Abstract
The thiourea isoxyl (thiocarlide; 4,4′-diisoamyloxydiphenylthiourea) is known to be an effective anti-tuberculosis drug, active against a range of multidrug-resistant strains of Mycobacterium tuberculosis and has been used clinically. Little was known of its mode of action. We now demonstrate that isoxyl results in a dose-dependent decrease in the synthesis of oleic and, consequently, tuberculostearic acid in M. tuberculosis with complete inhibition at 3 μg/ml. Synthesis of mycolic acid was also affected. The anti-bacterial effect of isoxyl was partially reversed by supplementing growth medium with oleic acid. The specificity of this inhibition pointed to a Δ9-stearoyl desaturase as the drug target. Development of a cell-free assay for Δ9-desaturase activity allowed direct demonstration of the inhibition of oleic acid synthesis by isoxyl. Interestingly, sterculic acid, a known inhibitor of Δ9-desaturases, emulated the effect of isoxyl on oleic acid synthesis but did not affect mycolic acid synthesis, demonstrating the lack of a relationship between the two effects of the drug. The three putative fatty acid desaturases in the M. tuberculosis genome, desA1, desA2, and desA3, were cloned and expressed in Mycobacterium bovis BCG. Cell-free assays and whole cell labeling demonstrated increased Δ9-desaturase activity and oleic acid synthesis only in the desA3-overexpressing strain and an increase in the minimal inhibitory concentration for isoxyl, indicating that DesA3 is the target of the drug. These results validate membrane-bound Δ9-desaturase, DesA3, as a new therapeutic target, and the thioureas as anti-tuberculosis drugs worthy of further development.
The prevalence of tuberculosis, particularly in concert with human immunodeficiency virus infection and AIDS, has been well documented (1). An equally serious public health problem is increasing multi-drug-resistant tuberculosis (2). At present only a few alternative chemotherapeutic regimens are available, resulting in poor therapeutic outcomes and high mortality rates among multi-drug-resistant tuberculosis patients (3). There is an urgent need to develop new effective antituberculosis drugs with bactericidal mechanisms different from those of the presently available agents.
It is prudent to re-examine drugs that were formerly deemed effective against tuberculosis. Isoxyl (ISO)1 (thiocarlide) (Fig. 1) is a thiourea derivative that was successfully used in the 1960s to treat tuberculosis (4–7). Recently, ISO was shown to have considerable antimycobacterial activity in vitro and to be effective against various clinical isolates of multidrug-resistant strains of Mycobacterium tuberculosis in the range of 1–10 μg/ml (8). An early note reported that ISO, like isoniazid (INH) and ethionamide (ETH), strongly inhibits the synthesis of mycolic acids (9), a result since confirmed with the demonstration that all types of mycolic acids are affected (8). In addition it was noted that ISO also inhibited shorter chain fatty acid synthesis (8–11), suggesting inhibitory effects different from those of INH and ETH and raising the prospects of novel fatty acid biosynthetic targets exploitable for new drug development against multi-drug-resistant tuberculosis.
Fig. 1.
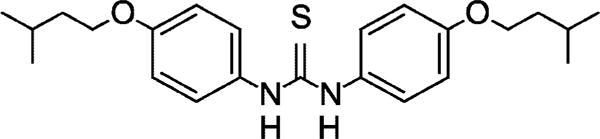
Structure of the thiourea known as thiocarlide or isoxyl, a 4,4′-diisoamyloxydiphenylthiourea.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Escherichia coli strain XL-1 Blue (Stratagene, La Jolla, CA), cultured on Luria-Bertani broth or agar medium (Invitrogen) and containing kanamycin at a concentration of 25 μg/ml, was used for generating recombinant clones. Mycobacterium bovis BCG strain 1173P2 was used for whole-cell labeling, MIC determinations, cell-free reactions, and expression of M. tuberculosis H37Rv desA1, desA2, and desA3 genes. Liquid cultures of M. bovis BCG and M. tuberculosis H37Rv (ATCC 27711) were grown in Sauton medium containing 0.025% tyloxapol (12). Recombinant M. bovis BCG clones were selected on Middlebrook 7H11 agar supplemented with oleic acid-albumin-dextrose-catalase (OADC) enrichment (Difco) (13) or on Sauton medium containing 20 μg/ml kanamycin. To determine whether the addition of this oleic acid supplement could override the effects of ISO, a series of 10-fold dilutions of M. tuberculosis was prepared from stock culture in a glycerol-alanine-salts medium with phosphate-buffered saline as diluent. An aliquot (5 μl) of each dilution was spotted on two types of agar plates, those with 7H11 agar medium supplemented with OADC or those with non-oleic acid-containing ADC (albumin-dextrose-catalase), prepared with the incorporation of ISO to a final concentrations of 0.1, 0.5, 1.0, 2.0, 3.0, 4.0, and 5.0 μg/ml. After inoculation, the plates were incubated at 37 °C for 21 days. The growth rates and bactericidal effects were scored by comparing size and number of colonies on plates.
Whole-cell Radiolabeling and Analysis of the in Vivo Effects of ISO and Sterculic Acid on Fatty Acid and Mycolic Acid Synthesis
Mycobacteria were grown in Sauton medium at 37 °C to A600 nm 0.250. ISO (a gift from Dr. P. Draper, National Institute of Medical Research, London, UK) was added followed by further incubation for 8 h before the addition of 1,2-[14C]acetate (110 mCi/mmol; PerkinElmer Life Sciences) to a final concentration of 1 μCi/ml. Cells were labeled for 24 h, harvested, washed, saponified with 15% tetrabutylammonium hydroxide at 100 °C overnight, methylated, and extracted (8). Extracts containing equal amounts of radiolabeled fatty acid methyl esters and mycolic acid methyl esters from control and ISO-treated cells were subjected to TLC on silica gel plates (silica gel 60 F254, Merck) that had been impregnated with 10% silver nitrate and activated (14). Plates were developed twice in petroleum ether-acetone (90:10), radioactive bands were located by autoradiography, and the oleic acid methyl ester standard was visualized by spraying with 10% sulfuric acid and anisaldehyde (15).
To analyze the effects of ISO on the synthesis of individual fatty acids, extracted methyl esters were treated with Trisil® (Pierce) to silate any free hydroxyl groups, and products were dissolved in hexane and injected onto a capillary HP-1 column (5 m × 0.53 mm inner diameter) (Supelco Inc., Bellefonte, PA) coupled to a Hewlett Packard HP 5890 Series II GC with a thermal conductivity detector attached to a GC-RAM™ radio detector (Inus Systems, Tampa, FL). The initial column temperature was 80 °C, which was increased to 185 °C at a rate of 30 °C/min, followed by an increase at the rate of 5 °C/min to a final temperature of 345 °C. The eluted peaks of labeled FAMEs were identified by comparison of their retention time with those of available fatty acid methyl ester standards. Analysis of the effects of sterculic acid, the Δ9-desaturase inhibitor, on oleic acid synthesis was performed by whole cell labeling with [1,2-14C]acetate followed by argentation TLC and GC analysis, as described above. The effects of sterculic acid and ISO on mycolic acid synthesis were compared by argentation TLC.
