Systemic acquired resistance and the associated priming response are regulated by a pipecolic acid/FMO1 signaling module that controls both salicylic acid-dependent and -independent defense pathways.
Abstract
We investigated the relationships of the two immune-regulatory plant metabolites, salicylic acid (SA) and pipecolic acid (Pip), in the establishment of plant systemic acquired resistance (SAR), SAR-associated defense priming, and basal immunity. Using SA-deficient sid2, Pip-deficient ald1, and sid2 ald1 plants deficient in both SA and Pip, we show that SA and Pip act both independently from each other and synergistically in Arabidopsis thaliana basal immunity to Pseudomonas syringae. Transcriptome analyses reveal that SAR establishment in Arabidopsis is characterized by a strong transcriptional response systemically induced in the foliage that prepares plants for future pathogen attack by preactivating multiple stages of defense signaling and that SA accumulation upon SAR activation leads to the downregulation of photosynthesis and attenuated jasmonate responses systemically within the plant. Whereas systemic Pip elevations are indispensable for SAR and necessary for virtually the whole transcriptional SAR response, a moderate but significant SA-independent component of SAR activation and SAR gene expression is revealed. During SAR, Pip orchestrates SA-dependent and SA-independent priming of pathogen responses in a FLAVIN-DEPENDENT-MONOOXYGENASE1 (FMO1)-dependent manner. We conclude that a Pip/FMO1 signaling module acts as an indispensable switch for the activation of SAR and associated defense priming events and that SA amplifies Pip-triggered responses to different degrees in the distal tissue of SAR-activated plants.
INTRODUCTION
In tissue inoculated by pathogenic microbes, plants are able to initiate a basal immune program that counteracts microbial infection. Plant basal resistance or pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) involves recognition of microbial structures by plant pattern recognition receptors, defense signal transduction, and transcriptional activation of defense-related gene expression (Boller and Felix, 2009). Yet, PTI can be overcome by well-adapted pathogen isolates. However, previous pathogen encounters can render plants significantly more resistant to a future challenge. For instance, systemic acquired resistance (SAR), a state of heightened resistance of the entire plant foliage to a broad spectrum of biotrophic and hemibiotrophic phytopathogens, is induced by a localized leaf inoculation with avirulent or virulent microbial pathogens (Mishina and Zeier, 2007; Fu and Dong, 2013). Plants with activated SAR exhibit enhanced systemic expression of antimicrobial PR proteins and other augmented immune responses (Sticher et al., 1997). In addition, biologically induced SAR conditions plants to react more quickly and vigorously to subsequent pathogen attack (Jung et al., 2009; Návarová et al., 2012), a phenomenon also designated as defense priming (Conrath, 2011).
The establishment of SAR is regulated by signal-active plant metabolites (Dempsey and Klessig, 2012; Shah and Zeier, 2013). From ∼1980 onwards, multiple studies have provided evidence that the phenolic defense hormone salicylic acid (SA) plays a pivotal role in SAR (reviewed in Vlot et al., 2009). The pathogen-induced biosynthesis of SA in Arabidopsis thaliana proceeds via isochorismate synthase-mediated conversion of chorismate to isochorismate (Nawrath and Métraux, 1999; Wildermuth et al., 2001). The Arabidopsis sid2-1 mutant, which is defective in ISOCHORISMATE SYNTHASE1 (ICS1), is unable to accumulate pathogen- and stress-inducible SA and is impaired in SAR activation (Nawrath and Métraux, 1999). Although SA was previously proposed as the mobile compound that travels from inoculated to distal leaves to induce SAR (Shulaev et al., 1995; Mölders et al., 1996), genetic studies support the notion that SA is not a SAR long-distance signal but that its isochorismate-derived de novo biosynthesis in systemic leaf tissue is required for proper SAR (Vernooij et al., 1994; Attaran et al., 2009). Since then, several other candidate long-distance signals have been suggested (reviewed in Shah and Zeier, 2013). A predominant portion of SA downstream responses is dependent on the transcriptional coactivator NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), which has been identified, in addition to its paralogs NPR3 and NPR4, as a bona fide SA receptor (Fu et al., 2012; Wu et al., 2012). NPR1 acts as a central regulator of SAR (Fu and Dong, 2013).
In addition to SA and NPR1, FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1) is a critical component of biologically induced SAR in Arabidopsis (Mishina and Zeier, 2006; Jing et al., 2011). The strongly pathogen-inducible FMO1 gene encodes a flavin monooxygenase that is activated in both locally inoculated and distal leaves (Bartsch et al., 2006; Koch et al., 2006; Mishina and Zeier, 2006). Notably, functional FMO1 is necessary for SA accumulation in the systemic, noninoculated leaves but dispensable for SA production in inoculated tissue (Mishina and Zeier, 2006). A biochemical characteristic of distinct FMOs from plants, insects, and mammals is that they are able to oxidize amino or sulfide groups within small metabolic substrates (Schlaich, 2007). We therefore previously hypothesized that an endogenously produced plant amine, amino acid, or S-containing metabolite might play a central function in SAR (Mishina and Zeier, 2006).
This hypothesis was confirmed by identifying the non-protein amino acid pipecolic acid (Pip; homoproline) as a critical SAR regulator (Návarová et al., 2012). Alongside with SA, Pip accumulates to high amounts in Arabidopsis leaves inoculated with SAR-inducing Pseudomonas syringae bacteria as well as in leaves distant from initial inoculation. Very specifically, and in contrast to SA and many other accumulating metabolites, Pip is enriched in phloem exudates collected from inoculated leaves, indicating specific transport of Pip out of inoculated leaves (Návarová et al., 2012). Pipecolic acid is a plant natural product with widespread occurrence throughout the angiosperms (Morrison, 1953; Zacharius et al., 1954), and its accumulation in leaves after inoculation with bacterial, fungal, or viral pathogens has been documented for rice (Oryza sativa), potato (Solanum tuberosum), tobacco (Nicotiana tabacum), soybean (Glycine max), and Arabidopsis (Pálfi and Dézsi, 1968; Návarová et al., 2012; Vogel-Adghough et al., 2013; Aliferis et al., 2014). In addition, Pip is overproduced in Arabidopsis autophagy mutants that exhibit stress-related phenotypes (Masclaux-Daubresse et al., 2014).
Feeding studies with isotope-labeled Lys demonstrated that, like animals (Broquist, 1991), plants synthesize l-Pip from Lys (Fujioka and Sakurai, 1997) and strongly suggested that Lys-to-Pip conversion in plants involves both a classical aminotransferase reaction removing the α-amino group from Lys and a subsequent reductase activity (Gupta and Spenser, 1969; Zeier, 2013). An aminotransferase with strong substrate specificity for Lys is encoded by AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) (Song et al., 2004a). Similar to FMO1 (Mishina and Zeier, 2006), ALD1 has been identified as an essential SAR component that is upregulated in both locally inoculated and distal leaf tissue (Song et al., 2004b). Our recent finding that ald1 knockout mutants are not able to biosynthesize and accumulate Pip after pathogen inoculation indicates that ALD1 is the aminotransferase required for the Lys-derived biosynthesis of Pip (Návarová et al., 2012). Moreover, the SAR defect of Pip-deficient ald1 can be rescued by exogenous supply of Pip in physiological doses, demonstrating that Pip accumulation is required for SAR activation (Návarová et al., 2012). Exogenous Pip is also sufficient to increase resistance to P. syringae in wild-type and ald1 plants to a similar degree as biological SAR (Návarová et al., 2012). Notably, Pip feeding is able neither to restore the SAR defect of fmo1 nor to increase pathogen resistance in this mutant, indicating that Pip requires functional FMO1 to activate SAR (Návarová et al., 2012). Like for P. syringae-induced SAR, Pip accumulation and FMO1 are also integral parts of the systemic immune response induced by local oviposition of insect eggs in Arabidopsis (Hilfiker et al., 2014).
The activation of SAR in noninfected distal tissue of pathogen-inoculated plants relies on the perception and amplification of endogenous plant signals (Shah and Zeier, 2013). Our current working model implies that Pip is a central player of a feedback amplification mechanism that realizes systemic SA accumulation and SAR establishment (Návarová et al., 2012; Zeier, 2013). Once established, SAR primes Arabidopsis plants for effective defense responses to future pathogen challenge. These responses include the accumulation of the phytoalexin camalexin and expression of a series of defense-related genes, including ALD1, FMO1, and PR1. Systemically elevated Pip during SAR is necessary and sufficient for the SAR-associated priming response (Návarová et al., 2012; Zeier, 2013).
In this study, we investigate the interplay of Pip and SA in Arabidopsis immune signaling and provide a detailed characterization of the systemic transcriptional response associated with SAR. Systemic transcriptional reprogramming upon localized P. syringae inoculation involves enhanced expression of microbial pattern recognition receptors, various defense signaling components, and specific transcription factor classes, as well as a massive downregulation of photosynthetic and growth-related genes. We show that Pip orchestrates SAR and virtually the whole transcriptional SAR response via SA-dependent and, less prominently, SA-independent activation pathways. Our data indicate that activation of a primed state in distal leaves of locally inoculated plants requires functional FMO1 downstream of Pip and that SAR priming of a subset of genes proceeds in SA-deficient sid2 to a similar extent as in the wild type. Our results therefore emphasize the significance of partially SA-independent signaling events during SAR establishment and the realization of SAR-associated defense priming. Moreover, they indicate that Pip and SA act both synergistically and independently from each other to mediate PR gene expression and plant basal resistance to P. syringae.
RESULTS
The SA and Pip Defense Pathways Provide Additive Contributions to Basal Resistance
To obtain deeper insights into the interplay between the immune signals Pip and SA in mediating Arabidopsis basal resistance, SAR establishment, and defense priming, we comparatively investigated resistance responses of the Col-0 wild type, Pip-deficient ald1 (Návarová et al., 2012), SA induction-deficient sid2-1 (sid2; Nawrath and Métraux, 1999), and a sid2 ald1 double mutant unable to generate both SA and Pip after pathogen inoculation. Whereas ald1 represents a T-DNA knockout line for ALD1 (Song et al., 2004b; Návarová et al., 2012), sid2 was previously obtained by ethyl methanesulfonate mutagenesis and carries a single base pair mutation in the ICS1 coding region that results in a premature stop codon and a full loss of ICS1 function (Nawrath and Métraux, 1999; Wildermuth et al., 2001). The sid2 ald1 double mutant was generated by screening progeny from a cross of the single mutants (Supplemental Figures 1 and 2).
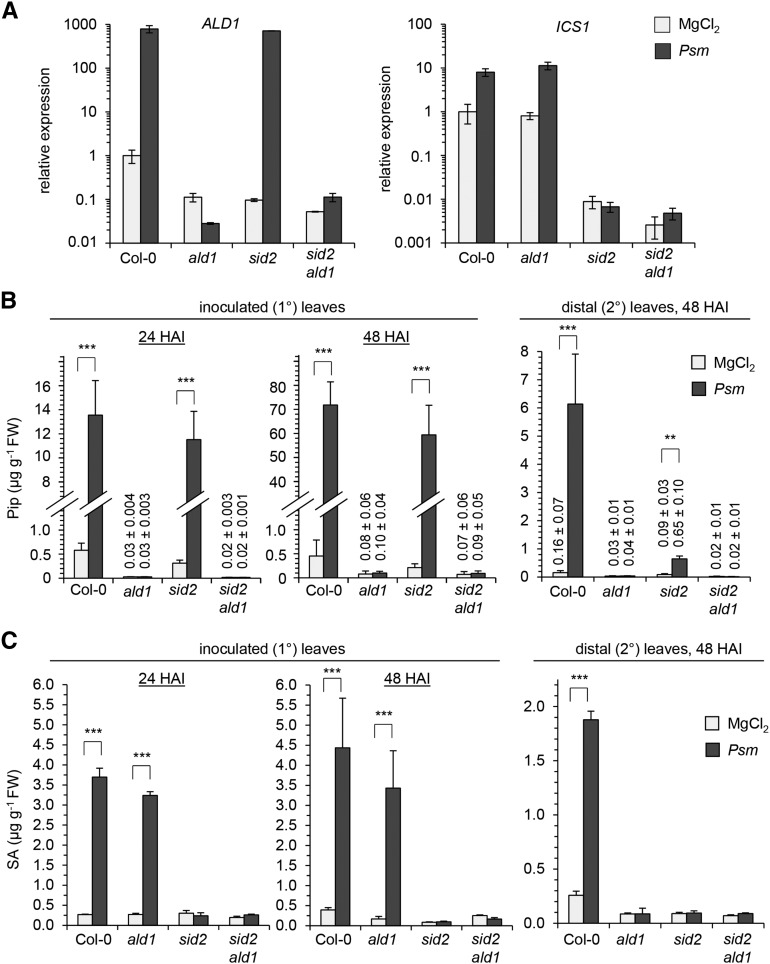
Upon leaf inoculation with the SAR-inducing bacterial strain Pseudomonas syringae pv maculicola ES4326 (Psm) (Mishina and Zeier, 2007; Attaran et al., 2009; Jing et al., 2011), we observed strong increases of ALD1 and ICS1 transcript levels in wild-type Col-0 plants, increases of ICS1 but not ALD1 in ald1, and elevations of ALD1 but not ICS1 in sid2. Moreover, sid2 ald1 lacked both basal and pathogen-induced expression of ALD1 and ICS1 (Figure 1A). On the metabolite level, Psm attack triggered a strong accumulation of both Pip and SA in inoculated (1°) leaves and in distal (2°), noninoculated leaves of Col-0 plants. By contrast, sid2 ald1 produced neither Pip nor SA after pathogen inoculation and contained only faint basal levels of the two immune-regulatory metabolites (Figures 1B and 1C). Consistent with our previous analyses (Návarová et al., 2012), ald1 completely lacked pathogen-induced Pip accumulation but was able to activate SA production in inoculated leaves, whereas sid2 showed a reciprocal accumulation pattern (Figures 1B and 1C).
Figure 1.
Pip and SA Biosynthesis in Local and Systemic Tissue of Wild-Type Col-0, ald1, sid2, and sid2 ald1 Plants Inoculated with SAR-Inducing Psm.
(A) Expression of ALD1 (left) and ICS1 (right) in Psm-inoculated leaves at 24 h after inoculation (HAI). Infiltration with 10 mM MgCl2 served as a mock control treatment. Transcript levels were assessed by qPCR analysis and expressed relative to the Col-0 mock control value. Data represent the mean ± sd of three biological replicate leaf samples from different plants. Each biological replicate consists of two leaves from one plant. Expression values for each biological replicate represent the mean of two technical replicates.
(B) and (C) Accumulation of Pip (B) and free SA (C) in Psm-inoculated (1°) leaves at 24 and 48 HAI (left) and in distal, noninoculated (2°) leaves (right) at 48 HAI. Data represent the mean ± sd of at least three biological replicate leaf samples from different plants. Each biological replicate consists of six leaves from two plants. Asterisks denote statistically significant differences between Psm and MgCl2 samples (***P < 0.001 and **P < 0.01; two-tailed t test). Numerical values for samples with very low metabolite contents are given above the respective bars.
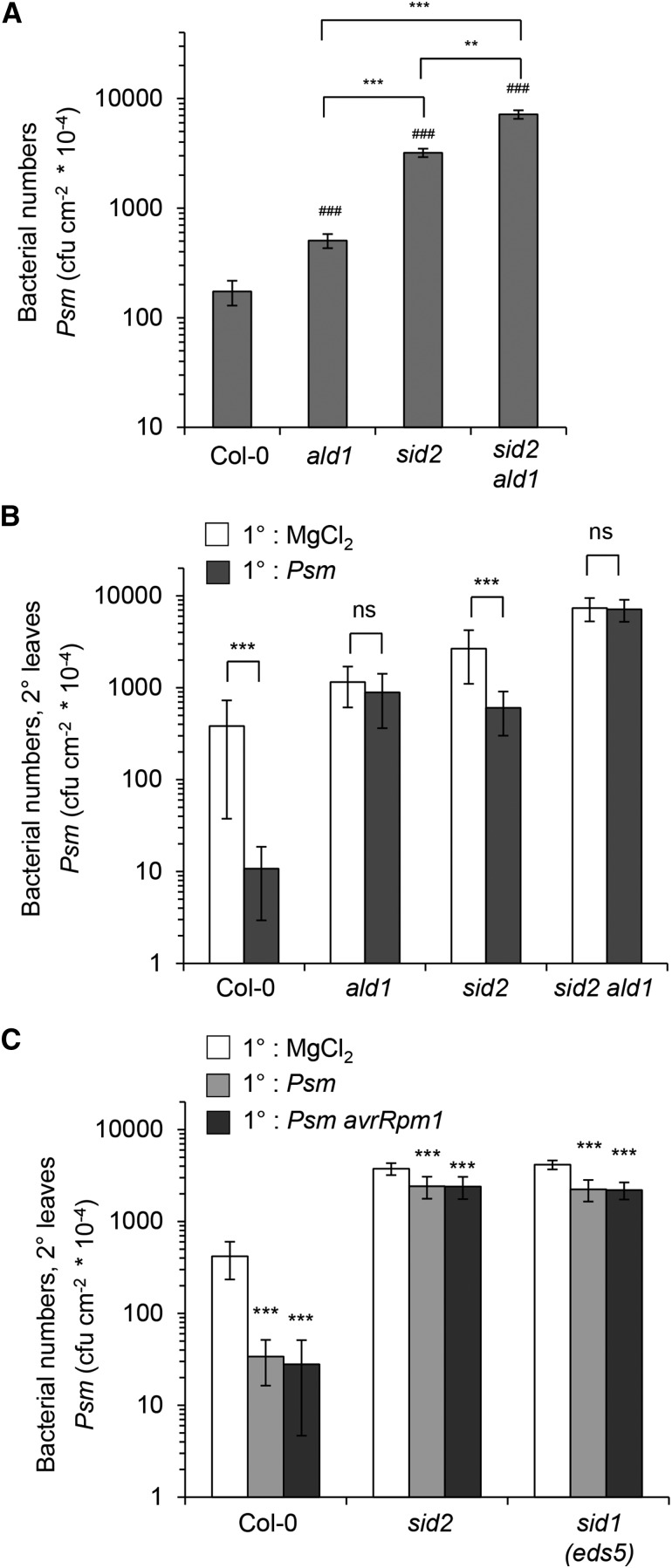
To assess basal resistance to bacterial attack, we compared the growth of the virulent Psm strain in leaves of Col-0, ald1, sid2, and sid2 ald1 3 d after inoculation. Both ald1 and sid2 allowed higher bacterial multiplication than the wild type (Figure 2A), confirming the previously suggested requirements for Pip and SA in the full basal immunity program at inoculation sites (Nawrath and Métraux, 1999; Návarová et al., 2012). The sid2 mutant permitted significantly higher bacterial multiplication than ald1, indicating that the relative contribution of SA to basal resistance is higher than that of Pip. Notably, leaf-inoculated sid2 ald1 showed the weakest resistance phenotype of all investigated lines and allowed a significantly higher bacterial multiplication than Col-0, ald1, and sid2 (Figure 2A). This indicates that SA and Pip provide additive contributions to Arabidopsis basal immunity against P. syringae.
