Abstract
Pluripotent cells give rise to distinct cell types during development and are regulated by often self-reinforcing molecular networks. How such networks allow cells to differentiate is less well understood. Here, we use integrative methods to show that external signals induce reorganization of the mouse embryonic stem cell pluripotency network and that a sub-network of four factors - Nac1, Oct4, Tcf3 and Sox2 – regulates their differentiation into the alternative mesendodermal and neuroectodermal fates. In the mesendodermal fate, Nac1 and Oct4 were constrained within quantitative windows, while Sox2 and Tcf3 were repressed. In contrast, in the neuroectodermal fate, Sox2 and Tcf3 were constrained while Nac1 and Oct4 were repressed. In addition, we show that Nac1 coordinates differentiation by activating Oct4 and inhibiting both Sox2 and Tcf3. Reorganization of progenitor cell networks around shared factors might be a common differentiation strategy and our integrative approach provides a general methodology for delineating such networks.
INTRODUCTION
Stem cells give rise to multiple cell types of an organism through progressive differentiation. While successive new fates are being specified, alternative fates are being restricted to create distinct cell lineages (Graf and Enver, 2009; Waddington, 1957). Cell-fate specifying information, in the form of spatial cues or inter-cellular signals, is processed through molecular networks whose causal regulations and dynamics ultimately define the final cellular outcome (Davidson, 2006). Understanding how such a network changes during cell fate choice is thus crucial to understanding development. Embryonic stem cells (ESC), which are both pluripotent and self-renewing (Evans and Kaufman, 1981; Martin, 1981; Nishikawa et al., 2007), represent a good model system for addressing this problem.
Mouse ESCs are regulated by an ensemble of transcription factors (TFs) including Pou5f1 (Oct4), Nanog, Sox2, Rex1, Nacc1 (Nac1), Klf4, cMyc and others (Figure S1A), which promote pluripotency by activating their own expression, and that of other pluripotency genes, and by suppressing genes required for differentiation (Cole and Young, 2008; Ng and Surani, 2011; Niwa, 2007; Silva and Smith, 2008). The key stem cell factor Nanog plays a central role in establishing the self-reinforcing pluripotency network through nested positive feedback and feed-forward regulations (Cole and Young, 2008; MacArthur et al., 2012). However, how the self-reinforcing regulations of the pluripotency network change as ESCs differentiate into alternative cell fates is not well understood.
Here, we used an integrative and quantitative approach to analyse how these regulations change as mouse ESCs exit pluripotency and choose between the alternative mesendodermal (ME) and neuroectodermal (NE) cell fates (Figures 1A) that act as precursors for germ layer specification during development (Gadue et al., 2005). We found that, during differentiation, the pluripotency network reorganises around four key TFs – Nac1, Oct4, Tcf3 and Sox2 – and that Nac1, a BEN and BTB (POZ) domain containing protein (Mackler et al., 2000), plays a coordinating role. Our findings suggest that pluripotency is a mutually balanced state among the differentiation-promoting factors, which then resolves during differentiation. Similar mechanisms may underlie the maintenance and differentiation of other progenitor and stem cells.
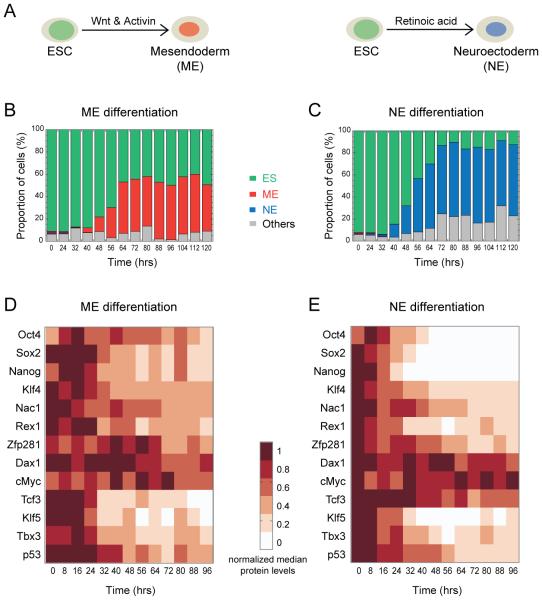
Figure 1. Differentiation-induced changes in the levels of pluripotency factors.
