Abstract
Cell-cell communication is also known as quorum sensing (QS) that happens in the bacterial cells with the aim to regulate their genes expression in response to increased cell density. In this study, a bacterium (L8A) isolated from dental plaque biofilm was identified as Citrobacter amalonaticus by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). Its N-acylhomoserine-lactone (AHL) production was screened by using two types of AHL biosensors namely Chromobacterium violaceum CV026 and Escherichia coli [pSB401]. Citrobacter amalonaticus strain L8A was identified and confirmed producing numerous types of AHL namely N-butyryl-L-homoserine lactone (C4-HSL), N-hexanoyl-L-homoserine lactone (C6-HSL), N-octanoyl-L-homoserine lactone (C8-HSL) and N-hexadecanoyl-L-homoserine lactone (C16-HSL). We performed the whole genome sequence analysis of this oral isolate where its genome sequence reveals the presence of QS signal synthase gene and our work will pave the ways to study the function of the related QS genes in this bacterium.
In the past few generations, dental scientists and microbiologists as well as microbial ecologists have researched on the identification of oral microbes which have led to oral diseases. Dental plaque has been reported as an important microhabitat for oral micro-organisms1. Dental plaque forms naturally on the teeth and aids in colonization of exogenous species, which disturbs the microbial homeostasis within oral cavity and becomes predisposed sites to the disease2. This diseased dental plaque influences the changes in environmental conditions and plays a vital role in the site specific diseases such as dental caries, gingivitis and periodontitis3. In addition, dental plaque which acts as a reservoir of Gram-negative bacteria and periodontium as a reservoir of inflammatory mediators indirectly play a role in systemic diseases such as bacteremia, cardio-vascular diseases, bacterial pneumonia, diabetes mellitus and low birth weight4. Previous researches show that a minimum number of 800 bacterial species are present in the oral cavity5,6 out of which total estimate of 415 species and 68 unseen species were reported from sub-gingival dental plaque5. The number of oral bacteria may be increasing with the advancement of biotyping techniques, for example mass sequencing technologies7,6, mass spectrometry analysis such as MALDI-TOF8 and next generation sequencing9.
On the other hand, quorum sensing (QS) has been well studied as bacteria genes regulation by signalling molecules termed autoinducers which will increase according to the cell density10,11. N-acylated-L-homoserine lactones (AHLs) and autoinducer AI-2 are the two common classes of autoinducers used by Gram-negative bacteria in their QS system12. With the availability of a numbers of AHL biosensors, this has greatly facilitated the screening of AHL production by using the lux, gfp or lacZ AHL biosensors reporter gene fusions or pigment induction13,14. AHL varies in acyl group length (C4 to C18), presence or absence of carbon-carbon double bond in the fatty acid chain and substitution of C3 (hydrogen, oxo- and hydroxyl group). To date, QS is an attractive approach for antimicrobial therapy in the treatment of infectious disease15 as it does not involve the use of antibiotics.
Citrobacter amalonaticus (formerly known as Levinea amalonaticus) is a Gram-negative rod-shaped facultative anaerobe with chemoheterotrophic metabolism16. The genus Citrobacter is described as an opportunistic human pathogen as it has the potential to cause neonatal meningitis17 and urinary tract infections18. It can be found in urine, sputum, soft tissue exudates and even water or food17,19. However, there is no report reveals the oral microbe associated Citrobacter species and the valuable information about the genomic study on Citrobacter amalonaticus is therefore, limited. To-date, next generation sequencing technology has revolutionized the genomic research by generating sequences data in hours or days. The aim of this study is to understand the occurrence of QS in C. amalonaticus isolated from dental plaque by studying the AHL production profile and insights gained from the annotated genomic features of C. amalonaticus strain L8A.
Results
Strain L8A identification
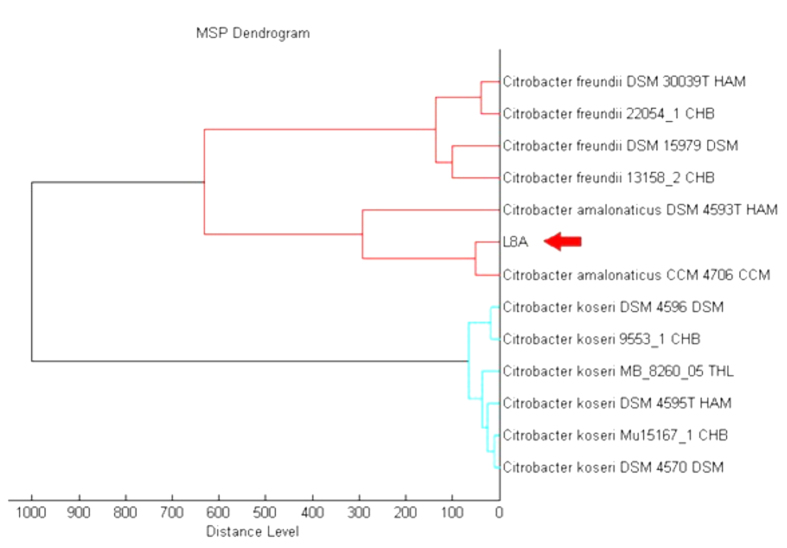
Initial microbial identification of strain L8A was done by Microflex MALDI-TOF Mass Spectrometry system. MALDI-TOF MS has been regarded as a reliable tool for microorganism identification20. A number of researchers reported the feasibility of MALDI-TOF MS analysis was able to classify and differentiate clinical isolates to species level21,22. Using MALDI-TOF mass spectrometry, strain L8A was identified at species level with score value of 2.390 and it was best matched to Citrobacter amalonaticus. The MALDI Biotyper MSP software was used as further support to generate the dendrogram of C. amalonaticus L8A with its closest match (Fig. 1).
