Abstract
When assayed in vitro, group IIC self-splicing introns, which target bacterial Rho-independent transcription terminators, generally fail to yield branched products during splicing despite their possessing a seemingly normal branchpoint. Starting with intron O.i.I1 from Oceanobacillus iheyensis, whose crystallographically determined structure lacks branchpoint-containing domain VI, we attempted to determine what makes this intron unfit for in vitro branch formation. A major factor was found to be the length of the helix at the base of domain VI: 4 base pairs (bp) are required for efficient branching, even though a majority of group IIC introns have a 3-bp helix. Equally important for lariat formation is the removal of interactions between ribozyme domains II and VI, which are specific to the second step of splicing. Conversely, mismatching of domain VI and its proposed first-step receptor in subdomain IC1 was found to be detrimental; these data suggest that the intron-encoded protein may promote branch formation partly by modulating the equilibrium between conformations specific to the first and second steps of splicing. As a practical application, we show that by making just two changes to the O.i.I1 ribozyme, it is possible to generate sufficient amounts of lariat intron for the latter to be purified and used in kinetic assays in which folding and reaction are uncoupled.
Keywords: group II intron, lariat intron, linear intron, self-splicing
INTRODUCTION
Group II introns are typically excised in the so-called “lariat” form, within which the first intron nucleotide and an internal adenosine residue are brought together by a 2′–5′ phosphodiester bond. This branched structure is essential for completion of “reverse splicing,” the process by which group II introns insert themselves into their DNA target with the help of the reverse transcriptase they encode (for review, see Lambowitz and Zimmerly 2011): retrotransposition of a linear intron is not only much less efficient than that of its branched counterpart (by some three orders of magnitude), but it is predominantly imprecise (Zhuang et al. 2009). The reason is that the second transesterification step of reverse splicing, in which the 3′ OH of the cleaved target DNA strand is ligated to the 5′ intron extremity, makes use of the 2′–5′ phosphodiester bond that was created at the initiation of splicing. Moreover, a branched structure facilitates the second step of forward splicing: branched intron–3′ exon intermediates of the yeast ai5γ group II intron carry out exon ligation much more efficiently (again by three orders of magnitude; Dème et al. 1999) than linear molecules.
In spite of the crucial role of the 2′–5′ bond in allowing efficient intron excision and retrotransposition, there exists an alternative group II processing pathway in which splicing is initiated by hydrolysis at the 5′ splice site, rather than by branch formation. This pathway, which results in linear intron products, was first identified in vitro, by using alternative reaction conditions (Jarrell et al. 1988) or molecules whose branchpoint, which normally coincides with a bulging adenosine residue on the 3′ side of secondary-structure domain VI, had been deleted or altered (van der Veen et al. 1987). Only later was it noticed that a relatively small number of natural group II introns happen to lack a canonical branchpoint. When representative members of this subclass were investigated in vivo (Vogel and Börner 2002) or in vitro (Li et al. 2011a), they were found to be excised exclusively as linear molecules. In keeping with the importance of a branched structure for retrotransposition, introns that lack a functional branchpoint do not encode a reverse transcriptase. Some of them, such as the trnV intron (Vogel and Börner 2002) which, like most other chloroplast introns, is strongly conserved in land plants, appear to have lost mobility altogether, while others belong to lineages that became associated with homing endonucleases (Toor and Zimmerly 2002; Mullineux et al. 2010; Li et al. 2011a; Salman et al. 2012); the latter have long been known to ensure mobility of many group I and some archaeal introns by a mechanism in which the intron RNA plays no part (Dujon 1989; Bell-Pedersen et al. 1990; Belfort and Roberts 1997).
Besides molecules that clearly miss a functional branchpoint, a growing number of group II introns that seemingly possess a normal domain VI (DVI) and branchpoint have been reported to fail to yield branched products during in vitro splicing (e.g., Nagy et al. 2013, for a recent publication). Most notorious among these are members of subgroup IIC, the vast majority of which target Rho-independent transcription terminators (Granlund et al. 2001; reviewed in Michel et al. 2007 and Lambowitz and Zimmerly 2011). RNA precursor molecules containing group IIC introns typically do not generate more than traces of branched forms under a diversity of in vitro conditions (Granlund et al. 2001; Toor et al. 2006, 2008). Nevertheless, most group IIC introns encode reverse transcriptases and many of them must be mobile, since they exist as multiple copies in their host genomes. Moreover, the potential for branch formation has clearly been retained, as it could be shown that ribonucleoprotein particles formed by incubating a group IIC precursor transcript with the intron-encoded protein (IEP) do generate lariats (Robart et al. 2007).
The purpose of this work was to pinpoint the structural peculiarities that prevent group IIC introns from using the branching pathway in the absence of the IEP. As a model system, we chose intron O.i.I1 from Oceanobacillus iheyensis. Multiple crystal structures have been published for the ribozyme component of this intron (Toor et al. 2008; Chan et al. 2012; Marcia and Pyle 2012), but none of them includes domain VI, apparently because this element gets degraded during RNA purification and/or crystallization of linear excised molecules (Toor et al. 2010).
In order to determine whether the catalytic core of the O.i.I1 ribozyme, or domain VI itself, was primarily responsible for the inability of the molecule to react by branching in vitro, we generated a diversity of constructs that differ not only in the length and sequence of the basal and distal sections of DVI, but also in intron subdomains that are known, or have been proposed to interact with that domain. We now report that some of these constructs generate a significant fraction of branched products and the latter may even become predominant under appropriate experimental conditions. Moreover, contrary to linear products and branched reaction intermediates, the excised intron lariat was found to be quite stable, which makes it possible to consider experimental approaches and setups that could not be contemplated with those constructs of the Oceanobacillus intron that had been available so far.
RESULTS
Chimeras derived from intron A.v.I2
In order to find out which components of the Oceanobacillus intron (O.i.I1) contribute to limit its ability to yield significant amounts of lariat by in vitro self-splicing, our initial strategy has been to generate chimeras between this intron and another group IIC molecule, A.v.I2 from Azotobacter vinelandii, which had been stated to be capable of self-splicing by branching (Toor et al. 2006). We first verified this point by incubating transcripts of an A.v.I2 construct under in vitro self-splicing conditions. Under optimal conditions (100 mM Mg2+, 55°C, see Materials and Methods), transcripts from our ORF-less construct with 46 and 58 nucleotides (nt), respectively, of the original A.v.I2 5′ and 3′ exons (see Materials and Methods) were found to react from 35% to 50% in 2 h (Fig. 1A). The precursor RNA actually initiates self-splicing by both transesterification and hydrolysis: one product migrates as expected for the linear intron and another one (20% to 30% of the total) as expected for the lariat in polyacrylamide gels (Fig. 1A).
FIGURE 1.
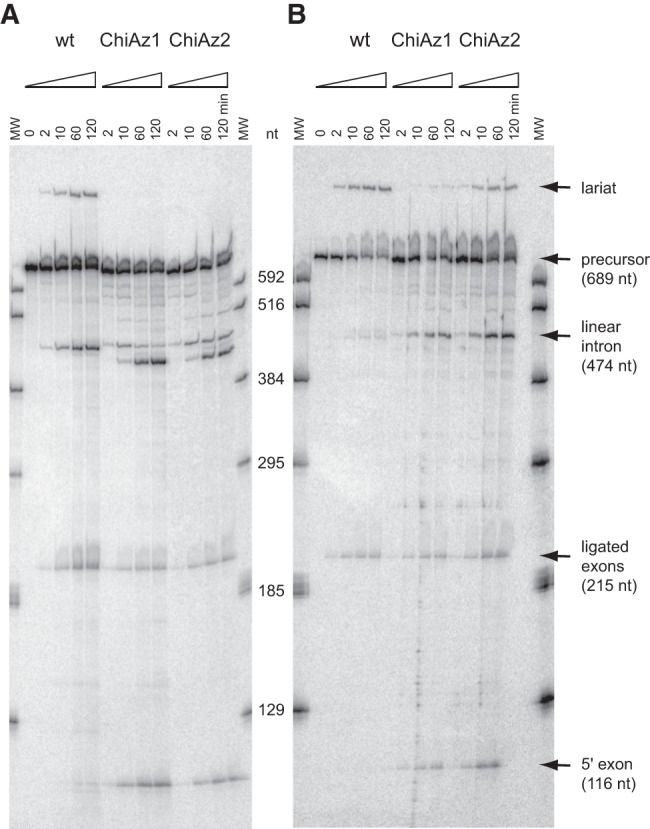
Time course of self-splicing at 55°C of intron A.v.I2 and A.v.I2-based chimeras ChiAz1 and ChiAz2. (A) Magnesium-containing buffer; (B) Manganese-containing buffer (see Materials and Methods for full experimental conditions). Bands corresponding to linear products were identified by reference to transcripts of known length (lanes MW).
