Analyses of structure-specific endonuclease mutants show that SEND1 plays a crucial backup role to MUS81 in maintaining genome stability and bring to light its unexpected role in telomere homeostasis.
Abstract
Structure-specific endonucleases act to repair potentially toxic structures produced by recombination and DNA replication, ensuring proper segregation of the genetic material to daughter cells during mitosis and meiosis. Arabidopsis thaliana has two putative homologs of the resolvase (structure-specific endonuclease): GEN1/Yen1. Knockout of resolvase genes GEN1 and SEND1, individually or together, has no detectable effect on growth, fertility, or sensitivity to DNA damage. However, combined absence of the endonucleases MUS81 and SEND1 results in severe developmental defects, spontaneous cell death, and genome instability. A similar effect is not seen in mus81 gen1 plants, which develop normally and are fertile. Absence of RAD51 does not rescue mus81 send1, pointing to roles of these proteins in DNA replication rather than DNA break repair. The enrichment of S-phase histone γ-H2AX foci and a striking loss of telomeric DNA in mus81 send1 further support this interpretation. SEND1 has at most a minor role in resolution of the Holliday junction but acts as an essential backup to MUS81 for resolution of toxic replication structures to ensure genome stability and to maintain telomere integrity.
INTRODUCTION
All living cells are exposed to different insults that can generate DNA double-strand breaks (DSBs). A key mechanism for repairing these lesions in both meiotic and mitotic cells is homologous recombination (HR). Repair of programmed DSB through HR in meiotic cells is essential for fertility, ensuring proper pairing and segregation of homologous chromosomes in the production of gametes. In somatic cells, HR acts to repair DNA damage-associated DSB or to restart stalled or collapsed replication forks (RFs). During the repair process, 3′-ended single-strand DNA generated through processing of the DSB invades an homologous DNA template and forms a joint recombination intermediate, the Holliday junction (HJ). Resolution of the covalently linked recombining DNA molecules is crucial for completion of the repair process. This is performed through dissolution by the BTR complex (BLM-TOPOIIIα-RMI1-RMI2) to yield non-crossover (NCO) products or through resolution by specialized nucleases, called resolvases, resulting in crossover (CO) or NCO products. The BTR complex (STR in the yeast Saccharomyces cerevisiae and RTR in plants) consists of a RecQ (recombination deficiency Q) family helicase, i.e., Sgs1 (Slow growth suppressor 1) in yeast, BLM (Bloom's syndrome helicase) in mammals, and RecQ4A in plants (Knoll et al., 2014), as well as a type IA topoisomerase and the structural protein RMI1 (RecQ-mediated genomic instability 1). This complex induces convergent migration of the double HJ to generate a singly linked DNA structure (hemicatenane), which is dissociated by the topoisomerase (Wu and Hickson, 2003, 2006). Mutation in the BLM gene in human leads to Bloom's syndrome disorder. Cell lines derived from these patients display genome instability and more than 10-fold increased sister chromatid exchanges (SCEs) (Bloom, 1954; Chaganti et al., 1974; German, 1993). Recently, it has been shown that these SCE arise through the action of three structure-specific endonucleases (i.e., SLX1-SLX4 [synthetic lethal of unknown function], MUS81-EME1 [MMS and UV-sensitive protein 81-Essential meiotic endonuclease 1], and GEN1 [Gen endonuclease homolog1] in human and SLX1-SLX4, Mus81-Mms4 [Methyl Methane Sulfonate sensitivity 4], and Yen1 [Holliday junction resolvase YEN1] in yeast), which resolve joint recombination intermediates (Wechsler et al., 2011; Castor et al., 2013; Garner et al., 2013; Wyatt et al., 2013). Mus81-Mms4 and Slx1-Slx4 were initially identified in yeast as essential proteins for viability of cells harboring a mutated SGS1 gene (ortholog of BLM) (Mullen et al., 2001). Mus81-Mms4 belong to the XPF (Xeroderma pigmentosum group F-complementing protein) endonuclease family, containing an Excision repair cross complementing 4 (ERCC4) nuclease domain and a tandem Helix-hairpin-helix (HhH) domain. In vitro studies have shown that this protein recognizes and processes branched DNA structures, such as 3′ flaps, RF structures, and nicked HJ (reviewed in Schwartz and Heyer, 2011). SLX1 is the catalytic subunit of the SLX1-SLX4 heterodimer and belongs to the GIY-YIG family of nucleases. It recognizes and cuts 5′ flap structures and has HJ resolvase activity (Fekairi et al., 2009; Muñoz et al., 2009; Svendsen et al., 2009). SLX1-SLX4 and MUS81-EME1 have recently been shown to interact at the G2/M-phase of the cell cycle, and biochemical analyses show that both nucleases cooperate to cleave HJ via an ordered nick and counter-nick mechanism. SLX1-SLX4 introduces the initial cut, and this nicked HJ is further processed by the MUS81-EME1 endonuclease (Matos et al., 2011; Wyatt et al., 2013; Matos and West, 2014). Thus, the SLX-MUS complex promotes asymmetric cleavage of the HJ to produce gapped and flapped intermediates that will require further processing before ligation. Resolvases that can process HJ by introducing symmetric nicks in two strands of the same polarity to generate products that can be directly ligated without the need for further processing have been isolated from bacteriophages T4 and T7, bacteria, and archaea (West, 1997; Lilley and White, 2001). The best-studied resolvase is the Escherichia coli protein RuvC (Crossover junction endoDNase RuvC). A nuclease promoting HJ resolution in the same manner as the bacterial nuclease has been identified in yeast (Yen1) and mammalian (GEN1) cells (Ip et al., 2008). These eukaryotic nucleases belong to the Rad2/XPG (Radiation-sensitive 2/Xeroderma pigmentosum group G-complementing protein) family of nucleases and contain XPG amino and internal nuclease domains and HhH DNA binding domains (Ip et al., 2008; Rass et al., 2010).
Mitotic mammalian cells exhibit very low levels of SCE, indicating that most HJs are resolved by the NCO-promoting pathway BTR. Mammalian cells show an elevated frequency of SCE in the absence of this pathway, which can be suppressed by depletion of MUS81, SLX1, or GEN1. Simultaneous depletion of both MUS81 and SLX1 results in similar levels of SCE reduction to that of the single mutants, indicating a cooperative role of these proteins in HJ resolution. However, depletion of GEN1 and SLX1 or MUS81 results in an additive reduction in SCE, indicating that the resolvase GEN1 is part of an independent pathway involved in the resolution of HJ (Wechsler et al., 2011; Wyatt et al., 2013; Sarbajna et al., 2014).
Loss of Mus81-Mms4/MUS81-EME1 sensitizes cells to agents causing replication stress, leading to RF stalling or collapse. Although yen1 mutant cells are not sensitive to these agents, the mus81 yen1 double mutant exhibits higher sensitivity than the mus81 single mutant. Interestingly, these mutant cells are more sensitive to treatments compromising replication than to agents inducing DSB, such as ionizing radiation, suggesting an essential function of these proteins in dealing with perturbed RF in mitotic cells (Blanco et al., 2010; Ho et al., 2010; Tay and Wu, 2010; Agmon et al., 2011). In the presence of exogenous DNA damage, mus81 yen1 mutant cells fail to segregate their chromosomes. Similarly, mammalian cells depleted of MUS81 and GEN1 and exposed to DNA damaging agents exhibit high frequencies of anaphase bridges and lagging chromosomes (Garner et al., 2013; Wyatt et al., 2013). This chromosome mis-segregation could result from the increase in toxic replication products or inappropriately resolved HJ resulting from the lack of processing of HR-dependent RF intermediates. Moreover, the reduction but not suppression of sister chromatid nondisjunction in mus81 yen1 mutant cells depleted of Rad51 (Radiation sensitive 51) suggests an essential role of these proteins in processing stalled RF rather than HR-dependent repair (Ho et al., 2010). Cells lacking both MUS81 and GEN1 are sensitive to replication inhibitors, activate the ATM/CHK2-mediated (Ataxia-telangiectasia mutated/Checkpoint kinase 2) DSB checkpoint response, show slow S-phase progression, and are impaired in RF movement (Sarbajna et al., 2014). Thus, Mus81-Mms4/MUS81-EME1 and GEN1/Yen1 are not only involved in processing HR intermediates but also play an essential role in resolving stalled replication forks.
