Significance
We demonstrate that forkhead box P1 (FOXP1) is a central transcriptional regulator of the most aggressive activated B cell (ABC) subtype of diffuse large B-cell lymphoma (DLBCL), the most prevalent non-Hodgkin’s lymphoma worldwide. We used a variety of methods to identify and functionally confirm FOXP1 target genes in DLBCL cell lines and primary clinical isolates. We found that FOXP1 target genes are sufficient to segregate ABC-DLBCL from the more indolent germinal center B-cell (GCB)-DLBCL subtype as well as to identify both hallmark and previously unidentified pathways underlying DLBCL pathology. Our findings extend the role of FOXP1 from a prognostic indicator of unknown mechanism to a driver of ABC-DLBCL neoplasia.
Keywords: FOXP1, DLBCL, lymphoma
Abstract
High expression of the forkhead box P1 (FOXP1) transcription factor distinguishes the aggressive activated B cell (ABC) diffuse large B-cell lymphoma (DLBCL) subtype from the better prognosis germinal center B-cell (GCB)-DLBCL subtype and is highly correlated with poor outcomes. A genetic or functional role for FOXP1 in lymphomagenesis, however, remains unknown. Here, we report that sustained FOXP1 expression is vital for ABC-DLBCL cell-line survival. Genome-wide analyses revealed direct and indirect FOXP1 transcriptional enforcement of ABC-DLBCL hallmarks, including the classical NF-κB and MYD88 (myeloid differentiation primary response gene 88) pathways. FOXP1 promoted gene expression underlying transition of the GCB cell to the plasmablast—the transient B-cell stage targeted in ABC-DLBCL transformation—by antagonizing pathways distinctive of GCB-DLBCL, including that of the GCB “master regulator,” BCL6 (B-cell lymphoma 6). Cell-line derived FOXP1 target genes that were highly correlated with FOXP1 expression in primary DLBCL accurately segregated the corresponding clinical subtypes of a large cohort of primary DLBCL isolates and identified conserved pathways associated with ABC-DLBCL pathology.
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin's lymphoma, striking ∼69,000 new patients annually in the United States (1). Although previously diagnosed and treated uniformly based on morphology and surface markers (2, 3), gene expression profiling (GEP) defined two major subtypes corresponding to the suspected B cell of origin (2, 3): the germinal center (GC) B cell for GCB-DLBCL (2) and the activated B cell (ABC) plasmablast (PB) for ABC-DLBCL (2). PBs exist transiently before terminal commitment to plasma cells (PC) and are proposed to be targeted for transformation in ABC-DLBCL (2, 4).
A hallmark of ABC-DLBCL is constitutive activation of the classical NF-κB pathway (4, 5). Activation of IKKβ and NF-κB signaling downstream of the B-cell receptor (BCR) (6) depends on the CBM complex, a signaling hub that includes CARD11, BCL10, and MALT1 (4). Roughly 10% of ABC-DLBCLs have CARD11 mutations (6). Another ∼10% harbor activating mutations in BCR components, including signal-transducing subunits CD79A and CD79B (6, 7). ABC-DLBCLs associated with chronic activation of BCR signaling (CABS) are specifically killed by shRNA targeting CBM components (4, 7). Another major route to NF-κB activation in ABC-DLBCL is via MYD88 (myeloid differentiation primary response gene 88), an adaptor protein whose mutation in ∼40% of ABC-DLBCL cases (8) up-regulates gene expression signatures of NF-κB, JAK-STAT, and type I IFN signaling (8).
Current multiagent chemotherapy achieves ∼80% 3-y survival for GCB-DLBCL, but only 45% for patients with ABC-DLBCL (1), and most ABC-DLBCL patients relapse with refractory disease (3, 9). GEP revealed genes associated with the length of survival (10). These “classifier” genes reflected biological features of the tumors that influenced the efficacy of chemotherapy (11). One such classifier gene is the transcription factor (TF) forkhead box P1 (FOXP1) (9). Implicated in development, proliferation, and differentiation in multiple contexts (12–16), FOXP1 is highly expressed in ABC-DLBCL relative to its levels in normal PB or in GCB-DLBCL (2, 3, 9, 17). Chromosomal loss, gain, or fusion of FOXP1 is associated with several B-cell malignancies, including a modest frequency in ABC-DLBCL (18–20). However, ABC-DLBCL tumors without such aberrations still show increased FOXP1 levels relative to GCB-DLBCL (20). The FOXP1 locus encodes multiple isoforms (13, 15, 17), and its shorter isoform 9 is up-regulated upon activation of nonmalignant B cells and overexpressed in ABC-DLBCL (17).
Here, we demonstrate that FOXP1 is a central regulator of ABC-DLBCL subtype distinction that directly or indirectly regulates hallmark DLBCL pathways, including repression of apoptosis, GCB cell identity, and tumor surveillance, while enforcing PB identity, hyper–NF-κB activity, and MYD88 signaling.
Results and Discussion
FOXP1 Expression Is Required for Viability of ABC-DLBCL but Not GCB-DLBCL Cell Lines.
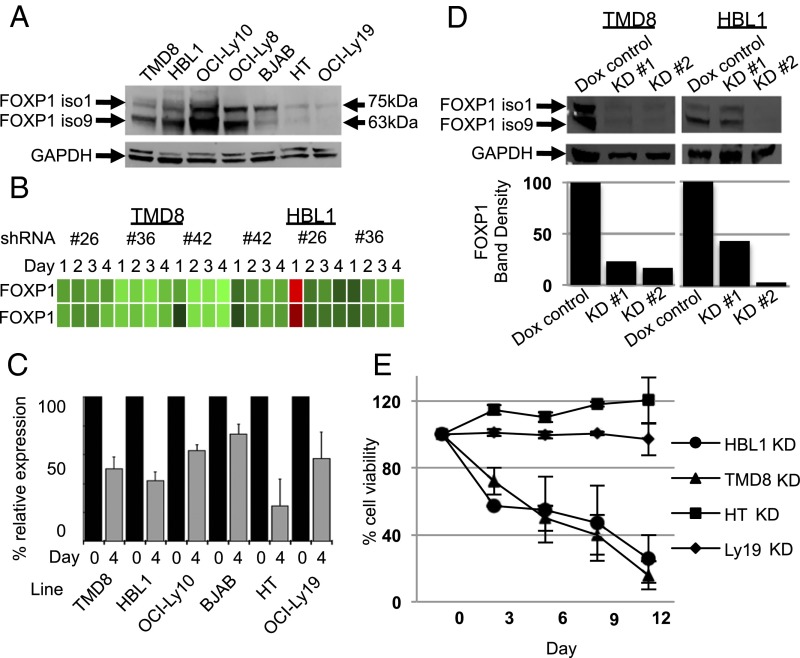
Comparison of FOXP1 protein levels in ABC-(TMD8, HBL1, and OCILy10), GCB-(BJAB, HT, and OCILy19), and OCILy8, considered to have an intermediate cell of origin, confirmed higher expression of both major FOXP1 isoforms 1 (∼75 kDa) and 9 (∼65 kDa) (17) in ABC lines (Fig. 1A and SI Appendix, Fig. S1). We used inducible shRNA knockdown (KD) to assess FOXP1 loss as a function of time. Optimized inducible KD (defined as ≥50% KD with multiple shRNAs at transcript and/or protein levels) (Fig. 1 B–D) was monitored by FACS to determine the percentage of live (GFP+) cells at indicated days (Fig. 1E). Nine to 12 d after shRNA induction, we observed ∼80% cell death of HBL1 and TMD8. Each line carries activating BCR mutations in CD79B ITAMs (Y196F; Y196H) and the MYD88 L265P activating mutation (7, 21). Singular loss of FOXP1, nonetheless, was sufficient for cell death. In contrast, equivalent KD in GCB-DLBCL lines HT and OCILy19 had no effect on viability (Fig. 1D). When we achieved at best ∼60% FOXP1 KD of FOXP1 in ABC line OCILy10, cell death was not observed. This observation was consistent with a recent report that also failed to observe FOXP1 KD-mediated cell death in OCILy10 (22). This finding could indicate that survival of all ABC lines are not dependent on FOXP1. Alternatively, because OC1Ly10 expresses FOXP1 at the highest levels of any in our panel of Fig. 1A, it is likely that the absolute—not relative—levels of FOXP1 determine sensitivity to death.
