Abstract
Background
B cells expressing IgE contribute to immunity against parasites and venoms, and are the source of antigen specificity in allergy, yet the developmental pathways producing these B cells in humans remain a subject of debate. Much of our knowledge of IgE lineage development derives from model studies in mice, rather than from human subjects.
Objective
We evaluate models for isotype switching to IgE in humans using Ig heavy chain (IGH) mutational lineage data.
Methods
We analyzed IGH repertoires in 9 allergic and 24 healthy adults using high-throughput DNA sequencing of 15,843,270 IGH rearrangements to identify clonal lineages of B cells containing members expressing IgE. Somatic mutations in IGH inherited from common ancestors within the clonal lineage are used to infer the relationships between B cells.
Results
Data from 613,641 multi-isotype B cell clonal lineages, of which 592 include an IgE member, are consistent with indirect switching to IgE from IgG- or IgA-expressing lineage members in humans. We also find that these inferred isotype switching frequencies are similar in healthy and allergic individuals.
Conclusions
We find evidence that secondary isotype switching of mutated IgG1-expressing B cells is the primary source of IgE in humans, with lesser contributions from precursors expressing other switched isotypes, and rarely IgM or IgD, suggesting that IgE is derived from previously antigen-experienced B cells, rather than naïve B cells that typically express low-affinity unmutated antibodies. These data provide a basis from which to evaluate allergen-specific human antibody repertoires in healthy and diseased individuals.
Keywords: IgE, isotype switching, direct, indirect, antibody, B cell, repertoire, high-throughput DNA sequencing
Introduction
B cells producing antibodies of distinct isotypes are the basis for humoral immune defenses, but are also the source of pathogenic antibodies such as allergen-specific IgE in allergic individuals. While mature but antigen-naïve cells exclusively express IgM and IgD, antigen stimulation and an appropriate microenvironment of interaction with T cells and cytokines can trigger B cell genomic rearrangements leading to the expression of IgG, IgA or IgE isotype subclasses, each of which have characteristic immune effector and modulatory functions, including activating the complement pathway, opsonizing antigens, binding to distinct Fc receptors, and, in the case of IgE, providing antigen specificity for mast cells and basophil responses1,2. In humans, debate continues over the extent to which IgE isotype switching occurs directly from IgM, or indirectly from other intermediate antibody isotypes, and even in tractable model organisms such as the mouse this question has not been resolved3-7. Recent studies employing IgE reporter mice have been interpreted to support predominantly direct or indirect switching pathways to IgE8-13. For example, He et al. 9 report evidence that IgE+ germinal center cells in mice derive from direct switching, while IgE+ plasma cells derive from sequential switching. Talay et al.10, 11, using a different reporter model, report that serum IgE in recall responses can be derived from IgE+ memory cells and IgG1+ memory cells, presumably by plasma cell differentiation in the first case, and sequential isotype switching and plasma cell differentiation in the latter case. Yang et al.12, using a third reporter mouse model, do not specifically address whether IgE derives from direct or indirect switching, but find IgE+ plasma cells with low mutation levels early in the immune response, when IgE+ germinal center B cells show elevated mutation levels, suggesting that these IgE+ cell types arise from different pathways. Studies in humans, reviewed by Davies et al.14, suggest that direct switching from IgM to IgE production occurs in incompletely organized germinal centers (GC) and results in minimally mutated IgE exhibiting low affinity for antigen. In contrast, indirect switching to IgE-production via other isotype intermediates is expected to result in IgE possessing a higher level of hypermutation and allergen affinity, and would be accompanied by shared patterns of hypermutation among B cells expressing IgE and members of the same clonal lineage expressing other isotype intermediates. Importantly, class switching is a one-directional process, because the genomic DNA between the upstream isotype and downstream isotype is excised during switching.
To obtain human data that could distinguish between the direct and indirect models of human B cell switching to IgE, and assess the extent to which direct and indirect switching are observed in healthy or allergic subjects, we performed deep sequencing of immunoglobulin heavy chain cDNA derived from peripheral blood B cells taken from 24 healthy individuals and 9 individuals with reported aero- or food allergies. We identified clonal lineages containing IgE-expressing B cells as well as lineage members expressing other Ig isotypes. Our data analysis takes advantage of the fact that antigen-stimulated B cells produce somatic mutations in their IGH V(D)J gene rearrangements at high rates, and these mutations are inherited by daughter cells, permitting reconstruction of relationships within clonal lineages based on the presence of shared mutations between different cells derived from a common ancestor. The results support a model for human IgE B cell development where isotype switching through intermediate isotypes is most common. In particular, we find that in both healthy and allergic individuals, IgE-encoding transcripts are most closely related in clonal lineages to IgG-encoding transcripts, particularly IgG1, and share extensive patterns of hypermutation with this isotype, with a smaller fraction appearing to switch directly from B cells expressing IgM or IgD. The data suggest that the pathogenic IgE antibodies in allergic individuals may differ from those in healthy subjects for reasons related to their antigenic specificities, affinities, or levels, rather than from major differences in the pathway of switching to IgE.