Functional plant homologs of BLM and Sgs1 have been identified in Arabidopsis thaliana (RecQ4A) and rice (Oryza sativa) (RecQL4). In both plants, loss of the RecQ helicase leads to hypersensitivity to DNA damaging agents and elevates the frequency of HR events (Bagherieh-Najjar et al., 2005; Hartung et al., 2007; Kwon et al., 2013). Arabidopsis mutants depleted for the two proteins partners of REQ4A in the RTR complex (top3A and rmi1 mutants) show increases in HR (Hartung et al., 2008). Simultaneous deletion of RECQ4A and MUS81 in Arabidopsis is lethal, and this lethality is suppressed by blocking the formation of recombination intermediates through inactivation of RAD51C (Hartung et al., 2007; Mannuss et al., 2010). Interestingly, mutants of RECQ4B, which shares conserved domains and high sequence similarity with RECQ4A, are not sensitive to DNA damage and are viable in a MUS81-deficient background. Thus, RECQ4B does not play a detectable role in somatic DNA repair. However, both RECQ4A and RECQ4B helicases have anticrossover activity in meiosis, with their simultaneous depletion inducing a 6-fold increase in CO frequency (Séguéla-Arnaud et al., 2015).
Much less is known about the structure-specific endonucleases that process HJ in plants. In Arabidopsis, MUS81 has been shown to cleave HJ in vitro (Geuting et al., 2009) and to act in HR, DNA repair, and processing of aberrant replication intermediates (Hartung et al., 2006, 2007; Mannuss et al., 2010). No SLX1-SLX4 complex has been characterized in plants, and although homologs of GEN1 have been identified, their function in planta has not been clarified. Most plants possess two homologs of GEN1/Yen1: GEN1 and SEND1 (SINGLE-STRAND DNA ENDONUCLEASE1) (Furukawa et al., 2003; Moritoh et al., 2005; Bauknecht and Kobbe, 2014). Biochemical analyses in rice and Arabidopsis showed that the two proteins have similar biochemical activities and are able to process 5′ flap, RF, as well as HJ structures (Furukawa et al., 2003; Yang et al., 2012; Bauknecht and Kobbe, 2014). In rice, GEN1 plays an essential role in pollen development (Moritoh et al., 2005), while SEND1 is preferentially expressed in proliferating tissues and is induced by UV and DNA damaging agents, suggesting a role in DNA repair (Furukawa et al., 2003).
Here, we describe the gen1 and send1 mutants of Arabidopsis. We show that GEN1 and SEND1, either alone or together, have no detectable role in DNA repair and meiosis. However, SEND1, but not GEN1, plays an essential overlapping role with MUS81. Growth of Arabidopsis plants lacking both MUS81 and SEND1 is severely affected, and these plants show high levels of spontaneous DNA damage enriched in S/early G2-phase nuclei, as well as chromosomal instability. We show that this chromosomal instability results, at least in part, from loss of telomere integrity. These phenotypes are not suppressed by the absence of RAD51, pointing to an essential role for SEND1 in replication in the absence of MUS81. Overall, our data identify SEND1 as the functional Arabidopsis homolog of GEN1/Yen1 and show that SEND1 plays a central role with MUS81 in the repair of toxic replication intermediates and telomere homeostasis.
RESULTS
Isolation and Molecular Characterization of GEN1 and SEND1 T-DNA Insertion Mutants
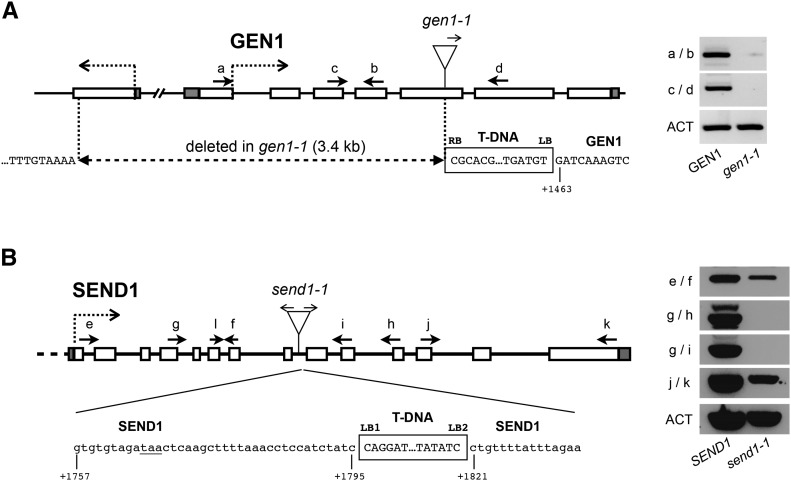
Two homologs of the human GEN1 and the budding yeast Yen1 are found in the Arabidopsis genome (https://www.arabidopsis.org; Furukawa et al., 2003; Bauknecht and Kobbe, 2014). These two proteins, GEN1 and SEND1, belong to the class IV of the RAD2/XPG family of nucleases and possess the characteristic N-terminal and internal XPG nuclease domains, as well as an HhH domain (Supplemental Figure 1). The highest sequence conservation is observed in the N-terminal region of the proteins, and both proteins contain catalytic residues that have been shown to be essential for nuclease activity (Supplemental Figure 1) (Ip et al., 2008; Bailly et al., 2010; Blanco et al., 2010; Lorenz et al., 2010; Rass et al., 2010; Bauknecht and Kobbe, 2014). Moreover, GEN1 and SEND1 have been shown to possess similar HJ resolvase activity in vitro, leading to the suggestion that plants, in contrast to yeast and mammals, have two functional canonical HJ resolvases (Bauknecht and Kobbe, 2014). Their function in planta remains unclear. We therefore searched for GEN1 and SEND1 mutants in public T-DNA insertion collections. We obtained and characterized GEN1 (FLAG_121A03) and SEND1 (SALK_135735) T-DNA insertion lines (Figure 1). Both insertions were verified by PCR and sequencing to determine the exact genomic position of the insertions (Figure 1). Homozygous mutant lines were analyzed by RT-PCR to confirm the absence of the respective transcripts (Figure 1). In gen1-1, the T-DNA is inserted in exon 5, and this insertion is associated with a large deletion (3.4 kb) of the DNA upstream of the T-DNA insertion site. Accordingly, no GEN1 transcripts could be detected in gen1-1 mutants (Figure 1A). In send1-1, the T-DNA is inserted in intron 8. This insertion is associated with a deletion of 25 bp and is flanked by two left borders in opposite orientations (Figure 1B). Although a transcript could be detected in send1-1 upstream of the T-DNA insertion, no transcript was detected with primers surrounding the T-DNA insertion site, confirming the absence of full-length transcript (Figure 1B). Sequence analysis of the T-DNA junction indicated that an in-frame stop codon is present in intron 8 before the T-DNA integration (Figure 1B) and a truncated protein of 228 amino acids (out of 600) could be expressed. This would contain the XPG N-terminal and internal nuclease domains as well as the catalytic residues essential for nuclease activity, but not the complete helix-hairpin-helix domain.
Figure 1.
Arabidopsis GEN1 and SEND1 T-DNA Insertion Mutants.
Structure of GEN1 (A) and SEND1 (B) and the gen1-1 and send1-1 T-DNA insertion mutant alleles. Boxes show exons (unfilled) and 5′ and 3′ untranslated regions (gray fill). The position of the T-DNA insertions is indicated with arrows showing the orientation of the left border and sequences of the T-DNA/chromosome junctions below. The gen1-1 T-DNA insertion is flanked by two left borders (LB1 and LB2) and accompanied by a 3396-bp deletion, which eliminates exons 1 to 4 and a large part of exon 5. A putative in-frame TAA STOP codon in send1-1 is underlined. Numbering under the sequences is relative to the GEN1 or SEND1 start codons. RT-PCR analyses of transcripts of gen1-1 (A) and send1-1 (B). Amplification of the actin transcript (ACT) was used as a control for RT-PCR. The positions and orientations of the PCR primers are shown in the diagrams.
To assess the involvement of GEN1 and SEND1 in DNA repair, we tested the sensitivity of the mutants to DNA damaging agents (Supplemental Figure 2). No hypersensitivity was observed to γ-rays or Mitomycin C in either single mutant or the double mutant (Supplemental Figure 2A). Similarly, no hypersensitivity to UV irradiation (Supplemental Figure 2C) or hydroxyurea (HU) (Supplemental Figures 2D and 2E) was observed, with the exception of a mild UV sensitivity of the gen1 send1 double mutant, suggesting that GEN1 and SEND1 are not essential for somatic DNA repair in Arabidopsis.
Cytogenetic analysis of meiosis in pollen mother cells further demonstrated normal meiotic progression in single and double mutants, indicating that GEN1 and SEND1 are also not required for wild-type meiosis (Supplemental Figure 3).
Arabidopsis SEND1 Plays an Overlapping Role with MUS81
Our results demonstrate that GEN1 and SEND1 are not essential for somatic DNA repair and meiosis. In mammalian cells, HJs formed during homologous recombination repair of DSBs are processed by the RTR complex, SLX-MUS resolvase complex, and GEN1 (Ip et al., 2008; Rass et al., 2010; Wyatt et al., 2013). We thus examined the possibility that GEN1 and/or SEND1 might act redundantly with other nucleases for the resolution of recombination intermediates. We crossed our gen1 and send1 mutants with the previously characterized mus81 (Hartung et al., 2006) and xpf (Dubest et al., 2002) nuclease mutants. Knockout of SEND1 in plants lacking XPF showed a wild-type phenotype. However, while the mus81 gen1 mutant developed normally, mus81 send1 double mutant plants were severely affected, showing strong growth retardation and impaired leaf and shoot development (Figures 2A and 2B). The triple mus81 send1 gen1 mutant has a similar developmental phenotype to the double mus81 send1 mutant (11.8 ± 2.7 seeds per silique, n = 40 siliques; Supplemental Figure 4).