Embryonic stem cells (ESC) exit pluripotency to choose between mesendodermal (ME) and neuroectodermal (NE) germ layer precursor fates, guided by Wnt and Activin, and retinoic acid respectively (A) (Gadue et al., 2006; Thomson et al., 2011; Ying et al., 2003). The proportion of cells in the pluripotent (ES), ME, NE and other, undetermined, states at the indicated time points during ESC (Sox1-GFP cell line) differentiation towards the ME (B) and NE (C) fate. Changes in the levels of indicated pluripotency factors during ME (D) and NE (E) differentiation. For each protein, median value (from single cell data) at the indicated time point was normalized to its maximum from the respective differentiation condition. See also Figure S1 and Supplemental Experimental Procedures.
RESULTS
Dynamic changes in TF levels as ESCs exit pluripotency
We studied the dynamic changes to the pluripotency network during mouse ESC differentiation into the ME and NE fates by systematically quantifying the TFs which regulate the ES state (Figures 1 and S1). In total, we measured thirteen TFs which included nine important members of the extended pluripotency network (Oct4, Sox2, Nanog, Klf4, cMyc, Nac1, Dax1, Rex1 and Zfp281) (Kim et al., 2008; Wang et al., 2006) and others (Tcf3, Klf5, p53 and Tbx3) which are thought to have various roles in regulating pluripotency (Cole et al., 2008; Ema et al., 2008; Han et al., 2010; Neveu et al., 2010). This set of TFs included the stem cell “trinity” of Oct4, Sox2 and Nanog (Silva and Smith, 2008), the Yamanaka reprogramming factors Oct4, Sox2, Klf4 and cMyc (Takahashi and Yamanaka, 2006), and the Wnt-responsive Tcf3, which modulates the balance between pluripotency and differentiation (Atlasi et al., 2013; Cole et al., 2008; Wray et al., 2011).
ESCs can be differentiated in-vitro into either the ME or NE fate: Chiron (CHIR99021, a Wnt agonist that inhibits glycogen synthase kinase 3β) plus Activin-A together promote the ME fate while retinoic acid promotes the NE fate (Figure 1A) (Gadue et al., 2006; Thomson et al., 2011; Ying et al., 2003). We employed these signals to induce the ME and NE fates from ESCs, and primarily focused on analysing the reorganization of the pluripotency transcriptional network during differentiation (Figure1, S1 and Supplemental Information). To examine the temporal response to the signals we followed cell populations for time periods of 0, 24, 32, 40, …, 120 hrs of ME and NE differentiation (Figure 1B and C). A limited mixture of ME- and NE-fates was observed under the pluripotency condition (0 hr), but these populations quickly diverged and by 40 hrs differentiation was specific and exclusive. By 72 to 80 hrs, maximal proportion of cells expressed the fate-specific T or Sox1 markers under the respective differentiation condition. We noticed a higher proportion of undifferentiated cells under ME than NE which might be due to Chiron, which is also known to promote pluripotency (Ying et al., 2008).
We then measured the protein levels for all thirteen TFs in a 96 hour time-course for each differentiation condition using quantitative immuno-fluorescence in wild-type ESCs (Figure S1F, G and Supplemental Information). Different cell populations were used for each immunostaining to measure up to 3 TFs and nuclear DNA. Oct4 was measured across all immunostainings as a consistency check. At a given time point and condition, protein measurements were fairly consistent across immunostainings (Figure S1H: representative single-cell distributions of Oct4). ME and NE differentiation signals regulated the TF protein levels differently (Figure S1I and J: representative single-cell distributions of Oct4 across all time points).
The majority of the median TF protein levels decreased over time (Figure 1D and E), with the decrease being more prominent for NE than for ME, particularly for the trinity of Nanog, Oct4 and Sox2. In contrast, Tcf3 decreased more rapidly for ME than for NE, as expected since the ME condition is induced by Chiron, a Wnt agonist, and Wnt signaling inhibits Tcf3 (Atlasi et al., 2013; Wray et al., 2011). Klf5, p53 and Tbx3 showed similar dynamics under both ME and NE conditions and Zfp281, cMyc and Dax1 showed the least overall changes. These temporal changes are largely consistent with previous observations involving both directed and undirected differentiation experiments (Lu et al., 2009; MacArthur et al., 2012; Pereira et al., 2006; Thomson et al., 2011). Our results captured the varying dynamics of the TFs as ESCs exit pluripotency and transit towards the ME or NE fate. These dynamic changes in TFs coincide with the onset of differentiation marked by T or Sox1 expression (Figure 1B and C). In order to understand how these dynamic changes in TF levels influence their causal regulations and the cell fate transition, and to identify the key regulators behind these changes, we turned to computational analysis.
Computational analysis reveals potential pluripotency factors involved in differentiation
We employed Principal Component Analysis (PCA) to extract the main features of the differentiation dynamics. PCA determines a sequence of orthogonal vectors which best capture the shape of the variance in a data set (Jolliffe, 2002). The contribution of individual factors towards the collective behavior of all factors within a network can be assessed using PCA. We represented the median levels of each TF as a multi-dimensional vector, at all 13 time points (samples every 8 hrs from 0 to 96 hrs) for both ME and NE conditions.
