Abstract
The human paracaspase MALT1 is a caspase homolog that plays a central role in NF-κB signaling. Over the past few years it has become clear that this is due to a combination of its scaffolding and proteolytic function. Knockout mice and mice expressing a catalytically dead variant of the protease have provided valuable information. This review aims to provide an overview of recent developments regarding the enzymatic mechanism and specificity of MALT1, its substrates discovered to date, different mouse models, as well as the role of MALT1 in NF-κB signaling downstream of a variety of different receptors.
Keywords: NF-κB signaling, Protease, Caspase, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1)
1. Introduction
1.1 MALT lymphoma
MALT (mucosa-associated lymphoid tissue) lymphoma is an extranodal malignancy of mucosa-associated lymphoid tissue caused by chronic inflammation [1]. The term was first coined in 1984 when Isaacson and Wright discovered that lymphomas of different sites were all of MALT origin [2, 3] – Figure 1. MALT lymphoma is the third most common form of non- Hodgkin lymphoma, and its incidence has shown a steady rise over the past two decades [1]. A large variety of sites can be affected including the stomach (70% of cases), lung (14%), ocular adnexa (12%), thyroid (4%), and small intestine (1%) [3].
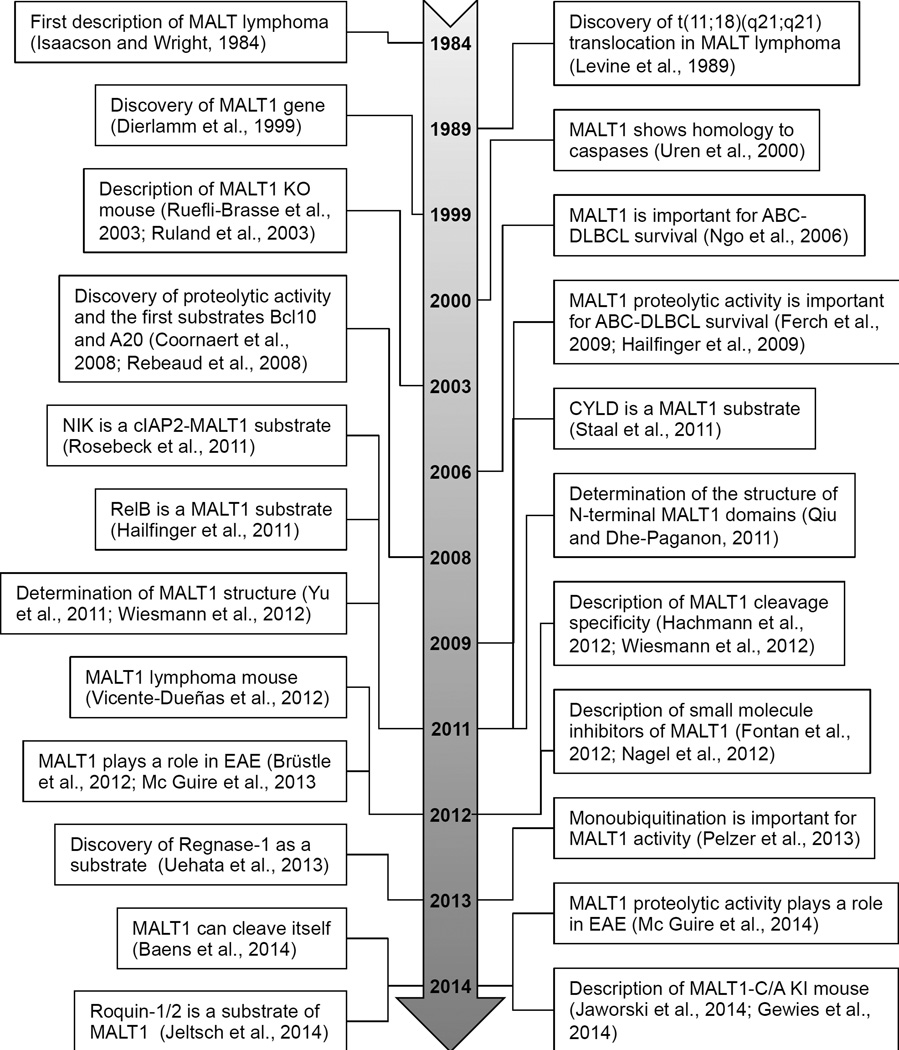
Figure 1. Three decades of MALT1 research.
See text for details. ABC-DLBCL, activated B-cell-like diffuse large B-cell lymphoma; EAE, experimental autoimmune encephalomyelitis (mouse model of multiple sclerosis); KI, knock in; KO, knockout.
MALT lymphoma can be caused both by bacterial infections like Helicobacter pylori (H. pylori) and autoimmune diseases like Sjögren’s syndrome and Hashimoto’s disease [1, 3]. Consistent presence of an infectious agent or autoimmune disease leads to chronic immune stimulation and inflammation with B cells trying to fight the infection stimulated by T cells. The accumulation of immune cells leads to the formation of acquired MALT and, under certain conditions, to MALT lymphoma. This usually indolent type of lymphoma can often be cured by stringent antibiotic therapy. However, at later stages genetic translocations often occur, and the treatment becomes more difficult [4].
The t(11;18)(q21;q21) translocation [5] is the most common chromosomal abnormality in lymphomas of mucosa-associated lymphoid tissue and occurs with frequencies varying widely from as high as 40% in the lung and 25% in the stomach, while being absent or rare in the salivary gland, thyroid, and skin. Other activating mutations include t(14;18)(q32;q21) and t(1;14)(p22;q32) [1].
1.2 Discovery of the MALT1 gene
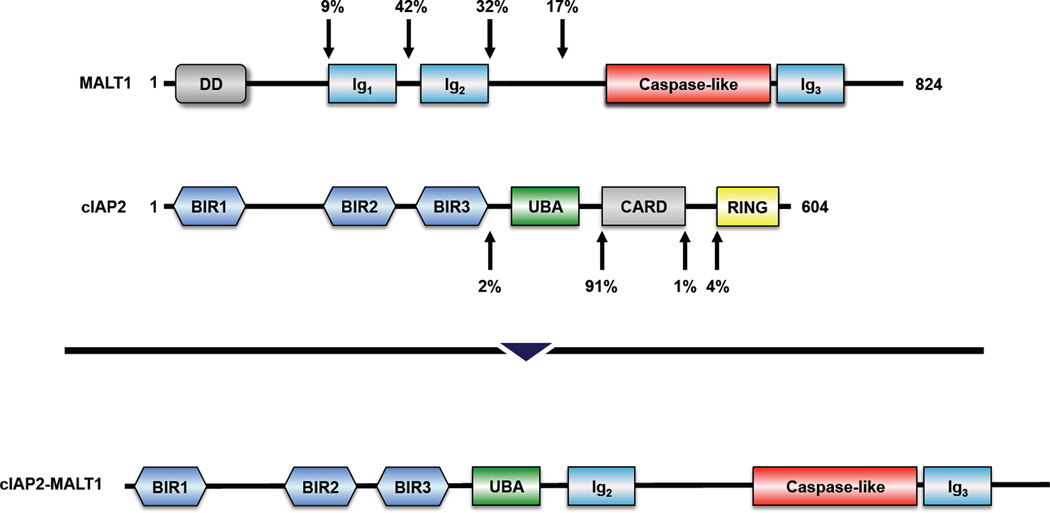
In 1999, it was discovered that the t(11;18)(q21;q21) translocation involves the gene of the cellular inhibitor of apoptosis protein 2 (cIAP2, also known as apoptosis inhibitor protein 2, API2) and an until then unknown gene named MLT (MALT lymphoma-associated translocation) [6], which was later renamed MALT1 (MALT lymphoma translocation protein 1) – Figure 2. cIAP2 was found to contribute the N-terminal part of the resulting cIAP2-MALT1 protein and MALT1 the C-terminal part [6].
Figure 2. Domain structure of MALT1, cIAP2, and the most common variant of cIAP2-MALT1.
The percentages represent the frequencies with which the genes break at these positions. The percentages are adapted from [3]. BIR, baculovirus inhibitor of apopotosis protein repeat domain; CARD, caspase recruitment domain; DD, death domain; Ig, immunoglobulin-like domain; RING, really interesting new gene domain; UBA, ubiquitin-associated domain.
1.3 MALT1 – a caspase homolog
Characterization of the MALT1 gene early on led to the seminal paper by Dixit and colleagues in 2000 [7] who discovered that the C-terminal part of the fusion contains a domain homologous to the caspases, and hence defined MALT1 as a “paracaspase”. In addition to the putative caspase domain MALT1 was revealed to contain a death domain (DD) and several immunoglobulin-like (Ig-like) domains. Based on sequence homology, paracaspase orthologs were found in different metazoans including humans, zebrafish, C. elegans, and the slime mold D. discoideum. More distantly related proteins categorized as metacaspases were found in several plant and fungal species as well as the protist P. falciparum. All proteins universally contain the cysteine and histidine residues belonging to the catalytic dyad of the caspase family [7] and were placed in the protease clan CD [8, 9], which includes evolutionarily related cysteine proteases (see the MEROPS database [8] for extensive details of phylogenetic relationships).
1.4 Discovery of MALT1 proteolytic activity
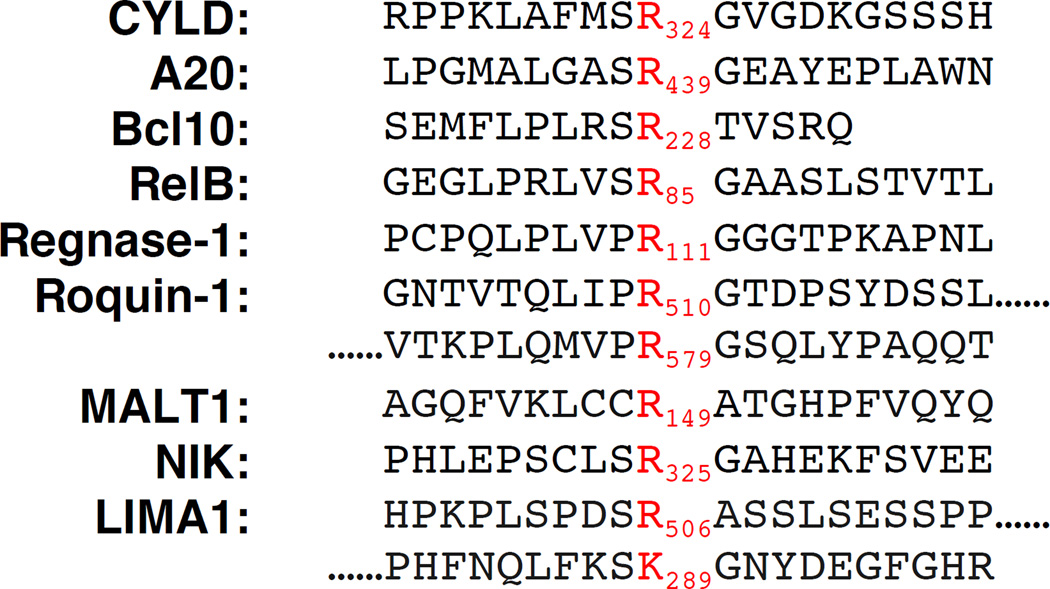
Despite its membership in the clan CD family, demonstrating proteolytic activity for MALT1 proved difficult. After initial unsuccessful attempts [10], it was not until 2008 that the first two substrates were discovered [11, 12], followed by additional substrates over the next few years. All substrates known to date are cleaved directly C-terminal of an arginine residue with the exception of LIMA1, which has been reported to be cleaved at two sites, one of which is Cterminal to a lysine residue – Figure 3.