Figure 2.
mus81 send1 Double Mutants Exhibit Strong Developmental Defects.
(A) Three-week-old plants. In contrast to single mutants, development of mus81 send1 plants is strongly affected and this can be complemented by expression of the wild-type SEND1 gene (a 2-cm scale bar is shown at the bottom left).
(B) Pictures of 6-week-old plants showing severe growth retardation and defects of mus81 send1 plants.
(C) Fertility is reduced in double mutants, which have short siliques and reduced number of seeds. Mean (±se) numbers of seeds per silique are shown in the upper parts of the image (wild type, n = 11 siliques; mus81 send1, n = 12 siliques).
To confirm that the phenotype of the mus81 send1 double mutant is due to the loss of the SEND1 gene, we performed genetic complementation. A 5.5-kb DNA fragment encompassing the full SEND1 genomic coding sequence (including introns) and 1.2 kb of upstream sequence was introduced into mus81/MUS81 send1/send1 plants. Four independent mus81 send1 transformants expressing the wild-type SEND1 construct (mus81 send1+SEND1) were selected and showed a wild-type phenotype (Figure 2A). This complementation strictly cosegregated with the SEND1 transgene in the following generation, confirming that the loss of SEND1 function is the cause of the defects observed in the mus81 send1 mutant.
Cell Cycle Defects and Cell Death in mus81 send1 Plants
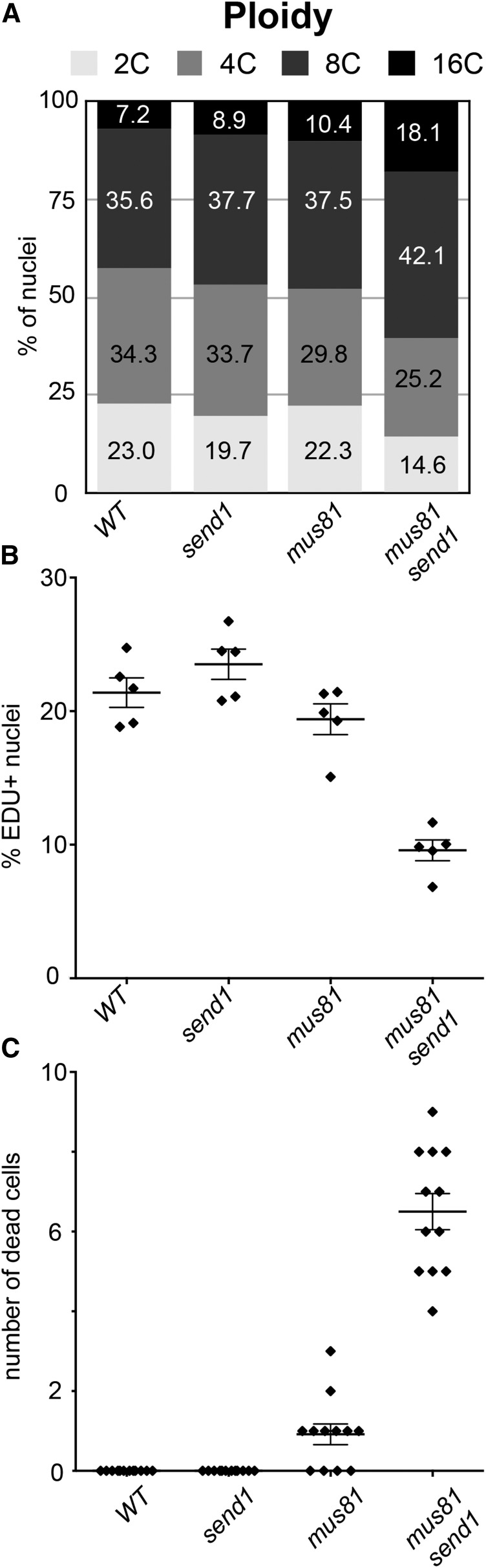
To determine the origin of these growth defects, we analyzed cell cycle progression and spontaneous cell death in mus81 send1 double mutants. Ploidy level increases when plants are subjected to highly stressful conditions, which is a good indicator of the presence of DNA damage (Adachi et al., 2011). Measurement of ploidy levels by flow cytometry in wild-type, send1, and mus81 mutants showed that the population of 2C and 4C nuclei represents more than 50% of the nuclei, while only 7 to 10% have C values of 16 (i.e., after two rounds of endoreduplication) (Figure 3A). In the mus81 send1 double mutant, however, 2C and 4C nuclei represent only 40% of the nuclei, while the proportion of 16C nuclei rose to 18% (Figure 3A). Accordingly, the mus81 send1 double mutant plants show increased endoreduplication, as seen in the higher endoreduplication index (EI; mean 163.6 ± 1.8 ; see Methods; Takahashi et al., 2008) compared with wild-type, mus81, and send1 single mutants (mean EI of 126.9 ± 4.5, 135.7 ± 1.4, and 136.1 ± 0.9, respectively).
Figure 3.
SEND1 and MUS81 Play Overlapping Roles in Preserving Genome Stability.
(A) Distribution of mitotic nuclear DNA contents in 1-week-old seedlings of wild-type, send1, mus81, and mus81 send1 mutant plants determined by flow cytometry. Percentage of nuclei in each category is indicated.
(B) and (C) Mean fraction (±se) of EDU+ nuclei (n = 5) (B) and mean (±se) number of dead cells per root tip (n = 12) (C) in wild-type, send1, mus81, and mus81 send1 mutant plants. Individual values are shown as filled diamonds. Cell cycle is affected in mus81 send1 mutant plants as shown by reduced EdU incorporation and spontaneous cell death.
We next examined cell cycle progression using 5-ethynyl-2′-deoxyuridine (EdU) labeling (Figure 3B). EdU is a thymidine analog that is incorporated into DNA during S-phase and can be detected cytologically with a fluorescence assay. One-week-old seedlings were incubated in a 10 µM EdU solution for 1 h and fixed. After 1 h of growth in the presence of EdU, 21% of wild-type root nuclei had detectable EdU incorporation. Similarly, 23% of send1 and 19% of mus81 nuclei were EdU positive (Figure 3B). EdU incorporation was significantly reduced in mus81 send1, with only 9% of the nuclei having detectable EdU incorporation (Figure 3B). These data indicate that cell cycle progression slows down in the mutant, consistent with the reduced growth rate of the mus81 send1 plants. Analysis of cell death by propidium iodide staining of root tips revealed elevated levels of cell death in root meristems of mus81 send1 plants, while few or no dead cells were observed in wild-type and the single mutant plants (Figure 3C).
SEND1 Is Not Essential for Somatic or Meiotic DNA Repair
MUS81 is known to play a key role in the resolution of recombination intermediates. SEND1 shows HJ resolvase activity in vitro and is thus reasonably expected to process HJ in vivo. Given this, we hypothesized that the growth and cell cycle phenotypes observed in the mus81 send1 mutant could result from accumulation of unresolved HJ produced during homologous recombination repair of DNA damage. Accordingly, suppressing the accumulation of HJ by inhibiting homologous recombination should suppress the mus81 send1 defects. To test this, we crossed our mus81 send1 mutant with rad51 mutant plants. Surprisingly, the developmental defects observed in mus81 send1 were not suppressed by the absence of RAD51 (Figure 4A). In agreement with this result, expression of the dominant-negative RAD51-GFP fusion protein (Da Ines et al., 2013) in mus81 send1 had no observable effect on development of the double mutant (Figure 4A).
Figure 4.
The mus81 send1 Phenotypes Are RAD51 Independent.
(A) The growth defects of mus81 send1 plants (left image) are not suppressed by the absence of RAD51 (middle image) nor by expression of the dominant-negative RAD51-GFP (right image). A 2-cm scale bar is shown at the bottom right.
(B) to (I) DAPI-stained meiotic nuclei. Normal meiotic progression can occur in mus81 send1 pollen mother cells, with complete synapsis at pachytene (B), five intact bivalents at metaphase I (C), two pools of five chromosomes at metaphase II (D), and tetrads with four balanced meiotic products (E). Meiotic chromosome segregation defects were occasionally observed in these plants (telophase I; [F]), leading to unbalanced pools of chromosomes at metaphase II (G) and aberrant telophase II tetrads with chromosome bridges (arrow; [H] and [I]). Bars = 10 µm.
