Abstract
RNAs must assemble into specific structures in order to carry out their biological functions, but in vitro RNA folding reactions produce multiple misfolded structures that fail to exchange with functional structures on biological time scales. We used carefully designed self-cleaving mRNAs that assemble through well-defined folding pathways to identify factors that differentiate intracellular and in vitro folding reactions. Our previous work showed that simple base-paired RNA helices form and dissociate with the same rate and equilibrium constants in vivo and in vitro. However, exchange between adjacent secondary structures occurs much faster in vivo, enabling RNAs to quickly adopt structures with the lowest free energy. We have now used this approach to probe the effects of an extensively characterized DEAD-box RNA helicase, Mss116p, on a series of well-defined RNA folding steps in yeast. Mss116p overexpression had no detectable effect on helix formation or dissociation kinetics or on the stability of interdomain tertiary interactions, consistent with previous evidence that intracellular factors do not affect these folding parameters. However, Mss116p overexpression did accelerate exchange between adjacent helices. The nonprocessive nature of RNA duplex unwinding by DEAD-box RNA helicases is consistent with a branch migration mechanism in which Mss116p lowers barriers to exchange between otherwise stable helices by the melting and annealing of one or two base pairs at interhelical junctions. These results suggest that the helicase activity of DEAD-box proteins like Mss116p distinguish intracellular RNA folding pathways from nonproductive RNA folding reactions in vitro and allow RNA structures to overcome kinetic barriers to thermodynamic equilibration in vivo.
Keywords: RNA, RNA folding, RNA chaperone, ribozyme, catalytic RNA, RNA structure, RNA-protein interaction, DEAD-box protein, Mss116p
INTRODUCTION
The ability of RNAs to adopt precise three-dimensional structures is critical for biological processes ranging from viral genome replication and protein synthesis to RNA processing, gene silencing, and chromosome maintenance. Although RNAs rapidly assemble into the correct functional structures in vivo, RNAs tend to adopt multiple nonfunctional structures in vitro with exchange kinetics that are slower than most RNA's biological lifetime (Fedor and Uhlenbeck 1990; Uhlenbeck 1995; Treiber and Williamson 1999; Woodson 2000). Our previous work has shown that the same RNAs that become kinetically trapped in stable misfolded structures during transcription in vitro rapidly adopt thermodynamically favored structures in vivo (Mahen et al. 2005, 2010). Results of whole-genome structure analyses and large-scale RNA folding studies have also revealed considerable differences between RNA folding outcomes in vivo, in vitro, and in silico (Ding et al. 2014; Yang and Zhang 2014) and highlight the importance of RNA structural dynamics (Kertesz et al. 2010; Underwood et al. 2010; Lucks et al. 2011; Li et al. 2012; Wan et al. 2012; Incarnato et al. 2014; Rouskin et al. 2014). Comparisons of RNAs designed to adopt specific alternative structures with precisely calibrated differences in thermodynamic stability suggest that a very narrow window of free energy limits conformational exchange (Mahen et al. 2010).
The mechanisms that ensure proper RNA folding in vivo are not well understood. ATP-dependent DEAD-box RNA helicases have been implicated in almost every aspect of RNA metabolism including transcription, mRNA and tRNA processing, protein synthesis, RNA nuclear export, and RNA degradation, and certain DEAD-box helicases have been shown to facilitate splicing and translation of a variety of RNAs in vivo and in vitro (Russell et al. 2013; Jarmoskaite and Russell 2014). DEAD-box proteins exhibit little substrate specificity in vitro, but are usually found in large macromolecular complexes in vivo, such as spliceosomes and degradosomes, which direct their activities to specific pathways. However, some DEAD-box helicases have been shown to mediate nonspecific RNA unwinding both in vitro and in vivo (Rössler et al. 2001; Yang and Jankowsky 2005; Uhlmann-Schiffler et al. 2006; Halls et al. 2007; Del Campo et al. 2009), suggesting that they might act generally to promote RNA rearrangements (Herschlag 1995; Schroeder et al. 2002; Rajkowitsch et al. 2007; Chu and Herschlag 2008; Russell 2008; Russell et al. 2013; Jarmoskaite and Russell 2014). The general increase in mRNA structure observed in yeast after ATP depletion (Rouskin et al. 2014) is consistent with the idea that ATP-dependent processes such as helix unwinding modulate RNA secondary structures.
We developed a system to investigate general features of RNA folding pathways directly and quantitatively in living cells using chimeric mRNAs with hairpin ribozyme inserts (Donahue and Fedor 1997; Donahue et al. 2000; Watson and Fedor 2009, 2011, 2012). We previously screened large numbers of hairpin ribozyme sequence variants to identify those with precisely tuned secondary structure stabilities that would enable us to monitor the kinetics and equilibria of well-defined steps in the assembly of a functional ribozymes (Donahue et al. 2000; Yadava et al. 2001, 2004; Mahen et al. 2005, 2010; Watson and Fedor 2009, 2011, 2012). In this approach, intracellular folding and cleavage rates of self-cleaving ribozymes are quantified by comparing the turnover rate of chimeric mRNA containing a self-cleaving ribozyme, which decays through cleavage and endogenous mRNA degradation pathways, to the turnover rate of the same mRNA with a mutationally inactivated ribozyme insert, which decays only through endogenous mRNA degradation pathways (Fig. 1A). While minimal and natural hairpin ribozymes catalyze the same chemical reaction (Fig. 1B), structural differences in the nature of an interhelical junction between the two forms lead to distinct kinetic mechanisms. Remarkably, our previous studies showed that base-paired RNA helices form and dissociate with virtually identical rates and equilibrium constants in vivo and in vitro provided that in vitro reaction conditions recapitulate an intracellular ionic environment (Donahue et al. 2000; Yadava et al. 2001, 2004). Despite the overall similarity between intracellular and in vitro folding pathways, however, certain secondary structure exchange steps reached thermodynamic equilibria much more quickly in vivo than in vitro (Mahen et al. 2005, 2010). Thus, some feature unique to the intracellular environment appears to alleviate kinetic traps by facilitating RNA secondary structure exchange.
FIGURE 1.
