Significance
Small multidrug resistance (SMR) transporters play an important role in the protection of prokaryotes from cytotoxic molecules. They exploit the proton electrochemical gradient to drive the transport of these molecules out of the cell against their concentration gradient. This work investigated how binding of protons power the conformational changes that enable substrate binding and subsequent alternating access of the Escherichia coli SMR transporter EmrE. The results show that protonation induces large-scale reconfiguration of the structure, including helical rotation and tilt and repacking of loops. A highly conserved charged residue primarily accounts for proton sensing, but other acidic residues control local structures. Our findings stimulate a structural model of transport, offering a novel perspective on proton-coupled multidrug transport.
Keywords: EmrE, SMR, EPR, DEER, multidrug transport
Abstract
The small multidrug transporter from Escherichia coli, EmrE, couples the energetically uphill extrusion of hydrophobic cations out of the cell to the transport of two protons down their electrochemical gradient. Although principal mechanistic elements of proton/substrate antiport have been described, the structural record is limited to the conformation of the substrate-bound state, which has been shown to undergo isoenergetic alternating access. A central but missing link in the structure/mechanism relationship is a description of the proton-bound state, which is an obligatory intermediate in the transport cycle. Here we report a systematic spin labeling and double electron electron resonance (DEER) study that uncovers the conformational changes of EmrE subsequent to protonation of critical acidic residues in the context of a global description of ligand-induced structural rearrangements. We find that protonation of E14 leads to extensive rotation and tilt of transmembrane helices 1–3 in conjunction with repacking of loops, conformational changes that alter the coordination of the bound substrate and modulate its access to the binding site from the lipid bilayer. The transport model that emerges from our data posits a proton-bound, but occluded, resting state. Substrate binding from the inner leaflet of the bilayer releases the protons and triggers alternating access between inward- and outward-facing conformations of the substrate-loaded transporter, thus enabling antiport without dissipation of the proton gradient.
Powered by the proton electrochemical gradient across the inner membrane of prokaryotes, small multidrug resistance (SMR) transporters extrude a spectrum of cytotoxic molecules that are primarily hydrophobic cations (1). The functional unit is typically a homodimer wherein each protomer consists of four hydrophobic transmembrane helices (TM). TM1–3 cradle a substrate binding pocket, whereas TM4 is involved in the contacts that stabilize the dimer. EmrE, the SMR transporter from Escherichia coli, has been a focal point of structural, spectroscopic, and mechanistic investigations (2–7). Seminal work from the Schuldiner laboratory over the last two decades has unlocked mechanistic principles of substrate–ion–antiport (1, 8–10). EmrE binds hydrophobic substrates in a membrane-embedded chamber coordinated by glutamate 14 (E14), an absolutely conserved, membrane-embedded, acidic residue in the middle of TM1. Coupling between substrate and proton arises from the principle of mutual exclusion between the two ligands at the binding site (8, 10).
In contrast to the elaborate understanding of the EmrE antiport mechanism, the conformational changes that enable binding and release of ligands have not been elucidated. Although the general framework of antiport is presumed to follow the principles of alternating access, the only structure available is of EmrE bound to the substrate tetraphenylphosphonium (TPP) (3). EM analysis of 2D crystals established an antiparallel orientation of the protomers in the dimer (2). This was later confirmed by the corrected crystal structure of TPP-bound EmrE trapped in an asymmetric state with an opening on one side of the transporter (3). This asymmetry arises from distinct conformations of each protomer in the dimer. Fleishman et al. (11) independently modeled EmrE on the basis of the EM structure and postulated the elegant notion that isomerization of the two protomers between the two conformations drives alternating access of the substrate-loaded transporter. Indeed solution NMR studies confirmed the isomerization of TPP-bound EmrE in bicelles and enabled the measurement of the time scale of the exchange between inward- and outward-facing states (6). Structurally, the inward- and outward-facing states of the dimer are related by a 180° rotation around an axis parallel to the plane of the membrane; that is, they are identical except for their orientations in the membrane.
In addition to alternating access of the substrate-bound EmrE, transport requires the protonation/deprotonation of E14 in the context of proton translocation from the extracellular milieu (8). It is typically assumed that the protonated intermediate has a similar structure to substrate-bound EmrE and undergoes isomerization by the isoenergetic exchange of the two protomers between two conformations (12). However, this model has not been challenged experimentally, and the structures of the protonated and apo states have not been determined. Thus, critical steps in the transport cycle remain structurally and dynamically unexplored.
To illuminate the structural and dynamic aspects of protonation in the transport mechanism, we present a global perspective on the ligand-induced conformational changes of EmrE. A systematic analysis of distances and distance changes between spin labels, site-specifically introduced across the dimer, reveals distinct structural rearrangements associated with protonation and substrate binding. These rearrangements reconfigure the backbone and side-chain orientations in the substrate binding chamber as well as modulate access to the bilayer. Protonation-induced movements are primarily dependent on E14, although residues E25 and D84 appear to influence the local conformation particularly in TM3, suggesting a departure from rigid body movement of the protomer. When integrated into the current biochemical and structural framework, these results provide a novel model of coupled transport by EmrE.
Results
General Methodology.
To investigate the EmrE structure under ligand conditions that are expected to promote transition between transport intermediates, we used our library of cysteine mutants (5), spanning almost the entire sequence of EmrE, to introduce single spin labels into the monomer leading to a doubly labeled dimer. Spin-labeled mutants, which did not show significant structural and functional perturbation in previous assays (5), were analyzed by Q-band (33.9 GHz) double electron electron resonance [DEER; also called pulsed electron--electron double resonance (PELDOR)] spectroscopy (13–15). Distance distributions were determined in n-dodecyl-β-d-maltopyranoside (β-DDM) detergent micelles, which maintain the structural and functional integrity of EmrE (16). The simplest model of EmrE transport satisfying energetics and coupling considerations entails at least three distinct intermediates: substrate-bound, proton-bound, and ligand-free or apo. It is likely that the apo state is only transiently populated, considering the high concentration of protons on the extracellular side and the obligatory exchange between protons and substrate (10). Therefore, DEER measurements were carried out at pH 5 to mimic the acidic environment of the periplasm and protonate acidic residues, at pH 8 to mimic the relatively higher pH of the cytoplasm and promote deprotonation, and at pH 8 in the presence of excess TPP to trap the substrate-bound conformation. For a limited number of mutants, we confirmed our interpretation in lipid bilayers through reconstitution of EmrE into nanodiscs.
Substrate Binding and Protonation Induce Distinct Conformations of EmrE.