Figure 2.
Analyses of Basal Resistance to Psm and SAR in Pip- and/or SA-Deficient Mutant Plants.
(A) Basal resistance of Col-0, ald1, sid2, and sid2 ald1 plants to Psm. Three leaves per plant were inoculated with a suspension of Psm (OD600 = 0.001) and bacterial numbers quantified 3 d later. Bars represent mean values (±sd) of colony-forming units (cfu) per square centimeter from at least seven biological replicate samples (n) derived from different plants. Each biological replicate consists of three leaf discs harvested from different leaves of one plant. Number signs denote statistically significant differences from the Col-0 wild-type value (#P < 0.05, ##P < 0.01, and ###P < 0.001; two-tailed t test). Asterisks designate statistically significant differences between indicated samples. To test whether the effects of ald1 and sid2 on bacterial proliferation are additive or synergistic, a linear model was used (log10 bacterial count ∼ ald1*sid2). No significant interaction of ald1*sid2 was detected both according to the F-test and according to Akaikes information criterion (P = 0.0508, AICsynergistic = −120 AICadditive = −122.7); hence, the effect is additive.
(B) SAR assay with Col-0, ald1, sid2, and sid2 ald1 plants. Lower (1°) leaves were infiltrated with either 10 mM MgCl2 or Psm (OD600 = 0.005) to induce SAR, and 2 d later, three upper leaves (2°) were challenge-infected with Psm (OD600 = 0.001). Bacterial growth in upper leaves was assessed 3 d after 2° leaf inoculation (n ≥ 7; as described in [A]). Asterisks denote statistically significant differences between Psm pretreated and mock control samples (***P < 0.001; ns, not significant, two-tailed t test).
(C) Biological SAR induction upon 1° leaf inoculation with compatible Psm and incompatible Psm avrRpm1 in Col-0, sid2, and sid1 plants (n ≥ 7; as described in [A] and [B]). Asterisks denote statistically significant differences between Psm or Psm avrRpm1 (OD600 = 0.005) pretreated and mock control samples (***P < 0.001; two-tailed t test).
Pip Regulates SAR via SA-Dependent and -Independent Activation Pathways
The 2° leaves of locally inoculated sid2 plants accumulated Pip to a moderate but significant extent, suggesting that pathogen-triggered systemic responses are not fully suppressed in sid2 (Figure 1B). On the contrary, ald1 was unable to elevate SA in 2° leaves, corroborating our previous results showing the necessity of Pip for the activation of SA biosynthesis and concomitant SA accumulation in distal leaves (Figure 1C; Návarová et al., 2012). To directly examine the SAR response in the lines under investigation, we inoculated plants with Psm in lower, 1° leaves to induce SAR or performed mock treatments with 10 mM MgCl2 to generate appropriate noninduced control plants and then challenge-inoculated upper, 2° leaves of both pathogen-inoculated and mock-treated plants with Psm 2 d after the primary treatment. Bacterial growth in upper leaves was assessed another 3 d later (Mishina and Zeier, 2007; Attaran et al., 2009; Jing et al., 2011; Návarová et al., 2012). In these assays, wild-type Col-0 plants exhibited a strong SAR response and the bacterial multiplication in challenge-infected leaves was generally attenuated by 95 to 98% as a consequence of SAR induction (Figure 2B). The Pip-deficient ald1 mutant was not able to activate any SAR upon attempted induction (Song et al., 2004b; Návarová et al., 2012), confirming the previously reported necessity of Pip accumulation in SAR establishment (Návarová et al., 2012). Remarkably, the SA-deficient sid2 mutant was able to significantly induce resistance upon localized Psm inoculation in distal leaves (Figure 2B). Although the observed SAR response in sid2 was small compared with wild-type SAR (a 50 to 80% reduction of bacterial growth), the effect was reproducible between experiments and occurred, besides in sid2, in sid1 (Figure 2C), another Arabidopsis mutant unable to activate stress-induced SA biosynthesis (Nawrath and Métraux, 1999), and in an ics1 ics2 double mutant (Supplemental Figure 3), which is not only blocked in induced SA production but also exhibits strongly diminished basal SA levels (Garcion et al., 2008). Moreover, the hypersensitive response-inducing Psm avrRpm1 strain triggered partial SAR activation in sid2 in a similar manner than the compatible Psm strain (Figure 2C). These results show that a moderate SAR response in plants can be triggered independently of inducible SA biosynthesis but not independently of Pip biosynthesis. However, SA accumulation upon pathogen encounter is required to realize a full SAR response. The residual, SA-independent SAR effect is absent in the sid2 ald1 double mutant, indicating that activation of this pathway and of the predominant, SA-dependent SAR pathway do both require Pip (Figure 2B). In sum, these data strongly suggest that Pip is a central regulatory metabolite for SAR that controls both SA-dependent and -independent SAR activation pathways.
Transcriptional Reprogramming in Distal Leaves of SAR-Activated Plants: Increased Readiness for Pathogen Defense Coupled with Decreased Photosynthesis, General Metabolism, and Growth
SAR establishment following 1° leaf inoculation involves increased expression of a whole battery of defense-related genes in the distal leaves (Ward et al., 1991; Gruner et al., 2013). To assess the contribution of Pip and SA to SAR on the transcriptional level, the transcriptional SAR response of Col-0 was characterized at the whole-genome level and compared with the responses in sid2 and ald1. Compatible Psm or hypersensitive response-inducing Psm avrRpm1 trigger SAR in Col-0 plants between days 1 and 2, and the full resistance response is apparent at 2 d after inoculation (Mishina et al., 2008; Návarová et al., 2012). We thus determined the transcriptional changes that occur in 2° leaves 2 d after Psm treatment of 1° leaves compared with 1° mock treatment by Illumina TruSeq RNA sequencing analyses. A first experimental set consisted of three independent, replicate SAR experiments (experiments 1 to 3) with Col-0 and sid2 plants, and a second analogous set involved Col-0 and ald1 (experiments 4 to 6). Principle component analysis (PCA) identified Psm treatment variation as the major variable between the samples, accounting for 58.0% of the variation. The second variable was experiment variation between the first and second experimental sets and accounted for 15.7% of the variation, indicating that experimental variation was small compared with treatment variation. To be conservative, the six Col-0 replicates were combined for analysis. The PCA also showed that in Col-0, the distance between mock and Psm treatment samples was widest. The distance between sid2 samples was small and the distance between ald1 samples was virtually nonexistent (Supplemental Figure 4).
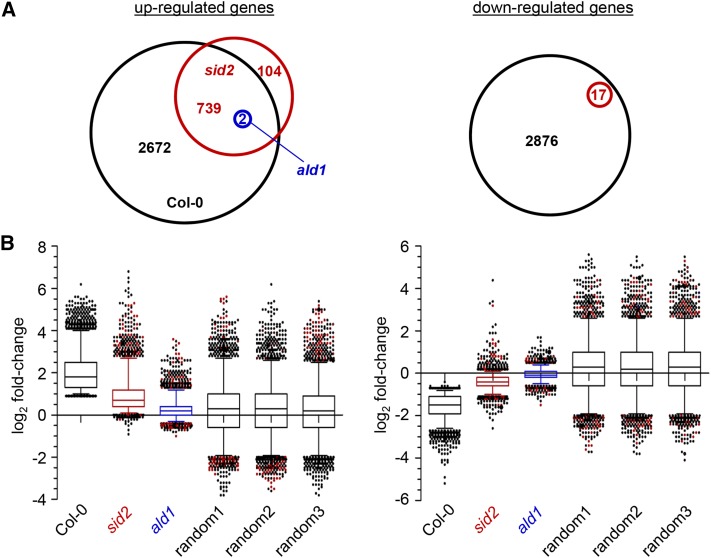
A threshold cutoff of expression values (reads per million) was defined that excluded genes with very low expression levels from the RNA-seq data set, i.e., genes that did not reach expression values of at least 5 in any of the mock or Psm samples. This reduced the set of 28,496 totally RNA-seq covered genes to a set of 15,239 expressed genes. To determine statistically significant changes in gene expression of Psm versus mock treatments for Col-0, sid2, and ald1, a false discovery rate (FDR) of 0.01 was assumed (Benjamini and Hochberg, 1995). Among the 15,239 investigated genes, 3413 were upregulated (designated as SAR+ genes) and 2893 were downregulated (SAR- genes) in a statistically significant manner in the Col-0 wild type (Figure 3A; Supplemental Data Set 1). To quantitatively assess the transcriptional changes between the SAR-induced and mock control state, we calculated log2-transformed ratios of the mean of expression values for Psm and mock samples (P/M-fold changes). The log2 P/M-fold changes for Col-0 averaged over all the SAR+ and SAR− genes were 2.05 and −1.57, respectively (Figure 3B).
Figure 3.
Transcriptional SAR Response in Distal Leaves of Plants Inoculated in Primary Leaves with Psm (OD600 = 0.005) at 48 HAI.
Six independent SAR assays for Col-0 and three independent SAR experiments for both sid2 and ald1 were performed. Gene expression was analyzed by RNA-seq analyses of the resulting replicate samples for Psm and mock treatments at the whole-genome level.
(A) Venn diagram depicting numbers of differentially regulated genes between Psm and mock treatments of Col-0 (black), sid2 (red), and ald1 (blue) (FDR < 0.01). Overlap of genes is indicated. Left: significantly upregulated genes (the Col-0 genes correspond to the SAR+ genes). Right: significantly downregulated genes (the Col-0 genes correspond to the SAR− genes). Note that only two genes are differentially regulated in ald1.
(B) Distribution of P/M-fold changes of SAR genes in Col-0, sid2, and ald1. Box plots depict log2-transformed P/M-fold changes. The distribution of log2 P/M-fold changes for three sets of randomly selected genes (left, 3413 genes; right, 2893 genes) is included (random a, b, and c). Left, SAR+ genes; right, SAR− genes.
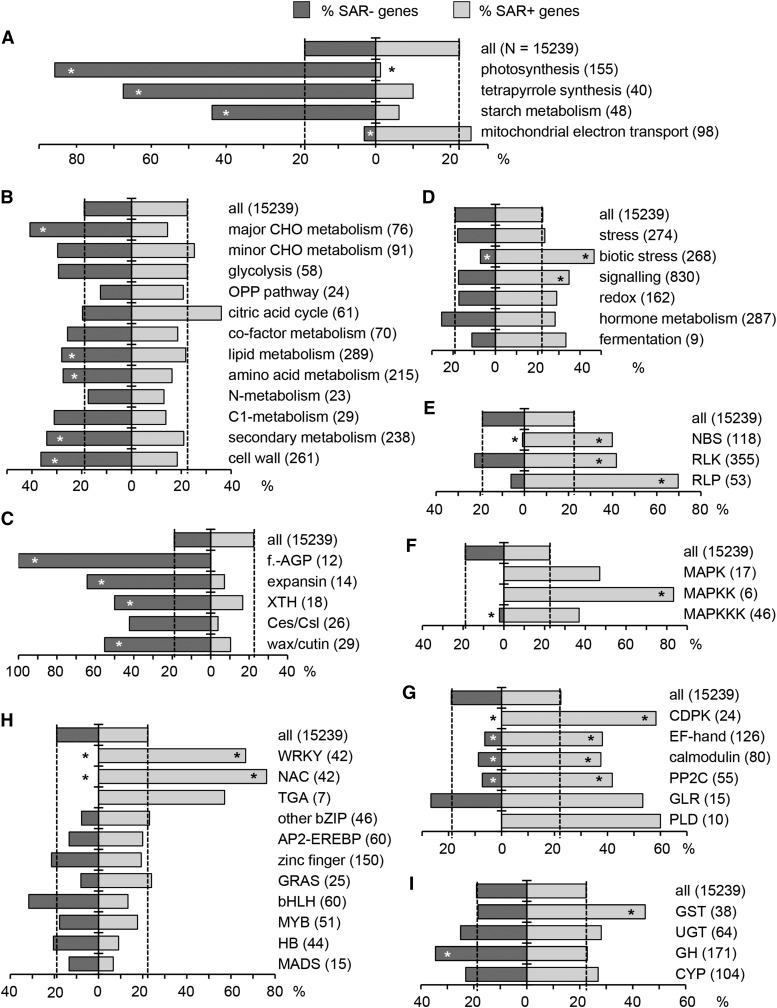
To identify and illustrate physiological and metabolic processes altered in Col-0 plants upon SAR establishment, we tested whether the up- and downregulated SAR genes are enriched in MapMan bins or particular Arabidopsis gene families (Thimm et al., 2004; http://www.arabidopsis.org/). Strikingly, 86% of the investigated genes annotated for an involvement in photosynthesis were significantly downregulated upon SAR induction in Col-0 (Figure 4A). Similarly, genes belonging to the MapMan categories photosynthetic light reactions, Calvin cycle, photorespiration, and tetrapyrrole biosynthesis were predominantly downregulated (Figure 4A, Supplemental Figures 5 to 7). Moreover, genes involved in starch metabolism and genes of the category major CHO metabolism were strongly overrepresented in the SAR− gene group (Figures 4A and 4B). To a lesser extent, this was also valid for genes of the categories lipid metabolism, amino acid metabolism, and secondary metabolism (Figure 4B). In addition, genes of the MapMan category cell wall were enriched among SAR− genes (Figure 4B). A closer look at the family level revealed that many genes coding for proteins involved in cell wall modification and growth (fasciclin-like arabinogalactan proteins, expansins, and xyloglucan endotransglucosylase/hydrolases) as well as genes associated with wax and cutin biosynthesis were strongly downregulated during SAR (Figure 4C). This suggests that in the distal leaves of SAR-activated plants, photosynthesis, several primary and secondary metabolic pathways, and growth processes are reduced compared with respective leaves of control plants. Gene Ontology (GO) term enrichment analysis confirmed the reduction in growth-related processes (Supplemental Data Set 2).
Figure 4.
Percentage of SAR+ and SAR− Genes in Defined Gene Groups Representing MapMan Metabolic Pathways, Functional Categories, or Arabidopsis Gene Families (http://www.arabidopsis.org).
Dashed vertical lines illustrate the percentage of SAR+ and SAR− genes in the whole, RNA-seq-covered transcriptome (28,496 genes) after threshold cutoff (15,239 genes). The number of genes in each category is given in parentheses. Asterisks on the bars indicate significant enrichment or depletion of gene categories in SAR+ (right) and SAR− (left) genes (Fisher’s exact test, P < 0.01).
(A), (B), and (D) MapMan metabolic pathways and functional categories.
(C) Gene families involved in cell wall remodeling and wax/cutin biosynthesis. f.-AGP, fasciclin-like arabinogalactan proteins; XTH, xyloglucan endotrans-glucosylase/hydrolases; Ces/Csl, cellulose synthase/cellulose synthase-like.
(E) Gene families involved in the perception of microbial structures and early defense signal transduction. NBS, nucleotide binding site-containing resistance proteins; RLP, receptor-like proteins.
(F) MAPK cascade members. MAPKK, MAPK kinase; MAPKKK, MAPK kinase kinase.
(G) Other gene categories involved in defense signaling. CDPK, calcium-dependent protein kinases; EF-hand, EF-hand-containing proteins; calmodulin, calmodulin binding proteins; GLR, glutamate receptor-like family; PLD, phospholipase D family.
(H) Main transcription factor families. WRKY, WRKY domain family; NAC, NAM-ATAF1,2-CUC2 transcription factors; TGA, TGACG motif binding factor; bZIP, basic leucine zipper; AP2-EREBP, APETALA2 and ethylene-responsive element binding proteins; zinc finger, zinc finger superfamily; GRAS, GRAS family; bHLH, basic helix-loop-helix; MYB, MYB family; HB, homeobox-leucine zipper; MADS, MADS box.
(I) Genes for different enzyme classes. GST, glutathione S-transferases; UGT, UDP-dependent glycosyltransferases; GH, glycosyl hydrolases; CYP, cytochrome P450 superfamily.
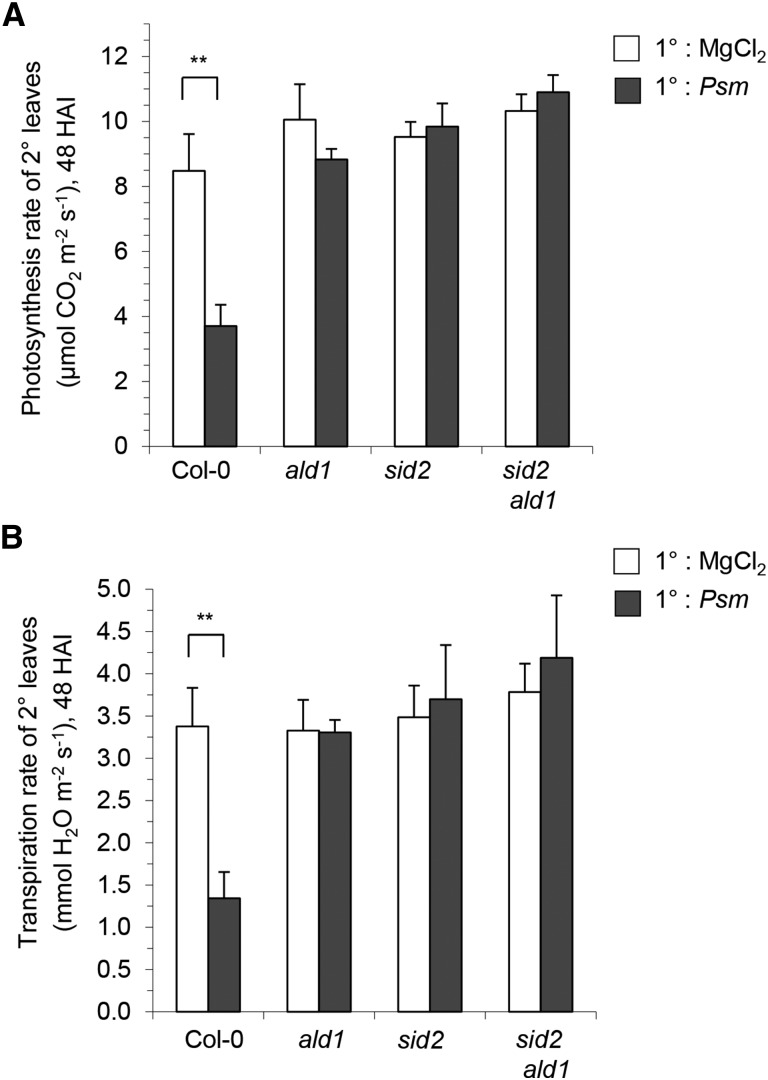
The massive downregulation of photosynthesis-associated genes upon SAR induction in Col-0 prompted us to comparatively investigate the photosynthetic rates of 2° leaves of plants infiltrated in 1° leaves 2 d earlier with Psm or mock solution. We measured the CO2 uptake of individual leaves using infrared gas analysis (IRGA) to determine the maximum rates of photosynthetic carbon assimilation (von Caemmerer and Farquhar, 1981). Upon SAR induction in Col-0, the rate of CO2 uptake of 2° leaves significantly dropped from ∼8 to 4 µmol m−2 s−1, indicating a marked decrease in photosynthetic rates (Figure 5A). Our data thus reveal an attenuation of photosynthesis in the noninoculated distal leaves of SAR-induced wild-type plants both at the transcriptional and the physiological level. Infrared gas analysis also showed decreased water loss from 2° leaves of SAR-induced Col-0 plants, suggesting a significant decline of leaf transpiration when SAR is established (Figure 5B). Decreased stomatal apertures may thus account for the attenuation of photosynthesis in SAR-induced plants.
