. Further enrichment was seen in sites annotated as potentially regulatory by RegulomeDB, but a deficit of effects was associated with known metabolic disease genes. The main result replicates in an independent sample of 75 individuals with RNA-seq and whole-genome sequence information. Three of four predicted large-effect sites were validated by CRISPR/Cas9 knockdown in K562 cells, but simulations indicate that effect sizes need not be unusually large to produce the observed burden. Unusually divergent low-frequency promoter haplotypes were observed at 31 loci, at least 9 of which appear to be derived from Neandertal admixture, but these were not associated with divergent gene expression in blood. The overall burden test results are consistent with rare and private regulatory variants driving high or low transcription at specific loci, potentially contributing to disease.
Introduction
In recent years, whole-exome sequencing has been used effectively to demonstrate that there is a burden of rare coding variants in individuals with a variety of neurological and developmental conditions.1, 2, 3, 4 Considering estimates that as many as 90% of disease-associated common variants are regulatory rather than structural,5, 6, 7 it is reasonable to assume that rare regulatory variants influencing the expression of causal genes might also be enriched in individuals with congenital abnormalities or common chronic diseases. Here we demonstrate that there is a burden of rare variants with gene expression itself, focusing on just the promoter regions of a targeted set of genes whose expression was measured by microarray analysis of peripheral blood samples.
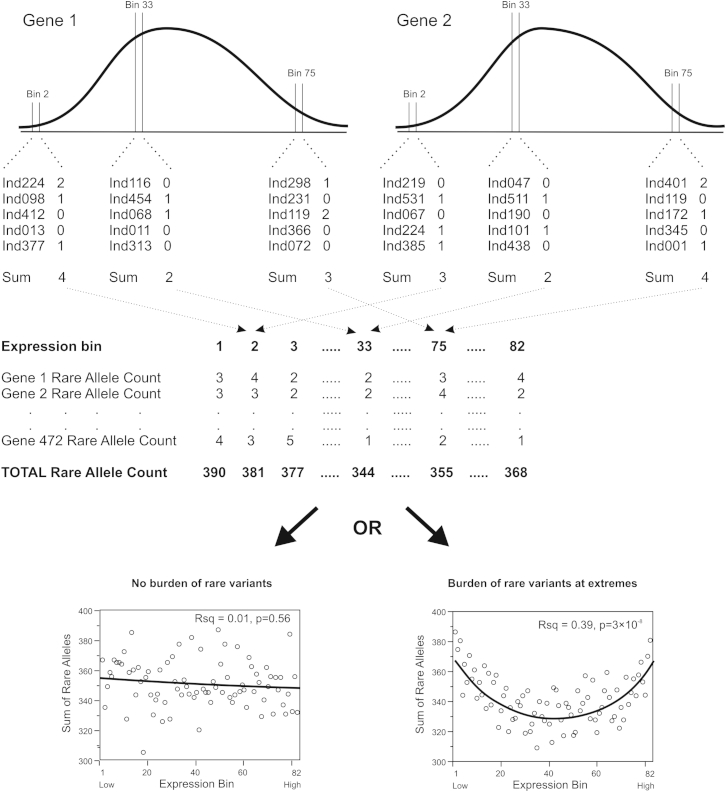
Our strategy, outlined in Figure 1, gains statistical power by pooling rare variant enrichments across the full range of expression of 472 genes measured in 410 individuals. This effectively generates almost 200,000 data points, but instead of focusing on just the most extreme individuals as required by burden tests designed for case-control comparisons,8, 9, 10, 11 we evaluate the shape of the distribution of cumulative counts of rare variants in equal sized bins of expression. For each gene in each individual, 2 kb of DNA sequence flanking the annotated transcription start site was sequenced after targeted capture of genomic DNA on custom beads.12 The count of rare variants with minor allele frequency less than 5% (or 1%) was assessed after alignment to the HuRef19 reference human genome with the Unified Genotyper in GATK.13 These counts were summed for 82 equal sized successive gene expression bins with 5 individuals each, and then tallied for all 472 genes.
Figure 1.
Schema Showing the Pooling Strategy to Evaluate Rare Variant Enrichment
For each gene, the normalized gene expression measures across all 410 individuals are sorted into 82 bins, resulting in somewhat normal frequency distributions shown in the top panels. Subsequently, the number of rare variants in the 2-kb promoter of each allele in that bin is tallied: for example, there are 2, 1, 0, 0, and 1 rare variants in the promoters of the 5 individuals (both alleles) in the second bin for gene 1, summing to 4, whereas the second bin for gene 2 has 3 rare variants. These expression bin rare allele counts are then summed over all 472 genes and plotted from lowest to highest bin to yield plots at the bottom of the figure that represent two alternative results. In the absence of a burden of rare variants at the extremes, there is neither a significant slope nor quadratic fit (left plot), whereas an excess of variants at both extremes produces a concave “smile” regression (right plot). If there were an excess at only the low or high expression, the linear slope would be significant.
Under the null hypothesis, there should be no relationship between rare variant count and gene expression and a plot of rare variant count on the y axis against expression bin on the x axis should yield a horizontal regression line. In the presence of rare variants that decrease expression, there should be larger counts in the low expression bins, toward the left in the plots in Figure 2, and similarly rare variants that increase expression should yield larger counts in the higher expression bins to the right. A general bias toward either effect would result in a significant linear slope term in a regression model. However, if both effects are present, a characteristic “smile” plot would ensue, the significance of which would be reflected in the quadratic term of a regression. We further assessed departure from the null by evaluating the significance of the complete quadratic model relative to 10,000 permutations of the full genotype and gene expression matrices, subsequently adjusting for various covariates to gain further insight into the nature of the burden of rare variants for extreme expression.
