Significance
Increasing evidence suggests that the exosomal messenger pathway warns neighboring cells against cellular stress and infection. Recent studies have shown that viruses and cancer cells exploit exosomes to transmit functional RNAs. Our studies reveal that a viral small RNA signal for innate immunity Epstein–Barr virus (EBV)-EBER1 is produced by latent EBV-infected B cells and recognized by noninfected dendritic cells activating an inflammatory response. We detected high amounts of EBV-EBER1 transcripts and EBV-microRNAs in inflamed skin lesions of autoimmune patients that are infiltrated with dendritic cells. Importantly, we found virtually no EBV-DNA present in these tissues, suggesting that continuous cell–cell EBER1 transmission via exosomes occurs in humans. We propose that innate sensing of latent EBV in predisposed individuals may be more harmful than previously thought.
Keywords: exosomes, EBV-EBER1, innate sensing, dendritic cells, skin inflammation
Abstract
Complex interactions between DNA herpesviruses and host factors determine the establishment of a life-long asymptomatic latent infection. The lymphotropic Epstein–Barr virus (EBV) seems to avoid recognition by innate sensors despite massive transcription of immunostimulatory small RNAs (EBV-EBERs). Here we demonstrate that in latently infected B cells, EBER1 transcripts interact with the lupus antigen (La) ribonucleoprotein, avoiding cytoplasmic RNA sensors. However, in coculture experiments we observed that latent-infected cells trigger antiviral immunity in dendritic cells (DCs) through selective release and transfer of RNA via exosomes. In ex vivo tonsillar cultures, we observed that EBER1-loaded exosomes are preferentially captured and internalized by human plasmacytoid DCs (pDCs) that express the TIM1 phosphatidylserine receptor, a known viral- and exosomal target. Using an EBER-deficient EBV strain, enzymatic removal of 5′ppp, in vitro transcripts, and coculture experiments, we established that 5′pppEBER1 transfer via exosomes drives antiviral immunity in nonpermissive DCs. Lupus erythematosus patients suffer from elevated EBV load and activated antiviral immunity, in particular in skin lesions that are infiltrated with pDCs. We detected high levels of EBER1 RNA in such skin lesions, as well as EBV-microRNAs, but no intact EBV-DNA, linking non–cell-autonomous EBER1 presence with skin inflammation in predisposed individuals. Collectively, our studies indicate that virus-modified exosomes have a physiological role in the host–pathogen stand-off and may promote inflammatory disease.
Viral nucleic acids are among the foreign molecules with pathogen-associated molecular patterns (PAMPs) that are recognized by innate immune host-sensors in the cytosol (1). Some RNA viruses establish a chronic productive infection: for example, hepatitis C virus (HCV), which evolved strategies to escape from innate immune sensors contributing to its pathophysiology (2). Nevertheless HCV, an enveloped RNA virus, can be recognized by sensory plasmacytoid dendritic cells (pDCs) (3). More recently, Hepatitis A virus, a nonenveloped RNA virus, was shown to hijack a host membrane, presumably to escape neutralizing antibodies while facilitating sensing by pDCs (4). How DNA viruses, and in particular latent herpes viruses, are recognized by the innate immune system and whether this causes or protects from pathogenesis remains unclear (5).
Chronic viral infections can be asymptomatic and only cause pathogenesis in immune-suppressed or genetically predisposed individuals, a prime example being infection by the large DNA herpesvirus Epstein–Barr virus (EBV) (6). Although there is no accurate animal model available, studies in human tissue and peripheral blood showed that EBV has a strong B-cell tropism and evolved a complex latency strategy to minimize transcriptional activity and avoid immune detection. Still, during persistence, EBV produces up to 105–6 copies of polymerase III (Pol III)-transcribed noncoding RNAs (EBERs) per infected B cell (7). EBERs consist of EBER1 and EBER2 molecules, although in latent-infected cells, EBER1 levels are generally higher. EBER1 expression is reduced in reactivated cells, implying a function in maintaining EBV latency, although in vivo EBER1 function remains unclear (8). EBER1 and EBER2 form stem-loop structures by intramolecular base pairing, which enables interaction with several cellular proteins. Both double-stranded loops and the uncapped 5′-triphosphate terminus of EBERs serve as ligands for pathogen recognition receptors (PRRs) (9–12). Because infected individuals (estimated at 90% of the world population) generally have no symptoms, it seems that EBER production in healthy individuals elude cell-intrinsic danger signaling. Currently, the recognition of herpesviruses during latent infection remains poorly understood (5).
Toll-like receptors (TLRs) -3, -7, and -8 are PRRs expressed in sensory dendritic cells that detect exogenous double- and single-stranded viral RNA in endosomes. Because the binding domains of TLRs face toward the lumen of endosomes, recognition of self-RNAs in the cytoplasm is avoided (13). Moreover, mRNAs are normally 5′-“capped” and bound by poly-A binding proteins, whereas tRNAs are enzymatically modified and ribosomal RNAs are masked as ribonucleoprotein complexes, preventing them from being recognized by cytosolic PRRs. Retinoic acid-inducible gene I (RIG-I) (14), IFN-induced proteins with tetratricopeptide repeats (IFITs) (15), and the double-stranded RNA-dependent protein kinase (PKR) can recognize 5′triphosphates (5′ppp)-bearing viral RNA in the cytoplasm (16). Antiviral signaling is also induced by modified self-RNAs under physiological stress conditions (17–19). Removal of proinflammatory self-RNAs, while maintaining the ability to selectively recognize virus-produced RNAs in the cell’s cytoplasm, is essential for proper regulation of antiviral response. Indeed, chronic activation of antiviral immunity by erroneous detection of extracellular RNA drives autoimmunity in predisposed individuals (20). Although binding of proinflammatory EBERs to cytoplasmic sensors has been reported in vitro (9–12), there is little evidence that this may occur in latently infected B cells in a physiologically relevant context (21), but there are multiple indications that EBERs are secreted from infected cells (9, 22, 23).
Here we determined a physiological effect of secreted EBER1 via exosomes, a subtype of extracellular vesicles of endosomal origin. We demonstrate that latently infected B cells sort 5′pppEBER1 molecules into exosomes that are taken up by DCs and trigger antiviral immunity. In a chronic setting, innate sensing of latent EBV by pDCs may pose an environmental risk factor for developing and driving nonresolving organ inflammation.
Results
Exosomes from Latently EBV-Infected Cells Induce Antiviral Immunity in Primary DCs.
We showed previously that latency III EBV-infected B cells influence gene expression in neighboring monocyte-derived DCs through exosome-mediated transfer of functional viral microRNAs (miRNAs) (24).
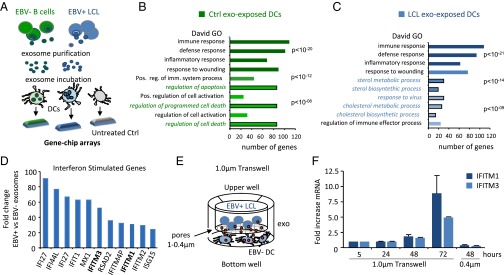
We wished to gain a global insight into the genes and pathways affected in DCs upon internalization of exosomes released by EBV-infected B cells. We purified exosomes from EBV-driven (latency III) lymphoblastoid cell lines (EBV+ LCLs) and noninfected (EBV−) B-cell lymphoma control cells (BJAB) that produce similar amounts of exosomes (25) (“LCL exosomes” and “Ctrl exosomes,” respectively). We incubated primary DCs with LCL and Ctrl exosomes and analyzed the gene expression profile of DCs with high-density arrays after 18 h of incubation (Fig. 1A). Gene ontology (GO) analysis (DAVID bioinformatics tool) showed that immune, defense, and inflammatory responses and “response to wounding” were the top four generally modulated processes in exosome-stimulated conditions (Fig. 1 B and C). Control exosomes also affected apoptosis-related pathways, but to a lesser extent. In contrast, incubation with LCL exosomes induced antiviral response-related pathways including sterol metabolic and biosynthetic processes in the DCs. Sterol-associated biochemistry was recently implicated in type I IFN-driven antiviral immunity induced by viral infection (26). In fact, type I IFN-stimulated genes (ISGs) were among the most strongly induced genes in DCs exposed to LCL exosomes. Among the most elevated genes in DCs exposed to LCL exosomes compared with Ctrl exosomes were interferon induced transmembrane protein 1 (IFITM1), which inhibits the entry of viruses to the cytoplasm, IFITM3, which disrupts intracellular cholesterol homeostasis (Fig. 1D), and the transcription factor STAT1 (27). Highly similar differences in the induction of IFITM1 and IFITM3 were observed when comparing DCs exposed to LCL exosomes or CD40L-stimulated primary B cell exosomes (Fig. S1 A and B). We conclude that LCLs exosomes but not control exosomes from EBV− B cells induce an antiviral transcription program in noninfected DCs.
Fig. 1.
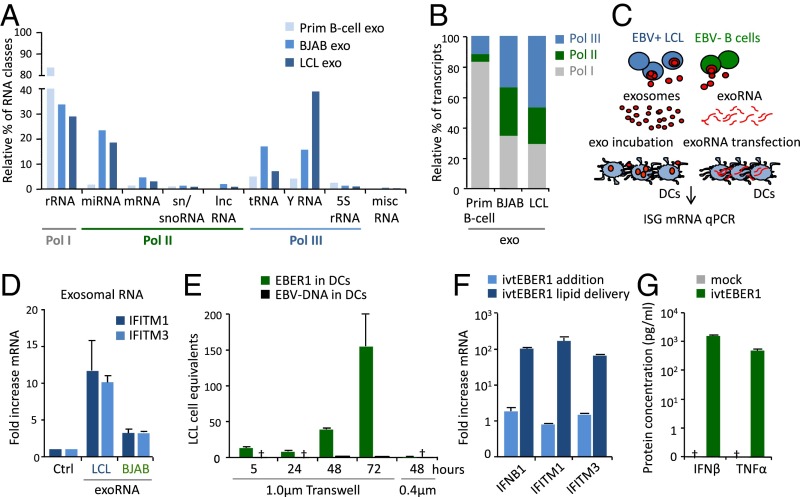
Exosomes released by latent EBV-infected LCLs trigger antiviral immunity in DCs. (A) Schematic representation of experimental design: primary DCs were incubated with exosomes derived from EBV+ LCLs or EBV− control B cells (BJAB), and a gene-expression profile was performed using high-density arrays after 18 h of incubation. (B and C) GO analysis performed with DAVID showing the top modulated pathways in DCs exposed to EBV− control B-cell (B) and EBV+ LCL (C) exosomes compared with untreated DCs. (D) Fold-difference of IFN-stimulated genes in DCs exposed to LCL exosomes compared with control exosomes. (E) Schematic representation of a transwell coculture system: exosome-producing B cells (upper compartment) and recipient DCs (lower compartment) are separated by a 1-µm pore size membrane, which allows the passage of exosomes. (F) Quantitative PCR (qPCR) for IFITM1 and IFITM3 in DCs at indicated time points during coculture using a transwell with either 1- or 0.4-µm membrane pores. Transcript levels are normalized to GAPDH and expressed as fold-increase relative to t = 0.