Figure 5.
Photosynthesis and Transpiration Rates in 2° Leaves of 1° Leaf-Inoculated Col-0, ald1, sid2, and sid2 ald1 Plants.
(A) CO2 uptake rates as a measure of photosynthetic capacity at 48 h after Psm inoculation or MgCl2 infiltration measured in distal, untreated leaves of Col-0, sid2, ald1, and sid2 ald1 plants, as determined by IRGA. Data represent the mean ± sd of four biological replicates (CO2 uptake rate of four distal leaves from different plants).
(B) Rates of transpiration (water loss) in distal leaves, determined as described above by IRGA.
Asterisks denote statistically significant differences between Psm-treated and mock control plants (**P < 0.01; two-tailed t test).
Analyses of the transcriptional SAR response also revealed that the categories biotic stress and signaling were overrepresented among the SAR+ genes (Figure 4D). GO term enrichment analysis of the SAR+ genes specified the biotic stress and signaling categories by identifying regulation of the hypersensitive response, SAR, and SA signaling and their respective parent terms as enriched processes. In addition, N-terminal protein myristoylation, Golgi-based protein targeting to membranes, and the endoplasmic reticulum unfolded protein response as well as their respective parent terms were enriched (Supplemental Data Set 3). Moreover, gene families typically involved in the perception of pathogen-derived elicitors, i.e., genes coding for nucleotide binding site (NBS)-containing resistance proteins (Tan et al., 2007), receptor-like protein kinases (RLKs; Shiu and Bleecker, 2001), and receptor-like proteins (Fritz-Laylin et al., 2005) were markedly enriched in the SAR+ gene group (Figure 4E). Notably, a preferential accumulation of members representing specific subfamilies of the large RLK family in the SAR+ group was apparent. For example, ∼70% of cysteine-rich protein kinases in the investigated gene set were consistently upregulated upon SAR induction (Supplemental Figure 8A). In addition, defense signaling components such as mitogen-activated protein kinase kinases, calcium-dependent protein kinases, EF-hand containing proteins, and calmodulin binding proteins were overrepresented among the SAR+ genes (Figures 4F and 4G). Among transcription factor families, a strong overrepresentation of WRKY- and NAC-type transcription factor genes was observed in the SAR+ gene group, whereas genes for transcription factor types such as MYB, bHLH, or bZIP were not enriched (Figure 4H). Finally, compared with other major types of enzyme classes, a prominent overrepresentation of glutathione S-transferases among the SAR+ genes was discernable (Figure 4I). Other, less prominent gene classes that were strongly overrepresented in the SAR+ group involved senescence-associated genes as well as genes coding for stomatin/prohibitin/flotillin/HflK/C (SPFH) domain-containing, FAD berberine-type, VQ motif-containing proteins, and plant U-box proteins (Supplemental Figure 8B). By contrast, the general expression patterns of genes from MapMan categories such as development, cell, transport, monolignol biosynthesis, or class III peroxidase genes were hardly or not at all changed upon SAR induction (Supplemental Figure 8C).
Overall, these characteristics suggest that SAR-activated plants prepare themselves for future pathogen attack by upregulating genes involved at different stages of defense signaling, such as elicitor perception, signal transduction, and transcriptional gene activation, and by downregulating photosynthesis and growth-associated processes.
The Transcriptional SAR Response Involves a Subset of Genes Whose Systemic Expression Is Partially SA Independent
The transcriptional SAR response in the mutants was compared qualitatively and quantitatively to the wild-type response (Figure 3). According to the statistical RNA-seq analysis, the transcript levels of 2672 out of the 3413 genes that were systemically upregulated in the Col-0 wild type following SAR induction were not elevated in sid2 in a statistically significant manner (Figure 3A). Therefore, a predominant part of the transcriptional SAR response is SA dependent. We created a list of all the SAR+ genes from this group and sorted them according to their mean P/M-fold change in sid2 in ascending order. Table 1 depicts the top 15 SAR+ genes from this list. Among the tightly SA-regulated SAR+ genes are PATHOGENESIS-RELATED GENE1 (PR1), a classical marker gene for an activated SA signaling pathway and SAR (Sticher et al., 1997), ACIREDUCTONE DIOXYGENASE3 (ARD3), and GLUTAREDOXIN13 (GRXS13).
Table 1. SAR+ Genes Tightly Regulated by SA (SA-Dependent SAR+ Genes).
| AGI Code |
Name |
Gene Name/Description |
Mean Expression Value |
Fold Change (Log2) |
P Value Log2 (P/M) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Col-0 M | Col-0 P | sid2 M | sid2 P | Col-0 P/M | sid2 P/M | Col-0 versus sid2 | |||
| At2g26400 | ARD3 | ACIREDUCTONE DIOXYGENASE3 | 17.5 | 1395.8 | 2.2 | 2.7 | 6.2* | 0.2 | 0.0115# |
| At2g14620 | XTH10 | Xyloglucan endotransglucosylase/hydrolase 10 | 3.1 | 156.4 | 0.9 | 1.6 | 5.3* | 0.4 | 0.0125# |
| At2g14610 | PR1 | PATHOGENESIS-RELATED PROTEIN1 | 98.7 | 3801.4 | 6.0 | 10.3 | 5.3* | 0.7 | 0.0243# |
| At3g22910 | ACA13 | Putative calcium-transporting ATPase 13 | 5.6 | 247.0 | 1.6 | 3.1 | 5.2* | 0.6 | 0.0160# |
| At3g28510 | – | AAA-type ATPase family protein | 11.0 | 411.5 | 1.8 | 2.7 | 5.1* | 0.4 | 0.0271# |
| At3g13950 | – | Unknown protein | 5.4 | 173.3 | 5.3 | 18.6 | 4.8* | 1.6 | 0.0357# |
| At4g10860 | – | Unknown protein | 1.6 | 57.2 | 0.1 | 0.3 | 4.5* | 0.2 | 0.0634 |
| At3g53150 | UGT73D1 | UDP-GLUCOSYLTRANSFERASE 73D1 | 1.4 | 49.9 | 0.1 | 0.6 | 4.4* | 0.6 | 0.1793 |
| At4g35180 | LHT7 | LYS/HIS TRANSPORTER7 | 4.2 | 105.9 | 0.7 | 1.9 | 4.4* | 0.7 | 0.0959 |
| At1g03850 | GRXS13 | GLUTAREDOXIN13 | 7.8 | 175.8 | 2.7 | 5.2 | 4.4* | 0.7 | 0.0138# |
| At4g01870 | – | TolB protein-related | 8.6 | 196.5 | 3.5 | 17.2 | 4.4* | 2.0 | 0.0716 |
| At1g65610 | GH9A2 | GLYCOSYL HYDROLASE 9A2 | 1.6 | 51.6 | 0.6 | 2.2 | 4.4* | 1.0 | 0.0274# |
| At3g61190 | BAP1 | BON ASSOCIATION PROTEIN1 | 2.1 | 58.6 | 1.2 | 5.8 | 4.2* | 1.6 | 0.1060 |
| At4g37530 | PER51 | Peroxidase superfamily protein | 12.4 | 241.7 | 8.0 | 20.6 | 4.2* | 1.3 | 0.0132# |
| At5g18270 | ANAC087 | NAC DOMAIN CONTAINING PROTEIN87 | 0.8 | 31.3 | 0.3 | 1.3 | 4.2* | 0.8 | 0.0191# |
RNA-seq analyses identified 2672 SAR+ genes with significant Psm-induced upregulation in the Col-0 wild type and no significant induction in sid2. RNA samples originate from distal leaves of Psm (P)-inoculated and mock (M)-treated Col-0 and sid2 plants at 48 HAI. Mean log2-transformed P/M ratios (fold changes) are depicted, and asterisks indicate significant changes between Psm and mock treatments (FDR < 0.01). The 2672 genes are listed in ascending order according to their sid2 P/M ratios. The top 15 SAR+ genes from this category (i.e., those with highest P/M-fold changes in Col-0) are shown. P values for differences in log2 fold changes in Col-0 and sid2, determined using a linear model framework [lm(log2(rpm) ∼ genotype*treatment)], are given. Number signs indicate significant differences (P < 0.05).
The quantitative RNA-seq assessment indicated that the median log2 P/M-fold change over all the SAR+ genes for sid2 accounted for 0.95 (Figure 3B). Although lower than the respective Col-0 value of 2.05, this value was markedly higher than the values calculated from different sets of randomly chosen genes from the total Arabidopsis genome (0.23 to 0.28). Therefore, sid2 exhibits an attenuated but still detectable transcriptional response to Psm in tissue distal from inoculation. Qualitatively, 845 genes were significantly upregulated in 2° leaves of sid2 upon 1° leaf inoculation, and 741 of them were induced in a significant manner also in the Col-0 wild type (Figure 3A). We sorted the SAR+ genes within this group according to their mean P/M-fold change values in sid2 in descending order. Table 2 lists 15 prominently expressed genes with strong upregulation in sid2.
Table 2. Partially SA-Independent SAR+ Genes.
| Pos. | AGI Code | Name | Gene Name/Description | Mean Expression Value | Fold Change (Log2) | P Value Log2 (P/M) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 M | Col-0 P | sid2 M | sid2 P | Col-0 P/M | sid2 P/M | Col-0 versus sid2 | ||||
| 1 | At2g24850 | TAT3 | TYROSINE AMINOTRANSFERASE3 | 55.6 | 2089.5 | 11.1 | 1302.5 | 5.2* | 6.8* | 0.8032 |
| 3 | At2g13810 | ALD1 | AGD2-LIKE DEFENSE RESPONSE PROTEIN1 | 12.1 | 378.6 | 0.8 | 140.2 | 4.9* | 6.3* | 0.1956 |
| 4 | At1g19250 | FMO1 | FLAVIN-DEPENDENT MONOOXYGENASE1 | 3.9 | 173.2 | 0.4 | 84.3 | 5.2* | 6.0* | 0.6120 |
| 5 | At2g43570 | CHI | Putative chitinase | 45.7 | 1523.2 | 3.7 | 286.4 | 5.2* | 5.9* | 0.7260 |
| 6 | At2g29460 | GST22 | GLUTATHIONE S-TRANSFERASE22 | 5.4 | 274.4 | 1.1 | 112.2 | 5.6* | 5.8* | 0.9231 |
| 7 | At3g09940 | MDAR3 | MONODEHYDROASCORBATE REDUCTASE3 | 4.9 | 126.6 | 0.8 | 93.8 | 4.8* | 5.7* | 0.8622 |
| 9 | At1g02930 | GSTF6 | GLUTATHIONE S-TRANSFERASE6 | 115.7 | 2530.5 | 14.7 | 596.6 | 4.1* | 5.3* | 0.4248 |
| 10 | At1g33960 | AIG1 | AVRRPT2-INDUCED GENE1 | 54.1 | 2285.9 | 7.6 | 325.5 | 5.4* | 5.3* | 0.7475 |
| 11 | At3g57260 | PR2 | PATHOGENESIS-RELATED PROTEIN2 | 179.9 | 3193.3 | 15.9 | 634.4 | 4.3* | 5.2* | 0.8218 |
| 12 | At3g22600 | LTPG5 | GPI-ANCHORED LIPID TRANSFER PROTEIN5 | 15.6 | 775.0 | 1.2 | 81.1 | 5.8* | 5.2* | 0.8393 |
| 13 | At3g26830 | PAD3 | PHYTOALEXIN DEFICIENT3 | 12.1 | 421.8 | 0.9 | 67.5 | 5.3* | 5.2* | 0.8512 |
| 14 | At2g38240 | DOXC46 | 2-Oxoglutarate-dependent dioxygenase superfamily | 0.2 | 20.0 | 0.2 | 41.4 | 4.8* | 5.1* | 0.7480 |
| 15 | At2g29350 | SAG13 | SENESCENCE-ASSOCIATED GENE13 | 27.4 | 1135.6 | 4.0 | 165.1 | 5.8* | 5.0* | 0.5042 |
| 16 | At1g57630 | – | Toll-Interleukin-Resistance domain family protein | 6.2 | 255.9 | 0.9 | 55.9 | 5.3* | 4.9* | 0.9346 |
| 17 | At2g04450 | NUDT6 | Nudix hydrolase homolog 6 | 35.7 | 623.0 | 2.7 | 102.0 | 4.1* | 4.8* | 0.9346 |
| 120 | At1g75040 | PR5 | PATHOGENESIS-RELATED PROTEIN5 | 274.8 | 5349.1 | 65.7 | 539.2 | 4.3* | 3.0* | 0.1159 |
| 350 | At4g28390 | AAC3 | ADP/ATP CARRIER3 | 15.7 | 165.0 | 5.9 | 25.6 | 3.3* | 2.0* | 0.0473# |
| 510 | At2g17720 | P4H5 | PROLYL 4-HYDROXYLASE5 | 28.2 | 188.0 | 18.5 | 58.3 | 2.7* | 1.6* | 0.0882 |
| 655 | At2g19190 | FRK1 | FLG22-INDUCED RECEPTOR-LIKE KINASE1 | 2.9 | 70.2 | 0.2 | 1.7 | 4.2* | 1.2* | 0.3649 |
SAR+ genes with significant Psm-induced upregulation in distal leaves of both Col-0 and sid2. The subgroup of SAR+ genes (741 genes in total) is listed in descending order according to their sid2 P/M ratios. Asterisks indicate significant changes (FDR < 0.01). The 15 genes with the highest P/M-fold change in sid2 and Col-0 log2 P/M values > 4.0 are depicted. Four selected genes from lower parts of the gene list are also shown. The position of each gene in the full SAR+ gene list is indicated in the first column. Note that the Pip pathway genes ALD1 and FMO1 occur at the top of the list. P values for differences in log2 fold changes in Col-0 and sid2 are given. Number values indicate significant differences (P < 0.05).
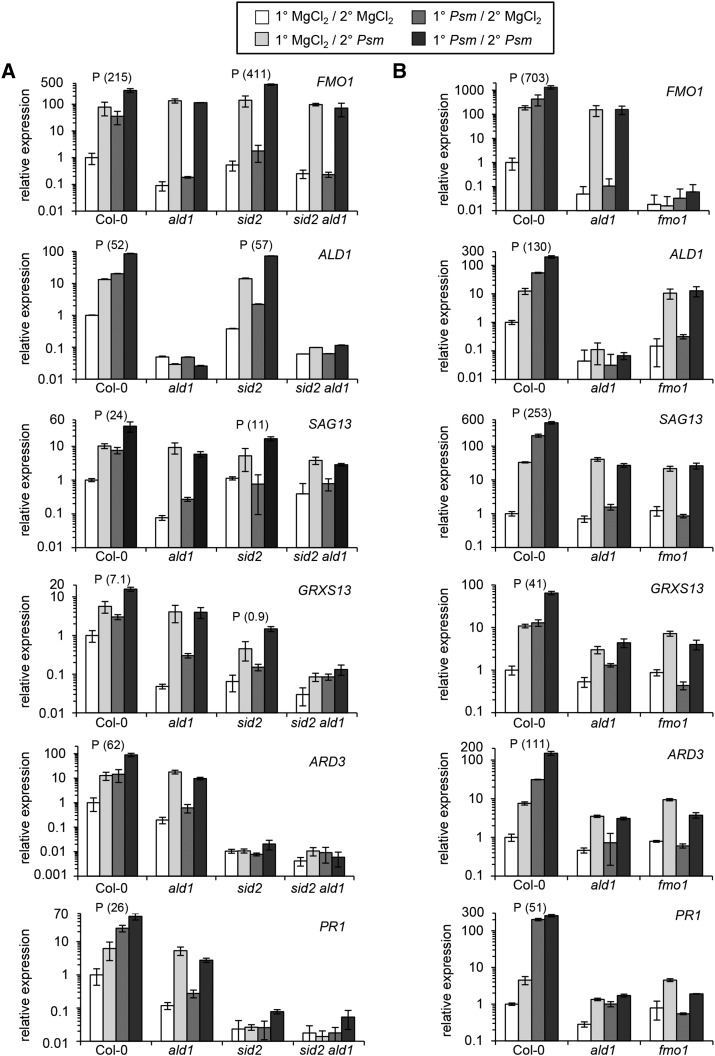
A common feature of these genes is that their P/M-fold changes (their inducibility) in sid2 are similar or even higher than in the wild type, but that their absolute expression in 2° tissue of mock- and pathogen-treated sid2 plants is markedly lower than in Col-0. This indicates that SA amplifies their expression under both inducing and basal conditions (Table 2; Supplemental Data Set 1 and Supplemental Figure 4), and we have therefore designated these genes as partially SA independent. Strikingly, ALD1 and FMO1 rank among the most strongly induced genes in the systemic tissue of sid2, demonstrating that the transcriptional activation of Pip biosynthesis and downstream signaling can be activated during SAR in a manner that is partially SA independent. Other paradigm examples of SAR+ genes that can be strongly upregulated in the absence of elevated SA are TYROSINE AMINOTRANSFERASE3 (TAT3), GLUTATHIONE S-TRANSFERASE22 (GST22), PHYTOALEXIN-DEFICIENT3 (PAD3), and SENESCENCE-ASSOCIATED GENE13 (SAG13) (Table 2). For the top members of the gene list, the high inducibility in sid2 resulted in expression values for Psm-treated sid2 samples that markedly exceeded those of mock-treated Col-0. Genes from the middle or bottom part of the gene list, however, generally showed a lower inducibility in sid2 than in Col-0, and the values from Psm-inoculated sid2 only moderately exceeded the Col-0 mock values (e.g., PR5, AAC3, and P4H5) or even stayed below (e.g., FRK1) (Table 2). Therefore, the absolute expression of the genes from the bottom part of the gene list such as FRK1 still very much depends on SA, although a slight (but statistically significant) upregulation exists in sid2.