These results strongly suggest that SEND1 is essential for the processing of aberrant replication intermediates in the absence of MUS81. Of course, this does not exclude the possibility that Arabidopsis SEND1 also plays a role in the resolution of HJ in vivo, as shown in vitro (Bauknecht and Kobbe, 2014). Analogously, the role of MUS81 in HJ resolution in somatic cells is only observed in the absence of the helicase activities involved in homologous recombination. Thus, severe developmental defects of Arabidopsis fancm and req4a helicase mutants in the absence of MUS81 (fancm mus81 and recq4A mus81) are suppressed by inhibition of homologous recombination (Mannuss et al., 2010; Crismani et al., 2012). The absence of developmental or fertility defects in fancm send1 and recq4A send1 double mutant plants, neither of which showed reduced fertility (Supplemental Figure 5), suggests that SEND1 does not have a similar role in the resolution of toxic recombination intermediates, although a cryptic role in the absence of other nucleases remains possible. Another test involved RNaseH2 (Ribonuclease H2)-deficient plants, which show increased homologous recombination and in which the absence of MUS81 (mus81 trd1 [triffid 1] double mutant) causes a severe inhibition of root and shoot growth (Kalhorzadeh et al., 2014). However, like the trd1 mutant (Kalhorzadeh et al., 2014), send1 trd1 plants grow normally (Supplemental Figure 6). Thus, while MUS81 plays an important role in HJ resolution in mitotic cells, SEND1 protein has at most a very minor role, and it is only in the absence of MUS81 that SEND1 becomes necessary.
The appropriate resolution of HJ is a key event in meiosis to ensure proper segregation of homologs during the production of gametes. The presence of unprocessed HJ hampers chromosome segregation and can lead to aneuploidy and infertility. The reduced fertility of mus81 send1 double mutants is seen in the low number of seeds per silique in these plants, with an average of 10 ± 0.9 seeds per silique (20% of the wild type; Figure 2C). Furthermore, Alexander staining of anthers showed that pollen viability is reduced, and toluidine blue staining showed that abnormal tetrads were frequent (up to 38%) in the mus81 send1 double mutant (Supplemental Figure 7). Although they need to be interpreted in the context of the severe growth and mitotic defects of these plants, these observations show possible meiotic defects in mus81 send1. We therefore checked meiotic progression in these plants in greater detail. Wild-type pairing and synapsis of homologs is seen as the synaptonemal complex at pachytene (Figure 3B), followed by further condensation and five bivalents at metaphase I (Supplemental Figure 3C). After segregation of homologous chromosomes to opposite poles, meiosis II gives rise to four haploid nuclei (Supplemental Figure 3D). All of these stages, and notably the presence of five bivalents at metaphase I (Figure 4C; n = 38), were also observed in mus81 send1 during meiosis, showing that normal meiotic progression can occur in the absence of MUS81 and SEND1 (Figures 4B to 4E). However, some meiotic defects were observed (Figures 4F to 4I), with 10% of telophase I/anaphase II nuclei showing segregation defects (Figures 4F and 4G; n = 40) and 20% of the telophase II tetrads being aberrant (Figures 4H and 4I; n = 59). However, we stress that given the strong mitotic and growth defects of mus81 send1 mutant plants, it is difficult to draw firm conclusions, and it remains very possible that these meiotic alterations result from the accumulation of toxic repair intermediates from the preceding mitosis or the premeiotic S-phase.
mus81 send1 Are Essential for Proper Telomere Replication
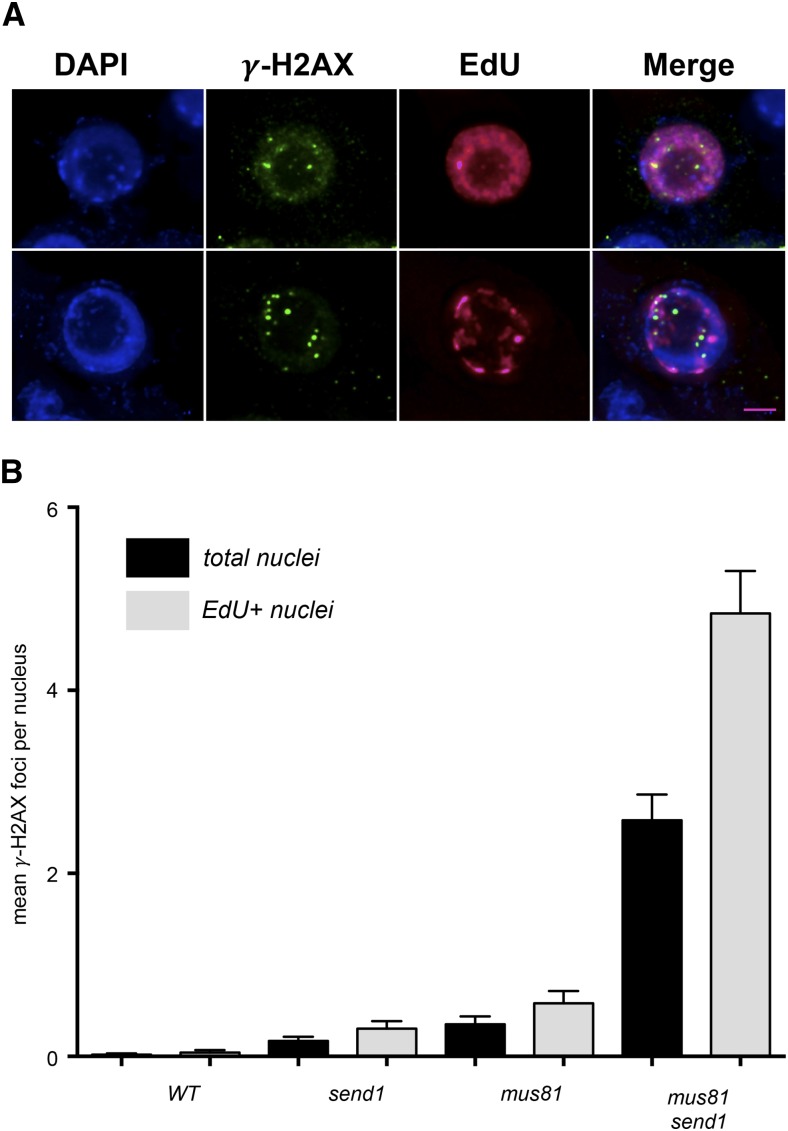
Plants lacking MUS81 and SEND1 exhibit severe cell cycle defects, increased polyploidy, and elevated levels of cell death, which are not suppressed by the absence of RAD51. The RAD51-independent origin of the structures requiring metabolism by SEND1 and/or MUS81 points to a role of these proteins in DNA replication. In mammalian cells, simultaneous depletion of MUS81 and GEN1 proteins results in impaired RF movement and prolonged fork stalling, which is proposed to lead to the formation of one-ended DSB (Sarbajna et al., 2014). This prompted us to investigate whether mu81 send1 mutants may accumulate toxic, unresolved DNA damage arising during replicative stress. To do so, we analyzed the presence of DSB in somatic cells by performing immunolocalization of γ-H2AX (histone H2AX). As expected, foci were only rarely detected in root tip nuclei of wild-type, send1, and mus81 plants (Figure 5). In contrast, a mean of 2.6 γ-H2AX foci/nucleus was observed in mus81 send1 mutant plants, showing the accumulation of DSBs in this double mutant (Figure 5). The question of whether these DSBs were associated with replication was tested by monitoring the presence of γ-H2AX foci in S-phase or early G2-phase nuclei. One-week-old seedlings were incubated in EdU solution for 1 h (after this time only replicating nuclei will have incorporated EdU), and EdU detection was then combined with immunolocalization of γ-H2AX. In accordance with a replicative origin of the DSB, we observed a significant enrichment of γ-H2AX foci in replicating cells of mus81 send1 mutant plants, with S/early G2-phase nuclei exhibiting an average of five foci (Figure 5B).
Figure 5.
mus81 send1 Double Mutants Accumulate DSBs, Which Are Enriched in Replicating Cells.
(A) γ-H2AX (green) foci in S- or early G2-phase (EdU+; magenta) mitotic nuclei of mus81 send1 plants. Bar = 2 µm.
(B) Mean (±se) numbers of γ-H2AX foci per nucleus in total nuclei (black fill) or EdU-positive nuclei (gray fill) of wild-type, send1-1, mus81, and mus81 send1 mutants (n = 100 for total nuclei and n = 50 for EdU-positive nuclei).