We first analyzed the extended pluripotency network with nine TFs: Oct4, Sox2, Nanog, Klf4, cMyc, Rex1, Nac1, Dax1 and Zfp281 (Kim et al., 2008; Wang et al., 2006). The first three principal components (PC) captured over 95% of the variance in the data (Figure 2A and B). PC1, which accounts for ~80% of the variance, captured the change in time; PC2 partially captured the difference between differentiation conditions and PC3 captured the changes within each condition. Although the two fates had different trajectories, they were indistinguishable along PC2 at several ME and NE time points (Figure 2A and B). This was in contrast to the mutually exclusive ME or NE choice observed (Figure 1B and C), suggesting that the nine factors were not sufficient to fully distinguish the two fate choices.
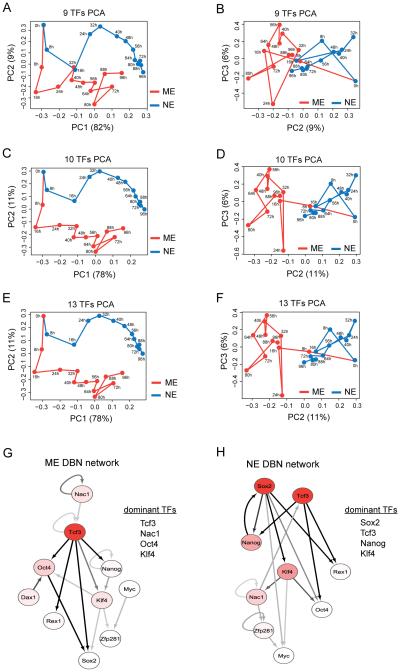
Figure 2. Identification of potential differentiation regulatory TFs by computational analysis.
Principal component analysis (PCA) using the median values of nine pluripotency factors - Oct4, Nanog, Sox2, Klf4, Rex1, Nac1, Zfp281, Dax1 and cMyc (A and B) and these nine plus Tcf3 (C and D) for the ME (red) and NE (blue) conditions. PCA with all 13 measured TFs (E and F). Principal component (PC) projections are shown as PC1 vs PC2 and PC2 vs PC3 plots. Dynamic Bayesian Network (DBN) analysis using median values of the ten TFs (above nine plus Tcf3) for ME (G) and NE (H) differentiation. The nodes are colored using a scale based on dominance scores which have been max-normalized for each network separately: white (0.5 and below), red gradient (0.5 to 1) and red (1). The edges are colored using a scale based on their posterior probability predictions with a threshold of 0.25: off-white (0.25), gray scale (0.25 to 0.5) and black (above 0.5). The extent of connectivity measured by dominance scores indicated the top four factors in the ME (G) and NE (H) networks. DBN networks are drawn using Cytoscape. See also Figure S2 and experimental procedures.
A second PCA analysis which included each of the additional TFs measured as the tenth TF revealed a distinct role for Tcf3. While there were no changes with Klf5, p53 or Tbx3, inclusion of Tcf3 surprisingly clearly separated the two differentiation trajectories (Figure 2C, D and Figure S2A - C). Tcf3 contributed significantly to PC2 (Figure S2M), suggesting that it played an important role in distinguishing the ME and NE fates. To further verify the role of each TF, we excluded them one at a time and repeated the analysis. While exclusion of Nac1, Oct4 or Sox2 had severe effects on the trajectory separation (Figure S2D - F), exclusion of the other TFs on the other hand had less effect (Figure S2G - L). In agreement with their role, the four TFs - Nac1, Oct4, Sox2 and Tcf3 were the dominant contributors to PC coefficients (Figure S2M). Furthermore, PCA on all thirteen proteins gave no further improvement (Figure 2E - F).
As a complementary approach to PCA we used Dynamic Bayesian Networks (DBN). Bayesian Networks have been widely used to infer causal relationships and to identify key regulators from complex biological data (Friedman et al., 2000; Pe'er, 2005) (Figure S2N). Non-homogeneous DBNs allow different model structures during each segment of a time-course, making it particularly applicable to processes like stem cell differentiation in which the underlying causal network is not necessarily static (Dondelinger, 2013). We assumed the initial DBN network to be random and incorporated in it the previously-known ME-promoting function of Oct4 through Sox2 regulation (Thomson et al., 2011) and the general differentiation-promoting function of Tcf3 through Nanog, Oct4 and Rex1 regulations (Cole et al., 2008; Pereira et al., 2006; Wray et al., 2011) (Supplemental Information).