Together, these expression analyses indicate that the extent of systemic upregulation for virtually all SAR+ genes is positively regulated by SA. For the majority of genes, this SA-mediated amplification is essential for a significant upregulation (SA-dependent SAR+ genes; Table 1) or a noticeable induction well above basal wild-type levels (bottom part of list of partly SA-independent genes, as exemplified by FRK1 in Table 2). However, the dependency on SA is lower for several hundred SAR+ genes, which therefore exhibit a marked systemic upregulation in sid2 (major part of the 741 partly SA-independent genes, as exemplified by the top 15 genes of Table 2). Notably, the SAR regulatory and pipecolic acid pathway genes ALD1 and FMO1 belong to the SAR+ genes with a pronounced SA-independent component of induction.
An Activated SA Pathway upon SAR Induction Suppresses Systemic Jasmonate Responses
The RNA-seq results show that 104 of the 845 genes systemically induced in sid2 were not upregulated or even significantly downregulated in Col-0 upon SAR activation (Figure 3A). We listed these genes according to their Col-0 mean P/M-fold change values in ascending order, and Table 3 exemplifies 15 of these specifically sid2 upregulated genes. Several jasmonate-responsive genes such as VEGETATIVE STORAGE PROTEIN2 (VSP2) or BENZOIC ACID/SALICYLIC ACID METHYLTRANSFERASE1 (BSMT1) belong to this group. We therefore tested whether this gene category would be actually enriched in jasmonic acid (JA)-responsive genes. A list of JA-responsive genes was taken from microarray data of Goda et al. (2008), studying the response of Arabidopsis to methyl jasmonate treatment. According to this list, the examined group of 15,239 Arabidopsis genes contained 3.2% of JA-responsive genes, and the genes systemically upregulated in Col-0 (SAR+ genes) showed a similar percentage of JA-regulated genes (3.7%; Table 4). By comparison, 12.8% of the genes systemically upregulated in sid2 were JA responsive, and the subgroup thereof containing only genes that were not upregulated in Col-0 showed by far the highest enrichment (48.1% JA-responsive genes). These findings show that a 1° leaf inoculation with Psm triggers a significantly higher expression of JA-responsive genes in 2° leaves of sid2 plants compared with Col-0 plants and indicate that an activated SA pathway in the SAR-induced wild type suppresses pathogen-inducible JA responses in systemic tissue.
Table 3. Genes Induced in Systemic Tissue of sid2 but Not in Col-0.
| Pos. | AGI Code | Name | Gene Name/Description | Mean Expression Value |
Fold Change (Log2) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Col-0 M | Col-0 P | sid2 M | sid2 P | Col-0 P/M | sid2 P/M | ||||
| 1 | At5g24770 | VSP2 | VEGETATIVE STORAGE PROTEIN2 | 2.9 | 7.6 | 10.4 | 1108.4 | 1.1 | 6.6* |
| 2 | At5g24780 | VSP1 | VEGETATIVE STORAGE PROTEIN1 | 0.8 | 1.9 | 1.8 | 260.4 | 0.7 | 6.5* |
| 3 | At4g16590 | CSLA01 | CELLULOSE SYNTHASE-LIKE A01 | 0.8 | 1.2 | 1.6 | 133.2 | 0.3 | 5.7* |
| 4 | At1g76790 | IGMT5 | INDOLE GLUCOSINOLATE O-METHYLTRANSFERASE5 | 0.6 | 0.4 | 1.8 | 116.5 | −0.2 | 5.4* |
| 5 | At4g17470 | – | α/β-Hydrolase superfamily protein | 1.1 | 2.6 | 2.7 | 148.8 | 0.8 | 5.3* |
| 6 | At2g24210 | TPS10 | TERPENE SYNTHASE10 | 0.1 | 0.5 | 0.5 | 54.8 | 0.4 | 5.3 |
| 7 | At3g11480 | BSMT1 | BENZOIC ACID/SALICYLIC ACID METHYLTRANSFERASE1 | 0.1 | 0.6 | 0.1 | 40.9 | 0.6 | 5.2* |
| 8 | At1g51780 | ILL5 | IAA-LEUCINE RESISTANT (ILR)-LIKE5 | 0.0 | 0.4 | 0.1 | 27.8 | 0.5 | 4.8* |
| 9 | At4g13410 | CSLA15 | CELLULOSE SYNTHASE LIKE A15 | 3.8 | 1.0 | 2.6 | 95.5 | −1.3 | 4.7* |
| 10 | At1g24070 | CSLA10 | CELLULOSE SYNTHASE-LIKE A10 | 1.3 | 1.0 | 1.4 | 55.0 | −0.2 | 4.5* |
| 11 | At3g28220 | – | TRAF-like family protein | 26.3 | 6.5 | 31.2 | 686.5 | −1.9* | 4.4* |
| 32 | At3g28290 | – | Sequence similarity to integrins | 2.4 | 0.5 | 1.9 | 28.9 | −1.2* | 3.4* |
| 33 | At3g28300 | – | Sequence similarity to integrins | 1.9 | 0.5 | 2.0 | 29.3 | −0.9* | 3.3* |
| 41 | At1g52000 | – | Mannose binding lectin superfamily | 28.5 | 6.6 | 33.4 | 306.8 | −2.0* | 3.2* |
| 70 | At2g43550 | – | Scorpion toxin-like knottin superfamily | 11.8 | 2.1 | 30.5 | 146.1 | −2.0* | 2.2* |
| 91 | At5g02940 | – | Protein of unknown function | 109.2 | 32.0 | 148.3 | 464.3 | −1.7* | 1.6* |
A total of 104 genes were identified with significant Psm-induced upregulation in distal leaves of sid2 but not Col-0 plants (Figure 3A). The genes were listed in descending order according to their sid2 P/M ratios. The 10 genes with highest P/M ratios in sid2 and all genes significantly downregulated in Col-0 (SAR− genes) from this group are depicted. The position of each gene in the list is indicated. Asterisks indicate significant changes between Psm and mock treatments (FDR < 0.01).
Table 4. Genes Systemically Induced in sid2 Are Strongly Enriched in JA-Responsive Genes.
| Gene Category | No. of Genes | JA-Inducible Genes (%)a | P Valueb |
|---|---|---|---|
| Whole gene setc | 15,239 | 3.2 | – |
| Col-0 up (SAR+) | 3,413 | 3.7 | 0.068 |
| sid2 up (total) | 845 | 12.8 | 9*10−38 |
| sid2 up/Col-0 not upd | 104 | 48.1 | 4*10−44 |
Percentage of JA-responsive genes in the total number of RNA-seq-analyzed genes and in different gene categories (as illustrated in the left Venn diagram of Figure 3A).
Percentage of genes determined to be JA inducible by Goda et al. (2008).
By Fisher’s exact test; indicates significance of enrichment.
After threshold cutoff.
Genes upregulated in sid2 but not in Col-0.
Pipecolic Acid Is a Central Regulator of the Systemic Transcriptional Reprogramming Response Associated with SAR
Col-0 plants responded to the 1° Psm inoculation with the upregulation of 3413 and the downregulation of 2893 genes in 2° leaves. Strikingly, for the Pip-deficient ald1 plants, only two genes, CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE6 (CRK6) and PLANT CADMIUM RESISTANCE1 (PCR1), were systemically upregulated in a statistically significant manner, and not a single gene was significantly downregulated (Figure 3A, Table 5). Moreover, albeit the statistical analyses identified CRK6 and PCR1 as upregulated in ald1, their absolute expression levels after Psm treatment were low and did not even exceed the values of the Col-0 mock control samples (Table 5). Therefore, the strong transcriptional reprogramming observed in 2° leaves of Col-0 plants after 1° leaf inoculation is essentially absent in ald1. This indicates that the accumulation of Pip upon pathogen inoculation is necessary for virtually the entire transcriptional SAR response. This also becomes evident when the MapMan heat maps of central metabolism for Col-0 and ald1 upon (attempted) SAR induction are compared (Supplemental Figures 5 and 7). Moreover, the mean P/M-fold change averaged over all the SAR+ genes for ald1 accounted for a value of 0.28, which was low compared with the Col-0 (2.05) or sid2 (0.95) values and in the range of the values for groups of randomly chosen genes (Figure 3B).
Table 5. The Transcriptional SAR Response Is Virtually Absent in ald1.
| AGI Code |
Name |
Gene Name/Description |
Mean Expression Value |
Fold Change (Log2) |
||||
|---|---|---|---|---|---|---|---|---|
| Col-0 M | Col-0 P | ald1 M | ald1 P | Col-0 P/M | ald1 P/M | |||
| At4g23140 | CRK6 | CYSTEINE-RICH RECEPTOR-LIKE KINASE6 | 52.4 | 636.3 | 5.3 | 26.1 | 3.6* | 2.1* |
| At1g14880 | PCR1 | PLANT CADMIUM RESISTANCE1 | 270.6 | 5673.3 | 7.2 | 29.0 | 4.4* | 1.9* |
The PCA showed that the transcriptome differences between the mock samples of Col-0 and those of ald1 were lower than the differences of mock values between Col-0 and sid2 (Supplemental Figure 4). This might indicate, as deduced before from the bacterial growth data (Figure 2A), that the contribution of Pip to basal resistance against P. syringae is lower than the contribution of SA. In addition, the SAR-associated downregulation of genes was not only blocked in ald1 but also severely compromised in sid2 (Figures 3A and 3B; Supplemental Figures 5 to 7). The RNA-seq results indicate that only 17 genes were significantly downregulated in sid2, whereas 2893 genes were repressed in Col-0 (Figure 3A). Therefore, a lack of induction of SA biosynthesis in plants seems to affect the pathogen-induced systemic gene repression to a broader extent than gene activation. Since the systemic downregulation of photosynthetic genes is one hallmark of the transcriptional SAR response in the wild type (Figure 4A), we expected that the observed induced systemic attenuation of photosynthetic rates in Col-0 would be severely affected in ald1 and sid2. IRGA analyses confirmed this assumption, since 1° leaf inoculation changed neither the photosynthetic rates nor the stomatal conductance in ald1, sid2, or sid2 ald1, indicating that both Pip and SA are required to mediate these responses (Figures 5A and 5B).
Pipecolic Acid Orchestrates SA-Dependent and -Independent Priming Responses upon SAR Activation
We have previously shown that the induction of SAR conditions Arabidopsis for timely and effective defense gene activation, SA biosynthesis, and camalexin accumulation. This state of defense priming becomes apparent upon a challenge infection of previously uninfected 2° leaves. Pip is a critical mediator of this SAR-associated priming response, because Pip-deficient ald1 plants completely lack this phenomenon. Moreover, exogenously applied Pip is sufficient to promote both Col-0 and ald1 plants into a primed, SAR-like state (Návarová et al., 2012). To investigate a possible interplay between Pip and SA in the activation of SAR-associated defense conditioning, we directly compared the abilities of Col-0, ald1, sid2, and sid2 ald1 plants to realize biologically induced defense priming.
For this purpose, the plants were infiltrated with Psm or mock solution in their lower (1°) leaves, and 2 d later, the distal (2°) leaves were challenged with Psm or mock-infiltrated obtaining four combinations: (1°/2°) mock/mock, Psm/mock, mock/Psm, and Psm/Psm (Supplemental Figure 9A). The magnitude of defense in the challenged 2° leaves was determined for the four combinations at 10 h after inoculation (Návarová et al., 2012). We defined a particular defense response as primed if the differences between the responses of SAR-induced plants to 2° Psm and 2° mock treatments (Psm/Psm – Psm/mock), respectively, were significantly larger than the same differences in noninduced plants (mock/Psm – mock/mock) (Supplemental Figure 9B). Moreover, to estimate quantitative differences in the strength of priming between genotypes with activated priming, we calculated the prgain (response gain due to priming) value (see Supplemental Figure 9C for details).
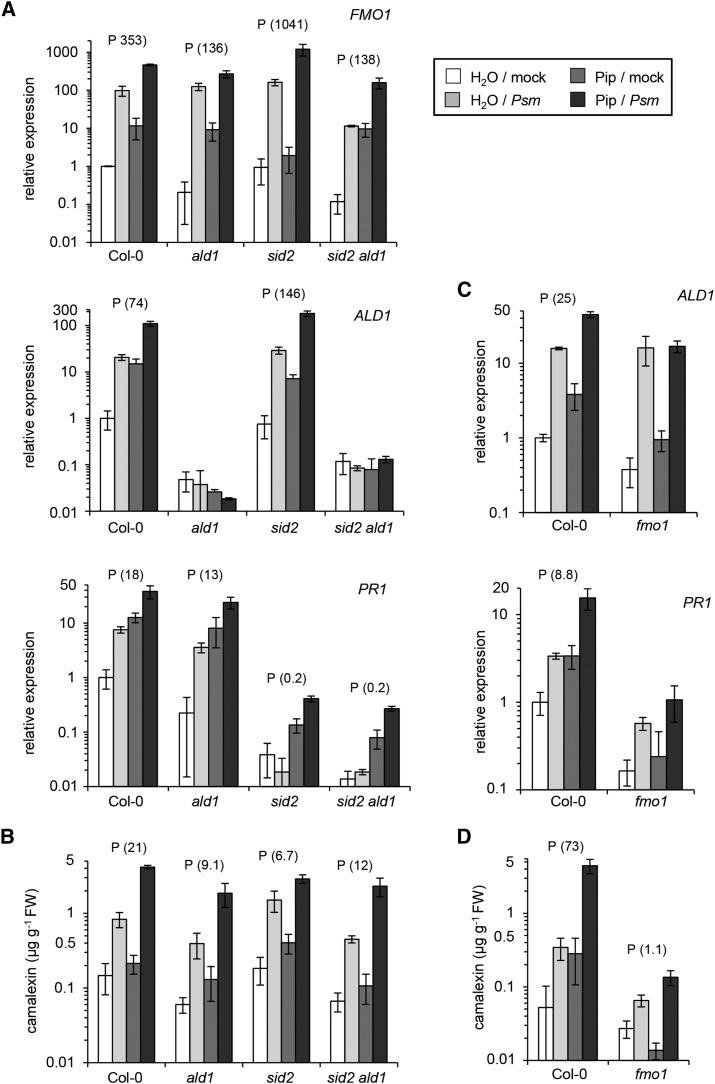
We first monitored the expression of ALD1, FMO1, and SAG13 (partially SA-independent SAR+ genes; Table 2) and of GRXS13, ARD3, and PR1 (SA-dependent SAR+ genes; Table 1) as defense outputs of the SAR priming assay. SAR induction significantly primed Col-0 wild-type plants for enhanced expression of all these genes (Figure 6A; Supplemental Figure 10A). This conditioning of gene expression was completely absent in both ald1 and sid2 ald1 for all the examined genes (Figure 6A; Supplemental Figure 10A), corroborating the previously identified central role for Pip in the activation of SAR-associated defense priming (Návarová et al., 2012). For sid2 plants, by contrast, the outcome of the priming assay depended on the nature of the investigated response. FMO1 and ALD1 expression were primed in sid2 to a higher or similar extent, respectively, than in Col-0. The expression of SAG13 was also significantly primed in sid2, but this response was markedly lower than in Col-0. Moreover, only a weak priming effect was observed for GRXS13 expression, and priming for both ARD3 and PR1 expression was completely abolished in sid2 (Figure 6A; Supplemental Figure 10A). Therefore, SAR induction primes sid2 plants for enhanced expression of three members of the partially SA-independent cluster of SAR+ genes, whereas priming is weak or fully absent with respect to expression of the three SA-dependent SAR+ genes. In addition to gene expression, we measured priming at the metabolite level. As previously shown (Návarová et al., 2012), we found that an activated SAR state primes Col-0 plants for enhanced camalexin and SA biosynthesis upon pathogen inoculation in a Pip-dependent manner (Figure 7A; Supplemental Figure 11A). Priming of camalexin accumulation was also absent in sid2, indicating that both Pip and SA are required for conditioning of camalexin biosynthesis during biological SAR (Figure 7A; Supplemental Figure 11A).
Figure 6.
SAR-Associated Priming of Defense-Related Gene Expression Fully Depends on a Functional Pip/FMO1 Module but Is Only Partially SA Dependent.
(A) SAR priming assays for Col-0, ald1, sid2, and sid2 ald1 plants.
(B) SAR priming assays for Col-0, ald1, and fmo1 plants (independent experiment).
The priming assay consisted of an inductive Psm inoculation or mock (MgCl2) treatment of 1° leaves, followed by a Psm challenge or mock treatment of 2° leaves 48 h later. Gene expression in 2° leaves was assessed 10 h after the second treatment (Supplemental Figure 9A). A particular defense response was defined as primed if the differences between the (1°-Psm/2°-Psm) and the (1°-Psm/2°-MgCl2) values were significantly larger than the differences between the (1°-MgCl2/2°-Psm) and the (1°-MgCl2/2°-MgCl2) values (two-sided Mann-Whitney U test, α = 0.005) (Supplemental Figure 9B). A P above the bars for a particular genotype indicates priming. Expression of three partially SA-independent SAR+ genes (FMO1, ALD1, and SAG13; Table 1) and three SA-dependent SAR+ genes (GRXS13, ARD3, and PR1) were monitored. Transcript levels were assessed by quantitative real-time PCR analysis and are given as means ± sd of three biological replicates. Each biological replicate involves two technical replicates. The transcript levels are expressed relative to the respective Col-0 mock control value. Note that the graphs use a base 10 logarithmic scale for the y axes to ensure recognizability of both high and low values. Graphs with a linear scale for the y axes, which more clearly illustrate differences between challenge-infected 1° mock-treated and 1° Psm-induced plants, are depicted in Supplemental Figure 10. As a measure of the gain of a response due to priming, we calculated the prgain (response gain due to priming) for each genotype with activated priming according to the formula given in Supplemental Figure 9C. prgain values are given in parentheses behind the priming indicator P and allow estimates about quantitative differences of the strength of priming between genotypes. The higher the prgain value, the stronger the priming. The data sets depicted in (A) and (B) are derived from independent experiments.
Figure 7.
SAR-Associated Priming of Camalexin and SA Biosyntheses Requires a Functional Pip/FMO1 Module, and SAR Priming of Camalexin Production Is SA Dependent.