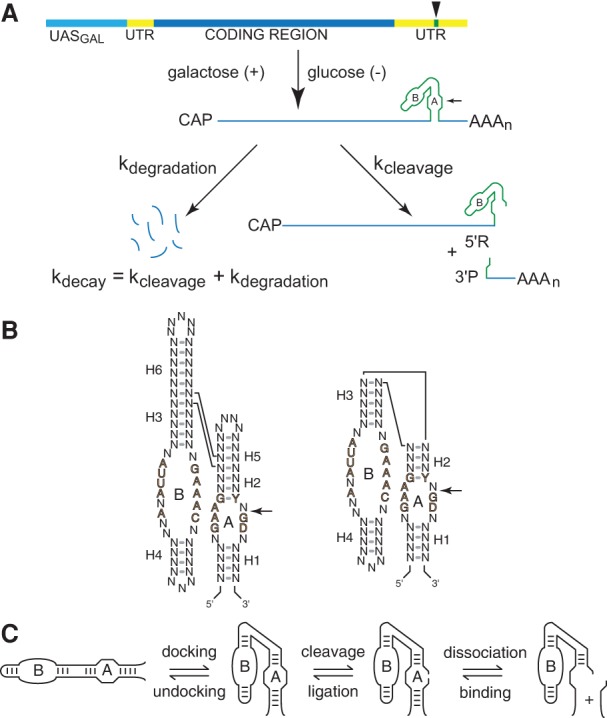
Chimeric ribozyme mRNAs as reporters of intracellular RNA folding. (A) Quantitative analysis of RNA folding in yeast. Ribozyme sequences (green) are inserted into the 3′ UTR (yellow) of the yeast PGK1 gene and transcribed under the control of the GAL1-10 upstream activation sequence, UASGAL (aqua), to allow measurement of chimeric mRNA decay kinetics after glucose inhibition. Self-cleaving mRNA decays both through self-cleavage (kcleavage) and through the normal mRNA degradation pathway (kdegradation), so self-cleavage accelerates decay by an amount that corresponds to the intracellular cleavage rate. (B) The two forms of hairpin ribozymes. Natural ribozymes (left) consist of two helix-loop-helix elements that assemble in the context of a four-way helical junction, whereas minimal ribozymes (right) assemble in the context of a two-way helical junction. Reversible cleavage of the reactive phosphodiester bond in Loop A is indicated by arrows. Defined nucleotides represent conserved sequences important for catalytic activity. (C) The hairpin ribozyme self-cleavage pathway. The minimal hairpin ribozyme (left) has two helix-loop-helix domains, A and B, that dock to form the active site, which catalyzes reversible cleavage of a specific phosphodiester bond in Loop A. Cleavage product dissociation occurs through unwinding of the intermolecular H1 helix. Diagrams adapted from Watson and Fedor (2009) and Mahen et al. (2010).
We have now made use of this approach to investigate whether the action of DEAD-box RNA helicases on kinetically trapped folding intermediates might account for this distinction between intracellular and in vitro folding outcomes. We focused on Mss116p, a DEAD-box helicase found in yeast mitochondria that has been particularly well characterized in vitro (Huang et al. 2005; Tijerina et al. 2006; Del Campo et al. 2007; Chen et al. 2008; Liu et al. 2008; Del Campo and Lambowitz 2009; Markov et al. 2009; Karunatilaka et al. 2010; Henn et al. 2012; Mallam et al. 2012; Russell et al. 2013; Jarmoskaite et al. 2014; Pan et al. 2014). Our findings are entirely consistent with previous studies of the effects of Mss116p on RNA folding reactions in vitro and suggest that DEAD-box helicase activity might account for the unique discrepancy in the kinetics of secondary structure exchange exhibited between hairpin ribozyme pathways in vitro and in yeast.
RESULTS
Overexpression of Mss116p in Saccharomyces cerevisiae does not affect RNA folding nonspecifically
Chimeric mRNAs containing ribozymes were expressed in yeast under the control of a glucose-repressible promoter, which enables quantification of RNA turnover rates after transcription inhibition (Fig. 1A). The Mss116p coding sequence was cloned under the control of the constitutive TEF1 promoter with a Flag-tag at the C terminus to facilitate Western blot analyses. Its mitochondrial targeting sequence (mts) was deleted to allow Mss116p colocalization with chimeric self-cleaving mRNAs. ATP binding is required for RNA unwinding by DEAD-box proteins, whereas ATP hydrolysis is primarily required for enzyme turnover (Chen et al. 2008; Liu et al. 2008). To determine whether unwinding activity was required for any observed effects on chimeric mRNA folding behavior, we compared the effects of the functional Δmts Mss116p DEAD-box helicase to that of the protein with an inactivating mutation (E268Q) in motif II that disrupts the DEAD sequence. In a related DEAD-box protein, Dbp5, this mutation abolishes ATPase activity, but does not affect ATP binding (Hodge et al. 2011).
Wild-type Mss116p, the mutant lacking the mitochondrial localization sequence (Δmts), and the D-E-A-D motif mutant (Δmts E268Q) were expressed at similar levels in yeast, evidence that the modifications used to ensure cytoplasmic localization and disrupt ATPase activity, respectively, did not interfere with Mss116p synthesis or turnover (Fig. 2). Mss116p overexpression did not affect yeast growth or viability; yeast harboring Mss116p expression vectors displayed similar growth curves, with a doubling time of ∼4 h (data not shown). Thus, Mss116p overexpression had no deleterious effects on RNA metabolism or any other pathways required for yeast viability. Coexpression of wild-type and mutant proteins with chimeric mRNAs had no detectable effect on protein expression levels (shown for yeast carrying HP44 and Mss116p Δmts E268Q in Fig. 2). Furthermore, Mss116p overexpression did not affect chimeric mRNA expression levels or alter the rate at which chimeric mRNAs were degraded through endogenous mRNA degradation pathways (Fig. 3). Thus, our analyses of the effects of Mss116p on ribozyme folding and catalysis were not complicated by nonspecific effects of Mss116p overexpression on RNA metabolism or yeast viability.
FIGURE 2.
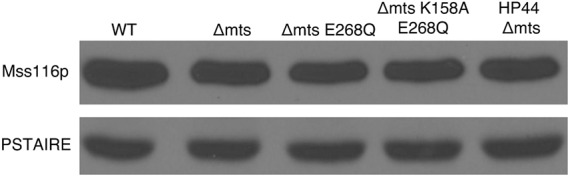
Wild-type and mutant Mss116 proteins were expressed at similar levels in yeast. Immunoblot of Flag-tagged wild-type (WT) Mss116p, Mss116p lacking the mitochondrial targeting sequence (Δmts), Mss116p lacking the mitochondrial targeting sequence with an inactivating mutation in the DEAD motif (ΔmtsE268Q), and Mss116p lacking the mitochondrial targeting sequence with an inactivating mutation in the P-loop (ΔmtsK158A). Coexpression of chimeric self-cleaving mRNA (HP44) with Mss116p (Δmts) did not affect Mss116p expression. After normalization using PSTAIRE protein loading controls, results obtained with different samples of the same yeast culture and with different yeast cultures varied less than twofold.
FIGURE 3.
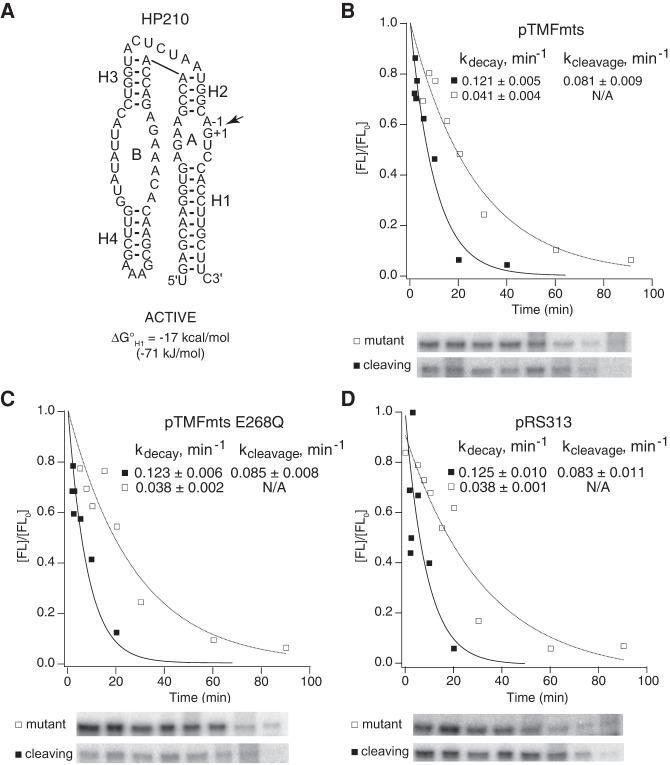
Mss116p did not affect chimeric mRNA stability or self-cleavage activity nonspecifically in yeast. (A) Secondary structure of HP210, which does not contain a complementary insert with the potential to form an alternative helix. Chimeric HP210 mRNAs were coexpressed in yeast with (B) active Mss116p (Δmts), (C) catalytically inactive Mss116p (ΔmtsE268Q), or (D) empty vector. Plots display results from a representative pair of experiments with functional (▪) and mutationally inactivated (□) chimeric HP210 mRNAs. Reported values represent the mean and standard deviation obtained from two or more pairs of experiments.