Methods
Collection of clinical samples from healthy individuals
Peripheral blood samples were obtained from the Stanford Ellison influenza vaccination cohort, consisting of 19 healthy individuals (8 males and 11 females, aged 21-88) and 8 allergic individuals (2 males and 7 females, aged 24-84) and consisted of longitudinal samples from 3 time points from each of 2 consecutive years, collected as part of an ongoing vaccination study. Peripheral blood samples from an additional 5 healthy subjects (4 males, 1 female, aged 28-44) and one allergic female individual were also included in the analysis. Patient recruitment, informed consent, and sample collection were carried out according to protocols approved by the Institutional Review Board at Stanford University. The clinical trial of influenza vaccination involving these patients is registered at ClinicalTrials.gov #NCT01827462.
cDNA production, IGH amplification and high-throughput DNA sequencing
Isolation of peripheral blood mononuclear cells (PBMCs), RNA extraction and preparation of isotype-specific IGH libraries was performed using methods previously described, modified for Illumina instrument sequencing15,16. Separate barcoded, isotype-specific Ig libraries for IgM, IgD, IgG, IgA and IgE were prepared from each sample15. Briefly, cDNA was PCR-amplified using multiplexed 5'- primer sets against the IGHV FR1 framework regions, together with 3'-primers specific for exon 1 of the constant regions for each isotype. Isotype primers consisted of a gene-specific sequence, a nucleotide sequence barcode, and an additional randomized 4-base sequence (Supplemental Table 1). To prevent cross-isotype chimeric PCR product formation, separate amplifications were carried out for each isotype. After this initial PCR step, a second PCR reaction was performed to complete the Illumina linker sequences at the 5’ and 3’ ends of the Ig amplicons, and to ensure that final PCR reactions were not amplified to saturation. PCR products were gel extracted, pooled and sequenced with an Illumina MiSeq instrument. Paired end reads were merged using FLASH17, then identical reads were collapsed to a single representative. Sequences can be accessed via dbGAP (http://www.ncbi.nlm.nih.gov/gap) with accession phs000666.v1.p1.
Annotation and pre-processing of Ig sequences
Variable, diversity, and joining gene identity for each immunoglobulin sequence were identified using IgBLAST18 following sequence demultiplexing and primer trimming. Isotype and subtype annotation required exact sequence match to non-primer-encoded sequence regions. The FR3-CDR3 boundary was obtained from IgBlast output, while the CDR3-FR4 boundary was subsequently identified using a position weight matrix search trained to identify the WGQG motif and sequence variants at this position. Sequences lacking V segment regions, or showing evidence for chimerism derived from PCR amplification were filtered and excluded from further analysis. Isotype identity for each sequence was confirmed by local sequence alignment of non-primer-encoded portions of the observed constant region.
Identification of clonally-related immunoglobulin sequences
IGH sequences from each participant were grouped into clusters where cluster members possessed a) the same variable gene segment, excluding allele information, b) identical CDR3 length and c) a minimum of 80% CDR3 nucleotide sequence similarity. The CDR3 similarity threshold was chosen based on comparisons of CDR3 sequence distances within individuals (where true clonally-related sequences are present), compared to distances between CDR3 sequences from different individuals (where clonal relationships are not possible, but rare similar sequences may be produced independently during receptor rearrangement). Clustering based on nucleotide sequence similarity was accomplished using cd-hit-est19. For clustered sequences sharing the same randomized 4-mer ID, putative PCR errors derived from the second PCR step of library preparation, as well as sequencing errors, were filtered out by selecting the sequence containing the lowest number of mutations with respect to the germline V and J segment for the rearrangement.
Identification of nearest-isotype-neighbor events
Sequences within each cluster were considered to be clonally-related members of the same B cell lineage. To identify sequences within a lineage that share somatic mutations, providing evidence of descent from a shared precursor, we counted the number of shared mutation positions between pairs of sequences of different isotypes. While all members of a lineage, by definition, had the same variable gene segment, excluding allele information, differences in allele calling (for variable gene segments) and occasionally diversity and joining gene annotation were observed, likely owing to the inherent difficulty in annotating more heavily mutated IGH sequences. Therefore, we began by identifying the consensus variable, diversity and joining genes for each lineage, which we defined as the most common V and J gene assignment for the unique sequences within that lineage. Next, mutated positions and the resultant base for each sequence were identified using pairwise alignments of germline V and J gene segments. We then identified the nearest isotype neighbor for each isotype within a lineage by by identifying the isotype possessing the greatest number of shared mutations between any one of its members and the members of the isotype in question. In cases where an isotype was equidistant from multiple isotypes within a cluster, all such relationships were reported. Given that alternative splicing within a single B cell can give rise to both IgM and IgD transcripts, in cases where the nearest isotype neighbor of IgM was IgD, or vice versa, that relationship was reported along with that of the second-closest isotype. As a second approach to determining relationships between isotypes within a lineage, we determined the minimum Hamming distance between members of each isotype and members of the other isotypes within a cluster. We first quantified the Hamming distance between pairs of sequences by performing a pairwise alignment of sequences of a particular isotype (excluding the constant region portion of the sequence) using the bio2align module of BioPython (www.biopython.org). For each isotype (the ‘query’ isotype) within a lineage, the nearest neighbor isotype was identified by finding the sequence of a different isotype with the minimum Hamming distance to a sequence in the query isotype.
Quantification and normalization of nearest-isotype-neighbor events by sequencing depth
We tallied the number of nearest neighbor events between each isotype and IgE (Ni), the total number of IgE nearest isotype neighbor events (Nt), the number of reads obtained per isotype amplification post-processing (Ri) and the total number of reads obtained post-processing (Rt). We define the probability that IgE has a nearest neighbor event with a given isotype as:
where (Ni/Nt) indicates the proportion of total IgE nearest neighbor events that involve a given isotype, (Rt/Ri) accounts for differences in sequencing coverage of each isotype amplification, and k is a scaling factor such that the sum of all isotype probabilities equals one.