(A) SAR priming assays for Col-0, ald1, sid2, and sid2 ald1 plants. Camalexin levels and total SA levels were determined as defense outputs. Values represent the mean ± sd of three biological replicates from different plants. Each biological replicate consists of six leaves from two plants. A P above the bars for a particular genotype indicates priming in this genotype. The prgain values are given in parentheses. Details of the priming assessments are described in the legends of Figure 6 and Supplemental Figure 9.
(B) SAR priming assays for camalexin and total SA production in Col-0, ald1, and fmo1 plants, as described in (A).
Note that the graphs use a logarithmic scale for the y axes. The same graphs with linear scaling are depicted in Supplemental Figure 11. The data sets depicted in (A) and (B) originate from independent experiments.
Together, these results indicate that Pip is able to prime plants for enhanced activation of a subset of defense responses such as ALD1 and FMO1 expression during biologically induced SAR independently of SA. However, for the priming of a second category of defense responses that involve ARD3 expression, PR1 expression, and camalexin accumulation, both Pip and SA are necessary. Notably, our data suggest overlapping regulatory principles of SAR activation and the realization of defense priming in challenge-infected plants: Priming of expression of partially SA-independent SAR+ genes can be achieved independently of SA, whereas priming of SA-dependent SAR+ genes requires SA (Tables 1 and 2, Figure 6A; Supplemental Figure 10A).
Arabidopsis plants exogenously supplied with 10 µmol Pip over the root system accumulate Pip in leaves to similar levels as 2° leaves of plants exhibiting P. syringae-induced SAR (Návarová et al., 2012). Pip applied in this way is sufficient to enhance resistance to P. syringae and induces defense priming to a similar extent as biological SAR (Návarová et al., 2012). To examine the role of SA in Pip-induced defense priming, we fed Col-0, sid2, ald1, and sid2 ald1 plants with 10 mL of 1 mM (=10 µmol) Pip, challenged leaves with Psm 1 d later and compared their defense responses 10 h after the challenge infection with those of unfed plants (supplied with 10 mL water). Similar to the SAR priming assay, we distinguished four treatments (soil/leaf): a control situation (water/mock), Pip treatment alone (Pip/mock), pathogen challenge alone (water/Psm), and Pip treatment with subsequent pathogen challenge (Pip/Psm). We defined defense priming by analogous criteria as for the SAR priming assay (Pip/Psm – Pip/mock > water/Psm – water/mock; Supplemental Figure 9). Exogenously supplied Pip markedly intensified the Psm-triggered expression of FMO1 and ALD1 in Col-0, and this priming response was even more pronounced in sid2 (Figure 8A; Supplemental Figure 12A). Pip feeding also primed Col-0 plants for enhanced PR1 expression during a Psm challenge infection, and here, the priming effect was almost fully suppressed in sid2 (Figure 8A; Supplemental Figure 12A). Pip-induced priming for camalexin production was also stronger in Col-0 than in sid2 (Figure 8B; Supplemental Figure 12B). These results overlap with those of the SAR priming assay and substantiate our conclusion that Pip regulates priming for certain responses in an SA-independent manner, whereas it requires SA to mediate priming of other responses. The responses triggered by exogenous Pip were generally somewhat lower in ald1 or sid2 ald1 than in Col-0 plants, indicating that endogenous Pip also amplifies the priming effects in these assays (Figures 8A and 8B; Supplemental Figures 12A and 12B).
Figure 8.
Exogenous Pip Confers Defense Priming in a FMO1-Dependent and Partially SA-Independent Manner.
(A) Pip-induced priming of gene expression (FMO1, ALD1, and PR1) in Col-0, ald1, sid2, and sid2 ald1 plants, as determined by qPCR analysis. Plants were supplied with 10 mL of 1 mM Pip (≡ dose of 10 µmol) or with 10 mL of water (control treatment) via the root system and leaves challenge-inoculated with Psm or mock-infiltrated 1 d later. Defense responses in leaves were determined 10 h after the challenge treatment. Values represent the mean ± sd of three biological replicates from different plants. Each biological replicate consists of two leaves from one plant and involves two technical replicates. A P above the bars for a particular genotype indicates defense priming in this genotype, as assessed in analogy to SAR priming. The prgain values are given in parentheses (see legend to Figure 6 and Supplemental Figure 9).
(B) Pip-induced priming for camalexin production in Col-0, ald1, sid2, and sid2 ald1 plants. Values represent the mean ± sd of three biological replicates from different plants. Each biological replicate consists of six leaves from two plants.
(C) Pip-induced priming assays in Col-0 and fmo1 plants, monitoring ALD1 and PR1 expression. Sampling as outlined in (A). The data sets depicted in (A) and (C) originate from independent experiments.
(D) Pip-induced priming assays in Col-0 and fmo1 plants, monitoring camalexin accumulation. Sampling is outlined in (B). The data sets depicted in (B) and (D) originate from independent experiments.
Note that the graphs use a logarithmic scale for the y axes. The same graphs with linear scaling are depicted in Supplemental Figure 12.
Pipecolic Acid Mediates SAR-Associated Defense Priming via FMO1
FMO1 is an essential component of biologically induced SAR (Mishina and Zeier, 2006). The finding that fmo1 mutant plants are unable to increase resistance upon Pip treatment indicates that FMO1 acts downstream of Pip in SAR signal transduction (Návarová et al., 2012). To investigate the role of FMO1 in defense conditioning, we subjected fmo1 plants to the priming assays described above. Like ald1, fmo1 plants were unable to intensify expression of partially SA-dependent or SA-independent SAR+ genes (Figure 6B; Supplemental Figure 10B), and to potentiate camalexin and SA production in challenge-infected 2° leaves upon a previous 1° pathogen inoculation (Figure 7B; Supplemental Figure 11B). In contrast to ald1, however, fmo1 was not capable of activating defense priming upon Pip feeding, because challenge-infected leaves of Pip-supplied plants were not or only very faintly able to intensify ALD1 expression, PR1 expression (Figure 8C; Supplemental Figure 12C), camalexin accumulation (Figure 8D; Supplemental Figure 12D), and SA biosynthesis (Supplemental Figure 13). This indicates that FMO1 is a critical mediator of Pip-activated conditioning events that determine SAR-associated defense priming.
Pip and SA Act Synergistically and Independently from Each Other to Induce PR Gene Expression and Disease Resistance
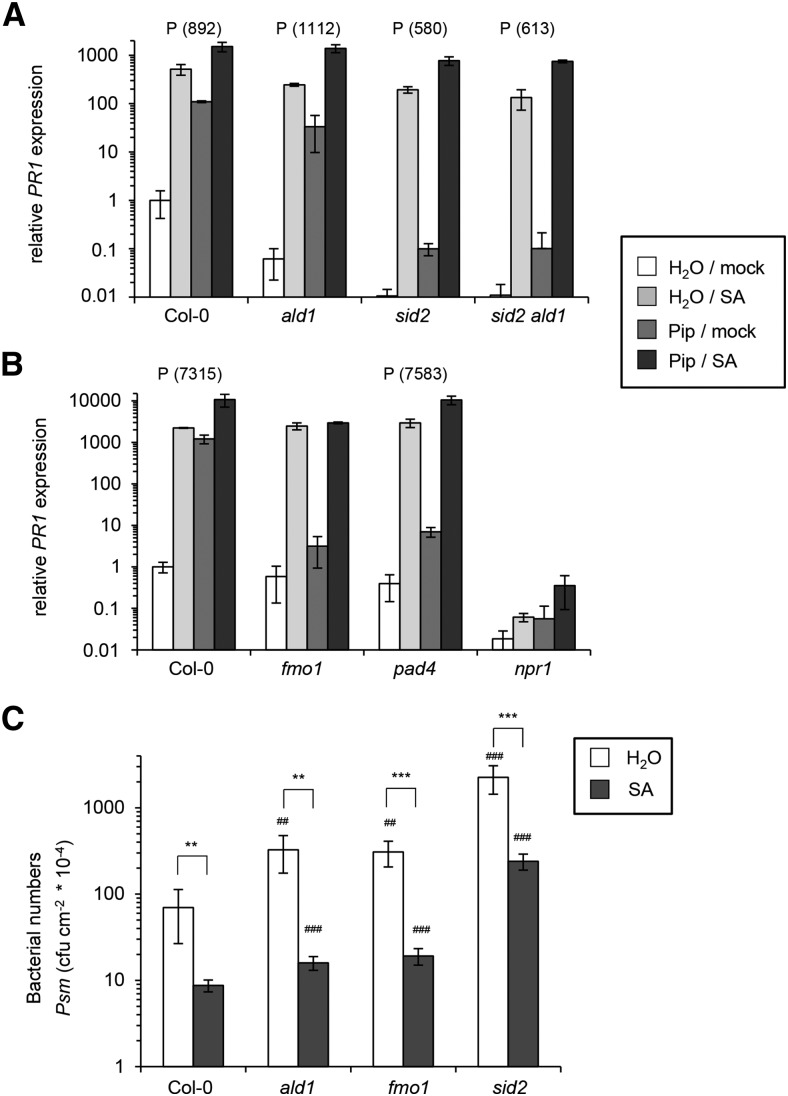
Exogenous SA is sufficient to induce expression of a set of defense-related genes and to confer enhanced plant resistance to different hemibiotrophic and biotrophic pathogens (Delaney et al., 1995; Sticher et al., 1997; Thibaud-Nissen et al., 2006). To examine a possible interplay of Pip and SA in the regulation of plant immune responses, we watered plants with 10 µmol Pip via the root system, subsequently infiltrated 0.5 mM SA into leaves, and determined the transcript levels of the classical SA-inducible gene PR1 4 h after SA treatment. Single Pip and SA applications as well as a control treatment were included, so that, as for the priming assays described above, four cases could be distinguished and priming assessed in an analogous manner (Pip/SA – Pip/mock > water/SA – water/mock; Supplemental Figure 9). SA alone induced strong expression of PR1 in Col-0, ald1, sid2, and sid2 ald1 plants, indicating that elevated SA levels are sufficient to trigger PR1 expression in the absence of Pip. However, this response was markedly fortified in all four genotypes when plants had been pretreated with Pip, indicating synergism between SA and Pip in the induction of PR1 (Figure 9A; Supplemental Figure 14A). Furthermore, the response to SA alone was higher in the Col-0 wild type than in the other lines, suggesting that the capacity of endogenously synthesizing Pip or SA positively affects the induction of PR1 by exogenous SA. Pip treatment alone caused increased expression of PR1 as well. This induction was almost absent in sid2 and in sid2 ald1, indicating that Pip-induced PR1 expression depends on an intact SA biosynthetic pathway (Figure 9A; Supplemental Figure 14A).
Figure 9.
Pip Amplifies SA-Induced PR1 Expression in a FMO1-Dependent Manner and Exogenous SA Enhances Basal Resistance in ald1, fmo1, and sid2.
(A) and (B) Plants were pretreated with Pip (or water) as described in the legend to Figure 8, and 1 d later 0.5 mM SA (SA) or water (mock) was infiltrated into leaves. PR1 expression in leaves was monitored 4 h after the SA (mock) treatment by qPCR analysis. Values represent the mean ± sd of three biological replicates from different plants. Each biological replicate consists of two leaves from one plant and involves two technical replicates. PR1 transcript levels are expressed relative to the water/mock value of Col-0. The data sets depicted in (A) and (B) originate from independent experiments. A P above the bars for a particular genotype indicates priming of SA responses in this genotype, as assessed in analogy to SAR priming. The prgain values are given in parentheses (see legend to Figure 6 and Supplemental Figure 9). Note that the graphs use a logarithmic scale for the y axes. The same graphs with linear scaling are depicted in Supplemental Figure 14.
(A) Col-0, ald1, sid2, and sid2ald1.
(B) Col-0, fmo1, pad4, and npr1.
(C) Basal resistance to Psm infection of Col-0, ald1, fmo1, and sid2 plants is enhanced by exogenous SA. Three leaves per plant were preinfiltrated with 0.5 mM SA or water, and 4 h later, the same leaves were challenged with Psm (OD600 = 0.001). Bacterial growth was assayed 3 d later as outlined in the legend to Figure 2A. Asterisks denote statistically significant differences between SA- and water-pretreated plants (**P < 0.01 and ***P < 0.001; two-tailed t test). Number signs above bars of mutant values denote statistically significant differences from the respective Col-0 wild-type value (##P < 0.01 and ###P < 0.001; two-tailed t test).
In addition to functional FMO1, intact PAD4 and NPR1 genes are required for a strong resistance induction by exogenous Pip (Návarová et al., 2012). To get deeper mechanistic information about the synergistic interplay of Pip and SA in PR1 induction, we examined the behavior of fmo1, pad4, and npr1 mutant plants in our assay. SA alone triggered a strong, wild-type-like expression of PR1 in fmo1. However, in contrast to the wild type, we observed neither significantly increased PR1 expression upon Pip treatment alone nor the Pip-triggered intensification of SA-induced PR1 expression in fmo1 (Figure 9B; Supplemental Figure 14B). This indicates that FMO1 acts downstream of Pip but upstream of SA in inducible PR1 expression. Moreover, FMO1 is required for the intensification of SA-induced PR1 expression by Pip. Similar to FMO1, PAD4 is not essential for SA-induced PR1 expression but is required for Pip-induced PR1 expression, indicating that PAD4 is also positioned between Pip and SA in the signaling pathway leading to PR1 induction. In contrast to fmo1, however, pad4 was not impaired in the Pip-mediated intensification of PR1 expression, suggesting that PAD4 is not involved in the synergistic interplay between SA and Pip (Figure 9B; Supplemental Figure 14B). Finally, the npr1 mutant did not show discernible expression of PR1 after any of the treatments, indicating that NPR1 functions downstream of both Pip and SA in the induction of PR1 (Figure 9B; Supplemental Figure 14B).
Exogenous Pip treatment results in a significant resistance induction in Col-0, ald1, and sid2 but not in fmo1 plants (Návarová et al., 2012). Complementarily, we now tested the abilities of Col-0, ald1, fmo1, and sid2 to augment basal resistance to Psm in response to exogenous SA (Figure 9C). SA treatment strongly increased resistance to Psm in all the genotypes. In fact, the degree of resistance induction was even higher in ald1 and fmo1 than in Col-0, since bacterial growth was attenuated by exogenous SA to 93 to 95% in the mutants but only to 88% in the wild type (Figure 9C). This indicates that exogenous SA can induce both PR gene expression (Figure 9A; Supplemental Figure 14A) and disease resistance (Figure 9C) past Pip/FMO1 signaling and points to a redundant mode of action of Pip and SA. Nevertheless, the above-described amplification of SA signaling by Pip/FMO1 (Figure 9A; Supplemental Figure 14A) was still apparent in this resistance assay, because exogenous SA restricted bacterial growth to lower absolute numbers in Col-0 than in ald1 or fmo1 (Figure 9C). Yet more strikingly, both Pip and SA pretreatments failed to reduce bacterial multiplication in sid2 to the same absolute values as in the wild type (Figure 9C; Návarová et al., 2012), indicating the necessity for endogenous SA production for full resistance induction. Together, the response patterns observed here for exogenous SA and by Návarová et al. (2012) for exogenous Pip treatments place FMO1 downstream of Pip but upstream of SA in resistance induction. Exogenous SA can trigger a strong but not a complete immune response without functional Pip signaling. Reciprocally, Pip can induce a marked but not a full resistance response independently from SA biosynthesis. Together, our findings show that Pip and SA exhibit synergistic, independent, and redundant modes of action in plant immunity.
DISCUSSION
This study provides insights into the interplay of two key SAR regulatory plant metabolites: the phenolic SA and the non-protein amino acid Pip (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Návarová et al., 2012), in SAR establishment, SAR-associated defense priming, and basal plant immunity. Key Pip and SA biosynthetic and signaling genes exhibit strong transcriptional activation upon SAR induction throughout the plant (Figure 1A; Song et al., 2004b; Mishina and Zeier, 2006; Attaran et al., 2009; Návarová et al., 2012), and both metabolites accumulate systemically in the foliage of Arabidopsis plants locally leaf-inoculated with P. syringae, whereby the initial rise of Pip in the systemic tissue timely precedes the increase of SA (Návarová et al., 2012). Similar to many other studies (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Vlot et al., 2009), we assessed here resistance responses of the SA induction-deficient sid2 mutant, which is defective in the pathogen-inducible SA biosynthesis gene ICS1, to investigate the function of SA in basal immunity and SAR. By analogy, we used ald1 plants defective in Pip accumulation to deduce the immune responses that are regulated by Pip. Taken together, isotope labeling, biochemical, and metabolite studies strongly suggest that Pip is derived from Lys by a two-step mechanism and that ALD1 catalyzes a first aminotransferase step therein (Gupta and Spenser, 1969; Song et al., 2004a; Návarová et al., 2012; Zeier, 2013). The direct involvement of ALD1 in Pip biosynthesis and the fact that exogenous Pip can complement ald1 resistance defects (Návarová et al., 2012) suggest that, just as sid2 is useful for SA-related research, ald1 is a suitable genetic tool to assess the function of Pip in plants. Given the above-mentioned lines of evidence, the possibilities that metabolites other than SA and Pip (or direct derivatives thereof) substantially contribute to the sid2 and ald1 phenotypes, respectively, are unlikely in our opinion. Whereas the recent identification of the bona fide SA receptors NPR1, NPR3, and NPR4 (Fu et al., 2012; Wu et al., 2012) has validated that free SA is an active signal, it is not clear yet whether unmodified Pip or a direct, possibly FMO1-generated Pip derivative is signaling active (see further discussion below regarding FMO1; Zeier, 2013). In addition to the respective single mutants, we generated a Pip- and SA-deficient sid2 ald1 double mutant as one of the tools to study the interplay between Pip and SA in basal and systemic immunity (Supplemental Figures 1 and 2).