Intracellular ribozyme folding and self-cleavage rates were calculated from the difference between decay kinetics of self-cleaving and mutationally inactivated chimeric mRNAs determined using decay time course experiments and from measurements of chimeric mRNA abundance taken at steady state, as described previously (Yadava et al. 2001; Mahen et al. 2005, 2010; Watson and Fedor 2009). The intracellular cleavage rate for a given ribozyme equals the difference between decay rates of uncleaved self-cleaving mRNAs and chimeric mRNAs with a G+1A mutation that inactivates self-cleavage (Fig. 1A). At steady state, both self-cleaving and inactive chimeric mRNAs are transcribed at the same rate, but the self-cleaving version decays both through cleavage and through intrinsic degradation pathways, whereas its inactive counterpart only decays through intrinsic degradation pathways. When intrinsic degradation rates are known, therefore, self-cleavage rates can also be calculated from the relative abundance of self-cleaving and mutationally inactivated chimeric mRNAs at steady state. Comparison of cleavage rates determined using both methods provides further assurance that self-cleavage rate measurements are not complicated by experimental artifacts.
Mss116p promotes exchange between adjacent secondary structure elements in vivo
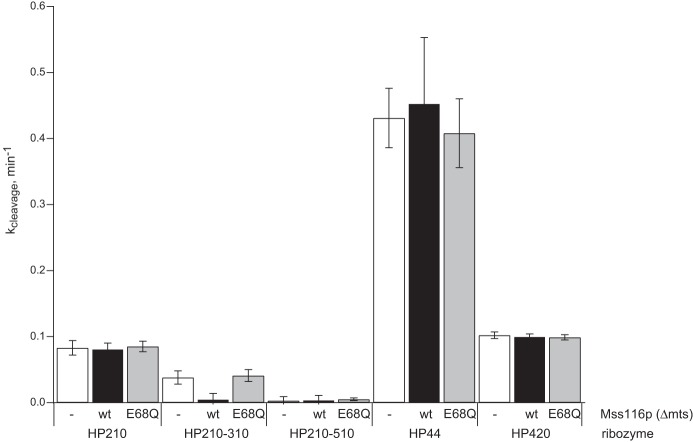
Intracellular cleavage of chimeric mRNAs was examined in yeast expressing various forms of Mss116p to reveal the role of this DEAD-box protein in RNA folding. In minimal ribozymes, the A and B helix-loop-helix elements form the arms of a two-way helical junction and dock in a noncoaxial orientation to create the active site (Fig. 1C). Self-cleavage within loop A gives 5′ and 3′ products that associate through complementary base pairs in helix H1 (Fig. 1C). The nature of the helical junction and the length and sequences of the base-paired helices are not essential for catalytic activity and can be manipulated to probe specific RNA structure–function relationships. HP210, a minimal hairpin ribozyme construct, cleaved at the same rate in yeast with or without Mss116p overexpression (Figs. 3, 4). We also observed similar self-cleavage rates for this ribozyme in yeast carrying wild-type or mutationally inactivated Mss116p or an empty vector (Figs. 3B–D, 4). The rate of ∼0.08 min−1 measured for HP210 self-cleavage agreed well with the rate of 0.082 min−1 reported previously for chimeric HP210 mRNA in yeast with no protein expression plasmid (Mahen et al. 2010). These results are consistent with our previous evidence that no feature of the intracellular environment, including endogenous DEAD-box helicase activity, alters folding kinetics or equilibria of simple hairpin ribozyme cleavage reactions relative to reactions carried out in vitro under conditions that approximate intracellular ionic conditions (Donahue et al. 2000; Yadava et al. 2001, 2004).
FIGURE 4.
Comparison of the effects of Mss116p overexpression on intracellular self-cleavage kinetics.
In our previous work, we introduced competing structures into minimal ribozymes to probe thermodynamic and kinetic determinants of partitioning among alternative RNA folding outcomes in vivo (Mahen et al. 2005, 2010). Specifically, a sequence that complements one strand of the essential H1 helix was inserted adjacent to the ribozyme sequence to create the potential to form an alternative H1 helix (AltH1) that inhibits cleavage by preventing the assembly of a functional ribozyme. Sequences needed to form AltH1 helices were located either downstream from the ribozyme (3′AltH1, Fig. 5A), where they were transcribed only after the entire ribozyme sequence had an opportunity to fold, or upstream of the ribozyme (5′AltH1, Fig. 6A), where the nonfunctional AltH1 had the potential to fold before the complete ribozyme sequence was transcribed. By systematically varying the relative thermodynamic stabilities of H1 and AltH1, we defined a narrow threshold of thermodynamic stability that determines whether RNAs adopt the thermodynamically favored structure or become kinetically trapped in the first helix that can form during transcription. These studies revealed that RNAs with adjacent H1 and AltH1 helices with calculated thermodynamic stabilities near −15 kcal/mol (62.8 kJ/mol) can undergo exchange and adopt the most thermodynamically favored structure, whereas RNAs with helices with calculated thermodynamic stabilities near −17 kcal/mol (71.1 kJ/mol) did not exchange and remained trapped with the 5′ helix that folds first during transcription (Mahen et al. 2010). These results define a very narrow range, on the order of 2 kcal/mol (8.4 kJ/mol), that limits the ability of RNA secondary structures to exchange within the biological lifetime of the chimeric mRNA. This folding behavior of chimeric mRNAs in vivo contrasts strongly with the folding outcomes we observed for transcription reactions in vitro where a helix with a calculated thermodynamic stability of −15 kcal/mol (62.8 kJ/mol) was sufficiently stable to prevent any thermodynamic equilibration with downstream structures (Mahen et al. 2005).
FIGURE 5.