Results
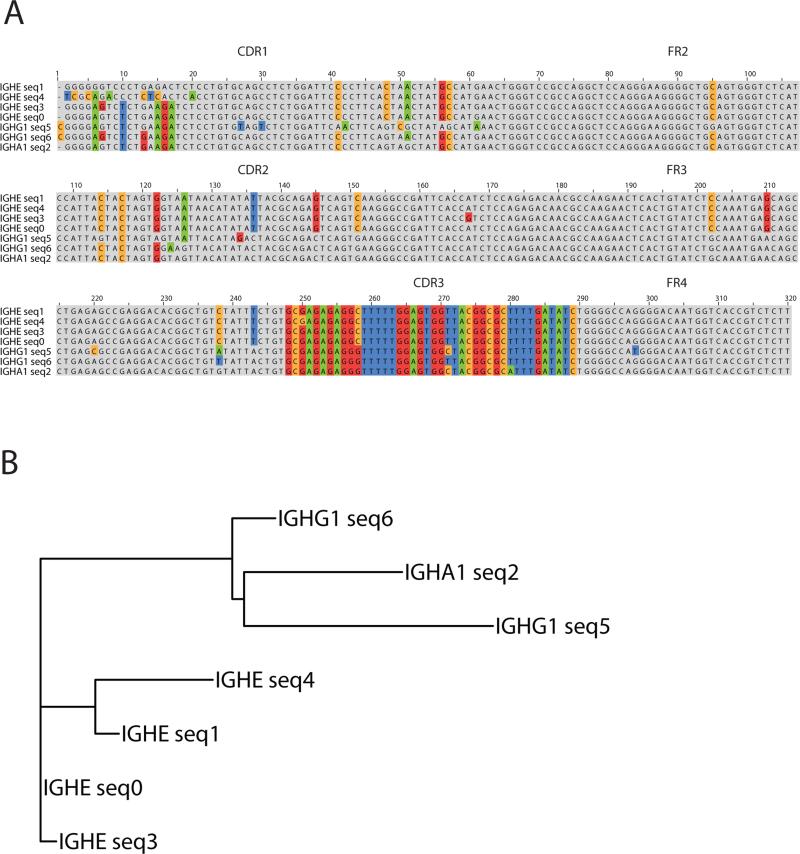
Expressed immunoglobulin heavy chain (IGH) sequences were measured from peripheral blood B cells of 24 healthy (non-allergic) individuals and 9 individuals with history of allergy to food or aeroallergens. We obtained 40,049,427 DNA sequence reads using Illumina MiSeq sequencing of IGH PCR amplified from cDNA template with multiplexed primers in the V segment and with separate PCR reactions for each isotype. 15,843,270 reads remained after sample demultiplexing, and sequence quality filtering. IGHV, IGHD and IGHJ segment identities, and the complementarity-determining region 3 (CDR3) that is encoded by the junctional region of the joined V, D and J segments were annotated. To identify candidate clonally-related lineages of B cells within each participant, we clustered reads that expressed the same variable gene segment, had CDR3 regions of the same length, and possessed at least 80% CDR3 nucleotide identity. Lineages contained either sequences of a single isotype or multiple isotypes. A total of 613,641 multi-isotype lineages were identified in all participants (average per individual 18,048 +/− 1172 standard error) of which 592 included an IgE sequence (Table 1). We were able to identify IgE-containing multi-isotype lineages in each participant. Notably, allergic individuals tended to possess approximately twice as many IgE-containing lineages as healthy individuals (average 119 vs 269 IgE lineages per 10K IgE reads, p=0.01, two-sided Student's t-test; Supplemental Figure 1A). As a distinguishing metric for allergic disease, measurement of IgE lineages compared favorably to ImmunoCap250 total IgE measurement from plasma (95 vs 105 kU/L for healthy and allergic individuals respectively, p=0.78, Student's two-sided t-test; Supplemental Figure 1B) although there was a weak positive correlation between the two metrics (Pearson's correlation =0.21, Supplemental Figure 1C, Table 2). The limited correlation of ImmunoCap250 total IgE with atopy status is consistent with prior literature20-25. Demographic and allergy history for the study cohort as well as measurements of IgE are provided in Table 2. For lineages containing IgE and at least one other isotype, we sought to identify the nearest isotype neighbor of IgE sequences in each lineage by comparing the number of shared mutations over the IGH variable and joining regions with antibodies of other isotypes in the lineage. An example case is presented in Figure 1. Nearest-neighbor isotypes for each individual are presented in Figure 2. We also performed an independent complementary analysis using Hamming distances (i.e., total number of sequence differences) calculated over the entire IGH sequence excluding the constant region, to identify nearest neighbors. Comparison by Hamming distances allows one to take into account sequence differences over the CDR3 region, where exonucleotide chewback and addition of non-templated nucleotides usually precludes the identification of mutated positions with respect to the original VDJ rearrangement in the clone. The two approaches yielded nearly identical results (Figure 2A; Supplemental Figure 2).
Table I.
Isotype composition of immunoglobulin sequence clusters derived from sequencing of peripheral blood in 24 healthy and 9 allergic individuals. Cluster members possess 80% or greater CDR3 nucleotide identity, and have identical variable gene annotations, excluding allele calls, and CDR3 lengths.
| Isotype | Total reads | Total lineages | Total multi-isotype lineages | Multi-isotype lineages containing IgE |
|---|---|---|---|---|
| IGHM | 3258527 | 2008310 | 269652 | 90 |
| IGHD | 3701739 | 2454754 | 256878 | 56 |
| IGHG3 | 288228 | 86418 | 52244 | 170 |
| IGHG1 | 2492494 | 478919 | 145877 | 407 |
| IGHG2 | 1598238 | 243873 | 122417 | 272 |
| IGHA1 | 2982500 | 554461 | 175730 | 193 |
| IGHG4 | 144221 | 32192 | 19663 | 93 |
| IGHE | 175584 | 3906 | 592 | 592 |
| IGHA2 | 1201739 | 269061 | 151506 | 131 |
Table II.