Fig. S1.
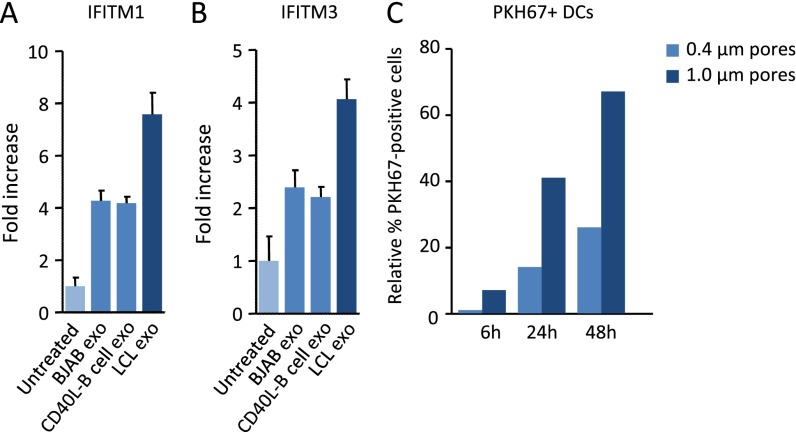
Induction of IFITM1 and IFITM3 by LCL exosomes compared with CD40L-stimulated B-cell exosomes, and efficiency of exosome transfer in transwell coculture systems. (A and B) qPCR analysis of IFITM1 (A) and IFITM3 (B) in DCs upon treatment with LCL exosomes, control BJAB, and CD40L-stimulated primary B-cell exosomes. (C) PKH67-labeled exosome-producing LCLs are placed in the upper compartment of a transwell system with 0.4- or 1-µm pore size membrane. Recipient DCs are placed in the lower compartment of the transwell and analyzed for PKH67-exosome uptake at indicated time points. Graph shows the relative percentage of PKH67+ DCs as analyzed by FACS.
To investigate whether the continuous release of exosomes in culture could reproduce the type I IFN-response signature induced by purified exosomes, we cocultured latently infected LCLs with primary DCs and measured IFITM1 and IFITM3 mRNA levels as markers for antiviral response activation (Fig. 1E). We detected a steady increase in IFITM1 and IFITM3 mRNA levels reaching twofold induction after 48 h and eightfold induction after 72 h (Fig. 1F), indicative of antiviral response activation. Interestingly, when we decrease exosome-transfer by separating the LCLs from DCs with a 0.4-µm porous membrane (instead of an exosome-permissive 1.0-µm membrane), we did not observe any induction of IFITM1 and IFITM3 after 48 h, suggesting that soluble factors are not involved in early activation (Fig. 1F and Fig. S1C). Thus, latently EBV-infected B cells secrete exosomes that can induce antiviral, proinflammatory behavior in sensory immune cells.
Latent EBV-Infected Exosomes Do Not Display an Antiviral Proteome Signature.
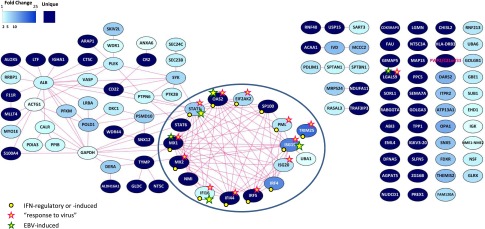
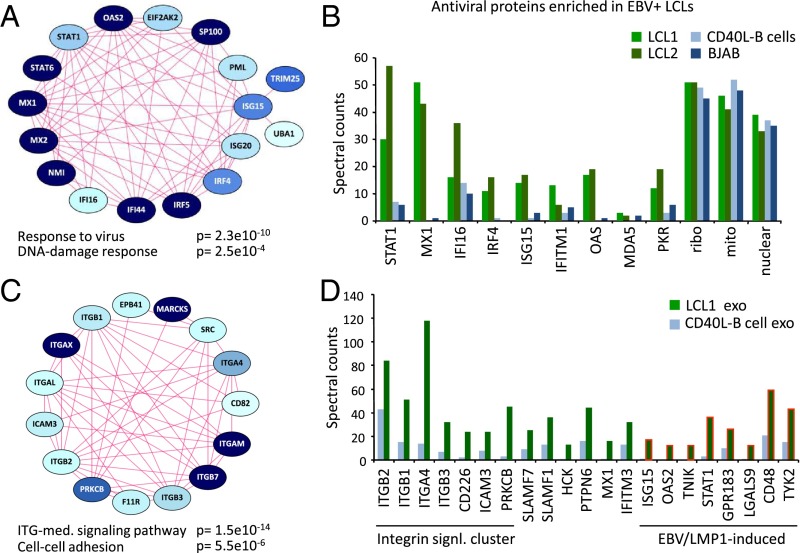
Recent studies in IFN-exposed DCs suggested that exosomes incorporate and transfer antiviral proteins (28). To investigate the possibility that latent EBV-infected cells release antiviral proteins via exosomes, we performed label-free mass spectrometric proteomics on LCLs whose proliferation is EBV-driven and, among others, CD40L-stimulated uninfected B cells with identical genetic background as a physiologically relevant control (Dataset S1). Differential analysis comparing CD40L-driven EBV− primary B cells (prim B-cells) with LCL cells driven by latent EBV, and subsequent GO mining of 113 proteins that were significantly more abundant in LCLs (P < 0.05) showed enrichment of the term “response to virus” (corrected P < 0.05) (Fig. S2 and Dataset S1). Proteins linked to this term were encompassed within a major cluster of protein–protein associations retrieved from the STRING database (29) (Fig. 2A and Fig. S2). Moreover, the majority of the proteins in this cluster are linked to type I IFN signaling and function. When comparing mass-spectral counts for selected proteins in two distinct LCL lines versus CD40L-driven and EBV− control B cells (BJAB) (Fig. 2B), the counts showed a consistent difference, indicating that the antiviral signature is related to EBV infection in human B cells.
Fig. S2.
Proteomics analysis on EBV-driven LCLs compared with CD40L-stimulated B cells shows an enrichment in the pathway “response to virus.” The circle highlights the antiviral protein cluster.
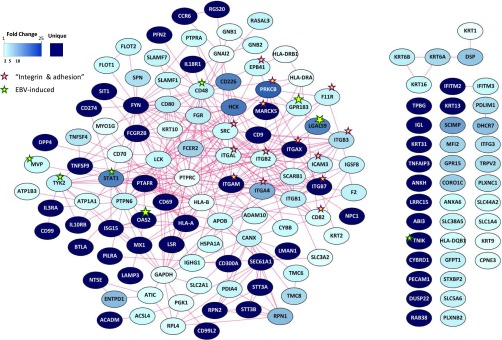
Fig. 2.
Latent EBV-infected cells display an antiviral proteomic signature that is not conveyed to their exosomes. (A and B) Proteomics analysis on EBV-driven LCLs compared with CD40L-stimulated B cells reveals enrichment in the pathway “response to virus” (Fig. S2) and “DNA-damage response,” with a dominant cluster of 17 antiviral proteins (A). Spectral counts of LCL cell lines, CD40L-stimulated B cells and BJAB cells showing association of the antiviral protein signature with EBV infection. Ribo: 40S ribosomal protein S3; mito: Stress-70 protein, mitochondrial; nuclear: Splicing factor 3B subunit 1 (B). (C and D) Proteomics analysis of exosomes from EBV-driven LCLs compared with CD40L-stimulated B cells (Fig. S3) reveals enrichment of “integrin-mediated signaling pathway” and “cell–cell adhesion” terms (C). Spectral counts showing enrichment of integrin signaling proteins and EBV/LMP1-induced proteins in LCL exosomes (D).
In parallel, we analyzed the proteome of exosomes released by EBV-driven LCL and exosomes from CD40L-driven B cells, all purified by ultracentrifugation on a sucrose cushion, as described previously (30). We detected 3,000+ proteins in the exosomes (Dataset S2). Interestingly, apart from immune-response terms, proteins enriched in the LCL exosomes relative to their EBV− counterparts (144 differential proteins in total, P < 0.05) were associated with “integrin-mediated signaling” and cell-adhesion terms (Fig. 2C and Fig. S3). Supportively, we identified characteristic EBV-LMP1 (latent membrane protein 1)–induced proteins enriched in LCL exosomes, including EBI2/GPR183, STAT1, and CD48/BLAST-1 proteins (Fig. 2D), consistent with a latent EBV signature. The most enriched antiviral protein in LCL exosomes was IFITM3, consistent with its subcellular localization at endosomal membranes (31). Generally, the spectral counts for antiviral proteins in both the LCL exosomes and EBV− B-cell exosomes were low. We conclude that the restricted antiviral protein signature in EBV-driven LCL is not selectively conferred to exosomes. This finding is distinct from exosomes released by cells that are exposed to IFN-α, which carry an elaborate antiviral proteome (28).
Fig. S3.
Proteomics analysis of exosomes from EBV-driven LCLs compared with exosomes from CD40L-stimulated B cells shows “immune system process” as the most represented pathway, and an enrichment in “integrin-mediated signaling” and “cell adhesion” terms.
EBER1 Sorting and Transmission via Exosomes Induces Antiviral Immunity in Primary DCs.
We previously found that the small RNA content of LCL exosomes includes functional Pol II-transcribed EBV-miRNAs (25). Bioanalyzer analysis of purified exosomal RNA indicated that latently infected LCLs and (CD40L-stimulated) EBV− primary B cells produce exosomes enriched in small RNA species, ranging from 15 to 200 nucleotides. RNA-seq analysis showed that exosomes carry multiple RNA classes, although their relative distribution is different (Fig. 3A). We classified the RNA sequences in Pol I, II, and III transcripts and found that almost 50% of all transcripts were Pol III-transcribed in LCL exosomes, against the 12% in the EBV− primary B-cell–derived counterparts (Fig. 3B). This observation is consistent with elevated levels of BDP1 and TFIIIC transcription factors in latently EBV-infected LCLs that stimulate host Pol III transcription (32).
Fig. 3.