Figure 1. Score-orientated dendrogram of strain L8A.
Bacterial strain L8A was clustered hierarchically based on the protein mass spectra patterns with distance value normalized to maximum value of 1,000.
QS signaling molecules production in C. amalonaticus L8A
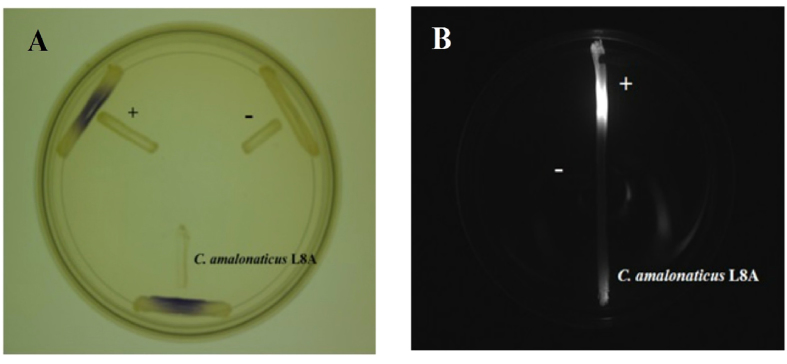
Production of QS activity by strain L8A was detected when it was cross-streaked with C. violaceum CV026 and E. coli [pSB401] AHL biosensors. Both purple violacein pigmentation (Fig. 2A) and bioluminescence activities (Fig. 2B) were induced by strain L8A at the meeting point between two strains. In particular, L8A showed positive result with both bioreporters used in this study.
Figure 2. Preliminary AHL screening with cross streak bioassay.
(A) Short chain AHL screening of strain L8A with CV026. E. carotovora PNP22 (negative control “−”) and E. carotovora GS101 (positive control “+”) were included for comparison. (B) AHL screening of strain L8A with E. coli [pSB401]. E. carotovora PNP22 (negative control “−”) and E. carotovora GS101 (positive control “+”) were included for comparison.
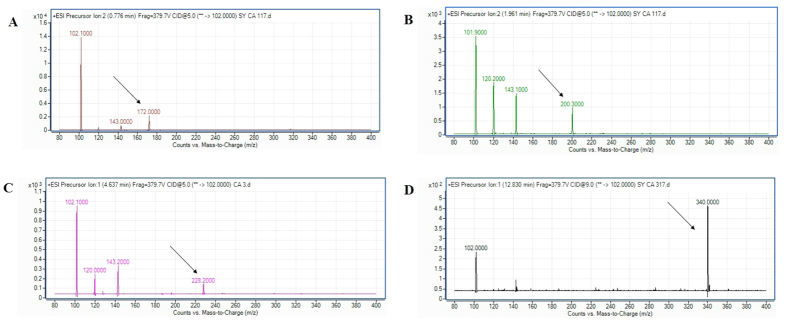
The extracted AHL from spent culture supernatant of strain L8A was analyzed using the high resolution triple quadrupole LC/MS mass spectrometry (MS) system and confirmed that C. amalonaticus L8A produced four different types of AHL molecules; namely N-butyryl-L-homoserine lactone (C4-HSL) (Fig. 3A), N-hexanoyl-L-homoserine lactone (C6-HSL) (Fig. 3B), N-octanoyl-L-homoserine lactone (C8-HSL) (Fig. 3C) and N-hexadecanoyl-L-homoserine lactone (C16 HSL) (Fig. 3D).
Figure 3. Mass spectrometry analysis of spent supernatants extract C. amalonaticus strain L8A.
(A) Mass spectra of C4-HSL (m/z 172.0000) (marked by arrow). (B) Mass spectra of C6-HSL (m/z 200.3000) (marked by arrow). (C) Mass spectra of C8-HSL (m/z 228.2000) (marked by arrow). (D) Mass spectra of C16-HSL (m/z 340.0000) (marked by arrow).
Data analysis on genomic features of strain L8A
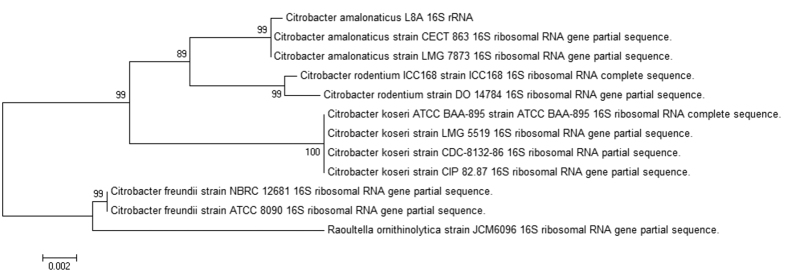
De novo genome assembly of C. amalonaticus L8A using CLC Genomics Workbench generated 110 contigs with the genome size of 5,273,145 bp and average 40-fold coverage. This genome has average GC content of 54.4% and encodes 4972 putative DNA-coding sequences. With RNAmmer23 and tRNAscan-SE24 tool, 16S, 23S and 5S rRNA genes and 70 tRNA genes were predicted on the chromosome of C. amalonaticus L8A. The 16S rRNA gene was found in contig 18 of the genome with the length of 1,542 bp. The 16S rRNA maximum likelihood phylogenetic tree was rooted using Raoultella ornithinolytica JCM6096 as outgroup species. Phylogenetic relationship inferred the highest 16S rRNA gene sequence 99% identical to Citrobacter amalonaticus (Fig. 4).