We used a Markov Chain Monte Carlo algorithm to find optimal and robust non-homogeneous DBNs with the median values of the ten TFs considered above (Oct4, Sox2, Nanog, Klf4, cMyc, Rex1, Nac1, Dax1, Zfp281 and Tcf3) at all 13 time points, for ME and NE conditions separately (Figure 2G and H). The inferred ME and NE networks were distinct from each other, suggesting that the causal network of pluripotency may undergo major reorganization during differentiation. Change-point analysis further suggested that the initial network changes the most within the first 32 hrs of differentiation (Figure S2O and P), and with no significant changes after 72 hrs. These dynamic changes to the pluripotency network precede the emergence of the ME and NE fate markers (Figure 1B and C). In order to identify the dominant regulators in the predicted causal networks, we assigned a score for each protein to estimate the degree of its connectedness (Supplemental Information). These dominance scores indicated the most important proteins that determine the inferred ME and NE networks: Tcf3, Nac1, Oct4 and Klf4 for ME; and Sox2, Tcf3, Nanog and Klf4 for NE (Figure 2G and H).
Since PCA analysis had also highlighted the potential differentiation roles for Sox2, Oct4, Nac1 and Tcf3, we pursued the hypothesis that these proteins, plus Nanog and Klf4, may play a key role in regulating the ME and NE fate choices. To explore this we turned to quantitative single-cell experiments.
Nac1 and Oct4 promote the ME fate, and Tcf3 and Sox2 promote the NE fate
We first examined the expression ranges and the correlations between protein levels of the above six TFs in pluripotent cells maintained in the presence of LIF and BMP4 (ES condition). Protein levels were measured in single cells using quantitative immuno-fluorescence. Max-normalized protein levels indicated that all factors, except Tcf3, occupied all three ranges of expression; low, medium and high (Figure S3A - F). In addition, their expression levels correlated highly with each other (Figure S3G), consistent with the self-reinforcing nature of the pluripotency network (Cole and Young, 2008; MacArthur et al., 2012; Niwa, 2007; Silva and Smith, 2008). Tcf3 levels on the other hand remained low in comparison to their maximum under the NE condition and poorly correlated with Nac1, Oct4 and Nanog.
Next, to delineate the quantitative patterns for these six proteins; we measured their levels in single ME and NE cells at 72 hrs into differentiation. Although the measurements at earlier time points might be informative, especially to address heterogeneity in response, at 72 hrs differentiation has just reached its maximal level, with reduced variability, allowing us to collect data from a statistically-significant number of ME or NE positive cells (Figure 1 and S1). This time point was also well suited for assessing the impact of the perturbation experiments reported below. The cell-fate marker signal was normalized across both ME and NE conditions to the maximum of its single-cell level in the respective condition: ME for T and NE for Sox1. Similarly, each TF level, except that of Tcf3, was normalized across both conditions to its single-cell maximum under the ES condition. Since Tcf3 was highest under NE, it was normalized to its maximum under the NE condition (Supplemental Information). Each normalized protein value was plotted against the normalized ME or NE fate marker (Figure 3 and Figure S3J - M). As observed earlier (Figure 1B and C), the differentiation was mutually exclusive, with no NE fate visible under ME and no NE fate visible under ME (Figure S3H and I).
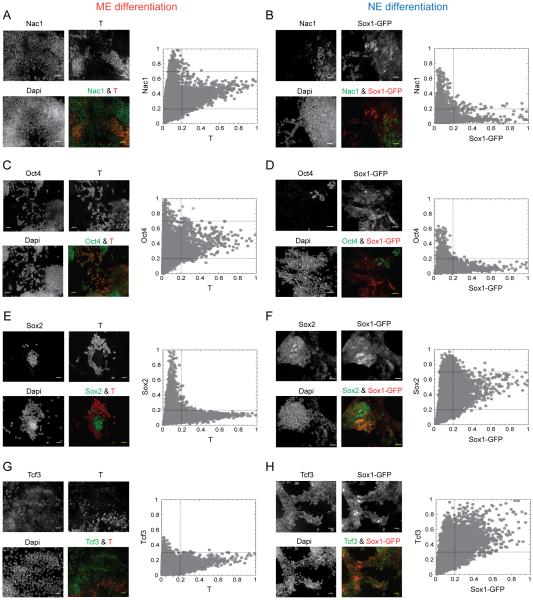
Figure 3. Differential regulation of potential TFs in the ME and NE cells.