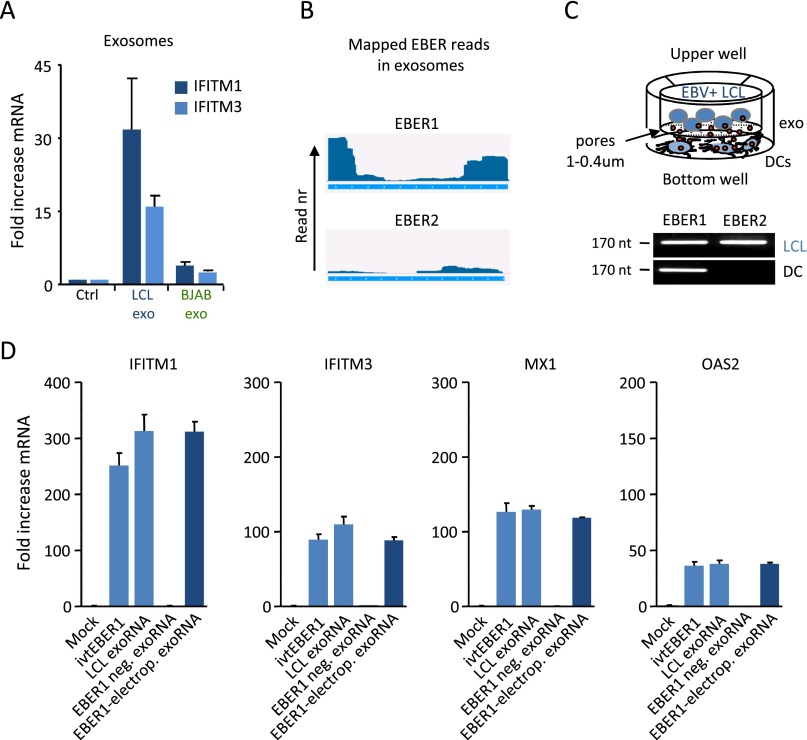
Exosome-mediated transfer of EBER1 triggers an antiviral response in recipient DCs. (A and B) Small RNA class distribution in exosomes from EBV+ LCLs and EBV− BJAB and primary B cells. Data are expressed as percentage relative to the total amount of sequencing reads (A). Relative percentage of Pol I, II, and III transcripts in LCL and control exosomes (B). (C and D) DCs were either incubated with exosomes from EBV+/EBV− cells or transfected with matching amounts of exosomal RNA (exoRNA), and the expression of ISGs was assessed by qPCR after 18 h of incubation (C). Exosomal RNA from EBV+ LCLs, but not from EBV− BJAB cells induced IFITM1 and IFITM3 expression (D). (E) qPCR analysis of EBER1 RNA and EBV-DNA in DCs at specific time points during the coculture period, using transwells with 1- or 0.4-µm membrane pores. qPCR data are expressed as EBV-infected LCL cell equivalents. (F) qPCR analysis of IFITM1, IFITM3, and IFNB1 in DCs upon transfection or direct addition of 5′pppEBER1. Transcript levels are normalized to GAPDH and expressed as fold-increase relative to experimental controls. (G) IFN-β and TNF-α production by DCs transfected with 5′pppEBER1; protein concentration was assessed by ELISA on culture supernatants.
Depending on their sequence, structure, and modifications, small RNAs may or may not trigger specialized viral sensors that recognize PAMPs, such as small double-stranded RNA loops or a 5′ppp moiety (14). To investigate whether exosome-incorporated small RNAs trigger viral sensors, we purified and transfected exosomal RNA (exoRNA) directly into DCs (Fig. 3C). Interestingly, exoRNA from latently infected LCLs strongly induced IFITM1 and IFITM3 transcription, compared with exoRNA from EBV− control (BJAB) cells (Fig. 3D), in agreement with the outcome of the array analysis in Fig. 1. Incubation of DCs with exoRNA or matched amounts of intact exosomes determined a similar fold-induction of IFITM1 and IFITM3 (Fig. S4A), suggesting that the small RNA content in LCL exosomes is modified by latent EBV infection and contains a trigger for antiviral gene expression.
Fig. S4.
EBER1 and EBER2 transfer to DCs after coculture with LCLs, and ISG expression in DCs after transfection with RNA isolated from EBER1-electroporated exosomes. (A) Transfection with exosomal RNA (see Fig. 3D) and incubation with exosomes had very similar effects on IFITM1 and IFITM3 expression. (B) Sequence coverage of EBER1 and EBER2 in cells and exosomes; y axis indicates the normalized counts (rpm). (C) Schematic representation of a transwell coculture system (Upper). Detection of full length EBER1 and EBER2 in LCLs and DCs after 48h of coculture (Lower). (D) ISG expression in DCs 24 h after transfection with RNA isolated from LCL exosomes, BJAB exosomes and BJAB exosomes electroporated with 5′pppEBER1.
EBV-encoded EBERs have proinflammatory properties and may reach up to 106 copies per latently infected B cell, with EBER1 being present at 10-fold higher levels than EBER2 (7). This finding is consistent with our small RNAseq analysis, indicating that EBER1-mapped reads outnumber those derived from EBER2 (Fig. S4B). To explore whether EBV+ LCL exosomes package full-length EBERs while avoiding unwanted codetection of EBV-DNA, we designed stem-loop RT-PCR assays. Full-length EBER1, but not EBER2, was detectable in recipient DCs after coculture with LCLs (Fig. S4C). EBER1 RNA levels increased in the recipient cells over time, suggesting that exogenously delivered EBER1 molecules are not immediately degraded (Fig. 3E). We ruled out the possibility that DCs were initially or subsequently infected with EBV, as EBV-DNA was not detected by DNA-PCR, indicating that the EBER1 molecules were most likely of exogenous origin. We deemed transfer via EBER1–protein complexes released by dying cells unlikely, because use of small (0.4 μm) pore size that obstructs LCL exosome passage blocked all EBER1 transfer (Fig. 3E).
To test whether EBER1 is detected by physiologically relevant cells expressing viral sensors (33), we generated EBER1 molecules by in vitro transcription (ivtEBER1) and introduced them into primary DCs by lipid transfection. Both IFITM1 and IFITM3, as well as interferon beta 1 (IFNB1) mRNA levels, were dramatically induced upon lipid delivery of EBER1 transcripts (Fig. 3F), whereas this was not observed by adding identical amounts of EBER1 directly to the culture dish. Finally, ELISAs demonstrated that EBER1 delivery stimulates the production of the inflammatory cytokines IFN-β and TNF-α by DCs (Fig. 3G). Thus, delivery of exogenous 5′pppEBER1 via lipid vesicles triggers antiviral gene expression and inflammatory cytokines production in primary DCs.
EBER1 Sensing Is Dependent upon Its 5′Triphosphate Moiety.
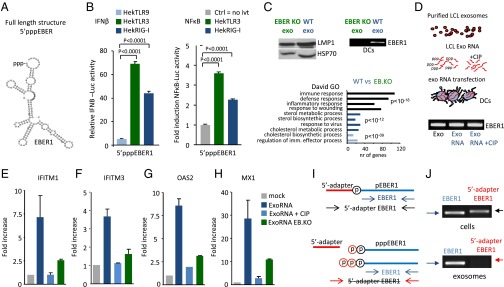
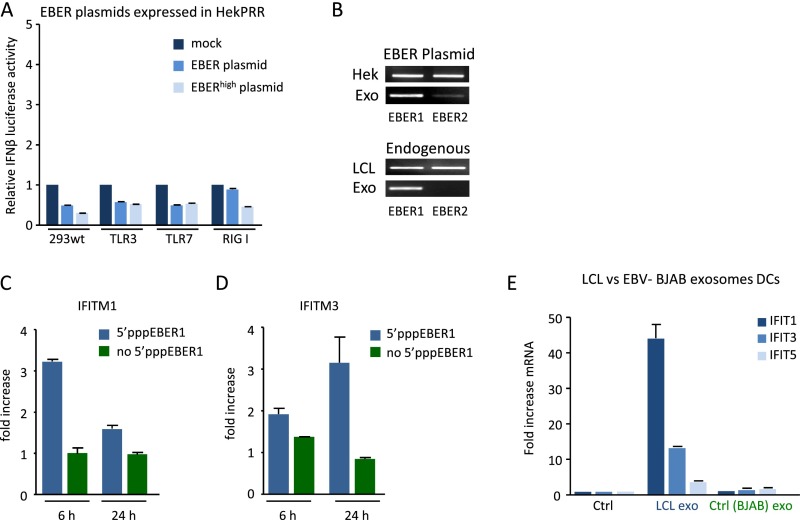
Sensor/PRR activation relies on the RNA structure, posttranscriptional modifications, protein-binding affinity, and subcellular localization of the activating RNA (1). We investigated 5′pppEBER1 (Fig. 4A) recognition in Hek293 cells overexpressing TLR9, TLR3, or induced RIG-I, by measuring IFN-β or NF-κB promoter activation with luciferase-reporters. IFN-β and NF-κB promoters were activated by 100 ng of in vitro transcribed 5′pppEBER1 in TLR3- and RIG-I–expressing cells, but not in TLR9-expressing or parental Hek cells (Fig. 4B). To formally establish that exosome-mediated transfer of EBER1 activates an antiviral response in noninfected DCs, we isolated exosomes from LCLs driven by an EBER-deficient mutant EBV strain (34). We performed microarray analysis of DCs that were incubated with EBER1-deficient (EB.KO) or EBER1-containing (wild-type) LCL exosomes. In a direct comparison we found that EBER1-containing exosomes were superior in their capacity to activate antiviral genes compared with EBER1-deficient LCL exosomes (Fig. 4C).
Fig. 4.
The 5′triphosphate moiety of EBER1 is required for antiviral response activation. (A) Predicted secondary structure of EBER1 (RNAfold web server). (B) IFN-β and NF-κB promoter activation (assessed with luciferase reporters) in response to 5′pppEBER1 (ivt) transfection in Hek293 cells expressing TLR9, TLR3, or RIG-I (cells were “primed” with rhIFN-α). Firefly luciferase activity is normalized to Gaussia luciferase activity and data are expressed relative to control. (C) Western blot analysis of LMP1 and Hsp70 in exosomes isolated from LCLs infected with EBER-deficient mutant (EB.KO) or wild-type (wt) EBV (Upper Left). DCs were incubated with EBER1-deficient or EBER1-containing exosomes and the EBER1 levels were assessed by qPCR (Upper Right). Microarray analysis (Lower) shows that antiviral genes are strongly overrepresented in DCs exposed to wild-type exosomes compared with DCs incubated with EBER1-deficient exosomes. (D–H) Exosomal RNA was incubated with CIP to remove the phosphate moieties, and DCs were transfected with CIP-treated or untreated exosomal RNA. EBER1 levels were comparable in all experimental conditions (D). qPCR analysis shows that treatment with CIP completely abrogates the ability of exosomal RNA to induce ISGs. Transcript levels are normalized to GAPDH and expressed as fold-increase relative to mock control (E–H). (I and J) LCL cellular and exosomal RNA were incubated with adapters that can only ligate 5′monophosphate RNAs; qPCR was performed using primers specific for the 5′adapter and 3′EBER1 (I). Full-length EBER1, but not 5′adapter ligated-EBER1, is detectable in LCL exosomes (J).