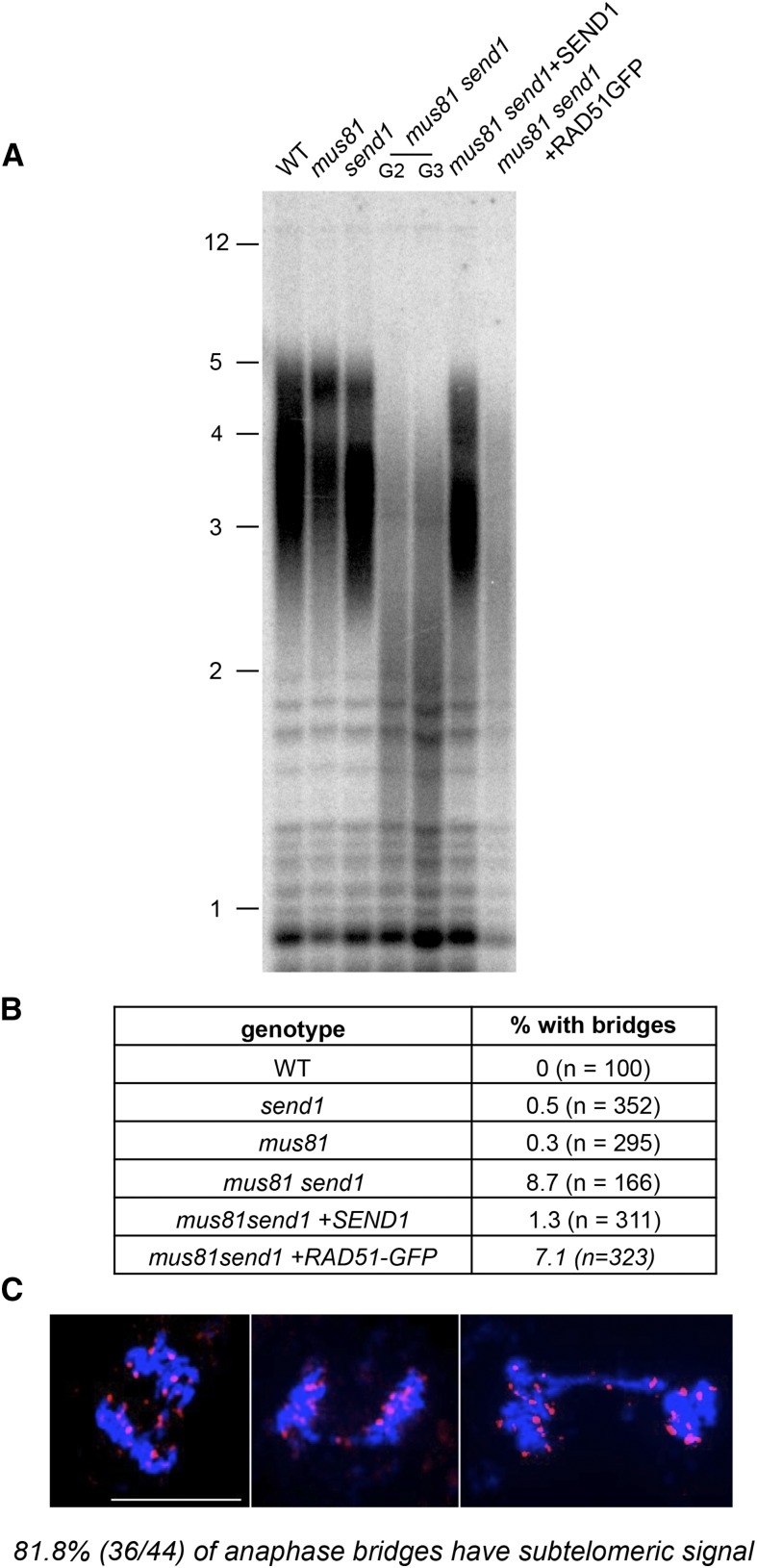
Given that high fidelity replication is also critical for maintaining telomere stability and in particular for preventing rapid telomere loss, we investigated telomere stability with terminal restriction fragment (TRF) analyses to determine bulk telomere length in the mutants. As expected, wild-type, mus81, and send1 single mutant plants exhibited a wild-type telomere length of ∼3 to 5 kb. In contrast, a striking loss of telomeric repeats was observed in the mus81 send1 double mutant, with TRF signals detected as a much shorter, broader smear (Figure 6A). This rapid, generalized loss of telomeric repeats is also seen in mus81 send1 plants expressing the dominant-negative RAD51-GFP protein but is absent in complemented mus81 send1+SEND1 transformants (Figure 6A).
Figure 6.
Telomere Instability and Chromosome Fusions in mus81 send1.
(A) TRF analysis of bulk telomere length in genomic DNA using the telomeric repeat probes. G2 and G3 indicate second and third mutant generations.
(B) Percentages of mitotic anaphases with at least one visible chromosome bridge in wild-type and mutant plants (n = number of anaphases counted).
(C) Examples of mus81 send1 mitotic anaphases showing chromosome bridges analyzed by FISH with a pool of nine subtelomeric BAC fluorescent probes (magenta). DNA is counterstained with DAPI (blue). Bar = 10 µm.
Dramatic loss of telomeric repeats is thus directly associated to the combined absence of MUS81 and SEND1. Furthermore, the fact that absence of RAD51 catalytic activity does not restore telomere length in mus81 send1 points to a role of the nucleases in proper telomere replication. The loss of telomeric repeats could generate dicentric chromosomes in mus81 send1 mutant plants, resulting from the repair of nonfunctional telomeres being recognized as DSB. Cytological analysis revealed elevated levels of mitotic anaphases with chromosome bridges in the mus81 send1 double mutant plants (8.7% of the mitotic anaphases; Figure 6B). In accordance with the TRF analyses, inactivation of RAD51 in these plants (mus81 send1+RAD51-GFP) had little effect, with 7.2% of anaphases with bridges. In contrast, anaphase bridges were rare or absent in the wild type, the two single mutants, and the complemented mus81 send1+SEND1 plants (Figure 6B). Fluorescence in situ hybridization (FISH) analysis was performed using the mixture of nine subtelomeric BAC probes to confirm that the anaphase bridges in mus81 send1 plants are the result of end-to-end chromosome fusions. As shown in Figure 6C, 80% of the anaphase bridges present a subtelomeric signal, indicating that at least one chromosome end is implicated in the generation of the dicentric chromosome. These data are in agreement with a role of SEND1 and MUS81 in the repair of toxic replication intermediates to avoid loss of telomeric repeats.
DISCUSSION
Most plants possess two putative GEN1/Yen1 homologs, GEN1 and SEND1 (Furukawa et al., 2003; Moritoh et al., 2005; Bauknecht and Kobbe, 2014), and in vitro studies of the Arabidopsis and rice proteins show that they can cleave HJ (Furukawa et al., 2003; Moritoh et al., 2005; Yang et al., 2012; Bauknecht and Kobbe, 2014). We thus isolated Arabidopsis mutants lacking GEN1 and/or SEND1 and analyzed their role in DNA repair and meiosis. Our data show that absence of GEN1 and/or SEND1 has no detectable effect on meiosis or on sensitivity to a variety of DNA damaging agents. This is in accordance with data in budding yeast and mammals showing that yen1/gen1 mutants are not sensitive to DNA-damaging agents and have no defects in meiosis (Blanco et al., 2010; Ho et al., 2010; Agmon et al., 2011). mus81 yen1 (or gen1 in animals) mutants show increased DNA damage sensitivity and cell cycle and meiotic defects with respect to mus81 single mutants, leading to the hypothesis that Mus81 is the primary resolvase and that Yen1 acts as a backup (Blanco et al., 2010; Ho et al., 2010; Agmon et al., 2011; Matos et al., 2011). To test this hypothesis, we crossed Arabidopsis mus81 mutant with gen1 and send1 mutants. While the mus81 gen1 double mutant shows no visible defects, the mus81 send1 double mutant exhibits strong growth retardation and severe genome instability, reminiscent of observations in yeast and mammalian cells. Furthermore, the triple mus81 send1 gen1 mutant shows a similar phenotype to that of the mus81 send1 double mutant. Our data thus indicate that GEN1 is not able to compensate for the absence of SEND1 and that GEN1 and SEND1 play nonredundant functions in vivo, with Arabidopsis SEND1 being the functional homolog of human GEN1 and budding yeast Yen1 (see also below).
Both GEN1 and SEND1 possess the characteristic XPG N-terminal and internal nuclease domains, as well as the HhH DNA binding domain, and have identical biochemical activity in vitro (Bauknecht and Kobbe, 2014). Although the functional differences between SEND1 and GEN1 and the in vivo role of GEN1 remain to be determined, our data strongly suggest that in plants, SEND1 plays a major role in DNA damage repair similar to that of GEN1/Yen1. However, comparative analysis of protein domains does bring to light a difference between the two proteins, with SEND1 but not GEN1 harboring a chromodomain-like motif potentially involved in DNA and histone recognition. This domain is not present in yeast Yen1 or mammalian GEN1, and although its function is not known, we hypothesize that it may be essential in plants for SEND1 function through binding to specific proteins or DNA structures.