When DVI of the A.v.I2 intron was replaced by its homolog in the O.i.I1 intron (chimera ChiAz1), the molecule remained reactive, but the lariat band was no longer visible. Moreover, ligation was less efficient, part of the ligated exon product being replaced by the 5′ exon, and the linear intron product was seen to get slowly converted into a molecule ∼30 nt shorter (Fig. 1A; the segment removed, the identity of which was not investigated, could correspond to DVI).
Substitution of manganese for magnesium has been reported to accelerate specifically branch formation in the self-splicing reaction of the yeast ai5γ model group II intron (Dème et al. 1999). Likewise, replacement of magnesium by manganese in self-splicing experiments with intron B.h.I1, a close relative of the Oceanobacillus intron, which normally reacts by hydrolysis, results in significant amounts (6% of total products) of a molecule migrating as a lariat (Toor et al. 2006). We found that substitution of magnesium has comparable effects on A.v.I2 self-splicing which, in the presence of manganese, was observed to occur predominantly by transesterification (Fig. 1B): the ratio of initial rates of branching versus hydrolysis (Rbr/hy, see Materials and Methods) was increased more than 10-fold, from 0.43 to 5.2 (Table 1). Moreover, when magnesium was replaced by manganese in the ChiAz1 reaction, branch formation became detectable (Fig. 1B), although hydrolysis remained largely predominant (Rbr/hy = 0.061, Table 1).
TABLE 1.
Ratios of initial rates of branching over hydrolysis in buffers containing magnesium and manganese
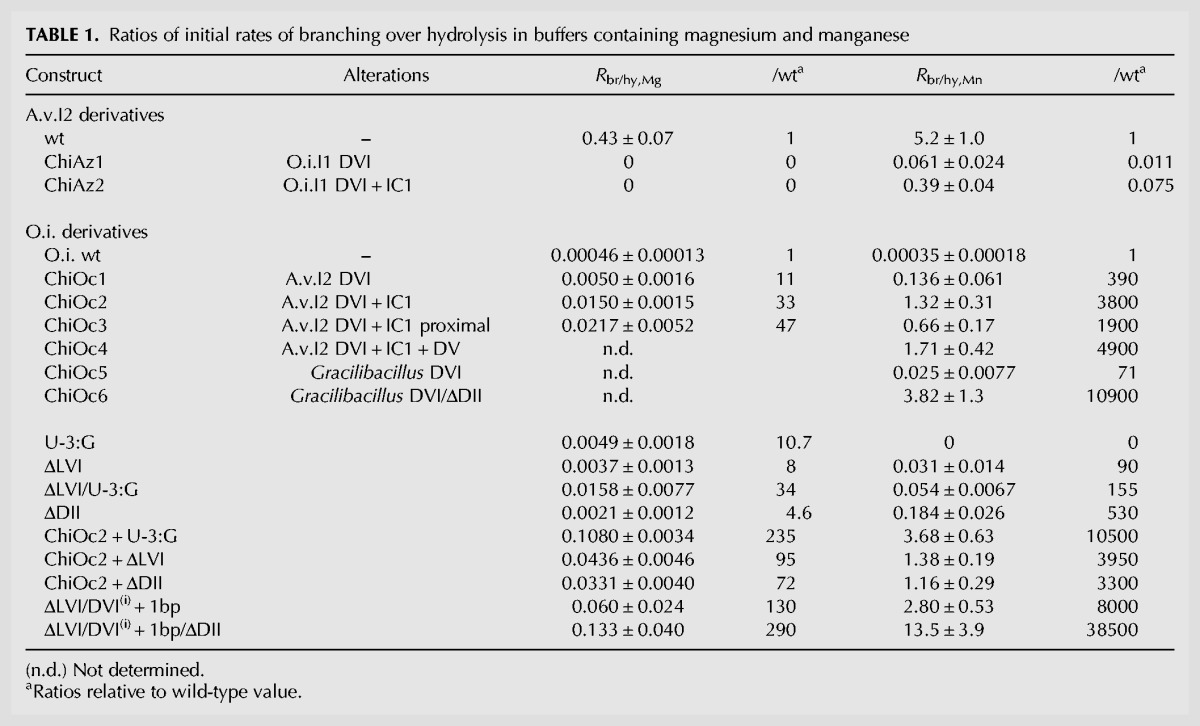
As shown in Figure 2, the A.v.I2 and O.i.I1 introns belong to different subdivisions of subgroup IIC and their so-called non-core components have diverged substantially, both in sequence and predicted secondary structure (Fig. 3). As substructure IC1 of domain I was shown to bind DVI during branch formation (Li et al. 2011b), mismatching of IC1 and DVI in construct ChiAz1 could contribute to the poor branching performance of that RNA. Replacement of subdomain IC1 from A.v.I2 by its homolog in O.i.I1 markedly improved indeed the fraction of branched molecules, with Rbr/hy increasing from 0.061 to 0.39 in the presence of manganese (chimera ChiAz2, Fig. 1B). Still, the latter value is an order of magnitude below that for a wild-type A.v.I2 transcript (Table 1): Even upon substitution of IC1, domain VI from O.i.I1 remains somewhat suboptimal in the A.v.I2 molecular context.
FIGURE 2.
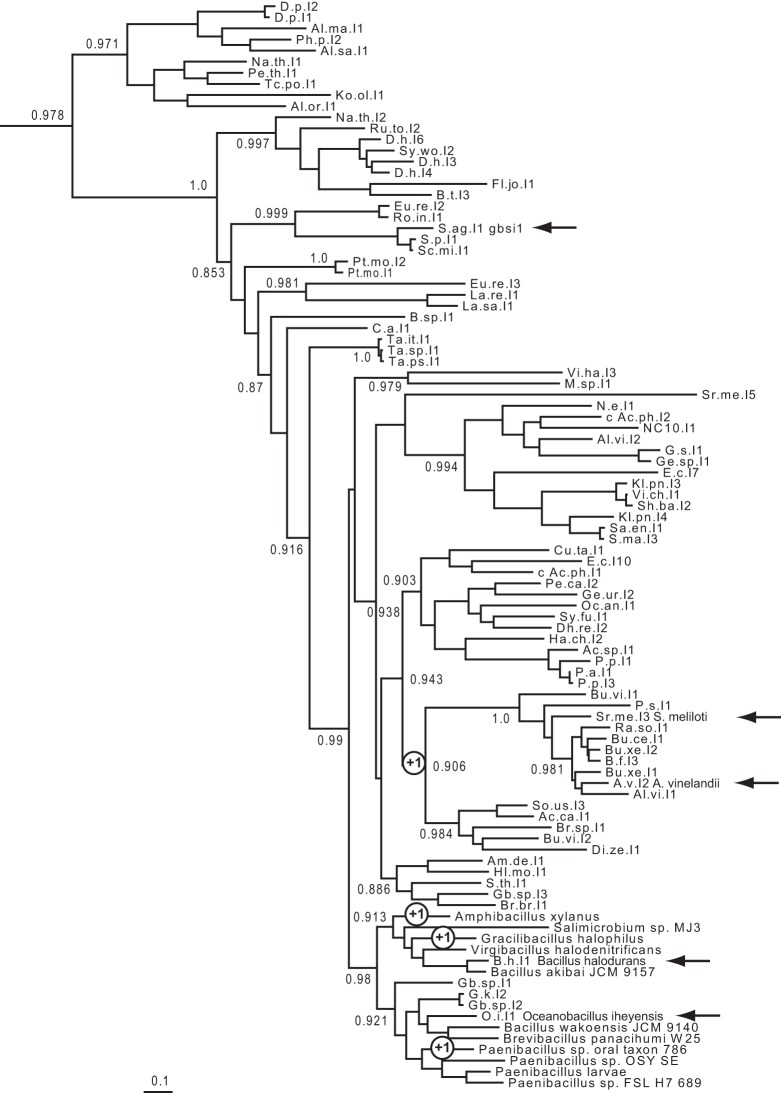
Phylogenetic tree of bacterial class C introns based on alignment of intron-encoded protein sequences (see Materials and Methods). Whenever possible, intron names are abbreviated as in previous works (Candales et al. 2012; Toro and Martínez-Abarca 2013). Arrows point to introns whose self-splicing reaction was examined in vitro (Granlund et al. 2001; Toor et al. 2006, 2008, 2010; this work), circled +1 signs indicate clades whose intron members have acquired an additional (fourth) base pair in their DVI basal helix (according to secondary structure models in the Database for bacterial group II introns of Candales et al. [2012] and F Michel [data not shown]).
FIGURE 3.
