This work describes cytogenetic and transcriptome analyses of repeated WGD events and genome reshuffling supporting preferential retention of stress-responding duplicates showing a signature of strong purifying selection.
Abstract
Whole-genome duplication (WGD) is usually followed by gene loss and karyotype repatterning. Despite evidence of new adaptive traits associated with WGD, the underpinnings and evolutionary significance of such genome fractionation remain elusive. Here, we use Buckler mustard (Biscutella laevigata) to infer processes that have driven the retention of duplicated genes after recurrent WGDs. In addition to the β- and α-WGD events shared by all Brassicaceae, cytogenetic and transcriptome analyses revealed two younger WGD events that occurred at times of environmental changes in the clade of Buckler mustard (Biscutelleae): a mesopolyploidy event from the late Miocene that was followed by considerable karyotype reshuffling and chromosome number reduction and a neopolyploidy event during the Pleistocene. Although a considerable number of the older duplicates presented signatures of retention under positive selection, the majority of retained duplicates arising from the younger mesopolyploidy WGD event matched predictions of the gene balance hypothesis and showed evidence of strong purifying selection as well as enrichment in gene categories responding to abiotic stressors. Retention of large stretches of chromosomes for both genomic copies supported the hypothesis that cycles of WGD and biased fractionation shaped the genome of this stress-tolerant polypolyloid, promoting the adaptive recruitment of stress-responding genes in the face of environmental challenges.
INTRODUCTION
Multiple rounds of whole-genome duplication (WGD) have shaped genome evolution throughout eukaryotes (Lynch and Conery, 2000; Wolfe, 2001; Van de Peer et al., 2009). In particular, WGDs have greatly contributed to plant evolution and speciation (Wood et al., 2009; Jiao et al., 2011). However, to what extent WGD leads to the origin of novel adaptive traits and fosters species radiation remains controversial (Ohno, 1970; Arrigo and Barker, 2012; Soltis et al., 2014). Comprehensive understanding of the molecular and evolutionary processes underlying the preservation and diversification of duplicated genes may thus be central to an integrative consideration of organismal diversity (McGrath and Lynch, 2012; Nei, 2013).
The importance of WGD as a major source of variation has been increasingly recognized with the documentation of various molecular mechanisms operating to restore structurally and functionally diploid states within a few million years (i.e., diploidization; Lim et al., 2007; Doyle et al., 2008; Freeling, 2009; Mandáková et al., 2010; Tayalé and Parisod, 2013). However, numerous duplicated genes escape deletion or other forms of pseudogenization (i.e., a phenomenon collectively referred as fractionation) and are stably retained over the long term (Freeling et al., 2012). Such retained duplicates contribute greatly to the genomes of paleopolyploids or younger mesopolyploids, and several nonmutually exclusive models have been proposed to account for their maintenance after WGD (Flagel and Wendel, 2009; Innan and Kondrashov, 2010). In particular, the overarching gene balance hypothesis posits that alteration of the stochiometry among interacting gene products is detrimental, fostering stable transmission of duplicates through purifying selection (Birchler and Veitia, 2012). Such a central role of gene dosage may explain the preferential retention of highly expressed genes following WGD (Garsmeur et al., 2014; Woodhouse et al., 2014). On the other hand, hypotheses related to change of function assume that one of the duplicated genes explores innovative functions (i.e., neo- or subfunctionalization leading to new or partitioned tasks) through neutral processes and positive selection (McGrath and Lynch, 2012). Various mechanisms certainly support the long-term retention of duplicates, but no consensus has yet emerged and their impact on species diversification remains elusive.
Analysis of plant genomes provided evidence of independent WGD events close to the Cretaceous-Paleogene boundary, supporting the origin of several paleopolyploids during the major extinction crisis that occurred approximately 65 million years ago (MYA; Fawcett et al., 2009; Vanneste et al., 2014b). The β-WGD event preceding the diversification of core Brassicales was postulated to be a result of this burst of polyploidy, but a recent study on this clade estimated this WGD to have occurred 90 MYA (Edger et al., 2015). The radiation of all Brassicaceae clades took place after a subsequent α-WGD event that is congruently dated around 30 MYA by independent studies (Edger et al., 2015; Hohmann et al., 2015). In addition to these multiple rounds of WGD having shaped the paleopolyploid crucifer genomes, transcriptome analysis in 23 Brassicaceae species recently revealed the origin of several mesopolyploids during the late Miocene (i.e., 10 to 5 MYA; Kagale et al., 2014). Despite apparently nonrandom distribution of WGD in time, the mechanisms promoting the radiation of polyploids remain poorly understood (Parisod et al., 2010; Vanneste et al., 2014a).
The genus Biscutella (Brassicaceae) comprises ∼50 species covering the whole Mediterranean basin and is mostly diversified in Southwestern Europe (Jalas et al., 1996). It apparently diverged relatively early from other Brassicaceae (Couvreur et al., 2010) and shows remarkable genomic variation, comprising species with chromosome numbers of either x = 6, x = 8, or x = 9 (Olowokudejo, 1986). Only specific taxa with x = 9 extended northward of the Mediterranean area and reached high elevation in central Europe. Accordingly, the Buckler mustard species complex (Biscutella laevigata) was postulated as case study of adalpine taxon having colonized the Alps in relation with autopolyploidy (i.e., WGD; Schönfelder, 1968; Favarger, 1971). This taxon indeed currently occurs as diploid populations (2n = 2x = 18) in thermophilous habitats of never-glaciated areas of central Europe, whereas multiple autotetraploid lineages (2n = 4x = 36) have established in a wide range of alpine habitats during the glacial cycles of the late Quaternary (Manton, 1937; Tremetsberger et al., 2002; Parisod and Besnard, 2007). Here, we use comparative chromosome painting in diploids and transcriptomics in autopolyploids of the Buckler mustard species to infer recurrent WGD events. Biased fractionation resulted in the retention of duplicates that show evidence of selection and that are preferentially involved in responses to ecological stressors. How genetic and environmental constraints may interact during WGD fractionation cycles and shape genomes is discussed.