Confirmation of Position of Double Band in Mono-unsaturated Fatty Acid
Fatty acids were derived from harvests of M. tuberculosis H37Rv wild type, M. bovis BCG wild type, and M. bovis BCG overexpressing M. tuberculosis desA3 and converted to the methyl esters (8). Methyl esters (0.5 mg) were dissolved in 50 μl of a pyridine-dioxane mixture (1:8) followed by the addition of 500 μg of OsO4 in 50 μl of dioxane (16). The solution was allowed to stand for 2 h at room temperature followed by another hour after the addition of 1.5 ml of a suspension of 16% Na2SO3 in water-methanol (1:3:4). The mixture was gradually acidified to pH 3.0 with HCl and extracted twice with n-hexane. The combined hexane extracts were dried under nitrogen and silated as described above. The osmium-oxidized-silated products were dissolved in n-hexane, and 1-μl aliquots were analyzed by GC/mass spectroscopy (Thermo-Quest gas chromatograph and mass spectrometer) under the following operating conditions: 30-m by 0.25-mm capillary column (Supelco); oven temperature, 60 to 170 °C at 30 °C for 1 min, then from 170 to 300 °C at 4 °C min−1; carrier gas, He; total running time, 32 min. The following fatty acid methyl esters, C18:1 Δ6, C18:1 Δ7, C18:1 Δ9, C18:1 Δ11, C18:1 Δ12, C18:1 Δ15, purchased from Supelco Inc. (Bellefonte, PA), were likewise oxidized and analyzed.
Cell-free Reaction for Analysis of the in Vitro Effect of ISO on Oleic Acid Synthesis
The wild type M. bovis BCG strain was grown in Sauton medium supplemented with FeSO4 (20 mg/liter) (17) and 0.025% Tyloxapol. Cells were harvested by centrifugation, resuspended in 0.25 M sucrose, pulse-disrupted by probe sonication, and centrifuged at 27,000 × g for 30 min at 4 °C. The Δ9-stearoyl-CoA desaturase activity was assayed as described with minor modifications (18) in a reaction mixture containing 1 μmol of NADPH (Sigma) in 0.1 mM potassium phosphate buffer, pH 7.2, and 2 mg of the M. bovis BCG 27,000 × g supernatant in a total volume of 1 ml. Mixtures were preincubated in the presence or absence of ISO (dissolved in 20 μl of a solution in 50% dimethyl sulfoxide and 1% Tween 20) at 37 °C for 20 min before the addition of 25 nmol (0.25 μCi) of [1-14C]stearoyl-CoA (10 μCi/μmol in 0.01 M 1:1 sodium acetate:ethanol; American Radiolabeled Chemicals, St. Louis, MO). Purified recombinant six-histidine-tagged protein (3.7–22 μg) produced in M. bovis BCG was added to some assays. After further incubation at 37° for 30 min, reactions were terminated by the addition of 2 ml of tetrabutylammonium hydroxide solution, saponified, and methylated. The radiolabeled products were resolved by argentation-TLC in petroleum ether:acetone (90:10, two developments), and the incorporation of radioactivity into oleic acid was analyzed using a Bioscan System 2000 Imaging Scanner (Bioscan Inc., Washington, D.C.).
Detection of M. tuberculosis desA3 Transcript
M. tuberculosis H37Rv was maintained in Sauton broth for RNA isolation and subsequent RT-PCR. After the addition of lysozyme and proteinase K to a cell suspension in RNase-free water, total RNA was isolated with Trizol reagent according to the manufacturer’s instructions (Invitrogen). The purified RNA was used subsequently in RT-PCR to detect desA3 transcripts. Reverse transcription and PCR were carried out sequentially in a single tube using Ready-To-Go RT-PCR beads (Amersham Biosciences). Each room-stable bead contained Moloney murine leukemia virus reverse transcriptase, RNase inhibitor, buffer, nucleotides, Taq DNA polymerase, and 30 μl of RNA template in RNase free water. The RT-PCR reaction was conducted in a programmable master-gradient thermal machine (Eppendorf, Germany) using specific desA3 primers 5′-GGT GAC CGA CTT AGC CAT ACG CTT-3′ (upper primer) and 5′-AGC ATC ACC TCT ATC CGG AC TGC C-3′ (lower primer). The thermal program involved reverse transcription at 55 °C for 30 min followed by the initial denaturation at 94 °C for 5 min. The amplification was performed for 35 cycles of denaturation at 94 °C for 1 min, annealing, and extension at 72 °C for 1 min. The final extension was set at 72 °C for 10 min. Negative controls contained all reaction components except the RNA template. To determine whether transcribed mRNA or contaminating DNA was amplified, purified nucleic acids were subjected to DNase and RNase treatment before cDNA synthesis and PCR. The amplicons obtained from RT-PCR were analyzed for the 341-bp fragment specific for M. tuberculosis desA3 by 1.0% agarose gel electrophoresis (19) in Tris borate-EDTA buffer (0.089 M Tris-HCl, pH 8.0, 0.044 M boric acid, and 0.001 M EDTA) at 100 V for 1 h. Gels were stained with ethidium bromide and visualized by UV transillumination.
Cloning and Overexpression of the M. tuberculosis desA Gene Family in M. bovis BCG
Two primers were designed to correspond to the 5′ and 3′ ends of each open reading frame previously identified as M. tuberculosis desA1 and desA2 (Rv0824c and Rv1094, respectively) (20). A 1034-base pair fragment of desA1 was PCR amplified with the primers 5′-CTCTGAATTCTCACCGGGCTAACGAC-3′ and 5′-CAGCCAAACTGACCGACCTGCA-3′, sense and antisense, respectively. A similar strategy was used to amplify a 895-base pair fragment of desA2 with the primers 5′-CACAAAACCTGTTGCTGAAGCCGCTGACCC-3′ and 5′-GCTAGAATTCGAGTGCGACGCTACTCCG-3′. Underlined sequences indicate the introduced EcoRI restriction sites. The PCR-amplified desA1 and desA2 genes were gel-purified, cut with EcoRI, and cloned into BalI-EcoRI sites of pMV261 under the control of the transcriptional and translational signals of the heat shock protein 60 (hsp60) promoter (21). The desA3, Rv3329c, was overexpressed in pVV16, a derivative of pMV261, allowing the production of C-terminal six-histidine-tagged fusion proteins under the control of the phsp60 promoter (22). The desA3 gene was amplified using the primers 5′-GTCGAGCAAAGCTTGGCTGCCAGATCGTC-3′ and 5′-AGGGAGAAGCATATGGCGATCACTGACGTC-3′ (underlined sequences correspond to HindIII and NdeI restriction sites, respectively). The 1360-base pair PCR product was digested with NdeI and HindIII and cloned into the similarly digested pVV16. The resulting expression vectors, pMV261desA1, pMV261desA2, and pVV16desA3, were electroporated into M. bovis BCG competent cells as described (23), and the production of recombinant DesA1, DesA2, and DesA3 proteins was then analyzed by immunoblotting (DesA1 and DesA3) or SDS-PAGE stained with silver nitrate and Coomassie Brilliant Blue (DesA2). Total protein extracts from each strain were loaded, separated on 0.1% SDS-polyacrylamide gels, and transferred to nitrocellulose (Schleicher & Schuell). The production of recombinant DesA1 and six-histidine-tagged DesA3 proteins was revealed using primary anti-DesA1 antibodies raised in a mouse (a gift from Dr. Brigitte Gicquel) and primary mouse anti-His-tagged antibody (Qiagen, Valencia, CA), respectively. Alkaline phosphatase-conjugated secondary mouse antibodies (Sigma) were then used, and color detection for proteins was performed using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Promega, Madison, WI). To locate DesA3 within the bacterial cell, Western blotting was performed using subcellular fractions prepared from the DesA3-His-tagged recombinant strain. The cytosol was recovered as the supernatant after centrifugation of cell lysate at 200,000 × g for 1 h, and the pellet represented the cell membrane fraction. Aliquots (100 μg) of protein from each fraction were analyzed by immunoblotting as described above. Alignment of amino acid sequences was achieved with Multalign (www.toulouse.inra.fr/multalin.html).