In mitotic cells, repair of DSB mainly generates NCO products by the action of helicases, which process HJ through dissolution and synthesis-dependent strand annealing; this process does not involve a double HJ intermediate. Thus, in mitotic cells, the resolvases act as a backup pathway to the action of helicases, and their role in crossing over can only be detected in the absence of the helicases activity. Thus, mammalian cells depleted of BLM helicase show increased SCE, which is dependent on the action of the resolvase activity of MUS81 or GEN1. Loss of the different pathways ensuring genome stability leads to high levels of mortality observed in cells depleted simultaneously of BLM, MUS81, and GEN1 (Wyatt et al., 2013). Analogously, growth is strongly affected in Arabidopsis plants lacking MUS81 and the helicases FANCM (for Fanconi anemia group M protein) or RECQ4A, suggesting accumulation of toxic unprocessed HJ in these cells (Mannuss et al., 2010; Crismani et al., 2012). In contrast to the essential role of MUS81 as a backup pathway to the action of the helicases in Arabidopsis, our observation of wild-type development of send1 recq4A or send1 fancm double mutants argues against any significant role of SEND1 as a resolvase in the presence of MUS81 in mitotic cells.
The strong developmental defects of mus81 send1 plants could thus be the result of accumulation of unprocessed HJ, with SEND1 only being required in the absence of MUS81. Inhibition of HR prevents the formation of HJ, and defects in mutants accumulating HJ can thus be alleviated in this way. Thus, absence of RAD51 suppresses the strong developmental defects of the Arabidopsis mus81 recq4A and mus81 fancm double mutants depleted for HJ processing helicase activity or synthesis-dependent strand annealing helicase and resolvase activities (Mannuss et al., 2010; Crismani et al., 2012). We show that this is not the case for the mus81 send1 double mutant, as neither the developmental defects nor the chromosomal instability of mus81 send1 double mutants were suppressed by deletion of RAD51 or expression of the dominant-negative RAD51-GFP fusion protein (Da Ines et al., 2013). This conclusion is consistent with data from budding yeast showing that defects observed in mus81 yen1 double mutants are not (or are only partially) suppressed by deletion of either Rad52 or Rad51 (Blanco et al., 2010; Ho et al., 2010; Agmon et al., 2011). Although we cannot exclude the possibility that mus81 send1 plants accumulate unresolved recombination intermediates, our data show that the developmental defects and chromosomal instability of Arabidopsis mus81 send1 mutants are due to the presence of RAD51-independent structures. Notwithstanding the presence of abnormalities in meiosis in some cells, presumably due to premeiotic events, meiosis can progress normally in mus81 send1 mutants. This supports the idea that SEND1 has no discernable role in the resolution of HJ formed during recombination, although the growth and mitotic defects of mus81 send1 plants make any conclusion concerning possible meiotic roles subject to considerable caution. Again, this is analogous to the only very slightly effects on CO formation and HJ resolution in budding yeast mus81 yen1 mutants compared with mus81 alone, notwithstanding strong reductions in spore formation and viability (Agmon et al., 2011; Matos et al., 2011; De Muyt et al., 2012; Zakharyevich et al., 2012). In the same way, mus81 yen1 mutants in the yeast Kluyveromyces lactis showed no further reduction in spore formation over mus81 single mutants (Chen and Aström, 2012). Arabidopsis SEND1 thus plays at most a minor role in the processing of mitotic and meiotic recombination intermediates.
Arabidopsis MUS81 plays a role in processing HJ as a backup pathway to the action of helicases in mitotic cells. However, as discussed above, we do not have any evidence that SEND1 functions in resolving RAD51-dependent (HR) joint molecules. This clearly points to a replication-dependent origin for the strong developmental defects and chromosomal instability of the mus81 send1 plants, presumably in the processing of stalled or collapsed replication forks. The involvement of MUS81 in the processing of replication intermediates is well established both in yeast and mammalian cells. The mus81 mutant in the yeast S. cerevisiae shows sensitivity to agents causing replication stress and chromosome mis-segregation, which is not exclusively due to the role of Mus81 in processing HJ, as demonstrated by only partial suppression of the phenotype in the absence of Rad51 (Ho et al., 2010; Agmon et al., 2011). In mammalian cells, the generation of MUS81-dependent double-strand DNA breaks after inhibition of replication by HU or aphidicolin suggests that collapsed or stalled replication fork are processed into DNA DSB by MUS81 (Hanada et al., 2007; Pepe and West, 2014). HeLa cells depleted in the RecQ helicase WRN (Werner syndrome ATP-dependent helicase) accumulate DSB in a MUS81-dependent manner after HU treatment. However, simultaneous depletion of WRN and RAD51 does not suppress these DSBs, suggesting that MUS81 operates upstream of RAD51 in the processing of toxic replication intermediates (Franchitto et al., 2008; Murfuni et al., 2012; Murfuni et al., 2013). Furthermore, MUS81 also plays a role in the cleavage of RFs remaining at later (G2/M) stages of the cell cycle at common fragile sites, and its absence leads to an increase in anaphase bridges due to improper disjunction of sister chromatids (Naim et al., 2013; Ying et al., 2013; Pepe and West, 2014). Much less is known about the potential role of GEN1/Yen1 in facilitating replication of difficult to replicate regions, although both yeast and mammalian cells lacking MUS81 and GEN1/Yen1 show increased genome instability compared with the corresponding single mutants (Ho et al., 2010; Agmon et al., 2011; Wyatt et al., 2013). Also, it has recently been shown that HeLa cells depleted in both MUS81 and GEN1 proteins show genome instability, delayed S-phase progression, and impaired replication fork movement (Sarbajna et al., 2014).
Here, we show that mus81 send1 Arabidopsis cells exhibit cell cycle defects, increased polyploidy, and elevated levels of cell death. These phenotypes are accompanied by mitotic anaphase bridges and accumulation of DSB at S-phase, suggesting replication fork collapse. These phenotypes are not observed in the corresponding single mutants. The requirement for the action of least one of the two proteins at replication is also seen in our observation of telomere loss in the double mutant plants. Sequences beyond the most distal replication origins at the ends of linear chromosomes, including telomeres, have the particular property of being replicated by one outward-moving replication fork, making these regions of the genome particularly sensitive to replication fork collapse, which can cause catastrophic telomere loss. The resulting shortened telomeres can be recognized as DSB, and their repair generates dicentric chromosomes detected as mitotic anaphase bridges. In agreement with this, we observe loss of telomeric repeats sequences in the double mutant and not in the single mutant plants. Furthermore, 80% of anaphase bridges in mus81 send1 plants include a subtelomeric FISH signal, indicative of end-to-end chromosome fusions. Our results suggest that MUS81 and SEND1 facilitate full telomere replication and thus contribute to the maintenance of functional telomeres. It is not known whether mammalian cells deficient for the MUS81 and GEN1 nucleases show loss of telomeric repeats. However, depletion of MUS81 in ALT (alternative lengthening of telomeres) cells, in which telomeres are maintained by HR, results in reduction of specific telomere recombination, telomere loss, and proliferation arrest (Zeng et al., 2009; Wan et al., 2013; Wilson et al., 2013). In contrast, no telomere loss was observed in telomerase-positive cells depleted of MUS81 (Pepe and West, 2014). Whether GEN1 participates in telomere recombination in ALT cells remains an open question.
In conclusion, here, we present an analysis of the in vivo roles of the two Arabidopsis Gen1/Yen1 homologs, GEN1 and SEND1. Arabidopsis gen1, send1, and gen1 send1 mutants grow normally, are fully fertile, and show no detectable hypersensitivity to DNA damaging agents. No visible phenotype was seen in mus81 gen1 mutants, which develop normally and are fertile. In striking contrast, the absence of MUS81 and SEND1 results in chromosomal instability, cell cycle defects, increased polyploidy, elevated levels of cell death, and severe developmental defects. These phenotypes are not suppressed by the absence of RAD51, pointing to roles of SEND1 and MUS81 in DNA replication rather than DNA break repair. A role in dealing with toxic replication intermediates is further supported by the enrichment of mitotic S-phase γ-H2AX foci nuclei and a striking loss of telomeric DNA in mus81 send1 plants. The strong growth and mitotic phenotypes make interpretation of the partial infertility of these plants subject to caution; however, we do show that mus81 send1 plants can carry out apparently normal meiosis and produce viable seed. Thus, while Arabidopsis SEND1 appears to have only a minor role in the resolution of HJ in mitosis and meiosis, MUS81 and SEND1 are essential for resolving toxic replication structures in order to ensure genome stability, telomere integrity, and faithful segregation of chromosomes in mitotic cells.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana plants used in this study were in the Columbia background, except gen1-1, which is of the Wassilewskija ecotype. Seeds of gen1-1 (FLAG_121A03) and send1-1 (SALK_135735) T-DNA insertion mutants were obtained through the INRA Versailles and the Nottingham Arabidopsis Stock Centres, respectively. For other mutants, we used the following alleles: mus81-2 (SALK_107515; Hartung et al., 2006), rad51-1 (GABI-KAT_134A01; Li et al., 2004), fancm-9 (SALK_120621; Crismani et al., 2012), recq4a-4 (Hartung et al., 2007), and RAD51-GFP (Da Ines et al., 2013). The RNaseH2 mutant, trd1-1 (Kalhorzadeh et al., 2014), was kindly provided by Lieven de Veylder.