RESULTS
Karyotype of Buckler Mustard
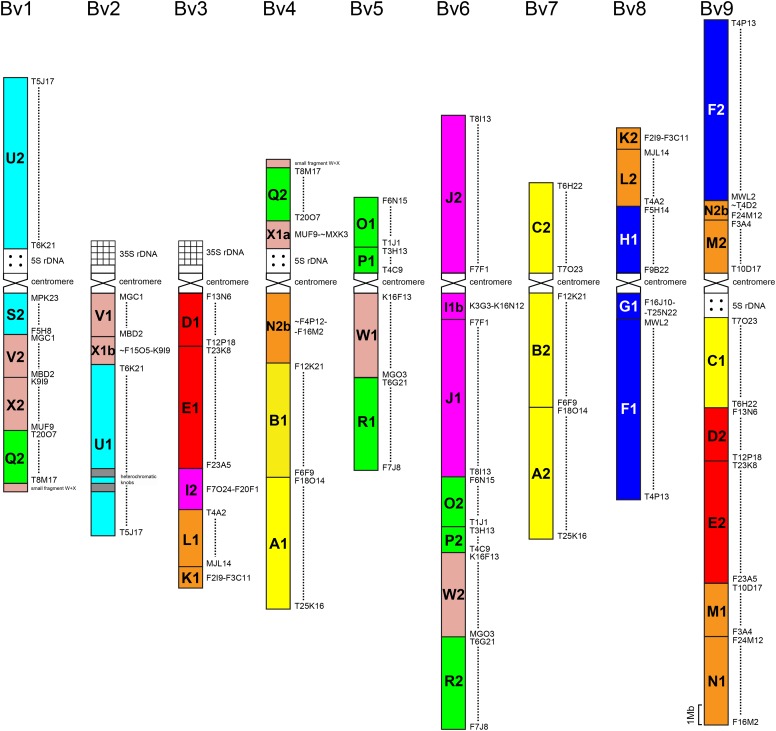
Chromosome painting in diploid Buckler mustard (B. laevigata subsp varia, 2n = 2x = 18) revealed a diploid-like karyotype, with nine bivalents in diakinesis and metaphase I (Figure 1). In addition to distinct domains of pericentromeric heterochromatin, two interstitial heterochromatic knobs were detected on the bottom arm of chromosome Bv2. Two chromosome pairs bear terminal 35S rDNA (Bv2 and Bv3), whereas 5S rDNA loci were observed in the pericentromeric region on three chromosomes (Bv1, Bv4, and Bv9). Given the lack of genetic map, the ancestral crucifer karyotype (ACK) with 24 genomic blocks (GBs) on eight chromosomes (AK1-8; Schranz et al., 2006) served as a reference for the construction of the B. laevigata karyotype using comparative chromosome painting (CCP; Figure 1). Although six copies of the shortest GBs (G2, H2, part of I1 = I1a, S1, T1, and T2) could not be placed into the map, all the remaining GBs (87.5%) were found duplicated. Homoeologous Arabidopsis thaliana BAC contigs indeed hybridized to two different chromosomes within haploid pachytene complements of diploid Biscutella (only both copies of GB J hybridized to the same chromosome). The GB duplicates slightly but consistently differed in length and fluorescence intensity; the longer and brighter copy was labeled as #1 (Figure 1).
Figure 1.
Karyotype of Buckler Mustard.
Cytomolecular comparative map of B. laevigata subsp varia (2n = 2x = 18) based on chromosome painting analyses. Colors of 48 GBs (A to X) reflect their position on the eight chromosomes AK1 to AK8 in the ACK. The longer and more fluorescent GB duplicates are labeled as #1 and the shorter and less fluorescent copies as #2. Six GBs (G2, H2, I1a, S1, T1, and T2) cannot be localized and are not considered. The size of GBs proportionally corresponds to the size of homoeologous blocks in the Arabidopsis genome, as delimited by A. thaliana BAC clones (Supplemental Table 4).
Several ancestral GB associations matching the ACK with eight chromosomes have been detected in the Biscutella karyotype. The characteristic GB association D/E of ancestral chromosome AK2 was located on chromosomes Bv3 and Bv9, whereas AK1-derived blocks A and B (and C) were together on Bv4 and Bv7. Similar to the ancestral chromosome AK4, genomic copies of J (J#1 and #2) as well as a part of I (I#1b) were on chromosome Bv6, whereas AK3-derived GBs F/G/H were together on Bv8. In contrast, chromosomes Bv5 and Bv6 possessed the O/P/W/R association that is diagnostic for ACK-derived proto-Calepineae karyotype (PCK) with n = 7 (Mandáková and Lysak, 2008). The origin of chromosome Bv6 apparently involved two centromere inactivation/deletion events preserving the collinearity of ancestral chromosomes PCK4 (AK4) and PCK6 (AK6/8). Furthermore, Bv1 and Bv4 presented PCK-like association of blocks Q and X. These O/P/W/R and Q/X block combinations thus link the ancestral Biscutella genome to an eight-chromosome precursor of PCK (Supplemental Figure 1).
Inference of WGDs with Transcriptomics
Genome dynamics involving WGD fractionation cycles were confirmed by the analysis of the Buckler mustard transcriptome that was iteratively assembled from long 454 reads and short SOLiD reads from leaf and root tissues of B. laevigata subsp laevigata (2n = 4x = 36; the autopolyploid relative of B. laevigata subsp varia; Supplemental Tables 1 and 2). After removal of perfectly duplicated contigs as well as contigs smaller than 150 bp, the transcriptome yielded 102,998 contigs (mean length, 413.50 bp; N50, 573 bp; for a total of 42,589,854 bp). BLASTx against the TAIR10 peptide database revealed 72,273 (70.1%) and 64,645 (62.7%) contigs annotated at e-values of e-5 and e-10, respectively. These contigs represented 16,389 unigenes (49% of genes in Arabidopsis) distributed among all GBs (Supplemental Figure 2), highlighting the representativeness and high quality of the inferred transcriptome. Genic contigs were translated into their reading frame, yielding 57,783 valid ESTs (thereafter called transcripts), among which 26,815 were grouped into 3141 families with at least two members. PAML estimated divergence between pairs of transcripts in each family for both nonsynonymous (Ka) and synonymous (Ks) substitutions.
Mixture models fitted four significant normal distributions on the distribution of Ks between 20,890 pairs of transcripts larger than 450 bp showing Ks < 2 (Table 1; Supplemental Figure 3), indicating large-scale gene duplications in the past of Buckler mustard. The oldest significant peak at mean Ks = 1.678 likely corresponds to the β-WGD event specific for the core Brassicales (Bowers et al., 2003; Lysak and Koch, 2011), whereas the peak at Ks = 0.886 matches the α-WGD event at the basis of the Brassicaceae. Two significant peaks characteristic of the clade analyzed here were further highlighted and strongly supported: (1) a WGD event (called Bl-m-WGD, referring to a mesopolyploidy event in the past of B. laevigata) that yielded duplicates with a mean Ks = 0.235 and that was also apparent in the karyotype of diploid B. laevigata subsp varia (Figure 1) and (2) a WGD event (called Bl-n-WGD, referring to a neopolyploidy event in the past of B. leavigata) at Ks = 0.032 that was only apparent in the transcriptome of autotetraploid B. laevigata subsp laevigata. As a number of duplicates from this latter n-WGD event may have collapsed into consensus sequences, it was not further investigated here. Using the α-WGD as calibration point (Hohmann et al., 2015), coarse dating of those WGD events pointed to ∼8.6 MYA for the Bl-m-WGD event and 1.2 MYA for the Bl-n-WGD event (Table 1). Such dating is consistent with evidence of mesopolyploidy events in other species (Kagale et al., 2014) and autopolyploidy in B. laevigata (Manton, 1937), respectively.
Table 1. Mixture Model of Synonymous Substitution (Ks) Distribution among Pairs of Transcripts Identifying Significant Peaks Indicative of WGD Events in Buckler Mustard (B. laevigata subsp laevigata; 2n = 4x = 36).
| WGD Event | Bl-n | Bl-m | α | β |
|---|---|---|---|---|
| Mean Ks ± sd | 0.032 ± 0.023 | 0.235 ± 0.121 | 0.886 ± 0.366 | 1.678 ± 0.198 |
| Estimated age ± sd (MYA)a | 1.17 ± 0.84 | 8.59 ± 4.42 | 32.42 | 61.36 ± 7.24 |
Means and sd of Ks-based age of duplicates were computed by 100 bootstraps and used to estimate the age of WGD events.
Calibration according to the α-WGD event that occurred 32.42 MYA following Hohmann et al. (2015).