Figure 4. Maximum likelihood phylogenetic tree of C. amalonaticus L8A 16S rRNA sequence and other closely related sequences.
The phylogenetic analysis of 16S rRNA of the strain L8A was hierarchical clustered under C. amalonaticus using MEGA 5.1.
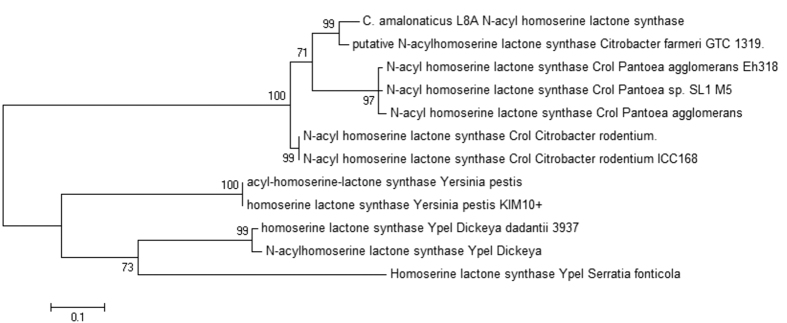
AHL-mediated QS system consists of three fundamental components namely LuxI-type autoinducer synthase (signal generator), AHL ligand (the signal itself) and LuxR-type receptor (cognate receptor)25,26,12. LuxI-type autoinducer synthase gene was determined and extracted from the genome sequence of C. amalonaticus L8A for further phylogenetic study. The 212 bp of putative protein-coding gene of C. amalonaticus L8A N-acylhomoserine lactone synthase was grouped with C. farmeri GTC1319 QS synthase gene (Fig. 5). C. amalonaticus L8A QS gene was further annotated using RAST server. The genome feature of QS homoserine lactone synthase gene and QS transcriptional activator gene were found in contig 12. The synteny analysis of annotated QS features were illustrated in Fig. 6 using Easyfig27 version 2.2.
Figure 5. Phylogenetic tree based on QS synthase gene in L8A.
In silico study of QS synthase gene of genome strain L8A was determined and represented in maximum likelihood phylogenetic tree.
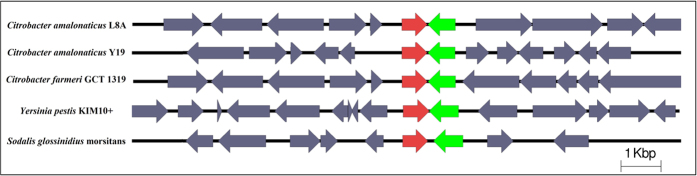
Figure 6. QS synthase gene and regulatory gene in L8A.
C. amalonaticus L8A QS genes were synteny analyzed using Easyfig (red arrow indicated the QS synthase gene and green arrow indicated the QS transcriptional regulator gene).
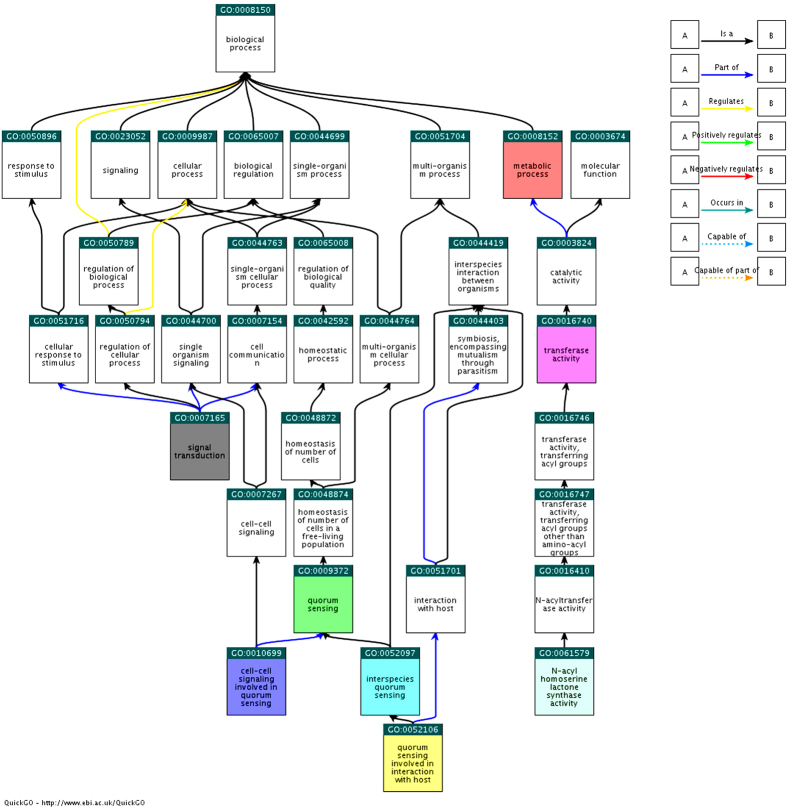
Association between these AHL synthase related terms and gene products is given in Fig. 7. As displayed in the comparison chart, interspecies QS and QS involved in interaction with host as well as implication of cell-cell signaling were predicted to happen in subclasses of QS which was classified as a type of homeostasis of number of cells within a population of free-living cells such as oral microbiome under biological process. Symbiotic interaction inclusive of interaction with host could be encompassed by genome L8A in oral cavity through mutualism or parasitism. In molecular function, mechanism of N-acylhomoserine lactone synthase activity in L8A was simulated and found to be associated with transferase activity. Occurrence of this transferase activity in genome L8A involved in catalysis of transferring acyl group to nitrogen atom in AHL. The term ‘N-acylhomoserine lactone synthase activity’ was linked to QS pathway via metabolic process under biological process.