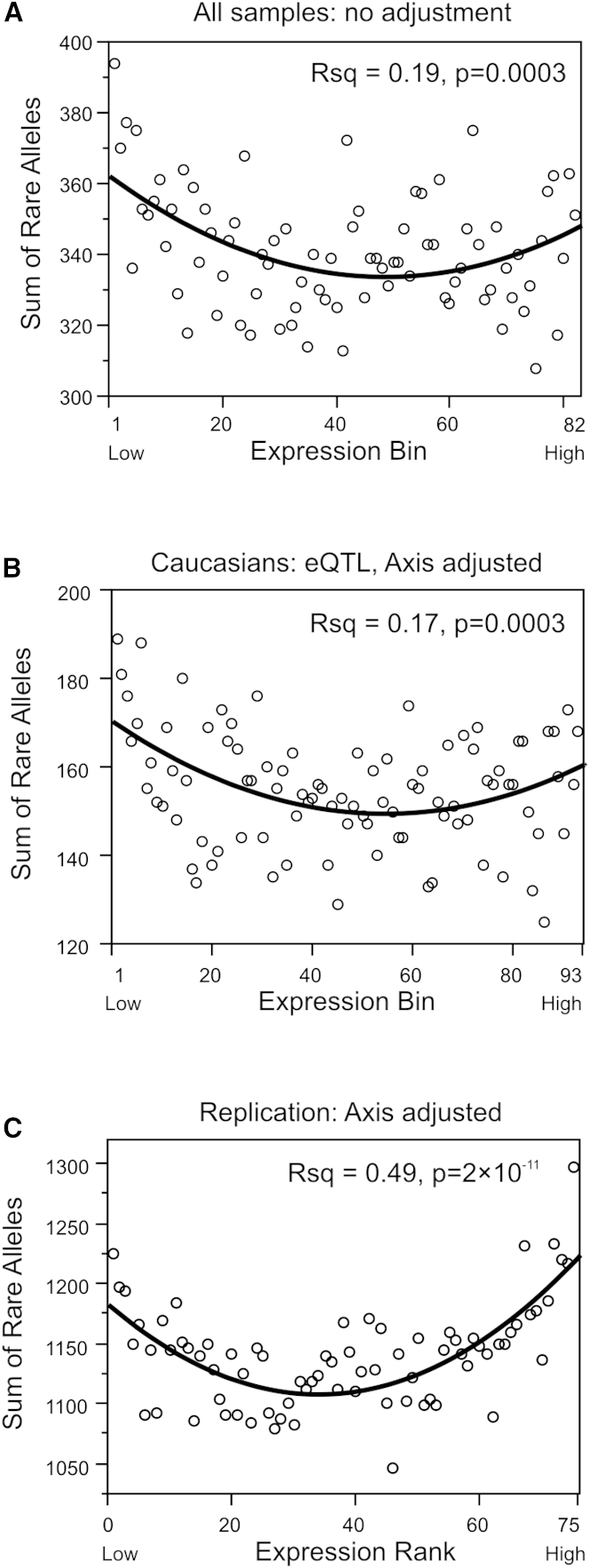
Figure 2.
Relationship between Rare Variant Counts and Transcript Abundance
Each plot shows the cumulative number of rare variants in equal sized bins across the indicated number of genes and individuals, with lowest expression bin to the left and highest to the right. Lines indicate the best fit quadratic model.
(A) 472 genes in 410 individuals of mixed ethnicity (5 individuals per bin), gene expression data normalized by SNM with variance adjustment (whole model R2 = 0.19, p = 0.0003).
(B) 472 genes in 279 Europeans (93 bins of 3 individuals) after removing effects of common eQTLs and conserved axes of covariation (R2 = 0.17, p = 0.0003).
(C) 4,633 genes in 75 replicates after removing effects of conserved axes of covariation and PC1 (R2 = 0.49, p = 2 × 10−11).
Although deleterious coding variants are generally loss of function, deleterious regulatory variants have similar a priori probabilities of increasing or decreasing transcript abundance. In fact, evolutionary studies14, 15 imply that gene expression is generally subject to moderate stabilizing selection, which acts to maintain transcript levels close to an optimal level. Thus, it is unlikely that many large effect mutations remain in the gene pool for extended periods of time, because purifying selection ensures that only moderate effect alleles persist. One class of haplotype that is particularly interesting in this regard is the few percent of alleles that can be traced to introgression from Neandertal or Denisovan populations.16, 17, 18 Although divergent at the genotype level and occasionally associated with extreme traits such as high-altitude adaptation,19 the maintenance of such haplotypes over several tens of thousands of generations leads to the expectation that they are not likely to collectively influence gene expression, a proposition that we also test here.
Material and Methods
Targeted Promoter Sequencing
DNA was obtained from 410 participants in the Atlanta CHDWB study,20, 21 under approval of the Emory University and Georgia Tech IRBs for genetic studies, and including written informed consent. The 410 samples comprised 274 females and 136 males, and included 297 individuals of European ancestry, 95 of African ancestry, and 18 of Asian ancestry. The age at entry into the program and initial sampling of blood spanned from 19 to 83 with a mean of 50.
A set of 500 genes was initially chosen for targeted promoter sequencing, but 28 of these located in the HLA complex were removed due to irregular read counts in the complex, resulting in a set of 472 genes that were included in the study. The full list is included as Table S1, and metrics of sequence quality along with a list of all promoter variants is available as additional material at the author’s website (see Web Resources). Among the 472 genes, 207 have common cis-eQTL, 244 are represented on the Metabochip, and 220 are represented on the Immunochip, 145 of which are on both chips.
Whole-genomic DNA was isolated from buffy coats using Flexigene DNA kits (QIAGEN). The location of the major transcription start site (TSS) of each gene was extracted from the UCSC Genome Browser (accessed May 2012), and oligonucleotide probes were designed with the Illumina Design Studio so as to pull down 1 kb upstream and 1 kb downstream of the major TSS for each of the 472 genes. 50-mer oligonucleotide probes were designed to ensure that the percentage of the total length of all regions targeted for enrichment was not less than 90%. Sequence capture libraries were generated and pooled with Illumina TruSeq DNA Sample Preparation Kits and TruSeq Custom Enrichment Kits. Paired end 100 bp sequencing was then performed on an Illumina HiSeq 2500 at Georgia Tech. Approximately 75% of the aligned reads with BWA mapped to the 2-kb promoter regions of the 472 genes, indicating good enrichment to the targeted regions. The average read depth across the dataset was more than 600×, with more than 90% of the reads in the 2-kb promoter regions having more than 20× read depth.
Variant Calling
After short read alignment, a variety of different strategies for variant calling were evaluated and contrasted, leading to the decision to use the output of the GATK Unified Genotyper algorithm13 applied to all samples in a pooled analysis. First, the BWA aligner was used to align fastq files to HuRef19. The total number of aligned reads per sample ranged from approximately 2 million to 40 million, with a mean of 14 million and standard deviation of 5 million. The mapped reads proportion was between 94.1% and 99.9% with a mean of 99.3% and a standard deviation of 1.05%. The percentage of concordantly paired reads ranged from 92.9% to 98.9% with a mean of 97.9% and a standard deviation of 1.2%. The GATK VQSR tool was used for variant filtering using the Illumina Omni chip array based on the 1000 Genomes Project as the training data with the highest-confidence SNPs from the 1000 Genomes Project’s call set used to validate the SNPs, utilizing a machine learning approach to optimize cutoffs for QD (quality by depth ratio), FS (Fisher’s exact test of strand bias), MQ (mapping quality metric), HaplotypeScore, MQRankSum (Mann-Whitney rank sum test for mapping qualities), and ReadPosRankSum (Mann-Whitney rank sum test for the distance of reads with the alternate allele to the end of the read).