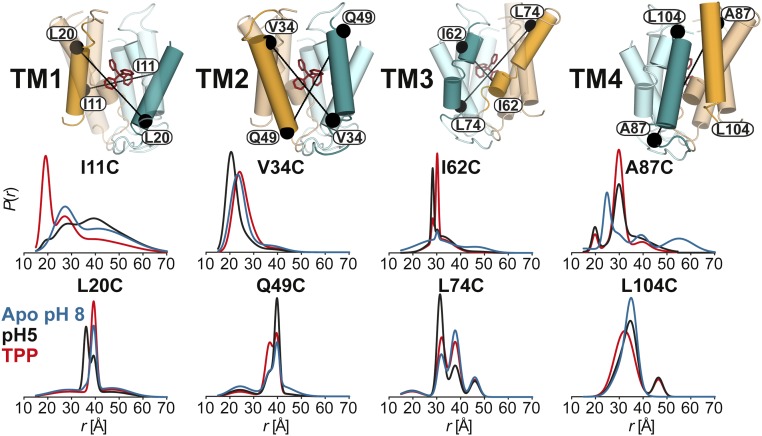
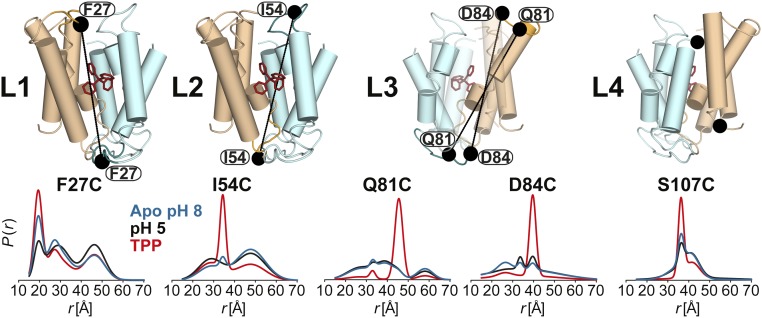
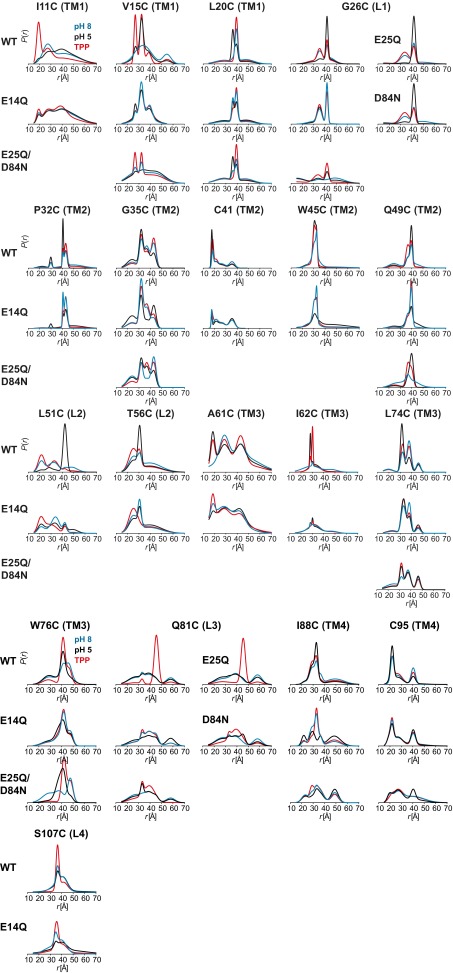
The DEER dataset (Figs. 1 and 2 and SI Appendix 1) was transformed into distance distributions characterizing the spatial relationships between pairs of TMs in the dimer as described in Methods. Overall, the distributions reveal a number of trends consistent with three distinct conformations corresponding to the proton-bound, substrate-bound, and apo EmrE intermediates. First, we observed changes in the average distances as well as the width of the distributions between the three conditions, unequivocally demonstrating extensive conformational rearrangements (Fig. 1 and SI Appendix 1). Second, the shape of these distributions suggests that the TPP-bound state is ordered in stark contrast with the highly dynamic apo state. Specifically, we observed broad distributions in TM1–3 at pH 8. At a number of sites, the shape of the distributions and the changes induced by different ligand conditions suggested equilibrium between multiple states. Finally, there are extensive, ligand-dependent rearrangements in the structure of the loops (L1–L4; Fig. 2 and SI Appendix 1), particularly L1 and L3 connecting TM1 and 2 and TM3 and 4, respectively, suggesting an important role for these segments in the mechanism of transport (17, 18). Notably, binding of substrate increases order in these loops, whereas protonation/deprotonation typically leads to broad distance distributions (Fig. 2).
Fig. 1.
Ligand-dependent conformational changes of EmrE in the transmembrane regions (TM1–4). Distance distributions depicting the probability of a distance P(r) versus distance (r) between identical positions in the doubly labeled dimer. Distance distributions for each pair were obtained in the ligand-free (blue; Apo pH 8), proton-bound (black; pH 5), and TPP-bound (red; TPP) intermediates.
Fig. 2.
Ligand-dependent conformational changes of EmrE in the loop regions (L1–4). Residue 107 is not resolved in the X-ray structure. Distance distributions were obtained in the ligand-free (blue), proton-bound (black), and TPP-bound (red) intermediates.
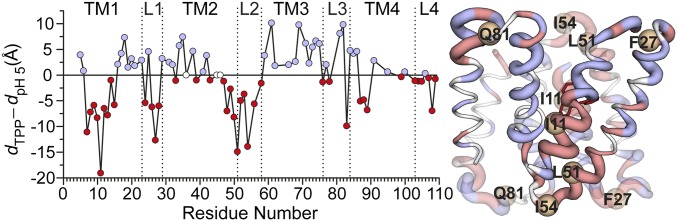
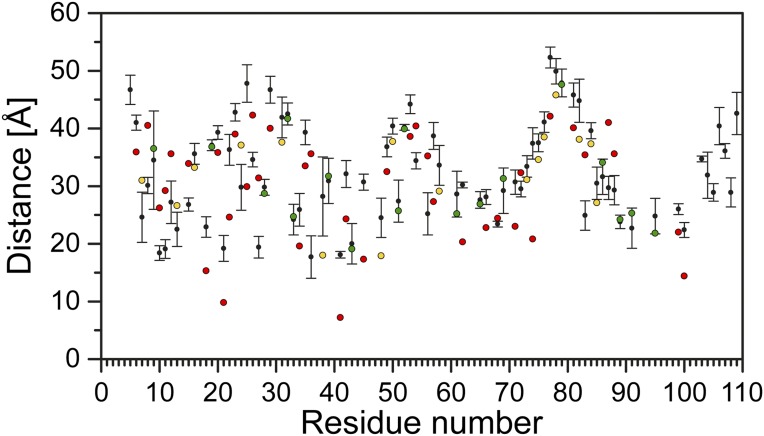
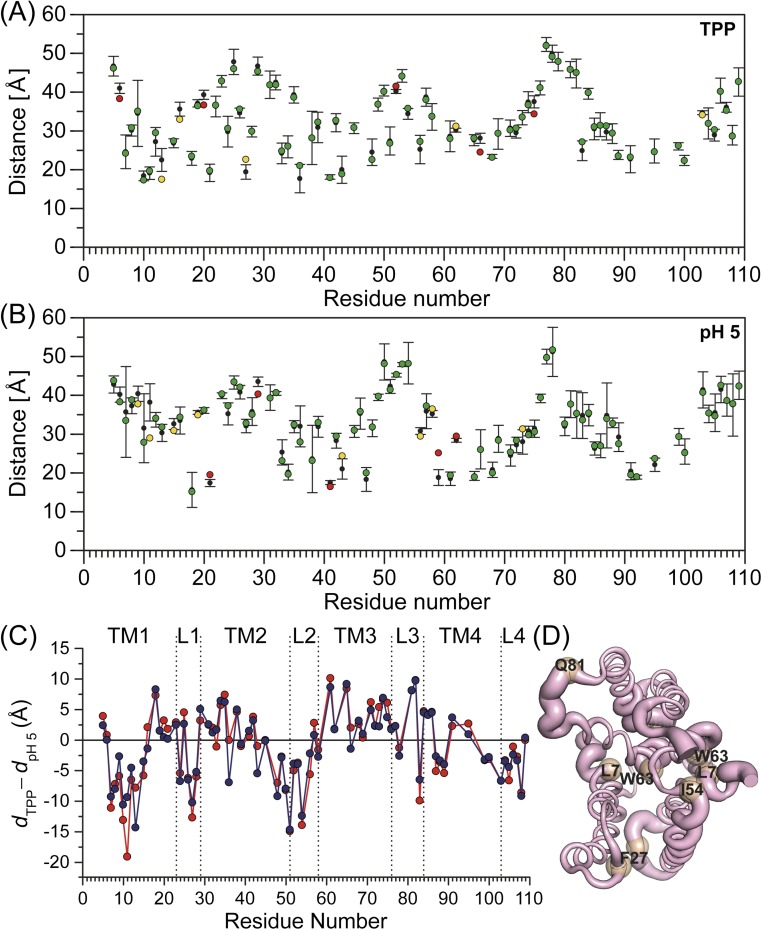
To provide a global perspective on the structural rearrangements, we plotted the change in the distance as a function of residue number (Fig. 3 and Fig. S1). This is necessarily an oversimplification, as many distributions particularly for the apo conformation are broad and cannot be rigorously characterized by a unique distance (see Fig. S1). Nevertheless, this representation allows the qualitative visualization of the regions of conformational changes, thereby identifying a complex web of structural rearrangements focused on TM1–3 and the loops as highlighted by the width of the ribbon representation of EmrE in Fig. 3.