Fig. 1.
FOXP1 is highly expressed in ABC-DLBCL cell lines, and its loss results in their death by apoptosis. (A) Anti-FOXP1 Western blot of whole cell lysates prepared from DLBCL cell lines shows stronger expression of FOXP1 isoforms 1 and 9 in ABC- than in GCB-DLBCL lines. (B) Heat map of FOXP1 probes after FOXP1 knockdown in HBL1 and TMD8 with indicated shRNAs and at indicated time points (microarray accession no. GSE64586). (C) Assessment of FOXP1 knockdown by RT-qPCR normalized to GAPDH in the indicated cell lines after 4 d of doxycycline (dox) (1–10 μg/mL) induction of shRNA. Data include a minimum of four biological repetitions; error bars are SE of the mean. (D) Western blot of FOXP1 KD after 4 d of dox induction in TMD8 and HBL1 cell lines confirms reduction in FOXP1 protein level with two different shRNAs; bar graphs are normalized to GAPDH and compared with nonspecific shRNA control (dox control). (E) Loss of cell viability of ABC- but not GCB-DLBCL lines after 12 d of inducible FOXP1 KD as assessed by constitutive GFP levels independently encoded by the FOXP1 shRNA vector pRSMX.
Direct FOXP1 Transcriptional Target Genes Are Highly Correlated Among ABC-DLBCL Lines and Overlap only Modestly with GCB-DLBCL Target Genes.
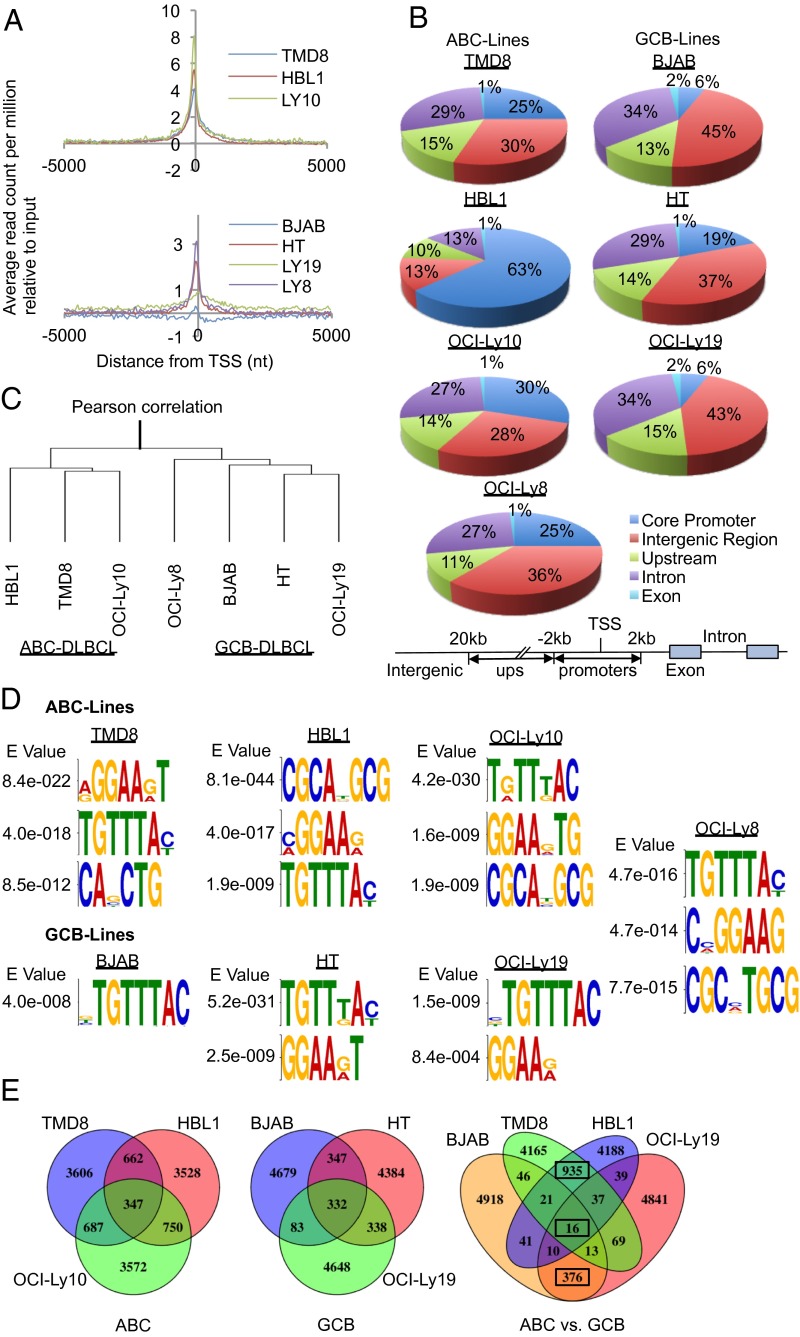
We next assessed global GEP of TMD8 and HBL1 after FOXP1 KD with several independent shRNAs across multiple time points before the advent of cell death; 553 of 19,526 genes were up-regulated and 563 of 19,526 were down-regulated upon FOXP1 KD (see Materials and Methods for modulation cutoffs), and high GEP concordance was observed among cell lines (SI Appendix, Fig. S2). To identify directly deregulated transcript expression, we performed FOXP1 ChIP-seq on ABC-(HBL1, TMD8, OCILy10), GCB-(HT, BJAB, OCILy19), and the ABC/GCB intermediate, OCILy8, lines. Several of these targets were confirmed by ChIP endpoint PCR (SI Appendix, Fig. S3). Read profiles generally identified stronger FOXP1 enrichment at transcriptional start sites (TSSs) in ABC relative to GCB lines (Fig. 2A). To eliminate bias from different peak numbers or cutoff levels between ChIP-seq datasets, the same number (5,452) of top peaks was used for downstream analyses, with the assumption that GCB peak sets contained more false positives. As shown in Fig. 2B, FOXP1 binding sites within proximal “core” promoters (±2 kb of TSS) were substantially higher in ABCs and were readily distinguished from GCBs by Pearson correlation of peak scores on target positions (Fig. 2C).
Fig. 2.