Purification of Recombinant His-tagged M. tuberculosis DesA3
The recombinant M. bovis BCG containing pVV16/desA3 were grown to mid-log phase (A600 0.7) at 37 °C in Sauton medium containing 20 μg/ml kanamycin and 20 mg/liter FeSO4. Cells were harvested and washed once in 50 mM MOPS buffer, pH 8. The cell pellet (20 g wet weight) was subsequently resuspended in 30 ml of buffer A (50 mM MOPS, pH 8, 10 mM MgCl2, 10% glycerol, and 500 mM NaCl) and sonicated for 10 cycles (60 s on; 90 s off) on ice. After the cell lysate was centrifuged at 27,000 g for 30 min, the resulting supernatant was collected and loaded onto a 1-ml metal BD-Talon™ resin-chelating column (BD Biosciences Clontech, Palo Alto, CA) previously equilibrated with 20 ml of buffer A. Column washing was performed with 25 ml of buffer A followed by 25 ml of buffer A containing 5 mM imidazole. Proteins were eluted using buffer A containing 50, 100, or 200 mM imidazole. The elution fractions were checked by SDS-PAGE (12% gel) and Western blot. The 200 mM imidazole fraction, which contained His-tagged M. tuberculosis DesA3, was subsequently adjusted to a volume of 2.5 ml with buffer B (10 mM MOPS pH 8, 1 mM MgCl2, 10% glycerol), desalted using a PD-10 column (Amersham Biosciences), and concentrated, and the protein concentration was measured before the determination of the enzymatic function of the purified protein.
Determination of the Effect of Overexpression of desA3 on the MIC of ISO
The microplate alamar blue assay as modified from Yajko et al. (24) was used to determine the MIC of ISO. Inocula were prepared from M. bovis BCG and the recombinant strain harboring the pVV16desA3 construct. Cells were grown in Sauton medium containing 0.025% Tyloxapol to a turbidity equal to that of a No. 1 McFarland standard (~2.0 × 107 colony-forming units/ml). Serial dilutions of ISO from a stock solution in Me2SO were prepared in Sauton medium. Duplicated treatments with different concentrations of ISO were conducted. Four controls were included that contained medium-only, culture-only, medium plus Me2SO, and culture plus Me2SO. At the outset, 10× alamar blue solution was added to the controls. After incubation overnight at 37 °C in a moisture control chamber, alamar blue was added to all treated wells. Plates were further incubated, and the color in each treated culture was recorded. MIC was determined as the lowest concentration of ISO for which the blue dye in the treated culture did not turn pink.
RESULTS
ISO Inhibits the Synthesis of Unsaturated Fatty Acids and Mycolic Acids
Previous studies with [1,2-14C]acetate as a precursor of fatty acid synthesis showed that ISO inhibited the synthesis of both mycolic acids and shorter chain fatty acids in M. tuberculosis (8). In this latter respect, ISO differed from INH and ETH, suggesting a different mode of action of ISO. To evaluate the effects of ISO on the synthesis of saturated fatty acids, unsaturated fatty acids, and mycolic acids, extracts containing 14C-labeled fatty acid and mycolic acid methyl esters were prepared from untreated and ISO-treated M. tuberculosis cultures and analyzed by argentation-TLC. In the presence of ISO at concentrations as low as 1 μg/ml, there was an apparent decided decrease in the synthesis of unsaturated fatty acids concomitant with a partial increase in the synthesis of saturated fatty acids (Fig. 2). Measurement of the radioactive counts in the relevant bands established that ISO at 1.0 μg/ml inhibited unsaturated fatty acid synthesis by 60%. The effect of ISO on mycolic acid synthesis, reported previously (8), can also be seen by this simple analysis; clearly, ISO inhibited the incorporation of [1,2-14C]acetate into all types of resolved mycolic acids (Fig. 2).
Fig. 2. Analysis of the effect of ISO on fatty acid and mycolic acid synthesis in M. tuberculosis.
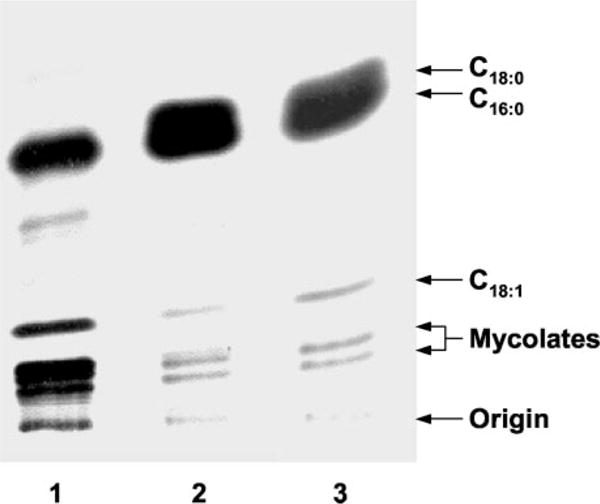
The inhibitory effect of ISO on the synthesis of fatty acids and mycolic acids was assayed by whole-cell labeling with [1,2-14C]acetate in the presence and absence of ISO. Labeled FAMEs and MAMEs were extracted from control and ISO-treated cultures, and equal counts of radiolabel were applied to TLC plates, which had been impregnated with a 10% aqueous AgNO3 solution. Autoradiograms were produced after the plate was developed twice in petroleum ether and acetone (90:10) and exposed to Kodak X-Omat AR film. Lane 1, untreated culture; lane 2, culture treated with 1.0 μg/ml ISO; lane 3, culture treated with 2.0 μg/ml ISO.
The cellular fatty acids synthesized under present conditions by M. tuberculosis and M. bovis BCG are palmitic acid, stearic acid, oleic acid, and tuberculostearic acid (Fig. 3, A and C), in accord with earlier reports (25, 26). In M. tuberculosis and M. bovis BCG treated with ISO at 2.0 μg/ml, the incorporation of [1,2-14C]acetate into oleic acid and tuberculostearic acid was almost totally eliminated as determined by radio-GC (Fig. 3, B and D). Similar results were obtained at 1 μg/ml ISO.
Fig. 3. Effects of ISO on in vivo synthesis of oleic acid and tuberculostearic acid in M. tuberculosis and M. bovis BCG.