IgE lineages, ImmunoCap total IgE and demographic and allergy history for study cohort.
| BFI | IgE lineages | Plasma Total IgE | Aeroallergen or Food Allergy Y/N | Comment in Cumulative Medical History | Age | Sex |
|---|---|---|---|---|---|---|
| BFI-0000372 | 161 | 770.0 | No | 24 | F | |
| BFI-0000374 | 132 | 19.8 | No | 61 | F | |
| BFI-0000376 | 91 | 2.7 | No | 66 | F | |
| BFI-0000377 | 178 | 80.9 | No | 65 | F | |
| BFI-0000378 | 95 | 33.0 | No | 75 | M | |
| BFI-0000379 | 51 | 14.5 | No | 62 | M | |
| BFI-0000380 | 114 | 18.1 | No | 61 | M | |
| BFI-0000381 | 55 | < 2 | No | 26 | F | |
| BFI-0000382 | 44 | 13.8 | No | 24 | M | |
| BFI-0000383 | 93 | 77.1 | No | 85 | M | |
| BFI-0000384 | 101 | 33.5 | No | 83 | F | |
| BFI-0000385 | 69 | 2.3 | No | 84 | F | |
| BFI-0000387 | 22 | 4.6 | No | 73 | F | |
| BFI-0000388 | 329 | 73.1 | No | 30 | M | |
| BFI-0000393 | 96 | 52.0 | No | 88 | F | |
| BFI-0000394 | 178 | 97.6 | No | 21 | M | |
| BFI-0000396 | 110 | 2.3 | No | 23 | M | |
| BFI-0000397 | 186 | 5.7 | No | 23 | F | |
| BFI-0000398 | 281 | 358.0 | No | 84 | F | |
| BFI-0000841 | 181 | 37.3 | No | 35 | M | |
| BFI-0002450 | 31 | 4.9 | No | 31 | M | |
| BFI-0002451 | 77 | 314.0 | No | 36 | M | |
| BFI-0002452 | 81 | 199.0 | No | 42 | F | |
| BFI-0002453 | 98 | 52.2 | No | 28 | M | |
| BFI-0000373 | 74 | 11.0 | Yes | Seasonal allergy to pollen | 84 | F |
| BFI-0000375 | 411 | 78.6 | Yes | Hay fever since childhood | 24 | M |
| BFI-0000386 | 237 | 308.0 | Yes | Macadamia nuts | 84 | M |
| BFI-0000389 | 73 | 10.5 | Yes | Seasonal allergies | 78 | F |
| BFI-0000390 | 192 | 122.0 | Yes | Pollen allergies and allergic rhinitis | 61 | F |
| BFI-0000391 | 275 | 50.3 | Yes | Dust | 69 | F |
| BFI-0000392 | 412 | 20.9 | Yes | Animal dander and hay fever since childhood | 26 | F |
| BFI-0000395 | 406 | 98.7 | Yes | Seasonal allergies | 27 | F |
| BFI-0001755 | 338 | 249.0 | Yes | Seasonal allergies | 34 | F |
Figure 1.
(A) Sequence alignment of clonally-related antibody heavy chains identified in B cells expressing different isotypes. Highlighted bases indicate positions of somatic mutation in V and J gene segments. The CDR3 region encoded by the VDJ junctions is also highlighted. (B) Tree representation of the clonal lineage. Sequences are labeled according to isotype. Tree building and visualization was performed using the ape package in R and the distance matrix obtained by comparing sequences using the ape “raw” method.
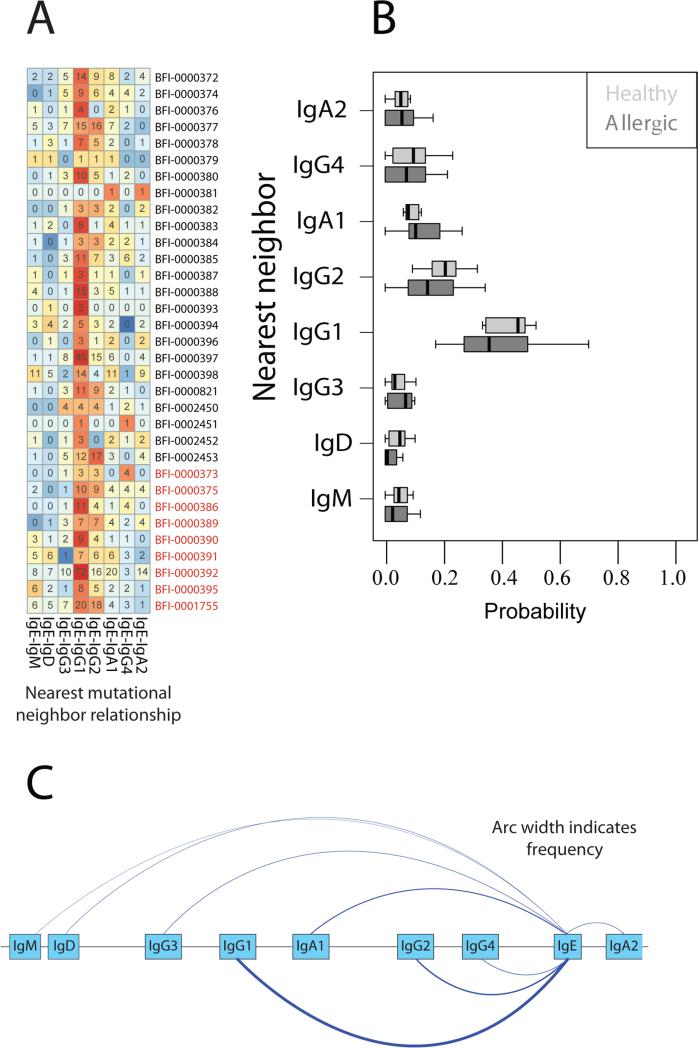
Figure 2.