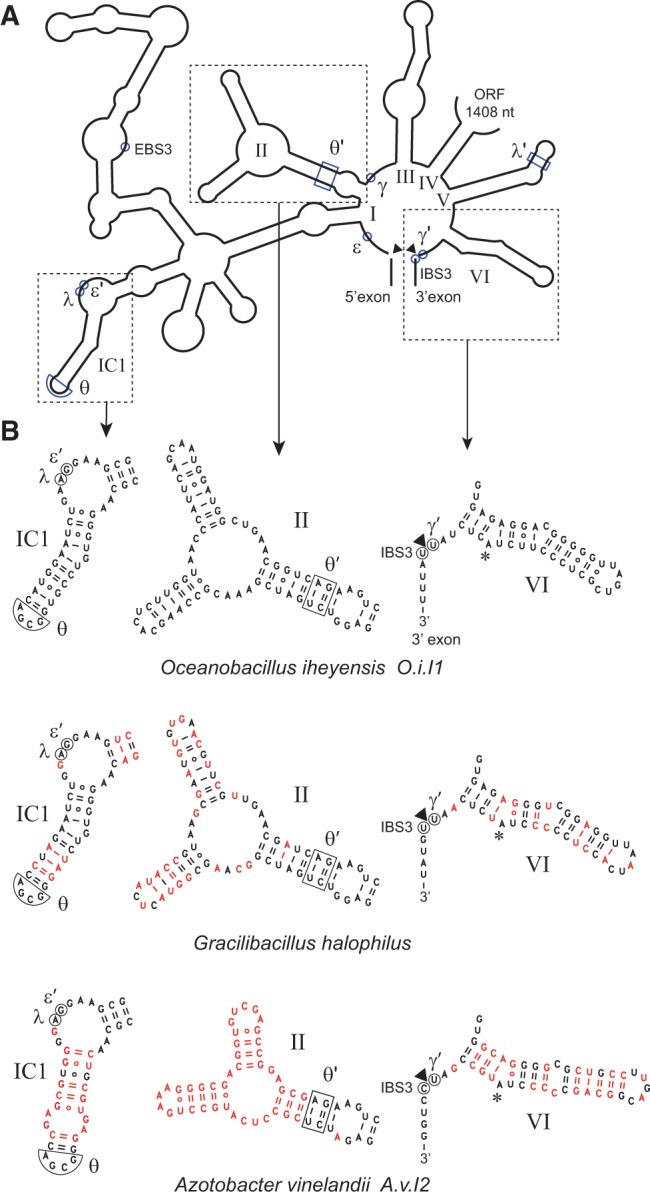
(A) Organization of secondary structure domains I to VI of intron O.i.I1 (from Oceanobacillus iheyensis). (B) Detailed secondary structure of domains IC1, II, and VI in introns O.i.I1, A.v.I2, and Gracilibacillus halophilus. Nucleotides that differ from their counterparts in O.i.I1 are in red. The asterisk marks the predicted branchpoint and an arrowhead points to the expected intron–3′ exon junction. Greek letters, EBS3, and IBS3 indicate nucleotides that participate in tertiary interactions (Costa et al. 2000).
Chimeras based on the Oceanobacillus intron
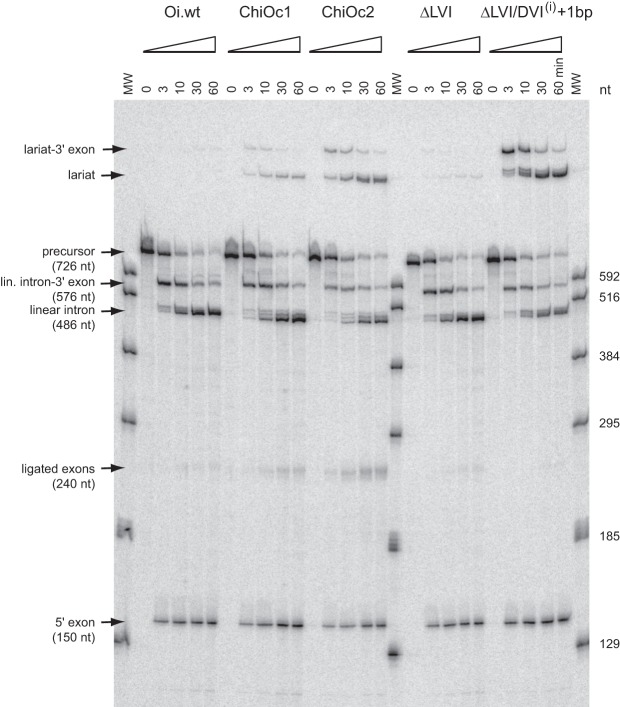
As previously reported (Toor et al. 2008), self-splicing of O.i.I1 transcripts in a magnesium-containing buffer (see Materials and Methods) was found to occur almost exclusively by 5′ splice site hydrolysis; only traces (∼0.2%) of lariat were observed at the end of the reaction (fraction of precursor reacted 0.89 ± 0.03; not shown). Moreover, the fraction of ligated exons is a slight one (0.085 of 5′ exon-containing reaction products). After replacement of O.i.I1 DVI by A.v.I2 DVI (chimera ChiOc1 in Fig. 4), the final fractions of lariat and ligated exons were found to have increased somewhat (to 0.6% and 14%, respectively; not shown), but remained low in absolute values. However, when the same substitution was tested in a manganese-containing buffer, we observed a marked increase in the yield of branched products, as measured by Rbr/hy; the latter was found to have changed by more than two orders of magnitude when O.i.I1 and its ChiOc1 derivative were compared (Table 1; Figs. 4, 5).
FIGURE 4.
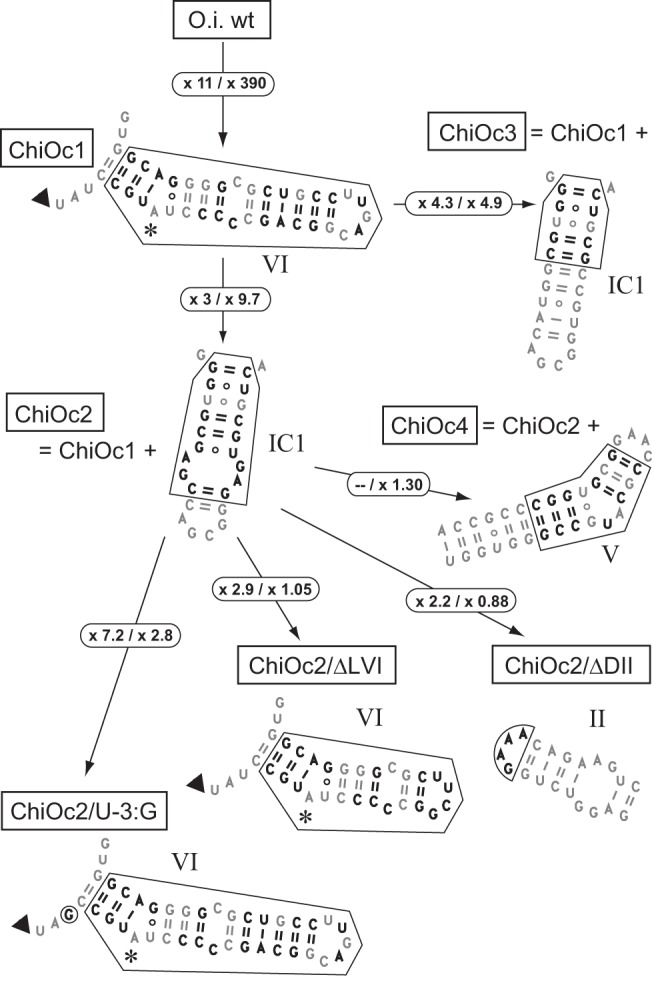
Chimeras based on the Oceanobacillus intron. Changes introduced with respect to the O.i.I1 sequence are in bold type. Arrows indicate the genealogy of constructs, e.g., chimera ChiOc2 was derived from ChiOc1 through replacement of O.i.I1 IC1 by its counterpart in A.v.I2. Circled numbers are increases in the ratio of initial rates of branching over hydrolysis (Table 1) that result from the molecular alterations shown in the figure (values for magnesium- and manganese-containing buffers are to the left and right side of the slash, respectively).
FIGURE 5.
Time course of self-splicing of O.i.I1-based chimeras and mutants (see Figs. 4, 6) in manganese-containing buffer (see Materials and Methods). Bands corresponding to linear products were identified by reference to transcripts of known length (lanes MW). Multiple bands visible at short reaction times just above the lariat intron of construct ΔLVI/DVI(i) + 1 bp were assumed to have been generated by the same process that results in multiple linear intron forms: Based on the migration of the latter, those bands were tentatively attributed to the use of surrogate 3′ splice sites after positions +7 and +14 of the 3′ exon.