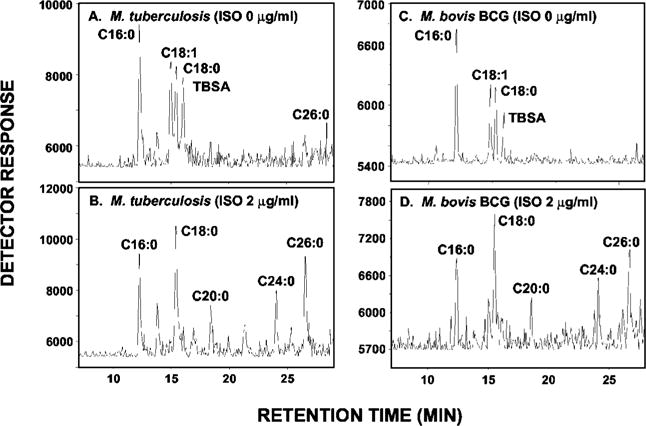
FAMEs (and MAMEs) of [1,2-14C]acetate-labeled M. tuberculosis and M. bovis BCG were prepared from untreated and ISO-treated cultures, and equal counts were subjected to radio-GC coupled to the Lablogic GC-RAM radio detector under conditions that resolve only the FAMEs. A, untreated M. tuberculosis culture; B, ISO-treated (2.0 μg/ml) M. tuberculosis culture; C, untreated M. bovis BCG culture; D, ISO-treated (2.0 μg/ml) M. bovis BCG culture. TBSA, tuberculostearic acid.
The Effects of Oleic Acid Supplement on Partial Rescue of M. tuberculosis from ISO
The observations that ISO inhibited oleic acid synthesis in M. tuberculosis and M. bovis BCG led to the concept that supplementation of the growth medium with oleic acid should rescue cells from the killing effect of ISO. The addition of oleic acid to the 7H11 agar medium in the form of the OADC partially reversed the bactericidal effect of ISO; there was comparable growth on the OADC-supplemented medium with or without ISO; however, the albumin-dextrose complex without oleic acid could not support growth in the presence of ISO (Fig. 4).
Fig. 4. Effects of an oleic acid supplement in reversing the bactericidal effect of ISO.
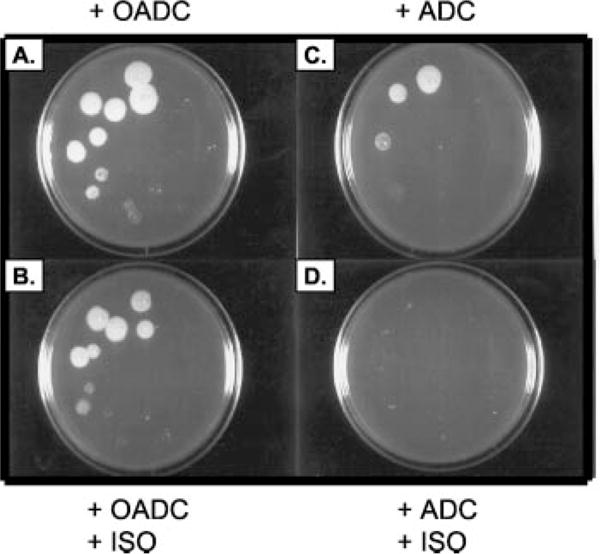
A series of 10-fold dilutions of M. tuberculosis were prepared in medium. An aliquot (5 μl) of each dilution was spotted on 7H11 agar supplemented with OADC or ADC with or without ISO. A, 7H11 medium and OADC; B, 7H11 with OADC and ISO at 0.5 μg/ml; C, 7H11 medium with ADC; D, 7H11 with ADC and ISO at 0.5 μg/ml.
Demonstration of Δ9 Desaturase Activity in Cell-free Extracts and the Effects of ISO
The specific inhibition of oleic acid synthesis by ISO suggested that the drug target is the aerobic desaturation system responsible for the synthesis of oleic acid from stearoyl-CoA in mycobacteria (17, 18, 27, 28). To test this hypothesis M. bovis BCG was grown in iron-supplemented medium, sonicated, and centrifuged at low speed, and both the cell wall pellet and the supernatant containing both cytosol and plasma membrane were assayed for Δ9 fatty acid desaturase activity. The assay was performed as originally described by Fulco and Bloch (18) with minor modifications. For instance, ferrous ions were added to the culture medium rather than to the reaction mixture itself because iron was shown to be required at the terminal stage of synthesis and/or assembly of the desaturase system (17). Only the supernatant allowed the conversion of stearoyl-CoA to the oleic acid derivative in the presence of atmospheric oxygen and NADPH. Consistent with the findings of Fulco and Bloch (18), in the absence of NADPH in the assay mixture, no oleic acid was detected. ISO consistently inhibited the incorporation of radioactivity into the oleic acid, but not into stearic acid, by 7% at 0.1 μg/ml and 61% at a concentration of 1.0 μg of ISO/ml in one typical experiment.
Overexpression of M. tuberculosis desA1, desA2, and desA3 Genes in M. bovis BCG and Evidence That desA3 Encodes a Fatty Acid Desaturase
Three open reading frames in the genome of M. tuberculosis are annotated as putative fatty acid desaturases: desA1, desA2, and desA3 (20). All three open reading frames of desA were amplified by PCR, cloned in-frame into the mycobacterial expression vector pMV261 or pVV16, and constitutively expressed under the control of the hsp60 promoter in M. bovis BCG. Production of all three proteins was demonstrated by SDS-PAGE and staining with silver and Coomassie Brilliant Blue (Des A2) or by immunoblotting (DesA1, DesA3) (results not shown). To assess the cloned open reading frames for Δ9-desaturase activity, the three overexpression strains and M. bovis BCG wild type were labeled with [1,2-14C]acetate, and their derived fatty acid methyl esters were analyzed by argentation-TLC. Quantitation of the production of [14C]oleic acid was performed by scraping and counting the bands corresponding to the methyl ester of oleate. The desA1 and desA2 overexpression strains synthesized oleic acid ~0.2-fold less than the M. bovis BCG wild type in contrast to a 2-fold increase in oleic acid production by M. bovis BCG overexpressing the desA3 gene, suggesting that the putative acyl-CoA desaturase gene desA3, but not the putative acyl-ACP desaturase genes desA1 or desA2, encodes the Δ9-desaturase responsible for the synthesis of oleic acid. Analysis of the radiolabeled fatty acid methyl esters by radio-GC confirmed this conclusion (Fig. 5). Consistent with these results, comparison over time of the Δ9-desaturase activity exhibited by cell-free extracts of M. bovis BCG wild type and the M. bovis BCG/pVV16desA3 clone over time showed an almost 50% increase in enzyme activity in the latter strain at the longer incubation periods (Fig. 6).
Fig. 5. GC analysis of the effects of the overexpression of the desA genes on oleic acid synthesis.