Fig. 3.
Ligand-dependent changes in the distance as a function of residue number for TPP to proton-bound intermediates (dTPP – dpH5). The absolute value of the change in the distance between the two states is displayed by the ribbon thickness on the TPP-bound crystal structure. Residues with positive and negative distance change are colored blue and red, respectively. Unchanged residues and those where no data were obtained are colored white. See SI Methods for more details (Fig. S1).
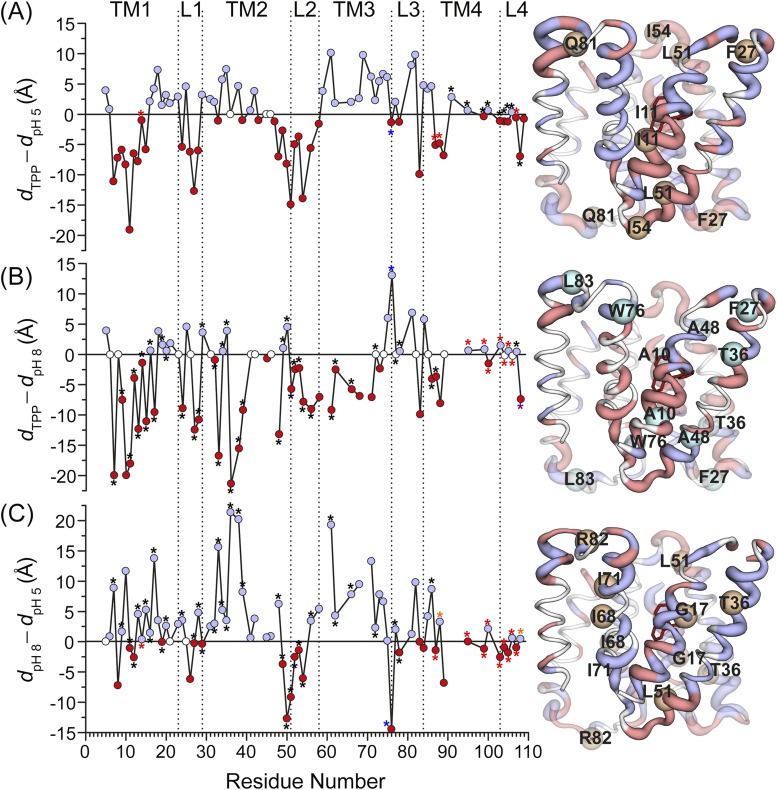
Fig. S1.
Ligand-dependent changes in the distance as a function of residue number. The absolute value of the change in the distance between the two states is displayed by the ribbon thickness on the TPP-bound crystal structure. Residues with positive and negative distance change are colored blue and red, respectively. Unchanged residues and those where no data were obtained are colored white. (A) TPP to proton-bound (dTPP – dpH5). (Residues 14, 87, 88, and 107; red asterisk) For the protonated (pH 5) state, weighted average distance of the distribution is used, and for the TPP-bound state, the center of the Gaussian component corresponding to this state is used. (Residues 91, 95, 99, 100, 103–106, and 108; black asterisk) For both states, weighted average distance of the distribution is used. (B) TPP-bound to apo (dTPP – dpH8). (Residues 7, 9, 11–17, 19, 20, 24, 27–29, 32–36, 38, 39, 48–54, 56, 61, 62, 66, 72, 77, 78, 86, 87, and 107; black asterisk) For the apo (pH 8) state, weighted average distance of the distribution is used, and for the TPP-bound state, the center of the Gaussian component corresponding to this state is used. (Residue 108; purple asterisk) For the TPP-bound state, weighted average distance of the distribution is used, and for the apo state, the center of the Gaussian component corresponding to this state is used. (Residues 95, 99, 100, and 103–106; red asterisk) For both states, weighted average distance of the distribution is used. (C) apo to proton-bound (dpH8 – dpH5). (Residues 7, 9, 11–13, 15–17, 19, 20, 24, 27–29, 32–36, 38, 39, 48–54, 56, 61, 62, 66, 72, 77, 78, and 86; black asterisk) For the apo (pH 8) state, weighted average distance of the distribution is used, and for the protonated (pH 5) state, the center of the Gaussian component corresponding to this state is used. (Residues 88 and 108; yellow asterisk) For the protonated (pH 5) state, weighted average distance of the distribution is used, and for the apo state, the center of the Gaussian component corresponding to this state is used. (Residues 14, 87, 95, 99, 100, and 103–107; red asterisk) For both states, weighted average distance of the distribution is used. In all panels, for the rest of the residues, the center of the Gaussian components corresponding to each state is used. For residue 76 (blue asterisk), the data for the reconstituted sample in nanodiscs are used. For residues where data were obtained for the E14Q mutant, the assignment of the distance components was based on the fact that this mutation favors the pH 5 component.
The TPP-Bound Conformation.
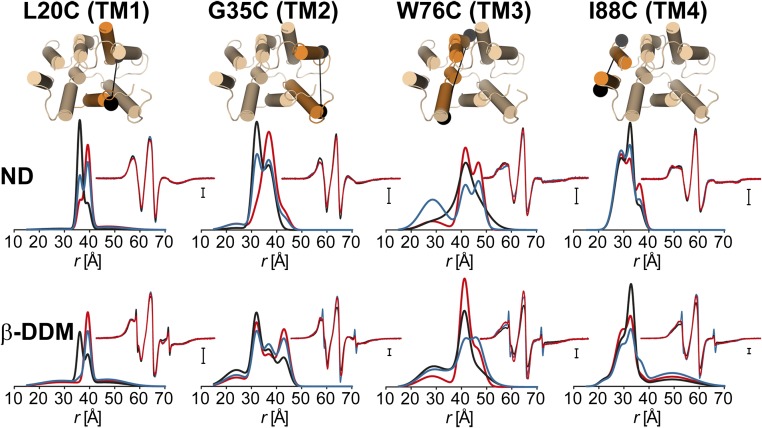
Because the current structural data are exclusively for the TPP-bound state, we will consider it as a reference for the purpose of interpreting the distance distributions in a structural context. Previous work from our laboratory provided a complete view of the accessibility and mobility profile of spin labels in this state (5). The data pointed to extensive disagreement between the crystal structure and the conformation in liposomes. The nature of these disagreements and the low resolution of the structure lead to the conclusion that there are substantial issues with the orientation of helices in the crystal structure (5).