Genome-wide analyses reveal direct transcriptional targets and pathways regulated by FOXP1 in DLBCL. (A) Binding profile of FOXP1 at core promoters in ABC lines (Top) and GCB lines (Bottom) indicates higher read counts in ABC lines. (B) Pie chart representation of the distribution of FOXP1 binding sites in six genomic regions of each DLBCL cell line tested. Core promoters are ±2 kb from the transcriptional start site (TSS); upstream, 2–20 kb upstream from the TSS; and intergenic, regions not included as a promoter, upstream region, intron, or exon. ABC lines show a higher proportion of proximal promoter binding than that of GCB lines. (C) Pearson correlation of ChIP-seq peaks (n = 5,452) distinguishes ABC-DLBCL lines from GCB-DLBCL lines. (D) De novo motif analysis from FOXP1 ChIP-seq in DLBCL cells. Analysis of the top 1,000 ChIP-seq peaks from each line using MEME identified the consensus TGTTT FOXP1 binding motif as significantly enriched in each line. (E) Venn diagrams of ABC lines, GCB lines, and two ABC with two GCB lines. ABC lines shared more peaks than GCB lines, indicating high concordance among ABC target loci. Targets among two ABC and two GCB lines revealed only 16 overlapped peaks in all four lines, with 935 overlapping in ABC lines and 376 in GCB lines, further indicating high correlation among ABC lines but low correlation between ABC and GCB lines.
FOXP1 motif enrichment using MEME (23) revealed FOXP1 consensus binding sites within proximal promoters and putative intronic and long-range enhancers in all lines tested (Fig. 2D). That the consensus motif was common among both ABC- and GCB-DLBCLs suggested that FOXP1 acts through similar pathways. Indeed, gene ontology (GO) analyses of TGTT bound genes (SI Appendix, Table S1) identified numerous significantly overlapping pathways (∼80) and many (∼22) with strong P values (<10−4), including the following: regulation of transcription from RNA polymerase II promoter (P = 6.55 × 10−5), positive regulation of transcription (P = 2.38 × 10−4), and apoptosis (P = 3.11 × 10−4). We also identified many unique pathways in ABC (121 significant; 8 with P < 10−4) and in GCB (105 significant; 10 with P < 10−4). Venn diagrams revealed 2,446 of 5,452 overlapped peaks in at least two ABC lines compared with 1,100 in at least two GCB lines (Fig. 2E). This indicated high concordance among ABC target loci, in agreement with GEP data (SI Appendix, Fig. S2). Comparison of FOXP1 targets among two ABC and two GCB lines identified only 16 overlapped peaks in all four lines, but 935 between ABC lines and 376 between GCB lines (Fig. 2E). Thus, direct FOXP1 targets are highly correlated among ABC lines, but less so between ABC and GCB lines.
FOXP1 Target Genes Distinguish ABC- from GCB-DLBCL and Underlie ABC-DLBCL Pathology.
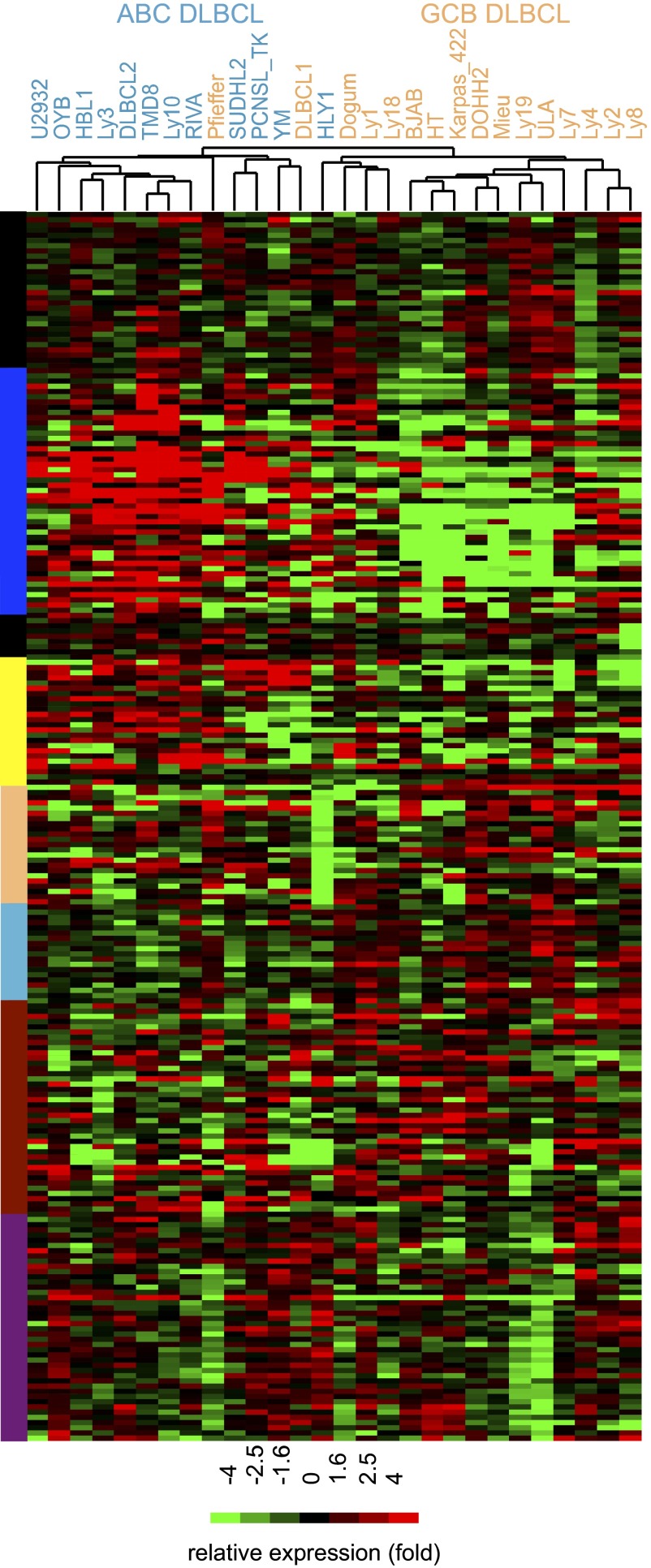
We next compared transcripts directly activated or repressed by FOXP1 in TMD8 and HBL1 ABC lines with transcripts expressed in an extended panel of ABC- and GCB-DLBCL lines (see Fig. 3 and SI Appendix, Fig. S4 for readable color-coded genes). Clustering based on correlation of expression patterns segregated ABC- and GCB-DLBCL types with high concordance. Notably, GCB-DLBCL lines with high absolute levels of FOXP1 (e.g., Ly1 and Karpas_422) were still accurately clustered. Thus, direct FOXP1 transcriptional targets are sufficient to distinguish ABC-DLBCL, regardless of their transforming mutations.
Fig. 3.
Expression of direct FOXP1 target genes is sufficient to differentiate ABC- from GCB-DLBCL. Direct FOXP1 target genes whose expression was modulated in TMD8 and HBL1 cells lines (qualifying genes reached log2 values of ±0.3 change in 5 of 12 microarray KD samples) were used for unbiased clustering of their corresponding endogenous expression levels in a panel of 11 ABC (blue) and 16 GCB lines (orange; top). Targeted genes are identifiable in order by color code left of heat map in SI Appendix, Fig. S5.