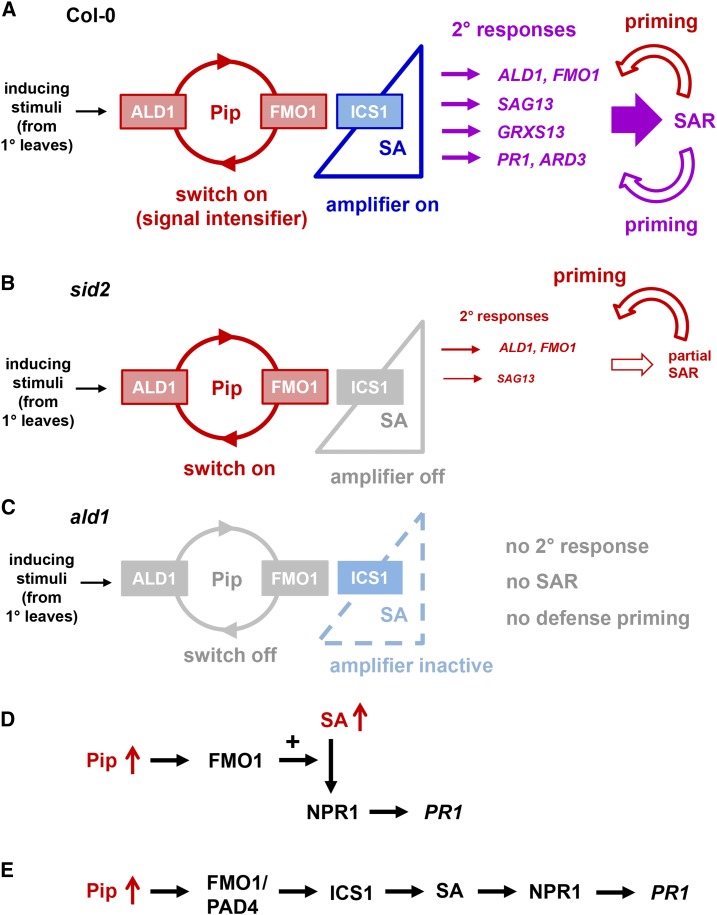
We showed that SAR establishment in Arabidopsis is characterized by a strong transcriptional reprogramming of the systemic leaf tissue that involves robust activation of more than 3400 SAR+ genes and suppression of nearly 2900 SAR- genes. Along with systemic resistance induction (Figure 2B; Song et al., 2004b; Návarová et al., 2012), this massive transcriptional SAR response is virtually absent in ald1, indicating that transcriptional reprogramming during SAR depends on the ability of plants to biosynthesize Pip after pathogen inoculation (Figures 1B, 3A, and 3B; Supplemental Figures 5 and 7; Návarová et al., 2012). Since knockout of ALD1 alone is sufficient to fully abrogate the transcriptional reprogramming in distal leaves, we consider it unlikely that analogous RNA-seq analyses with the sid2 ald1 double mutant, which we have not performed in this study, would provide mechanistic information about the transcriptional SAR response beyond that acquired for ald1, although we cannot fully rule out this possibility. However, in addition to ALD1, SAR and the transcriptional response associated with SAR fully require intact FMO1 (Mishina and Zeier, 2006; Gruner et al., 2013). Furthermore, elevation of Pip levels by exogenous application induces a SAR-like resistance response in a FMO1-dependent manner (Návarová et al., 2012). These findings indicate that the establishment of biological SAR and the associated transcriptional reprogramming of systemic leaf tissue are regulated by a master module that involves accumulating Pip and its downstream-acting component FMO1 (Figure 10A).
Figure 10.
Summary of the Roles of Pip, FMO1, and SA in Arabidopsis SAR and Associated Defense Priming, and Modes of the Synergistic Interplay between Pip and SA.
(A) to (C) Regulation of the SAR transcriptional response, SAR establishment, and SAR-associated defense priming by Pip, FMO1, and SA.
(A) The situation in wild-type plants. The full SAR and priming responses are established by elevated levels of ALD1-generated Pip, the action of FMO1 downstream of Pip, and ICS1-synthesized SA in 2° leaf tissue. The Pip/FMO1 module acts as an indispensable switch for SAR activation, and SA amplifies Pip-dependent responses to different degrees.
(B) The situation in sid2 mutant plants. In the absence of functional ICS1 and elevated SA, the Pip/FMO1 module is sufficient to induce a set of partially SA-independent responses to a certain level and trigger a moderate SAR response. Notably, an intact Pip/FMO1 module is also capable to prime plants for enhanced activation of partially SA-independent responses in the absence of SA elevations (bent red arrow).
(C) The situation in ald1 and fmo1 mutant plants. Functional ICS1 alone is not sufficient for SAR activation. Without Pip elevations or functional FMO1, SA biosynthesis is not activated. Failure of both Pip and SA elevations prevents the establishment of a primed state. Without the Pip/FMO1 module, inducing stimuli from 1° leaves are either not transduced into a meaningful response in 2° leaves or too weak to induce a noticeable response.
(D) and (E) Two modes of synergistic interplay between the immune signals Pip and SA.
(D) Elevated Pip levels amplify SA-induced PR1 expression. This amplification response is mediated by FMO1 but does not depend on PAD4.
(E) Elevated Pip levels induce PR1 expression via ICS1-triggered SA production. This signaling mode of Pip depends on both FMO1 and PAD4.
Whereas the critical function of Pip for SAR has been recognized only recently (Návarová et al., 2012), it has been known for more than two decades that SA functions as another key SAR player (Vernooij et al., 1994; Nawrath and Métraux, 1999). This study confirms this assessment and shows that the extent of SAR and of the transcriptional response in systemic tissue is strongly attenuated in sid2 plants (Figures 2B, 2C, 3A, and 3B). Our findings indicate that the degree of upregulation of the vast majority of the SAR+ genes in systemic tissue is positively influenced by SA (Tables 1 and 2). However, our study adds an important aspect to the current viewpoint of SAR regulation. We show that in the absence of induced SA production, a moderate SAR and a partial transcriptional response in systemic tissue occurs upon primary bacterial inoculation (Figures 2B, 2C, 3A, and 3B). This response not only becomes apparent in the ICS1-defective sid2 plants, but also in another SA induction-deficient mutant, sid1, which bears a defect in the multidrug and toxin extrusion-like transporter ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5) (Figure 2C). EDS5 is required for the export of chloroplast-synthesized SA from this organelle, and SA accumulation in sid1/eds5 is supposedly inhibited by autoinhibitory feedback (Serrano et al., 2013). Furthermore, the partial SAR response is established in ics1 ics2, in which both isochorismate synthase isoforms of Col-0 are inactive (Supplemental Figure 3). The ics1 ics2 double mutant not only is impaired in stress-inducible SA biosynthesis but also displays strongly reduced basal SA levels (Garcion et al., 2008). Therefore, our study reveals the existence of an SA-independent signaling pathway that is able to activate a comparatively small but significant SAR immune response in Arabidopsis. We show that this pathway is induced upon inoculation with two inherently different bacterial pathogen types, compatible (virulent) Psm and incompatible (avirulent) Psm avrRpm1, which, in contrast to Psm, causes a hypersensitive cell death response in Col-0 leaves (Bisgrove et al., 1994). SAR assays conducted with the sid2 ald1 double mutant indicate that the major, SA-dependent SAR activation pathway and the minor, SA-independent activation pathway are both regulated by Pip (Figure 2B). Therefore, the Pip/FMO1 module is able to switch on a moderate but clearly discernable SAR response in the absence of inducible SA biosynthesis (Figure 10B).
From our expression analyses we deduce that, after accumulating in systemic leaf tissue in a Pip/FMO1-dependent manner (Song et al., 2004b; Mishina and Zeier, 2006; Návarová et al., 2012), SA amplifies the expression of different SAR+ genes to varying degrees (Tables 1 and 2). Systemic expression of the predominant portion of the SAR+ genes shows a strict SA dependency, which signifies that these genes are not upregulated in a statistically significant manner in distal leaves of sid2 (Figure 3A, Table 1). Paradigm examples for these strictly SA-dependent SAR+ genes are PR1, a classical marker for the SA signaling pathway in plants and SAR establishment (Sticher et al., 1997), ARD3, an Arabidopsis gene with sequence similarity to a rice acireductone dioxygenase involved in the Yang cycle (Sauter et al., 2005), and GRXS13, encoding a glutaredoxin facilitating infection of Arabidopsis with the fungal necrotroph Botrytis cinerea (La Camera et al., 2011). However, about one-quarter of SAR+ genes were significantly upregulated in the systemic leaf tissue of sid2 upon 1° inoculation and showed higher absolute expression values in Psm-inoculated sid2 mutants than mock-treated Col-0 plants (Figure 3A, Table 2). These genes require SA for their full expression in systemic tissue but can be induced to different degrees without SA elevation. Notably, the two SAR regulatory and Pip pathway genes ALD1 and FMO1 rank among the SAR+ genes with the strongest SA-independent expression characteristics (Table 2). This suggests that the Pip signaling pathway can function separately from SA to switch on a subset of systemic resistance responses to a certain level. Nevertheless, for a full and strong SAR response, accumulation of SA is required to potentiate these partially SA-independent responses that include Pip production and signaling and to turn on other, strictly SA-dependent responses such as PR1 expression (Figures 10A and 10B).
Functional ICS1 is not a sufficient prerequisite for pathogen-induced SAR, as exemplified by the full SAR defects of ald1 and fmo1 (Figure 2B; Song et al., 2004b; Mishina and Zeier, 2006; Návarová et al., 2012). This is the case because Pip biosynthesis and signal transduction via FMO1 are required to switch on virtually every response in distal leaves of locally inoculated plants, including the expression of ICS1, SA biosynthesis, and SA downstream responses (Figures 1B, 1C, 3B, and 10A to 10C; Mishina and Zeier, 2006; Gruner et al., 2013). However, once SA levels elevate in leaves, significant SA responses can be activated in the absence of a functional Pip/FMO1 module. This is experimentally revealed in ald1 and fmo1 plants exogenously supplied with SA because these plants exhibit markedly enhanced expression of PR-1 and elevated resistance to Psm (Figure 9). This aspect is relevant for defense processes in inoculated leaves because here, in contrast to distal leaves, pathogen-induced ICS1 expression and SA accumulation are still active in ald1 and fmo1 (Figures 1A and 1C; Song et al., 2004b; Mishina and Zeier, 2006; Bartsch et al., 2006). Our findings that SA-deficient sid2 plants exhibit a more pronounced defect in local resistance to Psm than ald1 or fmo1 (Figures 2A and 9C) indicate that the contribution of the SA pathway to basal immunity exceeds that of the Pip/FMO1 pathway. A largely Pip/FMO1-independent activation of the SA defense pathway at pathogen inoculation sites might explain why defects in Pip biosynthesis and FMO1 result in comparatively moderate decreases of basal immunity. In fact, the importance of FMO1 for local plant immune responses is variable and depends on the nature of the attacking pathogen, with particular significance of FMO1 for local immunity to pathogens that activate EDS1-dependent and SA-independent branches of plant defense (Bartsch et al., 2006; Koch et al., 2006). Interestingly, EDS1 and its interacting partner PAD4, both master regulators of basal immunity (Feys et al., 2001), have also been identified as necessary SAR components (Mishina and Zeier, 2006; Jing et al., 2011; Rietz et al., 2011; Breitenbach et al., 2014). PAD4 positively regulates the transcription of a whole battery of defense genes including the SAR-relevant genes ALD1, FMO1, and ICS1 (Zhou et al., 1998; Jirage et al., 1999; Song et al., 2004b; Bartsch et al., 2006). Therefore, overlapping regulatory principles of basal immunity and SAR exist. However, the relevance of the Pip/FMO1 module for SA pathway activation seems to be a main distinguishing feature of the resistance modes at sites of Psm attack and in distal tissue (SAR). Whereas the Pip/FMO1 module possesses an obligate switch function for SAR activation, it seems to play a supportive but less rigorous role for resistance induction at inoculation sites.
Notably, our resistance assays with sid2 ald1 reveal that the SA and Pip defense pathways provide additive, independent contributions to Arabidopsis basal resistance toward Psm infection (Figure 2A). This is further corroborated by the findings that exogenous Pip is able to enhance resistance of sid2 to Psm (Návarová et al., 2012) and that exogenous SA increases resistance of ald1 to Psm (Figure 9C). Moreover, Bartsch et al. (2006) have shown that a sid2 fmo1 double mutant is more susceptible to the Hyaloperonospora arabidopsidis isolate Cala2, an avirulent oomycete pathogen that is recognized by the RPP2 resistance protein in the Arabidopsis accession Col-0 than either sid2 or fmo1 single mutants. This indicates additive contributions of the SA and Pip/FMO1 pathways also in resistance gene-mediated responses.
However, our data also show a synergistic relationship between Pip and SA signaling on several levels. First, application of Pip intensifies PR1 expression induced by exogenous SA in a FMO1-dependent manner (Figures 9A, 9B, and 10D; Supplemental Figures 14A and 14B). This enhancement, which also takes place in SA-deficient sid2 (Figure 9A), indicates that the Pip/FMO1 module amplifies SA downstream signaling events. Second, elevated Pip levels trigger PR1 expression via the induction of SA biosynthesis (Figure 10E), as indicated by both the shortcoming of sid2 to induce PR1 upon exogenous Pip feeding (Figure 9A) and the ability of exogenous Pip to moderately induce SA accumulation in Arabidopsis (Návarová et al., 2012). Pip therefore acts both upstream of SA to trigger PR1 expression and downstream of SA to fortify SA-induced PR1 expression (Figures 10D and 10E). The SA-mediated, PR1-inducing action of Pip as well as its function as an SA signal intensifier share overlapping mechanisms because both functions require functional FMO1 and operate upstream of NPR1. However, both operation modes of Pip also differ mechanistically because the PR1-inducing but not the intensifying mode requires PAD4 (Figures 9B, 10D, and 10E). Complementary to these positive effects of Pip on SA biosynthesis and signaling, SA feeding alone is sufficient to trigger small but significant elevations of ALD1 expression and Pip levels (Návarová et al., 2012). These findings exemplify that, in addition to their above-described independent modes of action, the Pip and SA pathways mutually reinforce each other in the regulation of plant immunity and SAR.
Crosstalk between different stress-related plant signals is well established (Derksen et al., 2013). For example, synergism between JA and ethylene signaling ensures effective induction of plant defenses directed against necrotrophic pathogen attack (Memelink, 2009). Moreover, the JA/ethylene and SA signaling pathways negatively influence each other to prioritize specific defense outputs in plant-pathogen interactions (Koornneef and Pieterse, 2008; Zander et al., 2014). JA biosynthesis and signaling is strongly induced by P. syringae infection in inoculated leaves. However, activation of the JA pathway is variable and strongly correlates with necrotic tissue disruption but not with systemic responses or SAR (Mishina and Zeier, 2007; Zoeller et al., 2012). With the inoculation conditions to induce SAR used in this study, JA biosynthesis and expression of JA-responsive genes are not elevated in systemic tissue of Col-0 wild-type plants (Table 4; Gruner et al., 2013). Although a considerable systemic JA response in Arabidopsis early after inoculation with high titers of avirulent P. syringae has been detected and associated with SAR (Truman et al., 2007), genetic analyses argue against a role of the JA pathway in Arabidopsis SAR establishment (Chaturvedi et al., 2008; Attaran et al., 2009). Notably, our comparative RNA-seq analyses reveal that distal leaves of locally inoculated sid2 plants express JA-responsive genes to a significantly higher extent than Col-0 plants. In particular, the genes that were upregulated in sid2 but not changed or downregulated in Col-0 are markedly enriched in JA-inducible genes (Table 4). This indicates that the accumulation of SA during SAR in wild-type plants suppresses systemic JA responses and directly illustrates the occurrence of SA/JA antagonism within SAR. The apparent prevalence of SA over JA signaling in SAR-induced plants might explain why SAR provides protection particularly against pathogens with a (hemi)biotrophic lifestyle (Sticher et al., 1997). Potential crosstalk of Pip with plant stress signals and hormones other than SA is not yet established. However, a recent study reports that treatment of Arabidopsis seedlings with methyl jasmonate decreases ALD1 expression and Pip levels in roots, indicating negative influences of JA on Pip biosynthesis (Yan et al., 2014).
SAR can condition plants to induce defense responses more quickly and efficiently after pathogen challenge (Jung et al., 2009; Návarová et al., 2012), a phenomenon designated as defense priming (Conrath, 2011). Within our experimental setup, we defined a particular defense response as primed, if the difference between the pathogen and mock response of SAR-activated (or Pip-stimulated) plants is significantly larger than the same difference of nonactivated plants (Supplemental Figure 9B), resulting in the strongest absolute response level in pathogen-challenged, SAR-activated (or Pip-stimulated) plants (Figures 6 to 8; Supplemental Figures 10 to 12). This does not mean that the pathogen-triggered fold change of the response in SAR- (Pip-) activated plants is larger than the respective fold change in control plants. For example, in the SAR-priming assay shown in Figure 6A (Supplemental Figure 10A), the quotient between the averaged 1°-Psm /2°-Psm to the averaged 1°-Psm /2° mock expression value for GRXS13 is 5.2 for the Col-0 wild type, whereas the quotient between the 1°-mock/2°-Psm to the 1°-mock/2°-mock value is 5.7. Therefore, an important determinant of the response gain due to priming is the marked increase of the response level caused by the inducing stimulus alone (SAR activation and Pip stimulation). A further increase of a strongly preelevated response level due to 2° challenge will ultimately lead to a much stronger absolute response level than a similar increase from a very small level present in noninduced plants. This mechanistic aspect of priming is consistent with a general model of the priming phenomenon proposed by Bruce and colleagues for two subsequent stress exposures (Bruce et al., 2007).
Between independent SAR or Pip induction experiments, the observed priming patterns in wild-type plants are reproducible and can be tested with our proposed algorithm (Supplemental Figures 9B and 9C). However, absolute or relative measurement values resulting from one of the four treatment cases (e.g., mock/mock, mock/Psm, Psm/mock, or Psm/Psm) might quantitatively vary between experiments (for example, compare Figures 6A with 6B, 7A with 7B, or Figures 8A/8B with 8C/8D). In this context, we performed linear model-based analyses of the SAR-associated priming responses (Figures 6 and 7), the Pip-induced priming responses (Figure 8), and the Pip-mediated intensification of SA-inducible PR1 expression (Figures 9A and 9B) in the Col-0 wild type using combined data of three independent experiments to estimate treatment and experimental effect terms (Supplemental Tables 1 to 3). In doing so, we assumed that priming in our assays corresponds to the interaction of a pretreatment (treatment 1) on the response of a second treatment (treatment 2). These analyses confirm that, although experimental variation significantly influences the individual treatment responses and the interaction term (priming) for most measured values, the individual treatment responses and the priming response are stable over different experiments (Supplemental Tables 1 to 3).