Figure 3. Cleavage sites of the human MALT1 substrates reported to date.
1.5 The structure of MALT1
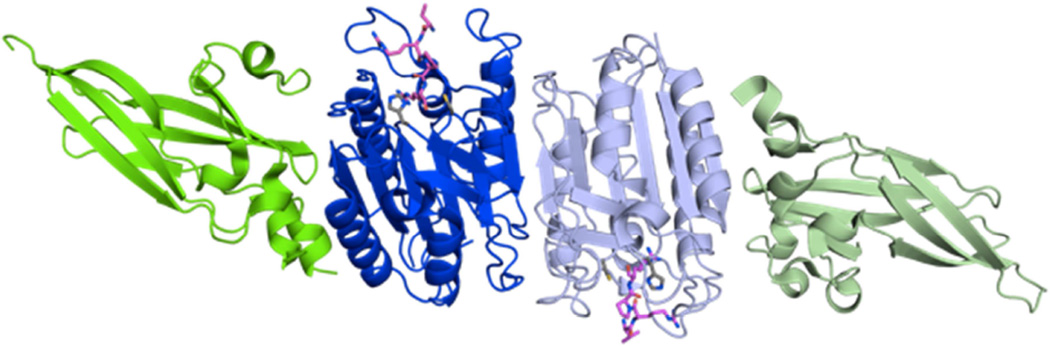
The crystal structure of MALT1 was solved in the absence as well as presence of the inhibitor Z-VRPR-FMK by two groups simultaneously [13, 14] – Figure 4.
Figure 4. Crystal structure of MALT1 in the presence of the inhibitor Z-VRPR-FMK.
MALT1 (aa 329–719) crystallizes as a dimer comprising the catalytic domain (dark and pale blue) as well as the C-terminal Ig3 domain (dark and pale green). The inhibitor Z-VRPR-FMK is shown in magenta, and the active site residues are shown in gray. PDB: 3UO8.
As expected from the dependence of MALT1 on kosmotropic salts for in vitro activity [15], the enzyme crystallized as a dimer. The overall structure of the catalytic domain displayed high similarity to the caspases with the typical six-stranded β-sheet surrounded by five α-helices.
In order to obtain crystals, it was necessary to express a construct containing both the catalytic domain and the adjoining Ig3 domain. A large hydrophobic contact area was observed between the Ig3 and protease domains rendering the catalytic domain alone unstable and relying on the Ig3 domain for stabilization. This large hydrophobic patch helps explain the high tendency of the catalytic domain alone to aggregate and the fact that only a small fraction of this construct was found to be active [13, 15].
In addition to the catalytic and Ig3 domain, the structures Ig1 and Ig2 domains as well as the CARD domain were also solved [16].
1.6 MALT1 activation mechanism
Interestingly, despite its dimeric state the enzyme crystalized in an inactive conformation in the absence of inhibitor. Several structural elements prevent its catalytic activity. The L2 loop harboring the active site is in a disordered position, and the sulfur ion of the catalytic Cys464 is not in the catalytically competent position and therefore cannot form the catalytic dyad with His415. The L3 loop, which is essential for the formation of the catalytic groove, is flexible and positioned in a way that blocks the entrance of substrates into the S1 pocket. Inhibitor binding triggers reordering, especially of the L2 and L3 loops, causing a stabilized L2 loop and formation of the active site and catalytic dyad [13].
The fact that a catalytically competent conformation is obtained only in the presence of inhibitor is noteworthy. Caspases are either activated via intersubunit linker cleavage of a preassembled inactive dimer (executioner caspases) or through dimerization with subsequent optional stabilization through cleavage of the intersubunit linker (initiator caspases)[17]. For MALT1, no cleavage in the region corresponding to the intersubunit linker is observed in the crystal structure or after incubation in activating kosmotropic buffer [13, 15], yet dimerization alone does not cause it to assume its active state. At least in vitro MALT1 seems to represent a third activation mechanism requiring dimerization and additional stabilization either through substrate/inhibitor binding or kosmotropic salts, which are most likely responsible not only for the dimerization of MALT1 but also the reordering and stabilization of active-site loops.
1.7 MALT1 active site
The interactions of the inhibitor with the subsite pockets explain the substrate specificity preferences in the P4-P1 positions (Schechter and Berger nomenclature [18]), which was determined to be LVSR or LISR, respectively, using positional scanning libraries [13, 15]. The S1 pocket contains three negatively charged residues that can optimally interact with Arg, whereas Lys with a carbon atom in the place of a nitrogen atom would not have the ability to undergo any of these interactions. The direct comparison of the P1 pocket of MALT1 with that of its caspase relatives shows that it is not a simple charge reversal that explains the diametrically opposite specificity in this position. The much larger arginine side chain compared to the aspartate preferred by caspases necessitates a larger S1 pocket, and consequently the substrate charges are countered by amino acids in structurally different positions [13].
The S2 pocket is relatively small, corresponding to the requirement for small amino acids in this position. The S3 pocket makes no specific contacts with the inhibitor, supporting the substrate library scanning results showing little influence of this position on the specificity of MALT1. The S4 pocket is hydrophobic, leading to a preference for similarly uncharged and hydrophobic residues in the P4 position of the substrate, which is exemplified by the interactions of the hydrophobic Val of the inhibitor with this pocket.
A systematic mapping of the prime positions C-terminal to the cleavage site has not been undertaken so far. However, the natural substrates discovered to date (Figure 3) suggest that small amino acids are preferred, glycine over all, as the majority of the substrates contain this amino acid in the P1’ position – a property shared with caspases [19].
2. NF-κB signaling
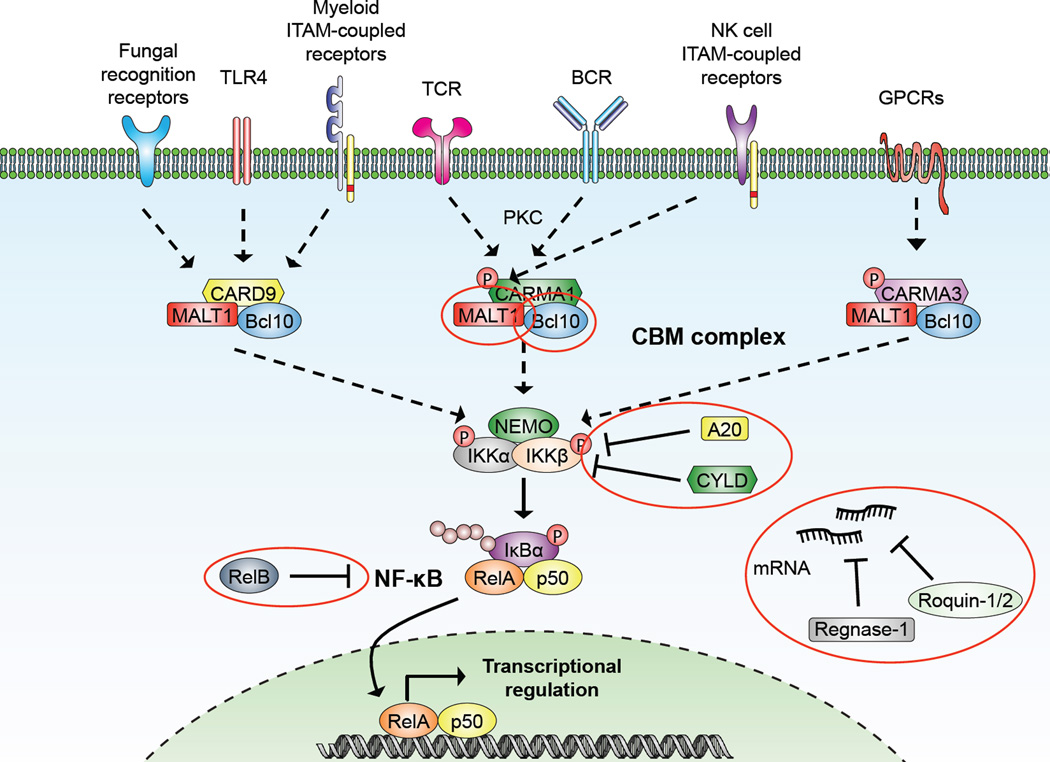
Immediately upon its discovery, evidence surfaced that MALT1 might play a role in NF-κB signaling [7] – Figure 5. The transcription factor NF-κB is a crucial regulator of lymphocyte activation, survival, and proliferation that signals through many different receptors including the T cell antigen receptor (TCR) and B cell antigen receptor (BCR), the tumor necrosis factor receptor (TNFR), and the B-cell-activating factor (BAFF) receptor. Depending on the receptor involved, the pathway is generally divided into the canonical and noncanonical pathway, both of which involve a signaling cascade that culminates in the relocation of transcription factors of the NF-κB family to the nucleus and the regulation of target gene transcription. The readers are referred to a number of extensive reviews on this subject [20–22]
Figure 5. MALT1 involvement in canonical NF-κB signaling downstream of different receptors.
See main text for explanations, abbreviations, and references. Dashed arrows represent additional signaling events and substrates are shown in red circles. The substrates NIK and LIMA1, which are exclusively cleaved by cIAP2-MALT1 are not depicted.