After variant calling, we detected 17,584 raw SNPs in total, but these were reduced to 10,451 SNPs passing the filters, that lie within the 2-kb promoter regions of 472 genes. 8,833 of the SNPs are rare (defined as MAF < 0.05 in our dataset, which is concordant with 1000 Genomes frequencies) and 1,618 are common (with MAF ≥ 0.05), which averages 1.5 rare variants per promoter per individual. Approximately 60% of these rare variants are private, meaning that they were observed in only a single individual. The number of rare, common, and private SNPs per gene, along with an estimate of the polymorphism rate (π) per gene in the 2-kb sequenced region, as well as a matrix of rare allele counts per gene in each individual are available at the author’s website.
Verification by Sanger Sequencing and Genotyping
To verify the accuracy of the high-throughput sequencing, we Sanger sequenced 500 bp segments of two genes, TRAF3IP3 (OMIM: 608255) and HSPA8 (OMIM: 600816). The sequenced region of TRAF3IP3 was chr1: 209,929,132–209,929,708, in which two rare SNPs and four common SNPs were observed in the 410 samples. All of these were included in 96 samples that were Sanger sequenced, and all were validated. The sequenced region of HSPA8 was chr11: 122,932,665–122,933,158, which contained 18 rare and 5 common SNPs in the 96 sequenced samples, which were again verified by Sanger sequencing. A handful of other individuals were nominally positive at some of the rare sites, but manual inspection of the traces revealed poor-quality sequence toward the ends of the reads in those individuals suggesting a false positive rate that in any case would be less than 0.5%. No other variants that were not present in the GATK analysis were called with high confidence by Sanger sequencing, whereas all common variants were also validated by the Sanger sequencing.
Whole-genome genotypes either from Illumina OmniExpress or Core+Exome arrays, imputed onto 1000 Genomes with Impute2,22 were also available for the majority of individuals. Extremely high concordance was observed. These genotypes were thus used for common variant eQTL analysis, which will be described in detail elsewhere. We also interrogated whether rare SNPs lie within the Illumina probes, but found just three examples in two genes, so these do not explain enrichment of rare variants downstream of the promoter with gene expression.
Gene Expression Profiling
Transcript abundance measures were generated in two batches using Illumina-HT12 human gene expression arrays. RNA was prepared from whole-blood samples collected and stored in Tempus tubes (Life Technologies), according to manufacturer-recommended protocols, and quality was confirmed with an Agilent Bioanalyzer such that all samples had RIN numbers greater than 8. The first batch of samples was processed for hybridization and bead intensity extraction by Expression Analysis and the second by HudsonAlpha. The raw data are available at the Gene Expression Omnibus (GEO) but additional data processing steps were employed for this study to account for batch effects that might have skewed the rare variant association statistics.
Raw expression data in the form of average bead intensities from the Illumina Genome Studio were first transformed to log2 values and then processed with two standard approaches for removing surrogate variables. For PEER analysis23 of the Europeans only (Figure S1A), batch effects were removed with ComBat,24 and then age and gender were fit in a general linear model, before fitting the PEER algorithm with 20 factors selected, 6 of which were notably stronger than the remainder. As an additional mode of analysis, surrogate variable analysis (SVA),25 we considered the full dataset, after removing batch effects with COMBAT, then used SVA with age as the biological variable, fitting a single surrogate variable identified by the open source R code. Because ethnicity still explained 9% of the variance and age and gender each approximately 0.5%, we removed these as linear terms, then extracted just the 279 European-ancestry samples for rare variant enrichment tests as described below, generating very similar results as with PEER. However, for most analyses reported below, we used normalization based on supervised normalization of microarrays (SNM) algorithm in R,26 without fitting the surrogate variables. We fit age as the biological variable and removed effects of batch and ethnicity by including these as adjustment variables with the rm = True option. Individual effects were accounted for as the intensity-dependent variable.
We next extracted the 472 genes for which we have promoter genotypes and averaged the estimates for 172 genes that are represented by two (132 genes) or more (40 genes) probes in the Illumina-HT12 arrays. Each gene expression distribution was then transformed to the same scale, namely to z-scores, which are standard normal distributions with a mean of 0 and standard deviation of 1. To ensure that there was no overall batch effect on the variances (namely, that individuals from one batch are not, for technical reasons, more likely to have extreme values), we fit the z-scores by batch and combined them into a single gene expression dataset that was used to generate all of the results reported here. Quantile normalization27 was also performed in parallel (Figure S1B) because it is commonly used in the literature. Quantile normalization ensures that the distribution of abundance estimates of the entire gene expression profile of each individual is the same but does not ensure adjustment of covariates influencing the variance (or average expression) of each gene. It was applied to the raw log2 distributions of each individual, and probes for the same gene were again averaged prior to assigning genes to expression bins for the regression on rare variant counts.
Rare Variant Burden Test
In order to evaluate whether there was a relationship between transcript abundance and number of rare variants in the promoter, for each gene, each individual was placed in one of 82 equal-sized bins of five individuals based on the rank of the batch-adjusted z-scores. We then tallied the number of rare variants in the promoter regions of those five individuals and subsequently summed the rare variant counts across all 472 genes to achieve statistical power to detect the overall burden. With a MAF cutoff of 0.05, only 1 homozygote is expected per gene, but because most rare variants are rarer than this, the actual number of homozygotes is too small to impact the sums, but they were counted twice in the tally at the gene level. The bin size was chosen as a compromise between smooth fitting of the quadratic regression and compensating for noise in individual gene expression measures assessed by microarray. However, additional analyses were performed with 41 bins of size 10 individuals (Figure S1E) or with MAF < 0.01 (Figure S1F), neither of which had a meaningful effect on the conclusions because both remain highly significant for enrichment at both extremes. We then evaluated the deviation of the distribution from the null hypothesis of no relationship by fitting a quadratic model where the linear term captures bias toward enrichment for either higher or lower expression and the quadratic term captures the effect of bias at both extremes simultaneously.
The significance values of the two terms were observed to be very similar to the empirical p values obtained by permuting the sum counts against the bin number. We also performed a more robust permutation to shuffle the genotype and gene expression vectors, keeping the full vector of promoter counts within each individual (and the full vector of expression ranks) constant so as to preserve any biological covariance. With the appropriately normalized gene expression data, such permutations generally resulted in flat regressions of allele count on expression bin, with non-significant linear and quadratic terms. We then evaluated the significance of the actual data by documenting how many permutations out of 10,000 have a more significant overall model fit, which is just a few cases, strengthening support for the inference (1) that the normalization has removed systematic biases and (2) that there is a true burden of rare variants at either extreme of the transcript distribution, averaging across 472 transcripts.