Genome Restructuring and Retention of Duplicates
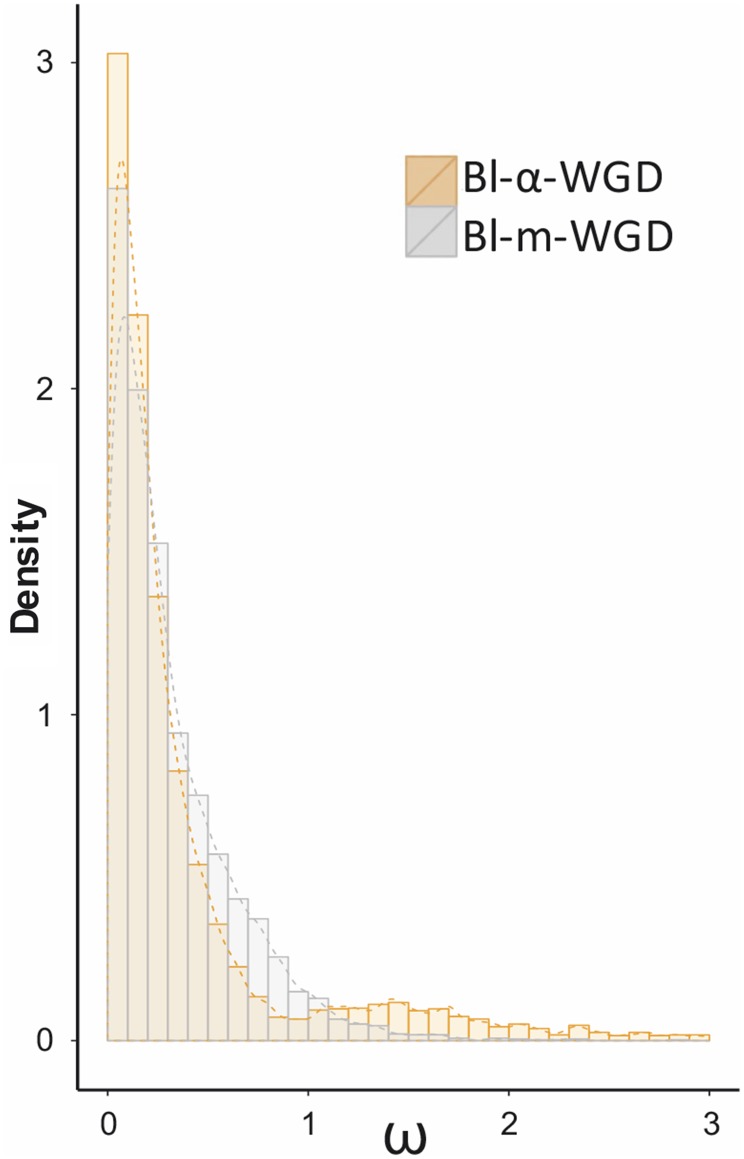
Both the α-WGD and the Bl-m-WGD events provided enough resolution to assess the evolutionary forces involved in the retention of duplicated transcripts. The Ka/Ks ratio (i.e., ω) between the 5126 pairs of transcripts having their origin after the α-WGD event (i.e., α-duplicates showing Ks within the 95% confidence interval of the peak) and between the 3933 pairs from the Bl-m-WGD event (i.e., Bl-m-duplicates with Ks within the 95% confidence interval of the peak) highlighted most duplicates with ω lower than 1 (Figure 2). Such depletion of nonsynonymous substitutions indicates that purifying selection is likely prevalent among retained duplicates. In contrast, only 592 pairs of α-duplicates (11.5%) and 150 pairs of Bl-m-duplicates (3.8%) presented evidence of positive selection with ω higher than 1. The α-WGD event thus yielded a significantly higher proportion of duplicated transcripts with evidence of positive selection than the younger Bl-m-WGD event (two-sided proportion test, z = 12.68, P < 0.01).
Figure 2.
Evidence of Selection among Duplicates.
Ratio (ω) of nonsynonymous and synonymous substitutions between duplicated transcripts having originated at the α-WGD and the Bl-m-WGD events (colored according to the panel) in Buckler mustard.
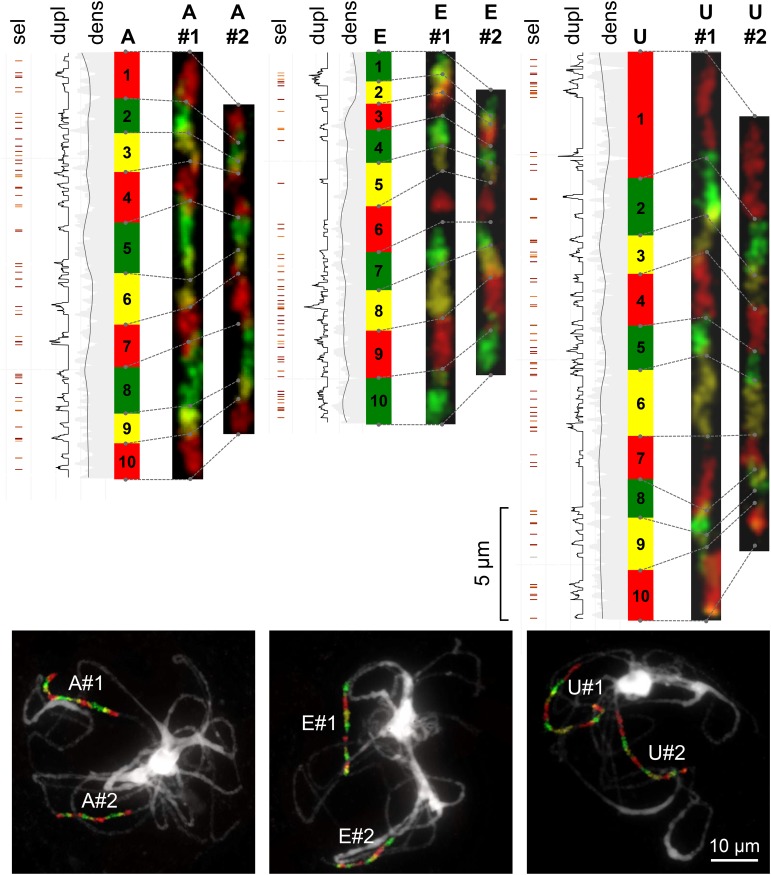
Comparative chromosome painting showed that Bl-m-duplicated GBs differed slightly but consistently in their physical length and corrected total fluorescence (see Figure 3 for examples). The longer and more fluorescent copy (labeled #1) of GBs A, B, E, J, O, P, R, and U was on average 1.55 times longer and presented 1.53 times higher fluorescence than the other copy (labeled #2; Table 2), indicating preferential fractionation of one copy. Whether the same subgenome is consistently more fractionated cannot be assessed with available data and must be further evaluated through phylogenomic approaches. Assuming conservation of the gene order within BACs between Arabidopsis and Biscutella, transcripts were mapped on the karyotype to gather additional insights on the mechanisms underlying fractionation. The proportion of duplicated transcripts among GBs strongly correlated with neither the ratio of fluorescence (robust regression, P = 0.54) nor the ratio of physical length between the longer and the shorter duplicated GB (robust regression, P = 0.047, R2 = 0.52). Despite uniform density of transcripts along those GBs, retained duplicates appeared mostly as stretches of nearby loci conserved under purifying selection (Supplemental Figure 2). Duplicated chromosome segments retained along GBs A, E, and U finely colocalized with inferred sections rich in duplicated transcripts (Figure 3). Mechanisms responsible for uneven reorganization of duplicates across the genome and particularly shorter and less fluorescent regions remain elusive and must be assessed with transcriptome and genome sequencing.