3. Knockout mice
3.1 Importance of CARMA1 and Bcl10
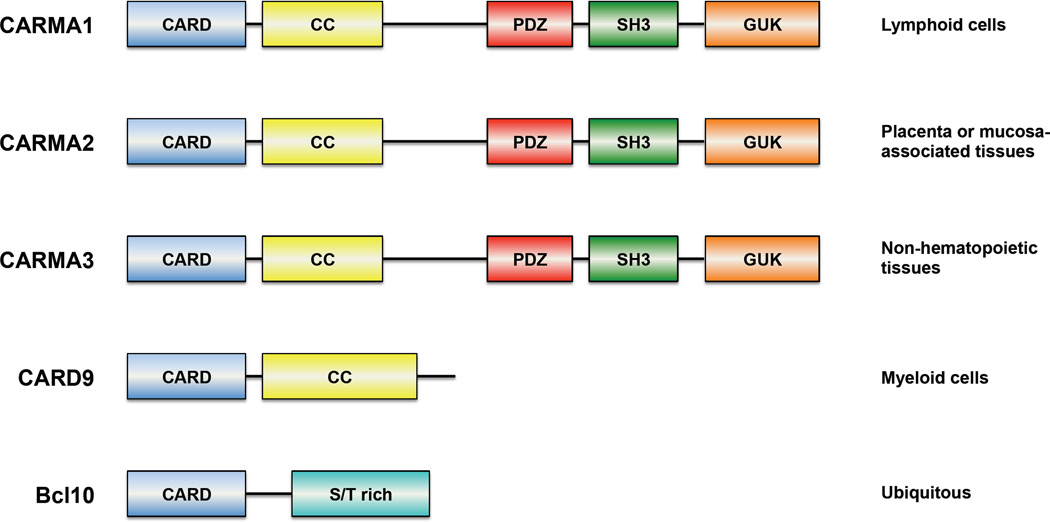
The activation of canonical NF-κB signaling in in T and B cells involves the activation of PKC-θ and PKC-β, respectively, which leads to the subsequent phosphorylation of the adaptor protein CARMA1 (CARD-containing MAGUK protein 1, also known as CARD11) [23], which next to a CARD domain contains a PDZ (PSD95-Dlg1-zonula occludens-1 protein), an SH3 (Src homology 3) and a GUK (guanylate kinase) domain as well as coiled-coil domain – Figure 6. Activation of CARMA1 leads to the recruitment of the CARD-containing adaptor protein Bcl10 through homotypic CARD-CARD interactions [24]. The importance of CARMA1 and Bcl10 in NF-κB signaling became evident from the CARMA1−/− and Bcl10−/− mice. Besides gross developmental problems of the Bcl10−/− mouse including a neural tube closure defect in a subset of animals leading to embryonic lethality, surviving animals displayed similarly impaired NF-κB signaling downstream of the TCR and BCR as the CARMA1−/− mouse [25–27]. Bcl10 and CARMA1 interaction was shown to be essential for NF-κB signaling through the TCR to the downstream IKK complex [25, 28, 29]. Since the association between Bcl10 and MALT1 had been discovered early on [7], it was speculated that MALT1 might provide the missing link to downstream effector proteins of the pathway and gene ablation studies were carried out.
Figure 6. CARD-containing members of the different CBM complexes and their tissue distributions.
CARD, caspase recruitment domain; CC, coiled coil domain; GUK, guanylate kinase; PDZ, PSD95-Dlg1- zonula occludens-1 protein; SH3, Src homology domain 3; S/T rich, serine and threonine-rich domain. See main text for references.
3.2 The MALT1−/− phenotype
In 2003, two independent groups published their findings on the MALT1−/− mouse [30, 31]. The mice were born at the expected Mendelian ratios, were viable and fertile, and appeared to be healthy with normal numbers and frequencies of B cells found in the bone marrow [30, 31]. The total number of T cells and the distribution of the CD4 and CD8 subsets in the spleen, lymph nodes, and thymus were found to be comparable between MALT1+/+ and MALT1−/− mice [30, 31], but a decrease of activated T cells in the periphery was observed [31]. Natural killer (NK) cells and natural killer T cells were not affected by the MALT1 knockout (KO) as opposed to the CARMA1−/− mouse, in which their development was impaired [26].
3.2.1 B cell development in MALT1−/− mice
Upon examination of splenic B cells, one research group observed no change in the ratio of IgD and IgM expressing B cells [31], while the other detected decreased numbers of IgMhighIgDlow B cells [30]. This B cell subtype can represent either transitional T1 B cells, marginal zone (MZ) B cells, or B1 B cells. Both groups determined in further examinations that the CD21highCD23low B cell subtype representing MZ cells was completely absent in the spleen. This corresponded with the finding that the MZ B cell area was barely visible in histological sections of in the spleen [30, 31]. Additionally, no B1 B cells were detected in the peritoneum, which matched the findings of the CARMA1−/− mouse [26, 27] as well as those of mice expressing inactivating mutant forms of CARMA1 [32, 33] and c-Rel−/− and NF-κB1−/− double KO mice [34], respectively. Taken together, these findings suggest that MALT1 is required for the development of MZ and B1 B cells but is dispensable for the development of normal B2 B cells [30, 31].
3.2.2 Antigen receptor signaling in MALT1−/− mice
When challenged with a T cell-dependent antigen, IgM and IgG1 production was strongly decreased [30] or completely absent [31] in MALT1−/− mice. The same downregulation was observed upon challenge with a T cell-independent antigen. Since this type of immune response depends mainly on MZ and B1 cells [35, 36] – the cell population decreased in the MALT1−/− mouse – the impaired immunoglobulin production was not surprising. Taken together, the findings point to defective antigen receptor signaling.
3.2.3 Immunoglobulin levels in MALT1−/− mice
Interestingly, basal serum immunoglobulin levels were decreased in MALT1−/− mice for all subtypes with IgM and IgG3 showing the most pronounced reduction [30, 31]. The decrease of these classes of immunoglobulins correlates well with the low numbers of B1 B cells, which have been reported to be the predominant producer of IgM and IgG3 [37].
3.2.4 T cell development in MALT1−/− mice
Detailed evaluation of the immature CD4−CD8−double negative (DN) thymocyte subset revealed variations in the developmental stages. While less DN3 CD25+CD44low cells were found, the population of DN4 CD25−CD44low was increased. Additionally, the subset of thymocytes expressing a rearranged TCRβ chain was increased with some cells even displaying a rearranged TCRα chain [30]. This pointed to premature maturation of the DN thymocytes since a rearranged TCRα chain is usually not displayed until the double positive CD4+CD8+ stage, which follows the DN stage [38, 39]. This phenotype was also observed in the Bcl10−/− mouse [31], providing further evidence that Bcl10 and MALT1 are part of the same pathway.
3.2.5 T cell proliferation in MALT1−/− mice
Stimulation of the TCR with anti-CD3 and anti-CD28 or a combination of PMA (phorbol 12-myristate 13-acetate) and ionomycin, which bypasses the receptor and directly activates PKC, led to decreased proliferation of MALT1−/− T cells compared to MALT1+/+ cells [30, 31]. Additionally, while interleukin-2 (IL-2) production was strongly enhanced in WT T cells, its upregulation was starkly decreased [30] or completely abrogated [31] in MALT1−/− cells. In line with this, WT T cells showed upregulated expression of CD25, the α chain of the IL-2 receptor, as well as CD44 and CD69, while this was abrogated in MALT1−/− cells [30, 31]. This phenotype could not be rescued by treatment with IL-2 [30, 31]. Furthermore, the viability of stimulated MALT1−/− cells was found to be decreased compared to MALT1+/+ cells, and the cells were smaller, which pointed to a proliferation block at an early time point before the initiation of cell growth [30]. Since PMA/ionomycin had the same effect as TCR stimulation in all of the described criteria, it was concluded that the proliferation defect was downstream of PKC [30, 31]. In addition, the MALT1 KO did not cause any appreciable effect on upstream signaling events downstream of the BCR or TCR, as evident from the fact that MALT1+/+ and MALT1−/− cells did not show a difference in total tyrosine phosphorylation. Hence, MALT1 was deduced to play a role further downstream in the pathway [30, 31]. Interestingly, some of the reported findings were dependent on T cell subtype, with CD4+ T cells showing a greater dependence on MALT1 than CD8+ T cells [40].
3.2.6 B cell proliferation in MALT1−/− mice
While both KO studies came to similar conclusions with regard to T cells, several differences were observed when elucidating B cell signaling. While Ruefli-Brasse et al. found the proliferation of splenic MALT1−/− B cells to be strongly impaired upon stimulation with various stimuli including anti-IgM and lipopolysaccharide (LPS), the latter of which signals through Tolllike receptors (TLR), Ruland et al. detected only minor differences [30, 31]. Interestingly, Bcl10−/− B cells show a proliferation defect after anti-IgM but not LPS stimulation [25], while the results for CARMA1−/− mice are ambivalent [26, 27].
3.2.7 NF-κB signaling after TCR and BCR stimulation in MALT1−/− mice
Upon examination of the effect on downstream signaling events, important differences were again observed between the two groups. While Ruefli-Brasse et al. found the degradation and phosphorylation of IκBα as well as DNA binding of NF-κB to depend on MALT1 with impairments seen in MALT1−/− B as well as T cells upon anti-IgM, anti-CD3/anti-CD28, or PMA and ionomycin treatment [30], Ruland et al. saw this impairment in MALT1−/− T cells but not in B cells [31], while the Bcl10−/− mouse showed defects in both cell types [25]. A closer look at BCR signaling revealed a complex picture in which the effects of MALT1 depend on the type of NF-κB involved. NF-κB signaling downstream of the BCR utilizes predominantly p65 (RelA) and c-Rel-containing dimers. Whereas MALT1 is essential for signaling involving c-Rel, p65 (RelA)-containing dimers continue to signal effectively in its absence [41].
3.2.8 Additional downstream signaling events after TCR and BCR signaling in MALT1−/− mice
Furthermore, Ruefli-Brasse et al. found c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) phosphorylation to be unaffected in MALT1−/− cells upon stimulation of TCR or BCR with anti-CD3/anti-CD28, anti-IgM, or PMA and ionomycin, suggesting that the mitogen-activated protein kinase (MAPK) and activator protein 1 (AP-1) signaling pathways are independent of MALT1 [30]. This was supported by normal levels of DNA binding observed for the AP-1 transcription factor after BCR and TCR stimulation [31]. The findings of Ruland et al. supported the results concerning ERK signaling, but in contrast showed JNK phosphorylation to be impaired in MALT1−/− T cells after PMA and ionomycin treatment. In addition, they found p38 phosphorylation to be similarly compromised under these conditions [31]. No impairment after stimulation of MALT1−/− T cells with the proinflammatory cytokines TNF-α [30, 31] or IL-1 [31] was observed, pointing to a receptor-specific role for MALT1.
3.2.9 Effect of the MALT1 KO on the development of experimental autoimmune encephalomyelitis
Recently, two independent groups described an important role for MALT1 in the development of experimental autoimmune encephalomyelitis (EAE), the main mouse model of human multiple sclerosis [42, 43]. While WT mice developed EAE in the experimental model, MALT1−/− mice were completely immune to the disease. No symptoms like demyelination, axonal degradation, or infiltration of inflammatory cells in the spinal cord were seen [43]. T helper 17 (TH17) cells along with the IL-17 and granulocyte-macrophage colony-stimulating factor (GM-CSF) cytokines produced by these cells have been reported to play an important role in EAE development, and all of these were reduced in MALT1−/− mice [42]. The importance of MALT1 for EAE development was substantiated by the fact that TH17 cells [42] or splenocytes [43] from MALT1−/− mice treated with EAE inducers and transplanted into the WT background failed to cause EAE in these mice. In contrast, TH17 cells or splenocytes from WT mice treated with these inducers and then transplanted into MALT1−/− mice did induce EAE [42, 43].
4. Different CBM complexes
The KO studies showed the importance of CARMA1, Bcl10, and MALT1 in NF-κB signaling. MALT1 and Bcl10 are expressed ubiquitously [44], but the expression of CARMA1 is mostly restricted to immune tissues [24]. It was discovered that while CARMA1 is the primary CARD-containing interactor of MALT1 and Bcl10 in B and T cells, other cell types and receptors utilize other scaffolding structures of the CARD family – Figure 6.