Figure 7. Comparison chart of eight QS-associated Gene Ontology terms using QuickGO (showing in highlight).
Different relationships between the terms are indicated by unique colour of arrows as depicted in the right panel.
Multiple genomes comparison
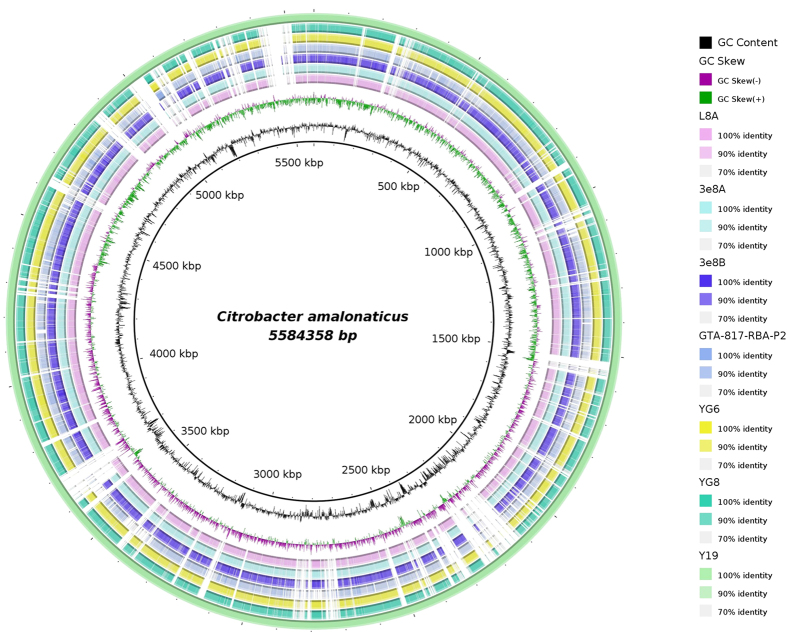
To date, there are only seven whole genome sequences publicly available from NCBI database inclusive L8A genome. Genome comparison of these seven C. amalonaticus genomes with clinical isolates (n = 3), environmental (n = 2), food (n = 1) and industrial (n = 1) was performed using BRIG (Fig. 8) in circular visualization. Among these seven genomes, six were draft genomes and one was complete genome which was used as reference genome. Average genome size was 5.139 Mb with GC content of 53.35%.
Figure 8. BRIG visualization of multiple C. amalonaticus genomes available from NCBI database.
The innermost rings show GC skew (purple/green) and GC content (black). The third innermost ring shows L8A genome and followed by strains 3e8A (CDQV01) and 3e8B (CDQX01) isolated from undernourished Malawian children’s gut. The remaining rings represent the ground beef isolate GTA-817-RBA-P2 (LAMY01) and environmental strains YG6 (LIGA01) and YG8 (LIGB01) prior the outermost ring with complete genome strain Y19.
Nucleotide sequence accession number
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JMQQ00000000. The version described in this paper is version JMQQ01000000.
Discussion
Citrobacter sp. is an opportunistic pathogen that infrequent infects neonates and in immunocompromised adults or older children28 who constantly follow the prophylactic β-lactam therapy29. However, C. amalonaticus infection is rare and only few cases have been described worldwide. In 2005, a 53 years old American male tourist was admitted to hospital and diagnosed with enteric fever syndrome caused by C. amalonaticus infection. He had initial cefriaxone treatment followed by co-trimoxazole for a period of 23 days30. Other case reports responded to bacterial infection with this species mostly happened in young pregnant female with underlying conditions such as urinary tract infection, vesico vaginal fistula and acute pyelonephritis31,32,33.
Bacteria in Citrobacter genus are currently divided into 11 species19. They are genotypically and biochemically diverse. In order to decrease the possibility of misidentification, molecular method and whole genome sequencing of strain L8A was performed. Both MALDI-TOF mass spectrometry and 16S rRNA gene results confirmed strain L8A is C. amalonaticus. Compared to the Human Oral Microbiome Database (HOMD)34 as part of Human Microbiome Project (HMP) and other literatures, surprisingly this strain has not been reported to be associated with oral cavity. When the 16S rRNA gene of C. amalonaticus L8A was searched (BLASTn) against the HOMD 16S rRNA reference sequences, the results returned the highest percentage identical to Enterobacter hormaechei references is only 97.7%, followed by Kluyvera ascorbata (97.5%) and E. coli (97.4%). This finding disclosed the existence of C. amalonaticus in the human oral cavity.
In general, Gram-negative bacteria communicate via expression of exogenous small diffusible molecules known as N-acyl homoserine lactone (AHL) to mediate gene expression in a cell density dependent manner. Frias and colleagues reported that QS in Porphyromonas gingivalis play a role in controlling the production of its virulence factor in oral cavity35. Besides, previous research suggested that AHL gene in Citrobacter rodentium has an unexpected role in affecting the mouse’s virulence factors in the manner of attaching and effacing lesions through QS36.
Typically, LuxI-like autoinducer synthase and LuxR-like receptor are clustered in pair. There are some articles reported that functional LuxI/LuxR pairs are not located on common bacterial chromosome or plasmids37,38 or absence of LuxI autoinducer synthase in bacterial QS circuit39. In order to identify the QS signal synthase and its cognate receptor genes, we performed the whole genome sequencing on C. amalonaticus L8A. Upon analysis of its genome sequence, we found that LuxI/LuxR functional pair of C. amalonaticus L8A is genetically adjacent and clustered on the same chromosome. The detected QS activity in this strain could be an alternative solution for treatment as most Gram-negative bacteria depend on QS to regulate virulence factors expression40.