Figure 3.
Biased Fractionation following the Bl-m-WGD Event in Buckler Mustard.
Fine-scale CCP along GBs A, E, and U is associated with the distribution of retained Bl-m-duplicates in the transcriptome. Transcriptome data are presented on the left of each GB, with the density of transcripts (dens) and of retained duplicates (dupl) as well as evidence of weak (light red) to strong (dark red) purifying selection (sel) along retained duplicates. Using CCP, the longest and most fluorescent GB copy was labeled as #1, and pachytene chromosomes (shown at the bottom) were straightened and displayed alongside the transcriptome data according to differentially painted sub-blocks (Supplemental Data Set 1).
Table 2. Length, Corrected Total Fluorescence, and Duplicated Genes among Selected GBs in Buckler Mustard.
| Average Length (μm)a |
Average CTFa |
Transcriptomeb |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GB | #1 | #2 | Ratio | #1 | #2 | Ratio | Unigenes | Duplicated Loci | Retained (%) |
| A | 20.5 | 14.6 | 1.40 | 4.7E-04 | 3.0E-04 | 1.58 | 988 | 54 | 5.46 |
| B | 16.8 | 17.8 | 1.06 | 5.9E-04 | 3.8E-04 | 1.54 | 696 | 31 | 4.45 |
| E | 19.6 | 15.6 | 1.25 | 5.0E-04 | 3.4E-04 | 1.48 | 818 | 50 | 6.11 |
| J | 22.0 | 17.4 | 1.26 | 8.0E-04 | 3.7E-04 | 2.19 | 911 | 45 | 4.94 |
| O | 8.0 | 4.0 | 2.00 | 4.1E-04 | 3.5E-04 | 1.18 | 237 | 11 | 4.64 |
| P | 6.0 | 2.0 | 3.00 | 9.4E-05 | 1.0E-04 | 0.93 | 135 | 5 | 3.70 |
| R | 16.0 | 11.0 | 1.45 | 3.7E-04 | 1.6E-04 | 2.22 | 1116 | 57 | 5.11 |
| U | 24.8 | 22.0 | 1.13 | 5.8E-04 | 5.3E-04 | 1.09 | 1330 | 75 | 5.64 |
Averaged measure from painting of multiple pachytene chromosomes, with #1 representing the longer and more fluorescent copy. CTF, corrected total fluorescence.
Percentage retained as the proportion of unigenes in the transcriptome that were located in the corresponding GB and showed evidence of Bl-m-duplicates.
Functionally Biased Retention
Transcripts corresponding to 12,848 unigenes did not group into a family. Although transcriptome data can hardly offer conclusive evidence, they were enriched in five Gene Ontology (GO) categories (i.e., DNA recombination, DNA dependent replication, DNA repair, response to DNA damage stimulus, and DNA metabolic process; Supplemental Table 3) compared with transcripts showing evidence of retained duplicates. These functional categories show consistent restoration of single copy states in flowering plants (De Smet et al., 2013).
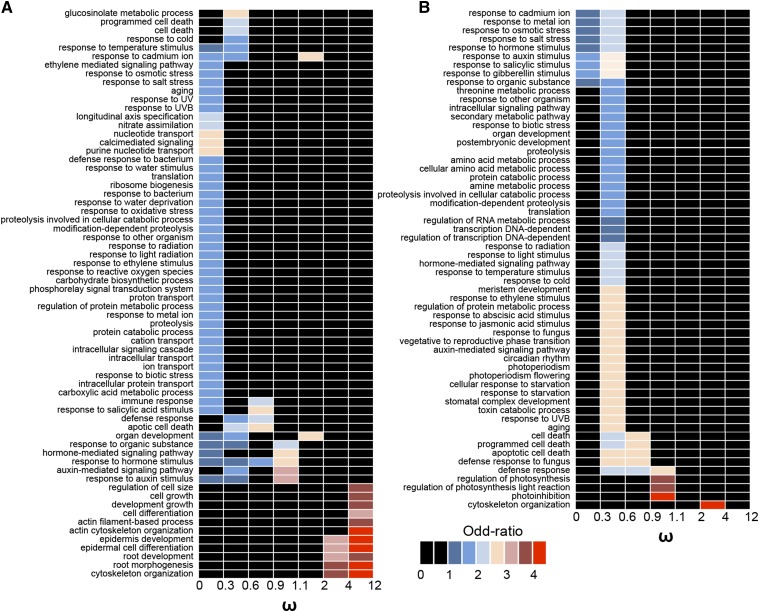
Using high-quality GO annotations from Arabidopsis as source of functional information for the related Buckler mustard, retained α-duplicates and Bl-m-duplicates with evidence of neutral divergence or weak selection (ω between 0.6 and 2) showed significant overrepresentation for only a few functional categories (Figure 4). In contrast, duplicates showing evidence of strong selection were significantly enriched in particular GO categories, providing insights on the selective agents having played an important role for gene retention. Duplicates retained under strong positive selection (ω higher than 2) after both the α-WGD and the m-WGD events were related to cytoskeleton organization, whereas duplicates showing evidence of strong purifying selection were significantly enriched in GO functions related to responses to ecological stressors. In particular, the vast majority of Bl-m-duplicates retained under strong purifying selection (i.e., ω lower than 0.6) were chiefly enriched for GO categories related to environmental stresses, such as temperature, light, or soil chemistry (Figure 4).
Figure 4.
Overrepresented GO Categories of α-Duplicates and Bl-m-Duplicates Retained under Different Selection Regimes in Buckler Mustard.
α-Duplicates (A) and Bl-m-duplicates (B). The strength of selection between duplicates is considered according to ω (i.e., Ka/ Ks ratio): strong purifying selection (ω < 0.6) versus weak selection or neutral divergence (ω between 0.6 and 2) versus strong positive selection (ω > 2). Significantly overrepresented categories are shown by colors according to the panel and corresponding to odd ratios quantifying the enrichment of the particular GO term (i.e., effect size), whereas categories in black are not significant at corrected α = 0.05.
DISCUSSION
Recurrent WGDs during Environmental Crises
Comparative chromosome painting in the Buckler mustard highlighted arrangements of GBs characteristic of both the ACK (n = 8; Schranz et al., 2006) and the more derived PCK (n = 7; Mandáková and Lysak, 2008). Presence of PCK-specific GB associations (O/P/W/R and Q/X) but absence of distinctive PCK5-like association (V/K/L/Wa/Q/X) indicate divergence of the ancestral Biscutella karyotype from the ACK before the establishment of the PCK genome. Phylogenetic connection of the Biscutelleae tribe to those of the extended lineage II remains to be clarified (Couvreur et al., 2010), but CCP supports early divergence of an ancestral Biscutella genome with eight chromosomes. Accordingly, the Bl-m-WGD event likely established a tetraploid genome with 16 chromosomes that underwent genome fractionation, with massive chromosome reshuffling and chromosome number reduction toward n = 9 in B. laevigata and n = 8 and 6 in other Biscutella species (Supplemental Figure 1). Future phylogenomic work will address the progenitor(s) of this mesopolyploid lineage, but we note that related genera present high chromosome numbers (e.g., Lunaria with n = 14, 15; Couvreur et al., 2010), suggesting that the Bl-m-WGD event may predate the diversification of the Biscutella genus.