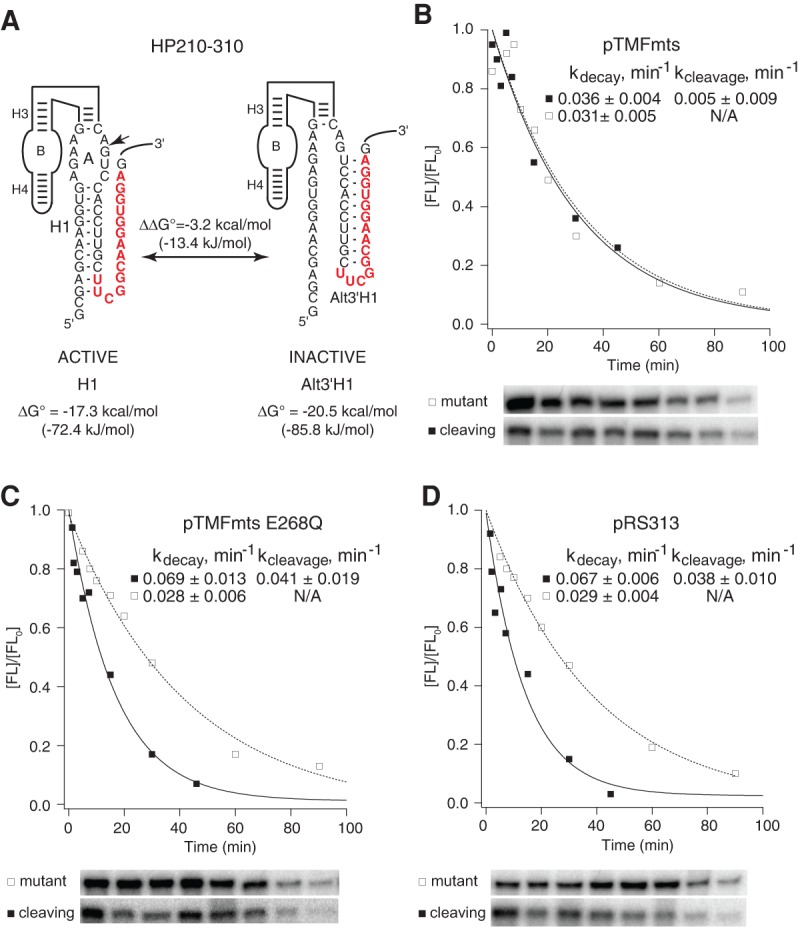
Mss116p promoted equilibration between H1 and AltH1 helices in vivo. (A) HP210-310 has the potential to adopt a functional structure with an H1 helix that has a calculated free energy of −17.3 kcal/mol (72.4 kJ/mol) or a nonfunctional structure with a 3′AltH1 that has a calculated free energy of −20.5 kcal/mol (85.8 kJ/mol). Chimeric mRNAs with HP210-310 were coexpressed in yeast with (B) active Mss116p (Δmts), (C) catalytically inactive Mss116p (ΔmtsE268Q), or (D) empty vector. HP210-310 chimeric mRNAs in yeast with active Mss116p exhibited eightfold lower intracellular cleavage rates than HP210-310 chimeric mRNAs that were coexpressed with catalytically inactive Mss116p (ΔmtsE268Q) or empty vector. Plots display results from a representative pair of experiments with functional (▪) and mutationally inactivated (□) chimeric HP210-310 mRNAs. Reported values represent the mean and standard deviation obtained from two or more pairs of experiments.
FIGURE 6.
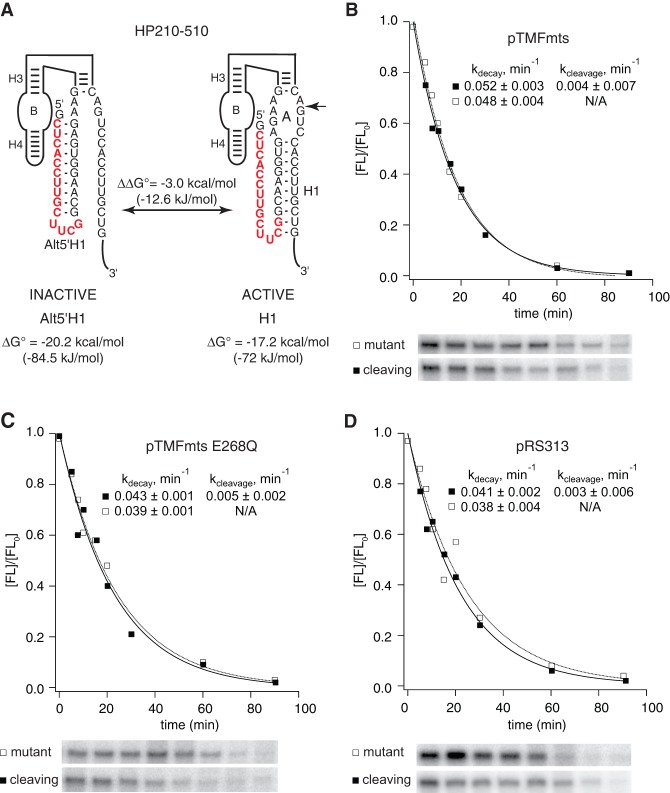
Mss116p did not alter RNA folding nonspecifically. (A) HP210-510 has the potential to adopt a functional structure with an H1 helix that has a calculated free energy of −17.2 kcal/mol (72.0 kJ/mol) or a nonfunctional structure with a 5′AltH1 that has a calculated free energy of −20.2 kcal/mol (84.5 kJ/mol). Chimeric mRNAs with HP210-510 were coexpressed in yeast with (B) active Mss116p (Δmts), (C) catalytically inactive Mss116p (ΔmtsE268Q), or (D) empty vector. Mss116p expression had no detectable effect on HP210-510 self-cleavage activity. Plots display results from a representative pair of experiments with functional (▪) and mutationally inactivated (□) chimeric HP210-510 mRNAs. Reported values represent the mean and standard deviation obtained from two or more pairs of experiments.
In order to test whether the Mss116p DEAD-box helicase promotes conformational equilibration of otherwise stable RNA secondary structures in vivo, we examined the effect of Mss116p overexpression on the folding behavior of a ribozyme with an upstream H1 helix that is normally too stable to allow equilibration with a nonfunctional downstream AltH1 helix that has greater thermodynamic stability. HP210-310 is a minimal ribozyme variant with an H1 helix that has a calculated thermodynamic stability of −17.3 kcal/mol (72.4 kJ/mol), which is located upstream of an AltH1 helix that has a calculated thermodynamic stability of −20.5 kcal/mol (85.8 kJ/mol) (Fig. 5A; Mahen et al. 2010). The ability of the HP210-310 ribozyme variant to adopt a functional structure with the H1 helix despite competition from a downstream AltH1 helix with greater stability was reflected in its intracellular self-cleavage rate that was just twofold below the rate observed for HP210, its counterpart with no competing helices (Figs. 3, 4; Mahen et al. 2010). This evidence that HP210-310 was kinetically trapped in the functional structure contrasts with the behavior of ribozyme constructs with intermolecular helices that had lightly lower thermodynamic stabilities, which did undergo thermodynamic equilibration with stable downstream helices (Mahen et al. 2010).
HP210-310 self-cleavage rates decreased eightfold in yeast expressing the active Mss116p DEAD-box helicase (Figs. 4, 5B). This loss of self-cleavage activity suggests that Mss116p overexpression allowed the AltH1 helix to compete with the upstream H1 helix to reach the catalytically inactive structure that is more thermodynamically favorable. HP210-310 exhibited virtually the same intracellular self-cleavage rate in yeast expressing mutationally inactivated Mss116p and the empty expression vector as reported previously in experiments with no Mss116p (Figs. 4, 5C,D). Thus, Mss116p overexpression reduced the thermodynamic threshold that limits exchange by ∼2 kcal/mol (8.4 kJ/mol) so that the H1 helix in HP210-310, with a calculated thermodynamic stability of −17.3 kcal/mol, was able to exchange with a more stable helix located downstream.