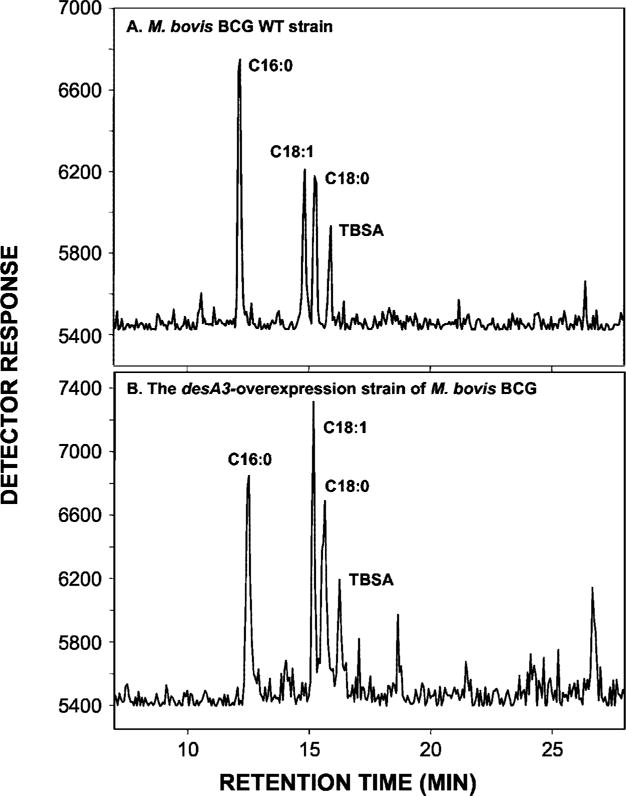
14C-Labeled FAMEs and MAMEs were extracted from M. bovis BCG wild type (WT) and the M. bovis BCG/pVV16desA3. Equal counts of the mixtures of FAMEs and MAMEs from each strain were silated and applied to a Hewlett Packard HP 5890 GC equipped with a capillary HP-1 column and the Lablogic GC-RAM detector. Peaks were identified by comparing the retention times with those of FAME standards.
Fig. 6. Comparison of the levels of Δ9-desaturase activity in M. bovis BCG wild type and M. bovis BCG/pVV16desA3.
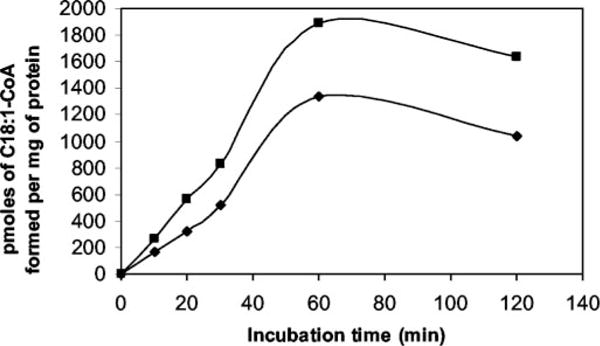
A final volume of 1 ml of reaction mixture contained crude cell lysate of M. bovis BCG wild type (diamonds) or BCG/pVV16desA3 (squares), 20 μl of solution containing 50% Me2SO and 1% Tween 20, 1 μmol of NADPH in 0.1 mM potassium phosphate buffer, pH 7.2, and 25 nmol of [1-14C]18: 0-CoA as substrate. Duplicate assays were performed. After the indicated incubation times, the reactions were stopped with 1 ml of 15% tetrabutylammonium hydroxide. FAMEs were extracted, methylated, and analyzed for[1-14C]C18:1 by argentation TLC and the Bioscan System.
That the C18 unsaturated product formed in the whole cells corresponded to oleic acid (C18:1Δ9) was checked by subjecting the methyl esters to OsO4 oxidation, trimethylsilylation, and GC-mass spectrometry analysis (16). The characteristics of the EI-mass spectrometry spectrum of C18:1Δ6 are a major fragment at m/z 257 and minor fragments at m/z 217 and 185. However, C18:1Δ9 yields typical fragments at m/z 259 and 217. The sole product identified in whole cells of wild type M. bovis BCG and M. bovis BCG/pVV16desA3 was C18:1Δ9 (data not shown).
Because the overexpression of desA3 in whole BCG cells could have indirectly stimulated the production of oleic acid, recombinant DesA3 purification was achieved (Fig. 7A). Although the purified enzyme itself showed no activity in the desaturase assay, presumably due to missing cofactors or components of the desaturase system (e.g. cytochrome c reductase or ferredoxin, flavoprotein) (18, 28), the addition of the purified enzyme to the standard Δ9 cell-free assay resulted in an approximate 2-fold increase in specific activity of the enzyme (Fig. 7B). The enzyme activity was dependent on the amount of recombinant DesA3 added to the assay mixture (results not shown) and the incubation time (Fig. 7B). These results confirmed the function of the DesA3 desaturase as responsible for the synthesis of oleic acid. Finally, a desA3 transcript was detected by RT-PCR in M. tuberculosis H37Rv during the exponential growth (data not shown), confirming the expression of this gene in vivo and emphasizing the importance of the enzymatic product and function.
Fig. 7. Effect of adding purified DesA3 on endogenous Δ9-desaturase activity.
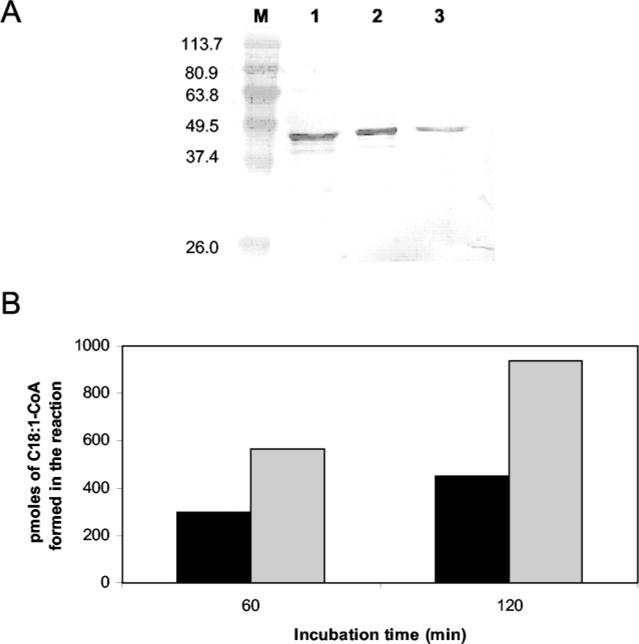
A, SDS-PAGE and Western blot of the purified DesA3 enzyme. M, Bench-Mark™ prestained protein ladder (Invitrogen). Lane 1, DesA3 eluted from a 1 ml of metal BD-Talon™ resin chelating column (BD Biosciences) with 50 mM imidazole. Lane 2, DesA3 as eluted with 100 mM imidazole. Lane 3, pure DesA3 as eluted with 200 mM imidazole. Other details and staining of the gel are described under “Experimental Procedures.” B, DesA3-purified protein was added to the enzymatic reaction to determine the effect of purified DesA3 on Δ9 desaturation. A final volume of 1 ml of reaction mixture contained crude cell lysate of M. bovis BCG wild type (0.55 mg), 20 μl of a solution containing 50% Me2SO and 1% Tween 20, 1 μmol of NADPH in 0.1 mM potassium phosphate buffer, pH 7.2, and 25 nmol of [1-14C]18:0-CoA as substrate. No purified DesA3 was added to the assay (black bars), or purified DesA3 was added in the amount of 4.8 μg (gray bars). Duplicate assays were performed. After 1 or 2 h of incubation at 37 °C, the [1-14C]C18: 1-CoA was quantified as described in Fig. 6.
Overexpression of the M. tuberculosis desA3 Increased the Resistance of M. bovis BCG to ISO
The MIC of ISO against the parent M. bovis BCG as determined by the microplate alamar blue assay was 3.0 μg/ml compared with 6.0 μg/ml for the M. bovis BCG/pVVdesA3-overexpression strain. Thus the terminal enzyme of the Δ9 desaturation system encoded by desA3 is very likely to be the target of the drug.