The isomerization between inward- and outward-facing conformations detected by NMR is not expected to lead to changes in the distance distributions, as the packing of the two protomers in the asymmetric dimer is identical (6). Therefore, we compared the experimental distances to those predicted from the crystal structure and found discrepancies that extend across the entire structure (Fig. S2), further confirming the conclusion deduced from the accessibility and mobility analysis regarding the accuracy of the structure. Nevertheless, the crystal structure [Protein Data Bank (PDB) ID code 3B5D] is presented in the figures to provide a general reference for the location of the labels. A model of the TPP-bound conformation, refined to agree with the experimental distances, is presented in Discussion.
Fig. S2.
Comparison of the obtained experimental distances for the TPP-bound state with distances predicted from the crystal structure. The average distances (colored circles) predicted by the X-ray structure were calculated using the MMM 2013.2 software package (27). The color code indicates whether distances are matched (green, within the SD of the specified experimental distance), poorly matched (yellow, within twice the SD), or not matched (red, deviating by more than twice the SD). Only experimental distances between residues 6 and 100 are compared, as the rest are not resolved for both monomers in the structure.
Conformational Changes Induced by Protonation.
Comparison of the pH 5 and TPP-bound distance distributions reveals extensive structural changes upon protonation. These are primarily observed at residues in TM1 and 3, which are directly involved in substrate binding, and TM2, which borders the bilayer. In contrast, minor rearrangements along the interface of TM4 are reported by the spin labels. Large changes in the average distances and the width of the distance distributions are observed along the N-terminal part of TM1 (residues 8–15; SI Appendix 1, A and B), with the largest changes occurring at sites with restricted mobility previously assigned to helix/helix interfaces (i.e., residues 11 and 15) (19). Therefore, we interpret these distance changes as reflecting a degree of rotation rather than a simple tilt of the helices. Rotation is expected to alter spin label mobility at helical interfaces and consequently lead to large distance changes due to the repacking of spin label side chains. In contrast, helix tilt would be expected to yield smaller amplitude distance changes and to be observed concurrently at lipid-facing sites.
Conformational changes at pH 5 include the C-terminal end of TM1 predominantly in the form of helix tilt. The different nature of rearrangements along the two segments of TM1 presumably necessitates a hinge point or an unwinding in the helix. Consistent with this conclusion, the distance distributions at 12 and 13 are broad, suggesting flexibility of the backbone (SI Appendix 1, B).
Large amplitude distance changes are observed in loop 1 (Figs. 2 and 3 and SI Appendix 1, C and D), indicating extensive ligand-dependent repacking. Although the exact nature of the underlying structural rearrangements is difficult to infer from the data, they suggest that the loop plays a central role in the occlusion/exposure of the substrate binding site. Notable is residue 27, where a short distance component is observed in the apo and in the TPP-bound state, whereas the pH 5 distribution suggests a highly disordered conformation. The penetration of this residue toward the binding site, implied by the short distance component, presumably stabilizes the bound substrate.
The N-terminal segment of TM2, consisting of residues 31–35, undergoes a closing motion upon protonation, evidenced by the shift in the average distance (Fig. 1 and SI Appendix 1, D and E). A residue-by-residue analysis of the 37–44 stretch is hindered by the close proximity of spin labels (13) (<20 Å; SI Appendix 1, E). However, the distributions at sites 41 and 42, where DEER decays can be analyzed, show the persistence of the closing trend. This movement is attenuated near the end of the helix, although changes in the distance distributions are detected at residues 48 and 49 (Fig. 1 and SI Appendix 1, F). As TM2 merges into loop 2, we observed evidence of ligand-induced changes in order, manifested by large changes in the width of the distributions at residues 50 and 51 (SI Appendix 1, F).
Except for residues 52 and 53, most residues in loop 2 were characterized by broad distributions, indicating a highly dynamic backbone (Figs. 2 and 3 and SI Appendix 1, G). The distributions consist of a well-defined component and a broad underlying component, which we interpret as reflecting the existence of one conformation wherein the loop backbone is rigid. This conformation is stabilized by ligand binding and is reduced in the apo intermediate.
The N terminus of TM3 is in direct contact with the bound substrate in the crystal structure. Moreover, previous mutagenesis studies implicated residues in this helix in substrate binding (20). Distributions at residues 62 and 68 and continuous wave (CW)-EPR spectrum at residue 64 suggest that the N-terminal part of TM3 undergoes repacking between proton- and TPP-bound states, but a quantitative interpretation is hindered by the broad distributions and the close distances (Fig. 1 and SI Appendix 1, H). Beginning at residue 71, protonation invariably leads to a distinct short distance component not observed in the TPP-bound state, suggesting a closing movement in this region of the transporter (Fig. 1 and SI Appendix 1, I). This movement is likely facilitated by the GVG motif in TM3, which displays changes in its dynamics in the absence of substrate (5, 21).
The conformational changes at the C-terminal half of TM3 propagates to the loop, linking it to TM4 (Figs. 2 and 3 and SI Appendix 1, J). Remarkably, we observed distinct evidence of short components in the pH 5 state (e.g., W76, Q81, R82, and D84) that imply a large-scale closing movement of the loop indicative of an occlusion of the substrate binding cavity in the absence of substrate.
Although noticeable ligand-induced changes in the distributions are observed at several sites in TM4 (Fig. 1 and SI Appendix 1, K and L), they are generally smaller in magnitude and no discernable pattern was evident from comparison of changes at successive sites. Given that this TM is involved in dimer formation, it is not unexpected that the rearrangements at TM1 and 3 necessitate repacking at the dimer interface.
The Apo State.
The profile of the apo state that emerges from the distributions at pH 8 is that of a highly dynamic conformation (Fig. S1 and SI Appendix 1). Broad distributions indicative of conformational sampling are observed along the N-terminal parts of TM1 and 3 as well as in the loops (Fig. 2 and SI Appendix 1, A, B, G, and H). However, there are notable exceptions that occur at functionally important residues. Specifically, the apo state distributions at residues 14 and 18 (SI Appendix 1, B) are narrower than in the ligand-bound state, suggesting that the substrate binding site may become occluded in the absence of ligands.
Unexpected short distance components at residues in loop 3 (Fig. 2 and SI Appendix 1, I and J) are indicative of a large amplitude excursion for this segment, which would be at either side of the membrane in an antiparallel structure. Such movement may be associated with an occlusion of the substrate binding region near TM3 similar to what is observed at pH 5. These results rationalize previous accessibility data that revealed simultaneous large exposure of spin labels in loop 3 to nickel(II)–ethylenediamine-N,N′-diacetic acid (NiEDDA) and molecular oxygen (5).
Conformational Dynamics in Lipid Bilayers.