Additionally, the pathway analyses of Gene Ontology terms had conveyed a simplified biological insight for large proteomic and genomic datasets. Multiple GO terms could be used for a single gene annotation manually or electronically with references. The QS pathway showing that interspecies QS and QS interplay with host occurred in L8A genome. Interspecies signaling between bacterial species via small diffusible AHL molecules has been suggested by Eberl and Tümmler41; it happened in patients with cystic fibrosis between Pseudomonas aeruginosa and Burkholderia cepacia. QS involves in interaction with host, in particular, a process applied by community of single-cell L8A microorganism living in intimate contact with host and controlling the population density with the concentration of signaling molecules. The host here referred to a larger organism which responded to symbiotic interaction42,43. In this case, strain L8A is able to use their available proteins to adhere to each other and form layer of biofilm. However, the information about the possible role of AHL-regulated QS of C. amalonaticus is still a mystery. Therefore, it is vital to link the possible proteins to signaling pathway for later experiment design and evaluation and future work in finding out anti-QS-based solution to control the oral cavity diseases.
In addition to this, overview of sequence feature information is able to be visualized in the context of sequence analysis results generated through the comparative genomics tool. The sole complete genome Citrobacter amalonaticus Y19 (CP011132) was assigned as reference genome in this comparative study. This strain was reported with capability to produce hydrogen from oxidizing toxic carbon monoxide44. The output image shows differences and similarity between the query genomes and reference genome, where BLAST matches are indicated as coloured sliding scale at side. From the comparison study, we can predict the possible antimicrobial resistance genes, important genomic traits for industrial application and other genomic features.
In summary, for the first time, we have profiled the unusual various types of AHLs produced by C. amalonaticus strain L8A isolated from dental plaque. Coupling with next generation sequencing technology and systematic bioinformatics analysis, our study has provided insights into C. amalonaticus genome features and invaluable information about this bacterial strain notably on the QS system. Besides this, the genome C. amalonaticus L8A reported in this work can be exploited as reference genome for future comparative, functional and clinical study of Citrobacter genus. The genome sequence may be useful for research on any related genes through its nucleotide sequences analysis, orientation, upstream and downstream regulatory elements and gene products, as well as its miscellaneous genomics features. By elucidating the QS genes in C. amalonaticus, this will pave the way to understand the QS regulation in this bacterium. In addition, this work also illustrated that oral cavity that could be potential reservoir for QS pathogen and hence more intense research should be carried out.
This is the first documentation reported C. amalonaticus possesses QS activity, whole genome sequencing, and its AHL production namely C4-HSL, C6-HSL, C8-HSL, and C16-HSL.
Materials and Methods
Ethics statement
All of the procedures were carried out in accordance with guidelines approved by the Medical Ethics Committee Faculty of Dentistry, University of Malaya (DFRD-1302/0033-L). Informed consent was obtained from all subjects prior to commencement of this study.
Sample Collection and Bacterial Strains Isolation
Dental plaque biofilms samples were collected from healthy patients who were scheduled to have their teeth extracted due to deep caries at the Primary Care Unit, Faculty of Dentistry. Soft and loose dental plaque around the buccal and proximal surfaces of the carious teeth was gently curated using sterile Gracey curettes. The collected samples were placed into a sterile microcentrifuge tube with PBS buffer concentration of 0.01 M at pH 7.4, maintained at 4 °C and transported to the laboratory for further analysis within 24 hours. Briefly, saline sample was vortex-homogenized for 1 min prior to 100 μL in aliquots spread on Lysogeny Broth (LB) medium and aerobically cultured overnight at 37 °C. The isolated bacterial strains from dental plaque samples were repeatedly streaked on LB agar to obtain the pure colony.
MALDI-TOF application for strain identification
MALDI-TOF mass spectrometry (MS) was used to identify the bacterial strains. Briefly, pure colony of interested strain was smeared thinly onto each well of MSP 96 MALDI Target plate prior to adding 1μl of HCCA MALDI matrix and allow the mixture to co-crystallize45. The target plate was then subjected to automated MALDI Biotyper bench-top mass spectrometer using FlexControl software version 3.3. Bruker MALDI Biotyper Real Time Classification (RTC) (Version 3.1) software was implemented to analyze the generated spectra by standard pattern matching with default settings and identification was provided with score value46. In addition, the score-oriented dendrogram was constructed using MALDI Biotyper MSP creation method.
Preliminary screening of AHLs
Two different short chain AHLs bacterial biosensors namely Chromobacterium violaceum CV026 and Escherichia coli [pSB401] were employed for the preliminary AHLs screening of bacterial isolates. Erwinia carotovora GS101 and E. carotovora PNP22 were used as positive and negative controls, respectively45,47. These AHLs biosensors and controls were cultivated overnight in LB medium at 28 °C. Bacterial isolates were then screened for its AHLs production by cross-streaking with AHL biosensors. Purple violacein pigmentation formed in C. violaceum CV026 and intensity of bioluminescence of E. coli [pSB401] will increase if exogenous short chain AHL was detected48,49. The bioluminescence image was electronically documented by Hamamatsu Photonics photon camera (Hamamatsu, Japan).
Extraction of AHLs
The QS positive strain was grown overnight in LB broth buffered with 50 mM 3-(N-morpholino) propanesulfonic acid (MOPS) (pH 5.5) in 37 °C incubator shaker with 200 rpm. The acidified organic solvent (0.1% v/v glacial acetic acid) ethyl acetate was used to extract the supernatant culture twice as reported previously50. The extracted supernatant was evaporated to complete dryness in fume hood. The dried extract was resuspended in 200 μL of acetonitrile (HPLC grade) and vortex vigorously to dissolve the extracted AHLs. The mixture was then centrifuged and any insoluble residue was discarded. The dissolved sample (100 μL) was inserted into sample vials for mass spectrometry analysis.