Comparative Effects of ISO and the Known Δ9 Desaturase Inhibitor, Sterculic Acid
The naturally occurring cyclopropene fatty acid, sterculic acid (9,10-cyclopropenoid acid), is a potent and specific inhibitor of the Δ9 desaturation of stearic to oleic acid (29–32) and, therefore, provided a useful means to compare the effects of ISO to those of an established inhibitor of acyl-CoA Δ9-desaturase. The effect of sterculic acid on the growth of M. bovis BCG was determined by the broth dilution technique (8) at concentrations of 10, 25, 50, and 100 μg/ml. Consistent with the fact that oleic acid is synthesized by a stearoyl-CoA utilizing Δ9-desaturase in mycobacteria, M. bovis BCG was found to be sensitive to sterculic acid. Sterculic acid at 50 μg/ml was sufficient to inhibit the growth of the cells by about 80%, and complete inhibition of ~1.0 × 106 colony-forming units/ml was obtained at a concentration of 100 μg/ml. Under present growth conditions employing the broth dilution technique, sterculic acid had a MIC value between 50 and 100 μg/ml. The inhibitory effect of sterculic acid on the Δ9-desaturase activity was also evaluated in intact cells of M. bovis BCG by measuring the incorporation of [1,2-14C]acetate into oleic acid and analyzing the extractable lipids by radio-GC. At 50 μg/ml, there was complete inhibition of the synthesis of oleic acid (Fig. 8), and the effects of ISO and sterculic acid on oleic acid synthesis were similar in this respect. However, at this concentration, sterculic acid, unlike ISO, had no effect on mycolic acid synthesis, whereas under these conditions, 1 μg ISO/ml resulted in inhibition of synthesis of α- and ketomycolates by 64 and 27%, respectively (results not shown). This comparison of the effects of ISO and sterculic acid leads to the conclusion that the production of oleic acid and mycolic acids is not linked, a conclusion reinforced by the fact that sterculic acid, unlike ISO, does not result in the accumulation of long chain fatty acids (C24:0 to C26:0) (Fig. 8). This latter phenomenon is also seen under conditions of INH treatment (10), suggesting that long chain fatty acid accumulation is a consequence of the inhibition of mycolic acid synthesis and provides further proof that the effects of ISO on mycolic acids are distinct from those on oleic acid synthesis.
Fig. 8. Comparative effects of sterculic acid and ISO on the synthesis of oleic and tuberculostearic acids from [1,2-14C]acetate in M. bovis BCG.
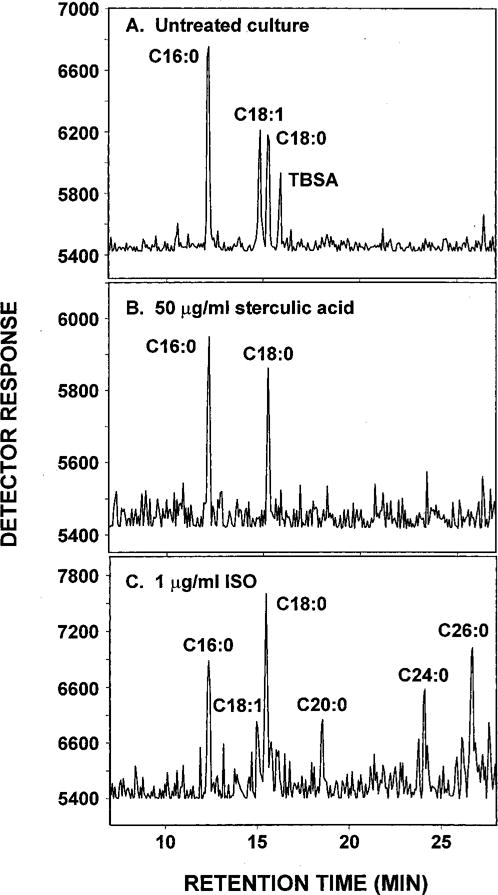
Conditions are described in Fig. 3. A, untreated M. bovis BCG; B, M. bovis BCG treated with 50 μg/ml sterculic acid; C, M. bovis BCG treated with 1.0 μg/ml ISO.
DISCUSSION
Previous observations demonstrated that ISO inhibits the synthesis of both mycolic acids and shorter chain fatty acids (8–10). In contrast, both INH and ETH inhibit mycolic acid synthesis (10, 11) and result in the accumulation of fatty acids (10). Cross-resistance between ISO and ETH suggests that these two drugs share a common step in their action, and a recent genetic and chemical study revealed that cross-resistance to ISO and ETH is associated with the activation of the thiocarbonyl moiety (Fig. 1) shared by these two drugs (33, 34). The product of the resistance-conferring gene was identified as a putative regulator controlling the expression of a flavin-containing monooxygenase that catalyzes the S-oxidation of the thiocarbonyl to S-oxide electrophilic intermediates (34, 35).
The present work serves to characterize the site of action of this activated form of ISO. Following the earlier observation of an inhibition of short chain fatty acids, efforts were focused in this area of metabolism, resulting in the conclusion that ISO is unique in its ability to inhibit the synthesis of oleic acid and tuberculostearic acid (Figs. 2 and 3). Oleic acid is the most abundant unsaturated fatty acid in Mycobacterium spp. (36) and is a vital constituent of mycobacterial membrane phospholipids (37, 38), where it apparently plays an essential role in membrane physiology (39). At physiological temperatures, phospholipids containing only saturated fatty acids cannot form a lipid bilayer, but the introduction of the appropriate unsaturated fatty acids decreases the transition temperature from gel to the liquid crystalline phases and provides membranes with the necessary fluidity for physiological function (40–43). The vital functions of oleic acid lead to the conclusion that inhibition of its synthesis results in cell death, and thus, the enzymes involved in oleic acid synthesis are probably lethal targets for drug therapy.
ISO also had a dramatic effect on the synthesis of the C19-monomethyl-branched stearic acid, tuberculostearic acid. Typically, mycobacteria contain a large amount of methyl-branched fatty acids (44, 45), mainly tuberculostearic acid and multimethyl-branched acids such as phthienoic (2,4,6-trimethyltetracos-Δ2-enoic acid) (46, 47) and mycocerosic (2,4,6,8-tetramethyloctacosanoic) acids (48). The mycocerosic acids are confined to the phthiocerol-containing cell wall lipids and, although apparently implicated in the pathogenesis of M. tuberculosis (49, 50), are not essential for viability. However, tuberculostearic acid is a key ingredient of membrane phospholipids. It appears likely that the inhibition of tuberculostearic acid by ISO is a consequence of its effect on oleic acid synthesis, since tuberculostearic acid arises by direct methylation of oleic acid by S-adenosylmethionine once oleic acid is first esterified in the form of oleoylphosphatidylethanolamine (51, 52). The indirect effect of ISO on tuberculostearic acid probably also extends to the synthesis of the phosphatidylinositol mannosides and eventually lipomannan and lipoarabinomannan, since tuberculostearic acid is a major constituent of all of the mycobacterial phosphoinositides (53).