We next investigated whether matching domains VI and IC1, as had been done for A.v.I2-based chimera ChiAz2, would improve the ability of O.i. ChiOc1 to initiate splicing by transesterification at the 5′ splice site. As shown in Figures 4 and 5, this was found to be the case: In manganese, Rbr/hy (Table 1) is ∼10-fold higher for construct ChiOc2 when compared to ChiOc1. Moreover, the yield of ligated exons (Fig. 5) also markedly improved, from 6% to 22% of reaction products.
Since the binding site for domain VI was proposed to be confined to the proximal section of the IC1 distal helix (Li et al. 2011b), only the first 5 bp of stem IC1 were replaced by their A.v.I2 counterparts in chimera ChiOc3 (Fig. 4). As expected, this construct behaves not too differently from chimera ChiOc2: Rbr/hy was found to be about twofold lower in Mn buffer, but somewhat higher in Mg buffer (Fig. 4; Table 1).
In addition to peripheral sections of the ribozyme, the middle part of domain V, which, together with its basal section and bound metal ions, forms the catalytic center of the molecule (Toor et al. 2008), exhibits divergent sequences in introns A.v.I2 and O.i.I1. Still, the Rbr/hy ratio in Mn buffer of transcript ChiOc4, in which domain V of O.i.I1 was replaced by that of A.v.I2, is very similar to that of transcript ChiOc2 (Fig. 4; Table 1).
Mutant derivatives of the Oceanobacillus intron and of Oceanobacillus-based chimeras
Disruption of interactions specific to the second step of splicing was reported to improve the yield of branched over linear products in intron S.c.cox1, a member of subgroup IIA (Costa et al. 1997a). We sought to determine whether altering presumably homologous components of the O.i.I1 and A.v.I2 introns would have similar effects. Deletion of the tip of DVI which, in subgroup IIB introns, contacts domain II (DII) to form the η–η′ second-step interaction (Chanfreau and Jacquier 1996), does increase Rbr/hy, by nearly two orders of magnitude (in Mn buffer), when carried out in our O.i.wt construct (Fig. 6; Table 1), but not in chimera ChiOc2 (Fig. 4). Deletion of the entire distal section of DII was found to have an even larger effect (Fig. 6; Table 1), but again not in chimera ChiOc2. In the latter construct, DVI and DII, which come from rather divergent introns (Fig. 3), may not bind efficiently and that could explain why subsequent attempts to disrupt interactions in this context by modification of either of these components had no significant effect.
FIGURE 6.
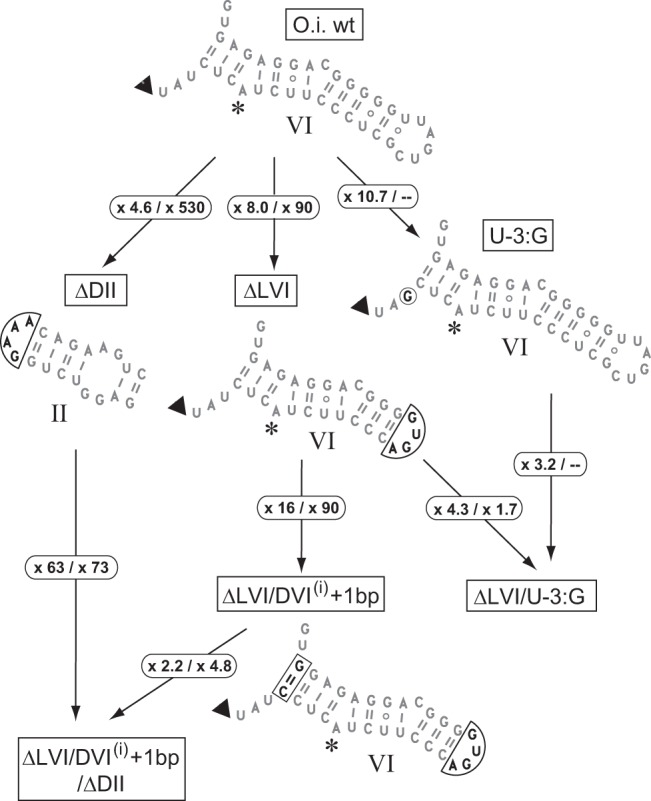
Modifications made to the Oceanobacillus intron. Changes are boxed; circled numbers as in Figure 4.
In a previous investigation of the ai5γ group IIB model molecule, the efficiency of branching was found to depend critically on the length of the basal helix of domain VI (Chu et al. 2001). In intron A.v.I2, that helix comprises 4 bp, instead of only 3 in the Oceanobacillus intron. In order to find out whether this difference could contribute to the better ability of A.v.I2 DVI to participate in branching, whether in its natural context or after transplantation into the O.i.I1 molecule, we introduced an additional base pair in the DVI basal helix of our ΔLVI O.i.I1 construct (mutant ΔLVI/DVI(i) + 1 bp, Fig. 6). The effect was a striking one, with Rbr/hy increasing 90-fold and 16-fold, respectively, in Mn-containing and Mg-containing buffers (Fig. 6; Table 1). Admittedly, changing the length of the basal helix of DVI could alter the interaction of DII with its distal section even when the latter is somewhat truncated. Nevertheless, an estimate of the specific contribution of adding 1 bp to the DVI basal helix may be obtained by introducing the DVI(i) + 1 bp mutation in the ΔDII context—deletion of DII is assumed to remove any second-step interaction between DVI and the rest of the intron—and that contribution turns out to be very large (63- and 73-fold in Mg and Mn solutions, respectively, see Fig. 6).
The basal helix of domain VI is composed of 3 bp not only in the Oceanobacillus intron, but in most members of subgroup IIC (Fig. 2). Except for A.v.I2 and its close relatives, only three introns, which all happen to be closely related to O.i.I1 (Figs. 2, 3), possess a 4-bp DVI basal helix, which they probably acquired independently. In order to confirm the connection between the length of the DVI basal helix and efficient branching, we created chimera ChiOc5 by replacing DVI of O.i.I1 by its homolog in the Gracilibacillus intron (Fig. 3B). Despite Rbr/hy having increased 70-fold as a consequence of this substitution (Table 1), the amount of branched products generated during in vitro self-splicing of construct ChiOc5 in Mn-containing buffer remained small. However, when substitution of DVI was combined with the deletion of the peripheral sections of domain II (chimera ChiOc6), branched products became predominant, the ratio of initial rates of branching over hydrolysis having increased 150-fold compared to construct ChiOc5.
Finally, we changed the ante-penultimate intron nucleotide (position “-3,” immediately 3′ of DVI), which is a U in O.i.I1, to G, as found in A.v.I2 and in fact, the vast majority of subgroup IIC introns. This move, which targeted a nucleotide whose possible structural role remains unknown, was found to improve the yield of branched products whatever the molecular context (wt, ChiOc2, or ΔLVI; Figs. 4, 6; Table 1). It is noteworthy that effects of this substitution were consistently larger in magnesium-containing buffer, whereas the converse was found to be true of most other changes, see Figure 7.
FIGURE 7.

Ratios of initial rate of branching over hydrolysis in manganese- (Rbr/hy,Mn) versus magnesium-containing (Rbr/hy,Mg) buffers. Constructs are depicted in Figures 4 and 6. Bars indicate standard errors (see Materials and Methods). The distribution of data points was tentatively fitted to a log-log linear equation (line).
Kinetic analysis of branched products from a mutant derivative of the Oceanobacillus intron
As reactions of precursor transcripts containing full-length group II introns are typically started by addition of splicing buffer, the accumulation of products reflects the complex interplay of RNA folding and reaction processes. By contrast, purified lariat molecules, which are expected to be chemically stable, may be renatured separately before being characterized kinetically through incubation with the 5′ exon (or an oligonucleotide counterpart of the latter), which will result in a partial debranching reaction (Chin and Pyle 1995; Costa et al. 1998; Dème et al. 1999).