Previously defined classifier genes (2, 24) were analyzed for perturbation after FOXP1 KD (SI Appendix, Fig. S5). Consistent with the above results, the majority of ABC classifiers were down-regulated, and the majority of GCB classifiers were up-regulated. That some classifiers were modulated in an inconsistent direction underscores the well-known heterogeneity among primary isolates (20, 25, 26). GO and pathway analyses (10, 27) predicted highly significant deregulation of signatures associated with B-cell/leukocyte activation and regulation of apoptosis/cell death in ABC lines (SI Appendix, Fig. S6). Also significantly reduced by FOXP1 KD were DLBCL-related signatures composed of genes within MYD88, JAK2, and NF-κB pathways (P = 2.25 × 10−58, 1.06 × 10−45, and 7.36 × 10−27, respectively) (SI Appendix, Table S2). In contrast, FOXP1 KD in GCB lines up-regulated MYD88 and JAK pathway transcripts typically repressed in ABC-DLBCL (P = 4.58 × 10−44 and 5.67 × 10−18) and pathways typically up-regulated in GCB-DLBCL, including the BCL6 (B-cell lymphoma 6)-transformed (GCB-DLBCL) and BCL6-untransformed GCB cell gene signatures (P = 1.26 × 10−10, 7.99 × 10−14, and 2.21 × 10−10, respectively) (SI Appendix, Table S2). Pathway analysis of direct FOXP1 target genes, identified by merging GEP and ChIP-seq, were enriched for the same signatures (SI Appendix, Table S2). These global data, along with the viability consequences of FOXP1 KD in ABC but not GCB lines, suggest that high FOXP1 expression is vital to ABC-DLBCL pathology and that its singular loss results in a GCB-like GEP phenotype.
FOXP1 Enforces Hallmark ABC-DLBCL Survival/Proliferative Pathways.
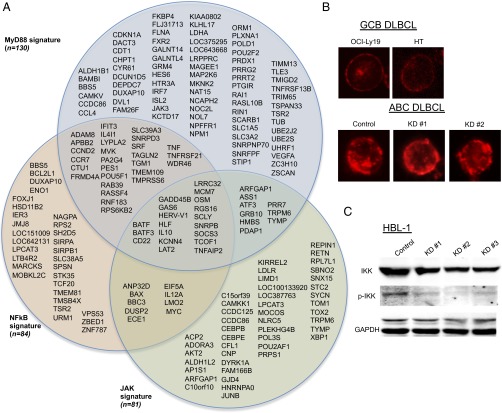
In ABC-DLBCL, MYD88 up-regulates NF-κB, JAK-STAT, and type I IFN signaling by virtue of the L265P mutation shared among the ABC lines analyzed in this study (8). Within the MYD88Up signature totaling 271 ABC-DLBCL–activated genes, 130 were down-regulated in HBL1 and 80 in both HBL1 and TMD8 upon FOXP1 KD (Fig. 4A and SI Appendix, Table S2). The MYD88Up signature, along with the NF-κB and JAK-STAT signatures, whose component members often overlap, were the most perturbed upon FOXP1 KD (Fig. 4A and SI Appendix, Table S2).
Fig. 4.
FOXP1 activates hallmark ABC-DLBCL pathways. (A) Venn diagram of FOXP1 up-regulated genes in three hallmark ABC-DLBCL pathways (4, 5). MyD88 (P = 2.25 × 10−58), NF-κB (P = 7.36 × 10−27), and JAK (P = 1.05 × 10−45) signatures were significantly down-regulated upon FOXP1 loss. n, the number of genes up-regulated by FOXP1 in the HBL1 cell line. (B) Chronic BCR activation, as assessed by mIgM clustering, is not perturbed by FOXP1 loss 4 d post KD in ABC line HBL1. Indicated GCB lines were used as controls. (C) Loss of FOXP1 blocks constitutive NF-κB signaling in ABC line HBL1. After 4 d of FOXP1 KD, signaling was assessed by Western blotting for phospho-IKKα/β and IKKβ.
Type I IFN signaling is a consequence of MYD88 activation in ABC-DLBCL (28). Perturbations of IFN pathways were detected by loss of TNF and IL-10 transcripts upon FOXP1 KD (Fig. 4A). MYD88 also promotes JAK-STAT signaling in ABC-DLBCL, and, consistent with this data, a positive modulator of that pathway, IL-6 (29), was directly repressed by FOXP1 (Fig. 6 A and B). Through its direct repression of IL-6, FOXP1 may also block PC differentiation by maintaining the PB phenotype of ABC-DLBCL.
Fig. 6.
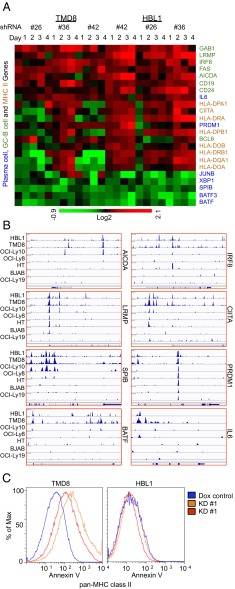
FOXP1 represses germinal center B-cell gene and MHC Class II expression in favor of partial plasma cell differentiation. (A) Heatmap of GCB and plasmablast (PB)/plasma cell (PC)-related genes after FOXP1 KD via shRNAs at indicated times in TMD8 and HBL1 ABC lines. GCB genes (left, green) are repressed by FOXP1, whereas PB/PC genes (left, blue) are activated, with the exception of PRDM1, encoding BLIMP-1, the master regulator of PC differentiation. Typically high GCB expression of MHC class II structural (HLA) and master regulator CIITA (red) are repressed. (B) FOXP1 is recruited to regulatory loci of GCB and PB/PC-related genes. ChIP-seq peaks from seven DLBCL cell lines are shown as read density tracks. In general, many loci had aligned peaks across all seven lines, whereas others had aligned peaks only in ABC lines, indicating different binding patterns between the DLBCL types. (C) MHC class II expression is restored after FOXP1 loss. Pan-MHC class II expression as measured by flow cytometry was markedly increased 4 d after FOXP1 KD in ABC cell line HBL1 but not in TMD8.
CABS is another defining characteristic of ABC-DLBCL that engages NF-κB (8) via the CBM complex (7). Modulated genes within a BCR-dependent signature were virtually all activated by FOXP1 (SI Appendix, Fig. S7). However, 21 of 23 of these genes also fall within the MYD88, NF-κB, and JAK-STAT pathways (Fig. 4 A and B). After FOXP1 KD in ABC lines, BCR clustering was not decreased (Fig. 4B). Thus, FOXP1 contributes to CABS-related gene expression, but only minimally to CABS-directed NF-κB activation. Consistent with these pathway disruptions upstream of NF-κB, IKK phosphorylation was reduced upon FOXP1 KD (representative data shown in Fig. 4C). Thus, FOXP1 directly regulates multiple, independent pathways underlying constitutively active NF-κB.
Notable, in addition to previously identified hallmark pathways, was direct FOXP1 activation of c-MYC (Fig. 4B and SI Appendix, Fig. S8). Most DLBCLs, including lines analyzed here, express high levels of MYC transcripts—an indication of worse outcomes after R-CHOP therapy (30)—but lack amplification or translocations of the MYC locus (4, 31). Thus, ABC-DLBCL must achieve c-MYC overexpression by other means, and FOXP1 likely contributes. c-MYC in normal GCB cells is repressed by BCL6, the quintessential marker of GCB-DLBCL (31), which is addressed in FOXP1 Indirectly Antagonizes BCL6.
FOXP1 Represses Apoptosis While Promoting Cell Survival.