This evidence suggested at once that ISO acts by inhibiting stearoyl desaturase, the enzyme responsible for the insertion of the double bond at carbon 9 of stearic acid. The existence of such a desaturation system catalyzing the formation of oleic acid in mycobacteria has actually been demonstrated using Mycobacterium phlei as a model strain and shown to involve a particulate terminal desaturase enzyme that desaturates stearoyl-CoA to form octadec-9-enoic acid (oleic acid) in the presence of NADPH as the reducing agent, NADPH-cytochrome c reductase, iron for electron flow, and oxygen as the final acceptor (17, 18, 27, 28). A desaturase assay based on that described by Fulco and Bloch (18) and conducted with cell-free extracts of M. bovis BCG in the presence or absence of ISO clearly demonstrated the ISO inhibits Δ9-desaturase activity in vitro. Furthermore, the Δ9-desaturase of the present system appears to be encoded by the putative acyl-CoA desaturase gene, desA3, as overexpression of the M. tuberculosis desA3 gene in M. bovis BCG, but not those of the other putative desaturase genes of the genome, desA1 or desA3, resulted in increased synthesis of oleic acid and Δ9-desaturase activity in the recombinant strain (Figs. 5 and 6). Our inability to associate Δ9-desaturase activity with the purified Des A3 enzyme is not surprising in light of its membranous nature and complex cofactor requirements. Instead, we had to rely on demonstrating a boosting effect on the endogenous activity of the wild type strain (Fig. 7). The overexpression of desA3 in M. bovis BCG also resulted in increased MIC of ISO in BCG/pVV16desA3 as compared with the parent strain, strongly suggesting that the terminal desaturase enzyme of the desaturation system is the target of the drug. Incidentally, Obukowicz et al. (54) show that a urea-derived inhibitor with a structure not unlike that of ISO selectively inhibits the stepwise Δ5 desaturase synthesis of arachidonic acid in rat and offers promise as an anti-inflammatory agent. This work further underlines the role of urea-based products as inhibitors of membrane and CoA requiring desaturases.
Consistent with its demonstrated membrane localization and affinity for acyl-CoA substrates, DesA3 shares signature motifs of membrane acyl-lipid and acyl-CoA desaturases characterized by three regions of primary conserved sequences, HX(3 or 4)H, HX(2 or 3) HH, and HX(2 or 3)HH, designated as region Ia, Ib, and II (39, 55). However, the alignment of the conserved “membrane desaturase” regions of DesA3 with some representative membrane Δ9 acyl-CoA desaturases from rat, yeast, and mouse revealed certain distinctions of some variable amino acids. In region Ia, all representative Δ9-desaturases contain the sequence HX4H, whereas M. tuberculosis DesA3 contains the sequence HX3H. In region Ib, the representatives of the Δ9-desaturases contain the sequence HX2HH, whereas M. tuberculosis DesA3 has HX3HH. In region II, the same representative Δ9-desaturases have a second occurrence of HX2HH, whereas DesA3 from M. tuberculosis has a second occurrence of HX3HH (Fig. 9). In these respects it is interesting that DesA3 shares closer sequence identity with the Δ6 acyl-lipid of cyanobacteria and higher plants than with Δ9 acyl-CoA and acyl-lipid desaturases of animals, yeasts, higher plants, and cyanobacteria (39). The hydropathy analysis of the secondary structure of DesA3 provided further evidence that DesA3 contains up to three hydrophobic domains, sufficient to span the membrane bilayer several times. Also, computer-assisted predictions (www.ch.embnet.org/software/TMPREDform.html) suggest the presence of at least two transmembrane domains starting at positions 104 and 194, and the three His regions are within hydrophilic loops. All of the His motifs (amino acids 90–94, 125–130, and 303–308) are located in the hydrophilic regions of the primary sequences as is the case in all desaturases, suggesting that the catalytic site of DesA3 is located on the cytosolic face of the membrane (55). Altogether, the presence of His conserved motifs and transmembrane domains supports the conclusion that DesA3 is a membrane desaturase, a conclusion reinforced by Western blotting, demonstrating directly that DesA3 is localized in the bacterial membrane fraction (results not shown).
Fig. 9. Multiple alignment of the putative membrane desaturase regions of the M. tuberculosis Δ9-desaturase, DesA3, and other related proteins.

Comparative amino acid sequence analysis of the various regions of the various membrane desaturases and the Δ9 and CoA-requiring DesA3 of M. tuberculosis (M. tb.) is shown. The eight conserved His residues are indicated by shading (two in region Ia, three in region Ib, and three in Region II). Identical amino acid residues are in bold letters. Gaps are introduced to facilitate the sequence alignment. Representatives of membrane Δ9-desaturases are from Rattus norvegicus (Rat), Saccharomyces cerevisiae (Yeast), and Mus musculus (Mouse).
The use of sterculic acid proved to be invaluable in demonstrating the parallel between its action as a known inhibitor of oleic acid synthesis and that of ISO. First, this comparison demonstrated that the inhibition of oleic acid synthesis is sufficient to kill M. bovis BCG and, thus, that the enzymes involved in the synthesis of this fatty acid are indeed lethal targets in mycobacteria. Second, the use of sterculic acid demonstrated that there is likely no relationship between the two effects of ISO on oleic acid and mycolic acid synthesis, i.e. mycolic acid probably does not originate in oleic acid. Rather, the results raise the possibility that ISO exerts an additional effect, possibly on the specialized enzymes, dehydratases and desaturases, that provide precursors for mycolic acid synthesis (56). Although ISO inhibits the formation of both oleic acid and mycolic acids, the data from this study support the concept that its inhibition of the growth of mycobacteria results primarily from effects on oleic acid rather than mycolic acid synthesis. This conclusion is drawn primarily from the observation of the rescue of ISO-treated M. tuberculosis with oleic acid and the high MIC value of BCG expressing desA3. The mechanism by which ISO inhibits mycolic acid synthesis has yet to be determined, but clearly it is not merely an effect on oleic acid synthesis.
Acknowledgments
We thank Drs. J. T. Belisle, S. Mahapatra, and V. Vissa for scientific advice and discussion and Marilyn Hein for preparing the manuscript.
Footnotes
This work was funded by NIAID, National Institutes of Health Grants AI-18357 and AI-38087 (a Research Project Agreement from the National Cooperative Drug Discovery Groups for the Treatment of Opportunistic Infections in AIDS).
The abbreviations used are: ISO, isoxyl; ETH, ethionamide; INH, isoniazid; FAME, fatty acid methyl ester; MAME, mycolic acid methyl ester; MIC, minimal inhibitory concentration; ADC, albumin-dextrose complex; OADC, oleic acid-albumin-dextrose complex; GC, gas chromatography; RT, reverse transcription; MOPS. 4-morpholinepropanesulfonic acid.