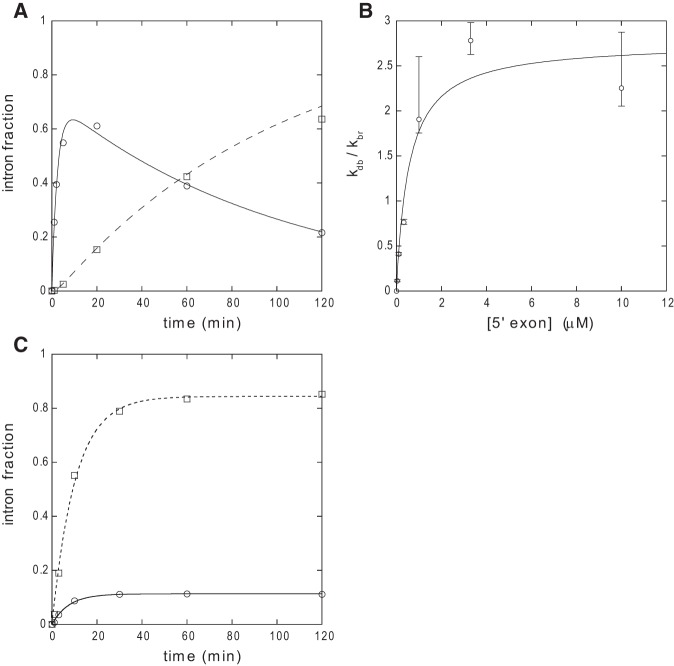
Debranching of gel-purified ΔLVI/DVI(i) + 1 bp lariat by an oligonucleotide substitute of the 5′ exon was observed to be a complex process (Fig. 8A). During a first phase, linear E5–intron molecules rapidly accumulate, after which their concentration begins to slowly decrease. This is a consequence of these molecules undergoing hydrolysis at the 5′ splice site, which results in a steadily growing fraction of slightly shorter linear intron (as a control, aliquots of purified, renatured lariat were verified to be fully stable throughout the duration of experiments; data not shown). Despite this secondary reaction, analytic equations that account for product accumulation may be derived (see Materials and Methods), and these make it possible in turn to estimate the affinity of intron constructs for E5 oligonucleotides by varying the concentration of the latter (Fig. 8B).
FIGURE 8.
Analysis of reaction kinetics. (A) Time course of debranching of purified ΔLVI/DVI(i) + 1 bp lariat molecules by 10 μM oligonucleotide Oceano 14-6 (see Materials and Methods). Circles and full curve, E5–intron linear form; squares, intron linear form. Reaction conditions: 45°C, 2 M NaCl, 40 mM Na-HEPES, pH 7.0 (37°C), 10 mM MnCl2, 0.01% SDS, 20 nM lariat. Debranched and linear fractions were fitted with Kaleidagraph 3.6 to Equations 1 and 2 in Materials and Methods, respectively, with 1/τ1 = 0.410 ± 0.046 min−1, 1/τ2 = 0.0098 ± 0.0011 min−1, and kdb = 0.285 ± 0.024 min−1 (Pearson's R was 0.9947 and 0.9967, respectively). (B) Debranching equilibrium of the ΔLVI/DVI(i) + 1 bp lariat as a function of the concentration of oligonucleotide Oceano 14-6. Reaction conditions: 45°C, 2 M NH4Cl, 50 mM Tris-HCl, pH 7.5 (37°C), 10 mM MnCl2, 0.01% SDS, 10 nM lariat. Bars indicate standard errors. See Materials and Methods for calculation of kdb and kbr. Data were fitted with Kaleidagraph 3.6 to (kdb,max/kbr)/(1 + Kd/[E5]), with kdb,max/kbr = 2.7 ± 0.03 and Kd = 0.56 ± 0.22 μM. (C) Reaction kinetics of precursor transcript O.i.I1 ΔDII in manganese-containing buffer. Data were fitted with Kaleidagraph 3.6 to (v0/k) [1−exp(−kt)], where k is the rate constant and v0 the initial rate of reaction. Circles and full curve, fraction of branched intron products (k = 0.133 ± 0.011 min−1, v0 = 0.015 ± 0.001 min−1); squares and dashed curve, fraction of linear intron products (k = 0.097 ± 0.007 min−1, v0 = 0.082 ± 0.006 min−1).
The Kd we estimated at 45°C for the ΔLVI/DVI(i) + 1 bp:Oceano 14-6 intron:oligonucleotide pair—0.56 ± 0.22 μM (Fig. 8B)—is markedly above the molar concentration (10 nM) that was used for reactions of precursor transcripts, which, moreover, were carried out at 55°C. This could explain in part why coupling of exon ligation with reaction at the 5′ splice site was typically poor (e.g., 22% for chimeric construct ChiOc2 in manganese buffer, Fig. 5); under such conditions, complexes between the 5′ exon and intron-3′ exon that happened to undergo dissociation should not be able to reform efficiently. However, we observed that coupling between the two splicing steps was even far worse for some constructs in which DVI came from the Oceanobacillus intron. As an extreme example, ligated exons were virtually undetectable among reaction products of the ΔLVI/DVI(i) + 1 bp precursor transcript (Fig. 5), in which the distance between the branchpoint and the 3′ splice site has been increased from 6 to 7 nt due to the insertion of an additional base pair in the basal helix of DVI. In this particular case, multiple bands are visible at short reaction times not only around the expected location of the linear intron, but also just above the lariat intron. These additional products, which we tentatively attribute to the use of surrogate 3′ splice sites at positions +7 and +14 of the 3′ exon (see legend to Fig. 5), just 3′ of the sequence AUUUAU, get slowly converted into a slightly shorter molecule that migrates as does the lariat of other constructs. The transient presence of those molecules must reflect inefficient use of the expected 3′ splice site, which accounts in turn for the absence of ligated exons.
Branched and linear intron–3′ exon reaction intermediates were also gel purified after partial reaction of ΔLVI/DVI(i) + 1 bp precursor transcripts in order to analyze the kinetics of their reaction with an oligonucleotide substitute of the 5′ exon. To our surprise, however, both products proved unstable. When incubated alone at 45°C in manganese-containing buffer, they get converted into lariat and linear intron, respectively, with indistinguishable rate constants (0.039 ± 0.008 min−1 and 0.037 ± 0.010 min−1) which correspond to half-reaction times of <20 min.
Instability of the ΔLVI/DVI(i) + 1 bp lariat-3′ exon reaction intermediates explains why self-splicing of this construct generates abundant intron lariat despite a very low yield of ligated exons (Fig. 5). It remains to be established whether such instability of the lariat-3′ exon, which we never observed before, will turn out to be a general property of O.i.I1-derived constructs (and, perhaps, of a majority of group IIC introns) or is specific instead to this particular molecule and experimental conditions. Whatever the answer to this point, it is remarkable that contrary to linear intron forms, which rapidly get trimmed and lose domain VI (Toor et al. 2010), lariat introns from our O.i.I1-derived constructs proved fully stable (at least judging from their migration on denaturing polyacrylamide gels). Thanks to the protection against degradation afforded by a branched structure, it has now become feasible to obtain sizeable amounts of molecules that stably include domain VI, while differing from crystallizable versions of the Oceanobacillus intron by only a limited number of sequence changes.
DISCUSSION
Structural requirements for branch formation by the Oceanobacillus intron
Among possible explanations for the inability of transcripts containing the Oceanobacillus intron to splice by branching in vitro, a plausible one was that domain VI is not positioned correctly into the catalytic center of the O.i.I1 ribozyme in the absence of the intron-encoded protein. We have now shown indeed that by modifying DVI on the one hand, and two intron components (IC1 and DII) that are known or have convincingly been proposed to interact with DVI on the other, the (initial) ratio of branched over linear products in magnesium-containing (Rbr/hy,Mg) and manganese-containing (Rbr/hy,Mn) solutions can be increased up to 290-fold and 38,000-fold, respectively (these figures correspond to the ratios of R values for construct ΔLVI/DVI(i) + 1 bp/ΔDII and the O.i.wt molecule, Table 1). In fact, our best-performing constructs self-splice primarily by branching in the presence of manganese and generate a sizeable fraction of branched products (∼10% for mutant ΔLVI/DVI(i) + 1 bp/ΔDII), even when magnesium is the only available divalent cation.
The data we gathered indicate that two major factors determine the ability of a molecular construct to make efficient use of DVI and initiate self-splicing by branching rather than by 5′ splice-site hydrolysis, the latter being the default mode of reaction in the absence of a functional branchpoint (van der Veen et al. 1987). One of these factors is the length of the DVI basal helix: adding a fourth base pair to the O.i.I1 3-bp helix was found to increase RMg and RMn 16- and 90-fold, respectively (compare constructs ΔLVI and ΔLVI/DVI(i) + 1 bp in Fig. 6 and Table 1).
The second factor is the extent to which DVI and DII can interact. In intron members of subgroup IIB1, which includes model molecules ai5γ (van der Veen et al. 1987) and Pl.LSU/2 (Costa et al. 1997b), two interactions that are known to be specific to the second step of splicing (i.e., exon ligation) bind DVI to the distal part of DII: η–η′ connects the tip of DVI to a specific receptor in DII (Chanfreau and Jacquier 1996; Robart et al. 2014), while the basal part of DVI is contacted by the terminal loop of stem DIIB (π–π′ interaction, Robart et al. 2014). Assuming that homologous interactions exist in group IIC introns (Fig. 9), their disruption should displace the conformational equilibrium that governs the position of DVI (Li et al. 2011b; Chanfreau and Jacquier 1996) in favor of the step 1 configuration (branching). In actual fact, deletion of the tip of DVI alone was found to increase RMg and RMn eight- and 90-fold, respectively, while deletion of the distal part of DII, which is predicted to remove any interaction between DVI and DII, has an even larger effect, resulting in a 530-fold increase in RMn (Fig. 4; Table 1).