AHL Profiling by High Resolution Tandem Liquid Chromatography Mass Spectrometry (MS)
The extracted AHLs profile of the bacterial strain was analyzed using high resolution MS namely Agilent 1290 Infinity LC system and Agilent 6490 Triple Quadrupole LC/MS system (Agilent Technologies Inc., Santa Clara, CA, USA) associated with Agilent ZORBAX Rapid Resolution High Definition SB-C18 Threaded Column. Mobile phases A and B used in this study were water and acetonitrile (both mobile phases added with 0.1% v/v formic acid), respectively with an initial ratio of 80:20. The parameters included flow rate, temperature were set as described previously46,51. The precursor ion scan mode will scan through m/z value from 150 to 400 and target the m/z 102 [M+H]+ product ion which is the characteristic of the lactone ring moiety and indicates the presence of AHL. The generated EI mass spectra were then compared to the standard AHL profile based on the retention index using the Agilent MassHunter software.
Library preparation for next generation sequencing
Total DNA of the strain was extracted using MasterPureTM Complete DNA and RNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA), performed as recommended in the manufacturer protocol. The quality and quantity of DNA were examined using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA), respectively, prior to library preparation using Illumina Nextera DNA Sample Preparation Kit (Illumina, USA). The quality control of the DNA library template was analyzed on Agilent 2100 Bioanalyzer system using High Sensitivity DNA Analysis Kit (Agilent Technologies, USA) whereas the concentration was determined using Illumina Eco qPCR machine, as described in the KAPA Library Quantification Kits for Illumina sequencing platforms (KAPA BioSystems, Boston, MA, USA). By calculating from qPCR reading, library template was diluted to 2 nM in Tris-HCl buffer (10mM, pH 8.5 with 0.1% v/v Tween 20). The DNA library was denatured and further diluted to 10 pM before loaded into cartridge for MiSeq (Illumina, USA) next generation sequencing platform, guided by the MiSeq Control Software (MSC) interface.
Genome assembly and annotation
The raw reads generated by MiSeq were first quality evaluated using FastQC analyzer52 prior to trimming the bad reads based on their base quality, filtering according to read length and ambiguous nucleotides were excluded using CLC Genomics Workbench53 version 7.0.4. In this genome data analysis, raw reads with average quality value lower than Q20 were excluded together with ambiguous nucleotide. In order to obtain the best assembly result, de novo genome assemblies were performed multiple times using different word size parameter from post-filtered reads with minimum read-length of 15 nucleotides.
A rapid prokaryotic genome annotation system (Prokka)54 was used to annotate the draft genome of strain L8A. All predicted genes by Prodigal55 version 2.60 were searched against the databases of known sequences (NCBI-NT/NR databases) using BLAST tool in order to assign appropriate functional annotation. In addition, the genome of strain L8A was subject to automatic SEED-based annotation using Rapid Annotation Subsystems Technology (RAST) server56. Easyfig version 2.2 was implemented for downstream synteny analysis of QS synthase and regulator genes.
Phylogenetic analysis
The predicted protein coding sequences of 16S rRNA and QS synthase gene sequences were extracted from strain L8A whole genome sequence with the aim to study their phylogenetic relationships with the related gene sequences available in public databases. Molecular Evolutionary Genetic Analysis version 5.1 (MEGA)57 tool was used to construct maximum likelihood (ML) phylogenetic trees with GTR nucleotide substitution model and 1,000 bootstrap values.
In-silico discovery QS system
The analysis of Gene Ontology terms or physical interaction network associated with amino acid regulated genes based on their involvement in cellular component, biological process and molecular function. With the accession number of N-acyl-homoserine lactone synthase of strain L8A (A0A083ZD14) which provided by UniProt Knowledgebase (UniProtKB), pathway was determined by fast web-based GO browser, QuickGO58. Five AHL synthase related GO terms coupled with three child terms from the term of ‘quorum sensing’ were used to construct the comparison chart.
Comparative genome studies of strain L8A
To perform genomes comparison for strain L8A, the assembled contigs were ordered and re-oriented by aligning them to a closely-related reference genome using Mauve59 and concatenated to produce a pseudo-molecule. The comparison files between the query pseudo-molecule genome sequence and reference genome sequence were created using the local BLASTn program to show the percentage of match hit between them. The structural similarities and differences between the genomes sequences were then visualized using BLAST Ring Image Generator (BRIG)60 software.
Additional Information
How to cite this article: Goh, S.-Y. et al. Quorum sensing activity of Citrobacter amalonaticus L8A, a bacterium isolated from dental plaque. Sci. Rep. 6, 20702; doi: 10.1038/srep20702 (2016).
Acknowledgments
This work is supported by the University of Malaya, High Impact Research Grants, Ministry of Education, Malaysia (no. H-50001-A000027 and A-000001-50001) awarded to Kok-Gan Chan.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.Y.G. and W.F.Y. conducted the experiments. S.A.K. and N.H.A.K. obtained the dental sample, applied and obtained the medical ethics approval. S.Y.G., S.A.K., K.K.T., N.H.A.K., K.G.C. and W.F.Y. analysed the data and contributed to the writing of the manuscript. K.G.C. and N.H.A.K. conceived the idea, K.G.C. executed the project, obtained funding and prepared the manuscript. All authors reviewed and approved the final manuscript for publication.