A further adjustment was made to account for unequal total read counts among individuals or in specific genes as follows. For the analyses involving mixed races, we performed a haplotype burden analysis by collapsing all multi-SNP promoters down to a count of 1, instead of the actual number of rare variants. This should be conservative because it will tend to underestimate the contributions of two or more variants in a single promoter. We also fit a “joint” analysis (Figure S1C) where instead of binning gene expression solely within individuals, we generated 82 bins of 2,100 gene expression measures (5 × 420), based in the ranked z-scores of all genes in all individuals, and performed the regression on the summed allele counts associated with the 2,100 measures. In this procedure, each gene contributes slightly disproportionately to each bin, yet the overall result was again retained.
Adjustment of Rare Variant Burden Test for Covariates
A variety of biological factors could mask the effect of rare variants by increasing the variance of gene expression. Two obvious effects are the contribution of common eQTLs, which will tend to cause individuals with the less active polymorphism to be in lower expression bins, and trans-acting sources of gene expression covariance. Because peripheral blood preserved in Tempus tubes is a complex mixture of leukocytes (residual red blood cell and platelet gene expression is thought not to contribute strongly to observed transcript abundance), an obvious source of covariance is cell counts. Cell counts (lymphocytes, monocytes, neutrophils, erythrocytes, and platelets) explain on average just 6.0% of the variance of each transcript abundance measure in our dataset, which is actually one quarter of the amount explained by seven empirically determined common axes of covariance (average 23.8%: compare Figures S2A and S2B for distributions of variance explained). These axes probably reflect a mixture of the contributions of cell counts and coordinate gene regulation for example by interferon or other systemic factors. The seven axes are defined by the first principal component of the expression of ten “blood informative transcripts (BITs)” per axis, where the BITs have been defined by comparison of multiple blood gene expression datasets.28 Note that fitting PC1 to the 5 or 100 most correlated transcripts in each axis results in almost identical axis scores.
Simple linear regression was used to fit both eQTL and co-expression axes. For the eQTL adjustment, we first performed whole-genome cis-eQTL analysis on the full CHDWB dataset and identified significant eQTLs located within 5 kb of the TSS or in the gene body for 207 of the 472 genes at p < 10−4, observing more than 70% overlap with the blood eQTL browser variants derived from meta-analysis of more than 5,000 samples.29 For approximately one fifth of the genes, multiple additional cis-eQTLs were observed conditioned on the primary eQTL. We used stepwise linear regression to fit these empirical eQTLs in our dataset, also including age and sex as covariates in the model (although neither age nor sex account for more than a few percent of the variance of any of the 472 genes). The residuals from the eQTL fit were then used for axis adjustment, for which we computed the seven PC1 scores from the full SNM normalized gene expression matrix, and then identified which axis was most strongly correlated with the expression of each gene (8,654 were influenced by an axis at p < 10−5, including 398 of the 472 genes included in the enrichment analyses). Univariate linear regression was then used to fit the relevant axis for each gene, yielding gene expression residuals that were taken forward to the adjusted burden test.
Partitioning the Sources of Rare Variant Burden on Gene Expression
Several potential modifiers of the rare variant contribution were evaluated by dividing the total European ancestry dataset into subsets and comparing the model fit. For example, to evaluate whether suspected regulatory sites are more likely to harbor rare variants, we downloaded the RegulomeDB30 assignments for each SNP and contrasted sites with scores in the ranges 1–4 (likely regulatory) or 5–7 (weak or no evidence for regulatory potential). Similar analyses reported in Table S1 contrast SNPs upstream and downstream of the TSS; SNPs in genes in the upper or lower halves of the overall average transcript abundance spectrum; SNPs in genes in the upper or lower halves of the average promoter polymorphism distribution; SNPs in genes with or without common eQTLs; and SNPs in genes represented on the Metabochip,31 Immunochip,32 or neither. For each comparison, we report the significance of the quadratic term and the linear term as well as the overall model fit alongside the average number of rare variants per gene in the two samples being compared.
Assessment of Archaic Origins of Haplotypes
Because 289 of the promoter sequences in the European ancestry samples (0.2%) have three or more substitutions relative to the reference human genome, we asked whether they might be derived from archaic genomes. For each of the 472 genes, vcftools v.3.033 was used to query the online sequences of the Neandertal individual from an Altai Mountain cave34 and the Denisovan individual from the same cave.35 62 of the genes were not covered in the sequence. For the remaining 410 genes, we identified 195 positions where one of the 7,779 polymorphisms in our European individual promoter sequence set matched either of the archaic genomes rather than HuRef19. 75 genes had haplotypes with 3 or more sites different from HuRef19; of these, 18 had haplotypes matching a Neandertal haplotype. 31 genes had haplotypes with 4 or more sites different from HuRef19; of these, 8 had haplotypes matching a Neandertal haplotype. In several cases, a Denisovan haplotype was similar to the Neandertal one, but the Neandertal matched the human sequences, so as expected all archaic alleles identified in our dataset are most likely of Neandertal origin. Table S2 shows the genotypes of the 22 individuals with archaic haplotypes in the 8 genes with 4 or more divergent sites.
The expected proportion of Neandertal alleles genome-wide is between 1% and 2%, which is an order of magnitude greater than the proportion observed in our data. A very conservative lower bound on the proportion of divergent Neandertal promoters is 67 of 289 haplotypes, or 0.06% of all promoters. Including promoters with just 2 non-reference sites, approximately 10% of which match a Neandertal allele, this proportion rises to 0.1%. Single divergent sites at least double this proportion again. But owing to the possibility of recurrent mutation and recombination, we chose not to include such sites in analysis of association of Neandertal alleles with gene expression, because they need to be assessed by reference to long-range archaic haplotype blocks. It should also be noted that the Altai individual represents only a fraction of all archaic polymorphisms. Our observed proportion of Neandertal-derived alleles in the divergent promoters, at least 20%, can be considered to be within the expected range given that some alleles are also related to rare haplotypes observed in the African-ancestry individuals and might have been retained from the out-of-Africa dispersal.
Replication Dataset
In order to replicate the rare variant enrichment on a completely independent dataset generated with different gene expression and genotyping technologies, we identified a small cohort of 75 individuals with whole-genome sequence and whole-blood RNA-seq at the Duke Center for Human Genome Variation (D.B. Goldstein, P.I.). Most of these individuals are from a schizophrenia study. Permission to perform genetic analysis was obtained under IRB approval of Duke University, affirmed by the Georgia Tech IRB, and written informed consent was obtained from all study subjects, their parent/guardian, or legally authorized representative. Analysis of the principal components of the genotypes indicated that the sample includes 49 individuals of African ancestry, 24 of European ancestry, and 2 of Asian ancestry.