FIGURE 9.
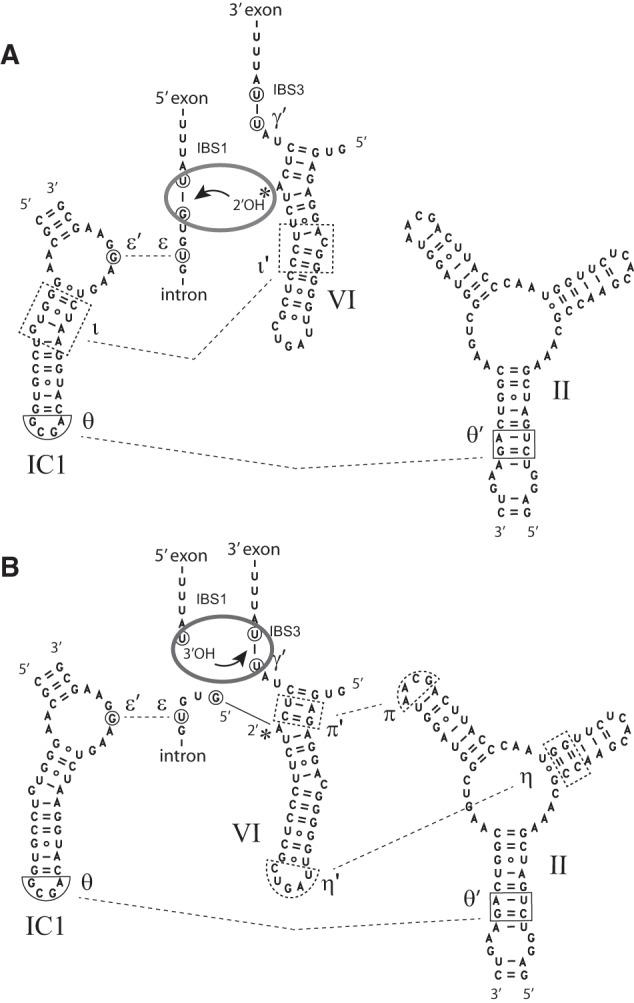
Postulated displacement of domain VI between the two splicing steps. (A) First-step conformation. (B) Second-step conformation. The θ–θ′ and ε–ε′ interactions (Costa et al. 1997a; Toor et al. 2008) are presumed to persist throughout the splicing process, whereas ι–ι′, η–η′, and π–π′ are step-specific interactions, whose location in the Oceanobacillus intron is tentatively inferred from studies on other group II introns (Li et al. 2011b; Robart et al. 2014). A gray ellipse symbolizes the catalytic center of the group II ribozyme and the arrow indicates nucleophilic attack of a phosphodiester bond. Movement of domain VI between its two preferred locations is assumed to occur irrespective of the presence of the 5′ exon (see text).
One advantage of the use of chimeric constructs is that it has allowed us to confirm the existence of an interaction specific to branch formation (ι–ι′, Li et al. 2011b), between DVI and the basal section of stem IC1 (Fig. 9). Interaction of these two components was suggested based on phylogenetic and mutational analyses, and strongly supported by the fact that a defective ribozyme unable to splice by branching was rescued by binding an oligonucleotide that brings stems DVI and IC1 together. However, nucleotide substitutions with compensatory effects could not be designed, for want of an atomic-resolution model. Fortunately, the considerable divergence exhibited by introns A.v.I2 and O.i.I1 not only in DVI, but also in IC1 (Fig. 3B), has now made it possible to carry out such a test. Replacement of the resident IC1 by a version that matches the introduced DVI significantly improves the ability of O.i.I1-based chimera ChiOc1 to perform branching, as the Rbr/hy,Mn ratio for matched combination ChiOc2 is 10-fold higher than for its mismatched counterpart. Moreover, some compensation was observed as well with A.v.I2-based chimera ChiAz2 (Fig. 1B).
Finally, substitution of O.i.I1 antepenultimate U by G (the base most commonly encountered at this position in subgroup IIC introns) stands apart in that contrary to all other changes we made, it has significantly larger effects in magnesium-containing than in manganese-containing buffers (Fig. 7; Table 1). Replacement of magnesium by manganese was reported to specifically accelerate branch formation (compared to 5′ splice site hydrolysis) by close to two orders of magnitude in subgroup IIB1 intron ai5γ (Dème et al. 1999); our own preliminary estimates for O.i.I1, based on the debranching kinetics of the ΔLVI/DVI(i) + 1 bp lariat in magnesium—data not shown—seem to indicate an even larger acceleration—by up to 400-fold at pH 8.0—and confirm the absence of effects on hydrolysis. The molecular basis for the action of manganese remains unknown, but must involve interaction of the ion with the branched form of DVI, at a site that could lie in the vicinity of intron position -3.
Functional and evolutionary implications
This is not the first time that truncating DVI is found to increase the fraction of branched molecules among in vitro self-splicing products. In yeast mitochondrial intron Sc.cox1/1, removal of the distal section of that subdomain was observed to decrease the share of linear intron forms from ∼35% to at most 10% (Costa et al. 1997a). That observation is of particular interest, because Sc.cox1/1 belongs to the highly divergent subgroup IIA, in which components of the η–η′ interaction have exchanged locations: at least in mitochondrial members of that subgroup, the GNRA loop tips DII, rather than DVI, and its receptor lies within the section of DVI that was severed in Costa et al. (1997a). This coincidence highlights the general occurrence, within group II, not merely of interactions between domains II and VI—those potentially persist even in introns that have lost the ability to initiate splicing by transesterification (Li et al. 2011a)—but of a toggling mechanism that allows DVI to swing permanently between its first-step and second-step binding sites both in branched forms (Chanfreau and Jacquier 1996; Costa et al. 2000) and precursor molecules. In many subgroup IIC and, to a lesser extent, subgroup IIA precursor transcripts, DVI may be too strongly bound to DII for branching to be possible under typical self-splicing conditions: the molecules nevertheless eventually react, but by 5′ splice-site hydrolysis, which is the default mode of splicing when a functional domain VI is not available.
Our finding that the length of the basal helix of DVI is a critical factor in determining a construct's ability to self-splice by branching and that four, rather than three, base pairs may be optimal (five was not tested) is also not without precedent. Chu and colleagues (Chu et al. 2001), who assessed the ability of a series of mutant constructs derived from intron ai5γ to initiate self-splicing by branching, already had concluded that the optimal number of base pairs in the DVI basal helix was four. What makes this coincidence truly remarkable, however, is not that introns ai5γ and O.i.I1 should belong to divergent group II subclasses, but that whereas the former molecule possesses the same, apparently optimal, 4-bp DVI basal helix as all fungal members of subgroup IIB1 (Li et al. 2011a), the latter shares its shorter 3 bp stem with most subgroup IIC introns, including the closely related intron B.h.I1 from Bacillus halodurans (Fig. 2), which was shown to generate lariats, but only when part of ribonucleoparticles (Robart et al. 2007), or else in the presence of manganese (Toor et al. 2006).
Is the length of the DVI basal helix generally correlated in group II introns with the ability to generate branched products under standard self-splicing conditions? The importance of the basal section of DVI and surrounding nucleotides is illustrated by the fact that in most group II intron subclasses, a fixed number of nucleotides, either 6 or 7, separate the branchpoint from the 3′ splice site (Michel et al. 1989). Still, among molecules tested for their ability to self-splice, a fair number of those with a 6-nt spacer (between the branchpoint and intron end), and 3-bp DVI basal helix were found to generate branched products in vitro (Adamidi et al. 2003; Ferat et al. 2003; Costa et al. 2006; Nagy et al. 2013). Within group IIC, on the other hand, a correlation between the length of the DVI basal helix and efficient branching in in vitro tests does seem to hold. Among molecules examined (Fig. 2), intron A.v.I2, with a 4-bp DVI basal helix, is the only one to self-splice by branching. Moreover, we have shown that domains VI of the A.v.I2 and Gracilibacillus intron, whose basal helices separately acquired a fourth base pair during the evolutionary diversification of subgroup IIC (Fig. 2), can both rescue to a large extent the ability to carry out the branching reaction when transplanted into the Oceanobacillus intron (Table 1).