References
- Nobbs A. H., Jenkinson H. F. & Jakubovics N. S. Stick to your gums mechanisms of oral microbial adherence. J Dental Res. 90, 1271–1278, doi: 10.1177/0022034511399096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8.2, 263–271, doi: 10.1177/08959374940080022001 (1994). [DOI] [PubMed] [Google Scholar]
- Liljemark W. F. & Bloomquist C. Human oral microbial ecology and dental caries and periodontal diseases. Crit. Rev. Oral. Biol. Med. 7.2, 180–198, doi: 10.1177/10454411960070020601 (1996). [DOI] [PubMed] [Google Scholar]
- Li X., Kolltveit K. M., Tronstad L. & Olsen I. Systemic diseases caused by oral infection. Clin. Microbiol. Rev. 13, 547–558, doi: 10.1128/CMR.13.4.547-558.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J. et al. Bacterial diversity in human subgingival plaque. Journal of Bacteriology 183, 3770–3783, doi: 10.1128/JB.183.12.3770-3783.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filoche S., Wong L. & Sissons C. H. Oral biofilms: emerging concepts in microbial ecology. J Dental Res. 89, 8–18 (2010). [DOI] [PubMed] [Google Scholar]
- Keijser B. J. F. et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dental Res. 87, 1016–1020, doi: 10.1177/154405910808701104 (2008). [DOI] [PubMed] [Google Scholar]
- Croxatto A., Prod’hom G. & Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36, 380–407, doi: 10.1111/j.1574-6976.2011.00298.x (2012). [DOI] [PubMed] [Google Scholar]
- Chan X. Y., Arumugam R., Choo S. W., Yin W. F. & Chan K. G. Metagenomic sequencing of prokaryotic microbiota from tropical surface seawater. Genome Announc. 1, e00540–13, doi: 10.1128/genomeA.00540-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooshangi S. & Bentley W. E. From unicellular properties to multicellular behavior: bacteria quorum sensing circuitry and applications. Curr. Opin. Biotechnol. 19, 550–555, doi: 10.1016/j.copbio.2008.10.007 (2008). [DOI] [PubMed] [Google Scholar]
- Vu B., Chen M., Crawford R. J. & Ivanova E. P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554, doi: 10.3390/molecules14072535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway W. R., Hodgkinson J. T., Bowden S. D., Welch M. & Spring D. R. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem.l Rev. 111, 28–67, doi: 10.1021/cr100109t (2010). [DOI] [PubMed] [Google Scholar]
- Lei Y., Chen W. & Mulchandani A. Microbial biosensors. Anal. Chim. Acta. 568, 200–210, doi: 10.1016/j.aca.2005.11.065 (2006). [DOI] [PubMed] [Google Scholar]
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938, doi: 10.1099/mic.0.2007/012856-0 (2007). [DOI] [PubMed] [Google Scholar]
- Cegelski L., Marshall G. R., Eldridge G. R. & Hultgren S. J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27, doi: 10.1038/nrmicro1818 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y. K., Kim H. J., Park S., Kim M. S. & Ryu D. D. Metabolic-flux analysis of hydrogen production pathway in Citrobacter amalonaticus Y19. Int. J. Hydrogen Energ. 33, 1471–1482, doi: 10.1016/j.ijhydene.2007.09.032 (2008). [DOI] [Google Scholar]
- Lipsky B. A., Hook E. W., Smith A. A. & Plorde J. J. Citrobacter infections in humans: experience at the Seattle Veterans Administration Medical Center and a review of the literature. Rev, Infect. Dis. 2, 746–760, doi: 10.1093/clinids/2.5.746 (1980). [DOI] [PubMed] [Google Scholar]
- Zhanel G. G. et al. A Canadian national surveillance study of urinary tract isolates from outpatients: comparison of the activities of trimethoprim-sulfamethoxazole, ampicillin, mecillinam, nitrofurantoin, and ciprofloxacin. Antimicrob. Agents Chemother. 44, 1089–1092, doi: 10.1128/AAC.44.4.1089-1092.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran T. I. The role of Citrobacter in clinical disease of children: review. Clin. Infect. Dis. 28, 384–394, doi: 10.1086/515106 (1999). [DOI] [PubMed] [Google Scholar]
- Keough T., Youngquist R. S. & Lacey M. P. A method for high-sensitivity peptide sequencing using postsource decay matrix-assisted laser desorption ionization mass spectrometry. Proc. Natl. Acad. Sci. USA 96, 7131–7136, doi: 10.1073/pnas.96.13.7131 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E., Maier T., Urban E., Terhes G. & Kostrzewa M. Species identification of clinical isolates of Bacteroides by matrix‐assisted laser‐desorption/ionization time‐of‐flight mass spectrometry. Clin. Microbiol. Infect. 15, 796–802, doi: 10.1111/j.1469-0691.2009.02788.x (2009). [DOI] [PubMed] [Google Scholar]
- Cherkaoui A. et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48, 1169–1175, doi: 10.1128/JCM.01881-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108, doi: 10.1093/nar/gkm160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 0955–964, doi: 10.1093/nar/25.5.0955 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. B. & Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296, 149–161, doi: 10.1016/j.ijmm.2006.02.005 (2006). [DOI] [PubMed] [Google Scholar]
- Geske G. D., O’Neill J. C. & Blackwell H. E. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chem. Soc. Rev. 37, 1432–1447, doi: 10.1039/B703021P (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K. & Beatson S. A. Easyfig: a genome comparison visualizer. Bioinformatics, 27, 1009–1010, doi: 10.1093/bioinformatics/btr039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J. G., Krieg N. R., Sneath P. H. A., Staley J. T. & Williams S. T. Group 5 facultatively anaerobic gram-negative rods in Bergey’s Manual of Determinative Bacteriology (9th ed. Williams & Wilkins) 177–178 (Baltimore, Md., 1994). [Google Scholar]