Mss116p overexpression did not affect folding or intracellular cleavage activity of HP210-510, a ribozyme variant with a very stable AltH1 helix (−20.2 kcal/mol) (84.5 kJ/mol) upstream of an H1 helix that has lower thermodynamic stability (−17.2 kcal/mol) (72.0 kJ/mol) (Fig. 6). HP210-510 exhibited no detectable self-cleavage activity with or without Mssp116p overexpression, evidence that it remained kinetically trapped in a nonfunctional structure with the more stable upstream AltH1 (−20.2 kcal/mol) (84.5 kJ/mol) (Figs. 4, 6B–D; Mahen et al. 2010). Thus, Mss116p overexpression facilitated thermodynamic equilibration of HP210-310, in which the downstream helix had lower thermodynamic stability, but it did not change the folding outcome in the HP210-510 variant in which the most stable helix was located upstream.
Mss116p does not accelerate product dissociation
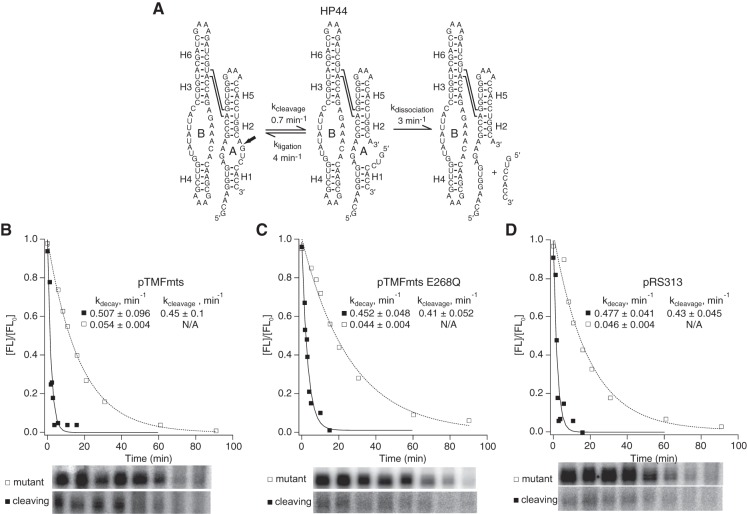
We examined the effect of Mss116p expression on helix dissociation kinetics using HP44, a variant of the natural form of the hairpin ribozyme (Fig. 7; Yadava et al. 2001). Relative to minimal ribozymes, natural hairpin ribozymes contain two additional helices and assemble in the context of a four-way helical junction (Fig. 1C). While minimal and natural ribozymes catalyze the same chemical reaction and have the same active sites (Fig. 1B), they exhibit distinct kinetic mechanisms (Fedor 1999, 2009). They exhibit higher cleavage rate constants compared to minimal ribozymes both in vitro and in yeast, and also strongly favor ligation of bound products relative to cleavage (Fig. 1B; Fedor 1999; Yadava et al. 2001). Observed intracellular cleavage kinetics for HP44 reflect partitioning between helix dissociation and re-ligation of bound products. Careful comparison of four-way junction ribozymes with many different H1 stabilities previously showed that re-ligation of bound products causes observed cleavage rates to decrease as H1 stability increases, both in vitro and in yeast, and cleavage products dissociate from four-way junction ribozymes more slowly than from minimal ribozymes (Fedor 1999; Yadava et al. 2001). HP44, with a carefully calibrated sequence of 4 base pairs (bp) in H1, exhibits product dissociation kinetics on the same order as the kinetics of re-ligation of bound products, on the order of 3 min−1 (Fig. 7A). Nearly equal partitioning between re-ligation and product dissociation reduces observed cleavage rates, by approximately twofold relative to four-way junction ribozymes with shorter or less GC-rich product RNAs that dissociate rapidly. This makes HP44 a particularly sensitive reporter of any effect of Mss116p overexpression on helix dissociation kinetics.
FIGURE 7.
Mss116p did not promote helix dissociation in vivo. (A) Kinetic mechanism of a natural ribozyme with 4 bp in H1, HP44, which partitions equally between dissociation and re-ligation of bound products (Donahue et al. 2000). Chimeric HP44 mRNAs were coexpressed in yeast with (B) active Mss116p (Δmts), (C) catalytically inactive Mss116p (ΔmtsE268Q), or (D) empty vector. Mss116p expression had no detectable effect on HP44 self-cleavage activity. Plots display results from a representative pair of experiments with functional (▪) and mutationally inactivated (□) chimeric HP44 mRNAs. Reported values represent the mean and standard deviation obtained from two or more pairs of experiments. Intracellular cleavage, ligation, and dissociation rate constants in the absence of Mss116p overexpression were reported previously (Yadava et al. 2001).
If Mss116p accelerated complete helix dissociation, intracellular cleavage rates for HP44 would increase, as rapid dissociation and dilution of ribozyme–product complexes would prevent re-ligation. Alternatively, observed cleavage rates might decrease if Mss116p accelerated helix annealing to drive assembly of ribozyme–product complexes that can undergo ligation. However, HP44 exhibited virtually the same self-cleavage kinetics in yeast overexpressing active Mss116p, mutationally inactivated Mss116p, or carrying an empty vector (Figs. 4, 7B–D). Observed intracellular self-cleavage rates agreed well with previous measurements (Yadava et al. 2001). Thus, Mss116p overexpression had no detectable effect on product dissociation kinetics, consistent with the possibility that Mss116p did not accelerate unwinding of the H1 helix of HP44 or that it affected winding and unwinding equally. These results are consistent with our previous evidence that no feature of the intracellular environment accelerates dissociation or annealing of base-paired RNA helices relative to ribozyme complex formation in vitro provided that in vitro reactions approximate intracellular ionic conditions (Donahue et al. 2000; Yadava et al. 2001, 2004).
Mss116p does not destabilize ribozyme tertiary structure
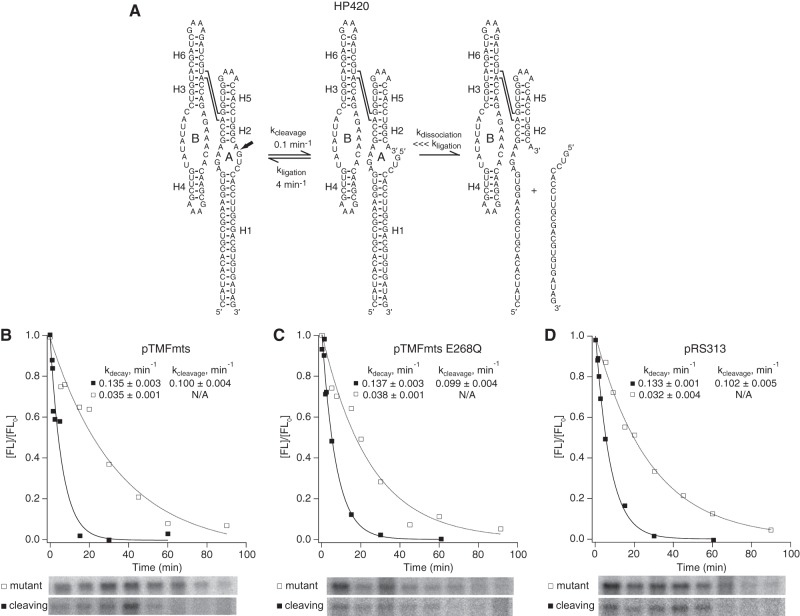
The enhanced ligation activity of a four-way junction ribozymes relative to a minimal ribozyme reflects its enhanced tertiary structure stability, which is likely to stabilize the positioning of cleavage product termini in the orientation needed for the ligation (Walter et al. 1999; Fedor 2009). To investigate whether Mss116p affects ribozyme tertiary structure stability, we examined the effect of Mss116p overexpression on HP420, a ribozyme with a four-way helical junction and 20 bp in its H1 helix (Fig. 8A). HP420 exhibits very slow intracellular self-cleavage kinetics because a ribozyme–product complex with 20 bp in the intermolecular H1 helix does not dissociate within the intracellular lifetime of HP420 chimeric mRNA (Yadava et al. 2001). Thus, HP420 cleavage kinetics reflects the internal equilibrium between cleavage and ligation of bound cleavage products, which reports on tertiary structure stability (Fig. 8A).