The transcriptome of Buckler mustard assembled by combining advantages of short and long reads into a set of long and framed transcripts largely confirmed cytogenetic conclusions and offered additional insights on the evolutionary processes underlying WGD fractionation cycles. Consistent peaks in the distribution of Ks between duplicated transcripts highlighted four WGD events in the history of Buckler mustard. In addition to evidence matching with the well-known core Brassicales-specific β-WGD event and the 30 million-year-old α-WGD event that predates the radiation of the Brassicaceae (Bowers et al., 2003; Lysak and Koch, 2011; Schranz et al., 2012), the Ks distribution pointed to two younger peaks (Bl-m-WGD at Ks = 0.235 and Bl-n-WGD at Ks = 0.032) indicative of recurrent WGD events in the more recent past of B. laevigata.
According to the dating of WGD events across angiosperms and Brassicaceae (Lysak and Koch, 2011; Hohmann et al., 2015), the mesopolyploidy Bl-m-WGD event (∼8 MYA) dates from the late Miocene era (5.3 to 11.6 MYA), when several other Brassicaceae species underwent WGD events (Kagale et al., 2014). This period was characterized by increasing seasonal aridity as well as major climatic changes such as the Messinian salinity crisis that strongly impacted Mediterranean vegetation (Blondel et al., 2010). It thus seems likely that environmental changes favored the Bl-m-WGD event among Biscutella ancestors. Evidence of the Bl-m-WGD event in the karyotype of the diploid B. laevigata subsp varia (2n = 2x = 18) indicates that the latter Bl-n-WGD event corresponds to the recent origin of autotetraploids B. laevigata subsp laevigata (2n = 4x = 36) that occurred during the Pleistocene ice ages (Manton, 1937; Parisod and Besnard, 2007). The n-WGD event thus also points to a period with major environmental changes across the Buckler mustard distribution range. Although Ks-based estimates offer rough dates to be taken with caution, recurrent WGD events in Buckler mustard appear consistent with data available from other species (Kagale et al., 2014; Vanneste et al., 2014b; Hohmann et al., 2015) and support the hypothesis that ecological challenges sustain evolution through polyploidy (Parisod et al., 2010; Vanneste et al., 2014a).
Recurrent WGDs followed by chromosome repatterning and descending dysploidy leading to diploid-like chromosome numbers within a few million years, as observed here, are fully congruent with genome dynamics reported in other Brassicaceae clades (Bowers et al., 2003; Mandáková et al., 2010; Chalhoub et al., 2014). Compared with other species having undergone a WGD event at the same time and still showing polyploid chromosome numbers (Kagale et al., 2014), Buckler mustard has gone through intense karyotype reshuffling. Similarly remarkable post-WGD genome evolution was also reported in the genus Brassica (Chalhoub et al., 2014). However, unlike the mesopolyploid Brassica genomes that involved genome triplication as well as subsequent allopolyploidy (Tang et al., 2012), B. laevigata underwent tetraploidization and autopolyploidy, ideally complementing investigation of the processes shaping polyploid genomes.
Biased Genome Fractionation over Time
The transcriptome of B. laevigata shed further light on the mechanisms underlying genome reorganization after recurrent WGDs by comparing duplicated transcripts that originated at the α- and Bl-m-WGD events. Pairs of transcripts analyzed here are predominantly derived from those WGD events (Supplemental Analysis) and, although specific loci may find their origin in other types of duplication, WGD appears as likely responsible for patterns observed in the genome of Buckler mustard. The Ka/Ks ratio (ω) highlighted that the majority of duplicates retained after both WGD events showed evidence of selection, matching predictions of biased fractionation over evolutionary times (Innan and Kondrashov, 2010; McGrath and Lynch, 2012). In particular, retained genes with evidence of positive selection (ω > 1) were significantly more frequent among older duplicates (i.e., retained after the α-WGD) than younger ones (Bl-m-WGD), matching a lag-time model whereby adaptive recruitment of innovative duplicates occurs long after their origin (Schranz et al., 2012; Tank et al., 2015). Remarkably, only duplicates related to cytoskeleton organization were retained under strong positive selection after both the α- and m-WGD events. With increased cell volume being one of the few constant alterations induced by polyploidy (Ramsey and Schemske, 2002), to what extent adaptive diversification of cytoskeleton genes may support the necessary scaling of intracellular structures after WGDs should be further examined (Hazel et al., 2013).
The vast majority of retained duplicates in B. laevigata presented evidence of purifying selection, matching the hypothesis that dosage effects are crucial in maintaining functional traits after WGDs (Birchler and Veitia, 2012). In the absence of a complete atlas of transcripts coupled with a high-quality genome sequence, only partial conclusions can be drawn. In particular, that specific duplicates have evolved partially different functions is not excluded, but high-quality GO annotations from Arabidopsis are expected to provide reliable insights in this non-model relative (Primmer et al., 2013). Remarkably, the relatively young Bl-m-duplicates with evidence of strong purifying selection were specifically enriched in functional categories related to stressing environments. Despite offering only candidate genes and genetic pathways of adaptive significance, such quantitative inference strongly supports that environmental constraints played an influential role in shaping genome fractionation in Buckler mustard. Accordingly, ploidy-dependent uptake of soil minerals and tolerance to salinity has recently been demonstrated (Chao et al., 2013). Here, a majority of the Bl-m-duplicates involved in responses to soil chemistry showed evidence of purifying selection, indicating that increased dosage may have enhanced tolerance of Biscutella to stressing edaphic conditions (Veitia et al., 2013), which is coherent with the species adaptation to toxic soils such as found on serpentine or mines (Gasser, 1986; Babst-Kostecka et al., 2014, and references therein). Several of the retained duplicates involved in responses to light and temperatures show a similar pattern and also represent strong candidates having been adaptively recruited during radiation of Buckler mustard across alpine habitats (Körner, 2003). Shifts from Mediterranean to alpine environments have indeed been postulated for several polyploid species (Favarger, 1971), matching the observed rounds of WGD fractionation in Buckler mustard. The underpinnings of the colonization of new environments remain elusive, but available insights suggest that adaptive retention of extra doses of functional duplicates likely supported establishment into otherwise harsh conditions. This hypothesis remains largely to be tested.
Cycles of WGD: Fractionation
Fractionation in Buckler mustard resulted in the retention of large chromosome segments interrupted by GB sections presenting decay of fluorescence (Figure 3). Molecular mechanisms driving such uneven genome reorganization are unknown, but likely involved DNA loss and dynamics of transposable elements such as regularly reported in this and other polyploids (Freeling et al., 2012; Woodhouse et al., 2014; Bardil et al., 2015). Recurrent WGD fractionation cycles point here to a balance between conservation of specific loci and high sequence turnover across other regions, matching predictions of Conant et al. (2014) that duplicates are initially preserved under purifying selection before some duplicates accumulate mutations leading to functional changes to be recruited under positive selection. The surmised impact of environmental constraints in shaping fractionation after the breaking of genetic constraints by WGD likely explains patterns of polyploid radiation and calls for additional studies addressing the contribution of ecological stresses during genome evolution (Wagner, 2011; Nei, 2013).