Whole-genome sequences were obtained on Illumina HiSeq2000 automated DNA sequencers and genotypes were called individually with the GATK algorithm. RNA-seq of whole blood preserved in Tempus tubes was performed also by paired end 100 bp sequencing on the Illumina platform. Raw read counts were log2 transformed and mean centered, and linear regression fitting each of the seven axes of variation (represented by PC1 of the blood informative transcripts) as well as the overall PC1 of gene expression variation (which is correlated with genetically determined ancestry). Subsequently, we assigned the rank of each gene in each individual and performed quadratic regression of the total number of individuals with at least one rare allele count for the gene (MAF < 0.05) in each of 75 ranks. That is, rather than pooling five individuals per bin, the analysis was essentially on bin sizes of 1, necessitated by the small sample of individuals.
Experimental Validation of SNP Effects by Genome Editing
Four SNPs were chosen for experimental validation by CRISPR/Cas9-mediated genome editing.36 Two (chr6: 24,667,167 in TDP2 [OMIM: 605764] and chr20: 33,999,719 in UQCC1 [OMIM: 611797]) were associated with loss of gene expression and two (chr5: 54,603,837 in DHX29 [OMIM: 612720] and rs182080358 in COMMD4 [HGNC: 26027]) with gain of gene expression in the CHDWB targeted sequencing analysis (Table S3). These four sites were all present as private alleles in one individual and were in the outlier set for an effect size greater than 2 SDs, visible in Figure 2A. For each SNP, we generated 11 or 12 independent approximately 10-cell K562 clones targeted by guide RNAs. Although K562 cells are erythroleukemic, rather than lymphoid or myeloid, because the variants are promoter proximal, we reasoned that they might have effects generally on transcript abundance and this cell line is well established for CRISPR experiments. Each promoter region in the cell line was re-sequenced to confirm that it is homozygous for the reference allele, also as reported for the publically available K562 sequence.
DNA oligonucleotides containing a G followed by 19-nt guide sequence were kinased, annealed, and ligated into pX330 (gift from Feng Zhang, Addgene plasmid #42230). The four guide RNAs were COMMD4, 5′-GCGCCAAGAAGCCAGGGCCC-3′; DHX29, 5′-GCTCTCACTGCTCCCAAAAA-3′; TDP2, 5′-GTGCGCAGGCGCCTGTGTCA-3′; and UQCC, 5′-GGTGAAGGAGTAATTTTCTA-3′. Once constructed, the plasmids were sequenced to confirm the guide strand region using the primer CRISPR_Seq (5′-CGATACAAGGCTGTTAGAGAGATAATTGG-3′). K562 cells (1 × 106) were transfected by nucleofection with 1 μg CRISPR plasmid construct for COMMD4, DHX29, TDP2, or UQCC and 300 ng of pmaxGFP, according to manufacturer’s recommended protocol (Lonza). GFP expression was analyzed 72 hr after transfection at which time DNA and RNA were prepared from pooled cells for preliminary analysis.
After a period of 4 to 10 days of growth, transfected cells were sorted by fluorescence activated cell sorting (FACS) and GFP-positive single cells or 10-cell colonies were sorted into 96-well plates. These were allowed to incubate for 2–4 weeks or until confluent and then subjected to T7E1 mutation detection assay37 in which the region of interest was amplified and 200 ng of purified PCR product was re-annealed and digested with T7 endonuclease 1. Cleavage was confirmed by the appearance of reduced molecular weight bands on 2% agarose gels that were quantified by ImageJ in order to estimate the fractional heterozygosity.
After confirming disruption of each relevant SNP by the T7E1 assay, five clones were chosen for each of the four genes with average heterozygosity between 16% and 23%. Droplet PCR,38 which is capable of detecting a 20% modulation of gene expression, was used with HPRT (OMIM: 308000) as a uniform control gene in all analyses, with UQCC as a second control for TDP2 and DHX29, and with TDP2 as the second control for UQCC and COMMD4 disrupted clones. In each experiment we contrasted the relative expression of the knocked out gene in the five clones to its average expression in five clones carrying a knockout of another gene (that is, TDP2 for DHX29 and vice versa; UQCC for COMMD4 and vice versa), performing a t test of the comparison. Differential expression was computed by formulating the ratio of each ddPCR count for the gene of interest to each reference gene, normalizing these ratios such that the average in the control cells was 1, then averaging the ratios to the two references. The average and significance of the change in expression is reported in Table S3 and Figure S3, which shows the proportional reduction or gain in signal for the five clones of each type, measured in two technical replicates of each clone.
Results
Figure 2A illustrates the core result that there is enrichment of rare variants for both increased and decreased gene expression in the full sample (model R2 = 0.19, p = 0.0003, permutation p = 0.0002). This is true for a variety of modes of normalization of the gene expression data detailed in the Material and Methods section, including a conservative strategy involving removal of batch effects with Combat24 followed by fitting age and gender followed by fitting PEER factors23 to just the European-ancestry individuals (Figure S1A), simple quantile normalization27 (Figure S1B), surrogate variable analysis,25 and an approach that first pooled the z-scores for all of the gene expression measures before assigning bins (Figure S1C). In subsequent analyses we employed supervised normalization of microarrays (SNM)26 because it optimally controls for confounding between technical and biological sources of variance. The full dataset consisted of 297 individuals of European ancestry, 18 of East Asian ancestry, and 95 of predominantly African ancestry, but because African ancestry is associated with mild differential expression of one third of the genes and an almost 4-fold elevation in rare variant counts in the promoter regions (consistent with HapMap estimates39, 40), we also considered each of the two larger population groups separately. The same trend was observed in both the European and African American samples but with only marginal significance in the latter due to the reduced sample size. Similarly, the result holds when we replaced total rare variant counts with rare haplotype counts, noting that some individuals carry alleles that differ from the reference allele at two or more sites. Because some genes show flatter distributions of expression in African ancestry samples, there is an excess of African ancestry at the extremes for the combined analysis, which we conservatively corrected by additionally standardizing the gene expression within each population group, which reduced the strength of the association with rare variant counts, particularly for increased expression (Figure S1D). Larger sample sizes will be required to explore whether there are population differences in rare variant contributions.