FIGURE 8.
Mss116p did not affect the internal equilibrium between cleavage and ligation of bound products. (A) Kinetic mechanism of a natural ribozyme with 20 bp in H1, which undergoes product dissociation much more slowly than re-ligation. Chimeric HP420 mRNAs were coexpressed in yeast with (B) active Mss116p (Δmts), (C) catalytically inactive Mss116p (ΔmtsE268Q), or (D) empty vector. Mss116p expression had no detectable effect on HP44 self-cleavage. Mss116p expression had no detectable effect on HP420 self-cleavage activity. Plots display results from a representative pair of experiments with functional (▪) and mutationally inactivated (□) chimeric HP420 mRNAs. Reported values represent the mean and standard deviation obtained from two or more pairs of experiments. Intracellular cleavage, ligation, and dissociation rate constants in the absence of Mss116p overexpression were reported previously (Yadava et al. 2001).
If Mss116p destabilizes ribozyme tertiary structure and shifts the internal equilibrium away from ligation to favor cleavage, we would expect to observe faster HP420 self-cleavage when Mss116p is overexpressed. However, observed HP420 cleavage rates were the same in yeast overexpressing active or mutant Mss116p or containing the empty vector (Figs. 4, 8B–D) and matched rates measured previously in yeast with no protein expression vector (Yadava et al. 2001). The absence of any change in the internal equilibrium between ligation and cleavage of bound products upon Mss116p overexpression suggests that Mss116p chaperone activity does not destabilize RNA tertiary structures nonspecifically. The absence of an effect of Mss116p overexpression on HP420 cleavage kinetics also supports the conclusion from experiments with HP44 that Mss116p does not accelerate complete helix dissociation. If Mss116p overexpression accelerated dissociation of the intermolecular H1 helix with 20 bp in HP420, faster product dissociation would have prevented re-ligation of bound products and increased observed cleavage rates.
DISCUSSION
Here, we have demonstrated the ability of the Mss116p DEAD-box helicase to promote rapid exchange between adjacent base-paired helices. While Mss116p might interact transiently and reversibly with RNA secondary and tertiary structures, we detected no overall effects on helix annealing or dissociation kinetics or on tertiary structure stability. Effects of Mss116p overexpression were limited to reactions in which a kinetically trapped structure could exchange with a more thermodynamically stable structure through a helical junction; Mss116p did not act broadly to completely unwind long RNA helices or reduce tertiary structure stability. Our ability to detect an effect of Mss116p overexpression only when unfolded RNA was able to adopt an alternative stable structure is consistent with our previous findings that the kinetics and equilibria of simple helix annealing and dissociation steps and the internal equilibrium between cleavage and ligation were virtually the same in vivo and in vitro despite the ubiquity of endogenous DEAD-box proteins (Donahue et al. 2000; Yadava et al. 2001, 2004; Mahen et al. 2005, 2010; Watson and Fedor 2009, 2011, 2012). Indeed, Mss116p only accelerated the kind of RNA conformational exchange reaction that we previously found to exhibit very different kinetic behavior in vitro and in yeast (Mahen et al. 2005, 2010). Thus, Mss116p can enable an RNA to adopt the lowest free energy structure within a biological timeframe by accelerating exchange of adjacent RNA helices, but it does not stabilize or destabilize RNA structures nonspecifically. The ability of DEAD-box RNA helicases, such as Mss116p, to facilitate equilibration among otherwise stable RNA secondary structures likely accounts, at least in part, for the differences in chimeric mRNA folding outcomes in vitro and in yeast that we reported previously (Mahen et al. 2005, 2010).
The shift we observed in the thermodynamic barrier to secondary structure exchange resulted from overexpression of Mss116p, but it is entirely consistent with the chaperone activities previously attributed to DEAD-box proteins under physiological conditions in vitro and in yeast deficient in these proteins (Russell et al. 2013; Jarmoskaite and Russell 2014). Mss116p, CYT-19, and Ded1 DEAD-box helicases have been shown to facilitate assembly of functional group I and II intron structures by disrupting non-native structures through a mechanism that does not require recognition of specific RNA structures or sequences (Huang et al. 2005; Tijerina et al. 2006; Del Campo et al. 2007; Potratz et al. 2011; Russell et al. 2013). Mss116p binds diverse RNA substrates and exhibits high RNA helicase activity in vitro (Mohr et al. 2006; Halls et al. 2007; Del Campo et al. 2009), supporting a model in which DEAD-box proteins serve as general RNA chaperones by unfolding RNA structures nonspecifically to give RNAs the opportunity to refold into their native states.
Our finding that Mss116p overexpression promoted exchange between adjacent secondary structures but did not accelerate RNA helix dissociation generally is consistent with the limited processivity characteristic of DEAD-box helicases. Unlike processive helicases that unwind duplexes through ATP-dependent translocation, DEAD-box helicases unwind just a few base pairs (Russell et al. 2013; Jarmoskaite and Russell 2014). Upon binding an RNA helix and a single molecule of ATP, DEAD-box proteins undergo a conformational change that results in strand separation (Henn et al. 2012; Russell et al. 2013). One ATP cycle may be enough to unwind short, relatively unstable helices, but not more stable structures. ATP hydrolysis is not necessary for unwinding, but it does promote release of the RNA, which may contribute to the protein's ability to unfold more complex structures. Our results support a model in which Mss116p facilitates exchange between adjacent secondary structures through a branch migration mechanism that lowers barriers to interhelical exchange through sequential melting of several base pairs at interhelical junctions (Fig. 9).
FIGURE 9.
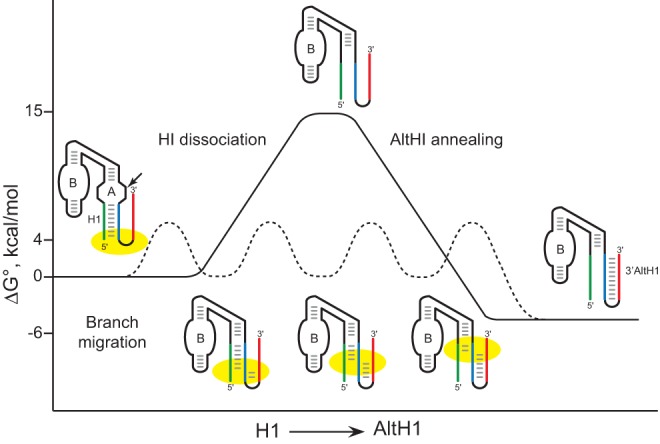
Free energy model of secondary structure exchange through RNA chaperone-assisted branch migration. RNA chaperone proteins, like Mss116p, that unwind 1–2 bp per ATP cycle could catalyze the exchange between adjacent RNA helices through small steps with low energy barriers to facilitate assembly of RNA structures with the lowest free energy. Diagram adapted from Mahen et al. (2010).