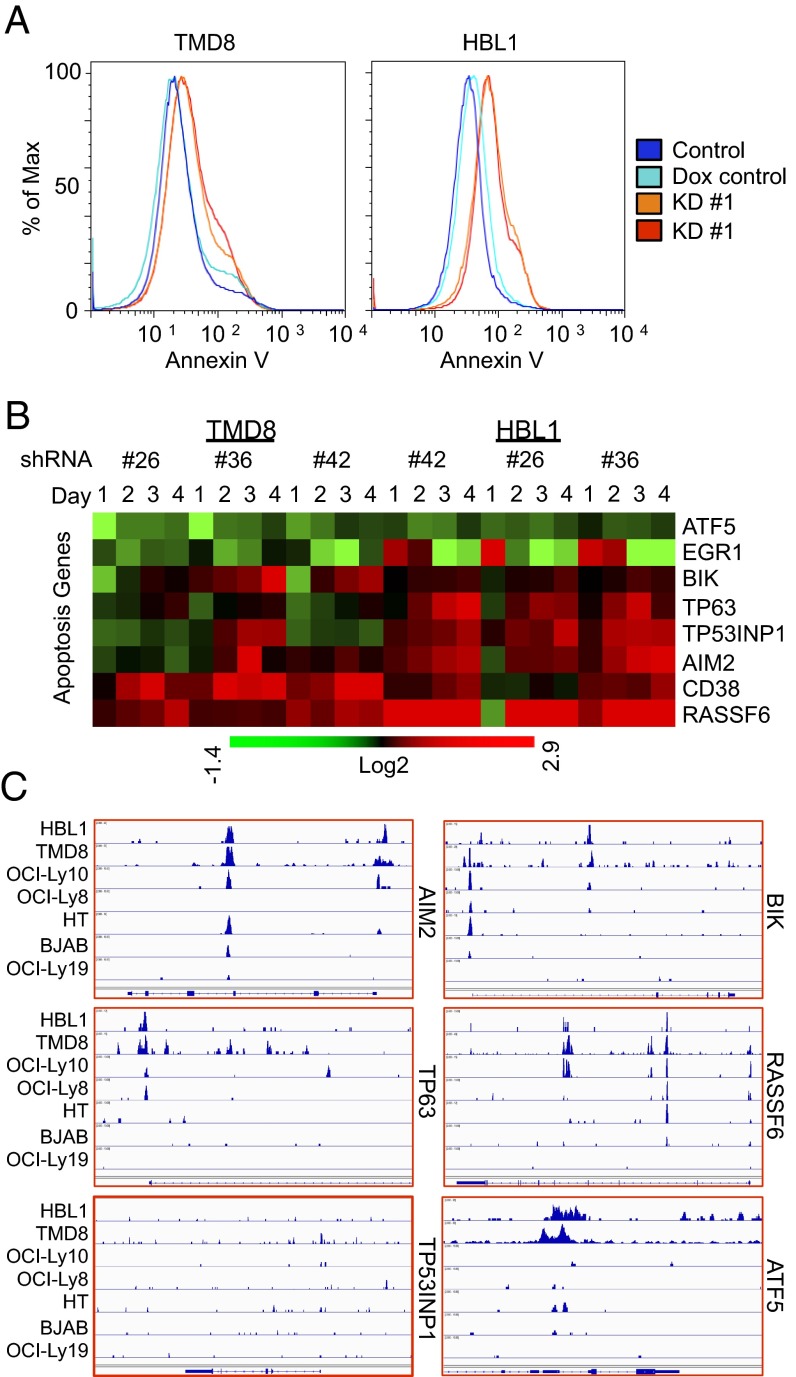
After FOXP1 KD, HBL1 and TMD8 showed increased annexin V staining (Fig. 5A), implicating apoptosis as the likely mechanism of cell death. Consistent with this phenotype, GO analyses identified significant deregulation of apoptotic signatures (SI Appendix, Fig. S6). Within these signatures, a number of proapoptotic genes were directly repressed in ABC lines (Fig. 5 B and C). Many of the same targets were identified in a recent study by van Keimpema et al. (22), including the following: Aim2, encoding a component of the canonical inflammasome; BIK, a proapoptotic BH3-only domain member of the BCL2 family; and TP63, a TP53/p53 homolog that promotes apoptosis or chemosensitivity in solid tumors (22). Factors encoded by two additional FOXP1-repressed proapoptotic target genes, TP53INP1 and RASSF6, along with TP63, encode p53-activating tumor suppressors whose expression is correlated with bad prognosis in DLBCL (22). Reciprocally, FOXP1 directly activated the antiapoptotic TF, ATF5 (32), and indirectly down-regulated the ATF5 target, EGR1 (33).
Fig. 5.
FOXP1 prevents ABC-DLBCL cell death. (A) FOXP1 loss results in apoptotic death of ABC-DLBCL lines as assessed by flow cytometric measurement of annexin V expression 4 d after dox induction of KD. (B) FOXP1 represses apoptosis-related genes and activates survival genes previously unassociated with DLBCL pathways. (C) FOXP1 is recruited to regulatory regions of apoptosis and survival-related genes. ChIP-seq data from seven DLBCL cell lines are shown as read density tracks. In general, numerous loci had aligned peaks across all seven lines, whereas many had aligned peaks only in ABC lines, indicating different binding patterns between DLBCL subtypes. Still other modulated genes had no significant peaks, as with TP53INP1 pictured here.
FOXP1 Indirectly Antagonizes BCL6.
Although both BCL6 and GCB gene signatures are repressed by FOXP1 (SI Appendix, Table S2 and FOXP1 Target Genes Distinguish ABC- from GCB-DLBCL and Underlie ABC-DLBCL Pathology), BCL6 itself is inconsistently repressed despite FOXP1 binding to its promoter region (Fig. 6A and SI Appendix, Figs. S3 and S8). These data support previous observations (34), suggesting that FOXP1 and BCL6 control many of the same GCB identity genes. We determined by GREAT analysis (35) that ∼50% of the 3,200 genes previously identified (31, 34) as bound by BCL6 were also bound by FOXP1 in ABC- or GCB-DLBCL lines (1,705 of 3,200 and 1,488 of 3,200, respectively) (SI Appendix, Fig. S9). Thus, FOXP1 not only directly activates the NF-κB pathway but also reinforces its activation by counteracting the negative effect of BCL6 on NF-κB (36).
FOXP1 Directly Represses Gene Signatures Associated with GCB Cell Identity.
As noted above, FOXP1 repressed signatures underlying GCB cell biology (Fig. 6 A and B and SI Appendix, Table S2). In particular, FOXP1 repressed AICDA, which encodes the master regulator of secondary antibody diversity, AID (Fig. 6 A and B). Reduction of AICDA transcripts would predict a reduction in somatic hypermutation (SHM). Consistent with this hypothesis, BCRs expressed by primary ABC-DLBCLs often are minimally mutated (37–39), and transgenic FOXP1-overexpressing mice show GCB cell deficiency (34).
Other notable GCB cell identity genes directly repressed after FOXP1 KD include (i) IRF8, which is highly expressed in normal GCB and GCB-DLBCL (40) and negatively regulates PC differentiation (41); (ii) GAB1, an adapter that recruits SH2 domain-containing proteins to the ligated BCR (42); and (iii) LRMP, a lymphoid-restricted membrane protein involved in Ag receptor assembly (43) whose overexpression is a GCB-DLBCL classifier (2, 43). Indirectly down-regulated is the GCB-DLBCL marker, Fas/CD95 (Fig. 6A).
These data suggest that appropriate timing of FOXP1 down-regulation is essential for GC function and formation. They further suggest (as addressed in the next section) that persistent FOXP1 expression contributes to B-cell lymphomagenesis by premature and sustained promotion of the normally transitory PB state.
FOXP1 Promotes the Partial Plasma Cell Differentiation Phenotype of ABC-DLBCL.