To investigate ligand-induced conformational changes in lipid bilayers, we carried out DEER measurements on representative spin-labeled EmrE mutants that have been reconstituted in lipid nanodiscs (Fig. 4 and SI Appendix 2). Comparison of the distance distributions demonstrates pH- and substrate-dependent rearrangements along the same structural elements as in detergents. More importantly, the sign of the distance changes is identical in detergent micelles and lipid bilayers, suggesting that similar conformations are stabilized by protonation and substrate binding in the two environments. However, we found that, for the majority of the residues investigated, the width of the distributions was narrower in lipid bilayers, indicating a more ordered/less dynamic structure. These were particularly notable for residues in the center of TM1 and 2 and the C-terminal part of TM3 (e.g., residue 12 in TM1; Fig. 4 and SI Appendix 2, A and B). In contrast, reconstitution in lipid bilayers either did not affect the disorder of the loops or, more notably, promoted fluctuations by loop 3, as evident by the increase in the population of the short component at residue 76 (Fig. 4 and SI Appendix 2).
Fig. 4.
Ligand-dependent conformational changes of EmrE in nanodiscs (ND) composed of E. coli polar lipids. The distributions in DDM micelles are shown for reference: Apo (blue), proton-bound (black; pH 5 for β-DDM, pH 6 for nanodiscs), and TPP-bound (red). The CW-EPR spectra are shown in the Insets.
The Proton Sensor of EmrE.
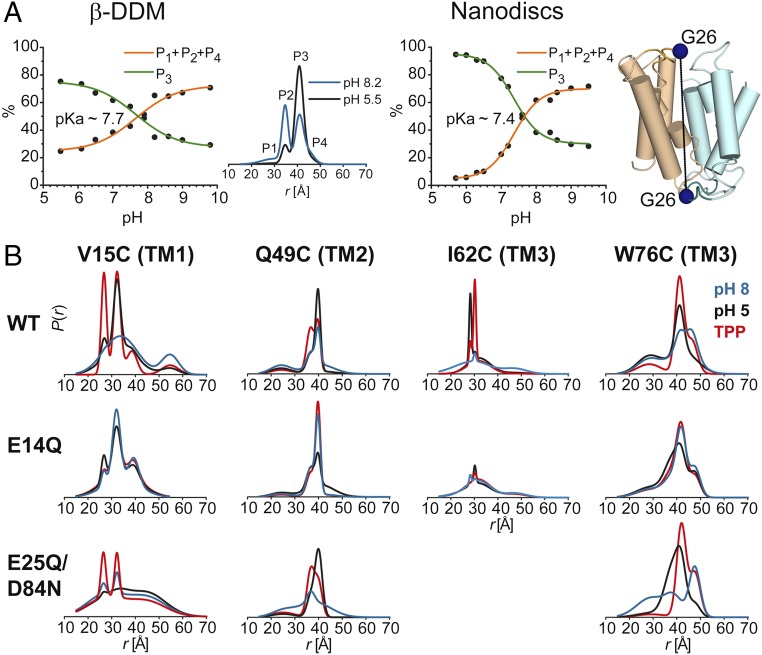
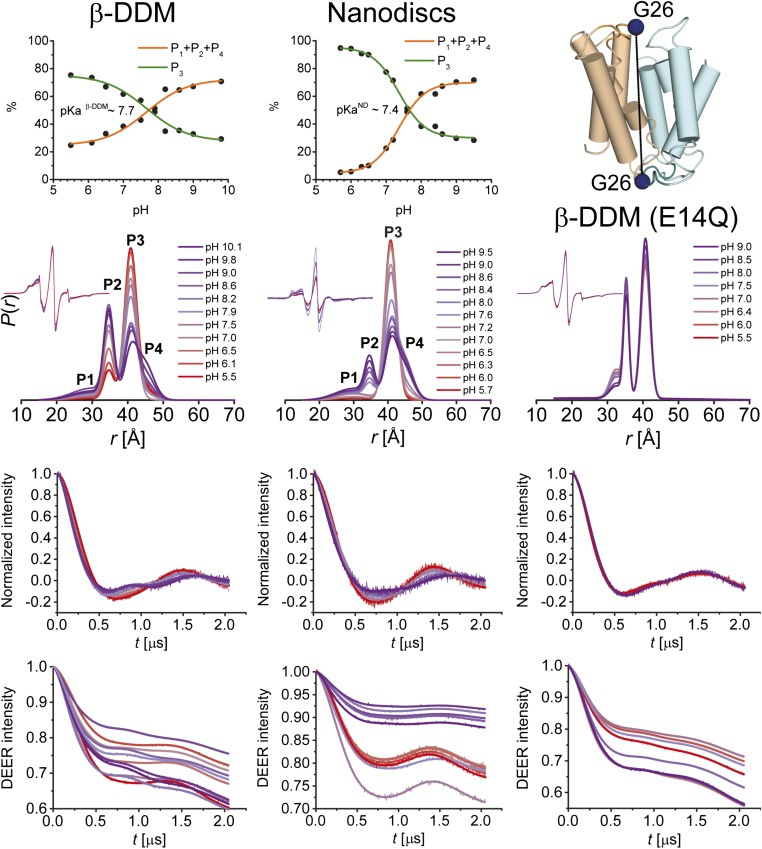
We determined the pKa of the conformational transition of ligand-free EmrE using G26 as a reporter (Fig. 5 and Fig. S3). A titration curve was constructed by determination of the amplitudes of the distance components in the pH 5–10 range. pKa values of ∼7.7 and 7.4 were obtained in detergent micelles and lipid bilayers, respectively, indicating that the environment of E14 was not substantially altered in lipids. Previous kinetic analysis yielded a pKa value for E14 of about 7.3, although a wider range was reported from a steady-state analysis (9). The similar values of the pKa suggest that the conformational changes detected by EPR are involved in the antiport mechanism of EmrE.
Fig. 5.
(A) pH-dependent conformational equilibrium of EmrE in nanodiscs and β-DDM micelles. Distance distributions of the G26C pair were obtained at different pH values ranging from 5.5–10.0 in β-DDM and 5.7–9.5 in nanodiscs (Fig. S3). The variation in population of rising (P1 + P2 + P4) or equally decreasing (P3) distance peaks (Middle panel) as a function of pH was used to estimate the pKa value for conformational changes in EmrE. (B) Effect of protonation–mimetic mutation of acidic residues on conformational states in equilibrium (β-DDM micelles). The single (E14Q) or double (E25Q/D84N) mutations were combined with the single-cysteine mutations (see Fig. S4 and SI Appendix 3).
Fig. S3.
pH-dependent conformational equilibrium of EmrE in nanodiscs and β-DDM micelles. Distance distributions of the G26C pair were obtained at different pH values ranging from 5.5–10.0 and 5.7–9.5 in β-DDM and nanodiscs, respectively. In addition, distance distributions were obtained for the protonation–mimetic mutant G26CE14Q in β-DDM at pH values ranging from 5.5–9.0. The variation in population of rising (P1 + P2 + P4) or equally decreasing (P3) distance peaks as a function of pH was used to estimate the pKa value for conformational changes in EmrE. From Bottom to Top, primary DEER traces with the corresponding fits, baseline-corrected and normalized DEER traces along with the fits, distance distributions and the CW-EPR spectra as Insets, and pH–titration curves are shown. The E14Q mutation abrogates the pH-dependent changes in the amplitude of the distance components.
Consistent with this conclusion, we found that the pH-induced distance changes in TM1 are primarily associated with the protonation of E14 (Fig. 5 and Fig. S3). Substitution of E14 with glutamine designed to mimic protonation abrogates the distance changes due to the shift from pH 8 to pH 5 in TM1 (e.g., residues 11, 15, and 20; Fig. 5, Fig. S4, and SI Appendix 3, A). More importantly, the distributions at pH 8 in the E14Q background are similar to those at pH 5 in the WT, suggesting that the E14Q mimics protonation of E14.