Given that few changes are required to convert a group IIC intron that cannot self-splice by branching into a molecule that generates fair amounts of branched products even in the absence of manganese (Table 1), why should evolution have resulted in the use of the branching pathway being dependent on the intron-encoded protein for intron B.h.I1 and, most likely, a majority of other group IIC introns, including its O.i.I1 close relative? It has already been pointed out (Toor et al. 2006) that since group IIC introns are inserted 3′ of transcription terminators, hydrolysis at the 5′ splice site may suffice in general to ensure that the presence of the intron does not perturb gene expression. Along this line of reasoning, expression of the IEP, which allows lariat formation and, as a result, not only efficient splicing (Jacquier and Jacquesson-Breuleux 1991), but efficient, accurate reinsertion into DNA by complete reverse splicing to take place (Lambowitz and Zimmerly 2011), would only occur under special circumstances, that make mobility an appropriate response. One might even speculate that the rather few group IIC introns whose domain VI basal helix is of appropriate length for efficient branch formation (and, in the case of A.v.I2, have actually been found to self-splice by branching) were selected for their ability to dispense with the IEP in order not only to excise themselves from, but reseal, the transcript that harbors them: this could be advantageous, should the integrity of that transcript be of functional importance to the host despite the fact that transcription resulted from leaky termination in the first place.
Somewhat less speculatively, our results also provide clues about how the IEP may enable the intron to be excised in branched form. It might have been thought that most subgroup IIC ribozymes were unable to perform branching because they lacked a first-step binding site for domain VI in subdomain IC1. However, since compensatory effects could be observed by matching DVI and IC1 in A.v.I2-based chimeras (Fig. 1; Table 1), the ι–ι′ interaction must be functional, at least to some extent, in the Oceanobacillus ribozyme. It is thus more likely that the protein acts by weakening interactions specific to the second step of splicing (Fig. 9B), so as to shift the conformational equilibrium in favor of a structure that supports the branching process; except that unlike the destabilization that resulted from deleting the tips of domains II and VI in our constructs, the effect of the protein should be only a transient one in order for the ensuing exon ligation to proceed efficiently. What makes this model especially attractive is that Prp8, which lies at the heart of the spliceosome and shares common ancestry with group II-encoded reverse transcriptases (Dlakić and Mushegian 2011; Galej et al. 2013), is also believed, based on genetic (Liu et al. 2007) and biochemical (Schellenberg et al. 2013) evidence, to participate in a conformational shift in between the two steps of nuclear pre-mRNA splicing.
Temporary destabilization of second-step-specific interactions cannot be the sole mode of action of the IEP, since our data imply that in the case of O.i.I1 and, presumably, other group IIC introns, the protein must also be capable of maximizing the catalytic efficiency of branch formation by positioning optimally the DVI substrate of the reaction, something that we have shown can be achieved (at least in part) in vitro by introducing an additional base pair in the DVI basal helix. Yet another, nonexclusive possibility is that the IEP is responsible for the specific recognition of Rho-independent terminators by the ribonucleoparticles it generates when it associates with intron B.h.I1 (Robart et al. 2007). The exon sequence targeted by the intron's 5′ exon-binding site (EBS1) is too short in the case of subgroup IIC introns to contribute significantly to the specificity of transposition and the use of surrogate 5′ splice sites during in vitro self-splicing reactions of intron B.h.I1 has been interpreted as reflecting an affinity for terminator-like sequences (Toor et al. 2006). The question of whether or not group IIC ribozymes specifically bind stem–loop structures when alone in salt solutions can now rigorously be addressed, by monitoring the debranching of purified intron lariat by predesigned 5′ exon oligonucleotides, as in Figure 8. In case the answer turned out to be a positive one, it should be of interest to find out how group IIC ribozymes, which are relatively small compared to those of other intron subgroups, manage to contact specifically an RNA double helix, and whether or not the substructures involved in that interaction may be regarded as counterparts of the second exon-binding site (EBS2) of other group II introns.
MATERIALS AND METHODS
Constructs
A.v.I2 intron
DNA segments corresponding to nt 4653472 to 4653038 and 4651647 to 4651510 of the Azotobacter vinelandii DJ genome were PCR amplified from strain UWR with oligonucleotide pairs 5′-CCATTTCGAATTCCGTGCTGAAAAAGCCCTGCCGG and 5′-AACTTAAGGTCTCCCTCTCGTTCGGTCCT, 5′-AACTTAAGGTCTCGAGAGTTCGCTCTCGAACCGCCCGGTG and 5′-GAAATTTAAGCTTGCTCCGGCCGAACCTCTC, respectively. The resulting PCR products were digested with EcoRI, BsaI, and HindIII (underlined sites in primer sequences) and ligated with a pBIIKS(−) vector previously cut with EcoRI and HindIII. After digestion of the resulting construct with Acc65I, T7 polymerase transcription generates a precursor RNA molecule with 5′ and 3′ exons of 116 and 99 nt (46 and 58 nt of which, respectively, come from A. vinelandii) and an ORF-less intron of 474 nt in which the 6-bp helix at the base of domain IV is capped by a UUCG terminal loop. In chimera ChiAz1, the 33-nt DVI of O.i.I1 was substituted for the 36-nt DVI of A.v.I2. Further replacement of the 24-nt IC1 hairpin of A.v.I2 by the 25-nt homologous structure in O.i.I1 gave rise to chimera ChiAz2.
O.i.I1 intron
DNA segments corresponding to nt 1841024 to 1840557 and 1839132 to 1838982 of the Oceanobacillus iheyensis HTE831 genome were PCR amplified with oligonucleotide pairs 5′-CTGCAGGAATTCGATAATTAGATGGTAAGGAAGTAAAT and 5′-CGCCTAGGTCTCACGAACGCCATACTTGTTCCGTCCTTCCCCTT, 5′-ATCAAAGGTCTCTTTCGCGCCTAAGCTTGAACCGCCGTATACCGA and 5′-GACTATATCGATAAGCTTGATCTAGGCGATGAAGCAGGAA, respectively. PCR products were digested with EcoRI, BsaI, and ClaI (underlined sites in primer sequences) and ligated with vector pBIIKS(−) that had been cut with EcoRI and ClaI. Digestion of the resulting construct (“O.i.wt” in Table 1) with ClaI, followed by T7 polymerase transcription generates a precursor RNA with 5′ and 3′ exons of 150 and 90 nt (77 and 77 nt of which come from O. iheyensis) and an ORF-less intron of 486 nt with the same DIV sequence (CAAGTATGGCGTTCGCGCCTAAGCTTG) as in Toor et al. (2008). Our chimeric and mutant constructs derived from O.i.wt are listed in Table 1, with corresponding base substitutions shown in Figures 4 and 6. All constructs were verified by sequencing.
Phylogenetic analyses
A continuous section of 330 amino acids of the protein encoded by intron O.i.I1 was aligned with homologous sequences from intron members of bacterial classes C, F, and g1 (Candales et al. 2012; Toro and Martínez-Abarca 2013). The tree in Figure 2 was generated with program PhyML3.1, using an SPR topology search and default options (e.g., an LG model of amino acid substitutions and four rate categories); branch support was estimated using approximate likelihood ratios. The following group II intron ORF sequences, which have been tentatively attributed to bacterial classes F and g1 (Toro and Martínez-Abarca 2013), were used as outgroup: UA.I4, c-Ku.st.I1, D.a.I1, Ni.ha.I1, Pe.ca.I3, Cl.kl.I2, S.ag.I2, So.us.I2, Ge.ur.I1, Pe.th.I2, My.va.I1, Fr.sp.I3, Rh.sp.I1, Mx.xa.I1, Sg.ce.I1, B.th.I3, W.e.I5, Ma.sp.I2, Rh.js.I1, Ca.ac.I1, Sp.li.I1, Sr.me.I2. In addition, 11 group II ORF sequences that are close relatives of the O.i.I1 IEP, but were not part of the Database of bacterial group II introns (Candales et al. 2012), were included in our analyses (species and strain names are followed by accession numbers and intron coordinates): Bacillus akibai JCM 9157, #BAUV01000115, 1884-2; Virgibacillus halodenitrificans, #ALEF01000009, 9-1890; Bacillus wakoensis JCM 9140, #BAUT01000148, 1-1883; Gracilibacillus halophilus, #APML01000087, 2314-429; Amphibacillus xylanus, #NC018704, 47424-49309; Salimicrobium sp. MJ3, #AMPQ01000013, 53882-55745; Paenibacillus sp. oral taxon 786, #ACIH01000268, 2-1887; Paenibacillus larvae subsp. larvae BRL-230010, #NZ_AARF01000332, 1149-3031; Paenibacillus sp. OSY-SE, #ALKF01000166, 874-2770; Paenibacillus sp. FSL H7-689, #ASPU01000054, 145387-143505; Brevibacillus panacihumi W25, #AYJU01000016, 649088-647210.