Representative qualitative images (left) and quantitative single cell measurements (right) for Nac1 (A & B), Oct4 (C & D), Sox2 (E & F) and Tcf3 (G & H) in ME and NE differentiation conditions. T and the indicated proteins were quantified by immuno-fluorescence in Sox1-GFP ES cell line. Mean fluorescence intensities of each protein from single cells, normalized as explained in the text, are plotted against the T (ME marker) and Sox1-GFP (NE marker) levels, also normalized, in each panel. Cells were fixed, stained and measurements were done at 72 hrs of differentiation. Dashed lines indicate the division of TF levels into low, medium and high and fate-markers into low and high ranges, as explained in the text. Scale bars represent 35 µm. See also Figures S3 and S4.
Normalized TF levels were divided into three ranges: low (0 – 0.2; 0 – 0.3 for Tcf3), medium (0.2 – 0.7) and high (0.7 −1.0) and fate markers into low (0 – 0.2) and high (0.2 – 1.0). Two characteristic shapes are observed from the single-cell plots of TF level versus fate marker (Figure 3 and Figure S3J – M): an L shape, indicating mutual exclusivity between ES (undifferentiated cells below the fate marker threshold) and ME (for Sox2, Tcf3, Nanog and Klf4) or NE (for Nac1, Oct4 and Nanog) cells; a V shape, in which the protein level can be high when the fate marker is low but is restricted to be medium if the fate marker is high (for Nac1 and Oct4 in ME and for Sox2 in NE cells). Tcf3 is somewhat similar; it exhibits a truncated L shape under ME, presumably due to its inhibition by Chiron, and a broader cone-like shape under NE in which its level is medium or high when the fate marker is high.
These results suggested that the central pluripotency factors such as Nanog and Klf4 are not required for ESC differentiation and were generally repressed (Figure S3J – M). The remaining four factors on the other hand were differentially regulated (Figure 3). Nac1 and Oct4 were constrained within a quantitative window, neither too high nor too low, in ME cells, but were repressed in NE cells (above the fate-marker threshold in Figure 3A - D). Similarly, Sox2 was constrained to intermediate levels and Tcf3 to intermediate-to-high levels in NE cells but they were repressed in ME cells (Figure 3E – H). Additionally, we tested four other TFs - Esrrb, Sall4, Smad1 and E2f1 - for their quantitative levels in ME and NE cells. These TFs are known to promote pluripotency (Chen et al., 2008; Dunn et al., 2014) but were not part of our original data set. We found that, in a similar way to Klf4 and Nanog, the levels of these factors are equally down-regulated in both ME and NE cells (Figure S4). Together, these distinct quantitative patterns suggested a requirement of Nac1 and Oct4 for the ME choice and Sox2 and Tcf3 for the NE choice.
To confirm this, we utilized siRNA-mediated perturbation to modulate their protein levels during differentiation. We first tested the siRNAs for their efficiency in down-regulating the specific TF and their effects on ESC survival. While scrambled siRNA pool was used as negative control, pools consisting of siRNAs against four sites of each target were used to down-regulate the TFs (Supplemental Information). Quantitative immuno-fluorescence of the target proteins at 72 hrs after siRNA transfection showed significant reduction in their expression while ES colonies were still largely intact (Figure S5A - G). Mock transfections with the scrambled siRNA pool (neg) did not alter the normal TF levels in ESCs (Figure S5G). However, after 6 days of transfection varying degree of effects was observed on ES colony survival (Figure S5H). Consistent with their central role, we found that Nac1, Oct4, Sox2 and Nanog down-regulation reduced ES survival by up to 80%. In contrast, Tcf3 and Klf4 had lesser effects, also consistent with their known functions. While Klf4 is redundant for the ES state (Jiang et al., 2008), Tcf3 down-regulation is known to stabilize the ES state (Pereira et al., 2006; Wray et al., 2011). Furthermore, mock transfection of ESCs with scrambled siRNA did not induce ME or NE fates at 72 hrs after transfection, thus ruling out the differentiation artifacts from transfection reagents alone (Figure S5I). Similarly the transfection reagents did not alter ESCs ability to differentiate into either the ME or NE cell fates (Figure S5G, J and K).
We then used the siRNAs to down-regulate Nac1 and Oct4 levels during ME, and Sox2 and Tcf3 during NE differentiation. The resulting single-cell protein levels, at 72 hrs of differentiation, were normalized as described above but with maximal levels taken from mock-transfected cells (negative control). Forcing Nac1 and Oct4 to relatively low levels compromised the ME choice (Figure 4A and B), more prominently with Nac1 perturbation. The NE choice on the other hand was equally compromised on reducing either Tcf3 or Sox2 levels (Figure 4C and D). Ectopic over-expression of these TFs in cells treated with siRNA largely rescued both the target TF levels as well as the respective cell-fate marker levels (Figure 4E – H). Although acute knockdown of these genes would cause defects in ESC maintenance (Figure S5H), these ME and NE lineage specific phenotypes at 72 hrs suggest that the general ESC differentiation is not compromised under our conditions. In addition to the specificity of siRNA pools used, the above results also confirm important role of some of the pluripotency TFs in promoting differentiation: ME by Nac1 and Oct4, and NE by Sox2 and Tcf3.