(A) Number of IgE nearest isotype neighbor events with each non-IgE isotype, observed in multi-isotype B cell clonal lineages. Numbers in the heat map grid indicate the number of times a given nearest-neighbor relationship was observed in the data. Coloring in the heat map indicates the proportion of nearest neighbor relationships to IgE for each isotype, with blue indicating low proportion and red indicating high proportion. Each row represents a different individual, IgG1 is usually the isotype with the closest mutational relationship to IgE. Patient identifier codes from allergic individuals are highlighted in red. (B) Nearest isotype neighbors for IgE. Probabilities indicate the relative frequencies for each other isotype to be the nearest neighbor to IgE within multiple-isotype lineages. (C) A model for frequencies of class switching to IgE in healthy individuals based on the data in Figure 2B. The width of each arc indicates the probability of a nearest mutational neighbor between each isotype pair.
The frequency at which any two isotypes are found to be nearest mutational neighbors within IgE-containing multi-isotype lineages is shown in Figure 2B. For both healthy and allergic individuals, the greatest fraction of IgE forms nearest-neighbor relationships with switched isotypes IgG or IgA (approximately 90% of IgE lineages) while a smaller fraction has nearest neighbors of IgM or IgD (5% and 3% respectively). Among switched isotypes, the nearest neighbors to IgE were, in decreasing order, IgG1 (38%), IgG2 (16%), IgA1 (11%), IgG4 (10%), IgA2 (7%), and IgG3 (6%). Although allergic individuals had on average twice the number of IgE positive lineages as healthy individuals (p=0.01, two-sided Student's t-test), inferred isotype switching frequencies were similar for both groups. A model of isotype switching to IgE based on the aggregate data is shown in Figure 2C. For a significant minority of IgE relationships, IgM/D and IgE were most closely related to each other, implying that IgM/D to IgE switching may also contribute to the human IgE-expressing B cell pool, at least in peripheral blood B cells. We note that in Figure 2B, the probability of nearest-neighbor relationships between IgM or IgD and IgE are calculated separately.
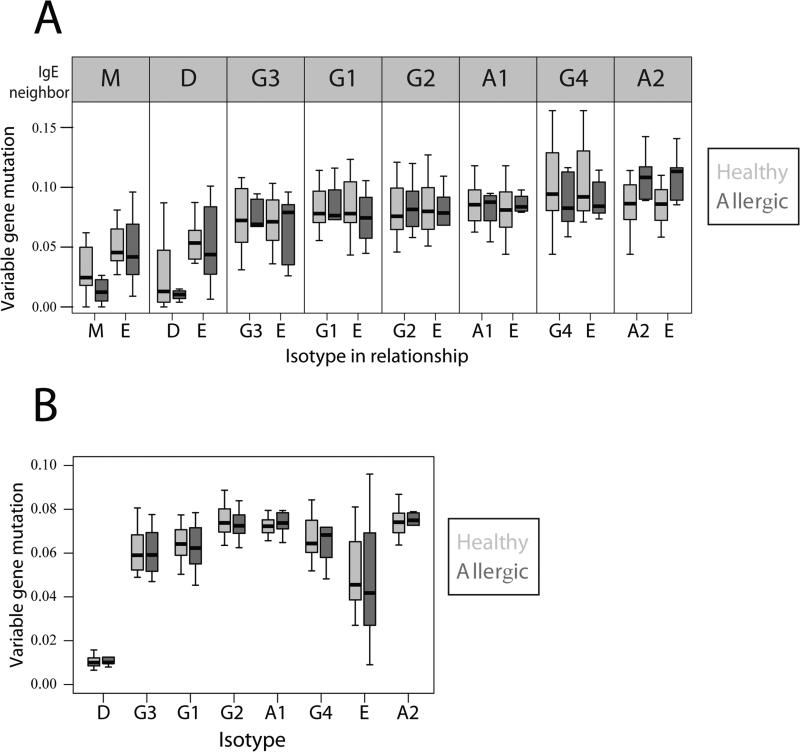
An important question in the study of allergic disease is the origin of highly mutated IgE. Studies in mice have supported different models, either that IgE expressing B cells may undergo somatic hypermutation, or else that highly-mutated IgE is derived from the IgG pool with little or no somatic hypermutation occurring once a cell has switched to IgE26. To examine the relationship between IgE mutation level and isotype switching origin, we calculated the average mutation level of the sequences involved in a nearest neighbor relationship with IgE, along with the mutation level of the cognate IgE. We find that IgE that is a nearest neighbor to IgM or IgD is significantly less mutated than IgE involved in a nearest neighbor relationship with switched isotypes (Figure 3A), such that IgE showing a somatic hypermutation rate of 7% or more almost always has an IgG or IgA nearest neighbor, while IgE having less than 7% mutation usually has either IgM/IgD as the nearest neighbor. Within the IgM/D-IgE relationships, the IgM/D is usually considerably less mutated than the cognate IgE, implying that IgE may have undergone additional hypermutation following direct switching from IgM/D. An alternative explanation would be that switching to IgE occurred via other unobserved isotype intermediates in these cases. While our data cannot rule out the possibility of unobserved intermediate isotypes, we note that IgG3, which is directly downstream from IgM/D in the genome, and must be the result of direct isotype switching from IgM/D, shows a similar mutational distance between the switched isotype and its nearest IgM neighbor in clonal lineages where both are observed, suggesting that significant accumulation of mutations may occur after isotype switching to IgG3 (Figure 3B). In contrast to the cases where IgE has a IgM/D nearest neighbor, IgE sequences whose nearest mutational neighbor is IgG or IgA show mutation levels very similar to the IgG or IgA sequences (Figure 3A), implying that little if any additional somatic hypermutation occurs after these switching events, as might be expected if switching to IgE occurred outside of the germinal center. We note that our data do not permit one to track the accumulation of somatic mutations in IgE-expressing B cells over time.
Figure 3.
(A) Average IGHV gene mutation frequency for members of IgE nearest neighbor relationships. Panels indicate the average mutation level for members of the indicated nearest-neighbor pair. (B) Average variable gene mutation frequency for sequences that are nearest neighbors to IgM. There are no significant differences between allergic and healthy individuals for these metrics after accounting for multiple hypothesis testing.