Apart from DEAD-box RNA helicases, additional intracellular factors certainly contribute to the ability of RNAs to avoid the kinetic traps that block assembly of functional RNA structures in vitro. RNA secondary structures with the potential to form within upstream sequences during transcription are clearly favored relative to structures that require interactions with downstream sequences that are transcribed later (Mahen et al. 2005, 2010), a process that is facilitated by sequence-specific transcriptional pausing (Pan et al. 1999; Pan and Sosnick 2006; Wong et al. 2007; Perdrizet et al. 2012). Interactions with specific RNA-binding proteins and small ligands have also been shown to “capture” certain RNA conformations (Weeks and Cech 1996; Haller et al. 2011). Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large and diverse class of proteins that interact with RNA polymerase II transcripts in nuclei (Dreyfuss et al. 1993; He and Smith 2009; Singh et al. 2015). Many hnRNPs exhibit RNA chaperone activity in vitro and in vivo suggesting that cotranscriptional binding of hnRNPs might modulate assembly of stable RNA structures within nascent transcripts (Karpel et al. 1982; Herschlag et al. 1994; Belisova et al. 2005). A second class of nonspecific nucleic acid–binding proteins with basic, intrinsically disordered domains destabilize RNA structures and have been reported to promote refolding of misfolded RNAs in vitro without the use of ATP (Tompa and Csermely 2004). Some members of this class, including ribosomal and HIV nucleocapsid proteins, have also been shown to rescue activity of misfolded self-splicing introns in vivo (Rajkowitsch et al. 2007; Chu and Herschlag 2008; Russell 2008). However, these basic, disordered proteins generally exhibit RNA unfolding activity over a narrow range of concentrations in vitro and many of them, such as histones and ribosomal proteins, have other primary functions. Future studies will be needed to establish whether these proteins also function to promote exchange among alternative RNA structures in a biological context.
MATERIALS AND METHODS
Plasmids
Plasmids containing the PGK1 gene and hairpin ribozyme sequence variants were used for chimeric ribozyme mRNA expression in yeast as described previously (Donahue et al. 2000; Yadava et al. 2001; Mahen et al. 2010). Briefly, the ribozyme sequence was inserted into the 3′ UTR of the yeast PGK1 gene in pGAL28, a pRS316 derivative in which the PGK1 gene is fused to the GAL1 promoter. Plasmids were propagated in Escherichia coli strain DH5α or in S. cerevisiae strain HYF114 (MATa ade2-1 his3-11,15 leu2-3 112 trp1-1 ura3-1 can1-100).
The Mss116p coding region was amplified from yeast genomic DNA (S. cerevisiae strain HYF114, MATa ade2-1 his3-11,15 leu2-3 112 trp1-1 ura3-1 can1-100) using primers BamHI-MSS116F and MSS116-Flag-Spe R (Table 1). The amplicon containing Mss116p, a Flag tag, and a triple stop codon was cloned into the pRS313 vector using BamHI and SpeI restriction sites. To express the protein constitutively, a truncated TEF1 promoter was amplified from yeast genomic DNA using primers TEF1XhoI and TEF1BamHI (Table 1) and cloned into the pRS313 plasmid using XhoI and BamHI restriction sites. The plasmid containing the full-length Mss116p coding region and TEF1 promoter was named pTMF. The Mss116p mitochondrial targeting sequence (amino acids 2–26) was deleted from pTMF using QuickChange mutagenesis (Stratagene) and oligonucleotides TEF1MSS116 -22 and TEF1MSS116 102 (Table 1) to create pTMFmts. A catalytically inactive version of Mss116p contained an E268Q mutation in the D-E-A-D motif. An E268Q mutation in the D-E-A-D motif was introduced using oligonucleotides MSS116 781 and MSS116 820 (Table 1) to create pTMFmts E268Q. All experiments were performed with Mss116p lacking the mitochondrial localization sequence to ensure cytoplasmic colocalization with chimeric HP mRNAs.
TABLE 1.
Oligonucleotides used for plasmid constructions

Plasmid templates for T7 RNA polymerase transcription of the 32P-labeled probes used to quantify yeast chimeric mRNAs contained sequences complementary to both chimeric PGK1 and actin mRNAs to maximize accuracy and precision. The ACT1 sequence was amplified using the primers DualActinF and DualActinR (Table 1) and Q5 High Fidelity 2X Master Mix (NEB), which added NdeI and NarI restriction sites to the 5′ and 3′ ends of the amplicon. The digested amplicon was inserted downstream from the chimeric PGK1 sequence into probe template plasmids used previously (Yadava et al. 2001; Mahen et al. 2010).
Ribozyme names reflect the important features of the ribozyme structure including the interdomain junction (two-way or four-way), the number of base pairs in H1, the location of the complementary insert, if any (5′ or 3′), and the number of base pairs in AltH1, if present. For example, HP210-310 is a ribozyme variant with a two-way helical junction that has 10 bp in H1 and a complementary insert located on the 3′ side of the ribozyme with the potential to form a 3′AltH1 with 10 bp, whereas HP420 is a variant with a four-way helical junction that has 20 bp in H1 and no AltH1.
Immunoblot analysis
Protein extracts were made by vortexing yeast with glass beads in lysis buffer (100 mM Tris pH 6.5, 2% SDS, 20% glycerol, 100 mM DTT, and 1 mM PMSF). After cells were boiled for 5 min, glass beads were added and the mixture was vortexed for 5 min. The glass beads were removed and the lysates were boiled again for 5 min. Cell debris was pelleted by brief centrifugation. Protein concentrations were determined by absorption at 280 nm. Extracts were fractionated using 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes by semidry blotting. Membranes were blocked with 10% nonfat dry milk in TBST (Tris-buffered saline, 0.25% Tween 20) and incubated in primary antibody (1:2000 in 5% nonfat dry milk in TBST) for 1 h to overnight followed by secondary antibody (1:5000 in 5% nonfat dry milk in TBST) for 1 h. Antibodies used were anti-Flag (M2, Sigma), anti-mouse IgG-horse radish peroxidase produced in goat (Sigma), and anti-PSTAIRE to control for protein loading (kindly provided by Curt Wittenberg, The Scripps Research Institute, La Jolla, CA) (Ma et al. 2014). Signals were detected on film by enhanced chemiluminescence (Super Signal West Dura, Pierce).
Intracellular cleavage kinetics
Intracellular cleavage rates were calculated from decay time course experiments and from measurements taken at steady state as described previously, except that hybridization probes contained sequences complementary to both chimeric PGK1 and actin mRNAs for improved accuracy (Yadava et al. 2001; Mahen et al. 2005, 2010; Watson and Fedor 2009). 2P-labeled RNAs used as hybridization probes were transcribed from linearized pGEM-4Z derivatives (Promega). Yeast cultures were grown at 30°C in galactose minimal medium to log phase overnight. Aliquots were taken at time zero before yeast were transferred to glucose minimal medium to inhibit transcription. After transfer, aliquots were taken at the indicated times and total RNA was extracted from yeast pellets. Ribozyme cleavage was quantified using RNase protection assays performed under acidic conditions at low temperature to suppress further self-cleavage activity during the analyses. Uncleaved 32P-labeled self-cleaving RNA was added to one yeast pellet and subjected to extraction and analysis procedures as a control experiment to exclude the possibility that self-cleavage occurred in vitro during extraction and analysis of yeast RNA rather than in yeast. In all reported experiments, <10% of the uncleaved ribozyme RNAs underwent cleavage, confirming that conditions used for RNase protection assays did not support ribozyme activity. Uncleaved chimeric mRNA levels were normalized to the abundance of ACT1 mRNA.