ABC-DLBCL primary isolates and cell lines demonstrate gene expression similar to PBs—the cell type most likely targeted for transformation (2, 4). Maintenance of the normal proliferative PB state is accompanied by down-regulation of prototypic B cell-specific TFs (e.g., BCL6, PAX5, and SPIB) balanced by up-regulation of PC TFs (e.g., IRF4, XBP1, and PRDM1) (44). Transcripts encoding TFs critical to PC differentiation are deregulated—both directly and indirectly—by FOXP1 KD in ABC lines (Figs. 4 A and B and 6A and SI Appendix, Fig. S3). BLIMP-1/PRDM1, considered the “master regulator” of terminal PC differentiation (45), is directly repressed by FOXP1 in our study. FOXP1 binds the PRDM1 proximal promoter in ABC lines, as well as within an alternative, internal promoter (PRDM1β), in both ABC- and GCB-DLBCL lines (Fig. 6 A and B and SI Appendix, Fig. S3). The PRDM1β isoform is missing much of the positive regulatory (PR) domain and has a diminished capacity for repressing target genes (46). Sagardoy et al. (34) previously identified FOXP1 as an activator of PRDM1 in OCILy1, a GCB line that expresses FOXP1 at relatively high levels (Fig. 3A). That FOXP1 peak binding overlaps in both studies at the β isoform promoter, but not at the promoter of the major isoform, suggests that the outcome of activation vs. repression is context-dependent.
Regulatory regions within SPIB, which encodes a prominent B-cell ETS family TF, were strongly bound by FOXP1 (Fig. 6 A and B and SI Appendix, Fig. S3). SPIB is often overexpressed in ABC-DLBCL via amplification or translocation (20) and directly represses PRDM1 while itself being repressed by BLIMP-1 (25, 44). A number of ABC-DLBCLs have eliminated functional BLIMP-1 expression via PRDM1 translocations, mutations, deletions, or epigenetic silencing (26). Our data indicate that FOXP1 may collaborate with SPIB to ensure PRDM1 repression-—a favorable situation for retention of PB identity and ABC biology. SPIB binds DNA cooperatively with either IRF4 or IRF8 at ETS-IRF composite “EICE” motifs (41). That the consensus ETS motif GGAA was enriched within FOXP1-associated chromatin in all lines tested (Fig. 2D) raises the possibility that SPIB may also collaborate with FOXP1 to further fine-tune transcription of a subset of FOXP1 target genes.
Two additional FOXP1 directly deregulated targets implicated in the GCB-PB-PC transition were the basic leucine zipper TFs BATF and BATF3 (Fig. 6 A and B and SI Appendix, Fig. S8). BATF is a direct target of NF-κB that is expressed transiently in activated B cells en route to PC differentiation and highly in ABC-DLBCL (24).
Thus, continual high expression of FOXP1 results in repression of GCB-cell genes and activation of genes that encourage exit from the GC, whereas full extinction of FOXP1 is necessary for terminal PC differentiation (47).
FOXP1 Down-Regulation of HLA Class II Expression May Contribute to Tumor Surveillance Failure in ABC-DLBCL.
Tumor cells must avoid immune surveillance, and, consequently, lymphomas accumulate genetic lesions in genes necessary for immune recognition. Loss of MHC class II expression, or HLAs, is considered a hallmark of ABC-DLBCL (10). HLAs are also lost upon normal plasma cell differentiation due to BLIMP-1 repression of CIITA, an essential regulator of HLA transcription (48). FOXP1 directly repressed CIITA in HBL1 cells (Fig. 6 A and B), whereas multiple HLA genes were derepressed post-FOXP1 KD (Fig. 6A). As predicted from the target deregulation data, rescue of HLA surface expression was observed after FOXP1 KD (Fig. 6C).
FOXP1 Target Genes Derived from Cultured DLBCL Lines Distinguish Primary ABC- and GCB-DLBCL Isolates.
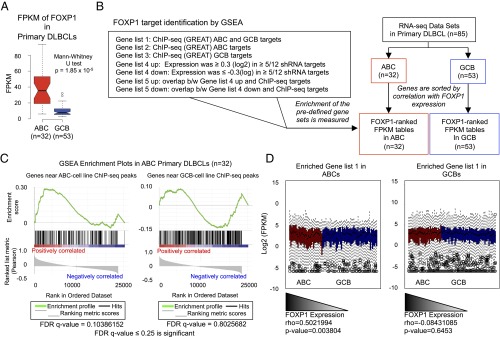
Recent studies indicate that only a small number of DLBCL cell line-derived “classifier transcripts,” including those often used for diagnosis, are shared among primary isolates (20, 25, 26). To investigate whether FOXP1 targets discovered above might better deconvolute this heterogeneity, we analyzed 32 ABC-DLBCL and 53 GCB-DLBCL primary isolate RNA-seq datasets from the Genotypes and Phenotypes database (dbGaP) (accession no. phs000235) (49, 50). Transcript abundance by FPKM (fragments per kilobase of an exon per million fragments mapped) showed that FOXP1 levels were significantly higher in ABC isolates compared with GCB isolates (Mann–Whitney U test, P = 1.85 × 10−5) (Fig. 7A). However, clustering the entire list of cell line-derived FOXP1-bound and/or modulated genes based on gene expression levels in primary isolates did not statistically segregate ABC subtypes vs. GCB subtypes. This finding was not unexpected based on previous primary DLBCL analyses (20, 25, 26), and it likely results from redundant transcriptional regulation mechanisms and/or modest target overlap of genes/pathways common across subtypes. Thus, we used gene set enrichment analysis (GSEA) to correlate expression of FOXP1 with its direct and indirect targets by generating gene sets derived from the following: (i) genes bound by FOXP1 in ABC cell lines and/or GCB cell lines; (ii) genes modulated by FOXP1 KD in the HBL1 and TMD8 cell lines; and (iii) genes bound and modulated by FOXP1 (Fig. 7B and SI Appendix, Table S3). GSEA revealed significant enrichment in multiple contexts (Fig. 7C and SI Appendix, Fig. S10). For example, ∼25% (50 of 215) of FOXP1-bound target genes in all three ABC cell lines [false discovery rate (FDR) q-value = 0.10] were significantly enriched in primary ABC isolates, but enrichment in targets bound in all three GCB cell lines was not (FDR q-value = 0.80) (Fig. 7C). When FOXP1-bound target genes in both ABC and GCB lines (gene list 1) were enriched by correlation with FOXP1 expression in ABC, as expected, the median expression of the targets showed a descending trend of expression in correlation with FOXP1 levels in ABC (Spearman’s rho = 0.502, P = 0.0038). In contrast, the same gene list enriched in GCB did not follow this trend (rho = −0.0843, P = 0.6453). Therefore, subtype-specific enrichment allowed us to specifically identify FOXP1 coexpression targets in corresponding subtypes (Fig. 7D).
Fig. 7.
FOXP1 target genes correlated with its expression are able to differentiate primary ABC- from GCB-DLBCL by their expression profiles. (A) Primary DLBCL FOXP1 transcript abundance was assessed by FPKM (fragments per kilobase of an exon per million fragments mapped). FOXP1 levels were significantly higher in ABC isolates (Mann–Whitney U test, value of P = 1.852e−05). (B) Flowchart for identification of GSEA-derived gene lists. Genes from ABC or GCB primary data were sorted by correlation with FOXP1 expression, and expression of each gene list was analyzed by GSEA to detect enriched gene sets. (C) An example of GSEA analysis. Target genes bound by FOXP1 in all three ABC cell lines (∼25%, 50/215) were significantly enriched and positively correlated with FOXP1 expression across ABC isolates (FDR q-value = 0.10). Conversely, direct FOXP1 target genes in all three GCB cell lines did not achieve enrichment in the same set of FOXP1 correlated genes in the ABC primaries (FDR q-value = 0.80). (D) Box plots of enriched gene lists show that median expression of FOXP1 targets is correlated with FOXP1 expression. Gene list 1 is used as an example, in primary FOXP1 (rho = 0.502 and P = 0.0038) and in GCB subtype (rho = −0.0843, P = 0.6453).