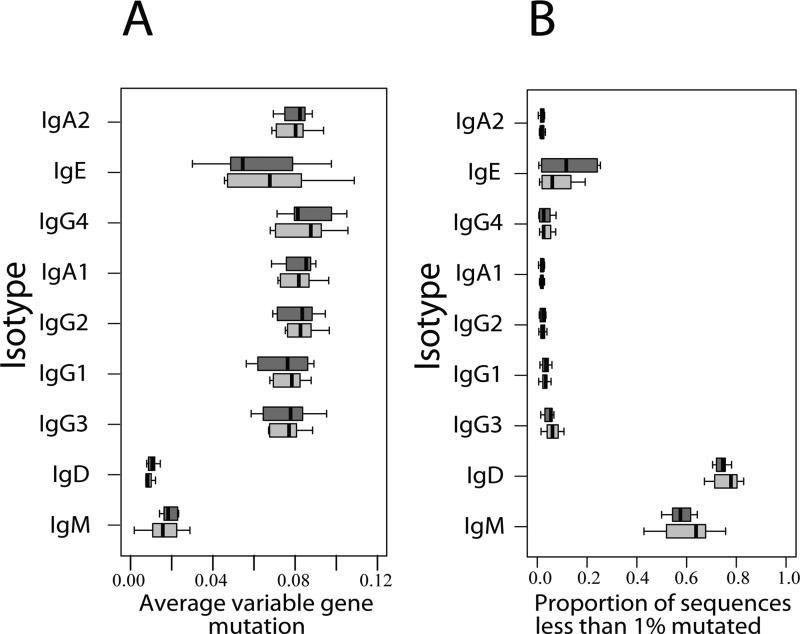
Finally, we sought to determine whether the clonal origins of IgE suggested by our analysis are supported by the overall level of variable gene mutation observed for each isotype. We find that mutation frequency generally increases in proportion to the distance of the isotype gene from the IgM/D locus (Figure 4A), suggesting that isotypes further downstream in the IgH locus are produced by cells that have undergone more rounds of cell division and somatic mutation than isotypes proximal to the IgM/D locus. As might be expected from this last result, we also find that the proportion of germline sequences of each isotype decreases in proportion to the distance of the isotype from the IgM/D locus (Figure 4B). The exceptions to these trends are IgE and, to a lesser extent, IgA2, which show lower levels of somatic mutation than upstream isotypes and, in the case of IgE, a higher proportion of germline sequences. This is a consistent with a scenario where the bulk of IgE arises indirectly from B cells expressing mutated IgG or IgA1, while a sizeable minority of the IgE pool is derived directly from IgM/D and consequently has undergone little or no somatic hypermutation.
Figure 4.
(A) Average IGHV gene mutation frequency per isotype. (B) Proportion of isotype sequences having less than 1% mutation in IGHV.
Discussion
At a molecular level, the frequency of switching between isotypes is determined by the chromatin state of the isotype switch regions, the distance between the switch regions (which is negatively correlated with switch synapse frequency), the DNA sequence of the switch regions themselves, and regulation of the enzymatic machinery that carries out switching27. To examine the origins of IgE expressing B cells in humans, we evaluated evidence for isotype switching events by identifying inherited patterns of somatic hypermutation among clonally related heavy chain Ig sequences expressed as different isotypes. The results suggest that indirect isotype switching to IgE from other switched isotypes, particularly IgG1, is the predominant pathway in healthy and allergic human peripheral blood B cells, and that there are not gross differences between allergic and healthy individuals in this aspect of IgE biology. It is currently unknown what fraction of an allergic patient's IgE repertoire is directly responsible for their allergic symptoms, and our results do not preclude the possibility that there may be individual antibody lineages important for disease pathogenesis that differ from the predominant patterns we describe.
Several observations in mice and humans further support the notion that indirect isotype switching to IgE predominates. Studies of Sμ-Sε switch circle junctions have identified fragments of IgG switch regions, or switch fragments from other isotype intermediates4, 6, 28, 29,30. While the literature also contains reports of primarily direct switching to IgE based on the analysis of switch junctions31, 32, it is important to note that only a fraction of excision recombination events from indirect isotype switching will leave evidence of the intermediate switch fragment, and indeed the precise frequency at which switch fragments remain following a switch event is not known. Therefore, while studies of switch regions can provide decisive evidence for indirect class switching, they are less suited to measuring the rate of direct switching. In addition, studies of IgE maturation in mice using fluorescence microscopy and flow cytometry indicate that IgE expressing cells are underrepresented in germinal centers, and that switching to IgE is antagonized by germinal center factors13, 33, 34. These findings have been supported by more recent studies employing IgE reporter mice, where observations suggest that IgE cells are rare and short-lived in germinal centers and that memory and germinal center IgG1 expressing B cells may contribute substantially to IgE recall responses11, 12, 35, 36. Our data from human peripheral blood B cells identifies a predominance of IgE with moderate to high levels of mutation ( >7% mutation). In these cases, the IgE is almost always closely related in mutational state to an IgG or IgA1 precursor, when members of the B cell clone expressing non-IgE isotypes are detected. This is consistent with a scenario where switching to IgE often occurs from affinity-matured isotype intermediates, outside of the germinal center, with little subsequent somatic mutation.