Further evidence that the regression models truly capture enrichment for rare variants at extremes of gene expression comes from the observation that the fit and significance improve after adjustment for covariates that are known to influence gene expression. Figure 2B shows the model fit for the Europeans (so as to avoid false positives due to population stratification) when gene expression bins were reassigned to the residuals after also fitting known common variant eQTL effects29 for each gene, as well as principal component scores for seven common axes of peripheral blood gene expression covariance.28 These seven axes collectively explain an average of 26% of the expression variance of the 472 genes (range 2% to 89%), which is approximately four times as much as the variance explained by the major cell types in whole blood (average 6%, range 0% to 44%; Figure S5). Fitting common eQTLs and the axis scores improves the model R2 from 0.05 (p = 0.07) to 0.17 (p = 0.0003, permutation p < 0.0001).
Next we asked whether the enrichment might be attributed to particular classes of gene or gene region, summarizing the results in Table S1 and Figure S4. First, categorizing all variants with respect to predicted regulatory potential according to RegulomeDB30 classifications confirms that variants that lie within features such as DNase hypersensitive sites or transcription factor binding sites (classes 1–4) are more enriched at the extremes than all other variants in classes 5–7 (p = 0.002 versus 0.08). Second, surprisingly, the enrichment was much stronger for variants located downstream of the transcription start site (p = 0.0007) versus upstream (p = 0.09). Third, we investigated whether there was a difference between overall low-abundance and high-abundance transcripts by dividing the dataset into two halves according to that parameter, but no difference was noted. Analyzing the genes separately in sets with low, intermediate, and high levels of promoter polymorphism suggests that the enrichment is observed across all levels of polymorphism. Notably as well, the burden remains when we reduced the minor allele frequency threshold to 0.01 (Figure S1F), reflecting the fact that the majority of rare variants are found in five or fewer individuals.
The most striking differential enrichment was observed with respect to gene function. The 472 genes were chosen for analysis in order to obtain approximately equal representation with respect to two criteria: whether or not they contain known common variant eQTLs, and whether or not they are thought to be associated with common chronic diseases. Genes not represented on the MetaboChip31 are significantly more likely to be enriched for rare regulatory variants (p = 9 × 10−6 versus 0.73 for metabolic disease-related genes). To confirm that this is significant, we compared the deviation in R-squared values with those of 1,000 random partitions of the European dataset keeping the number of individuals in the two sets constant, and observed that the Metabochip deviation is toward the tail (p = 0.007). A similar trend was observed for genes on the Immunochip,32 p = 0.005 versus 0.11, but this difference was not significant relative to the random partitions. These trends toward reduced rare regulatory SNP presence in promoters of disease-associated genes might be explained by relaxation of purifying selection on genes not associated with disease.
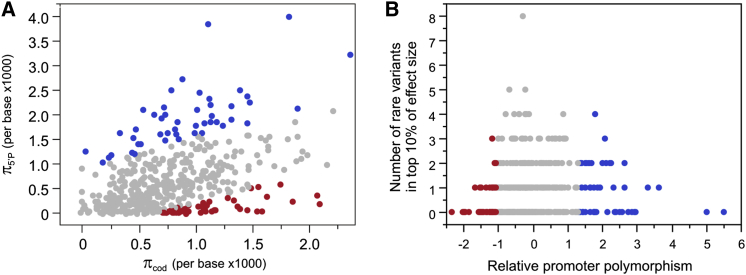
The possibility that some genes are more tolerant of regulatory variants is also implied by the observation that genes with common eSNPs are much more likely to harbor rare promoter-proximal variants affecting gene expression (p = 0.0006) than those without (p = 0.11). Furthermore, genes with low promoter polymorphism (π5′P) relative to coding region polymorphism (πcod) highlighted in Figure 3A are highly significantly depleted for rare variants in the top decile of effect sizes inferred from our data (Figure 3B; t test contrasting number in bottom 10% of low π5′P / πcod genes against the remainder, p = 3 × 10−5). The deficit is not due solely to low polymorphism because the contrast remains significant when π5′P is included as a covariate, and the bottom 10% of π5′P genes overall are as likely to harbor large effect rare variants as all other genes. This result raises the possibility that more extensive analyses of rare regulatory variant association with transcript abundance in different tissues might give rise to a measure of promoter region tolerance to functional mutation similar to the RVIS41 and constraint scores42 that classify genes with respect to intolerance to disruptive and pathogenic protein mutations.
Figure 3.
Genes with a Low Ratio of Promoter to Coding Polymorphism Are Intolerant to Large Effect Regulatory Variants
(A) Regression of π5′P on πcod highlighting the bottom 10% of genes with the largest negative residuals (lowest ratio, red) and top 10% (blue).
(B) Plot of relationship between the number of variants per gene in the top decile of estimated effect size from all 8,833 rare variants (MAF < 0.05) and residual from the regression in (A) as a measure of relative promoter polymorphism.
Another potential source of functional regulatory variation is divergent promoters derived from admixture with archaic humans. We identified 31 genes in the dataset that harbor haplotypes with 4 or more rare variants in Europeans, and for 8 of these genes (26%), the divergent haplotype matches informative Neandertal or Denisovan sequences. Similarly, 15 of 75 (20%) haplotypes with 3 or more rare variants appear to be archaic, whereas less divergent haplotypes are more likely to be generated by successive mutations in the human lineage. The remaining divergent haplotypes might represent Neandertal alleles not found in the Altai cave individual, as-yet-unrecognized archaic lineages, or fast-evolving alleles also present in the African gene pool. Two of the non-archaic divergent haplotypes, both also found in multiple African ancestry individuals in our sample, were likely to associate with elevated expression, in CDK10 (2 individuals, two-tailed t test p = 1.2−10) and NDUFB10 (12 individuals, two-tailed t test p = 0.0028). By contrast, there was no tendency for any of the archaic haplotypes to be associated with extreme expression in peripheral blood. Further analyses imputing ancestry based on long-range haplotypes from multiple Neandertal individuals, and assessments in multiple tissues, might nevertheless define roles for archaic promoter polymorphism in gene regulation.