To investigate whether the 5′ppp moiety of EBER1 serves as a PAMP in recipient DCs, we isolated exosomal RNA and treated this with calf intestinal phosphatase (CIP) to remove the phosphate groups. Subsequently we transfected CIP-treated and untreated exoRNA into DCs and measured ISG expression (Fig. 4D). Strikingly, CIP treatment completely impeded ISG induction (Fig. 4 E–H). EBER1 levels were nevertheless similar in DCs transfected with CIP-treated and nontreated exoRNA (Fig. 4D), excluding variation in intracellular levels as an explanation for the observed differences. Moreover, the exoRNA from EBER1-deficient LCLs induced ISG expression to a much lower extent compared with the exoRNA from wild-type EBV+ LCLs, suggesting that EBER1 is a major factor in the IFN response (Fig. 4 E–H). To confirm that exosome-transmitted EBER1 causes this effect, we electroporated 5′pppEBER1 transcripts into exosomes from EBER− control cells. We found that 5′pppEBER1 addition to EBER− exosomes fully recovered the induction of IFN response (Fig. S4D). These results, combined, suggest that EBER1 molecules in exosomes bear a 5′ppp motif and induce antiviral genes in DCs. To corroborate this finding, we ligated adapters to 5′monophosphate-bearing small RNA molecules and performed RT-PCR with a 5′adapter and 3′EBER1 specific primer set (Fig. 4I). Although we readily detected intracellular EBER1, we did not detect adapter-ligated EBER1 in the exosomes, yet with conventional EBER1 primers we confirmed the presence of EBER1 (Fig. 4J). Thus, the exosomal pool of EBER1 harbors minimally a di- or triphosphate group that both function as a viral PAMP in cultured monocyte-derived DCs.
Cytoplasmic 5′pppEBER1 in Latently Infected B Cells Evades Viral Sensors by Association with Lupus Antigen Protein.
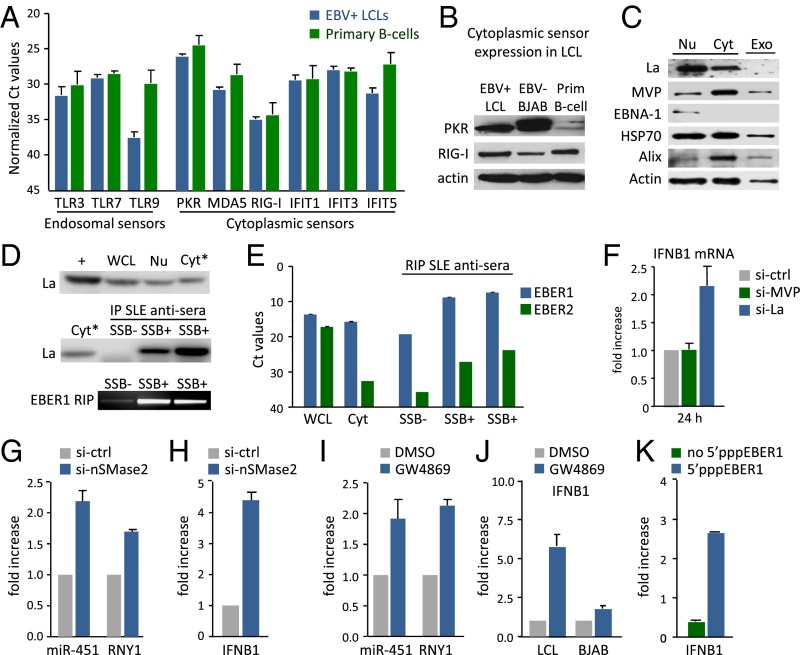
Because EBV-EBER1 fails to trigger inflammatory cytokine production in latently infected LCLs, we hypothesized that these cells may lack viral sensors to detect them. We profiled LCLs and primary B cells by RT-PCR for nine prominent sensors and found that they were all expressed at the transcriptional level (Fig. 5A). RIG-I and PKR, both of which can sense 5′pppRNA, were also detected at the protein level (Fig. 5B), making it unlikely that the lack of cytokine production is merely due to the absence of cytosolic sensors.
Fig. 5.
EBER1 evades viral sensors by association with La. (A) qPCR analysis of viral sensors in EBV+ LCLs (blue bars) and primary B cells (green bars). (B) Western blot analysis of PKR and RIG-I in EBV+ LCLs, EBV− BJAB cells and primary B cells. (C) Western blot analysis of La, MVP, and indicated proteins in LCL nuclear (Nu), cytosolic (Cyt), and exosomal (Exo) fraction. (D and E) RNA immunoprecipitation (RIP) detects EBER1–La complexes in the cytosolic fraction. Western blot analysis of La in whole-cell lysate (WCL), nuclear and cytosolic fractions (D, Top). La immunoprecipitation in the cytosolic fraction using SSB− control antiserum and two different SSB+ antisera (Middle). Detection of coimmunoprecipitated full-length EBER1 (D, Bottom). qPCR analysis of EBER1 and EBER2 in WCL, input (cytosol), and control (SSB−) and La (SSB+) immunoprecipitates (E). (F) siRNA knockdown experiments show that La- but not MVP-knockdown increases IFNB1 mRNA expression in LCLs. (G and H) Intracellular levels of miR-451 and RNY1 (G) and IFNB1 (H) in LCLs after treatment with siRNA against nSMAse2. (I and J) Intracellular levels of miR-451 and RNY1 (I) and IFNB1 (J) in LCLs after treatment with GW4869. (K) IFNB1 mRNA levels after LCL transfection with ivtEBER1 with or without 5′triphosphate.
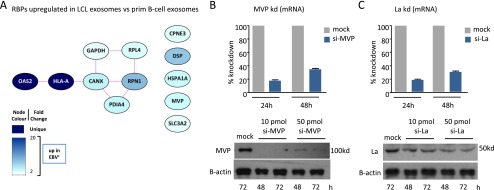
Some miRNAs may be sorted into exosomes through association with nuclear ribonucleoproteins (RNP) (35). To search for potential EBER1 transport proteins, we analyzed the proteomics data for all known RNA-binding partners. We found few Pol III RNA-binding proteins enriched in LCL exosomes compared with control exosomes from EBV− B cells (Fig. S5A). One of these, the major vault protein (MVP), associates with vRNA1-1 (36), which was enriched in LCL exosomes. However, we did not detect the lupus antigen (La/SSB) (Fig. 5C), the most commonly described binding-partner of EBERs. As expected, Hsp70, Alix, and β-Actin were detected in exosomes, whereas the nuclear protein EBNA1 was absent. To investigate whether EBER1 associates with cytoplasmic La before sorting into exosomes, we performed RNA immunoprecipitation experiments in the cytosolic fraction of latent EBV-infected LCLs. Strikingly, after confirming specific pull-down of the La protein with human La/SSB reactive sera, we detected full-length coimmunoprecipitated EBER1 as shown on gel (Fig. 5D). Moreover, when normalized to input with La/SSB nonreactive sera, we estimated a 1,000-fold enrichment of EBER1 with La/SSB reactive sera (Fig. 5E). In comparison, EBER2 pull-down was very low, consistent with the observation that EBER2 transcripts are relatively low in the cytosolic fraction (Fig. 5E) and not transferred via exosomes (Fig. S4C).
Fig. S5.
Differential representation of RNA-binding partners in exosomes, MVP and La knockdown efficiency. (A) Proteomics analysis showing Pol III RNA-binding partners enriched in exosomes from EBV-driven LCLs compared with primary B cells. (B and C) siRNA-mediated knockdown of MVP (B) and La (C) at mRNA (Upper) and protein levels (Lower) at indicated time points. Transcript levels are normalized to GAPDH and expressed as percentage relative to mock control.
To investigate whether the association of EBER1 with cytoplasmic La is physiologically relevant in that it shields its 5′ppp motif from sensor detection, we reduced La protein level by small-interfering RNA (siRNA) knockdown and analyzed IFNB1 expression. La-knockdown increased IFNB1 transcription in LCLs, whereas MVP-knockdown had no such effect (Fig. 5F and Fig. S5 B and C). Thus, EBERs may evade detection by cytosolic sensors in latently infected B cells by interacting with the La protein, as previously suggested for respiratory syncytial virus-produced small RNAs (37). We also tested this in HekPRR reporter cells that expressed EBERs from nuclear plasmids. Despite sorting and trafficking into exosomes, we did not observe an increase in IFN-β promoter activation (Fig. S6 A and B). Because EBER1 is sorted into exosomes without La, we wondered whether interference of small RNA sorting at endosomes, might disrupt EBER evasion from sensors. To investigate this, we treated LCLs with a small-interfering RNA against nSMase2, a protein implicated in both exosome release and small RNA secretion (38). As expected, siRNA treatment elevated intracellular levels of miR451 and Pol III RNY1 (Fig. 5G). Importantly, this effect was associated with a significant increase in IFNB1 transcription (Fig. 5H). Treatment of LCLs with GW4869, an inhibitor of nSMase2 function (39), yielded similar results (Fig. 5 I and J). Finally, transfection of 5′pppEBER1 directly into LCLs also increased IFNB1 and ISG (IFITM1 and IFITM3) transcription, confirming that endogenous sensors can sense 5′pppEBER1 molecules (Fig. 5K and Fig. S6 C and D). Thus, EBERs can be recognized in latently infected B cells, but interaction with La and sorting into exosomes prevent detection.
Fig. S6.
IFN response after transfection with EBER expression plasmids or in vitro-transcribed 5′pppEBER1. (A) IFN-β promoter activation (assessed with a luciferase-reporter construct) after transfection with EBER plasmid (1 EBER copy) or EBERhigh plasmid (10 EBER copies) in Hek293 cells expressing TLR3, TLR7, or RIG-I. Reporter firefly luciferase activity is normalized to Gaussia luciferase activity and data are expressed as fold-change relative to the mock control. (B) Detection of EBER1 and EBER2 in Hek293 cells transfected with EBER expression plasmids (Upper) and endogenous EBER1 and EBER2 levels in LCL cells and exosomes (Lower). (C and D) IFITM1 and IFITM3 expression in LCLs at indicated time points after transfection with in vitro transcribed 5′pppEBER1. (E) qPCR analysis of IFIT1, IFIT3, and IFIT5 in DCs after treatment with LCL or BJAB exosomes.
Exosomes from Latent EBV-Infected Cells Target Primary Tonsillar Plasmacytoid DCs.
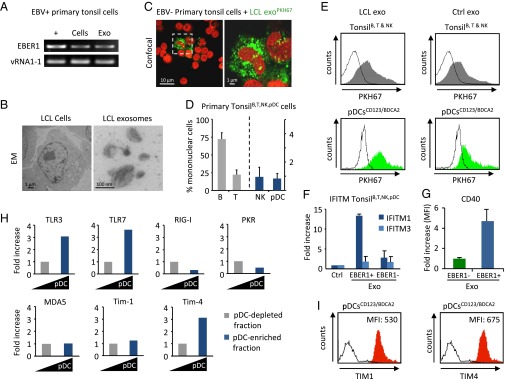
A prominent model of EBV persistence predicts that in healthy individuals newly infected naïve B cells follow a “normal” germinal center reaction in tonsillar tissue (40). Exosomes from infected B cells might transmit physiological messages to neighboring cells in these tissues. We cultured the primary tonsil cells in exosome-depleted medium and isolated the released tonsillar exosomes after 48-h culture. Interestingly, we could detect EBER1 transcripts in the exosomes of the tonsil cell population from EBV+ donors (as confirmed by EBNA1 Q-K DNA-PCR) (Fig. 6A).