RNA synthesis and purification
Synthesis and purification of precursor transcripts internally labeled with 32P were carried out essentially as described in Costa et al. (1997b). Templates for T7 polymerase transcription of A.v.I2- and O.i.I1-derived constructs were obtained by digestion of plasmid DNA with Acc65I and ClaI, respectively. Transcription was carried out in the presence of 20 mM Mg2+ (17.5 mM nucleotide triphosphates) in order to minimize self-splicing.
Kinetic analyses
Self-splicing reactions were started by addition of an equal volume of 2×-concentrated buffer to a solution of precursor transcript in water (at a final concentration of typically 10 nM). Reactions were stopped by addition of an equal volume of formamide loading buffer containing Na2EDTA (final concentration 20 and 110 mM, respectively, for manganese- and magnesium-containing splicing buffers). Samples were run on denaturing 4% polyacrylamide gels (50% urea w:v, 19:1 acrylamide:bis-acrylamide ratio), and bands generated by the precursor and reaction products were quantitated with a PhosphorImager (Molecular Dynamics).
Reactivity of both O.i.I1 and A.v.I2 precursor transcripts was found to be optimal at 55°C in 0.5 M NH4Cl, 100 mM MgCl2, 40 mM Tris-HCl (pH 7.5 at 37°C) (“magnesium-containing” buffer) or 2 M NH4Cl, 10 mM MnCl2, 50 mM Tris-HCl (pH 7.5 at 37°C) (“manganese-containing” buffer) and the same conditions were used for all precursor constructs examined in this work (lowering the temperature to 40°C improves somewhat the coupling between 5′ cleavage and exon ligation which, as measured by the molar ratio of ligated to 5′ exon, increases from 0.08 to 0.18 for our wild-type O.i.I1 construct, but results in a sharp drop in the reactivity of precursor transcripts—from above 80% to below 30%).
As was done in Li et al. (2011b), we chose to characterize competing branching and hydrolysis processes (a representative example is provided in Fig. 8C) by the ratio of initial reaction rates (Rbr/hy), a parameter that was found to ensure both reproducibility of measurements for any particular construct and optimal discrimination between different molecules. The standard error on Rbr/hy (Fig. 7; Table 1) was estimated by fitting kinetic data with Kaleidagraph 3.6 and adding relative errors for branching and hydrolysis.
Lariat debranching reactions were carried out at 45°C rather than 55°C in order to ensure increased affinity of oligonucleotide substitutes of the 5′ exon (E5) for the intron. Reactions were started by addition of the E5 oligonucleotide in reaction buffer to a solution of lariat that had been renatured by addition of an equal volume of 2× reaction buffer at 55°C followed by slow cooling (0.8°C min−1) to 45°C.
Lariat debranching is a reversible process (Chin and Pyle 1995; Costa et al. 1998). However, debranched E5–intron molecules may also react by hydrolysis of the 5′ splice site, which competes with branching and is an irreversible process:
where kdb, kbr, and khy are rate constants for debranching, branching, and hydrolysis reactions, respectively. At saturating E5 concentrations, the system becomes pseudo-first order (e.g., Fersht 1977) with
| (1) |
| (2) |
where [E5-I] and [I] are molar fractions of debranched and linearized intron molecules, respectively, and τ1 and τ2 are relaxation times estimated by fitting experimental data. Rate constants of individual reaction steps can then be obtained from the following equations:
In order to estimate Kd, the dissociation constant of the intron–E5 pair, reactions were set up at different E5 concentrations and for each of these, kdb,[E5] was determined by fitting experimental data (Fig. 8A), assuming that koff is much higher than kdb (as we had found indeed for another group II intron; Costa and Michel 1999; F Michel, unpubl.). As a 5′ exon substitute, we used 5′-AAAACCTTCCCTTTUGUUAU (Oceano 14-6); ribonucleotides (in bold type) are complementary to the intron-contained EBS1 sequence (as visible in the crystallographic structure of Toor et al. 2008) and the deoxyribonucleotide tail is destined to ensure separation of linear E5-I molecules from linear intron molecules on denaturing polyacrylamide gels.
ACKNOWLEDGMENTS
We thank Eric Westhof for critical reading of our manuscript. This work was supported by the French Agence Nationale de la Recherche (grant ANR-10-BLAN-1502 to F.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054643.115.
REFERENCES
- Adamidi C, Fedorova O, Pyle AM. 2003. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42: 3409–3418. [DOI] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res 25: 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Quirk S, Clyman J, Belfort M. 1990. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res 18: 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candales MA, Duong A, Hood KS, Li T, Neufeld RAE, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S. 2012. Database for bacterial group II introns. Nucleic Acids Res 40: D187–D190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A. 1996. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J 15: 3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Chan RT, Robart AR, Rajashankar KR, Pyle AM, Toor N. 2012. Crystal structure of a group II intron in the pre-catalytic state. Nat Struct Mol Biol 19: 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, Pyle AM. 1995. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA 1: 391–406. [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Adamidi C, Liu Q, Perlman PS, Pyle AM. 2001. Control of branch-site choice by a group II intron. EMBO J 20: 6866–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F. 1999. Tight binding of the 5′ exon to domain I of a group II self-splicing intron requires completion of the intron active site. EMBO J 18: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Dème E, Jacquier A, Michel F. 1997a. Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol 267: 520–536. [DOI] [PubMed] [Google Scholar]
- Costa M, Fontaine JM, Loiseaux-de Goër S, Michel F. 1997b. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J Mol Biol 274: 353–364. [DOI] [PubMed] [Google Scholar]
- Costa M, Christian EL, Michel F. 1998. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA 4: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E. 2000. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19: 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Molina-Sánchez MD, Martinez-Abarca F, Toro N. 2006. An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie 88: 711–717. [DOI] [PubMed] [Google Scholar]
- Dème E, Nolte A, Jacquier A. 1999. Unexpected metal ion requirements specific for catalysis of the branching reaction in a group II intron. Biochemistry 38: 3157–3167. [DOI] [PubMed] [Google Scholar]
- Dlakić M, Mushegian A. 2011. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. 1989. Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene 82: 91–114. [DOI] [PubMed] [Google Scholar]
- Ferat J-L, Le Gouar M, Michel F. 2003. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol Microbiol 49: 1407–1423. [DOI] [PubMed] [Google Scholar]
- Fersht A. 1977. Enzyme structure and mechanism. W.H. Freeman, Reading/San Francisco. [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K. 2013. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493: 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granlund M, Michel F, Norgren M. 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol 183: 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A, Jacquesson-Breuleux N. 1991. Splice site selection and role of the lariat in a group II intron. J Mol Biol 219: 415–428. [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS. 1988. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439. [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. 2011. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-F, Costa M, Bassi G, Lai Y-K, Michel F. 2011a. Recurrent insertion of 5′-terminal nucleotides and loss of the branchpoint motif in lineages of group II introns inserted in mitochondrial preribosomal RNAs. RNA 17: 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-F, Costa M, Michel F. 2011b. Linking the branchpoint helix to a newly found receptor allows lariat formation by a group II intron. EMBO J 30: 3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM. 2007. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol 14: 519–526. [DOI] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30. [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Doucet AJ, Ferat J-L. 2007. Specialized lineages of bacterial group II introns. Biochimie 89: 542–553. [DOI] [PubMed] [Google Scholar]
- Mullineux S-T, Costa M, Bassi GS, Michel F, Hausner G. 2010. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA 16: 1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V, Pirakitikulr N, Zhou KI, Chillón I, Luo J, Pyle AM. 2013. Predicted group II intron lineages E and F comprise catalytically active ribozymes. RNA 19: 1266–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Seo W, Zimmerly S. 2007. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci 104: 6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. 2014. Crystal structure of a eukaryotic group II intron lariat. Nature 514: 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman V, Amann R, Shub DA, Schulz-Vogt HN. 2012. Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria. Proc Natl Acad Sci 109: 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Wu T, Ritchie DB, Fica S, Staley JP, Atta KA, LaPointe P, MacMillan AM. 2013. A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nat Struct Mol Biol 20: 728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Zimmerly S. 2002. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA 8: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Robart AR, Christianson J, Zimmerly S. 2006. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res 34: 6461–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM. 2010. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Martínez-Abarca F. 2013. Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PLoS One 8: e55102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Kwakman JH, Grivell LA. 1987. Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J 6: 3827–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T. 2002. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J 21: 3794–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Mastroianni M, White TB, Lambowitz AM. 2009. Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation. Proc Natl Acad Sci 106: 18189–18194. [DOI] [PMC free article] [PubMed] [Google Scholar]