Seeds were stratified in water at 4°C for 2 d and grown on soil in a growth chamber or in a greenhouse. For in vitro culture, seeds were surface sterilized for 5 min with 75% ethanol and 0.05% SDS, rinsed with 95% ethanol for 5 min, and air-dried. Sterilized seeds were then sown on half-strength Murashige and Skoog medium, stratified at 4°C for 2 d, and placed in a growth cabinet. All plants were grown under 16-h-light/8-h-dark cycles (white fluorescent tubes, 100 to 130 µmol m−2 s−1) at 23°C and 40 to 60% relative humidity
Molecular Characterization of gen1-1 and send1-1 T-DNA Insertion Mutants
The gen1-1 mutant was genotyped using primers c and d to detect the wild-type loci and primers d and TAG6 (Versailles T-DNA left border-specific primer) to detect the T-DNA insertion allele. The send1-1 mutant was genotyped using primers c and d to detect the wild-type loci and primers d and TAG6 (Versailles T-DNA left border-specific primer) to detect the T-DNA allele. For the send1-1 mutant, genotyping was performed using primers i and l to detect the wild-type loci and primers i, l, and Lba1 (SALK T-DNA left border-specific primer) to detect the T-DNA allele. Sequences of primers used for genotyping are listed in Supplemental Table 1.
Cloning and Transformation of SEND1 Complementation Constructs
For complementation, a 5577-bp DNA fragment encompassing the full SEND1 genomic coding sequence (including introns) and 1.2 kb of upstream sequence was amplified by PCR (forward primer TGATTAGCATTTAGCGTCAAG and reverse primer ATCCTTGCAATCTGTTACACC) using Arabidopsis Columbia-0 wild-type genomic DNA as a template. The PCR product was inserted into pDONR221 (Invitrogen) and verified by sequencing. The complete SEND1 fragment was then transferred to the Gateway destination vector pB7FWG2 from which the 35S promoter was removed with a SacI/SpeI digest. Eventually, the plasmid was transformed into Agrobacterium tumefaciens C58C1, which was subsequently used to transform mus81/MUS81 send1/send1 mutant plants by the floral dip method (Bechtold and Pelletier, 1998; Clough and Bent, 1998). Seeds from the Agrobacterium-treated plants were sown on soil, and transformants were selected for BASTA resistance. Presence of the SEND1 construct and genotypes of BASTA-resistant plantlets were verified by PCR.
RT-PCR Analyses
For RT-PCR, total RNA was extracted from rosette leaves of wild-type, gen1-1, and send1-1 plants using TRizol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. After treatment with RQ1 RNase-free DNase (Promega) and reverse transcription using M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions, 80 ng was used for each PCR amplification. Primers used for PCR are indicated in Figure 1, and their sequences listed in Supplemental Table 1. Amplifications were performed three times for send1 and twice for gen1 (with multiple pairs of primers per repetition; see Figure 1).
TRF Analyses
TRF analysis of telomere length in MboI-digested genomic DNA was assayed as described previously (Gallego and White, 2001).
Sensitivity Assays
For all assays, seeds were surface-sterilized and sown onto solid medium containing half-strength Murashige and Skoog salts, 1% sucrose, and 0.8% agar, stratified in the dark for 2 d at 4°C, and transferred to a growth cabinet.
For the MMC assay, medium was supplemented with 40 µM Mitomycin C (Sigma-Aldrich), and plants were then grown for 2 weeks. For γ-irradiation, 5-d-old seedlings were irradiated with a dose of 100 Gy from a 137Cs source, returned to standard growth conditions, and further grown for 10 d. Sensitivity was then analyzed in 2-week-old seedlings by counting the number of true leaves as previously described (Bleuyard and White, 2004). Plants with more than three true leaves were considered to be resistant. For the HU sensitivity assay, seeds were germinated on vertical plates containing medium supplemented with 0.5 mM HU and allowed to grow for 12 d. Sensitivity was analyzed by measuring root growth between days 5 and 12. For UV sensitivity assays, seeds were germinated on vertical plates and allowed to grow for 5 d. Five-day-old seedlings were then treated with 300 J⋅m−2 UV-C radiation (GS Gene linker; Bio-Rad) and allowed to grow for 7 d in the dark to avoid photoreactivation, and final root length was measured. Relative root lengths were calculated as the final root length of a given plant over the mean final root length of the corresponding wild-type plants.
Cell Death Assay
For analysis of cell death in roots, 5-d-old seedlings grown on solid medium were immersed in a 5 µg/mL propidium iodide solution for 1 min and then rinsed three times in water. Roots were then transferred to microscope slides in a drop of water, covered with cover slips, and observed using a motorized Zeiss AxioImager.Z1 epifluorescence microscope.
Analyses of DNA Content with Flow Cytometry
Nuclei were prepared with the Cystain UV Precise P kit (Partec) according to the manufacturer’s protocol. Briefly, 20 1-week-old seedlings were chopped with a sharp razor blade in 200 mL of Cystain UV Precise P nuclei extraction buffer supplemented with 800 mL of Cystain UV Precise P staining buffer. Samples were filtered through a 30-µm nylon mesh, and supernatants were analyzed using an Attune Acoustic Focusing Cytometer (Life Technologies) following the manufacturer’s protocol. Results were analyzed using the Attune Cytometric Software version 1.2.5. The EI represents the average number of endocycles undergone by a typical nucleus (EI = 1*4C+2*8C+3*16C) (Takahashi et al., 2008).
EdU Incorporation
Five-day-old seedlings were transferred to half-strength Murashige and Skoog liquid medium containing 10 mM EdU and incubated for 1 h at room temperature. Seedlings were fixed for 15 min in 3.7% (v/v) formaldehyde in PBS, pH 7.4, rinsed twice with PBS/3% BSA, and placed in 0.5% (v/v) Triton X-100/PBS for 20 min. Seedlings were then incubated in Click-It reaction solution for 30 min with gentle agitation, rinsed in PBS/3% BSA, and eventually transferred to PBS. EdU detection was then performed using the Click-It EdU Alexa Fluor imaging kit following the manufacturer’s instructions (Molecular Probes).
DAPI Staining of Mitoses and FISH
Whole inflorescences were collected and fixed, and mitotic nuclei of flower pistils were squashed onto slides. Slides were then mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; 1.5 µg⋅mL−1) (Vector Laboratories). FISH with subtelomeric probes was performed as previously described (Vannier et al., 2009).
Immunostaining Using γ-H2AX Antibodies
Slide preparation, immunostaining, and quantification of γ-H2AX foci in root tip nuclei were performed as previously described (Amiard et al., 2011).
Analysis of Meiosis
Pollen viability and tetrad analysis were performed as described (Crismani and Mercier, 2013). Meiotic chromosome spreads were prepared according to Ross et al. (1996). Briefly, whole inflorescences were fixed in ice-cold ethanol/glacial acetic acid (3:1) for 3 × 30 min and stored at −20°C until further use. Immature flower buds were rinsed twice at room temperature in distilled water for 5 min followed by two washes in 1× citrate buffer for 5 min. Buds of appropriate size were selected under a binocular microscope and incubated for 3 h on a slide in 100 μL of enzyme mixture (0.3% [w/v] cellulase [Sigma-Aldrich], 0.3% [w/v] pectolyase [Sigma-Aldrich], and 0.3% cytohelicase [Sigma-Aldrich]) in a moist chamber at 37°C. Each bud was then softened for 1 min in 20 μL 60% acetic acid on a microscope slide at 45°C, fixed with ice-cold ethanol/glacial acetic acid (3:1), and air dried. Slides were then mounted in Vectashield mounting medium with DAPI (1.5 µg⋅mL−1) (Vector Laboratories).
Microscopy
All observations were made with a motorized Zeiss AxioImager.Z1 epifluorescence microscope (Carl Zeiss) using a PL Apochromat 100×/1.40 oil objective. Photographs were taken with an AxioCam Mrm camera (Carl Zeiss) and Zeiss filter sets adapted for the fluorochromes used: filter set 25HE (DAPI), filter set 38HE (Alexa 488), and filter set 43HE (Alexa 596, propidium iodide). Images were captured in three dimensions (x, y, z) and further processed with the Zeiss ZEN lite and Adobe Photoshop CS4 software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: GEN1 (AT1G01880), SEND1 (AT3G48900), MUS81 (AT4G30870), RAD51 (AT5G20850), FANCM (AT1G35530), RECQ4A (AT1G10930), and TRD1 (At2G25100).