Fig. 6.
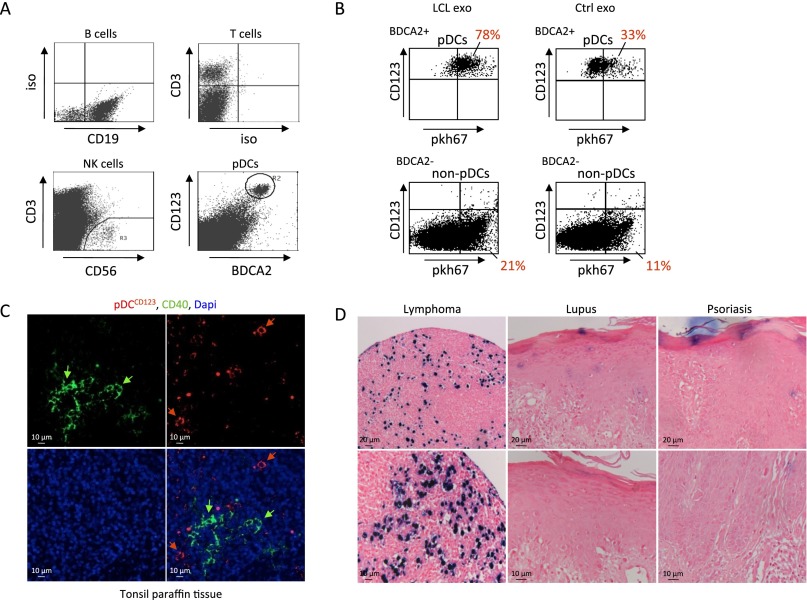
Exosomes from latent EBV-infected cells are internalized by Tim1/4, TLR3/7-expressing pDCs. (A) Full-length EBER1 and vRNA1-1 in tonsil cells from EBV+ donors and corresponding exosomes. (B) Electron microscopy micrograph of LCL cells and exosomes. (C) Confocal microscopy image of primary mononuclear tonsil cells (from EBV− donors) incubated with PKH67-labeled LCL exosomes. Nuclei are stained with TO-PRO-3. (D) Relative percentage of B, T, and NK cells and pDCs in the tonsil mononuclear cell population as analyzed by FACS (Fig. S7A). Graph shows the average percentage of cells from at least four different donors. (E) FACS analysis of tonsil B, T, and NK cells (Upper) and CD123+/BDCA2+ pDCs (Lower) incubated with PKH67-labeled LCL/BJAB exosomes. Tonsillar pDCs internalize LCL exosomes more efficiently compared with nonpDC. (F and G) IFITM1 and IFITM3 mRNA expression in tonsil cells (F) and CD40 (G) expression in the pDC subpopulation upon internalization of EBER1+ or EBER1− exosomes. (H and I) Receptor expression in the pDC subpopulation compared with the pDC-depleted fraction by qPCR (H), and expression of TIM1 and TIM4 exosome receptors at pDC surface as analyzed by FACS (I). MFI, mean fluorescence intensity.
To study exosome communication between EBV-infected B cells and surrounding cells, we first purified LCL exosomes by ultracentrifugation, yielding pure populations of vesicles ranging from 70 to 100 nm in size (Fig. 6B) and labeled these with PKH67, a green fluorescent dye. We incubated the PKH-exosomes with primary mononuclear tonsil cells from EBV− donors. We observed with confocal microscopy that only a small proportion of cells internalize the PKH-exosomes efficiently (Fig. 6C). FACS analysis revealed that tonsils harbor mainly B and T cells (up to 75% and 24%, respectively) and only minor populations of NK cells (CD3+/CD56+) and CD123+/BDCA2+ pDCs (Fig. 6D and Fig. S7A). Importantly, when incubating tonsil mononuclear cells with fluorescently labeled LCL or control BJAB exosomes, we found that CD123+/BDCA2+ pDCs internalize exosomes much more efficiently than the CD123−/BDCA2− cell population (Fig. 6E and Fig. S7B). Moreover, assuming exosomes are added in excess, we observed that LCL exosomes are more efficiently internalized by pDCs compared with EBV− control exosomes. Finally, we found that EBV+ LCL exosomes, but not control exosomes, induced IFITM1 (but not IFITM3) transcription in tonsil mononuclear cells (Fig. 6F). This result argues against the possibility that PKH67 particles or PKH67-induced clustering of exosomes have a role in ISG induction. In vivo, IFITM proteins have various physiological functions and expression patterns, possibly explaining why we did not detect IFITM3 induction in bulk tonsil cell populations (41, 42). As a clear sign of activation, CD40 surface expression was increased in pDCs upon interaction with unstained EBV+ LCL exosomes (Fig. 6G).
Fig. S7.
Exosome uptake by tonsil mononuclear cells, CD123/CD40 expression in tonsil tissue and EBER1 detection in skin tissues. (A) FACS analysis of tonsil mononuclear cells based on CD19, CD3, CD56, BDCA2, and CD123 surface markers. (B) PKH67-exosome uptake by CD123+/BDCA2+ pDCs and CD123−/BDCA2− tonsil cells. (C) Immunofluorescence staining for CD123 (red) and CD40 (green) on tonsil tissue sections reveals single CD40+ cells and CD123+ pDCs. Nuclei are stained with DAPI. (D) EBER1 in situ hybridization in SLE and inflamed skin tissues. EBV+ lymphoma tissue was used as positive control.
As expected, tonsillar pDCs express endosomal RNA sensors TLR7 and TLR3, more so than other tonsil cells, whereas RIG-I, PKR, and MDA5 transcripts are not enriched (Fig. 6H). T-cell immunoglobulin mucin (TIM) proteins 1/4 exosome receptors on tonsillar pDCs are highly enriched at the RNA (Fig. 6H) and protein (surface) level as determined by FACS (Fig. 6I). We propose that EBER1-sensing in pDCs is mediated by exosome capture, possibly through surface TIM1 or -4 exosomes receptors (43).
EBER1 Transfer to Noninfected Cells in Vivo.
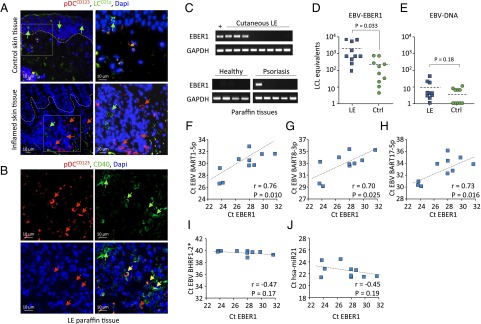
To evaluate whether EBER1 molecules are taken up by pDCs in vivo, we studied pDC-infiltrated skin lesions of lupus erythematosus (LE) patients that suffer from chronically elevated EBV loads (44). We examined paraffin sections of skin biopsies by immunofluorescent staining. Using CD123 as a marker, we detected infiltration of pDCs in inflamed and LE tissues (Fig. 7 A and B). Immunofluorescent staining for CD40 revealed activation of the infiltrating pDCs in LE patient skin (Fig. 7B and Fig. S7C). Next, we extracted RNA from these tissues and performed EBER1 RT-PCR. In three of eight LE tissues, but not in healthy control skin (zero of four), we detected the presence of EBER1 molecules (Fig. 7C).
Fig. 7.
EBER1 is present in pDC-infiltrated SLE skin lesion in the absence of EBV-DNA. (A) Immunofluorescence staining of paraffin-embedded inflamed and healthy skin tissue. CD123 is used as pDC marker and CD1a as Langerhans cell (LC) marker. Nuclei are stained with DAPI. (B) Immunofluorescence staining of cutaneous LE paraffin tissues shows CD123+/CD40+ cells. (C) qPCR of EBER1 in lupus erythematosus and control (healthy and psoriasis) tissues. (D and E) qPCR analysis of EBER1 (D) and EBV-DNA (E) in control and cutaneous LE frozen tissues. qPCR data are expressed as EBV+ LCL equivalents. (F–J) Correlation of EBV-miRNAs and hsa-miR-21 with EBER1 in skin tissues analyzed by qPCR.
Because archived paraffin material can yield low intact RNA yield, we analyzed 10 additional frozen LE biopsies. In these tissues we measured very high levels of EBER1 transcripts, underscored by representing EBER1 levels as “infected LCL B-cell equivalents” (Fig. 7D). EBER1 levels in the LE tissues were significantly higher (P = 0.033) than in the control tissues, whereas EBV-DNA was virtually absent (Fig. 7 D and E), suggesting that EBER1 signals are not derived from actual EBV-infected cells. In accordance with this conclusion, no EBER1 nuclear staining was detected on systemic lupus erythematosus (SLE) skin sections by in situ EBER hybridization (Fig. S7D). In addition, we found elevated EBV-BART miRNAs in the inflamed skin tissues, the expression levels of which positively correlated with that of EBER1, whereas no correlation was observed between hsa-miR21 and EBER1. In contrast, BHRF1-2 miRNAs expressed during EBV-lytic stage or in latency type III LCLs (45) were not detected, suggesting that the EBV transcripts in these lesions were derived from latently infected cells (Fig. 7 E–I).
Discussion
Latent EBV-infected cells continuously produce small noncoding EBERs that can be detected by PRRs and ignite proinflammatory responses (9, 46, 47). It remained unclear whether EBERs gain access to cytoplasmic compartments to instigate antiviral immunity (21). Here we show that cytoplasmic 5′pppEBER1 is selectively sorted into exosomes that are internalized by pDCs, triggering antiviral immunity. The exosomal messenger pathway of cellular stress and infection may be an evolutionarily conserved protective mechanism that may cause disease if not properly regulated.
EBERs can bind to viral (EBNA1) (48) and host RNA binding proteins (49). EBER1 and EBER2 associate with La, an abundant RNP in the nucleus of latently infected B cells (50) that binds nascent Pol III transcripts, protecting 3′ ends from exonucleases (51). The abundance of small RNAs in EBV-infected cells is linked to EBNA1 protein expression and the transcription factor TFIIB, which raises Pol III activity (32). Whereas EBER1 interacts with the La phosphoprotein in nuclei, our data show that a fraction of EBERs interact strongly with cytoplasmic La. Although the exact physiological function of this interaction is unclear, knock-down of La protein led to increased IFNB1 transcription (Fig. 5F), similar to what has been observed in respiratory syncytial virus-infected cells. Here, La associates with leader small RNAs (leRNA) and knock-down caused IFNB1 transcription in infected cells (37). EBERs do not acquire a 5′-triphosphate cap-structure, making them susceptible for detection by cytosolic sensors. Indeed, introducing “naked” 5′pppEBER1 directly into RIG-I–expressing latent-infected B cells triggers IFNB1 transcription (Fig. 5K), indicating that the endogenous pool of cytoplasmic 5′pppEBERs fails to activate cytosolic sensors.