METHODS
Plant Materials and Cultivation Conditions
Four individuals of Biscutella laevigata subsp laevigata (2n = 4x = 36) were collected in a previously described population (Parisod and Christin, 2008), grown outdoors for 1 year in a common garden (Botanical Garden of Neuchâtel, Switzerland), and analyzed there.
CCP
Whole inflorescences from individuals of B. laevigata subsp varia (2n = 2x = 18), used by Bardil et al. (2015), were fixed in freshly prepared ethanol:acetic acid fixative (3:1) overnight, transferred into 70% ethanol, and stored at −20°C until use. Selected inflorescences were rinsed in distilled water and citrate buffer (10 mM sodium citrate, pH 4.8) and digested in a 0.3% mix of pectolytic enzymes (cellulase, cytohelicase, and pectolyase; all from Sigma-Aldrich) in citrate buffer for ∼3 h. Individual anthers were disintegrated on a microscopic slide by a needle in a drop of water. The suspension was then softened by adding 20 μL of 60% acetic acid, spread by stirring with a needle on a hot plate at 50°C for 2 min, fixed by adding 100 μL of ethanol:acetic acid fixative (3:1), postfixed in 4% formaldehyde in water (10 min), and air-dried. Chromosome preparations were pretreated with RNase (100 µg/mL; AppliChem) and pepsin (0.1 mg/mL; Sigma-Aldrich) dissolved in 0.01 M HCl for 10 min, postfixed in 4% formaldehyde in 2× SSC (2× SSC is 3M sodium chloride and 300 mM trisodium citrate, pH 7.0) for 10 min, and dehydrated in an ethanol series (70, 80, and 96%).
The BAC clone T15P10 (AF167571) bearing 35S rRNA genes was used for in situ localization of 35S rDNA, and the clone pCT 4.2 corresponding to 500-bp 5S rRNA repeat (M65137) was used for localization of 5S rDNA loci. For CCP, a total of 547 Arabidopsis thaliana BAC clones were assembled to represent the 24 GBs of the ancestral crucifer karyotype (Schranz et al., 2006; Supplemental Table 4). To further characterize the orientation and internal structure of particular GBs, the respective BAC contigs were arbitrarily broken into smaller subcontigs and differentially labeled. DNA probes were labeled with biotin-dUTP, digoxigenin-dUTP, or Cy3-dUTP by nick translation as described by Mandáková et al. (2010). Labeled BAC DNAs were pooled, precipitated, and resuspended in 20μL of hybridization mixture (50% formamide and 10% dextran sulfate in 2×SSC) per slide. Labeled probes and chromosomes were denatured together on a hot plate at 80°C for 2 min and incubated in a moist chamber at 37°C for 16 to 72 h. Posthybridization washing was performed in 20% formamide in 2× SSC at 42°C. Immunodetection was performed as follows: biotin-dUTP was detected by avidin-Texas Red (Vector Laboratories) and signals amplified by goat anti-avidin-biotin (Vector Laboratories) and avidin-Texas Red; digoxigenin-dUTP was detected by mouse antidigoxigenin (Jackson ImmunoResearch) and goat anti-mouse-Alexa Fluor 488 (Molecular Probes). Cy3-dUTP-labeled probes were observed directly. Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole (2 μg/mL) in Vectashield (Vector Laboratories). Fluorescence signals were analyzed with an Olympus BX-61 epifluorescence microscope and CoolCube CCD camera (MetaSystems). Images were acquired separately for the four fluorochromes using appropriate excitation and emission filters (AHF Analysentechnik). The four monochromatic images were pseudocolored and merged using Adobe Photoshop CS2 software (Adobe Systems).
The length and fluorescence intensity of selected GBs were measured on average in 10 pachytene chromosomes using Image J software (National Institutes of Health). For determining the level of fluorescence, corrected total fluorescence (Burgess et al., 2010; Potapova et al., 2011) was calculated: corrected total fluorescence = integrated density – (area of selected chromosome segment × mean fluorescence of background readings). BAC painted chromosomes in Figure 3 were straightened using the plug-in “Straighten Curved Objects” (Kocsis et al., 1991) in Image J with sub-blocks differentially labeled according to Supplemental Data Set 1.
cDNA Library Preparation and Transcriptome Sequencing
Tissue from young leaves and roots of the four individuals of B. laevigata subsp laevigata (2n = 4x = 36) were simultaneously collected in liquid nitrogen, and total RNA was extracted using PureLink RNA Mini Kit (Life Technologies), following the manufacturer’s instructions. RNA quality was checked with the Agilent bioanalyzer. A 454 normalized library was prepared on poly(A)-enriched RNAs from root and leaf tissues of one individual that were pooled, fragmented, and single-end sequenced on a quarter of a plate using Roche GS FLX Titanium Series (service provided by Microsynth, following the manufacturer’s instructions). Six quantitative cDNA libraries (four individuals; two with pooled root and leaf; two with root and leaf separated; Supplemental Table 1) were prepared and bar-coded for SOLiD, mixed, and single-end sequenced on six lanes of SOLiD 5500xl (service provided by Microsynth, following the manufacturer’s instructions).
Transcriptome Assembly and Annotation
After trimming of adaptor sequences, reads of low quality or showing GC bias and reads of low complexity or size as well as exact duplicates were removed using PRINSEQ (Schmieder and Edwards, 2011) and FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/).
Transcriptome assembly was performed through six iterative steps using the hybrid mode of MIRA (Chevreux et al., 2004). As a first step, 454 reads were assembled with a SOLiD library to obtain a first set of assembled contigs as well as singletons. Contigs were then used together with the 454 library in a new round of hybrid assembly with a new SOLiD library. Previously assembled contigs that failed to be elongated at this specific step were retrieved from singletons and merged back with contigs for further elongation in a new round of hybrid assembly. This iterative procedure was followed until all SOLiD libraries were included. At each step, summary statistics were computed on the resulting set of contigs to follow improvement of the assembly (Supplemental Table 2). At the final step, singletons and exact duplicates as well as contigs shorter than 150 bp were filtered out to produce the working set of transcripts (i.e., transcriptome).
BLAST against various databases (e.g., nucleotide collection of NCBI) returned hits from various Brassicaceae for the vast majority of assembled contigs (data not shown). We decided to rely on the TAIR10 database and its accurate annotations from Arabidopsis (Lamesch et al., 2012) for subsequent analyses (Primmer et al., 2013). Assembled contigs were annotated by BLASTx (minimum e-values set at e-5 and e-10) against the TAIR 10 peptide database. Reading frames were identified and excised through comparison with the TAIR10 database using Exonerate (Slater and Birney, 2005).
Assuming that gene order within BACs is conserved between Arabidopsis and Biscutella, unigenes were assigned to GBs (Supplemental Table 4) according to their annotation (Schranz et al., 2006). Transcript coverage along GBs was then assessed using overlapping sliding windows of 40 loci from the Arabidopsis reference genome (increment of 10), visualizing the representativeness of the B. laevigata transcriptome. The distribution of retained duplicates along GBs was similarly assessed as the proportion of loci with transcripts showing evidence of Bl-m-duplicates. Coordinates of BAC clones used for GB painting (Supplemental Data Set 1) were used to determine encompassed genes and colocate transcriptome and cytogenetic data.