Our data suggest that direct IgM/D to IgE switching in humans occurs at a substantially lower frequency than IgG and IgA1 to IgE switching. In these cases, the cognate IgE is often more mutated than the IgM/D precursor, suggesting that affinity maturation of the IgE-expressing B cell may occur following a direct switching event. These instances of putative direct switching are associated with IgE that is less mutated than IgE inferred to have derived from indirect switching, consistent with the notion that the highest affinity IgE may be derived from IgG or IgA precursors37. We cannot fully exclude the possibility that apparent cases of IgM/D to IgE direct switching might have actually involved indirect switching via an unobserved isotype intermediate. Similarly, a conceivable but unlikely alternate scenario for apparent indirect isotype switching relationships would be that an unobserved highly mutated IgM-expressing B cell gave rise to daughter cells that switched to IgE, and others that switched to the apparent intermediate isotype. We do not see evidence of such highly mutated IgM clone members in the relatively deep sequencing coverage of IgM and other isotypes in this study. The data we have analyzed also do not directly address the origin of IgE B cell clonal lineages where no members of the clone expressing other isotypes were observed.
Our results indicate that indirect isotype switching from IgG4 to IgE contributes to the overall IgE repertoire, but is not the predominant source of IgE. This may seem counterintuitive given the conceptual links and reported correlations in serum levels of IgG4 and IgE proteins in the literature38. The contribution of any isotype to the IgE pool is determined by the frequency at which B cells of that isotype undergo switching to IgE, but also the overall abundance of B cells of that isotype within the body. In light of the rarity of IgG4-expressing B cells within the IgG pool (1.5% of total IgG-expressing B cells on average, in these data), their contribution to the IgE pool is significant, and may be of special importance in allergy or response to immunotherapy.
We note that our analysis is based on peripheral blood B cells. Cell-extrinsic signals regulate the tendency of switched B cells to reside in a particular tissue or migrate to distant tissues, which could affect the detection of isotype switching events in a given tissue source. For example, there is evidence that IgA and IgG expressing B cells can have trafficking properties that depend on both their site of induction and the presence of chemokines and adhesion/homing receptor engagement39. In this context, one important unresolved question is the extent to which frequencies of isotype switching to IgE differ among different tissues, particularly those such as the gut, respiratory tract, and skin, where allergen molecules are likely to be encountered. The results of the present study, favoring the indirect isotype switching route for most IgE in humans, provide a starting point for further investigations into the allergen-specific lineages whose IgE members are responsible for human allergic disease.
Supplementary Material
Key Messages.
Somatic mutation patterns in antibody heavy chain genes of B cell clones containing IgE-expressing members can be evaluated for evidence of direct (from IgM or IgD) versus indirect (from an intermediate switched isotype) isotype switching.
Both allergic and healthy individuals give support for indirect switching, particularly from IgG1, as the predominant pathway to IgE expression in humans.
Acknowledgements
The investigators thank our study volunteers for their participation, Sally Mackey for project, regulatory and data management, Research Nurses Sue Swope and Nancy Mastman, Research Assistants Ashima Goel and Thu Quan, and Phlebotomist Michele Ugur.
Funding sources: The project was supported by NIH NIAID grants 1U19AI0420901 (S. Boyd, PD), U19AI090019 (M. Davis, PI, S. Boyd, PD), and NIH/NCRR CTSA award UL1 RR025744.
Abbreviations
- PBMC
Peripheral Blood Mononuclear Cell
- Ig
immunoglobulin
- IGH
immunoglobulin heavy chain gene
- IGHV
Immunoglobulin Heavy Chain V
- IGHD
Immunoglobulin Heavy Chain D
- IGHJ
Immunoglobulin Heavy Chain J
- CDR3
Complementarity Determining Region 3
- FR3
Framework region 3
- FR4
Framework region 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, et al. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 2.Janeway Cea. Immunobiology. 5th ed Garland Science; New York: 2001. [Google Scholar]
- 3.Jabara HH, Loh R, Ramesh N, Vercelli D, Geha RS. Sequential switching from mu to epsilon via gamma 4 in human B cells stimulated with IL-4 and hydrocortisone. J Immunol. 1993;151:4528–33. [PubMed] [Google Scholar]
- 4.Mills FC, Thyphronitis G, Finkelman FD, Max EE. Ig mu-epsilon isotype switch in IL-4-treated human B lymphoblastoid cells. Evidence for a sequential switch. J Immunol. 1992;149:1075–85. [PubMed] [Google Scholar]
- 5.Stoep Nea Van der. In vivo and in vitro IgE isotype switching in human B lymphocytes: evidence for a predominantly direct IgM to IgE class switch program. Eurpoean Journal of Immunology. 1994;24:1307–11. doi: 10.1002/eji.1830240610. [DOI] [PubMed] [Google Scholar]
- 6.Cameron L, Gounni AS, Frenkiel S, Lavigne F, Vercelli D, Hamid Q. S epsilon S mu and S epsilon S gamma switch circles in human nasal mucosa following ex vivo allergen challenge: evidence for direct as well as sequential class switch recombination. J Immunol. 2003;171:3816–22. doi: 10.4049/jimmunol.171.7.3816. [DOI] [PubMed] [Google Scholar]
- 7.Baskin B, Islam KB, Evengard B, Emtestam L, Smith CI. Direct and sequential switching from mu to epsilon in patients with Schistosoma mansoni infection and atopic dermatitis. Eur J Immunol. 1997;27:130–5. doi: 10.1002/eji.1830270120. [DOI] [PubMed] [Google Scholar]