Prior studies showed that EBV-EBER transcripts produced from cytoplasmic AT-rich (EBV)DNA triggers RIG-I signaling (52, 53). Our studies imply that Pol III viral transcripts transferred through exosomes and not herpes virus DNA are detected by immunological guardian cells during persistence. It was previously shown that EBER1 complexed to RNPs is detected by TLR3, suggesting that double-stranded RNA loops act as PAMPs. This finding is distinct from exosome-delivered EBER1, where enzymatic removal of 5′ppp rendered EBER1 molecules noninflammatory (Fig. 4 E–H). Thus, 5′ppp-recognizing sensors (i.e., RIG-I, IFIT, or PKR) are more likely to have a role in the recognition of the exosome-delivered small RNA. Moreover, only EBER1 exosomes induced IFIT (interferon induced proteins with tetratricopeptide repeats) 1 and 3 RNA sensor mRNA expression in recipient DCs (Fig. S6E). This finding suggests that 5′pppRNA exosomes transport stress-related signals to neighboring cells, priming them for 5′ppp-RNA detection. Interestingly, breast cancer stroma cells under stress deliver endogenous 5′pppRNAs that are recognized by RIG-I in tumor cells (54). Thus, a conserved intercellular pathway exists in which stress signals in the form of small RNAs are transported between cells via exosomes.
Herpes simplex virus-1 DNA has been detected in the cytosol of infected cells, but its origin (i.e., nuclear, cytoplasmic, or endosomal) remained unclear (55). Upon viral entry, EBV-DNA in the cytosol may be transcribed by RNA pol III, producing cytoplasmic EBERs activating RIG-I (52, 53); although unlikely, we cannot exclude the possibility that some EBV-DNA accesses the cytosol of latently infected LCLs. We propose that by interaction with La and sorting into exosomes, nuclear 5′pppEBER1 eludes cytosolic detection in established latent-infected cells. In contrast, EBERs produced from incoming (cytosolic) EBV-DNA by Pol III, may be sensed by RIG-I in newly EBV-infected B cells. Possibly, resting primary B cells have lower amounts of cytosolic La protein, making them particularly prone to EBER sensing. Although EBER–La complexes may be released in a vesicle-independent manner, we did not detect La in exosomes by proteomics or by Western blotting (9). We used sucrose density-based purification of exosomes, which may explain why we do not detect La in exosomes preparations (23). In addition, in EBV-infected LCLs, but not in EBV− control cells, inhibiting small RNA sorting raises IFNB1 transcription (Fig. 5 G–J). Furthermore, EBER1, but not EBER2, was sorted and released via exosomes, consistent with EBER2 having a nuclear function (56).
Constitutive expression of TLRs and a predisposition for rapid and massive type I IFN release by pDCs pose a risk for developing autoimmunity (57). Exosomes from EBV-infected B cells could trigger inflammation directly or indirectly by elevating the expression of 5′pppRNA-sensors (IFIT proteins) (58) that can recognize self-RNA (Fig. S6E). Although a role for extracellular viral small RNAs has not been considered, pDC infiltration in skin lesions of autoimmune patients is frequently observed (57, 59, 60). There is evidence that the EBV-related herpes simplex virus is recognized by innate sensors during latency in skin-infiltrated pDCs (61, 62). Moreover skin inflammation can be a consequence of UV-stress, because damaged nuclear small RNAs from keratinocytes trigger TLR3, presumably in a non–cell-autonomous fashion (18). Because pDCs constitutively express TLR3, they are a likely candidate cell-type involved in tissue inflammation. In addition, antiviral immunity triggered by exosomal transfer of viral RNA to pDCs has been suggested in HCV infection (3). HCV persistence and pathology, depends on strategies evolved to avoid innate recognition in infected cells (2). It will become important to resolve how pathogenic (viral) RNAs are incorporated and transported via exosomes, as the packaging machineries of virion capsids are presumably lacking. Recently, we and others showed that B cells latently infected with EBV export functional miRNAs to uninfected cells via exosomes (25, 58).
One question that still needs addressing is whether bystander recognition of latent EBV-infected cells contributes to life-long persistence. Recent studies in humanized mice suggest that EBV-deficient in EBER1 is not impaired in establishing persistence (8), whereas EBER2 increases LMP2 transcription (56). One could imagine that EBER1-activated pDCs during primary EBV infection primes NK cells to restrict outgrowth (63). Indeed, failure to respond to primary EBV infection can be fatal in predisposed individuals suffering from X-linked lymphoproliferative syndrome (64). During persistence, EBER recognition may keep the pool of latent-infected memory B cells that do not overtly expose EBV-antigens, at low levels. The absence of detectable BHRF1 miRNAs and EBV-genomes, but the positive correlation between EBER1 transcripts and EBV-BART miRNAs in exosomes, strongly suggest a joint delivery mechanism from latently infected cells (45). Our findings in vitro were made with naturally infected latency III LCLs that grow rapidly and release well-defined exosomes (65), whereas the most dominant infected cell type in lymphoid tissues and circulation are presumably memory B cells (40). Latency I memory B cells have sustained expression of EBV-noncoding RNAs, except the BHRF1-miRNAs (45). Although resting, these cells may retain the ability to release exosomes that have incorporated EBER1. However, latency III cells have been detected in circulation of individuals under immune suppression (44), and may represent an additional source of EBER1+ exosomes. Although chronic sensing of latent EBV may lead to disease in predisposed individuals, EBER1 recognition during early stages of EBV infection is possibly protective for the host, as at this time adaptive immune responses are not yet functional.
In conclusion, we propose that EBER1-containing exosomes have a role in immune recognition of latently EBV-infected B cells, which is a normal feature of EBV persistence, but maybe exacerbated in EBV-linked inflammatory autoimmune diseases like lupus erythematosus and multiple sclerosis.
Methods
Exosome Isolation and Purification.
Exosomes were isolated by differential centrifugation, as previously described (30). When indicated, exosomal RNA was treated with CIP (New England Biolabs) to remove 5′-triphosphate. For uptake experiments, purified exosomes were labeled with PKH67 dye (Sigma-Aldrich), as described previously (25).
Microarray Analysis.
DC RNA quantity and quality were determined on a 2100 Bioanalyzer, and samples were subjected to Agilent high-density Microarrays according to the manufacturer’s protocol. Data were truncated for maximum intensities at 10,000 and filtered for minimum intensities (45). Fold-change was set higher than 1.5 at least in one sample per group. Quantile normalization was applied. The resulting 10.383 filtered genes were subjected to unpaired class comparison univariate test to select differentially expressed genes (α = 0.0005). Log2 ratio between the geometrical mean of intensities for each class were calculated. Genes with fold-change greater than 2 were selected. Analyses were performed using BRB-ArrayTools, developed by Richard Simon and the BRB-ArrayTools Development Team (linus.nci.nih.gov/BRB-ArrayTools.html).
Stimulation of DCs and Coculture.
The 5′pppEBER1 (100 ng) was added to or transfected in immature DCs using Lipofectamine 2000 (Life Technologies) for 24 h. Coculture experiments were performed as previously described (25).
All clinical samples and primary cell culture (i.e., dendritic cells and tonsil cells) were obtained with informed consent, evaluated by the Vrije Universiteit Medical Center ethical board committee and used in compliance with the Declaration of Helsinki.
Additional methods are included in SI Methods. See Table S1 for adapter, primers, and probes.
Table S1.
List of adapter, primers, and probes
| Target | Oligonucleotide | Sequence (5′- 3′) |
| Small noncoding RNA | ||
| EBER1 | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAACATGCG |
| Forward | AGGACCTACGCTGCCCTAGA | |
| Stem-loop Reverse | ATTCAGTTGAGAAAACATGCGG | |
| EBER2 | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATAGCG |
| Forward | AGGACAGCCGTTGCCCTAGTGGTTTC | |
| Stem-loop Reverse | ATTCAGTTGAGAAAAATAGCGG | |
| vtRNA1-1 | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAGGACTG |
| Forward | GGCTGGCTTTAGCTCAGCGG | |
| Stem-loop Reverse | ATTCAGTTGAGAAAAGGACTGG | |
| RNY1 | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAGACTAG |
| Forward | GCTGGTCCGAAGGTAGTGAG | |
| Stem-loop Reverse | ATTCAGTTGAGAAAAGACTAGT | |
| 5′-adapter-EBER1 | RA5 adapter | GUUCAGAGUUCUACAGUCCGACGAUC (Illumina #15013205) |
| Forward (adapter) | GTTCAGAGTTCTACAGTCCGA | |
| Reverse (EBER1) | AAAACATGCGGACCACC | |
| miRNA | ||
| EBV BART1-5p | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGCACG |
| Forward | ACACTCCAGCTGGGTCTTAGTGGAAGTGAC | |
| Probe | TTCAGTTGAGACAGCACG | |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCA | |
| EBV BART8-3p | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACGACC |
| Forward | ACACTCCAGCTGGGGTCACAATCTATGGGG | |
| Probe | TTCAGTTGAG CTACGACC | |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCA | |
| EBV BART17-5p | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTTGTATG |
| Forward | ACACTCCAGCTGGGTAAGAGGACGCAGGCA | |
| Probe | TTCAGTTGAGCTTGTATG | |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCA | |
| EBV BHRF1-2* | Stem-loop RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTATCTG |
| Forward | ACACTCCAGCTGGGAAATTCTGTTGCAGCA | |
| Probe | TTCAGTTGAGGCTATCTG | |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCA | |
| hsa-miR21-5p | TaqMan MicroRNA Assay ID 000397 | |
| hsa-miR451 | TaqMan MicroRNA Assay ID 001105 | |
| mRNA | ||
| IFN-β | Forward | CCTGAAGGCCAAGGAGTACA |
| Reverse | AAGCAATTGTCCAGTCCCAG | |
| IFITM1 | Forward | ATGTCGTCTGGTCCCTGTTC |
| Reverse | GTCATCAGGATGCCCAGAAT | |
| IFITM3 | Forward | ATGTCGTCTGGTCCCTGTTC |
| Reverse | GGGATGACGATGAGCAGAAT | |
| OAS2 | Forward | GGTGAACACCATCTGTGACG |
| Reverse | ACCATCGGAGTTGCCTCTTA | |
| MX1 | Forward | GATTTTGGGGCTTTCCAGTC |
| Reverse | GATGATCAAAGGGATGTGGC | |
| TLR3 | Forward | TCACTTGCTCATTCTCCCTT |
| Reverse | GACCTCTCCATTCCTGGC | |
| TLR7 | Forward | TGCTCTGCTCTCTTCAACCA |
| Reverse | AACCATCTAGCCCCAAGGAG | |
| TLR9 | Forward | ACACAGCTGCGCAAGCTTAAC |
| Reverse | TCGAGTGAGCGGAAGAAGATG | |
| PKR | Forward | GGGACCTTGGAACAATGGAT |
| Reverse | CCTTGTTCGCTTTCCATCAT | |
| RIG-I | Forward | ATATCCGGAAGACCCTGGAC |
| Reverse | GAGAAAAAGTGTGGCAGCCT | |
| IFIT1 | Forward | TACCTGGACAAGGTGGAGAA |
| Reverse | GTGAGGACATGTTGGCTAGA | |
| IFIT3 | Forward | CTTACGGCAAGCTGAAGAGT |
| Reverse | AATTGCCAGTCCAGAGGAGA | |
| IFIT5 | Forward | GGCCATTCTGTTGGAGTTAG |
| Reverse | GCTTCTTCAAGCTGGTCCAT | |
| MVP | Forward | ATCTGAGGCGTTTGTTGCAG |
| Reverse | GGATGCGGATGATGAACTCT | |
| La/SSB | Forward | TAATGAAAAGATGGCTGCCC |
| Reverse | AAGGTACCCAGCCTTCATCC | |
| TIM1 | Forward | CCAGTAGCCACTTCACCATCTTCAC |
| Reverse | CGGTGTCATTCCCATCTGTTGTG | |
| TIM4 | Forward | ACAGGACAGATGGATGGAATACCC |
| Reverse | AGCCTTGTGTGTTTCTGCG | |
| GAPDH | Forward | GAGTCAACGGATTTGGTCGTAT |
| Reverse | ATGGGTGGAATCATATTGGAAC | |
| EBV DNA | ||
| EBNA1 QK | Forward | GCCGGTGTGTTCGTATATGG |
| Reverse | CAAAACCTCAGCAAATATATGAG | |
| Probe 1 | TCTCCCCTTTGGAATGGCCCTG-fluorescein | |
| Probe 2 | LCRed640-ACCGGCCCACAACCTG | |
| Cloning | ||
| EBER1 | Forward | TGCAAGATCTTAATACGACTCACTATAGGAGGACCTACGCTGCCATAGA |
| Short Forward* | TGCAAGATCTTAATACGACTCACTATAGGAGGACC | |
| Reverse | ACGTTCTAGAAAACATGCGGACCACCAGC |
The T7 promoter sequence is underlined.