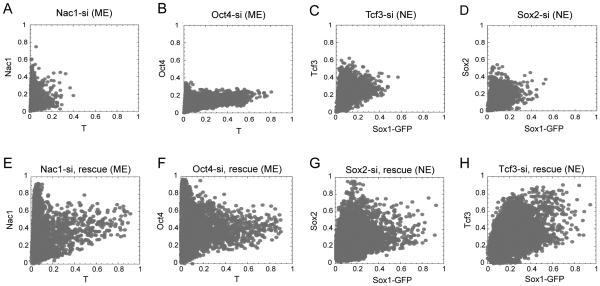
Figure 4. Differential requirement of potential TFs for the ME and NE fate choice.
Scatter plot for T with Nac1 (A) and Oct4 (B) in cells with siRNA mediated down-regulation of Nac1 and Oct4 respectively during ME differentiation. Scatter plot for Sox1-GFP with Tcf3 (C) and Sox2 (D) in cells with siRNA mediated down-regulation of Tcf3 and Sox2 respectively during NE differentiation. Rescue of Nac1 (E), Oct4 (F) during ME differentiation, and Sox2 (G) and Tcf3 (H) during NE differentiation, following their respective siRNA transfection. A 25 nM pool of siRNAs was used to down-regulate Nac1, Oct4, Tcf3 and Sox2 during the indicated condition. Plasmid bearing the individual TF was co-transfected with siRNA for the rescue experiments in E to H. Cells were transfected 12 hrs prior to addition of differentiation signal. Cells were fixed, stained and measurements were done at 72 hrs of differentiation. Normalization was performed as in Figure 3 but using the maximum levels from mock transfected cells using scrambled siRNA alone (for A to D) or with an empty plasmid (for E to H) under the same experimental conditions. See also Figure S5.
Nac1 activates Oct4 and inhibits Tcf3 and Sox2 to favor the ME fate
The repression of Nac1 and Oct4 in NE cells and their presence in ME cells indicated that they might oppose the NE choice (Figure 3A - D). Similarly, the repression of Sox2 and Tcf3 in ME cells and their presence in NE cells indicated that they might oppose the ME choice (Figure 3E - H). To gain further insights into the possible mechanisms that may explain the distinct quantitative patterns observed, we asked whether the regulations among these TFs were different in ME and NE cells. We hypothesized that in order to promote ME, Nac1 and Oct4 might repress NE by inhibiting Sox2 and Tcf3, and vice versa. To understand these, we systematically tested for the causal relationships among Nac1, Oct4, Sox2 and Tcf3 during both the ME and NE differentiation conditions using specific siRNA perturbations.
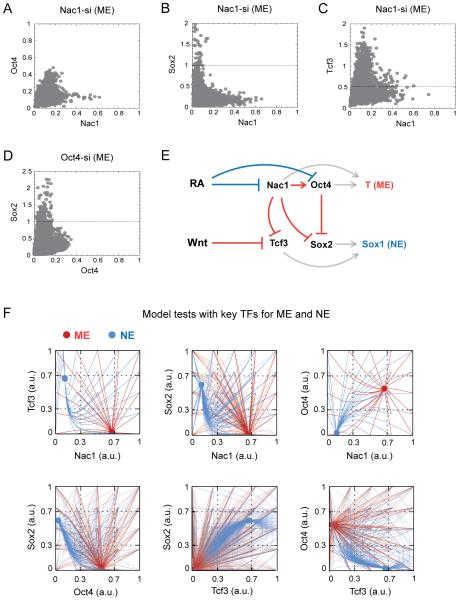
We examined the impact of down-regulating Nac1 or Oct4 during ME differentiation and Tcf3 or Sox2 during NE differentiation on changes in the levels of all four identified factors - Nac1, Oct4, Sox2, and Tcf3 – as well as Nanog (Figure 5A - D and Figure S6A - Q). Nac1 down-regulation during ME differentiation resulted in the decrease of Oct4 level but an increase in both the Sox2 and Tcf3 levels (Figure 5A – C and S6Q). Similarly, down-regulation of Oct4 during ME differentiation resulted in increased Sox2 levels (Figure 5D and S6Q) but had no effect on Nac1 and Tcf3 levels (Figure S6A and B). These results indicated that during ME choice, Nac1 favors Oct4 expression and inhibits both Tcf3 and Sox2, and that Oct4 inhibits Sox2. By inhibiting the NE-promoting TFs, both of these Nac1- and Oct4-mediated actions favor the ME choice and Nac1 in particular, may play a central role. In agreement with this, Nac1 down-regulation resulted in prominent reduction of the ME fate (Figure S6M, N, & Q)
Figure 5. Nac1 coordinates the ME fate selection through Oct4 activation and inhibition of Sox2 and Tcf3.