Fig. S4.
Effect of protonation–mimetic mutation of acidic residues on distance distributions in β-DDM micelles. The single (E14Q, E25Q, D84N) or double (E25Q/D84N) mutations were combined with the single-cysteine mutations.
In TM2, the E14Q substitution also attenuates the pH-dependent distance changes, but unlike TM1 the shape of the distribution at pH 5 is noticeably broader than in the WT background (Fig. S4 and SI Appendix 3, B and C). To identify the residues involved in modifying the effects of E14 protonation, we introduced the D84N/E25Q substitutions while monitoring selected sites (Fig. 5B, Fig. S4, and SI Appendix 3). We observed an increase in disorder primarily for the pH 8 conformation and to a lesser extent the pH 5 conformation in this background. Consistent with a role for the protonation of these residues in the transport mechanism, we found that although E14Q substitution attenuated the distance changes in TM3, it also introduced a large degree of disorder reflected in the width of the distance distributions (e.g., I62; Fig. 5B, Fig. S4, and SI Appendix 3, C and D). We interpret this result as reflecting the change in protonation state of D84 and E25. Importantly, conformational changes in loop 3 are primarily determined by the protonation of D84 rather than E14 (Fig. S4 and SI Appendix 3, D), consistent with previous reports (17). Together the data reveal that pH-induced structural changes, although primarily mediated by E14, are affected by other acidic residues such as D84.
Discussion
The results presented above reveal extensive, functionally relevant, conformational changes as a consequence of protonation/deprotonation of glutamate 14. Rotation and tilting of TM1–3, which together form the substrate/proton binding site, not only reconfigure access to the cavity but presumably modulate substrate affinity through reorientation of side chains. The interhelical loops emerge as central elements in this protonation switch, undergoing extensive repacking. The large, ligand-dependent distance changes cannot arise from the isoenergetic alternating access through simple conformational exchange of each protomer because the two resulting dimer structures are identical except for their orientation relative to the bilayer. Rather the distance changes reflect the population of novel conformations that are primarily stabilized by the binding/dissociation and protonation/deprotonation of E14. In addition, the data uncovered a contribution of E25 and/or D84 to the conformational switch primarily through extensive repacking of loop 3. Finally, deprotonation and substrate release appear to induce a high level of disorder, suggesting large amplitude equilibrium fluctuations in multiple structural elements, notably TM3. However, structural interpretation of the distance distributions in this apo state is hindered by the breadth of these distributions.
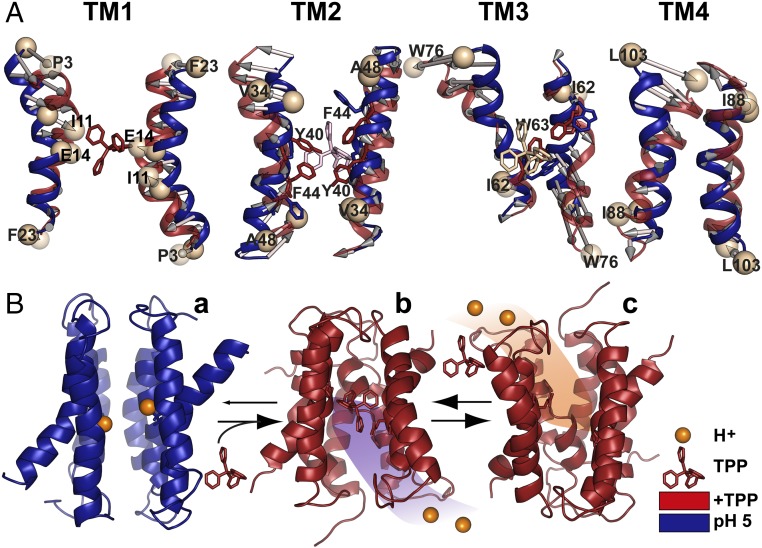
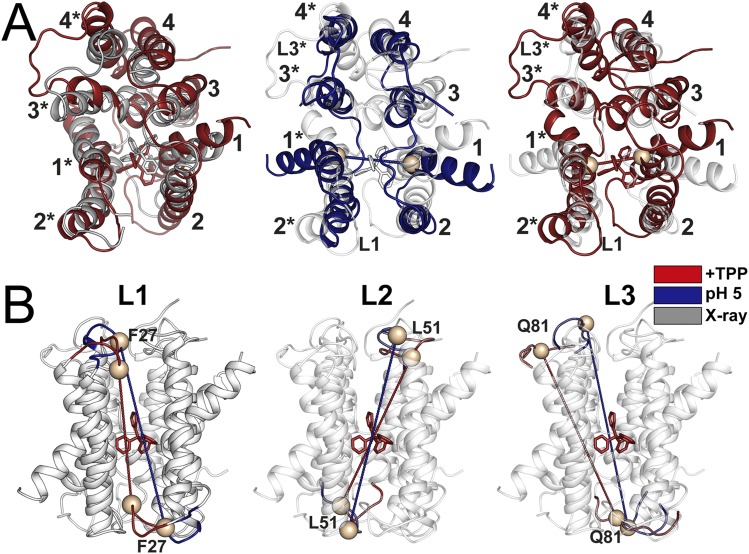
To highlight the conformational changes induced by protonation in a structural context, we carried out detailed de novo modeling of the pH 5 conformation using BCL::Fold and Rosetta (22–25). The conformational search was restrained by the experimental distances at pH 5 (22, 23). A twofold symmetry was imposed such that both protomers have similar conformations, in contrast to the crystal structure, where the two protomers have distinct conformations and form an asymmetric dimer. However, despite the extensive nature of the distance restraints and their coverage along the protein sequence, compatibility of the DEER data with an asymmetric dimer cannot be excluded—primarily due to the uncertainty in translation of measured distances between the spin labels into backbone structural restraints (23, 25). However, as discussed below, we consider the asymmetric dimer to be less mechanistically plausible for the protonated state. The TPP-bound crystal structure was refined using MODELLER (26) restrained by the distances obtained in the TPP-bound conformation following a protocol introduced by Jeschke and coworkers (Fig. 6 and Figs. S5 and S6) (27). To ensure that the spin label side chains were treated similarly in both models, the de novo pH 5 models were further refined using the MODELLER protocol (Fig. 6 and Figs. S5 and S6).
Fig. 6.
Model of EmrE transport derived from EPR data. (A) Conformational changes between protonated (pH 5) and TPP-bound intermediates. Overall alignment between respective TM pairs is shown; for TM3, alignment was based on residues 58–64. (B) The resting state is a protonated but water-occluded conformation of EmrE, represented here with the symmetric model generated by BCL::Fold/Rosetta and refined in MODELLER by pH 5 distance restraints (a). Subsequent binding of the substrate from inner membrane leaflet promotes release of the protons, yielding the refined TPP-bound crystal structure (b). Conformational exchange of the monomers enables alternating access to the extracellular milieu, and exchange of substrate with protons resumes the cycle (c).
Fig. S5.
Comparison of the obtained experimental distances for the TPP-bound and protonated states with distances predicted on the generated models. (A) Refined X-ray structure. (B) The symmetric protonated (pH 5) model. The average distances (colored circles) were predicted at 298 K using the MMM 2013.2 software package (27). The color code is described in Fig. S2. (C) Ligand-dependent changes (TPP-bound to protonated) in the experimental distances (red) versus predicted ones on the TPP-bound and protonated models (royal blue). (D) C-α rmsd between the refined TPP-bound X-ray structure and the symmetric protonated model is displayed by the ribbon thickness on the refined X-ray structure.