Besides microbial encounters, adverse abiotic conditions can prime plants for enhanced pathogen responses (Singh et al., 2014). For more than two decades, SA has been attributed an important role in defense priming. For instance, in leaf tissue directly adjacent to pathogen attack, SA has been implicated in the potentiation of expression of defense genes that are not directly SA responsive (Mur et al., 1996). Furthermore, exogenous SA or pretreatment with chemicals such as 2,6-dichloroisonicotinic acid or S-methyl-1,2,3-benzothiadiazole-7-carbothioate (BTH), which are often vaguely designated as SA analogs, are able to prime cultured plant cells or intact plants for enhanced defense responses (Kauss et al., 1992; Thulke and Conrath, 1998), and BTH-induced priming proceeds via the SA downstream regulator NPR1 (Kohler et al., 2002). Moreover, treatments with chemicals that inhibit SA glycosylation and thereby endogenously elevate free SA levels lead to priming responses in Arabidopsis (Noutoshi et al., 2012), and defense priming triggered by exogenous β-amino butyric acid or the bacterial quorum sensing compound N-acyl-homoserine lactone requires an intact SA signaling pathway (Zimmerli et al., 2000; Schenk et al., 2014). Our study reveals a differentiated role of SA in the defense priming responses associated with SAR: SA is required to prime plants for enhanced activation of certain defense responses but is dispensable for the potentiation of others. On the one hand, genes such as PR1, ARD3, and GRXS13, which we had classified as SA dependent according to their expression characteristics in systemic tissue upon SAR induction, also require SA accumulation for augmented expression during challenge infection (Figure 6A). On the other hand, the expression of the partially SA-independent SAR genes ALD1, FMO1, and SAG13 is potentiated in the SAR priming assays to a similar extent in Col-0 and sid2, and a primary inoculation thus primes plants for enhanced expression of these genes in an SA-independent manner in systemic tissue (Figure 6A).
Our previous work identified Pip as a critical regulator of SAR-associated defense priming in Arabidopsis (Návarová et al., 2012). Exogenous Pip also increases resistance of tobacco plants to P. syringae pv tabaci and primes plants for early SA accumulation, indicating a conserved role of Pip in plant defense priming (Vogel-Adghough et al., 2013). This study shows that Pip elevations in leaves are both required and sufficient for the priming of both SA-dependent and -independent SAR responses (Figures 6 to 8; Supplemental Figures 10 to 12). Moreover, our results provide evidence that, in addition to Pip, functional FMO1 is essential for the SAR-associated priming phenomenon (Figure 6B). In fact, FMO1 acts downstream of Pip in the activation of defense priming because the priming responses induced by exogenous Pip are virtually blocked in fmo1 (Figure 8B; Supplemental Figure 13). This finding is consistent with the critical role of FMO1 for SAR and resistance induction by Pip (Mishina and Zeier, 2006; Návarová et al., 2012; Gruner et al., 2013). Future work should focus on the biochemical function of the flavin monooxygenase FMO1. A conceivable scenario is that FMO1 catalyzes the oxidation of Pip or a Pip-derived metabolite for defense signal transduction (Zeier, 2013).
The existence of SA-independent defense priming responses associated with SAR parallel recent findings on silicon-induced resistance in Arabidopsis, in which increased silicon absorption primed plants for effective responses also in a sid2 background (Vivancos et al., 2015). Moreover, SA-independent SAR activation, which is moderate for Arabidopsis (Figures 2B and 2C), might be more pronounced in monocots, as activation of systemic immunity in barley by leaf inoculation with bacterial pathogens has been shown to proceed in an NPR1-independent manner (Dey et al., 2014). Which molecular components besides Pip and FMO1 could be involved in the SA-independent signaling pathway leading to partial SAR and defense priming in Arabidopsis? Recent data point to a role of mitogen-activated protein kinase (MAPK) signaling cascades in the activation of SA-independent resistance responses and SAR. On one hand, Tsuda and colleagues have shown that a sustained activation of mitogen-activated protein kinase 3 (MPK3) and MPK6 in leaves of transgenic Arabidopsis plants results in the induction of PR1 and other SA-responsive genes in sid2 and npr1, indicating that strong activation of MAPK signaling can induce defense gene expression independently of a functional SA pathway (Tsuda et al., 2013). PR1 induction in systemic leaf tissue of P. syringae-inoculated plants, however, is strictly SA dependent (Table 1), indicating differences between the SA-independent branch of biological SAR and the SA-independent signaling pathway activated by sustained MAPK expression (Tsuda et al., 2013). Nevertheless, Beckers et al. (2009) reported that priming of abiotic stress responses and SAR are compromised in a mpk3 mutant and attenuated in mpk6, indicating a role for MAPK3/6 signaling in SAR. In our SAR analyses, several MAPK, MAPK kinase, and MAPK kinase kinase genes are significantly upregulated upon SAR induction (Figure 4F), including MPK3 and MPK6. In sid2, however, systemic upregulation of MAPK cascade genes is only weakly induced, suggesting that transcriptional activation of these genes is largely SA dependent (Supplemental Figure 15). Future experiments should further dissect the role of MAPK signaling in SAR and, in particular, clarify its relationship to the Pip signaling pathway.
Our RNA-seq data suggest that SAR prepares plants for future pathogen attack by the transcriptional preactivation of different, consecutive stages of defense signaling. First, a widespread upregulation of genes putatively involved in pathogen recognition and early signal transduction occurs upon SAR establishment. For example, many Toll-interleukin-1 receptor-like domain (TIR)-NBS-leucine rich repeat (LRR) or coiled coil (CC)-NBS-LRR-type resistance proteins are upregulated upon SAR activation (Figure 4E; Supplemental Figure 15). These include VICTR, which encodes a TIR-NBS-LRR protein that associates with EDS1 and PAD4 in protein complexes and is required for small molecule-mediated crosstalk between abscisic acid and immune signaling (Kim et al., 2012). Furthermore, genes coding for receptor-like proteins and members of specific receptor-like kinase families prominently occur in the SAR+ gene group (Figure 4E). For instance, cysteine-rich protein kinase (CRK) genes are widely upregulated upon SAR establishment. Dexamethasone-inducible expression of CRK13 in Arabidopsis, one of the most strongly upregulated CRK genes during SAR (Supplemental Figure 15), resulted in increased SA-mediated resistance to P. syringae (Acharya et al., 2007). These results suggest that the increased, broad-spectrum expression of resistance proteins and pattern recognition receptors during SAR is an integral part of systemic resistance activation and defense priming. Moreover, the well-characterized pattern recognition receptor genes EFR, BAK1, and CERK1, but not FLS2 (Zipfel, 2014), are among the SAR+ genes (Supplemental Figure 15). SA has been shown to dynamically regulate the levels of FLS2 and BAK1 and to promote their upregulation at later signaling stages (Tateda et al., 2014). Consistent with the notion that SA positively regulates microbial pattern receptors, sid2 shows strongly reduced upregulation of these genes. However, a marked SA-independent (but Pip-dependent) component of receptor upregulation is discernable, as exemplified by increased systemic expression of several CRK genes in sid2 (Supplemental Figure 15).
Besides the above-discussed MAPK members, several other gene types involved in signaling steps downstream of pathogen perception are broadly activated in systemic tissue of SAR-induced plants. These include genes coding for Ca2+ signaling-related proteins such as calcium-dependent protein kinases, EF-hand-containing proteins, and calmodulin binding proteins (Figure 4G). Among those is CPK5 (Supplemental Figure 15), an Arabidopsis calcium-dependent protein kinase that phosphorylates the NADPH oxidase isoform RbohD (Respiratory burst oxidase homolog D). CPK5 is required for the rapid leaf-to-leaf propagation of defense responses upon PAMP perception, and reactive oxygen species-mediated cell-to-cell communication involving CPK5 and RbohD has been suggested as a basis for long-distance defense signal propagation (Dubiella et al., 2013). However, a direct function of CPK5 in pathogen-induced SAR has not yet been established.
Many immune-related signal transduction pathways culminate in the activation of transcription factors and cofactors in the nucleus and thereby trigger enhanced expression of defense-related genes. The transcriptional coactivator NPR1 acts as a central transducer of SA responses and is essential for biological SAR as well as for SAR-related transcriptional responses (Fu and Dong, 2013). For example, NPR1 is required for the expression of ∼98.5% of BTH-inducible genes (Wang et al., 2006). It is likely that NPR1 also largely regulates, besides the SA-dependent SAR activation pathway, also the Pip-dependent and partially SA-independent branch of SAR induction described here, because exogenous Pip induces a much weaker resistance response in npr1 than in sid2 (Návarová et al., 2012). NPR1 interacts with different members of the TGA subfamily of bZIP transcription factors to regulate PR gene expression (Fu and Dong, 2013). NPR1, its paralog NPR3, and three TGA genes (TGA5, TGA3, and TGA1) are SAR+ genes showing a moderate but significant degree of upregulation upon SAR induction (Supplemental Figure 15).
Remarkably, more than one-third of the WRKY family of transcription factors belong to the SAR+ group, suggesting that WRKY factors play a major role in the SAR transcriptional response (Figure 4H). For many WRKYs, positive or negative regulatory roles in distinct plant immune responses have been established (Pandey and Somssich, 2009). Among the SAR+ genes are several WRKY factor genes whose BTH-induced expression depend on NPR1 (Supplemental Figure 15; Wang et al., 2006). WRKY18, for example, controls about one-fourth of BTH-induced and NPR1-regulated genes in Arabidopsis, indicating that individual WRKY factors regulate expression of subgroups of the SAR transcriptome (Wang et al., 2006). Interestingly, the marked upregulation of WRKY genes correlates with an enrichment of VQ motif-containing genes in the SAR+ group (Supplemental Figure 8B). VQ motif-containing proteins have been shown to interact with WRKY proteins and appear to be part of transcriptional regulatory protein complexes in plant immunity (Cheng et al., 2012). Besides WRKYs and TGAs, NAC transcription factors are overrepresented in the SAR+ gene group, whereas other classes are not (Figure 4H). This suggests a dominant function of particular transcription factor families in mediating the SAR transcriptional response. Sequential waves of specific transcription factor activities have been suggested to orchestrate plant immunity (Moore et al., 2011), and this might be particularly true for SAR activation. For the initiation of this transcriptional program leading to SAR, the Pip/FMO1 module and the transcriptional coregulator NPR1 are indispensable.
Our results not only illustrate that SAR equips plants with an increased defense capacity at different signaling levels, but also show that the state of increased pathogen resistance is associated with a diminished photosynthetic rate systemically in the foliage and strongly decreased expression of photosynthesis-related genes (Figures 4A and 5A; Supplemental Figure 5 and Supplemental Data Set 2). Moreover, the transcriptional SAR response illustrates an overall decline of several anabolic pathways, secondary metabolism, cell wall remodeling, and wax and cutin biosynthesis (Figures 4A to 4C; Supplemental Data Set 2), suggesting that SAR is associated with reduced plant growth. The induced decline in photosynthetic rates is completely absent in both ald1 and sid2 (Figure 5A), indicating that the Pip-dependent accumulation of SA attenuates photosynthesis systemically in the plant. This interpretation is consistent with the observation that plants exhibiting constitutively high SA levels show decreased efficiency of several photosynthetic parameters (Mateo et al., 2006). Similarly, enhanced SA levels during SAR appear to trigger growth alleviation because transcriptional gene downregulation is strongly suppressed in sid2 (Figure 5A) and plants with high constitutive SA levels exhibit retarded growth (Bowling et al., 1997; Mauch et al., 2001). The decrease in photosynthesis (and growth) might be a consequence of reduced stomatal conductance in SAR-activated plants, as the decreases in transpiration rates indicate (Figure 5B). This would in turn limit CO2 uptake and carbon fixation. Since the transpiration rates do not systemically decrease in ald1 and sid2 (Figure 5B), and exogenous SA is able to induce stomatal closure in Arabidopsis (Zeng et al., 2011), accumulating SA during SAR also likely accounts for this phenomenon. A closure of stomata represents an integral part of the PAMP-induced immune response of plants to bacterial pathogen entry (Melotto et al., 2006), and our data suggest that the stomatal defense response is also preactivated systemically in the foliage upon localized pathogen encounter.
In conclusion, Arabidopsis SAR and the associated defense priming phenomenon are master-regulated by Pip accumulation and the action of FMO1 downstream of Pip. The Pip/FMO1 switch module activates SA-dependent and SA-independent responses that both contribute to SAR establishment and the execution of defense priming. SA amplifies Pip/FMO1-dependent responses to different degrees and thereby ensures strong SAR and priming reactions. This work further indicates that Pip and SA cooperatively interact but can also function independently from each other in the induction of plant immune responses.
METHODS
Plant Material and Cultivation
Arabidopsis thaliana plants were grown individually in pots containing a mixture of soil (Substrat BP3; Klasmann-Deilmann), vermiculite, and sand (8:1:1) in a controlled cultivation chamber with a 10-h-day (9 am to 7 pm; PFD 100 µmol m−2 s−1)/14-h-night cycle and a relative humidity of 70%. Day and night temperatures were set to 21°C and 18°C, respectively. Experiments were performed with 5- to 6-week-old, naïve plants exhibiting a uniform appearance.
The ald1 and fmo1 mutants represent the SALK lines SALK_007673 and SALK_026163, respectively (Mishina and Zeier, 2006; Návarová et al., 2012). sid2-1 (sid2, ics1; Nawrath and Métraux, 1999), ics1 ics2 (Garcion et al., 2008), npr1-2 (npr1, NASC ID: N3801), and pad4-1 (pad4; Jirage et al., 1999) were used in this study. All mutant lines are in the Col-0 background.
The sid2-1 ald1 (sid2 ald1) double mutant was generated by crossing of the sid2-1 (female parent) and ald1 (male parent) single mutants. From every fertilized silique, the F1 seeds were collected individually and dried before planting. About 110 sid2 ald1 candidate plants were analyzed in the F2 generation as outlined in Supplemental Figures 1 and 2. One positive F2 candidate (sid2 ald1 #6) was self-pollinated and used for experiments. The position of the ethyl methanesulfonate-generated mutation in sid2-1 results in a stop codon (TAA) at residue 449 instead of a glutamine (Wildermuth et al., 2001). To identify the ethyl methanesulfonate-generated mutation, site specific primers were designed and the specific annealing temperature of 64°C determined in a gradient PCR. To identify the homozygous T-DNA insertion of ald1 by PCR, the method described by Alonso et al. (2003) was applied, using gene-specific primers (Supplemental Table 4). Seeds of ics1 ics2 were sterilized and germinated on full Murashige and Skoog medium containing 2% sucrose (pH 5.7) before the seedlings were transferred to soil (Garcion et al., 2008).
Cultivation of Bacteria
Psm and Psm harboring the avrRpm1 avirulence gene (Psm avrRpm1) were cultivated at 28°C in King’s B medium to which appropriate antibiotics were added (Zeier et al., 2004). Overnight log phase cultures were diluted to different final optical densities at 600 nm (OD600) for leaf inoculations. As experimentally determined, OD600 = 1 corresponds to 3.2*108 colony-forming units per mL.
SAR Experiments, Defense Priming, and Plant Basal Resistance
To activate SAR, plants were infiltrated between 10 and 12 am into three lower (1°) leaves with bacterial suspensions of OD600 = 0.005. Infiltration with 10 mM MgCl2 served as the mock control treatment. Upper (2°) leaves were harvested 48 h after the primary treatment for the determination of systemic gene expression and metabolite contents, as well as for RNA-seq analyses. For systemic resistance assays, 2° leaves were inoculated with Psm (OD600 = 0.001) 2 d after the 1° treatment. The number of Psm bacteria in 2° leaves was assessed another three days later as described in Návarová et al. (2012). SAR-associated defense priming was assessed by challenging 2° leaves with either Psm (OD600 = 0.005) or infiltrating 10 mM MgCl2 as a mock control 2 d after the 1°-inducing or 1° mock treatments. The 2° leaves were collected 10 h later for analyses (Supplemental Figure 9). For the determination of basal resistance, suspensions of Psm (OD600 = 0.001) were infiltrated into three full-grown leaves of naive plants. Bacterial growth was assessed 3 d later (Návarová et al., 2012).
Exogenous Treatments with Pip and/or SA
Exogenous Pip was applied to plants by pipetting 10 mL of a 1 mM (10 µmol) d,l-pipecolic acid solution (S47167; Sigma-Aldrich) onto the soil substrate of individually cultivated plants. Control plants were supplemented with 10 mL of water instead. Pip feeding in this way served as the inducing treatment for Pip-priming assays. The bacterial challenge was performed 1 d after Pip application as described for the SAR priming assay. To determine synergism between Pip and SA, SA was infiltrated into leaves in a concentration of 0.5 mM 1 d after Pip feeding, and water infiltration served as a mock control. Leaf samples were collected 4 h after treatment.
Metabolite Determination
Determination of Pip levels in leaves was performed by a protocol detailed by Návarová et al. (2012), using gas chromatography-mass spectrometry-based analysis following propyl chloroformate derivatization. Free SA, conjugated SA, and camalexin were determined by a method based on vapor-phase extraction and gas chromatography-mass spectrometry analysis of metabolites as described by Attaran et al. (2009) and Návarová et al. (2012).
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qPCR) analysis to assess gene expression in leaves was performed as detailed by Návarová et al. (2012) with minor modifications. In brief, 1 µg total RNA extracted with PeqGOLD RNAPure reagent (PeqLab) was used for cDNA synthesis using the RevertAid RT reverse transcription kit from Thermo Fisher Scientific. Transcript levels were measured based on SYBR Green technology using Promega GoTaq qPCR master mix according to the manufacturer’s instructions. Data were analyzed using the ΔΔCT method. Primers used for qPCR analyses are given in Supplemental Table 4.
Assessment of Photosynthesis and Transpiration Rates by Gas Exchange
Maximum photosynthesis rates were determined by CO2 uptake (µmol CO2 m−2 s−1) measurements, and stomatal conductance was assessed via the determination of transpiration rates (mmol water m−2 s−1) in intact leaves at a PPFD of 1000 µmol photons m−2 s−1 using a LI-6400XT portable photosynthesis system (LI-COR Environmental). Before recording the maximum photosynthetic rates, leaves were incubated for ∼20 min in the measurement chamber until photosynthetic rates were stable. The temperature in the measuring cell was set to 22 to 23°C and the relative humidity was kept constant at 40% during measurements.
Reproducibility of Experiments and Statistical Analyses
The presented results are generally derived from one experimental data set, consisting of at least three biological replicates per treatment and genotype for metabolite and qPCR analyses, of seven biological replicates for bacterial growth assays, and of four biological replicates for IRGA analyses. For qPCR analyses, values for biological replicates were calculated as the mean of two technical replicates. The presented results were similar in three independent experiments. For the statistical evaluation of the qPCR and metabolite data by Student’s t test, log10-transformed relative expression and metabolite content values were analyzed. Bacterial growth data were analyzed by Student’s t test without prior log transformation.