Decay rates for intracellular chimeric mRNAs were determined by fitting a single exponential rate equation to the data. Cleavage rates calculated using both methods typically agreed within 30% and never varied more than twofold. Plots represent results of a single decay time course experiment. Reported values represent the mean and standard deviation obtained from two or more experiments.
ACKNOWLEDGMENTS
We thank Marisela Guaderrama for expert technical assistance with plasmid constructions and immunoblots. This work was supported by National Institutes of Health grant RO1 GM062277 to M.J.F.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055178.115.
REFERENCES
- Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, Steiner G. 2005. RNA chaperone activity of protein components of human Ro RNPs. RNA 11: 1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. 2008. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci 105: 20203–20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VB, Herschlag D. 2008. Unwinding RNA's secrets: advances in the biology, physics, and modeling of complex RNAs. Curr Opin Struct Biol 18: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Lambowitz AM. 2009. Structure of the yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell 35: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang QS, Jankowsky E, Russell R, Lambowitz AM. 2007. Do DEAD-Box proteins promote group II intron splicing without unwinding RNA? Mol Cell 28: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Mohr S, Jiang Y, Jia HJ, Jankowsky E, Lambowitz AM. 2009. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol 389: 674–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YL, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Donahue CP, Fedor MJ. 1997. Kinetics of hairpin ribozyme cleavage in yeast. RNA 3: 961–973. [PMC free article] [PubMed] [Google Scholar]
- Donahue CP, Yadava RS, Nesbitt SM, Fedor MJ. 2000. The kinetic mechanism of the hairpin ribozyme in vivo: influence of RNA helix stability on intracellular cleavage kinetics. J Mol Biol 295: 693–707. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321. [DOI] [PubMed] [Google Scholar]
- Fedor MJ. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. [DOI] [PubMed] [Google Scholar]
- Fedor M. 2009. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys Biomol Struct 38: 271–299. [DOI] [PubMed] [Google Scholar]
- Fedor MJ, Uhlenbeck OC. 1990. Substrate sequence effects on “hammerhead” RNA catalytic efficiency. Proc Natl Acad Sci 87: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. 2011. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400. [DOI] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. 2007. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 365: 835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YW, Smith R. 2009. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci 66: 1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Bradley MJ, De La Cruz EM. 2012. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys 41: 247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. 1995. RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. 1994. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J 13: 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev 25: 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. 2005. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci 102: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incarnato D, Neri F, Anselmi F, Oliviero S. 2014. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol 15: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2014. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83: 697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Bhaskaran H, Seifert S, Russell R. 2014. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc Natl Acad Sci 111: E2928–E2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel RL, Miller NS, Fresco JR. 1982. Mechanistic studies of RNA renaturation by a nucleic acid helix-destabilizing protein. Biochemistry 21: 2102–2108. [DOI] [PubMed] [Google Scholar]
- Karunatilaka KS, Solem A, Pyle AM, Rueda D. 2010. Single-molecule analysis of Mss116-mediated group II intron folding. Nature 467: 935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. 2012. Global analysis of RNA secondary structure in two metazoans. Cell Rep 1: 69–82. [DOI] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci 105: 20209–20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. 2011. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc Natl Acad Sci 108: 11063–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Han B-K, Guaderrama M, Aslanian A, Yates JR, Hunter T, Wittenberg C. 2014. Psy2 targets the PP4 family phosphatase Pph3 to dephosphorylate Mth1 and repress glucose transporter gene expression. Mol Cell Biol 34: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen EM, Harger JW, Calderon EM, Fedor MJ. 2005. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol Cell 19: 27–37. [DOI] [PubMed] [Google Scholar]
- Mahen EM, Watson PY, Cottrell JW, Fedor MJ. 2010. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol 8: e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. 2012. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature 490: 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. 2009. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast 26: 423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM. 2006. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci 103: 3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sosnick T. 2006. RNA folding during transcription. Annu Rev Biophys Biomol Struct 35: 161–175. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. 1999. Folding of a large ribozyme during transcription and the effect of the elongation factor nusA. Proc Natl Acad Sci 96: 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Potratz JP, Cannon B, Simpson ZB, Ziehr JL, Tijerina P, Russell R. 2014. DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol 12: e1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdrizet GA, Artsimovitch I, Furman R, Sosnick TR, Pan T. 2012. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci 109: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potratz JP, Del Campo M, Wolf RZ, Lambowitz AM, Russell R. 2011. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo. J Mol Biol 411: 661–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Starnpfl S, Sernrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4: 118–130. [DOI] [PubMed] [Google Scholar]
- Rössler OG, Straka A, Stahl H. 2001. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res 29: 2088–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. 2008. RNA misfolding and the action of chaperones. Front Biosci 13: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Jarmoskaite I, Lambowitz AM. 2013. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol 10: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Grossberger R, Pichler A, Waldsich C. 2002. RNA folding in vivo. Curr Opin Struct Biol 12: 296–300. [DOI] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. 2006. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci 103: 16698–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Csermely P. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. 1999. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol 9: 339–345. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC. 1995. Keeping RNA happy. RNA 1: 4–6. [PMC free article] [PubMed] [Google Scholar]
- Uhlmann-Schiffler H, Jalal C, Stahl H. 2006. Ddx42p—a human DEAD box protein with RNA chaperone activities. Nucleic Acids Res 34: 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. 2010. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 7: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG, Burke JM, Millar DP. 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol 6: 544–549. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Ouyang ZQ, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. 2012. Genome-wide measurement of RNA folding energies. Mol Cell 48: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. 2009. Determination of intracellular RNA folding rates using self-cleaving RNAs. Methods Enzymol 468: 259–286. [DOI] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. 2011. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol 18: 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. 2012. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol 8: 963–965. [DOI] [PubMed] [Google Scholar]
- Weeks KM, Cech TR. 1996. Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science 271: 345–348. [DOI] [PubMed] [Google Scholar]
- Wong TN, Sosnick TR, Pan T. 2007. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci 104: 17995–18000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. 2000. Compact but disordered states of RNA. Nat Struct Biol 7: 349–352. [DOI] [PubMed] [Google Scholar]
- Yadava RS, Choi AJ, Lebruska LL, Fedor MJ. 2001. Hairpin ribozymes with four-way helical junctions mediate intracellular RNA ligation. J Mol Biol 309: 893–902. [DOI] [PubMed] [Google Scholar]
- Yadava RS, Mahen EM, Fedor MJ. 2004. Kinetic analysis of ribozyme-substrate complex formation in yeast. RNA 10: 863–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. 2005. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44: 13591–13601. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J. 2014. Human long noncoding RNAs are substantially less folded than messenger RNAs. Mol Biol Evol 32: 970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]