The protein CARD9 is expressed in a variety of tissues including spleen, liver, lung, peripheral blood monocytes, brain, and placenta. In these, it serves as the CARD-containing interactor of Bcl10. CARD9 contains an N-terminal CARD and a C-terminal coiled-coil domain. The interaction between Bcl10 and CARD9 depends on the CARD domains of the two proteins [45]
CARMA3 (also known as Bimp1 or CARD10) is another interactor of Bcl10. The protein is expressed ubiquitously and was found in all tissues examined including heart, brain, lung, liver, skeletal muscle, kidney, testes, and spleen with the highest expression in heart and kidney. Like CARMA1 and CARMA2 the protein is made up of five domains, a CARD at the N-terminus, followed by coiled-coil, PDZ, SH3, and GUK domains. Essentially the same findings as for the other CARD proteins were made again: interaction with Bcl10 mediated by the CARD domains and NF-κB activation upon overexpression [46]. Similar to Bcl10−/− mice, about half of CARMA3−/− mice display neural tube closure defects and die perinatally [47], providing further evidence that the proteins are involved in the same pathway.
CARMA2 was originally found to be expressed exclusively in the placenta [24], although this has recently been questioned, and a mucosal expression pattern was reported [48]. As for the other CARD family proteins, interaction with Bcl10 and NF-κB activation upon overexpression were observed. CARMA2 has the same domain structure as CARMA1 and CARMA3.
5. Additional receptors
As described, the different CARD proteins are expressed in a variety of tissues besides B and T cells, and over the years they were found to couple the Bcl10-MALT1 duo to a host of different receptors besides the T and B cell receptors – Figure 5.
5.1 BAFF receptor and noncanonical signaling
While the results concerning the involvement of MALT1 in BCR signaling remain ambiguous [30, 31], further elucidation of MALT1−/− mice showed that MALT1 is required for efficient BAFF receptor signaling. Ligation of this receptor activates noncanonical NF-κB signaling in B cells, and it was observed that the relocation of the noncanonical NF-κB members RelB and p52 to the nucleus was decreased in MALT1−/− B cells. Additionally, MALT1 is required for BAFF-induced survival and upregulation of antiapoptotic genes in MZ B cells [49]. This established MALT1 as an important member of not only the canonical but also the noncanonical NF-κB pathway whose significance is cell- and receptor-dependent.
5.2 Toll-like receptors
The MALT1−/− and Bcl10−/− mice gave inconclusive results when it came to TLR signaling. While the proliferation of B cells after stimulation with LPS was found to be normal in Bcl10−/− mice [25], studies either described a strong dependence on MALT1 [30] or found it to play only a minor role [31]. In another study the role of the two proteins was elucidated further coming to the conclusion that both proteins are essential [50]. According to the report, upon LPS stimulation of macrophages Bcl10 is recruited to the TLR4 complex via the IL-1 receptorassociated kinase (IRAK1) – which it binds in a CARD-independent manner – where it is activated presumably via oligomerization. Subsequently, NF-κB is activated after a complicated chain of events involving MALT1 and other downstream effectors [50]. An additional study implicated a role for the CARD9-Bcl10-MALT1 complex in TLR4 signaling [51].
5.3 Fungal recognition receptors
The fungal recognition receptors dectin-1 and dection-2 are C-type lectins that recognize carbohydrates present in the cell wall of fungi. The stimulation with C. albicans, zymosan, or other glucans mimicking fungal carbohydrates has been reported to activate the receptors and lead to the assembly of a complex containing CARD9, Bcl10, and MALT1 [52]. The CARD9−/− mouse displays severe defects in fighting C. albicans infection, and bone marrow-derived macrophages as well as dendritic cells showed defective NF-κB signaling after exposure to zymosan or C. albicans [52].
In addition, a case of a family with several members affected by deleterious mutation in CARD9 has been reported. The patients suffered severe chronic fungal infections [53]. The defense against fungal infections is complex and only partially understood, but it is known to involve the activation of TH17 cells through cytokines produced by dendritic cells exposed to fungi. The expression of these TH17-inducing cytokines was found to depend on dectin-1 and dectin-2, the CARD9-Bcl10-MALT1 signalosome, and the selective activation of the c-Rel subunit of NF-κB. If MALT1 was not present, the expression of TH17-inducing cytokines was severely impaired [54]. These findings are especially interesting considering that next to the defense against fungi, TH17 cells play roles in many autoimmune diseases such as Crohn’s disease and ulcerative colitis as well as the development of allergies [55].
5.4 ITAM-coupled receptors in myeloid cells
When MALT1 and Bcl10 were found to play an essential function in mast cell signaling [56, 57], this represented the first reported role in nonlymphoid cells. Mast cells are myeloid cells that are mainly activated by binding of the Fc portion of immunoglobulins to Fc receptors, which couple to ITAM (immunoreceptor tyrosine-based activation motif)-containing adapter molecules such as FcRγ and DAP12 to transmit signals [58]. The main mast cell receptor is FcεRI, the high-affinity receptor for IgE. After ligand binding, several actions are responsible for the role of mast cells in fighting infections as well as the course of allergic reactions. These involve the release of proinflammatory molecules such as histamine from granular intracellular stores as an immediate reaction as well as the transcription of target genes such as the cytokines TNF-α and IL-6 [59]. While MALT1 does not seem to be involved in the immediate processes or mast cell killing [57], reports for Bcl10 are less clear [56, 57]. However, both proteins are clearly required for NF-κB signaling and cytokine production [56, 57].
A study by Hara et al. showed the involvement of MALT1 and Bcl10 in the signaling of further ITAM-coupled receptors of cells of the myeloid lineage including dendritic cells and macrophages. This painted a complex picture suggesting the participation of MALT1 and Bcl10 in NF-κB signaling downstream of receptors such as FcγR, OSCAR (osteoclast associated, immunoglobulin-like receptor), and TREM-1 (Triggering receptor expressed on myeloid cells 1). CARD9 was found to be the upstream interaction partner for these receptors as well as for dectin-1, while CARMA1 was dispensable [51].
5.5 ITAM-coupled receptors in NK cells
Natural killer (NK) cells are cells of the lymphoid lineage that are responsible for fighting viruses and tumor cells as part of the innate immune system. Like many other immune cells they express different receptors that couple to ITAM-containing adapter molecules, notably NK1.1, Ly49D, Ly49H, and NKG2D. It was found that just as in myeloid cells, signaling through these ITAM-coupled receptors depends on Bcl10 and MALT1 for NF-κB activation. In addition, a requirement for both proteins was described in JNK and p38 signaling. However, in contrast to myeloid cells, the signaling depends on CARMA1 instead of CARD9 [60].
5.6 GPCRs
G-protein coupled receptors (GPCRs) are a huge family of proteins regulating a vast number of diverse cellular processes [61, 62]. One known substrate is lysophosphatidic acid (LPA). It can bind to GPCRs of the endothelial differentiation gene (Edg) family and regulates the proliferation, migration, and survival of different cell types. In fibroblasts, LPA is able to activate NF-κB and subsequent chemokine production, and this was found to depend on PKC [63] as well as Bcl10 and MALT1 [64, 65]. Studies involving CARMA3−/− mice showed that CARMA3 in combination with Bcl10 is required for NF-κB signaling events downstream of LPAinduced GPCR signaling [47]. The involvement of MALT1 was not formally proven but given the combined information from these studies seems likely.
Further support for the involvement of MALT1 in GPCR signaling came from a study involving the type I angiotensin II receptor (AT1R), another member of the GPCR family [44]. Although angiotensin II is classically known as a blood pressure regulator, additional functions such as that of a proinflammatory mediator in various tissues are emerging [66, 67]. Stimulation of the AT1R by angiotensin II causes NF-κB signaling and is seen for example during liver injury. The study examined the role of CARMA3, Bcl10, and MALT1 after angiotensin II stimulation of hepatocytes and found the proteins to be required for NF-κB signaling [44].
Additional GPCRs have been reported to rely on CARMA3-Bcl10-MALT1 for signaling, including the thrombin receptor PAR-1, the IL-8/CXCL8 receptor CXCR2, and the CXCL12/SDF-1α receptor CXCR4 [68–70], suggesting that this represents a general mechanism of GPCR signaling through NF-κB.
5.7 Overall significance of MALT1 in NF-κB signaling
Overall, a picture emerges in which MALT1 and Bcl10 play essential roles downstream of many different types of receptors. In cells of the lymphoid lineage including B and T cells as well as NK cells they cooperate with CARMA1 to form the classical CARMA1-Bcl10-MALT1 (CBM) complex. In cells of the myeloid lineage the complex involves CARD9 instead of CARMA1, and in nonhematopoietic cells CARMA3 becomes the binding partner. All three types of CBM complexes serve the common goal of activating the transcription of NF-κB target genes [48].
6. Relevance of MALT1 proteolytic activity
Mounting evidence showed that the proteolytic activity of MALT1 contributes to NF-κB signaling, but in all of the studies only a partial impairment of the pathway was seen upon inhibition of MALT1 activity [11, 12]. Even though it was proven that MALT1 can cleave substrates, the question remained as to how important this aspect of its function was. To begin to elucidate this question, two independent groups took clues from a study involving diffuse large B-cell lymphoma (DLBCL) cell lines.
6.1 Diffuse large B cell lymphoma
DLBCL is the most common type of non-Hodgkin lymphoma consisting of enlarged B cells. The neoplasm presents either in the lymph nodes or extranodally at a variety of sites. It can arise de novo or develop from a less aggressive lymphoma. DLBCL is further categorized into subgroups, but a significant number does not easily conform to any specific group and is thus denominated as DLBCL, not otherwise specified (NOS) [71]. This group comprises 25–30% of all non-Hodgkin lymphoma cases in the western world with slightly higher incidences in developing countries [72]. It is further divided into two subtypes, activated B cell-like diffuse large B cell lymphoma (ABC-DLBCL) and germinal center B cell-like diffuse large B cell lymphoma (GCB-DLBCL) [73, 74], with both types making up about half of the cases. The division is made according to gene expression profiles, and as the name suggests, GCB cases are believed to originate from peripheral B cells of the germinal center, whereas ABC cases arise from peripheral activated B cells of post germinal center origin [73, 74]. One notable difference significant for the potential involvement of MALT1 is that ABC-DLBCL shows constitutive NF-κB activation whereas GCB-DLBCL does not [75, 76]. The disease affects mostly older patients with approximately 20,000 cases per year in the United States and a rising incidence [77, 78]. GCB-DLBCL has a better overall prognosis than ABC-DLBCL [73, 74], although treatment with the anti-CD20 antibody rituximab is promising and might level this difference [77].
6.2 The role of MALT1 in DLBCL
In 2006, Ngo et al. searched for genes whose knockdown would specifically decrease the survival or proliferation of lymphoma cells. They discovered that while CARMA1, Bcl10, and MALT1 were dispensable for GCB-DLBCL cell lines as well as another subtype of DLBCL called primary mediastinal large B cell lymphoma (PMBL), they were required for the survival of ABC-DLBCL cell lines [79].