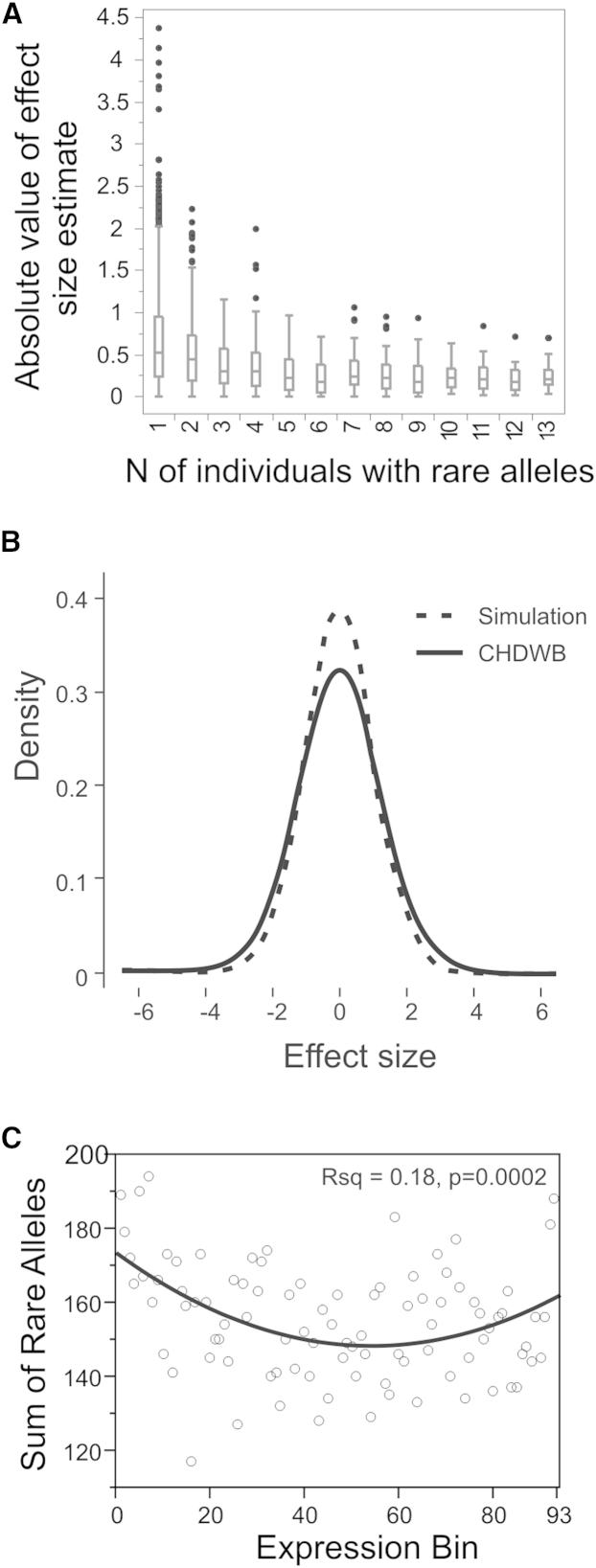
In order to estimate the frequency and magnitude of effects that would be consistent with the observed enrichments, we performed a simulation study where effects were assigned to the empirically determined genotypes and added to randomly generated and normally distributed gene expression values. Figure 4A shows the actual observed effects from the data, and this distribution is compared with those drawn from a gamma(1.5, 0.12) distribution (Figure 4B) that generates results comparable to the observed enrichment (Figure 4C). The estimated effect sizes under this model are somewhat smaller than those estimated from the data, probably a consequence of sampling overestimation in the experiment and the absence of technical noise in the simulated data. However, the result suggests that observed effect sizes are very rarely greater than 2 SDs and that most of the enrichment is driven by rare variants with influences comparable to those of common eQTLs, namely allelic substitutions in the range of 0.5 to 1 SD.
Figure 4.
Estimation of Effect Sizes of Rare Variants
(A) The distribution of absolute values of the estimated effect size from the CHDWB data as a function of the number of alleles in the sample. Boxes show mean and interquartile range with whiskers at 1.5 times IQR with outliers as single points.
(B) Comparison of estimated effect size distribution from the data (bold curve) and in the simulation (thin black curve) showing slight excess of larger effect variants in the observed data.
(C) Simulated model fit assuming a gamma distribution with gamma(1.5,0.12), for 472 genes in 279 individuals, showing excess of extreme expression as in the actual data (R2 = 0.18, p = 0.0002).
Next we replicated the result with an independent dataset consisting of 75 whole-genome sequences of mixed ancestry, linked to whole-blood RNA-seq profiles. Figure S5 shows that the same trend was observed with the 472 genes as in the Atlanta cohort, but owing to the small sample size of individuals the result is only significant at p = 0.008. However, increasing the analysis to include the same total number of comparisons as in the CHDWB, namely 4,633 genes, and adjusting for the axes of variation as well as a strong surrogate variable corresponding to ancestry provides clear replication of the rare variant enrichment (R2 = 0.49, p = 2 × 10−11, permutation p < 0.0001; Figure 2C).
To experimentally validate rare variant regulatory effects predicted from the statistical analysis, we used CRISPR/Cas9 to mutagenize four sites that had estimated effect sizes greater than 5-fold higher or lower than the population mean in a single individual, in K562 erythroleukemia cells.43 11 individual clones were grown for each disruption and once cleavage was confirmed via the T7E1 assay,37 indicating 16% to 23% average heterozygosity, 5 clones were chosen for RNA abundance measurement. Droplet digital RT-PCR,38 which quantifies relative abundance by counting the number of nanodroplets from a dilution of the RNA sample that yield PCR product, assessed relative to control genes, was used to demonstrate that disruption of three of the four sites resulted in reduced or increased transcript abundance (Table S3). Disruption of rs182080358 associated with increased expression of COMMD4 weakly increased transcript abundance in three of the four clones and greatly increased it in another. All five clones with disruptions in UQCC reduced expression almost by half, whereas disruption of a negative control site had no effect and four of five clones with disruptions in TDP2 reduced expression to varying degrees (Figure S3). Because our CRISPR protocol causes small deletions rather than targeted replacement of polymorphisms, it is expected that the disruptions remove binding sites for transcriptional activators in each case. This could cause loss or gain of expression depending on the nature of the transcription factor, but it is notable that in each case the effect of disruption was in the same direction as the nucleotide substitution. The difference in estimated effect size might be attributed to over-estimation of the effect from the microarray data and underestimation in the CRISPR clones that have incomplete heterozygosity because K562 cells are largely triploid.43
Discussion
These experiments demonstrate that the combination of gene sequencing with transcript profiling in peripheral blood has good potential as a screening approach to identify rare promoter-proximal variants that significantly disrupt the regulation of gene expression. Although the average effect sizes we estimate are not large, they are on a par with those estimated for disease-associated common eQTLs.29 That is to say, large-effect rare variants typically alter expression of a gene between one half and one standard deviation unit. For a transcript otherwise expressed in the inter-quartile range, namely in the middle half of the distribution, this will often be enough to cause it to shift it to the upper or lower decile where it would presumably be more likely to contribute to pathology or abnormality.