- 8.Aea Erazo. Unique maturation program of the IgE response in vivo. Immunity. 2007;26:191–203. doi: 10.1016/j.immuni.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J-S, Meyer-Hermann M, Xiangying D, Zuan LY, Jones LA, Ramakrishna L, et al. The distinctive germinal center phase of IgE(+) B lymphocytes limits their contribution to the classical memory response. The Journal of Experimental Medicine. 2013;210:2755–71. doi: 10.1084/jem.20131539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talay O, Yan D, Brightbill HD, Straney EE, Zhou M, Ladi E, et al. IgE(+) memory B cells and plasma cells generated through a germinal-center pathway. Nat Immunol. 2012;13:396–404. doi: 10.1038/ni.2256. [DOI] [PubMed] [Google Scholar]
- 11.Talay O, Yan D, Brightbill HD, Straney EEM, Zhou M, Ladi E, et al. IgE+ memory B cells and plasma cells generated through a germinal-center pathway. Nat Immunol. 2013;14:1302–4. doi: 10.1038/ni.2770. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Sullivan Brandon M, Allen Christopher DC. Fluorescent In Vivo Detection Reveals that IgE+ B Cells Are Restrained by an Intrinsic Cell Fate Predisposition. Immunity. 2012;36:857–72. doi: 10.1016/j.immuni.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361:2046–55. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies JM, Platts-Mills TA, Aalberse RC. The enigma of IgE+ B-cell memory in human subjects. J Allergy Clin Immunol. 2013;131:972–6. doi: 10.1016/j.jaci.2012.12.1569. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Liu Y, Xu LT, Jackson KJ, Roskin KM, Pham TD, et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J Immunol. 2014;192:603–11. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 20.Barbee RA, Halonen M, Lebowitz M, Burrows B. Distribution of IgE in a community population sample: correlations with age, sex, and allergen skin test reactivity. J Allergy Clin Immunol. 1981;68:106–11. doi: 10.1016/0091-6749(81)90167-6. [DOI] [PubMed] [Google Scholar]
- 21.Gergen PJ, Arbes Sj, Jr, Calatroni A, Mitchell HE, Zeldin DC. Total IgE levels and asthma prevalence in the US population: results from the National Health and Nutrition Examination Survey 2005-2006. J Allergy Clin Immunol. 2009;124:447–53. doi: 10.1016/j.jaci.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton RG. Proficiency survey-based evaluation of clinical total and allergen-specific IgE assay performance. Arch Pathol Lab Med. 2010;134:975–82. doi: 10.5858/2009-0518-OA.1. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton RG, Adkinson NF., Jr. Clinical laboratory assessment of IgE-dependent hypersensitivity. J Allergy Clin Immunol. 2003;111:S687–701. doi: 10.1067/mai.2003.123. 23. [DOI] [PubMed] [Google Scholar]
- 24.Sunyer J, Anto Jm, Castellsague J, Soriano JB, Roca J. Total serum IgE is associated with asthma independently of specific IgE levels. Int J Epidemiol. 1999;28:728–34. [Google Scholar]
- 25.Wittig HJ, Belloit J, De Fillippi I, Royal G. Age-related serum immunoglobulin E levels in healthy subjects and in patients with allergic disease. J Allergy Clin Immunol. 1980;66:305–13. doi: 10.1016/0091-6749(80)90026-3. [DOI] [PubMed] [Google Scholar]
- 26.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA, Lafaille JJ. Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med. 2012;209:353–64. doi: 10.1084/jem.20111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–22. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills FC, Mitchell MP, Harindranath N, Max EE. Human Ig S gamma regions and their participation in sequential switching to IgE. J Immunol. 1995;155:3021–36. [PubMed] [Google Scholar]
- 29.Xiong H, Curotto de Lafaille MA, Lafaille JJ. What is unique about the IgE response? Adv Immunol. 2012;116:113–41. doi: 10.1016/B978-0-12-394300-2.00004-1. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida K, Matsuoka M, Usuda S, Mori A, Ishizaka K, Sakano H. Immunoglobulin switch circular DNA in the mouse infected with Nippostrongylus brasiliensis: evidence for successive class switching from mu to epsilon via gamma 1. Proc Natl Acad Sci U S A. 1990;87:7829–33. doi: 10.1073/pnas.87.20.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Stoep N, Korver W, Logtenberg T. In vivo and in vitro IgE isotype switching in human B lymphocytes: evidence for a predominantly direct IgM to IgE class switch program. Eur J Immunol. 1994;24:1307–11. doi: 10.1002/eji.1830240610. [DOI] [PubMed] [Google Scholar]
- 32.Lin M, Du L, Brandtzaeg P, Pan-Hammarstrom Q. IgA subclass switch recombination in human mucosal and systemic immune compartments. Mucosal Immunol. 2014;7:511–20. doi: 10.1038/mi.2013.68. [DOI] [PubMed] [Google Scholar]
- 33.Erazo A, Kutchukhidze N, Leung M, Christ AP, Urban JF, Jr., Curotto de Lafaille MA, et al. Unique maturation program of the IgE response in vivo. Immunity. 2007;26:191–203. doi: 10.1016/j.immuni.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124:1289–302. e4. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oea Talay. IgE+ memory B cells and plasma cells generated through a germinal-center pathway. Nature Immunology. 2012;13:396–404. doi: 10.1038/ni.2256. [DOI] [PubMed] [Google Scholar]
- 36.He J-S, Narayanan S, Subramaniam S, Ho W, Lafaille J, Curotto de Lafaille M. Biology of IgE Production: IgE Cell Differentiation and the Memory of IgE Responses. In: Lafaille JJ, Curotto de Lafaille MA, editors. IgE Antibodies: Generation and Function. Springer International Publishing; 2015. pp. 1–19. [DOI] [PubMed] [Google Scholar]
- 37.Aalberse RC, Platts-Mills TA. How do we avoid developing allergy: modifications of the TH2 response from a B-cell perspective. J Allergy Clin Immunol. 2004;113:983–6. doi: 10.1016/j.jaci.2004.02.046. [DOI] [PubMed] [Google Scholar]
- 38.Rea Aalberse. Immunoglobulin G4: an odd antibody. Clinical and Experimental Allergy. 2009;39:469–77. doi: 10.1111/j.1365-2222.2009.03207.x. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel EJ, Butcher EC. Plasma-cell homing. Nat Rev Immunol. 2003;3:822–9. doi: 10.1038/nri1203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.