Fig. S6.
Structural comparison of the generated models and X-ray structure. (A) Overall structural alignments between X-ray structure and the refined model (Right) and refined X-ray structure and the symmetric protonated model (Middle and Left) are shown. (B) Conformational changes in the loop regions (L1–3) between TPP-bound and protonated models.
The conformational changes gleaned from comparing these models suggest plausible and previously unappreciated mechanistic elements of EmrE transport (Fig. 6A and Figs. S5D and S6). The flexibility of TM1, presumably a consequence of the two consecutive glycines at positions 8 and 9, enables large-scale reconfiguration upon concurrent substrate dissociation and protonation of E14. Not only does the distance between the two TM1s increase (Fig. S6A), but extensive rotation of the N-terminal part alters the side chain orientations in the substrate binding site (Fig. 6A). The rearrangements of the backbone and side chains of TM1 and 3 may provide the mechanism to lower the affinity to substrate in conjunction with the competition by protons for binding to E14 (Fig. S5D).
Although substrate access to the binding site is typically represented as occurring from the cytoplasm, a competing model posits that for hydrophobic substrates, such as those of EmrE, partitioning is likely to occur from the inner leaflet of the bilayer (28). Consistent with this model, we observed that rotation/tilting of TM2 (Fig. 6A) swings open a gate consisting of Tyr-40 and Phe-44, thereby enabling direct access to and from the bilayer to the binding site.
Substrate dissociation and protonation induces repacking of the N terminus of helix 3, which participates in the coordination of the substrate (Fig. 6A and Figs. S5 C and D and S6A). In concordance with the conclusion from accessibility data (5), the C-terminal part of TM3 undergoes large amplitude movement coupled to extensive repacking of loop 3 (Fig. 6A and Fig. S6). This movement is controlled by the protonation state of E25 and/or D84, suggesting a degree of decoupling of this loop from the protonation state of E14.
How may these conformational changes enable coupled transport? Because of the high concentration of protons on the extracellular side, we envision that in the absence of substrate, E14 is protonated and the transporter is in the protonated conformation (corresponding to pH 5 here; Fig. 6B). Measured values of E14’s pKa would imply proton leakage unless the structure is proton-occluded, which for EmrE would require a symmetric conformation wherein E14 is not exposed to the pH of the cytoplasm. Therefore, we propose that the protonated conformation does not undergo the isoenergetic alternating access and thus is symmetric, as shown in Fig. 6B. Substrate binding to this conformation, which occurs from the inner leaflet of the bilayer through the TM2 fenestration, releases the two protons by stabilizing the asymmetric TPP-bound conformation. Through its isoenergetic alternating access (6, 11), this state exposes the substrate to the extracellular milieu, at which point protons displace the substrate, enabling a new cycle of transport (Fig. 6B).
Methods
Mutagenesis, Expression, Purification, Labeling, and Reconstitution of EmrE.
This study uses the previously generated constructs of single cysteine mutants of EmrE (5). Specific functional regions of EmrE, including residues in the GG7 dimerization motif, located in TM4, were excluded from the analysis. Further functional mutants (E14Q, E25Q, D84N, E25Q/D84N) were introduced on the same constructs using site-directed mutagenesis. EmrE mutants were expressed, purified in 1.5% (wt/vol) β-DDM, and spin-labeled using the same protocol as previously described (5). Purified EmrE was concentrated with Amicon Ultra-10 kDa centrifugal filter units (Millipore). Samples for DEER spectroscopy were prepared in the 100–200 μM protein concentration range. A final concentration of glycerol of 30% (wt/vol) was used in all samples as a cryoprotectant. The TPP-bound state was obtained by addition of sixfold molar excess of the substrate TPP. For pH 5 and apo pH 8 states, respectively, a calibrated volume of 1 N HCl or 1 N NaOH was added to samples in EmrE size exclusion chromatography buffer (50 mM sodium phosphate monobasic, 50 mM NaCl, 0.02% β-DDM, and 0.02% NaN3, pH 7.2). For pH titration experiments, an Orion 9810BN micro pH electrode (Thermo Scientific) was used to adjust the pH values. See SI Methods for reconstitution of EmrE in nanodiscs (29).
CW-EPR and DEER Spectroscopy.
For CW-EPR, spin-labeled EmrE samples were loaded in capillaries, and spectra were collected on a Bruker EMX spectrometer using a 10-mW microwave power level and a modulation amplitude of 1.6 G. DEER spectroscopy was performed on an Elexsys E580 EPR spectrometer (Super QFT bridge with ELDOR; Bruker) operating at Q-band frequency (33.9 GHz) with the dead-time free four-pulse sequence at 83 K (13). Primary DEER decays were analyzed using home-written software operating in the Matlab environment (29). Briefly, the software carries out global analysis of the DEER decays obtained under different conditions for the same spin-labeled position. The distance distribution is assumed to consist of a sum of Gaussians, the number of which is determined based on a statistical criterion.
Refinement of X-Ray Structure and Modeling the EmrE Structure at pH 5.
The X-ray structure of EmrE in a TPP-bound state (PDB ID code 3B5D) was refined in several iterations using MODELLER version 9.10 (26). A previously built complete atomistic model of dimeric EmrE was used (5). In silico spin labeling of the protein structure using a rotamer library approach was performed using the MMM 2013.2 software package (27). See SI Methods for more details on refinement using MODELLER. The stereochemical quality of the generated models was evaluated using PROCHECK (Table S1) (30).
Table S1.
Quality assessment of the generated models
| Models/Structure | GA341 score* | Ramachandran plot statistics,† % residues in most favored regions | Ramachandran plot statistics,† % residues in allowed regions | Main-chain parameters,† overall G-factor | Cα rmsd between two chains |
| pH 5, H+-bound | 0.84 | 73.1% | 94.0% | −0.1 | 0.7 |
| Refined X-ray, +TPP | 0.99 | 72.0% | 95.6% | −0.2 | 4.7 |
| X-ray | 4.1 |
A model is predicted to be reliable when the GA341 score is higher than a prespecified cutoff (0.7). A reliable model has a probability of the correct fold that is larger than 95% (26).
The stereochemical quality of the generated models was evaluated using PROCHECK (30). G-factor values below –0.5 are considered unusual.
The initial model of the EmrE structure at pH 5 was predicted by a two-step approach using BCL::Fold (22, 23) to assemble the secondary structure elements (SSEs) in 3D space and Rosetta (24, 25) to construct loop regions and predict side-chain conformations. See SI Methods for more details on BCL/Rosetta modeling. The generated models using this approach were further refined in MODELLER.
SI Methods
Reconstitution of EmrE in Nanodiscs.