Added after 20 rounds of amplification to increase PCR efficiency and yield.
SI Methods
Clinical Specimens.
Serum samples containing autoantibodies against La/SSB were obtained from patients with Sjögren’s syndrome and SLE. Paraffin-embedded and frozen skin tissues from cutaneous lupus erythematosus patients and controls were obtained from the Department of Pathology and from the Department of Dermatology of the Vrije Universiteit Medical Center (VUmc).
All clinical samples were obtained with informed consent, evaluated by the VUmc ethical board committee, and used in compliance with the Declaration of Helsinki.
Cell Culture.
EBV+ LCLs IB4, RN, EBER-knockout LCL AM58, wtEBV LCL IK140508, and EBV− B-cell line (BJAB) were cultured in RPMI-1640 supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin sulfate, and 2 mM glutamine. CD40L stimulated EBV− (RZL) and EBV+ LCL (RZB), were cultured as described previously (66). Hek293, HekTLR3, HekTLR7, and HekTLR9 were cultured in DMEM. DCs were generated from peripheral blood mononuclear cells from healthy donors, as described previously (25). Tonsils were obtained from patients undergoing routine tonsillectomies for obstructed breathing disorders at the ENT department of the VUmc. Tonsil mononuclear cells were isolated as described previously (67). All primary cell cultures were obtained with informed consent, evaluated by the VUmc ethical board committee, and used in compliance with the Declaration of Helsinki.
RNA and DNA Isolation and qPCR.
Total cellular and exosomal RNA and genomic DNA were isolated using TRIzol Reagent (Life Technologies), according to the manufacturer’s protocol. Cytosolic RNA was extracted using TRIzol LS (Life Technologies). RNA from paraffin-embedded skin tissue was isolated using RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Ambion). The quality of exosomal RNA was assessed on a 2100 Bioanalyzer system (Agilent).
EBNA1 QK DNA-PCR was performed as described previously (25). mRNA analysis was performed by SYBR Green qPCR. The detection of EBV-encoded miRNAs was performed as described previously (30). The expression of small noncoding RNAs was performed by adapting the stem-loop method. Stem-loop RT primers specific for the 3′-end of the noncoding RNA of interest were used in a multiplex reaction according to the TaqMan MicroRNA RT protocol (Life Technologies). Detection was performed by SYBR Green qPCR using a target-specific forward primer and a reverse primer spanning the target-stem-loop junction (30). For the detection of 5′adapter ligated-EBER1, a 5′adapter and 3′EBER1-specific primer set was used. Adapter, primers, and probes are listed in Table S1.
Cloning and in Vitro Transcription of EBER1.
Full-length EBER1 (NC_007605.1) was obtained from RN cDNA, and the T7 promoter sequence was introduced by PCR. BglII and XbaI were used to clone T7-EBER1 in pcDNA3, plasmids were linearized and in vitro transcription performed with T7 RNA polymerase (Promega). ivtEBER1-RNA was treated with DNase I and optionally with CIP (New England Biolabs). Primers are listed in Table S1.
Luciferase Reporter Assay.
Hek293, HekTLR, or “primed” hek293 cells were transfected with an IFN-β promoter reporter (pGL3-p125) or with a NF-κB promoter reporter (p3x-κB-Luc) using Lipofectamine 2000 (Life Technologies); a GLuc plasmid was used for normalization. Cells were cotransfected with 100 ng ivtEBER1, or 1 µg EBER-expression plasmids BSAII-EBER and EKS10. Poly(I:C) (Sigma) and imiquimod (Invivogen) were used as positive controls. Luciferase activity was measured after 24 h.
Exosome Electroporation.
BJAB exosomes were electroporated using the EasyJet Plus instrument (EQUIBIO). Briefly, BJAB exosomes were mixed in 400 μL electroporation medium (Cell Projects), added with 100 ng ivtEBER1, and electroporated at 400 V and 150 μF in a 4-mm cuvette. RNA isolation was performed using TRIzol Reagent.
Stimulation of pDCs.
Mononuclear tonsil cells were characterized by FACS (BD FACS Calibur) using antibodies against CD19 (FITC, clone HIB19), CD3 (PE, clone SK7), CD56 (FITC, clone MOC-1), CD123 (PECy5, clone 9F5), BDCA2 (FITC, clone AC144), TIM1 (clone 1D12), and TIM4 (clone 9F4). When indicated, pDCs were enriched using CD304/BDCA4 microbeads (Miltenyi Biotec). Total tonsil cells were incubated with PKH67-labeled LCL or BJAB exosomes. Exosome uptake was analyzed by FACS using antibodies against CD123 (PECy5) and BDCA2 (PE, clone AC144). Cells were incubated overnight with wtEBV LCL and EBV− BJAB exosomes, followed by CD40 (clone MAB89) FACS analysis or RT-PCR.
Stimulation and Transfection of Wild-Type EBV LCL.
Wild-type EBV LCL (5E5) were microporated (Neon Transfection System, Life Technologies) with ivtEBER with/without 5′ppp. Cells were incubated for 6–24 h and harvested for RNA analysis. For knockdown experiments, 10–50 pmol si-RNA against SSB, MVP, nSMase2, or cyclophillin B (Dharmacon) were used. Cells were incubated 24–72 h before RNA and Western blot analysis.
Nuclear/Cytosolic Fractionation and Western Blotting.
EBV+ IB4 cells were resuspended in hypotonic buffer (10 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, pH 7.5), disrupted using a Dounce homogenizer and centrifuged at 500 × g for 5 min at 4 °C. The nuclei-containing pellet was resuspended in TRIzol or lysis buffer. The supernatant (cytosolic fraction) was then centrifuged for 10 min at 20,000 × g at 4 °C and TRIzol LS was added for RNA isolation.
Cells and exosomes were lysed with RIPA buffer. Lysates were run on a 10% SDS gel and blotted on a nitrocellulose membrane. Primary antibodies against RIG-I (clone Alme-1, Adipogen), PKR (clone K-17, Santa Cruz), Hsp70 (clone W27, Santa Cruz), Alix (clone 3A9, Cell Signaling), La/SSB (clone OTI9D6, Origene), MVP (clone MVP37), LMP1 and EBNA1 (clones OT21C and OT1X, respectively, provided by J.M.M.), and β-Actin (clone C4, Santa Cruz) were used.
RNA Immunoprecipitation.
RN cells were cross-linked with 0.8% formaldehyde for 15 min. The cross-linking reaction was quenched with glycine 0.4M. Cells were resuspended in lysis buffer (20 mM Tris⋅HCl 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% Nonidet P-40) containing protease inhibitor mixture (Roche) and RNase OUT (Life Technologies), and incubated 15 min on ice. Cell lysates were centrifuged at 10,000 × g for 15 min and the supernatant (cytosolic fraction) was collected in a new tube. The nuclei-containing pellet was solubilized in urea lysis buffer (50 mM Tris⋅HCl pH 6.8, 6 M urea, 2% SDS) and sonicated. Two milligrams of cytosolic fraction were incubated for 4 h at 4 °C with Protein A dynabeads precoupled with SSB/La+ sera or SSB/La− control serum. Ten percent of the cytosolic fraction was used as input. Before protein analysis, samples were boiled 10 min in loading buffer containing freshly added b-mercaptoethanol. For RNA analysis, de-cross-linking was performed by incubating the samples in 250 µL of elution buffer containing 10 mM Tris⋅HCl pH 7.5, 1% SDS, 10 mM EDTA, and 0.2 mg/mL Proteinase K (Roche), for 15 min at 65 °C. Eluted RNA was resuspended in 750 µL of TRIzol LS (Life Technologies).
Proteomics Analysis.
Proteomics analysis was performed as previously described (68). Proteins that were determined to be significantly differential (P < 0.05) according to a β-binomial–based test were uploaded as the corresponding gene symbol in the web-based STRING tool (v9.1) (29) to retrieve protein–protein associations. The latter were imported into Cytoscape (v2.8.3) and visualized as networks. Enriched GO terms were retrieved using the BiNGO plugin for Cytoscape (v2.44). Subclusters were extracted using the ClusterONE plugin for Cytoscape (v0.93).
Small RNA Sequencing.
Small RNA libraries were generated as previously described (69) and analyzed using a pipeline based on the sRNAbench analysis package (70). Briefly, reads were aligned by means of the Bowtie algorithm to a pooled index of the human (GRCh37 patch release 5; downloaded as hg19 from the University of California, Santa Cruz) and EBV genome (GenBank accession no. NC_007605). To provide annotations for RNA elements that mapped to human and EBV genomes, several databases were used: (i) miRBase v19; (ii) National Center for Biotechnology Information Reference Sequences (RefSeq) (at February 6, 2013); (iii) genomic tRNA database; (iv) RepeatMasker algorithm (University of California, Santa Cruz); (v) piwiRNA (piRNA) sequences were downloaded from the National Center for Biotechnology Information nucleotide database. For those annotations in fasta format, we obtained the genome coordinates by mapping the sequences to the genome. Read counts were normalized using all genome mapped reads (RPMM). For classification of RNA Pol III transcripts, we refer to Dieci et al. (71).