Inference of WGD from Transcriptome
After having filtered out mitochondrial and plastid genes as well as transposable element containing genes, framed contigs showing higher than 30% sequence similarity over at least 450 bp were grouped into gene families (custom script). Transcripts from clusters with at least two members were aligned using the codon-aware MACSE algorithm (Ranwez et al., 2011). Corresponding phylogenetic trees were inferred with FastTree (Price et al., 2009) to infer synonymous and nonsynonymous substitutions (Ks and Ka, respectively) in PAML using the F3X4 model (Yang, 2007).
The fixation rate of synonymous substitutions being relatively constant, Ks can be used to evaluate the time of duplication between two sequences (Maere et al., 2005). When all genes are duplicated simultaneously during WGD, the Ks distribution of resulting pairs of transcripts is approximately Gaussian (Blanc and Wolfe, 2004). Accordingly, mixture models can infer WGD events by fitting normal distributions to the data (Barker et al., 2009). After removal of identical pairs of contigs (i.e., Ks = 0) as well as pairs with Ks > 2 that proved less reliable for inference of WGDs (Vanneste et al., 2013), normal peaks in the distribution of Ks between members of gene families were accordingly inferred using the package mixtools in R cran (Benaglia et al., 2009). Both Ks and root-squared Ks values were analyzed and gave similar results. The most likely number of normal distributions in the observed distribution of Ks among pairs of transcripts was tested by a parametric bootstrap analysis (100 bootstraps) comparing the likelihood ratio of k versus k + 1 in between k = 1 and k = 10. Then, the iterative “expectation-maximization” algorithm (i.e., normalmixEM procedure) converged to the positions and standard deviations of four normal distributions that were further evaluated by 100 bootstraps.
Converting Ks time equivalents into real time using the α-WGD event (mean Ks = 0.896) as a calibration point offered rough estimates of the time of duplication events. Following Hohmann et al. (2015), we used 32.42 million years as the minimum age of α-WGD event (Lysak and Koch, 2011). A constant accumulation of synonymous mutations among simultaneously duplicated transcripts with Ks < 2 was assumed and used to scale the mean Ks and sd of normal peaks into time since WGD events.
Biased Retention of Duplicated Transcripts
Overrepresented GO categories among framed contigs larger than 150 bp that were not grouped in gene families compared with those having been included in a group (i.e., retained duplicates) was assessed using FatiGo (Al-Shahrour et al., 2004).
Duplicated pairs of transcripts (i.e., contigs grouped in a family) were assigned to their WGD event of origin according to their Ks value caught between the 5% and the 95% quantiles of the corresponding peak. For the α-WGD event (mean Ks = 0.896), this interval was artificially truncated to α-duplicates with Ks ranging from 0.41 to 0.90, avoiding possible inclusion of duplicates from the β-WGD event. For the Bl-m-WGD event (mean Ks = 0.235), duplicates with Ks ranging from 0.085 to 0.37 were considered as Bl-m-duplicates. Difference in the proportion of retained duplicates after the α- versus Bl-m-WGD events was assessed with a two-sided nonparametric proportion test and evolutionary forces underlying their retention were inferred by computing their Ka/Ks ratios (i.e., ω) using PAML (Yang, 2007). The proportion of retained duplicates after the Bl-m-WGD event was associated with ratio of the physical length and painting intensity between duplicated GBs A, B, E, J, O, P, R, and U, using robust regressions (FastLTS approach) with P values assessed through permutation.
Using high-quality annotations of unigenes from the Arabidopsis reference, overrepresented GO categories of duplicated transcripts showing evidence purifying selection (i.e., ω below 0.9: strong from 0 to 0.6 or weak from 0.6 to 0.9), neutral divergence (i.e., ω of 0.9 to 1.1), or positive selection (i.e., ω higher than 1: weak from 1.1 to 2, strong from 2 to 12) compared with the whole transcriptome data set were assessed separately using FatiGo (Al-Shahrour et al., 2004). Significance was assessed according to Fisher’s exact tests and adjusted P values corrected for multiple testing with the false discovery rate procedure (Benjamini and Hochberg, 1995). Odds ratio, quantifying the probability that the list is enriched in a particular term, further provided insights on the strength of the association (i.e., effect size). Significantly overexpressed GO terms at the 0.05 level were retrieved and summarized by their GO classification (ftp://ftp.arabidopsis.org).
Accession Numbers
Sequence data and the assembled transcriptome are under the GenBank BioProject PRJNA302858.
Supplemental Data
Supplemental Figure 1. Scenario of genome evolution in Biscutella from the ancestral crucifer karyotype.
Supplemental Figure 2. Transcriptome data along each genomic block.
Supplemental Figure 3. Distribution of synonymous substitutions between pairs of duplicated transcripts.
Supplemental Table 1. Sequenced cDNA libraries.
Supplemental Table 2. Iterative hybrid assembly of cDNA libraries.
Supplemental Table 3. Overall retention bias of transcripts.
Supplemental Table 4. Genomic blocks of the ancestral crucifer karyotype.
Supplemental Analysis. Phylogenetics of gene pairs under whole-genome duplications.
Supplemental Data Set 1. Genomic sub-blocks used for fine-scale comparative chromosome painting.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for valuable comments on a previous draft of the manuscript and S. Zoller and A. Bardil for their help with bioinformatic analyses. Analyses were performed using facilities of the Genetic Diversity Centre, ETH Zurich. N.A. is funded by a grant from the Swiss National Science Foundation. Work in the Lysak lab was supported by the Czech Science Foundation (Grant 13-10159S) and by the European Social Fund (Projects CZ.1.07/2.3.00/30.0037 and CZ.1.07/2.3.00/20.0189). Work in the Parisod lab was supported by the Velux Stiftung (Project 705) and the Swiss National Science Foundation (Grant PZ00P3-131950).
AUTHOR CONTRIBUTIONS
C.G. and C.P. completed the transcriptome assembly, analysis, and interpretation, with assistance from N.A. T.M. and M.A.L. completed and interpreted the cytogenetics work. C.P. and T.M. integrated transcriptome and cytogenetics data. C.G., M.A.L., and C.P. wrote the manuscript. All authors commented and approved the final manuscript.