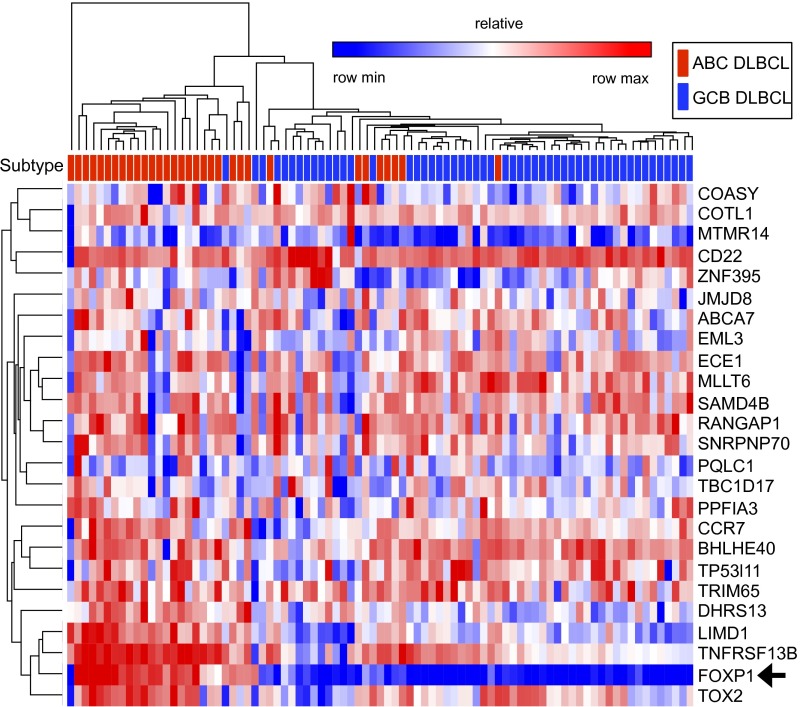
Hierarchical clustering of enriched gene subsets by Pearson correlation was capable of segregating ABC- and GCB-DLBCL primary isolates to varying degrees (Fig. 8 and SI Appendix, Fig. S11). For example, genes that were both directly bound and down-regulated upon FOXP1 loss in TMD8 and HBL1 clustered 21 of 32 ABC primary isolates as well as a majority of GCB primary isolates, as indicated by ABC (red) and GCB (blue) designation above the heatmaps in Fig. 8. Genes bound by FOXP1 in both ABC lines and GCB lines provided appropriate subtype classification when correlated with primary isolate gene expression (SI Appendix, Fig. S11). FOXP1 target gene expression also identified GCB subtypes, conceivably because its lower abundance in GCB-DLBCL tumors (Fig. 1A) restricts binding to higher affinity targets. The level of ABC vs. GCB segregation achieved was equivalent to FOXP1 subtype prediction in DLBCL cell lines (Fig. 3A). Collectively, the data demonstrate that, unlike many other classifiers (20, 25, 26), the prognostic value of high FOXP1 expression can accurately extend to primary subtype prediction.
Fig. 8.
FOXP1 gene sets segregate primary ABC- from GCB-DLBCL by their expression profiles. Hierarchical clustering of enriched gene subsets defined in Fig. 7 by Pearson correlation was capable of segregating ABC- and GCB-DLBCL primary isolates to varying degrees. Gene list 5 down as defined in Fig. 7B is used as an example.
Correlation of genes expressed in ABC-DLBCL primary isolates with those down-regulated after FOXP1 KD in ABC lines revealed a number of shared pathways. Among these pathways are several BCL-6 signatures (31, 51), as well as signatures associated with activation of ABC-hallmark NF-κB, MYD88, and JAK2 pathways (P = 8.6 × 10−11, P = 1.52 × 10−14, and P = 4.34 × 10−17, respectively) (SI Appendix, Table S4). FOXP1 target genes within these signatures were identified as key discriminators within the gene lists (e.g., BCL2L1, CCR7, LAT2, MYC within “NF-κB”; CCDC86, RUNX1, TNFRSF13B, and GADD45B within “MYD88”; and ECE1, TOX2, TRIM28, and TNF within “JAK2” signatures) (SI Appendix, Fig. S12). Further, genes up-regulated by FOXP1 KD that were also preferentially expressed in GCB primary tumors constituted the same major signatures but with reciprocal magnitudes: i.e., repression of MYD88 and JAK2 signatures (P = 1.61 × 10−13 and P = 2.34 × 10−10, respectively) (SI Appendix, Table S4). Finally, GO analysis confirmed previously identified signatures related to cell death, proliferation, and lymphocyte/B-cell activation (SI Appendix, Table S5).
Overall, FOXP1 gene targets in DLBCL cell lines, when applied to primary DLBCLs, are able both to segregate ABC and GCB subtypes and to reassemble essential DLBCL pathway signatures.
Conclusions
In this study, we report that FOXP1 is a central regulator of DLBCL survival and subtype distinction by direct and indirect transcriptional regulation of hallmark pathways that repress apoptosis and GCB cell identity while enforcing plasmablast identity and NF-κB signaling via the MYD88 and JAK-STAT pathways.
It is important to note that the CABS/MYD88/constitutively activated NF-κB model is based primarily on ABC-DLBCL cell lines—the majority of which carry activating mutations. However, singular knockdown of FOXP1 in cell lines harboring such mutations was capable of overcoming these survival pathways and led to cell death. In primary tumors, direct and indirect targets of FOXP1 that correlated with FOXP1 expression segregated ABC and GCB subtypes. Further analysis of these FOXP1 targets reinforced signatures identified in ABC-DLBCL cell lines, including up-regulation of NF-κB, MYD88, JAK-STAT, and proliferative signatures and down-regulation of BCL6 signatures that enforce germinal center B-cell identity.
Considering the limited treatment options currently available for ABC-DLBCL and the poor prognosis for patients with recurrent disease, new therapeutics and diagnostics are urgently required. Accurate diagnosis between DLBCL subtypes, as well as between DLBCL and Burkitt lymphoma, carries significant clinical relevance. We contend that further functional understanding of FOXP1 and other master TFs corrupted by ABC-DLBCL is essential for designing better prognostic and therapeutic approaches.
Materials and Methods
shRNA Knockdown.
Cells were stably transduced with a retrovirus expressing the bacterial tetracycline repressor (TETR) and the blasticidin resistance gene and then retrovirally transduced with Phoenix-E packaged pRSMX-PG TETR-inducible vectors (21) containing shRNA targeted to FOXP1 or a scrambled shRNA control (dox control). Cells were harvested for RNA isolation at multiple time points. FOXP1 KD was considered sufficient for analysis when it reached 50% or lower relative expression compared with controls by reverse transcription quantitative real-time PCR (RT-qPCR). SYBR green real-time qPCR was performed after high capacity cDNA reverse transcription, both following manufacturer’s protocols (cat. nos. 4385618 and 4368814; Applied Biosystems). Sequences of shRNAs and primers are in SI Appendix.
Flow Cytometry.
Flow cytometry was performed on a Fortessa flow cytometer (BD Biosciences) and analyzed with FloJo software (TreeStar). Antibodies used are in SI Appendix.