ELISA.
The human IFN-β (PBL IFN Source 41410–1A) and human TNF-α (Sanquin M1920) ELISA kit were used according to the manufacturer’s protocol.
Immunohistochemical Analysis and in Situ Hybridization.
For immunofluorescent staining, slides were incubated overnight with a combination of CD1a (1:50, IgG1 κ, clone 1CA04; Neomarkers) and CD123 (1:100; Abcam, ab154402) after standard EDTA antigen retrieval. AlexaFluor-labeled goat anti-rabbit-A546 and goat anti-mouseIgG1-A488 (Life Technologies) were incubated 1 h at room temperature. For the combined CD40/CD123 staining on tonsil and skin tissues, the rabbit anti-human CD40 (1:50; Abcam, ab13545) and the mouse anti-human CD123 (1:25; clone 7G3) were used. The primary antibodies were detected with a combination of AlexaFluor-labeled goat anti-rabbit A647 and AlexaFluor-labeled goat anti-mouse IgG2a-A546. Slides were mounted using VectaShield mounting medium containing DAPI (Vector Laboratories). Images were acquired with an LSM700 confocal laser-scanning microscope equipped with an LCI Plan-Neofluar 25×/0.8 ImmKorr DIC M27 objective (Zeiss).
The presence of EBV in skin tissues was evaluated by EBER-RISH using the commercial PNA-Based Hybridization Kit (Dakocytomation).
Statistical Analysis.
A Kolmogorov–Smirnov test was used to check the Gaussian distribution, followed by unpaired two-tailed t test or two-tailed Pearson correlation. All analyses were performed using Graphpad Prism 4 software.
Supplementary Material
Acknowledgments
We thank W. Stoorvogel (University Medical Center Utrecht) for the provision of RN cells; A. Moosmann for CD40L-stimulated Epstein–Barr virus (EBV−) (RZL) and EBV+ lymphoblastoid cell lines (LCLs) (RZB); R. Khanna and M. Gandhi (Queensland Institute of Medical Research) for established and early passage spontaneous wild-type EBV LCL lines; M. Holwerda, R. Bakker, and E. Abels for their assistance in constructing the EBER in vitro transcription constructs, EBER-stem loop PCR, and patient sample analysis; Douwe Buma and Sander Spiekstra for preparing the tissue sections; and Dr. Richard Maraia (National Institutes of Health) for his assistance in the cytoplasmic lupus antigen pull-down experiments. The pGL3-p125 carrying the IFN-β promoter was a kind gift from N. Kato (University of Tokyo) and the p3x-κB-Luc plasmid carrying the NF-κB promoter was a kind gift from E. Kieff (Harvard Medical School). The BSAII-EBER was provided by Dr. I. Ruf and EKS10 by K. Takada. D.K.L. was supported by grants from the Dutch Arthritis Foundation (13-2-401), Worldwide Cancer Research (11-0157), and the Dutch Cancer Society (KWF) VU2012-5510 (to D.M.P.). F.J.M.v.K. is supported by Netherlands Organization for Scientific Research 700.59.007. M.A.J.v.E. and E.S.H. were supported by KWF VU2007-3775/3776 (to J.M.M.), and S.R.B. was supported by the Italian Association for Cancer Research (AIRC) and Marie Curie Actions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518130113/-/DCSupplemental.
References
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19(7):879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreux M, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12(4):558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Z, et al. Human pDCs preferentially sense enveloped hepatitis A virions. J Clin Invest. 2015;125(1):169–176. doi: 10.1172/JCI77527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol. 2011;11(2):143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Münz C, Lünemann JD, Getts MT, Miller SD. Antiviral immune responses: Triggers of or triggered by autoimmunity? Nat Rev Immunol. 2009;9(4):246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78(2):805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregorovic G, et al. Epstein-Barr viruses (EBVs) deficient in EBV-encoded RNAs have higher levels of latent membrane protein 2 RNA expression in lymphoblastoid cell lines and efficiently establish persistent infections in humanized mice. J Virol. 2015;89(22):11711–11714. doi: 10.1128/JVI.01873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwakiri D, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med. 2009;206(10):2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samanta M, Iwakiri D, Takada K. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene. 2008;27(30):4150–4160. doi: 10.1038/onc.2008.75. [DOI] [PubMed] [Google Scholar]
- 11.Ruf IK, Lackey KA, Warudkar S, Sample JT. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J Virol. 2005;79(23):14562–14569. doi: 10.1128/JVI.79.23.14562-14569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quan TE, Roman RM, Rudenga BJ, Holers VM, Craft JE. Epstein-Barr virus promotes interferon-alpha production by plasmacytoid dendritic cells. Arthritis Rheum. 2010;62(6):1693–1701. doi: 10.1002/art.27408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 14.Bowie AG, Fitzgerald KA. RIG-I: Tri-ing to discriminate between self and non-self RNA. Trends Immunol. 2007;28(4):147–150. doi: 10.1016/j.it.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 16.Nallagatla SR, et al. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318(5855):1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 17.Eckard SC, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol. 2014;15(9):839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard JJ, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18(8):1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malathi K, et al. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16(11):2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stetson DB. Connections between antiviral defense and autoimmunity. Curr Opin Immunol. 2009;21(3):244–250. doi: 10.1016/j.coi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tycowski KT, et al. Viral noncoding RNAs: More surprises. Genes Dev. 2015;29(6):567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jochum S, Ruiss R, Moosmann A, Hammerschmidt W, Zeidler R. RNAs in Epstein-Barr virions control early steps of infection. Proc Natl Acad Sci USA. 2012;109(21):E1396–E1404. doi: 10.1073/pnas.1115906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed W, Philip PS, Tariq S, Khan G. Epstein-Barr virus-encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV-transformed cells. PLoS One. 2014;9(6):e99163. doi: 10.1371/journal.pone.0099163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegtel DM, van de Garde MD, Middeldorp JM. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta. 2011;1809(11-12):715–721. doi: 10.1016/j.bbagrm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanc M, et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 2011;9(3):e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol. 2013;14(8):793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 29.Franceschini A, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue) D1:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verweij FJ, van Eijndhoven MAJ, Middeldorp J, Pegtel DM. Analysis of viral microRNA exchange via exosomes in vitro and in vivo. Methods Mol Biol. 2013;1024:53–68. doi: 10.1007/978-1-62703-453-1_5. [DOI] [PubMed] [Google Scholar]
- 31.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6(8):610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felton-Edkins ZA, et al. Epstein-Barr virus induces cellular transcription factors to allow active expression of EBER genes by RNA polymerase III. J Biol Chem. 2006;281(45):33871–33880. doi: 10.1074/jbc.M600468200. [DOI] [PubMed] [Google Scholar]
- 33.Guiducci C, et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. J Exp Med. 2013;210(13):2903–2919. doi: 10.1084/jem.20131044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88(4):1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amort M, et al. Expression of the vault RNA protects cells from undergoing apoptosis. Nat Commun. 2015;6(May):7030. doi: 10.1038/ncomms8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bitko V, Musiyenko A, Bayfield MA, Maraia RJ, Barik S. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J Virol. 2008;82(16):7977–7987. doi: 10.1128/JVI.02762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: Diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. J Virol. 2009;83(8):3968–3976. doi: 10.1128/JVI.02609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M, et al. Expression profile and histological distribution of IFITM1 and IFITM3 during H9N2 avian influenza virus infection in BALB/c mice. Med Microbiol Immunol (Berl) 2015;204(4):505–514. doi: 10.1007/s00430-014-0361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feeley EM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 44.Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. EBV and systemic lupus erythematosus: A new perspective. J Immunol. 2005;174(11):6599–6607. doi: 10.4049/jimmunol.174.11.6599. [DOI] [PubMed] [Google Scholar]
- 45.Qiu J, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7(8):e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25(18):4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002;21(5):954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snudden DK, Hearing J, Smith PR, Grässer FA, Griffin BE. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 1994;13(20):4840–4847. doi: 10.1002/j.1460-2075.1994.tb06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conrad NK, Fok V, Cazalla D, Borah S, Steitz JA. The challenge of viral snRNPs. Cold Spring Harb Symp Quant Biol. 2006;71:377–384. doi: 10.1101/sqb.2006.71.057. [DOI] [PubMed] [Google Scholar]
- 50.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173(3):319–325. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fairley JA, et al. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc Natl Acad Sci USA. 2005;102(51):18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ablasser A, et al. RIG-I–dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boelens MC, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160(4):607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guiducci C, et al. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J Exp Med. 2010;207(13):2931–2942. doi: 10.1084/jem.20101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bordon Y. Innate immunity: An inducible RNA sensor? IFITs the bill. Nat Rev Immunol. 2011;11(7):440–441. doi: 10.1038/nri3011. [DOI] [PubMed] [Google Scholar]
- 59.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL. Plasmacytoid dendritic cells (natural interferon- alpha/beta-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol. 2001;159(1):237–243. doi: 10.1016/s0002-9440(10)61689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollenberg A, et al. Plasmacytoid dendritic cells: A new cutaneous dendritic cell subset with distinct role in inflammatory skin diseases. J Invest Dermatol. 2002;119(5):1096–1102. doi: 10.1046/j.1523-1747.2002.19515.x. [DOI] [PubMed] [Google Scholar]
- 61.Donaghy H, et al. Role for plasmacytoid dendritic cells in the immune control of recurrent human herpes simplex virus infection. J Virol. 2009;83(4):1952–1961. doi: 10.1128/JVI.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA. 2014;111(46):E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strowig T, et al. Tonsilar NK cells restrict B cell transformation by the Epstein-Barr virus via IFN-gamma. PLoS Pathog. 2008;4(2):e27. doi: 10.1371/journal.ppat.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hislop AD, et al. Impaired Epstein-Barr virus-specific CD8+ T-cell function in X-linked lymphoproliferative disease is restricted to SLAM family-positive B-cell targets. Blood. 2010;116(17):3249–3257. doi: 10.1182/blood-2009-09-238832. [DOI] [PubMed] [Google Scholar]
- 65.Wubbolts R, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278(13):10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 66.Wiesner M, et al. Conditional immortalization of human B cells by CD40 ligation. PLoS One. 2008;3(1):e1464. doi: 10.1371/journal.pone.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hochberg D, et al. Acute infection with Epstein-Barr virus targets and overwhelms the peripheral memory B-cell compartment with resting, latently infected cells. J Virol. 2004;78(10):5194–5204. doi: 10.1128/JVI.78.10.5194-5204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piersma SR, et al. Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: Method evaluation, differential analysis, and verification in serum. J Proteome Res. 2010;9(4):1913–1922. doi: 10.1021/pr901072h. [DOI] [PubMed] [Google Scholar]
- 69.Koppers-Lalic D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Reports. 2014;8(6):1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 70.Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: An update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39(Web Server issue):W132–W138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.