Scatter plots for Nac1 with Oct4 (A), Sox2 (B) and Tcf3 (C), and Oct4 with Sox2 (D) in cells with siRNA-mediated down-regulation of Nac1 and Oct4 respectively during ME differentiation. siRNA transfection, differentiation and normalization were performed as in Figure 4. Dashed lines indicate the expected maximum for a given TF in mock-transfected cells under the ME condition. (E) Schematic of the mathematical model based on experimental data for ESC differentiation into the ME and NE fates. Arrows indicate activation; blunt ends indicate inhibition. Regulations that are shown in red favor ME and those shown in blue favor NE fate choice. While T (ME fate marker) expression was defined by Nac1 and Oct4 levels, Sox1 (NE fate marker) expression was defined by Tcf3 and Sox2 levels (gray arrows). (F) The model simulation results, with chosen parameter values, on varying initial concentrations for each TF and their resulting steady-state levels for ME (red trajectories) and NE (blue trajectories) differentiation. Results are projected as pair-wise combinations among Nac1, Oct4, Sox2 and Tcf3. See also Figure S6 and Supplemental Experimental Procedures.
In analyzing the NE fate, while Tcf3 and Sox2 down-regulation equally reduced the NE fate (Figure S6O – P), we found no evidence that either Tcf3 or Sox2 inhibits either Nac1 or Oct4 (Figure S6E, F, I and J). It is known that Tcf3 inhibits Oct4 and Nanog under pluripotency condition (Cole et al., 2008; Pereira et al., 2006) but this may not continue in the NE condition. However, both Nac1 and Oct4 are down-regulated in NE (Figure 3B and D), suggesting their indirect regulation either through retinoic acid signaling itself or other factors, rather than one of the specific factors considered here. Similarly, a role for the ME differentiation signals to inhibit Tcf3 was observed (Figure 3G).
Down-regulation of Nac1 and Oct4 under ME and Sox2 during NE differentiation led to enhanced reduction in undifferentiated ESCs (as measured by Nanog levels) (Figure S6C, D and H). Similarly, while Oct4 and Nac1 were limited from their high range (0.7 – 1) of expression under ME (Figure 5A and S6A), they were restricted to the low range (0 – 0.3) in cells with down-regulated Sox2 under NE condition (Figure S6E and F). On the other hand, Tcf3 down-regulation had no effects on Oct4, Nac1, Sox2 and Nanog levels (Figure S6I – L). These results are consistent with the Nac1, Oct4 and Sox2 mediated activation of Nanog to maintain pluripotency (Figure S1A) (Kim et al., 2008), and further suggest that these positive regulations are lost in ME and NE positive cells (Figure S3J and K).
Nac1 modulates the extent of NE fate selection through Tcf3 inhibition
Together, the above data suggested that the self-reinforcing regulations among a subset of TFs – Nac1, Oct4, Tcf3 and Sox2 – in ESCs are modified so as to favor the ME choice over the NE choice. To reveal further functional outcomes that might be concealed in our data, we constructed a mathematical model using parsimonious assumptions to describe how the levels of Nac1, Oct4, Sox2 and Tcf3 adjust to each other during either the ME or NE differentiation (Figure 5E) (Supplemental Procedures). The model was formulated to regulate ME choice through repression of Tcf3 by Nac1, and Sox2 by both Nac1 and Oct4; or NE choice through repression of Nac1 and Oct4, mediated by either the NE differentiation signal itself or by some as-yet unknown factors. The model was set up to exhibit a single steady state for each differentiation regime (Figure S6R), thereby reflecting the mutually exclusive choice between ME and NE fates (Figure 1B - C). The model's parameter values were chosen manually to be consistent with our experimental findings, so that the eventual steady-state levels of Nac1, Oct4, Sox2 and Tcf3 fell within the ranges for cells expressing high levels of the respective ME or NE fate marker. We also performed parameter sensitivity analysis by varying each of the model parameter by ±25% and found that the model is robust to changes in parameter values (data not shown). Of the various TF concentrations and their combinations tested, only high levels of Nac1 and Oct4 favored the ME fate, and high levels of Tcf3 and Sox2 favored the NE fate (Figure 5F).