Western Blots.
Western blots were performed by common procedures; FOXP1 band densities were quantified with ImageJ (52). Antibodies used are in SI Appendix.
Microarray.
Total RNA was extracted using TRIzol reagent (Invitrogen) followed by RNAeasy kit cleanup (Qiagen), and labeled cRNA was prepared as described for Agilent human 4 × 44 microarrays. Gene expression was determined by comparing signal passing confidence criteria provided by the manufacturer from dox-induced cells with an ineffective control shRNA (Cy3) to dox-induced cells bearing FOXP1 targeting shRNAs (Cy5) at each time point. Data are shown as log2 ratio (Cy5/Cy3). Genes with log2 values of ±0.3 change in at least 5 of 12 microarray KD samples per cell line were considered modulated.
ChIP-Seq.
ChIP-seq assays were performed as described previously (53). ChIP pulldowns that were performed with either an IgG1 monoclonal FOXP1 antibody (15) or an anti-FOXP1 rabbit polyclonal antibody (54) were optimized by Endpoint ChIP-PCR with a previously determined FOXP1 target, the enhancer of RAG (Erag) (SI Appendix, Fig. S3) (54). IP for ChIP-seq was performed with the anti-FOXP1 rabbit polyclonal antibody. DNA was analyzed by Illumina deep sequencing technology.
Data Analysis.
Detailed data analysis is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Chhaya Das and Maya Ghosh for help with ChIP and cell culture. Anti-FOXP1 rabbit polyclonal antisera were kindly provided by Dr. Edward Morrissey. Library preparation and Illumina ChIP-seq were performed at the NGS core of the MD Anderson Cancer Center. Access to the datasets from the Genotypes and Phenotypes database (dbGaP) (accession no. phs000235) was essential for this study. The dbGaP data were generated by the Cancer Genome Characterization Initiative (CGCI). The Texas Advanced Computing Center (TACC) at The University of Texas at Austin was vital for providing computing resources that have contributed to this manuscript’s results (https://www.tacc.utexas.edu/). Support for this work was provided by the Intramural Research Program of the National Institutes of Health (NIH) National Cancer Institute, Center for Cancer Research (L.M.S., A.L.S., and H.K.); Lymphoma Research Foundation Fellowship 300463 (to J.D.D.); NIH Grant R01CA31534; Cancer Prevention Research Institute of Texas (CPRIT) Grants RP120348 and RP120459; and the Marie Betzner Morrow Centennial Endowment (H.O.T.). This work was also supported in part by NIH Grants R01CA130075 and R01CA95548 and CPRIT Grants RP120194 (to V.R.I.) and RP100612 (to G.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (ChIP-seq accession no. GSE63257 and microarray accession no. GSE64586).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524677113/-/DCSupplemental.
References
- 1.Howlader NNA, et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; Bethesda, MD: 2013. Based on November 2012 SEER data submission. Available at seer.cancer.gov/archive/csr/1975_2010/. Accessed March 7, 2015. [Google Scholar]
- 2.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, et al. Lymphoma/Leukemia Molecular Profiling Project The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 7.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banham AH, et al. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11(3):1065–1072. [PubMed] [Google Scholar]
- 10.Staudt LM, et al. 2007. Methods for identifying, diagnosing, and predicting survival of lymphomas. US Patent US20070105136 A1.
- 11.Shaffer AL, et al. Signatures of the immune response. Immunity. 2001;15(3):375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- 12.Feng X, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115(3):510–518. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabut M, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147(1):132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Shi C, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112(12):4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278(27):24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131(18):4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 17.Brown PJ, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111(5):2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 18.Wlodarska I, et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia. 2005;19(8):1299–1305. doi: 10.1038/sj.leu.2403813. [DOI] [PubMed] [Google Scholar]
- 19.Sagaert X, et al. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J Clin Oncol. 2006;24(16):2490–2497. doi: 10.1200/JCO.2006.05.6150. [DOI] [PubMed] [Google Scholar]
- 20.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441(7089):106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 22.van Keimpema M, et al. FOXP1 directly represses transcription of proapoptotic genes and cooperates with NF-κB to promote survival of human B cells. Blood. 2014;124(23):3431–3440. doi: 10.1182/blood-2014-01-553412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Care MA, et al. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PLoS One. 2013;8(2):e55895. doi: 10.1371/journal.pone.0055895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 26.Mandelbaum J, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18(6):568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21(6):723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19(21):2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 30.Kluk MJ, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS One. 2012;7(4):e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ci W, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113(22):5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8(3):225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- 33.Liu DX, Qian D, Wang B, Yang JM, Lu Z. p300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2011;31(18):3906–3916. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagardoy A, et al. Downregulation of FOXP1 is required during germinal center B-cell function. Blood. 2013;121(21):4311–4320. doi: 10.1182/blood-2012-10-462846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. BCL-6 negatively regulates expression of the NF-kappaB1 p105/p50 subunit. J Immunol. 2005;174(1):205–214. doi: 10.4049/jimmunol.174.1.205. [DOI] [PubMed] [Google Scholar]
- 37.Sebastián E, et al. Molecular characterization of immunoglobulin gene rearrangements in diffuse large B-cell lymphoma: Antigen-driven origin and IGHV4-34 as a particular subgroup of the non-GCB subtype. Am J Pathol. 2012;181(5):1879–1888. doi: 10.1016/j.ajpath.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 38.Cox MC, et al. Clinicopathologic characterization of diffuse-large-B-cell lymphoma with an associated serum monoclonal IgM component. PLoS One. 2014;9(4):e93903. doi: 10.1371/journal.pone.0093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruminy P, et al. The isotype of the BCR as a surrogate for the GCB and ABC molecular subtypes in diffuse large B-cell lymphoma. Leukemia. 2011;25(4):681–688. doi: 10.1038/leu.2010.302. [DOI] [PubMed] [Google Scholar]
- 40.Tinguely M, et al. IRF8 is associated with germinal center B-cell-like type of diffuse large B-cell lymphoma and exceptionally involved in translocation t(14;16)(q32.33;q24.1) Leuk Lymphoma. 2014;55(1):136–142. doi: 10.3109/10428194.2013.793324. [DOI] [PubMed] [Google Scholar]
- 41.Carotta S, et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014;211(11):2169–2181. doi: 10.1084/jem.20140425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingham RJ, et al. The Gab1 docking protein links the b cell antigen receptor to the phosphatidylinositol 3-kinase/Akt signaling pathway and to the SHP2 tyrosine phosphatase. J Biol Chem. 2001;276(15):12257–12265. doi: 10.1074/jbc.M010590200. [DOI] [PubMed] [Google Scholar]
- 43.Tedoldi S, et al. Jaw1/LRMP, a germinal centre-associated marker for the immunohistological study of B-cell lymphomas. J Pathol. 2006;209(4):454–463. doi: 10.1002/path.2002. [DOI] [PubMed] [Google Scholar]
- 44.Schmidlin H, et al. Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood. 2008;112(5):1804–1812. doi: 10.1182/blood-2008-01-136440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 46.Györy I, Fejér G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170(6):3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- 47.van Keimpema M, et al. The forkhead transcription factor FOXP1 represses human plasma cell differentiation. Blood. 2015;126(18):2098–2109. doi: 10.1182/blood-2015-02-626176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon HS, et al. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol. 2012;189(5):2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basso K, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115(5):975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ippolito GC, et al. Dendritic cell fate is determined by BCL11A. Proc Natl Acad Sci USA. 2014;111(11):E998–E1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu H, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7(8):819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.