- Underwood S. & Avison M. B. Citrobacter koseri and Citrobacter amalonaticus isolates carry highly divergent β-lactamase genes despite having high levels of biochemical similarity and 16S rRNA sequence homology. J. Antimicrob. Chemother. 53, 1076–1080, doi: 10.1093/jac/dkh235 (2004). [DOI] [PubMed] [Google Scholar]
- Suwansrinon K., Wilde H., Sitprija V. & Hanvesakul R. Enteric fever-like illness caused by infection with Citrobacter amalonaticus. J. Med. Assoc.Thai. 88, 837–840 (2005). [PubMed] [Google Scholar]
- Rizvi M., Khan F., Shukla I. & Malik A. & Shaheen. Rising prevalence of antimicrobial resistance in urinary tract infections during pregnancy: necessity for exploring newer treatment options. J. Lab. Physicians 3, 98–103, doi: 10.4103/0974-2727.86842 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabaya A. & Auta B. Microorganisms associated with the urinogenital system of Vesico Vaginal Fistula (VVF) patients in north western Nigeria. Science World J. 1, 37 (2007). [Google Scholar]
- Artero A., Alberola J., Eiros J. M., Nogueira J. M. & Cano A. Pyelonephritis in pregnancy. How adequate is empirical treatment? Rev. Esp. Quimioter. 26, 30–33 (2013). [PubMed] [Google Scholar]
- Chen T. et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, baq013, doi: 10.1093/database/baq013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias J., Olle E. & Alsina M. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69, 3431–3434, doi: 10.1128/IAI.69.5.3431-3434.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst S. J. et al. Quorum sensing has an unexpected role in virulence in the model pathogen Citrobacter rodentium. EMBO Rep. 8, 698–703, doi: 10.1038/sj.embor.7400984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S. & Venturi V. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385, doi: 10.1099/mic.0.026849-0 (2009). [DOI] [PubMed] [Google Scholar]
- Brameyer S., Kresovic D., Bode H. B. & Heermann R. Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. USA 112, 572–577, doi: 10.1073/pnas.1417685112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar A. V. & González J. E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33, 739–756, doi: 10.1111/j.1574-6976.2009.00163.x (2009). [DOI] [PubMed] [Google Scholar]
- Fuqua C. & Greenberg E. P. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695, doi: 10.1038/nrm907 (2002). [DOI] [PubMed] [Google Scholar]
- Eberl L. & Tümmler B. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 294, 123–131, doi: 10.1016/j.ijmm.2004.06.022 (2004). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056, doi: 10.1093/nar/gku1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainala S. K., Seol E. & Park S. Complete genome sequence of novel carbon monoxide oxidizing bacteria Citrobacter amalonaticus Y19, assembled de novo. J. Biotechnol. 211, 79–80, doi: 10.1016/j.jbiotec.2015.07.012 (2015). [DOI] [PubMed] [Google Scholar]
- Chen J. W., Koh C. L., Sam C. K., Yin W. F. & Chan K. G. Short chain N-acylhomoserine lactone production by soil isolate Burkholderia sp. strain A9. Sensors 13, 13217–13227, doi: 10.3390/s131013217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngeow Y. F., Cheng H. J., Chen J. W., Yin W. F. & Chan K. G. Short chain N-acylhomoserine lactone production by clinical multidrug resistant Klebsiella pneumoniae strain CSG20. Sensors 13, 15242–15251, doi: 10.3390/s131115242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong T. M. et al. Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian tropical montane forest. Sensors 12, 4846–4859, doi: 10.3390/s120404846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean K. H. et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143, 3703–3711, doi: 10.1099/00221287-143-12-3703 (1997). [DOI] [PubMed] [Google Scholar]
- Winson M. K. et al. Construction and analysis of luxCDABE‐based plasmid sensors for investigating N‐acyl homoserine lactone‐mediated quorum sensing. FEMS Microbiol. Lett. 163, 185–192, doi: 10.1111/j.1574-6968.1998.tb13044.x (1998). [DOI] [PubMed] [Google Scholar]
- Ortori C. A. et al. Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4 (1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal. Bioanal. Chem. 399, 839–850 (2011). [DOI] [PubMed] [Google Scholar]
- Lau Y. Y., Sulaiman J., Chen J. W., Yin W. F. & Chan K. G. Quorum sensing activity of Enterobacter asburiae isolated from lettuce leaves. Sensors 13, 14189–14199, doi: 10.3390/s131014189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Reference Source. (2010) Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Accessed: 9th December 2014).
- Chan X. Y., Chua K. O., How K. Y., Yin W. F. & Chan K. G. Global genome comparative analysis reveals insights of resistome and life-style adaptation of Pseudomonas putida strain T2-2 in oral cavity. The Scientific World J. 2014, doi: 10.1155/2014/930727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics, btu153, doi: 10.1093/bioinformatics/btu153 (2014). [DOI] [PubMed] [Google Scholar]
- Hyatt D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119, doi: 10.1186/1471-2105-11-119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, doi: 10.1186/1471-2164-9-75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739, doi: 10.1093/molbev/msr121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D. et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 25, 3045–3046, doi: 10.1093/bioinformatics/btp536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman A. I. et al. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25, 2071–2073, doi: 10.1093/bioinformatics/btp356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Zakour N. L. B. & Beatson S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402, doi: 10.1186/1471-2164-12-402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]