Glossary
- WGD
whole-genome duplication
- MYA
million years ago
- ACK
ancestral crucifer karyotype
- GB
genomic block
- CCP
comparative chromosome painting
- GO
Gene Ontology
- PCK
proto-Calepineae karyotype
References
- Al-Shahrour F., Díaz-Uriarte R., Dopazo J. (2004). FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580. [DOI] [PubMed] [Google Scholar]
- Arrigo N., Barker M.S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Babst-Kostecka A.A., Parisod C., Godé C., Vollenweider P., Pauwels M. (2014). Patterns of genetic divergence among populations of the pseudometallophyte Biscutella laevigata from southern Poland. Plant Soil 383: 245–256. [Google Scholar]
- Bardil A., Tayalé A., Parisod C. (2015). Evolutionary dynamics of retrotransposons following autopolyploidy in the Buckler Mustard species complex. Plant J. 82: 621–631. [DOI] [PubMed] [Google Scholar]
- Barker M.S., Vogel H., Schranz M.E. (2009). Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 1: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglia T., Chauveau D., Hunter D.R., Young D.S. (2009). mixtools: An R package for analyzing finite mixture models. J. Stat. Softw. 32: 1–29. [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Birchler J.A., Veitia R.A. (2012). Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel J., Aronson J., Bodiou J.-Y., Boeuf G. (2010). The Mediterranean Region: Biological Diversity in Space and Time. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbé J.-C., Lorca T., Castro A. (2010). Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA 107: 12564–12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Chao D.-Y., Dilkes B., Luo H., Douglas A., Yakubova E., Lahner B., Salt D.E. (2013). Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341: 658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B., Pfisterer T., Drescher B., Driesel A.J., Müller W.E.G., Wetter T., Suhai S. (2004). Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14: 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Birchler J.A., Pires J.C. (2014). Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 19: 91–98. [DOI] [PubMed] [Google Scholar]
- Couvreur T.L.P., Franzke A., Al-Shehbaz I.A., Bakker F.T., Koch M.A., Mummenhoff K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27: 55–71. [DOI] [PubMed] [Google Scholar]
- De Smet R., Adams K.L., Vandepoele K., Van Montagu M.C.E., Maere S., Van de Peer Y. (2013). Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 110: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Flagel L.E., Paterson A.H., Rapp R.A., Soltis D.E., Soltis P.S., Wendel J.F. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Edger P.P., et al. (2015). The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. USA 112: 8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favarger C. (1971). Relations entre la flore méditerranéenne et celle des enclaves à végétation subméditerranéenne d’Europe centrale. Boissiera 19: 149–168. [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Freeling M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Garsmeur O., Schnable J.C., Almeida A., Jourda C., D’Hont A., Freeling M. (2014). Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 31: 448–454. [DOI] [PubMed] [Google Scholar]
- Gasser M. (1986). Genetic-ecological investigations in Biscutella laevigata L. Veröff. Geobot. Inst. ETH. 86: 7–86. [Google Scholar]
- Hazel J., Krutkramelis K., Mooney P., Tomschik M., Gerow K., Oakey J., Gatlin J.C. (2013). Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann N., Wolf E.M., Lysak M.A., Koch M.A. (2015). A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27: 2770–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kondrashov F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Jalas J., Suominen J., Lampinen R. (1996). Atlas Florae Europaeae. (Helsinki, Finland: The Academic Bookstore; ). [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Kagale S., Robinson S.J., Nixon J., Xiao R., Huebert T., Condie J., Kessler D., Clarke W.E., Edger P.P., Links M.G., Sharpe A.G., Parkin I.A.P. (2014). Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis E., Trus B.L., Steer C.J., Bisher M.E., Steven A.C. (1991). Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J. Struct. Biol. 107: 6–14. [DOI] [PubMed] [Google Scholar]
- Körner C. (2003). Alpine Plant Life. (Heidelberg, Germany: Springer Verlag; ). [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.Y., Kovarik A., Matyasek R., Chase M.W., Clarkson J.J., Grandbastien M.A., Leitch A.R. (2007). Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 175: 756–763. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lysak, M.A., and Koch, M.A. (2011). Phylogeny, genome, and karyotype evolution of Crucifers (Brassicaceae). In Genetics and Genomics of the Brassicaceae, R. Schmidt and I. Bancroft, eds (New York: Springer-Verlag), pp. 1–31. [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Joly S., Krzywinski M., Mummenhoff K., Lysak M.A. (2010). Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A. (2008). Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton I. (1937). The problem of Biscutella laevigata L. II. The evidence from meiosis. Ann. Bot. (Lond.) 51: 439–465. [Google Scholar]
- McGrath C.L., Lynch M. (2012). Evolutionary significance of whole-genome duplication. In Polyploidy and Genome Evolution, Soltis P.S., Soltis D.E., eds (Berlin, Heidelberg, Germany: Springer-Verlag; ), pp. 1–20. [Google Scholar]
- Nei M. (2013). Mutation-Driven Evolution. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. (Berlin, Germany: Springer; ). [Google Scholar]
- Olowokudejo J.D. (1986). The infrageneric classification of Biscutella (Cruciferae). Brittonia 38: 86–88. [Google Scholar]
- Parisod C., Besnard G. (2007). Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae). Mol. Ecol. 16: 2755–2767. [DOI] [PubMed] [Google Scholar]
- Parisod C., Christin P.A. (2008). Genome-wide association to fine-scale ecological heterogeneity within a continuous population of Biscutella laevigata (Brassicaceae). New Phytol. 178: 436–447. [DOI] [PubMed] [Google Scholar]
- Parisod C., Holderegger R., Brochmann C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186: 5–17. [DOI] [PubMed] [Google Scholar]
- Potapova T.A., Sivakumar S., Flynn J.N., Li R., Gorbsky G.J. (2011). Mitotic progression becomes irreversible in prometaphase and collapses when Wee1 and Cdc25 are inhibited. Mol. Biol. Cell 22: 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer C.R., Papakostas S., Leder E.H., Davis M.J., Ragan M.A. (2013). Annotated genes and nonannotated genomes: cross-species use of Gene Ontology in ecology and evolution research. Mol. Ecol. 22: 3216–3241. [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D.W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33: 589–639. [Google Scholar]
- Ranwez V., Harispe S., Delsuc F., Douzery E.J.P. (2011). MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6: e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfelder, P. (1968). Chromozomenzahlen einiger Arten des Gattung Biscutella L. Österreichische Botanische Zeitschrift 115: 363–371.
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006). The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Mohammadin S., Edger P.P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Slater G.S., Birney E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Segovia-Salcedo M.C., Jordon-Thaden I., Majure L., Miles N.M., Mavrodiev E.V., Mei W., Cortez M.B., Soltis P.S., Gitzendanner M.A. (2014). Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (). New Phytol. 202: 1105–1117. [DOI] [PubMed] [Google Scholar]
- Tang H., Woodhouse M.R., Cheng F., Schnable J.C., Pedersen B.S., Conant G., Wang X., Freeling M., Pires J.C. (2012). Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank D.C., Eastman J.M., Pennell M.W., Soltis P.S., Soltis D.E., Hinchliff C.E., Brown J.W., Sessa E.B., Harmon L.J. (2015). Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol. 207: 454–467. [DOI] [PubMed] [Google Scholar]
- Tayalé A., Parisod C. (2013). Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenet. Genome Res. 140: 79–96. [DOI] [PubMed] [Google Scholar]
- Tremetsberger K., Konig C., Samuel R., Pinsker W., Stuessy T.F. (2002). Infraspecific genetic variation in Biscutella laevigata (Brassicaceae): new focus on Irene Manton’s hypothesis. Plant Syst. Evol. 233: 163–181. [Google Scholar]
- Van de Peer Y., Maere S., Meyer A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10: 725–732. [DOI] [PubMed] [Google Scholar]
- Vanneste K., Baele G., Maere S., Van de Peer Y. (2014b). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Maere S., Van de Peer Y. (2014a). Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Van de Peer Y., Maere S. (2013). Inference of genome duplications from age distributions revisited. Mol. Biol. Evol. 30: 177–190. [DOI] [PubMed] [Google Scholar]
- Veitia R.A., Bottani S., Birchler J.A. (2013). Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation. Trends Genet. 29: 385–393. [DOI] [PubMed] [Google Scholar]
- Wagner A. (2011). The Origins of Evolutionary Innovations. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Wolfe K.H. (2001). Yesterday’s polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333–341. [DOI] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Cheng F., Pires J.C., Lisch D., Freeling M., Wang X. (2014). Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. USA 111: 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.