Hailfinger et al. and Ferch at al. independently went on to investigate whether the dependence extended to the proteolytic activity of MALT1 [80, 81]. They used a fluoromethylketone tetrapeptide inhibitor (Z-VRPR-FMK) that was loosely based on the sequence of the discovered substrates to investigate the effect of MALT1 inhibition on different B cell lymphoma cell lines. The inhibition of MALT1 proteolytic activity led to decreased cell viability in cell lines derived from ABC-DLBCL. Cell lines derived from other types of B cell lymphomas like GCB-DLBCL, Burkitt’s lymphoma, and marginal zone lymphoma were not affected [80, 81]. These results suggested that it is not only the scaffolding function of MALT1 that is important for its function but also its proteolytic activity.
This was substantiated in a mouse model of MALT lymphoma, which had initially proven difficult to develop because mouse models overexpressing Bcl10 or cIAP2-MALT1 in B cells failed to recapitulate the MALT lymphoma phenotype [82, 83]. It wasn’t until 2012 that the groups of Isidro Sanchez-Garcia and Jose Martinez-Climent managed to produce a mouse strain, which overexpressed MALT1 specifically in hematopoietic stem cells leading to a MALT lymphoma phenotype [84]. Furthermore, in combination with p53 deletion this led to a phenotype resembling the activated B-cell-like subtype of DLBCL – more evidence that MALT1 plays a critical role in lymphoma development and cell survival. In addition, B cells isolated from the MALT1 and MALT1/p53 lymphoma mice showed increased susceptibility to apoptosis upon inhibition of MALT1 proteolytic activity by treatment with Z-VRPR-FMK providing further proof of the importance of MALT1’s proteolytic activity [84].
6.3 MALT1 inhibitors
To further study the importance of MALT1 proteolytic activity, two independent groups developed small molecule inhibitors for MALT1.
The MI-2 inhibitor described by Fontan et al. was reported to display kinetics consistent with an irreversible mode of binding and an IC50 of 5.84 µM. It most likely functions via covalent modification of the active site Cys [85].
Nagel et al. discovered the potential of several phenothiazine derivatives – which are either still approved for the use as antipsychotic or sedative drugs or have been in the past – to inhibit MALT1 [86]. The most effective compound was mepazine, which binds to an allosteric site at the interface between the catalytic domain and the Ig3 domain preventing the rearrangement of the inactive enzyme into the active conformation [87]. The reversible inhibition was reported to display an IC50 of 0.83 µM [86].
Both inhibitors caused a decrease in the amount of MALT1 substrate cleavage and decreased expression of NF-κB target genes. Growth inhibition, decreased viability, and increased apoptosis were observed in ABC-DLBCL but not GCB-DLBCL cells after inhibitor treatment. The size of ABC-DLBCL tumors decreased in a mouse model upon inhibitor treatment [85, 86]. Primary cells isolated from biopsies of two patients classified to have non- GCB DLBCL displayed decreased viability upon MI-2 treatment compared to non-treated cells, while no effects were seen in cells isolated from a third non-GDB DLBCL patient or cells from GCB-DLBCL biopsies [85].
The in vivo effectiveness of the mepazine inhibitor was substantiated by the fact that treatment of EAE mice with mepazine reduced symptoms both when administered before or after disease onset [88]. In contrast, the levels of Treg cells, which are an essential component of the immune system and which were found to be severely reduced in MALT1−/− mice [43], were normal after mepazine treatment [88].
6.4 MALT1 catalytic mutant mouse model
The recently published mouse models in which WT MALT1 was replaced by the catalytic mutant (knock in mouse, KI) dispersed any remaining doubts about the importance of MALT1 proteolytic activity [89–92]. While the studies did not reach the same conclusions for all aspects analyzed, the overarching message clearly supports a key role for MALT1 proteolysis. In the following section the mice will be referred to as MALT1 KI or WT regardless of homozygous or heterozygous status. The studies compared either MALT1mutant/mutant and MALT1+/+ [89, 91, 92] or MALT1mutant/− and MALT1+/− mice [90]. This may be responsible for some of the differences between the models.
While the studies found no impairment of IKK activation in the MALT1 KI mice, many other effects of the MALT1−/− mouse were directly phenocopied, with some of the observed effects slightly less extreme in the KI mouse. Besides the lack of MALT1 substrate cleavage, the B1 and MZ B cell subsets were severely reduced, T cell proliferation was decreased [89–92], serum levels of several but not all Ig subsets were downregulated, and the response to T-independent antigen stimulation was abrogated or reduced [89, 91, 92]. The T-dependent response was found to be slightly delayed [90] or strongly decreased [89, 91, 92]. The activation of both NK and dendritic cells downstream of a variety of receptors was decreased [89, 92]. As expected from the MALT1 inhibition studies in the EAE model [88, 91], the MALT1 KI mice were completely protected from EAE development [89, 91].
However, in contrast to MALT1 WT and MALT1−/− mice, over time the MALT1 KI mice displayed severely enlarged lymph nodes, decreased body weight, and autoimmune gastritis and other forms of inflammatory infiltrates. In addition, a Treg cell development defect was observed both in MALT1−/− and KI mice, which therefore must depend on the proteolytic activity of MALT1 [89–91]. This is in contrast to a study by Mc Guire et al. in which mepazine was found to have no effect on Treg levels [88].
In addition, in one of the studies ataxia and motor-coordination deficits were observed in KI but not MALT1−/− or WT mice. These effects depended on the CBM complex and were intrinsic to lymphocytes since double mutant mice obtained by crossing MALT1 KI mice with Bcl10−/− or Rag1−/− mice did not show any of these effects [90]. In addition, Gewies et al. found increased mRNA stability as well as serum levels of IFN-γ and other cytokines. Interestingly, many of the observed effects depended on IFN-γ since IFN-γ−/− and MALT1 KI double mutant mice did not display ataxia, weight loss, gastritis, or increased TNF levels. Other effects such as enlarged lymph nodes were unaffected in the double mutant mice [90].
The studies reached different conclusions with regard to colitis. While Jaworski et al. reported that MALT1 KO mice were completely protected from autoimmune colitis and KI mice developed it with lower penetrance [89], Gewies et al. found KI mice to be more susceptible to inflammatory bowel disease than WT mice [90]. However, it is difficult to directly compare the studies because different colitis models were used.
A complex picture emerged upon evaluation of the effect the different genotypes had on gene expression. While some genes were downregulated both in MALT1−/− and KI T cells, others were only affected in MALT1−/− T cells. Interestingly, a distinct set of genes including the IFN-γ gene (IFNg) as well as genes controlled by the cytokine were specifically upregulated in KI but not MALT1−/− T cells [90].
Overall, these data prove the importance of the proteolytic activity of MALT1 for its function and paint a complex picture for the application of MALT1 inhibitors in the treatment of disease. MALT1 seems to have both pro- and anti-inflammatory effects with the proteolytic activity largely responsible for the anti-inflammatory effects, but more research is needed to clarify this. Depending on the disease context and duration of treatment inhibitors might nonetheless prove useful. However, at the very least, it will be important to closely monitor the development of autoimmune reactions as well as the level of Treg cells as a result of MALT1 inhibition, which more closely mimics the KI than the KO model [89].
7. Is MALT1 implicated in other diseases?
Considering the importance of MALT1 in NF-κB signaling and the relevance of this pathway in a host of different settings, the implication of MALT1 in a variety of diseases is not surprising. As described, MALT1 is involved in lymphoma as well as multiple sclerosis. Furthermore, a recent report suggests that deregulated MALT1 levels are also observed in cases of acute myeloid leukemia (AML) [93] as well as pediatric T-cell acute lymphomblastic leukemia (ALL) [94], although more research is needed to substantiate these claims.
In addition, mutations in MALT1 have recently been described in several patients with combined immunodeficiency (CID) [95–97]. CID is characterized by severely impaired T and B cell functions, while total cell numbers remain unchanged. Although most cases of CID are spontaneous, in one of the reports the patients, one male and one female, were siblings and came from a first-cousin marriage from a family with numerous consanguineous marriages. Both patients showed frequent infections and died of respiratory failure at a young age (7 and 13 years, respectively). The mutation in MALT1 was determined to be Ser89Ile, which is within the core of the MALT1 DD. MALT1 mRNA was present, but no protein was detected, making it likely that the mutation destabilizes MALT1 and causes its degradation [95]. In the second report, the patient was a teenage girl also born from a first cousin marriage. Amongst other symptoms, she had chronic dermatitis, infections, and gastrointestinal infections since infancy. The mutation in MALT1 was Trp580Ser, which is localized between the catalytic domain and the Ig3 domain. mRNA levels were normal, and the MALT1 protein was decreased but not completely absent [96]. This can potentially explain some of the differences observed between the patients. In the third report, the patient came from a non-consanguineous family and developed a severe rash, irritable bowel syndrome as well as different bacterial and viral infections, alongside other symptoms. The mutations in MALT1 were heterozygous, one also present in the mother and affecting a splice acceptor site and one de novo leading to a frameshift after Tyr353 and truncation shortly after. No functional protein was detectable. At the age of 18 months the patient received a curative hematopoietic stem cell transplant followed later by an additional donor T cell infusion [97].
The analyses of lymphocytes performed throughout the patients’ lives showed largely normal numbers of B and T cells as well as immunoglobulin levels in two of the studies [95, 96]. The antibody responses of the patients varied, being poor in two of the studies [95, 97], while largely normal in the third [96]. Isolated peripheral blood mononuclear cells (PMBCs) showed decreased proliferation upon stimulation with anti-CD3 and other mitogens compared to WT cells [95, 97]. T cells isolated from the patients showed impaired IκBα degradation and IL-2 production upon stimulation with PMA and ionomycin [95–97], matching the mouse KO results [30, 31]. Reconstitution of MALT1−/− T cells with the Ser89Ile mutant gene did not rescue the IκBα and IL-2 defects, while the WT gene did, confirming that the Ser89Ile mutation renders MALT1 nonfunctional. The contrasting results seen with regard to immunoglobulin levels (normal in the patients compared to reduced in the MALT1−/− mice) were speculated to be due to the basal level of infections seen in the patients as opposed to the mice raised in a pathogenfree environment [95].
Since the cause of CID is mostly unknown, it would be interesting to see how common MALT1 mutations are in the disease and patient screening was recommended [95, 97]. Interestingly, two cases of CID containing CARMA1 mutations resulting in the loss of the protein have also been recently described [98, 99], substantiating the importance of the CBM complex in this disease.
8. Relevance of individual substrate cleavage events
Over the past years, several substrates have been discovered for MALT1; a brief summary is provided in Figure 7. In light of the fact that the proteolytic activity of MALT1 is important, the question arises which cleavage events are essential and which substrates represent innocent bystanders. To begin to elucidate this, it is important to distinguish between gain of function and loss of function cleavage events. In a gain of function cleavage the cleavage products take on a role not previously fulfilled by the full-length substrate. For example, the cleavage of NIK represents a gain of function cleavage because the C-terminal cleavage fragment contributes to uncontrolled noncanonical NF-κB signaling, while the fulllength protein is constantly downregulated by ubiquitination and degradation [100]. In contrast, in a loss of function situation the full-length protein is no longer present at sufficient concentrations to fulfill its function. A20 is an example of such a loss of function with the cleavage fragments unable to carry out the ubiquitin editing role of full-length A20 [11].