Supplemental Data
Supplemental Figure 1. Sequence alignment of GEN1/Yen1 proteins.
Supplemental Figure 2. Sensitivity of gen1, send1, and gen1 send1 to DNA damage.
Supplemental Figure 3. Pollen viability and meiosis in wild-type and mutant plants.
Supplemental Figure 4. Developmental defects in double and triple mutant plants.
Supplemental Figure 5. Fertility of fancm send1 and recq4a send1 mutants.
Supplemental Figure 6. Growth of trd1 and trd1 send1 mutants.
Supplemental Figure 7. Pollen viability and tetrad analysis in mus81 send1 mutants.
Supplemental Table 1. Primers used for characterizing gen1 and send1 T-DNA mutants.
Supplementary Material
Acknowledgments
We thank Lieven de Veylder, Raphael Mercier, and Holger Puchta for providing trd1, fancm, and mus81 and recq4a mutant seeds, respectively. This work was financed by a European Union research grant (FP7-KBBE-2008-227190), the Centre National de la Recherche Scientifique, the Université Blaise Pascal, the Université d’Auvergne, and the Institut National de la Santé et de la Recherche Médicale.
AUTHOR CONTRIBUTIONS
M.O., O.D.I., S.A., H.S., M.E.G., and C.I.W. conceived and designed the experiments. M.O., O.D.I., S.A., C.G., and M.E.G. performed the experiments. M.O., O.D.I., S.A., M.E.G., and C.I.W. analyzed the data. M.O., O.D.I., M.E.G., and C.I.W. wrote the article.
Glossary
- DSB
double-strand break
- HR
homologous recombination
- RF
replication fork
- HJ
Holliday junction
- NCO
non-crossover
- CO
crossover
- SCE
sister chromatid exchange
- HU
hydroxyurea
- EI
endoreduplication index
- EdU
5-ethynyl-2′-deoxyuridine
- TRF
terminal restriction fragment
- FISH
fluorescence in situ hybridization
- DAPI
4′,6-diamidino-2-phenylindole
References
- Adachi S., et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10004–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N., Yovel M., Harari Y., Liefshitz B., Kupiec M. (2011). The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 39: 7009–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S., Depeiges A., Allain E., White C.I., Gallego M.E. (2011). Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell 23: 4254–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherieh-Najjar M.B., de Vries O.M., Hille J., Dijkwel P.P. (2005). Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 43: 789–798. [DOI] [PubMed] [Google Scholar]
- Bailly A.P., Freeman A., Hall J., Déclais A.C., Alpi A., Lilley D.M., Ahmed S., Gartner A. (2010). The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet. 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauknecht M., Kobbe D. (2014). AtGEN1 and AtSEND1, two paralogs in Arabidopsis, possess holliday junction resolvase activity. Plant Physiol. 166: 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266. [DOI] [PubMed] [Google Scholar]
- Blanco M.G., Matos J., Rass U., Ip S.C., West S.C. (2010). Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Bleuyard J.Y., White C.I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. (1954). Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am. J. Dis. Child. 88: 754–758. [PubMed] [Google Scholar]
- Castor D., Nair N., Déclais A.C., Lachaud C., Toth R., Macartney T.J., Lilley D.M., Arthur J.S., Rouse J. (2013). Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol. Cell 52: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg S., German J. (1974). A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Aström S.U. (2012). A catalytic and non-catalytic role for the Yen1 nuclease in maintaining genome integrity in Kluyveromyces lactis. DNA Repair (Amst.) 11: 833–843. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crismani W., Mercier R. (2013). Identifying meiotic mutants in Arabidopsis thaliana. Methods Mol. Biol. 990: 227–234. [DOI] [PubMed] [Google Scholar]
- Crismani W., Girard C., Froger N., Pradillo M., Santos J.L., Chelysheva L., Copenhaver G.P., Horlow C., Mercier R. (2012). FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Da Ines O., Degroote F., Goubely C., Amiard S., Gallego M.E., White C.I. (2013). Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 9: e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., Dayani Y., Lichten M. (2012). BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubest S., Gallego M.E., White C.I. (2002). Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 3: 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S., et al. (2009). Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A., Pirzio L.M., Prosperi E., Sapora O., Bignami M., Pichierri P. (2008). Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Kimura S., Ishibashi T., Mori Y., Hashimoto J., Sakaguchi K. (2003). OsSEND-1: a new RAD2 nuclease family member in higher plants. Plant Mol. Biol. 51: 59–70. [DOI] [PubMed] [Google Scholar]
- Gallego M.E., White C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E., Kim Y., Lach F.P., Kottemann M.C., Smogorzewska A. (2013). Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Reports 5: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. (1993). Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 72: 393–406. [PubMed] [Google Scholar]
- Geuting V., Kobbe D., Hartung F., Dürr J., Focke M., Puchta H. (2009). Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol. 150: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R. (2007). The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14: 1096–1104. [DOI] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Knoll A., Wurz-Wildersinn R., Puchta H. (2008). Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Mazón G., Lam A.F., Symington L.S. (2010). Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 40: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S.C., Rass U., Blanco M.G., Flynn H.R., Skehel J.M., West S.C. (2008). Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Kalhorzadeh P., Hu Z., Cools T., Amiard S., Willing E.M., De Winne N., Gevaert K., De Jaeger G., Schneeberger K., White C.I., De Veylder L. (2014). Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell 26: 3680–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Schröpfer S., Puchta H. (2014). The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front. Plant Sci. 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.I., Abe K., Endo M., Osakabe K., Ohtsuki N., Nishizawa-Yokoi A., Tagiri A., Saika H., Toki S. (2013). DNA replication arrest leads to enhanced homologous recombination and cell death in meristems of rice OsRecQl4 mutants. BMC Plant Biol. 13: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D.M., White M.F. (2001). The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2: 433–443. [DOI] [PubMed] [Google Scholar]
- Lorenz A., West S.C., Whitby M.C. (2010). The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 38: 1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., Maslen S., Skehel J.M., West S.C. (2011). Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., West S.C. (2014). Holliday junction resolution: regulation in space and time. DNA Repair (Amst.) 19: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S., Miki D., Akiyama M., Kawahara M., Izawa T., Maki H., Shimamoto K. (2005). RNAi-mediated silencing of OsGEN-L (OsGEN-like), a new member of the RAD2/XPG nuclease family, causes male sterility by defect of microspore development in rice. Plant Cell Physiol. 46: 699–715. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz I.M., et al. (2009). Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35: 116–127. [DOI] [PubMed] [Google Scholar]
- Murfuni I., De Santis A., Federico M., Bignami M., Pichierri P., Franchitto A. (2012). Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis 33: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Murfuni I., Nicolai S., Baldari S., Crescenzi M., Bignami M., Franchitto A., Pichierri P. (2013). The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene 32: 610–620. [DOI] [PubMed] [Google Scholar]
- Naim V., Wilhelm T., Debatisse M., Rosselli F. (2013). ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15: 1008–1015. [DOI] [PubMed] [Google Scholar]
- Pepe A., West S.C. (2014). MUS81-EME2 promotes replication fork restart. Cell Reports 7: 1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U., Compton S.A., Matos J., Singleton M.R., Ip S.C., Blanco M.G., Griffith J.D., West S.C. (2010). Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 24: 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Jones G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4: 507–516. [DOI] [PubMed] [Google Scholar]
- Sarbajna S., Davies D., West S.C. (2014). Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 28: 1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E.K., Heyer W.D. (2011). Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla-Arnaud M., et al. (2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA 112: 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen J.M., Smogorzewska A., Sowa M.E., O’Connell B.C., Gygi S.P., Elledge S.J., Harper J.W. (2009). Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inzé D., De Veylder L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y.D., Wu L. (2010). Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 285: 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J.-B., Depeiges A., White C., Gallego M.E. (2009). ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 5: e1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B., et al. (2013). SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Reports 4: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler T., Newman S., West S.C. (2011). Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. (1997). Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31: 213–244. [DOI] [PubMed] [Google Scholar]
- Wilson J.S., Tejera A.M., Castor D., Toth R., Blasco M.A., Rouse J. (2013). Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Reports 4: 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. (2003). The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. (2006). DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40: 279–306. [DOI] [PubMed] [Google Scholar]
- Wyatt H.D., Sarbajna S., Matos J., West S.C. (2013). Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ishino S., Yamagami T., Kumamaru T., Satoh H., Ishino Y. (2012). The OsGEN-L protein from Oryza sativa possesses Holliday junction resolvase activity as well as 5′-flap endonuclease activity. J. Biochem. 151: 317–327. [DOI] [PubMed] [Google Scholar]
- Ying S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., Hickson I.D. (2013). MUS81 promotes common fragile site expression. Nat. Cell Biol. 15: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N. (2012). Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Xiang T., Pandita T.K., Gonzalez-Suarez I., Gonzalo S., Harris C.C., Yang Q. (2009). Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 11: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.