E. coli polar lipid extract (Avanti Polar Lipids) was dissolved in chloroform, evaporated to dryness on a rotary evaporator, and desiccated overnight under vacuum. Membrane scaffold protein (MSP1D1) was expressed and purified as described earlier (29). The lipids were hydrated in MSP buffer (50 mM Tris, 0.1 M NaCl, pH 7.5) containing 0.5% (wt/vol) β-DDM, filtered through 0.2 μm polycarbonate membrane (Whatman), and stored in small aliquots at –80 °C. For reconstitution into nanodiscs, purified spin-labeled proteins in β-DDM micelles were mixed with lipid, MSP1D1, and β-DDM in the following molar ratios: lipid:MSP1D1, 60:1; MSP1D1:EmrE, 8:1; β-DDM:lipid, 5:1. Mixtures were rocked at room temperature for 2 h. Biobeads (0.8 g/mL) were then added to the solution and incubated overnight at 4 °C with rocking. The nanodisc assembly solution was filtered using a 0.45-μm filter to remove biobeads. Full nanodiscs were separated from empty nanodiscs by size-exclusion chromatography (50 mM sodium phosphate monobasic, 50 mM NaCl, 0.02% NaN3, and 5% glycerol, pH 7.2). Nanodiscs were concentrated using Amicon Ultra-50 kDa centrifugal filters (Millipore). Proteo-nanodiscs were then characterized using SDS/PAGE to verify reconstitution. Concentration of spin-labeled mutants in nanodiscs was determined by comparing the intensity of the integrated CW-EPR spectrum to that of the same mutant in detergent micelles.
Refinement of X-Ray Structure Based on DEER Distance Constraints.
The X-ray structure of EmrE in the TPP-bound state (PDB ID code 3B5D) was refined in several iterations using MODELLER version 9.10 (26). A previously built complete atomistic model of dimeric EmrE was used (5). In silico spin labeling of the protein structure using a rotamer library approach was performed using MMM 2013.2 software package (27). In each iteration, the rotamer ensemble was first calculated at 298 K on the spin-labeled positions. A rotamer that best fits the mean N–O midpoint position of the whole ensemble was attached to the template structure that was supplied to MODELLER. The X-ray structure was refined using the center of the Gaussian components corresponding to the TPP-bound state. Secondary structure assignments deduced from our previous study were also considered as constraints (5). Only models with a GA341 score of 0.75 or higher were included in the ensemble. GA341 specifies a threshold, below which models should be rejected (Table S1). After importing models to MMM, rotamer ensembles were recalculated on them. The stereochemical quality of the generated models was evaluated using PROCHECK (Table S1) (30).
Modeling the EmrE Structure at pH 5.
The protein structure prediction protocol consisted of a two-step approach using BCL::Fold (22, 23) to assemble the SSEs in the 3D space and Rosetta (24, 25) to construct loop regions and predict side-chain conformations. In one of the models, a twofold symmetry was imposed such that both protomers have similar conformations. In another model, the symmetry was relaxed to obtain an asymmetric dimer. Over the course of the optimization, transformations are applied to the SSEs to sample different conformations, and the free energy of the sampled conformations was approximated using knowledge-based potentials evaluating properties like the burial of residues, residue–residue interactions, and steric interference between residues. The knowledge-based potentials were supplemented with a scoring function based on the cone model to quantify the agreement of the conformation with the EPR distance data (23). To achieve sufficient coverage of the conformational space, 100,000 models were sampled for the pH 5 state using this protocol. The resulting models were sorted by their pseudoenergy score, encompassing the scores from the knowledge-based potentials and the EPR agreement. The model with the most favorable score was selected for loop construction and refinement with Rosetta. The resulting model was refined using Rosetta relax (24) to improve agreement with the EPR data, and a cone mode-based scoring function was used to quantify the agreement. Upon construction of all loop regions, the model with the most favorable score was selected.
The generated models using this approach were further refined in MODELLER as described above.
Supplementary Material
Acknowledgments
This work is dedicated to Dr. Hanane A. Koteiche who initiated this research direction in the laboratory. We thank Dr. Sepan T. Amadi and Sanjay Mishra for their preceding studies on EmrE, which paved the way for this work. We thank Drs. Richard Stein and Derek Claxton for critical reading and editing of the manuscript. This work was supported by NIH Grants R01 GM077659 (to H.S.M.) and R01 GM080403 (to J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520431113/-/DCSupplemental.
References
- 1.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270(12):6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 2.Tate CG, Kunji ERS, Lebendiker M, Schuldiner S. The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 A resolution. EMBO J. 2001;20(1-2):77–81. doi: 10.1093/emboj/20.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104(48):18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehner I, et al. The key residue for substrate transport (Glu14) in the EmrE dimer is asymmetric. J Biol Chem. 2008;283(6):3281–3288. doi: 10.1074/jbc.M707899200. [DOI] [PubMed] [Google Scholar]
- 5.Amadi ST, Koteiche HA, Mishra S, Mchaourab HS. Structure, dynamics, and substrate-induced conformational changes of the multidrug transporter EmrE in liposomes. J Biol Chem. 2010;285(34):26710–26718. doi: 10.1074/jbc.M110.132621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison EA, et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481(7379):45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho MK, Gayen A, Banigan JR, Leninger M, Traaseth NJ. Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance. J Am Chem Soc. 2014;136(22):8072–8080. doi: 10.1021/ja503145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yerushalmi H, Schuldiner S. A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett. 2000;476(1-2):93–97. doi: 10.1016/s0014-5793(00)01677-x. [DOI] [PubMed] [Google Scholar]
- 9.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794(5):748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Schuldiner S. Competition as a way of life for H(+)-coupled antiporters. J Mol Biol. 2014;426(14):2539–2546. doi: 10.1016/j.jmb.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleishman SJ, et al. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J Mol Biol. 2006;364(1):54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 12.Henzler-Wildman K. Analyzing conformational changes in the transport cycle of EmrE. Curr Opin Struct Biol. 2012;22(1):38–43. doi: 10.1016/j.sbi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 14.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40(1):1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 15.Mchaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19(11):1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soskine M, Mark S, Tayer N, Mizrachi R, Schuldiner S. On parallel and antiparallel topology of a homodimeric multidrug transporter. J Biol Chem. 2006;281(47):36205–36212. doi: 10.1074/jbc.M607186200. [DOI] [PubMed] [Google Scholar]
- 17.Banigan JR, Gayen A, Cho MK, Traaseth NJ. A structured loop modulates coupling between the substrate-binding and dimerization domains in the multidrug resistance transporter EmrE. J Biol Chem. 2015;290(2):805–814. doi: 10.1074/jbc.M114.601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaubitz C, et al. 31P-CP-MAS NMR studies on TPP+ bound to the ion-coupled multidrug transport protein EmrE. FEBS Lett. 2000;480(2-3):127–131. doi: 10.1016/s0014-5793(00)01916-5. [DOI] [PubMed] [Google Scholar]
- 19.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 20.Yerushalmi H, Lebendiker M, Schuldiner S. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 1996;271(49):31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 21.Gayen A, Banigan JR, Traaseth NJ. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52(39):10321–10324. doi: 10.1002/anie.201303091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karakaş M, et al. BCL::Fold – de novo prediction of complex and large protein topologies by assembly of secondary structure elements. PLoS One. 2012;7(11):e49240. doi: 10.1371/journal.pone.0049240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer AW, et al. BCL:MP-fold: Membrane protein structure prediction guided by EPR restraints. Proteins. 2015;83(11):1947–1962. doi: 10.1002/prot.24801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirst SJ, Alexander N, Mchaourab HS, Meiler J. RosettaEPR: An integrated tool for protein structure determination from sparse EPR data. J Struct Biol. 2011;173(3):506–514. doi: 10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 27.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13(6):2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 28.Bolhuis H, et al. Multidrug resistance in Lactococcus lactis: Evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 1996;15(16):4239–4245. [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra S, et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife. 2014;3:e02740. doi: 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26(2):283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.