Figure 7. MALT1 substrates discovered to date and the effects of their cleavage.
CAP-Gly, cytoskeleton-associated protein glycine-rich; CARD, caspase recruitment domain; CC, coiled coil domain; DD, death domain; DUB, deubiquitinating enzyme domain; Ig, immunoglobulin-like domain; LZ, leucine zipper domain; OTU, ovarian tumor domain; Pro, proline-rich domain; RHD, Rel homology domain; S/T rich, serine and threonine-rich domain; T3-binding, tumor necrosis factor associated factor 3-binding domain; TAD, trans-activating domain; ZF, zinc finger domain. [11, 12, 100, 101, 104, 106–108]
The simplest way to test the importance of a substrate is to knock down its expression, and this has been done for many of the substrates discovered to date. If the cleavage event represents a loss of function, knockdown of the substrate could potentially have the same effect as cleavage. However, this approach is highly problem-prone because the cell might depend on the protein and not be able to function correctly without it. In addition, no information can be obtained for gain of function events. Another relatively simple approach is the expression of cleavage site mutants, which has been done for most of the reported MALT1 substrates. Depending on whether cleavage leads to a loss or gain of function, additional knockdown of the endogenous substrate might be necessary. To explore the effect of a gain of function cleavage, one possibility is the transfection of cleavage fragments into cells, which has been utilized in the investigation of the A20, NIK, and MALT1 cleavages. However, this strategy is problematic because the expression in these cells will be constitutive, whereas cleavage is a controlled event for which timing can be essential. A more efficient way is the targeted specific cleavage of the substrates of interest independent of other cleavage events that might be initiated by MALT1. One way would be to engineer the substrate of interest to include a TEV-cleavage site instead of the natural MALT1 cleavage site as previously described for caspases [110]. TEV protease has a highly selective cleavage specificity and is unlikely to mediate off-target cleavage events in mammalian cells. This strategy would allow to clearly differentiate between the relevance of the different substrates as well as between the proteolytic and scaffolding function of MALT1. However, in case the presence of a substrate in a certain signaling complex is required for the cleavage to have an effect and in case this constellation is only achieved as a consequence of signaling events involving the MALT1 scaffolding function, such events – and consequently a protease-dead background – would be required in order to judge the results. To date, no experiments involving TEV protease have been performed for any of the reported substrates of MALT1.
Nonetheless, since cleavage inhibition did not result in dramatic effects for any of the substrates discovered to date, it is tempting to speculate that it is either the combined effect of MALT1 on all of the substrates that explains the importance of MALT1 proteolytic activity or that the most relevant substrate(s) are yet to be discovered.
8.1 Autoproteolytic cleavage
Interestingly, Baens et al. reported autoproteolytic cleavage of MALT1 at R149 between the DD and the Ig1 domain. The study suggests that while the cleavage has no effect on the proteolytic activity of MALT1, the C-terminal cleavage fragment of 76 kDa can activate the expression of NF-κB target genes in a Bcl10-independent yet TRAF6-dependent manner. However, the cleavage site mutant does not affect phosphorylation of IκBα nor the relocalization of NF-κB subunits to the nucleus, and so the authors speculate that the C-terminal fragment exerts its effect on NF-κB signaling even further downstream after shuttling to the nucleus [108]. More work is needed to clarify this situation.
8.2 MALT1 activity-based probes
The need to distinguish between active and inactive MALT1 led to the development of activity-based probes (also known as active-site probes) directed against MALT1 [111, 112]. In addition to inhibiting MALT1, the main advantage of the probes is their ability to label and thereby visualize active MALT1 both in the recombinant in vitro setting and in the cellular context extending to endogenous MALT1.
As a general setup, activity-based probes contain a peptide scaffold or a linker as well as a label and an electrophilic warhead, which in combination leads to the selective inhibition and labeling of a target protease that can then be detected using a variety of techniques including Western blotting, microscopy, and mass spectrometry. The nature of the warhead depends on the protease in question. Epoxides, vinyl sulfones, and acyloxymethyl ketones (AOMKs) are commonly used for cysteine proteases. The label is chosen depending on the desired detection technique to be used. Biotin as well as fluorophores are common [113, 114]. The activity-based probes developed by the Krappmann group as well as our laboratory utilized the specificity of MALT1 combined with an AOMK warhead, which is highly selective for Cys proteases, as well as different labels leading to the general format label-L(V/R)SR-AOMK [111, 112].
Currently, the cleavage of endogenous MALT1 substrates is often used to answer the question whether MALT1 is active under certain cellular conditions. However, the incomplete cleavage and instability of the cleavage fragments observed for several of the substrates renders this difficult. Another approach that has been used in the literature to evaluate MALT1 cellular activity has been the ability of cellular lysates to cleave tetrapeptide substrates such as Ac-LRSR-AMC [12]. However, background cleavage of optimal MALT1 tetrapeptide substrates by unknown cellular proteases makes this approach similarly unsatisfying.
Instead, activity-based probes have the potential to be applied routinely to evaluate MALT1 activity. An example includes elucidation of the question whether posttranslational modifications of MALT1 such as monoubiquitination [115, 116] are important for its activity. Additionally, applications in inhibitor screens could be envisioned. After treating cells with potential inhibitors, cell lysates could be incubated with an activity-based probe to see if decreased probe binding is observed either in gel-based assays or in a proteomics-based approach.
9. Conclusions
Overall, a tremendous amount of progress has been made in MALT1 research over the last several years. Especially the development of MALT1 knock in mice has proven the relevance of MALT1 proteolytic activity. The development of the first MALT1 inhibitors will surely prove useful in further elucidating its role in different contexts including such different diseases as lymphoma, CID, and multiple sclerosis, and the next few years are sure to continue to shed light on the exact role MALT1 activity plays in distinct cellular processes.
Highlights.
MALT1 is a caspase homolog with proteolytic activity
MALT1 plays an important role in NF-κB signaling downstream of a variety of receptors
Mouse models demonstrated the importance of MALT1 proteolytic activity
MALT1 is implicated in a variety of diseases including lymphoma and CID
Acknowledgments
Supported by NIH grant 1R01GM099040 and a Corporate Sponsored Research Agreement from Genentech, Inc.
Abbreviations
- ABC
activated B cell-like
- cABC-DLBCL
activated B cell-like diffuse large B cell lymphoma
- Ac
acetyl
- ALL
acute lymphomblastic leukemia
- AMC
7-amino-4-methylcoumarin
- AML
acute myeloid leukemia
- AOMK
acyloxymethyl ketone
- AP-1
activator protein 1
- API2
apoptosis inhibitor protein 2
- AT1R
type I angiotensin II receptor
- BAFF
B-cell-activating factor
- Bcl10
B-cell lymphoma/leukemia 10
- BCR
B cell receptor
- Bimp1
Bcl10-interacting MAGUK protein 1
- CAP-Gly
cytoskeleton-associated protein glycine-rich
- CARD
caspase recruitment domain
- CARMA1
CARD-containing MAGUK (membrane-associated guanylate kinase) 1
- CBM
CARMA1-Bcl10-MALT1
- CC
coiled coil domain
- CD
cluster of differentiation
- cIAP2
cellular inhibitor of apoptosis protein 2
- CID
combined immunodeficiency
- CXCL
Chemokine CXC motif ligand
- CXCR
CXC chemokine receptor
- DAP12
DNAX-activation protein of 12 kDa
- DD
death domain
- DLBCL
diffuse large B cell lymphoma
- DN
double negative
- DUB
deubiquitinating enzyme domain
- EAE
experimental autoimmune encephalomyelitis
- Edg
endothelial differentiation gene
- ERK
extracellular signal-regulated kinase
- Fc
fragment, crystallizable (of an antibody)
- FcRγ
Fc receptor common gamma signaling chain
- FcγR
Fc-gamma receptor
- FcεRI
Fc-epsilon receptor I
- FMK
fluoromethylketone
- GCB
germinal center B cell-like
- GCB-DLBCL
germinal center B cell-like diffuse large B cell lymphoma
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- GPCR
G-protein coupled receptor
- GUK
guanylate kinase
- Ig
immunoglobulin or immunoglobulin-like domain
- IgG
immunoglobulin G
- IKK
IκB kinase
- IL
interleukin
- IRAK1
IL-1 receptor-associated kinase
- ILZ
leucine zipper domain
- ITAM
immunoreceptor tyrosine-based activation motif
- JNK
c-Jun N-terminal kinase
- KI
knock in
- KO
knockout
- LPA
lysophosphatidic acid
- LPS
lipopolysaccharide
- MAGUK
membrane-associated guanylate kinase
- MALT
mucosa-associated lymphoid tissue
- MALT1
mucosa-associated lymphoid tissue lymphoma translocation protein 1
- MAPK
mitogen-activated protein kinase
- MLT
MALT lymphoma-associated translocation
- MZ
marginal zone
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NIK
NF-κB-inducing kinase
- NK
natural killer
- NKG2D
natural killer group 2, member D
- NOS
not otherwise specified
- OSCAR
osteoclast associated, immunoglobulin-like receptor
- OTU
ovarian tumor domain
- PAR-1
protease-activated receptor-1
- PDZ
PSD95-Dlg1-zonula occludens-1 protein
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- PMBC
peripheral blood mononuclear cell
- PMBL
primary mediastinal large B cell lymphoma
- Pro
proline-rich domain
- RHD
Rel homology domain
- SDF-1α
stromal cell-derived factor-1α
- SH3
Src homology 3
- S/T rich
serine and threonine-rich domain
- T3-binding
tumor necrosis factor associated factor 3-binding domain
- TAD
trans-activating domain
- TCR
T cell receptor
- TEV
Tobacco Etch Virus
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- TNFR
tumor necrosis factor receptor
- TRAF
TNF receptor-associated factor
- TREM-1
triggering receptor expressed on myeloid cells 1
- WT
wild type
- Z
benzyloxycarbonyl
- ZF
zinc finger domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Du MQ. MALT lymphoma : recent advances in aetiology and molecular genetics. J Clin Exp Hematop. 2007;47:31–42. doi: 10.3960/jslrt.47.31. [DOI] [PubMed] [Google Scholar]
- 2.Isaacson P, Wright DH. Extranodal malignant lymphoma arising from mucosa-associated lymphoid tissue. Cancer. 1984;53:2515–2524. doi: 10.1002/1097-0142(19840601)53:11<2515::aid-cncr2820531125>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 3.Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat. Rev. Cancer. 2004;4:644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 4.Sugiyama T, Asaka M, Nakamura T, Nakamura S, Yonezumi S, Seto M. API2-MALT1 chimeric transcript is a predictive marker for the responsiveness of H. pylori eradication treatment in low-grade gastric MALT lymphoma. Gastroenterology. 2001;120:1884–1885. doi: 10.1053/gast.2001.25305. [DOI] [PubMed] [Google Scholar]