Assessment of the primed state was performed as described in Supplemental Figure 9 by comparing permutated differences of measured values by a two-sided Mann-Whitney U test (α = 0.005). To assess treatment effects, interaction terms between the two consecutive treatments, and between-experiment variation in the SAR-associated priming assays, the Pip-induced priming assays, and the Pip/SA coapplication assays, we assumed a linear model and performed ANOVA (Brady et al., 2015) with type II-sum of squares on Col-0 response data from three independent experiments using the R statistical package (https://www.r-project.org/) and the following command (see also Supplemental Tables 1 to 3):
 |
The statistical analysis of the RNA-seq data is described below.
Genome-Wide Transcriptional Analyses of the SAR State
Three biologically independent, replicate SAR induction experiments were performed as described in the paragraph above describing SAR experiments with Col-0 and sid2 plants (experimental set 1) and three other biologically independent experiments with Col-0 and ald1 plants (experimental set 2). In each SAR experiment, at least six upper (2°) leaves from six different plants pretreated in 1° leaves with Psm (MgCl2) were pooled for one biological Psm (mock control) replicate. In this way, three biologically independent, replicate samples per treatment and plant genotype were obtained within each SAR set.
RNA was extracted from leaf sample replicates with the Plant RNeasy extraction kit (Qiagen) and treated on-column (Qiagen) and in solution with RNA-free DNase (New England Biolabs). RNA integrity, sequencing library quality, and fragment size were checked on a 2100 Bioanalyzer (Agilent). Libraries were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina), and library quantification was performed with a Qubit 2.0 (Invitrogen). Leaf samples were multiplexed 12 libraries per lane, yielding ∼150 million reads per lane. All libraries were sequenced on the HiSeq 2500 Illumina platform. Libraries for the not stranded RNA-seq experiments were sequenced in the single end mode with length of 50 or 100 nucleotides.
Reads were demultiplexed and mapped to the Arabidopsis genome by the CLC bio Genomics Workbench with default parameters (alignment over at least 80% of the length of the read, up to three mismatches allowed) without alternative transcript detection. Differences in change ratios between mock and Psm treatment for the two independent Col-0 experimental sets were determined using a linear model framework [lm(log2(rpm) ∼ experiment*treatment)], resulting in no statistically different change ratios. The data for the two experimental sets were then combined for analyses. Differential expression between mock-treated and Psm-treated samples was calculated as a pairwise using edgeRs classic method on raw read counts as implemented in Bioconductor (Robinson et al., 2010). Genes are considered significantly differentially expressed if the FDR is below 0.01 (Benjamini and Hochberg, 1995). Reads per million (rpm) are reported for all transcripts. Reads per million were averaged for the replicates of each condition and the log2 of fold changes between Psm and mock values (=P/M-fold changes) were calculated by formally adding one read to all rpm values to account for transcripts with no expression detected in one or more samples before fold change calculation and logarithmic transformation (Bräutigam et al., 2011). This allowed us to keep genes with very low mock expression values but marked Psm-triggered expression in the data sets without significantly changing the actual logarithmized P/M ratios. Differences in change ratios between Col-0 and sid2 for all genes without 0 counts in any experiment were determined using a linear model framework [lm(log2(rpm) ∼ genotype*treatment)] with P values corrected (Benjamini and Hochberg, 1995). The complete RNA-seq data of the SAR experiments are provided in Supplemental Data Set 1. Arabidopsis transcripts are annotated with descriptions from TAIR10 (Swarbreck et al., 2008) and functional annotations from MapMan (Thimm et al., 2004). Overlapping and exclusive membership in the significantly changed genes were calculated with Linux functions. To determine the percentages of SAR genes in specific gene categories or families (http://www.arabidopsis.org/), selected gene sets (major MapMan categories [Thimm et al., 2004], categories with a presumed role in defense signaling, and categories with an evident enrichment in SAR+ and SAR− genes based on careful inspection by eye of the whole data set) were aligned to the RNA-seq data set using the Excel macro FIRe (Garcion et al., 2006). Fisher’s exact test (P < 0.01) corrected according to Benjamini and Hochberg (1995) was used to determine whether gene categories were significantly enriched or depleted in SAR+ and SAR− genes. GO term enrichment was analyzed and visualized using the GOrilla tool using the target and background gene list method (Eden et al., 2009).
Accession Numbers
RNA-seq data were deposited in the ArrayExpress database under the accession number E-MTAB-4151. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ALD1 (At2g13810), FMO1 (At1g19250), ICS1 (At1g74710), PR-1 (At2g14610), PAD4 (At3g52430), NPR1 (At1g64280), ARD3 (At2g26400), SAG13 (At2g29350), and GRXS13 (At1g03850).
Supplemental Data
Supplemental Figure 1. Identification of the sid2 ald1 (sid2-1 ald1) double mutant.
Supplemental Figure 2. Characterization of the sid2 ald1 double mutant.
Supplemental Figure 3. Systemic acquired resistance assay with Col-0 and ics1 ics2 plants.
Supplemental Figure 4. Principle component analysis of the normalized transcriptome data.
Supplemental Figure 5. MapMan visualization: the transcriptional SAR response in Col-0.
Supplemental Figure 6. MapMan visualization: a diminished transcriptional SAR response exists in sid2.
Supplemental Figure 7. MapMan visualization: the transcriptional SAR response is absent in ald1.
Supplemental Figure 8. Percentage of SAR+ and SAR− genes in additional gene classes/families.
Supplemental Figure 9. SAR-associated defense priming: assay and definition.
Supplemental Figure 10. Graphs of Figure 6 with a linear scale for the y axes instead of a log scale.
Supplemental Figure 11. Graphs of Figure 7 with a linear scale for the y axes instead of a log scale.
Supplemental Figure 12. Graphs of Figure 8 with a linear scale for the y axes instead of a log scale.
Supplemental Figure 13. Pip-induced priming of salicylic acid biosynthesis requires functional FMO1.
Supplemental Figure 14. Graphs of Figures 9A and 9B with a linear scale for the y axes instead of a log scale.
Supplemental Figure 15. The transcriptional SAR response: activation of multiple stages of defense signaling.
Supplemental Table 1. Linear model-based analysis of the SAR-associated priming response in Col-0 plants to estimate treatment and experimental effect terms.
Supplemental Table 2. Linear model-based analysis of the pipecolic acid-induced priming response in Col-0 plants to estimate treatment and experimental effect terms.
Supplemental Table 3. Linear model-based analysis of the amplification of salicylic acid-induced PR1 expression by pipecolic acid in Col-0 plants to estimate treatment and experimental effect terms.
Supplemental Table 4. List of primers used in this study.
Supplemental Data Set 1. Complete RNA-seq data of the SAR experiments with Col-0, sid2, and ald1.
Supplemental Data Set 2. GO term enrichment analysis of the SAR− gene group.
Supplemental Data Set 3. GO term enrichment analysis of the SAR+ gene group.
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation (DFG Grant ZE467/6-1 and Graduate Program IRTG 1525). We thank Anna Busch for excellent technical assistance, Wolfgang Kaisers (Center for Bioinformatics and Biostatistics, HHU Düsseldorf), Markus Kollmann, and Martin Lercher for support on statistical issues and Jean-Pierre Métraux for kindly providing ics1 ics2 seeds. We thank Michael Hartmann and Philippe Reymond for critically reading the manuscript.
AUTHOR CONTRIBUTIONS
F.B., A.-C.D., K.G., and S.S. performed the experiments. A.B., F.B., and J.Z. contributed to RNA-seq analyses and evaluation of expression data. F.B. and A.B. assisted J.Z. in manuscript preparation. J.Z. designed the research and wrote the manuscript.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- SAR
systemic acquired resistance
- SA
salicylic acid
- Pip
pipecolic acid
- Psm
Pseudomonas syringae pv maculicola ES4326
- PCA
principle component analysis
- FDR
false discovery rate
- GO
Gene Ontology
- IRGA
infrared gas analysis
- NBS
nucleotide binding site
- RLK
receptor-like protein kinase
- JA
jasmonic acid
- BTH
S-methyl-1,2,3-benzothiadiazole-7-carbothioate
- MAPK
mitogen-activated protein kinase
- rpm
reads per million
- HAI
hours after inoculation
References
- Acharya B.R., Raina S., Maqbool S.B., Jagadeeswaran G., Mosher S.L., Appel H.M., Schultz J.C., Klessig D.F., Raina R. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50: 488–499. [DOI] [PubMed] [Google Scholar]
- Aliferis K.A., Faubert D., Jabaji S. (2014). A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One 9: e111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Attaran E., Zeier T.E., Griebel T., Zeier J. (2009). Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers G.J., Jaskiewicz M., Liu Y., Underwood W.R., He S.Y., Zhang S., Conrath U. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bisgrove S.R., Simonich M.T., Smith N.M., Sattler A., Innes R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bowling S.A., Clarke J.D., Liu Y., Klessig D.F., Dong X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Burow M., Busch W., Carlborg Ö., Denby K.J., Glazebrook J., Hamilton E.S., Harmer S.L., Haswell E.S., Maloof J.N., Springer N.M., Kliebenstein D.J. (2015). Reassess the t test: Interact with all your data via ANOVA. Plant Cell 27: 2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach H.H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol. 165: 791–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquist H.P. (1991). Lysine-pipecolic acid metabolic relationships in microbes and mammals. Annu. Rev. Nutr. 11: 435–448. [DOI] [PubMed] [Google Scholar]
- Bruce T.J., Matthes M.C., Napier J.A., Pickett J.A. (2007). Stressful “memories” of plants: evidence for possible mechanisms. Plant Sci. 173: 603–608. [Google Scholar]
- Chaturvedi R., Krothapalli K., Makandar R., Nandi A., Sparks A.A., Roth M.R., Welti R., Shah J. (2008). Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54: 106–117. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhou Y., Yang Y., Chi Y.J., Zhou J., Chen J.Y., Wang F., Fan B., Shi K., Zhou Y.H., Yu J.Q., Chen Z. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159: 810–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16: 524–531. [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Friedrich L., Ryals J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92: 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey D.A., Klessig D.F. (2012). SOS - too many signals for systemic acquired resistance? Trends Plant Sci. 17: 538–545. [DOI] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207: 79–87. [DOI] [PubMed] [Google Scholar]
- Dey S., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166: 2133–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.P., Schulze W.X., Romeis T. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A., Parker J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., Krishnamurthy N., Tör M., Sjölander K.V., Jones J.D.G. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., Dong X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Fujioka S., Sakurai A. (1997). Conversion of lysine to L-pipecolic acid induces flowering in Lemna paucicostata 151. Plant Cell Physiol. 38: 1278–1280. [Google Scholar]
- Garcion C., Baltensperger R., Fournier T., Pasquier J., Schnetzer M.A., Gabriel J.P., Métraux J.-P. (2006). FiRe and microarrays: a fast answer to burning questions. Trends Plant Sci. 11: 320–322. [DOI] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J.P. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gruner K., Griebel T., Návarová H., Attaran E., Zeier J. (2013). Reprogramming of plants during systemic acquired resistance. Front. Plant Sci. 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.N., Spenser I.D. (1969). Biosynthesis of the piperidine nucleus. The mode of incorporation of lysine into pipecolic acid and into piperidine alkaloids. J. Biol. Chem. 244: 88–94. [PubMed] [Google Scholar]
- Hilfiker O., Groux R., Bruessow F., Kiefer K., Zeier J., Reymond P. (2014). Insect eggs induce a systemic acquired resistance in Arabidopsis. Plant J. 80: 1085–1094. [DOI] [PubMed] [Google Scholar]
- Jing B., Xu S., Xu M., Li Y., Li S., Ding J., Zhang Y. (2011). Brush and spray: a high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol. 157: 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. (2009). Priming in systemic plant immunity. Science 324: 89–91. [DOI] [PubMed] [Google Scholar]
- Kauss H., Theisinger-Hinkel E., Mindermann R., Conrath U. (1992). Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 2: 655–660. [Google Scholar]
- Kim T.H., Kunz H.H., Bhattacharjee S., Hauser F., Park J., Engineer C., Liu A., Ha T., Parker J.E., Gassmann W., Schroeder J.I. (2012). Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 24: 5177–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M., Vorwerk S., Masur C., Sharifi-Sirchi G., Olivieri N., Schlaich N.L. (2006). A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 47: 629–639. [DOI] [PubMed] [Google Scholar]
- Kohler A., Schwindling S., Conrath U. (2002). Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M.J. (2008). Cross talk in defense signaling. Plant Physiol. 146: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S., L’haridon F., Astier J., Zander M., Abou-Mansour E., Page G., Thurow C., Wendehenne D., Gatz C., Métraux J.-P., Lamotte O. (2011). The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 68: 507–519. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Clément G., Anne P., Routaboul J.M., Guiboileau A., Soulay F., Shirasu K., Yoshimoto K. (2014). Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26: 1857–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P.M., Karpinski S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57: 1795–1807. [DOI] [PubMed] [Google Scholar]
- Mauch F., Mauch-Mani B., Gaille C., Kull B., Haas D., Reimmann C. (2001). Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 25: 67–77. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Memelink J. (2009). Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570. [DOI] [PubMed] [Google Scholar]
- Mishina T.E., Griebel T., Geuecke M., Attaran E., Zeier J. (2008). New insights into the molecular events underlying systemic acquired resistance. In Biology of Plant-Microbe Interactions, Vol. 6, Lorito M., Woo S.L., Scala F., eds (St. Paul, MN: International Society for Molecular Plant-Microbe Interactions; ), paper 81. [Google Scholar]
- Mishina T.E., Zeier J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141: 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50: 500–513. [DOI] [PubMed] [Google Scholar]
- Mölders W., Buchala A., Métraux J.-P. (1996). Transport of salicylic acid in tobacco necrosis virus-infected cucumber plants. Plant Physiol. 112: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.W., Loake G.J., Spoel S.H. (2011). Transcription dynamics in plant immunity. Plant Cell 23: 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison R.I. (1953). The isolation of L-pipecolinic acid from Trifolium repens. Biochem. J. 53: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Grant N., Warner S.A.J., Sugars J.M., White R.F., Draper J. (1996). Salicylic acid potentiates defence gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 9: 550–571. [Google Scholar]
- Návarová H., Bernsdorff F., Döring A.-C., Zeier J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi Y., Okazaki M., Kida T., Nishina Y., Morishita Y., Ogawa T., Suzuki H., Shibata D., Jikumaru Y., Hanada A., Kamiya Y., Shirasu K. (2012). Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24: 3795–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálfi G., Dézsi L. (1968). Pipecolic acid as an indicator of abnormal protein metabolism in diseased plants. Plant Soil 29: 285–291. [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N., Vlot A.C., Feys B.J., Niefind K., Parker J.E. (2011). Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191: 107–119. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., Lorbiecke R., Ouyang B., Pochapsky T.C., Rzewuski G. (2005). The immediate-early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J. 44: 718–729. [DOI] [PubMed] [Google Scholar]
- Schenk S.T., Hernández-Reyes C., Samans B., Stein E., Neumann C., Schikora M., Reichelt M., Mithöfer A., Becker A., Kogel K.H., Schikora A. (2014). N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26: 2708–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich N.L. (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12: 412–418. [DOI] [PubMed] [Google Scholar]
- Serrano M., Wang B., Aryal B., Garcion C., Abou-Mansour E., Heck S., Geisler M., Mauch F., Nawrath C., Métraux J.-P. (2013). Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 162: 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Zeier J. (2013). Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V., Leon J., Raskin I. (1995). Is salicylic acid a translocated signal of systemic acquired resistance in tobacco? Plant Cell 7: 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Yekondi S., Chen P.W., Tsai C.H., Yu C.W., Wu K., Zimmerli L. (2014). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26: 2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.T., Lu H., Greenberg J.T. (2004a). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.T., Lu H., McDowell J.M., Greenberg J.T. (2004b). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40: 200–212. [DOI] [PubMed] [Google Scholar]
- Sticher L., Mauch-Mani B., Métraux J.P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35: 235–270. [DOI] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36: D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Meyers B.C., Kozik A., West M.A., Morgante M., St Clair D.A., Bent A.F., Michelmore R.W. (2007). Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C., Zhang Z., Shrestha J., Jelenska J., Chinchilla D., Greenberg J.T. (2014). Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26: 4171–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F., Wu H., Richmond T., Redman J.C., Johnson C., Green R., Arias J., Town C.D. (2006). Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 47: 152–162. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Thulke O., Conrath U. (1998). Salicylic acid has a dual role in the activation of defence-related genes in parsley. Plant J. 14: 35–42. [DOI] [PubMed] [Google Scholar]
- Truman W., Bennett M.H., Kubigsteltig I., Turnbull C., Grant M. (2007). Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 104: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Mine A., Bethke G., Igarashi D., Botanga C.J., Tsuda Y., Glazebrook J., Sato M., Katagiri F. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij B., Friedrich L., Morse A., Reist R., Kolditz-Jawhar R., Ward E., Uknes S., Kessmann H., Ryals J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos J., Labbé C., Menzies J.G., Bélanger R.R. (2015). Silicon-mediated resistance of Arabidopsis against powdery mildew involves mechanisms other than the salicylic acid (SA)-dependent defence pathway. Mol. Plant Pathol. 16: 572–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206. [DOI] [PubMed] [Google Scholar]
- Vogel-Adghough D., Stahl E., Návarová H., Zeier J. (2013). Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant Signal. Behav. 8: e26366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S., Farquhar G.D. (1981). Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387. [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahl-Goy P., Métraux J.P., Ryals J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J.Y., Boyle P., Wang Y., Brindle I.D., De Luca V., Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports 1: 639–647. [DOI] [PubMed] [Google Scholar]
- Yan H., Yoo M.J., Koh J., Liu L., Chen Y., Acikgoz D., Wang Q., Chen S. (2014). Molecular reprogramming of Arabidopsis in response to perturbation of jasmonate signaling. J. Proteome Res. 13: 5751–5766. [DOI] [PubMed] [Google Scholar]
- Zacharius R.M., Thompson J.F., Steward F.C. (1954). The detection, isolation and identification of L(-)-pipecolic acid in the non-protein fraction of beans (Phaseolus vulgaris). J. Am. Chem. Soc. 76: 2908–2912. [Google Scholar]
- Zander M., Thurow C., Gatz C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 165: 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J. (2013). New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 36: 2085–2103. [DOI] [PubMed] [Google Scholar]
- Zeier J., Pink B., Mueller M.J., Berger S. (2004). Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683. [DOI] [PubMed] [Google Scholar]
- Zeng W., Brutus A., Kremer J.M., Withers J.C., Gao X., Jones A.D., He S.Y. (2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L., Jakab G., Métraux J.-P., Mauch-Mani B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β -aminobutyric acid. Proc. Natl. Acad. Sci. USA 97: 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35: 345–351. [DOI] [PubMed] [Google Scholar]
- Zoeller M., Stingl N., Krischke M., Fekete A., Waller F., Berger S., Mueller M.J. (2012). Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.