References
- 1.World Health Organization. Strategic Framework to Decrease the Burden of TB/HIV Document WHO/CDS/TB/2002.296, WHO/HIV_AIDS/2002.2. World Health Organization; Geneva, Switzerland: 2002. [Google Scholar]
- 2.Dye C, Williams BG, Espinal MA, Raviglione MC. Science. 2002;295:2042–2046. doi: 10.1126/science.1063814. [DOI] [PubMed] [Google Scholar]
- 3.Espinal MA. Tuberculosis. 2003;83:44–51. doi: 10.1016/s1472-9792(02)00058-6. [DOI] [PubMed] [Google Scholar]
- 4.Titscher R. Prax Pneumol. 1966;20:202–206. [PubMed] [Google Scholar]
- 5.Urbancik B. Tubercle. 1966;47:283. doi: 10.1016/s0041-3879(66)80007-7. [DOI] [PubMed] [Google Scholar]
- 6.Urbancik B. Antibiot Chemother. 1970;16:117–123. doi: 10.1159/000386811. [DOI] [PubMed] [Google Scholar]
- 7.Lambelin G. Antibiot Chemother. 1970;16:84–95. doi: 10.1159/000386807. [DOI] [PubMed] [Google Scholar]
- 8.Phetsuksiri B, Baulard A, Cooper AM, Minnikin DE, Douglas JD, Besra GS, Brennan PJ. Antimicrob Agents Chemother. 1999;43:1042–1051. doi: 10.1128/aac.43.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winder FG, Collins PB. J Gen Microbiol. 1970;63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- 10.Winder FG. In: The Biology of Mycobacteria. Ratledge C, Stanford J, editors. Academic Press, Inc.; New York: 1982. pp. 353–438. [Google Scholar]
- 11.Winder FG, Collins PB, Whelan D. J Gen Microbiol. 1971;66:379–380. doi: 10.1099/00221287-66-3-379. [DOI] [PubMed] [Google Scholar]
- 12.Sauton B. C R Acad Sci (Paris) 1912;155:860–861. [Google Scholar]
- 13.Cohn ML, Waggoner RF, McClatchy JK. Am Rev Respir Dis. 1968;98:295–296. doi: 10.1164/arrd.1968.98.2.295. [DOI] [PubMed] [Google Scholar]
- 14.Morris LJ. J Lipid Res. 1966;7:717–730. [PubMed] [Google Scholar]
- 15.Dunphy PJ, Kerr JD, Pennock JF, Whittle KL, Feeney J. Biochim Biophys Acta. 1967;136:136–147. doi: 10.1016/0304-4165(67)90329-7. [DOI] [PubMed] [Google Scholar]
- 16.Evans JE, Dulaney JT. Biochim Biophys Acta. 1983;752:346–352. doi: 10.1016/0005-2760(83)90133-9. [DOI] [PubMed] [Google Scholar]
- 17.Kashiwabara Y, Nakagawa H, Matsuki G, Sato R. J Biol Chem. 1975;78:803–810. doi: 10.1093/oxfordjournals.jbchem.a130969. [DOI] [PubMed] [Google Scholar]
- 18.Fulco AJ, Bloch K. J Biol Chem. 1964;239:993–997. [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 20.Cole ST, Blosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Kinglmei K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krongh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Squares R, Sulston JE, Taylor K, Whitehead S, Barrel BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 21.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Benet LT, Bansal P, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Jr, Bloom BR. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 22.Jackson M, Crick DC, Brennan PJ. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- 23.Kremer L, Baulard A, Estaquier J, Content J, Capron A, Locht C. J Bacteriol. 1995;177:642–653. doi: 10.1128/jb.177.3.642-653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yajko DM, Madej JJ, Lancaster MV, Sanders CA, Cawthon VL, Gee B, Babst A, Hadley WK. J Clin Microbiol. 1995;33:2324–2327. doi: 10.1128/jcm.33.9.2324-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou S, Chedore P, Haddad A, Paul NR, Kasatiya S. J Clin Microbiol. 1996;34:1317–1320. doi: 10.1128/jcm.34.5.1317-1320.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou S, Chedore P, Kasatiya S. J Clin Microbiol. 1998;36:577–579. doi: 10.1128/jcm.36.2.577-579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fulco AJ, Bloch K. Biochim Biophys Acta. 1962;63:545–546. doi: 10.1016/0006-3002(62)90130-0. [DOI] [PubMed] [Google Scholar]
- 28.Kashiwabara Y, Sato R. J Biol Chem. 1973;74:405–413. doi: 10.1093/oxfordjournals.jbchem.a130260. [DOI] [PubMed] [Google Scholar]
- 29.Johnson AR, Pearson JA, Shenstone FS, Forgerty AC. Nature. 1967;214:1244–1245. doi: 10.1038/2141244a0. [DOI] [PubMed] [Google Scholar]
- 30.Jeffcoat R, Pollard MR. Lipids. 1977;12:480–485. doi: 10.1007/BF02535446. [DOI] [PubMed] [Google Scholar]
- 31.Quintana J, Barrot M, Fabrias G, Camps F. Tetrahedron. 1998;54:10187–10198. [Google Scholar]
- 32.Andrianaivo-Rafehivola AA, Gaydou EM, Rakotovao LH. Oleagineux. 1994;49:177–188. [Google Scholar]
- 33.Barry CE, III, Slayden RA, Sampson AE, Lee RE. Biochem Pharmacol. 2000;59:221–231. doi: 10.1016/s0006-2952(99)00253-1. [DOI] [PubMed] [Google Scholar]
- 34.DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE., III Proc Natl Acad Sci U S A. 2000;97:9677–9682. doi: 10.1073/pnas.97.17.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. J Biol Chem. 2000;275:28326–28331. doi: 10.1074/jbc.M003744200. [DOI] [PubMed] [Google Scholar]
- 36.Ratledge C. In: The Biology of Mycobacteria. Ratledge C, Stanford J, editors. Academic Press, Inc.; New York: 1982. pp. 53–93. [Google Scholar]
- 37.Okuyama H, Kankura T, Nojima S. J Biochem (Tokyo) 1967;61:732–737. doi: 10.1093/oxfordjournals.jbchem.a128607. [DOI] [PubMed] [Google Scholar]
- 38.Walker RW, Barakat H, Hung JGC. Lipids. 1970;5:684–691. doi: 10.1007/BF02531435. [DOI] [PubMed] [Google Scholar]
- 39.Los DA, Murata N. Biochim Biophys Acta. 1998;1394:3–15. doi: 10.1016/s0005-2760(98)00091-5. [DOI] [PubMed] [Google Scholar]
- 40.Stubbs CD, Smith AD. Biochim Biophys Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 41.Russell NJ. Trends Biochem Sci. 1984;9:108–112. [Google Scholar]
- 42.Hazel JR. Annu Rev Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- 43.Houslay MD, Gordon LM. Curr Top Membr Transp. 1984;18:179–231. [Google Scholar]
- 44.Kaneda T. J Biol Chem. 1963;238:1229–1235. [PubMed] [Google Scholar]
- 45.Campbell I, Nowal J. J Lipid Res. 1969;10:593–598. [PubMed] [Google Scholar]
- 46.Anderson RJ. J Biol Chem. 1929;83:169–175. [Google Scholar]
- 47.Carson J, Sumrell G. J Am Chem Soc. 1950;72:4837–4838. [Google Scholar]
- 48.Ginger LG, Anderson RJ. J Biol Chem. 1945;157:203–211. [Google Scholar]
- 49.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 50.Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 51.Lennarz WJ, Scheuerbrandt G, Bloch K. J Biol Chem. 1962;237:664–671. [PubMed] [Google Scholar]
- 52.Akamatsu Y, Law JH. J Biol Chem. 1970;245:701–708. [PubMed] [Google Scholar]
- 53.Besra GS, Morehouse CB, Rittner CM, Waechter CJ, Brennan PJ. J Biol Chem. 1997;272:18460–18466. doi: 10.1074/jbc.272.29.18460. [DOI] [PubMed] [Google Scholar]
- 54.Obukowicz MG, Welsch DJ, Salsgiver WJ, Martin-Berger CL, Chinn KS, Duffin KL, Raz A, Needleman P. J Pharmacol Exp Ther. 1998;287:157–166. [PubMed] [Google Scholar]
- 55.Shanklin J, Whittle E, Fox BG. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 56.Barry CE, III, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]