- 5.Levine EG, Arthur DC, Machnicki J, Frizzera G, Hurd D, Peterson B, Gajl-Peczalska KJ, Bloomfield CD. Four new recurring translocations in non-Hodgkin lymphoma. Blood. 1989;74:1796–1800. [PubMed] [Google Scholar]
- 6.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21)p6ssociated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- 7.Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JM, Rawlings ND, Stevens RA, Barrett AJ. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett. 1998;441:361–365. doi: 10.1016/s0014-5793(98)01574-9. [DOI] [PubMed] [Google Scholar]
- 10.Snipas SJ, Wildfang E, Nazif T, Christensen L, Boatright KM, Bogyo M, Stennicke HR, Salvesen GS. Characteristics of the caspase-like catalytic domain of human paracaspase. Biol. Chem. 2004;385:1093–1098. doi: 10.1515/BC.2004.142. [DOI] [PubMed] [Google Scholar]
- 11.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 12.Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, Fasel N, Thome M. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 13.Wiesmann C, Leder L, Blank J, Bernardi A, Melkko S, Decock A, D'Arcy A, Villard F, Erbel P, Hughes N, Freuler F, Nikolay R, Alves J, Bornancin F, Renatus M. Structural Determinants of MALT1 Protease Activity. J. Mol. Biol. 2012 doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M, Kasperkiewicz P, Drag M, Salvesen GS. Mechanism and specificity of the human paracaspase MALT1. Biochem. J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu L, Dhe-Paganon S. Oligomeric Structure of the MALT1 Tandem Ig-Like Domains. PloS one. 2011;6:e23220. doi: 10.1371/journal.pone.0023220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schechter I, Berger M. On the size of the active site in proteases. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 19.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 20.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 22.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Wang D, Lin X. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 25.Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001;104:33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 26.Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D'Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 27.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O'Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr. Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat. Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 29.Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat. Immunol. 2002;3:836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 30.Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 31.Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 32.Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, Domashenz H, Hong NA, Glynne RJ, Nelms KA, Goodnow CC. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751–762. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 33.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 2003;13:1247–1251. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 34.Pohl T, Gugasyan R, Grumont RJ, Strasser A, Metcalf D, Tarlinton D, Sha W, Baltimore D, Gerondakis S. The combined absence of NF-kappa B1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4514–4519. doi: 10.1073/pnas.072071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin F, Kearney JF. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a "natural immune memory". Immunol. Rev. 2000;175:70–79. [PubMed] [Google Scholar]
- 36.Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 2001;14:617–629. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 37.Tarakhovsky A. Bar Mitzvah for B-1 cells: how will they grow up? J. Exp. Med. 1997;185:981–984. doi: 10.1084/jem.185.6.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 39.von Boehmer H. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat. Rev. Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 40.Kingeter LM, Schaefer BC. Loss of protein kinase C theta, Bcl10, or Malt1 selectively impairs proliferation and NF-kappa B activation in the CD4+ T cell subset. J. Immunol. 2008;181:6244–6254. doi: 10.4049/jimmunol.181.9.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferch U, zum Buschenfelde CM, Gewies A, Wegener E, Rauser S, Peschel C, Krappmann D, Ruland J. MALT1 directs B cell receptor-induced canonical nuclear factor-kappaB signaling selectively to the c-Rel subunit. Nat. Immunol. 2007;8:984–991. doi: 10.1038/ni1493. [DOI] [PubMed] [Google Scholar]
- 42.Brüstle A, Brenner D, Knobbe CB, Lang PA, Virtanen C, Hershenfield BM, Reardon C, Lacher SM, Ruland J, Ohashi PS, Mak TW. The NF-kappaB regulator MALT1 determines the encephalitogenic potential of Th17 cells. J. Clin. Invest. 2012;122:4698–4709. doi: 10.1172/JCI63528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mc Guire C, Wieghofer P, Elton L, Muylaert D, Prinz M, Beyaert R, van Loo G. Paracaspase MALT1 Deficiency Protects Mice from Autoimmune-Mediated Demyelination. J. Immunol. 2013 doi: 10.4049/jimmunol.1201351. [DOI] [PubMed] [Google Scholar]
- 44.McAllister-Lucas LM, Ruland J, Siu K, Jin X, Gu S, Kim DS, Kuffa P, Kohrt D, Mak TW, Nunez G, Lucas PC. CARMA3/Bcl10/MALT1-dependent NF-kappaB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:139–144. doi: 10.1073/pnas.0601947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertin J, Guo Y, Wang L, Srinivasula SM, Jacobson MD, Poyet JL, Merriam S, Du MQ, Dyer MJ, Robison KE, DiStefano PS, Alnemri ES. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 2000;275:41082–41086. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 46.McAllister-Lucas LM, Inohara N, Lucas PC, Ruland J, Benito A, Li Q, Chen S, Chen FF, Yamaoka S, Verma IM, Mak TW, Nunez G. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-kappaB induction. J. Biol. Chem. 2001;276:30589–30597. doi: 10.1074/jbc.M103824200. [DOI] [PubMed] [Google Scholar]
- 47.Grabiner BC, Blonska M, Lin PC, You Y, Wang D, Sun J, Darnay BG, Dong C, Lin X. CARMA3 deficiency abrogates G protein-coupled receptor-induced NF-{kappa}B activation. Genes Dev. 2007;21:984–996. doi: 10.1101/gad.1502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blonska M, Lin X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tusche MW, Ward LA, Vu F, McCarthy D, Quintela-Fandino M, Ruland J, Gommerman JL, Mak TW. Differential requirement of MALT1 for BAFF-induced outcomes in B cell subsets. J. Exp. Med. 2009;206:2671–2683. doi: 10.1084/jem.20091802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong W, Liu Y, Peng J, Chen L, Zou T, Xiao H, Liu Z, Li W, Bu Y, Qi Y. The IRAK-1-BCL10-MALT1-TRAF6-TAK1 cascade mediates signaling to NF-kappaB from Toll-like receptor 4. J. Biol. Chem. 2006;281:26029–26040. doi: 10.1074/jbc.M513057200. [DOI] [PubMed] [Google Scholar]
- 51.Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, Inui M, Takai T, Shibuya A, Saijo S, Iwakura Y, Ohno N, Koseki H, Yoshida H, Penninger JM, Saito T. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat. Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 52.Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 53.Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, Jamal S, Manguiat A, Rezaei N, Amirzargar AA, Plebani A, Hannesschlager N, Gross O, Ruland J, Grimbacher B. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. The New England journal of medicine. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gringhuis SI, Wevers BA, Kaptein TM, van Capel TM, Theelen B, Boekhout T, de Jong EC, Geijtenbeek TB. Selective C-Rel activation via Malt1 controls anti-fungal T(H)-17 immunity by dectin-1 and dectin-2. PLoS Pathog. 2011;7:e1001259. doi: 10.1371/journal.ppat.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louten J, Boniface K, de Waal Malefyt R. Development and function of TH17 cells in health and disease. J. Allergy Clin. Immunol. 2009;123:1004–1011. doi: 10.1016/j.jaci.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Pappu BP, Zeng H, Xue L, Morris SW, Lin X, Wen R, Wang D. B cell lymphoma 10 is essential for FcepsilonR-mediated degranulation and IL-6 production in mast cells. J. Immunol. 2007;178:49–57. doi: 10.4049/jimmunol.178.1.49. [DOI] [PubMed] [Google Scholar]
- 57.Klemm S, Gutermuth J, Hultner L, Sparwasser T, Behrendt H, Peschel C, Mak TW, Jakob T, Ruland J. The Bcl10-Malt1 complex segregates Fc epsilon RI-mediated nuclear factor kappa B activation and cytokine production from mast cell degranulation. J. Exp. Med. 2006;203:337–347. doi: 10.1084/jem.20051982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamerman JA, Ni M, Killebrew JR, Chu CL, Lowell CA. The expanding roles of ITAM adapters FcRgamma and DAP12 in myeloid cells. Immunol. Rev. 2009;232:42–58. doi: 10.1111/j.1600-065X.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat. Rev. Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross O, Grupp C, Steinberg C, Zimmermann S, Strasser D, Hannesschlager N, Reindl W, Jonsson H, Huo H, Littman DR, Peschel C, Yokoyama WM, Krug A, Ruland J. Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NFkappaB and MAPK activation to selectively control cytokine production. Blood. 2008;112:2421–2428. doi: 10.1182/blood-2007-11-123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kobilka BK. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cummings R, Zhao Y, Jacoby D, Spannhake EW, Ohba M, Garcia JG, Watkins T, He D, Saatian B, Natarajan V. Protein kinase Cdelta mediates lysophosphatidic acid-induced NFkappaB activation and interleukin-8 secretion in human bronchial epithelial cells. J. Biol. Chem. 2004;279:41085–41094. doi: 10.1074/jbc.M404045200. [DOI] [PubMed] [Google Scholar]
- 64.Klemm S, Zimmermann S, Peschel C, Mak TW, Ruland J. Bcl10 and Malt1 control lysophosphatidic acid-induced NF-kappaB activation and cytokine production. Proc. Natl. Acad. Sci. U. S. A. 2007;104:134–138. doi: 10.1073/pnas.0608388103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, You Y, Lin PC, Xue L, Morris SW, Zeng H, Wen R, Lin X. Bcl10 plays a critical role in NF-kappaB activation induced by G protein-coupled receptors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:145–150. doi: 10.1073/pnas.0601894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benigni A, Cassis P, Remuzzi G. Angiotensin II revisited: new roles in inflammation, immunology and aging. EMBO molecular medicine. 2010;2:247–257. doi: 10.1002/emmm.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips MI, Kagiyama S. Angiotensin II as a pro-inflammatory mediator. Current opinion in investigational drugs. 2002;3:569–577. [PubMed] [Google Scholar]