Two other studies have also found indirect evidence for rare variant influences on gene expression in cell lines, albeit spread across extended cis-regulatory regions. Montgomery et al.44 performed eQTL analysis on the 1000 Genomes Project lymphoblast cell lines and observed that transcripts with rare instances of allele-specific expression (ASE) had a median of four perfectly concordant rare putative regulatory variants within 100 kb of the TSS, compared with three for control transcripts not exhibiting ASE. Interestingly, the enrichment was greater for non-synonymous than synonymous coding ASE variants and biased to lower expression, suggesting an epistatic interaction between regulatory and structural polymorphism. They also found that very rare alleles in conserved putative regulatory sites were more likely to be 2 SDs from the mean. Reanalyzing the Geuvadis lymphoblast cell line data, Zeng et al.45 focused on rare instances where the correlation between two transcripts was aberrant in a few individuals and described an enrichment of private variants specifically in the vicinity of enhancers in the 1 Mb region of the genes.
We do not observe any tendency for variants to be more likely to increase or decrease gene expression, which is consistent with the inference that globally, transcript abundance is under stabilizing selection.14, 15 Recent results from yeast46 also indicate that there is selection against noise promoted by regulatory variants, so it will be interesting in larger studies to evaluate whether genes with reduced promoter relative to coding polymorphism also tend to have reduced expression variability due to rare variants.
If our experimental strategy can be applied to tissues or cell types directly relevant to certain pathologies, such as neurons or cardiac cells, it could enhance efforts to infer whether rare variants in regulatory regions are functionally deleterious. It is possible that iPSC culture might be beneficial in this regard, particularly if the effects on transcription are greater in uniform cell culture than in mixed whole-blood cell populations from individuals who experience a wide range of environments. By extrapolation from our data, we estimate that the average individual probably carries several dozen such variants that cause gene expression to be toward the extreme, and consequently these cannot be ignored as a potential source of disease-related pathology. Determination of whether or not rare regulatory variants are sufficient to cause rare diseases in the same manner as structural variants are inferred to will require experimental designs targeted to individuals with particular congenital disorders.
Acknowledgments
This work was supported by NIH Program Project 1-P01-GM0996568 (Project 3, G.G. Principal Investigator), directed by Bruce Weir (University of Washington). We are grateful to Cathy Laurie and Sarah Nelson for assistance with imputation, Shweta Biliya for sequencing, Sahar Gelfman and David Goldstein for provision of the replication dataset (which was generated with the support of NIH grant RC2MH089915), Mary-Jane Chandler for assistance with the ancestral genome analysis, and Cathy Laurie, Joe Lachance, Patrick McGrath, and Fred Vannberg for comments on the manuscript. We thank all of the staff and participants at the CHDWB for their willing assistance, especially Ken Brigham for his vision for the Center.
Published: February 4, 2016
Footnotes
Supplemental Data include five figures, three tables, and an Appendix and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.023.
Accession Numbers
Short read sequences have been deposited at the SRA (accession number phs001021.v1.p1) with restricted access requiring approval by dbGaP under the terms of the consent provided by CHDWB participants. The raw data are available at the Gene Expression Omnibus as accession GEO: GSE61672.
Web Resources
The URLs for data presented herein are as follows:
HUGO Gene Nomenclature Committee, http://www.genenames.org/
Neandertal and Denisovan genome sequences, http://cdna.eva.mpg.de/neandertal/altai/
OMIM, http://www.omim.org/
Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra
UCSC Genome Browser, http://genome.ucsc.edu
Zhao et al. additional materials, http://cig.gatech.edu/supplementary/zhao-et-al-2016
Supplemental Data
References
- 1.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K., Bergen S.E., Kähler A. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt K.A., Mistry V., Bockett N.A., Ahmad T., Ban M., Barker J.N., Barrett J.C., Blackburn H., Brand O., Burren O. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–235. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgenthaler S., Thilly W.G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Chandrasekharappa S.C., Lach F.P., Kimble D.C., Kamat A., Teer J.K., Donovan F.X., Flynn E., Sen S.K., Thongthip S., Sanborn E., NISC Comparative Sequencing Program Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood. 2013;121:e138–e148. doi: 10.1182/blood-2012-12-474585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemos B., Meiklejohn C.D., Cáceres M., Hartl D.L. Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- 15.Hodgins-Davis A., Rice D.P., Townsend J.P. Gene expression evolves under a House-of-Cards model of stabilizing selection. Mol. Biol. Evol. 2015;32:2130–2140. doi: 10.1093/molbev/msv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 18.Qin P., Stoneking M. Denisovan ancestry in East Eurasian and Native American populations. Mol. Biol. Evol. 2015;32:2665–2674. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- 19.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brigham K.L. Predictive health: the imminent revolution in health care. J. Am. Geriatr. Soc. 2010;58(Suppl 2):S298–S302. doi: 10.1111/j.1532-5415.2010.03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabassum R., Cunningham L., Stephens E.H., Sturdivant K., Martin G.S., Brigham K.L., Gibson G. A longitudinal study of health improvement in the Atlanta CHDWB wellness cohort. J. Pers. Med. 2014;4:489–507. doi: 10.3390/jpm4040489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 25.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mecham B.H., Nelson P.S., Storey J.D. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Preininger M., Arafat D., Kim J., Nath A.P., Idaghdour Y., Brigham K.L., Gibson G. Blood-informative transcripts define nine common axes of peripheral blood gene expression. PLoS Genet. 2013;9:e1003362. doi: 10.1371/journal.pgen.1003362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voight B.F., Kang H.M., Ding J., Palmer C.D., Sidore C., Chines P.S., Burtt N.P., Fuchsberger C., Li Y., Erdmann J. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes A., Brown M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S.W., Kim S., Kim J.M., Kim J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 37.Mashal R.D., Koontz J., Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 1995;9:177–183. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- 38.Kiss M.M., Ortoleva-Donnelly L., Beer N.R., Warner J., Bailey C.G., Colston B.W., Rothberg J.M., Link D.R., Leamon J.H. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal. Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozzio B.B., Lozzio C.B. Properties of the K562 cell line derived from a patient with chronic myeloid leukemia. Int. J. Cancer. 1977;19:136. doi: 10.1002/ijc.2910190119. [DOI] [PubMed] [Google Scholar]
- 44.Montgomery S.B., Lappalainen T., Gutierrez-Arcelus M., Dermitzakis E.T. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Y., Wang G., Yang E., Ji G., Brinkmeyer-Langford C.L., Cai J.J. Aberrant gene expression in humans. PLoS Genet. 2015;11:e1004942. doi: 10.1371/journal.pgen.1004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzger B.P., Yuan D.C., Gruber J.D., Duveau F., Wittkopp P.J. Selection on noise constrains variation in a eukaryotic promoter. Nature. 2015;521:344–347. doi: 10.1038/nature14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.