- 68.Delekta PC, Apel IJ, Gu S, Siu K, Hattori Y, McAllister-Lucas LM, Lucas PC. Thrombin-dependent NF-{kappa}B activation and monocyte/endothelial adhesion are mediated by the CARMA3.Bcl10.MALT1 signalosome. J. Biol. Chem. 2010;285:41432–41442. doi: 10.1074/jbc.M110.158949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J. Biol. Chem. 2009;284:6038–6042. doi: 10.1074/jbc.C800207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rehman AO, Wang CY. CXCL12/SDF-1 alpha activates NF-kappaB and promotes oral cancer invasion through the Carma3/Bcl10/Malt1 complex. International journal of oral science. 2009;1:105–118. doi: 10.4248/IJOS.09059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissues. World Health Organization. 2008 [Google Scholar]
- 72.Anderson JR, Armitage JO, Weisenburger DD. Epidemiology of the non-Hodgkin's lymphomas: distributions of the major subtypes differ by geographic locations. Non-Hodgkin's Lymphoma Classification Project. Ann. Oncol. 1998;9:717–720. doi: 10.1023/a:1008265532487. [DOI] [PubMed] [Google Scholar]
- 73.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 74.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. P. Lymphoma/Leukemia Molecular Profiling, The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 75.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, Staudt LM. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin. Cancer. Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 77.Cultrera JL, Dalia SM. Diffuse large B-cell lymphoma: current strategies and future directions. Cancer control : journal of the Moffitt Cancer Center. 2012;19:204–213. doi: 10.1177/107327481201900305. [DOI] [PubMed] [Google Scholar]
- 78.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 80.Ferch U, Kloo B, Gewies A, Pfander V, Duwel M, Peschel C, Krappmann D, Ruland J. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baens M, Fevery S, Sagaert X, Noels H, Hagens S, Broeckx V, Billiau AD, De Wolf-Peeters C, Marynen P. Selective expansion of marginal zone B cells in Emicro-API2-MALT1 mice is linked to enhanced IkappaB kinase gamma polyubiquitination. Cancer Res. 2006;66:5270–5277. doi: 10.1158/0008-5472.CAN-05-4590. [DOI] [PubMed] [Google Scholar]
- 83.Li Z, Wang H, Xue L, Shin DM, Roopenian D, Xu W, Qi CF, Sangster MY, Orihuela CJ, Tuomanen E, Rehg JE, Cui X, Zhang Q, Morse HC, 3rd, Morris SW. Emu- BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-kappaB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphoma. Blood. 2009;114:4158–4168. doi: 10.1182/blood-2008-12-192583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vicente-Dueñas C, Fontan L, Gonzalez-Herrero I, Romero-Camarero I, Segura V, Aznar MA, Alonso-Escudero E, Campos-Sanchez E, Ruiz-Roca L, Barajas-Diego M, Sagardoy A, Martinez-Ferrandis JI, Abollo-Jimenez F, Bertolo C, Penuelas I, Garcia-Criado FJ, Garcia-Cenador MB, Tousseyn T, Agirre X, Prosper F, Garcia-Bragado F, McPhail ED, Lossos IS, Du MQ, Flores T, Hernandez-Rivas JM, Gonzalez M, Salar A, Bellosillo B, Conde E, Siebert R, Sagaert X, Cobaleda C, Sanchez-Garcia I, Martinez-Climent JA. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10534–10539. doi: 10.1073/pnas.1204127109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, Garcia M, Cerchietti L, Shaknovich R, Yang SN, Fang F, Gascoyne RD, Martinez-Climent JA, Glickman JF, Borden K, Wu H, Melnick A. MALT1 Small Molecule Inhibitors Specifically Suppress ABC-DLBCL In Vitro and In Vivo. Cancer Cell. 2012;22:812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nagel D, Spranger S, Vincendeau M, Grau M, Raffegerst S, Kloo B, Hlahla D, Neuenschwander M, Peter von Kries J, Hadian K, Dorken B, Lenz P, Lenz G, Schendel DJ, Krappmann D. Pharmacologic Inhibition of MALT1 Protease by Phenothiazines as a Therapeutic Approach for the Treatment of Aggressive ABC-DLBCL. Cancer Cell. 2012;22:825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 87.Schlauderer F, Lammens K, Nagel D, Vincendeau M, Eitelhuber AC, Verhelst SH, Kling D, Chrusciel A, Ruland J, Krappmann D, Hopfner KP. Structural analysis of phenothiazine derivatives as allosteric inhibitors of the MALT1 paracaspase. Angewandte Chemie. 2013;52:10384–10387. doi: 10.1002/anie.201304290. [DOI] [PubMed] [Google Scholar]
- 88.Mc Guire C, Elton L, Wieghofer P, Staal J, Voet S, Demeyer A, Nagel D, Krappmann D, Prinz M, Beyaert R, van Loo G. Pharmacological inhibition of MALT1 protease activity protects mice in a mouse model of multiple sclerosis. Journal of neuroinflammation. 2014;11:124. doi: 10.1186/1742-2094-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jaworski M, Marsland BJ, Gehrig J, Held W, Favre S, Luther SA, Perroud M, Golshayan D, Gaide O, Thome M. Malt1 protease inactivation efficiently dampens immune responses but causes spontaneous autoimmunity. EMBO J. 2014 doi: 10.15252/embj.201488987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gewies A, Gorka O, Bergmann H, Pechloff K, Petermann F, Jeltsch KM, Rudelius M, Kriegsmann M, Weichert W, Horsch M. Uncoupling Malt1 Threshold Function from Paracaspase Activity Results in Destructive Autoimmune Inflammation. Cell Reports. 2014 doi: 10.1016/j.celrep.2014.10.044. [DOI] [PubMed] [Google Scholar]
- 91.Bornancin F, Renner F, Touil R, Sic H, Kolb Y, Touil-Allaoui I, Rush JS, Smith PA, Bigaud M, Junker-Walker U, Burkhart C, Dawson J, Niwa S, Katopodis A, Nuesslein-Hildesheim B, Weckbecker G, Zenke G, Kinzel B, Traggiai E, Brenner D, Brustle A, St Paul M, Zamurovic N, McCoy KD, Rolink A, Regnier CH, Mak TW, Ohashi PS, Patel DD, Calzascia T. Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J. Immunol. 2015;194:3723–3734. doi: 10.4049/jimmunol.1402254. [DOI] [PubMed] [Google Scholar]
- 92.Yu JW, Hoffman S, Beal AM, Dykon A, Ringenberg MA, Hughes AC, Dare L, Anderson AD, Finger J, Kasparcova V, Rickard D, Berger SB, Ramanjulu J, Emery JG, Gough PJ, Bertin J, Foley KP. MALT1 Protease Activity Is Required for Innate and Adaptive Immune Responses. PloS one. 2015;10:e0127083. doi: 10.1371/journal.pone.0127083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi L, Chen S, Lu Y, Wang X, Xu L, Zhang F, Yang L, Wu X, Li B, Li Y. Changes in the MALT1-A20-NF-kappaB expression pattern may be related to T cell dysfunction in AML. Cancer cell international. 2013;13:37. doi: 10.1186/1475-2867-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arora M, Kaul D, Varma N, Marwaha RK. Cellular proteolytic modification of tumorsuppressor CYLD is critical for the initiation of human T-cell acute lymphoblastic leukemia. Blood cells, molecules & diseases. 2014 doi: 10.1016/j.bcmd.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 95.Jabara HH, Ohsumi T, Chou J, Massaad MJ, Benson H, Megarbane A, Chouery E, Mikhael R, Gorka O, Gewies A, Portales P, Nakayama T, Hosokawa H, Revy P, Herrod H, Le Deist F, Lefranc G, Ruland J, Geha RS. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J. Allergy Clin. Immunol. 2013;132:151–158. doi: 10.1016/j.jaci.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McKinnon ML, Rozmus J, Fung SY, Hirschfeld AF, Del Bel KL, Thomas L, Marr N, Martin SD, Marwaha AK, Priatel JJ, Tan R, Senger C, Tsang A, Prendiville J, Junker AK, Seear M, Schultz KR, Sly LM, Holt RA, Patel MS, Friedman JM, Turvey SE. Combined immunodeficiency associated with homozygous MALT1 mutations. J. Allergy Clin. Immunol. 2014;133:1458–1462. 1462, e1451–e1457. doi: 10.1016/j.jaci.2013.10.045. [DOI] [PubMed] [Google Scholar]
- 97.Punwani D, Wang H, Chan AY, Cowan MJ, Mallott J, Sunderam U, Mollenauer M, Srinivasan R, Brenner SE, Mulder A, Claas FH, Weiss A, Puck JM. Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation. J. Clin. Immunol. 2015;35:135–146. doi: 10.1007/s10875-014-0125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, Cohen S, Shachar I, Miosge LA, Schlesier M, Fuchs I, Enders A, Eibel H, Grimbacher B, Warnatz K. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J. Allergy Clin. Immunol. 2013;131:477–485. e471. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 99.Greil J, Rausch T, Giese T, Bandapalli OR, Daniel V, Bekeredjian-Ding I, Stutz AM, Drees C, Roth S, Ruland J, Korbel JO, Kulozik AE. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J. Allergy Clin. Immunol. 2013;131:1376–1383. e1373. doi: 10.1016/j.jaci.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 100.Rosebeck S, Madden L, Jin X, Gu S, Apel IJ, Appert A, Hamoudi RA, Noels H, Sagaert X, Van Loo P, Baens M, Du MQ, Lucas PC, McAllister-Lucas LM. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science. 2011;331:468–472. doi: 10.1126/science.1198946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, Gevaert K, Beyaert R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 2011;13:1437–1442. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 104.Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Decaillet C, Grau M, Dorken B, Lenz P, Lenz G, Thome M. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruben SM, Klement JF, Coleman TA, Maher M, Chen CH, Rosen CA. I-Rel: a novel rel-related protein that inhibits NF-kappa B transcriptional activity. Genes Dev. 1992;6:745–760. doi: 10.1101/gad.6.5.745. [DOI] [PubMed] [Google Scholar]
- 106.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S. Malt1-Induced Cleavage of Regnase-1 in CD4 Helper T Cells Regulates Immune Activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 107.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, Gloury R, Martin N, Lohs C, Lech M, Stehklein JE, Geerlof A, Kremmer E, Weber A, Anders HJ, Schmitz I, Schmidt-Supprian M, Fu M, Holtmann H, Krappmann D, Ruland J, Kallies A, Heikenwalder M, Heissmeyer V. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat. Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 108.Baens M, Bonsignore L, Somers R, Vanderheydt C, Weeks SD, Gunnarsson J, Nilsson E, Roth RG, Thome M, Marynen P. MALT1 auto-proteolysis is essential for NF-kappaBdependent gene transcription in activated lymphocytes. PloS one. 2014;9:e103774. doi: 10.1371/journal.pone.0103774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nie Z, Du MQ, McAllister-Lucas LM, Lucas PC, Bailey NG, Hogaboam CM, Lim MS, Elenitoba-Johnson KS. Conversion of the LIMA1 tumour suppressor into an oncogenic LMO-like protein by API2-MALT1 in MALT lymphoma. Nature communications. 2015;6:5908. doi: 10.1038/ncomms6908. [DOI] [PubMed] [Google Scholar]
- 110.Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–4407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- 111.Eitelhuber AC, Vosyka O, Nagel D, Bognar M, Lenze D, Lammens K, Schlauderer F, Hlahla D, Hopfner K, Lenz G, Hummel M, Verhelst SH, Krappmann D. Activity-Based Probes for Detection of Active MALT1 Paracaspase in Immune Cells and Lymphomas. Chem. Biol. 2014 doi: 10.1016/j.chembiol.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 112.Hachmann J, Edgington-Mitchell LE, Poreba M, Sanman LE, Drag M, Bogyo M, Salvesen GS. Probes to Monitor Activity of the Paracaspase MALT1. Chem. Biol. 2014 doi: 10.1016/j.chembiol.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 114.Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Curr. Opin. Chem. Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat. Immunol. 2013 doi: 10.1038/ni.2540. [DOI] [PubMed] [Google Scholar]
- 116.Hachmann J, Edgington-Mitchell LE, Poreba M, Sanman LE, Drag M, Bogyo M, Salvesen GS. Probes to Monitor Activity of the Paracaspase MALT1. Chem. Biol. 2015;22:139–147. doi: 10.1016/j.chembiol.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]