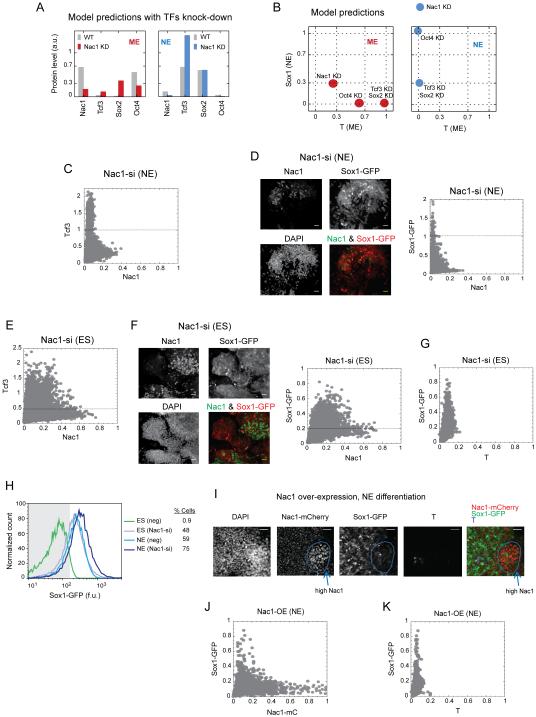
Next, we utilized this simple model to predict the effects of selective perturbation of Nac1, Oct4, Sox2 and Tcf3 on both the TF levels and the fate selection. For most part, simulated partial knock-down of these factors recapitulated experimental results with siRNA perturbations (Figure 6A, Figure S6S, Figure 5A - D and Figure S6A - P). However, the model also showed an increased level of Tcf3 (Figure 6A) and an enhanced NE fate selection when Nac1 was knocked-down in NE condition (Figure 6B). To confirm these unexpected predictions, we first down-regulated Nac1 during NE differentiation and consistently found twice the level of Tcf3 (Figure 6C). To test the fate selection predictions, we down-regulated each of the four TFs during both ME and NE differentiation, and measured the changes to both cell fates (Figure S7A – H). The results reconfirm the ME promoting role of Nac1 and Oct4, and NE promoting role of Sox2 and Tcf3. In addition, enhanced Sox1-GFP signals upon Nac1 down-regulation during NE differentiation confirmed the model prediction (Figures 6B, S7E and 6D). To examine the role of Nac1 further we conducted similar experiments in ESCs in the absence of any differentiation signals. Surprisingly, down-regulating Nac1 resulted in up to 4 times higher Tcf3 levels (Figure 6E). Nac1 down-regulation alone was sufficient to trigger higher Sox1-GFP expression and induce NE fate as the default choice in up to 48% of naïve ESCs (Figure 6F - H). Furthermore, ESCs with ectopic over-expression of Nac1 were blocked from NE fate selection during NE differentiation process (Figure 6I - K). These results suggested that Nac1 inhibits the NE-promoting Tcf3 in ES as well as in ME and NE cells.
Figure 6. Nac1 regulates the extent of NE choice and suppresses it in naïve ESCs through Tcf3 inhibition.
The model predictions for changes in Nac1, Tcf3, Sox2 and Oct4 levels on partial knock-down (KD) of Nac1 (A), and changes in the ME (T) and NE (Sox1) fate choice (B) upon partial knock-down of indicated protein, during the ME (red) and NE (blue) conditions. Nac1 down-regulation mediated changes in Tcf3, and Sox1-GFP levels during NE differentiation (C and D), under ES condition (E and F), and T and Sox1-GFP levels under ES condition (G). Images show the overlay of Nac1 and Sox1-GFP images at indicated condition. (H) Flow cytometry analysis for Sox1-GFP fluorescence in cells with (Nac1-si) and without (neg) Nac1 down-regulation during pluripotency (ES) and NE conditions. Grey area indicates the cut off used to measure percent of positive (% Cells) Sox1-GFP cells. (I) Images showing the ectopic over-expression of Nac1-mCherry and the extent of Sox1-GFP expression during NE differentiation. DAPI image and image overlays are also shown. Cells over-expressing Nac1-mCherry (Nac1-mC) are highlighted with a blue circle. Scatter plots showing quantitative changes in Nac1-mC and Sox1-GFP (J), and T and Sox1-GFP (K) levels during Nac1 over-expression and NE differentiation. Nac1 down-regulation and normalizations were performed as in Figures 4 & 5. Dashed lines indicate the expected maximum of Tcf3 or Sox1-GFP signal under the indicated condition in mock transfected cells. WT: wild type; neg: negative control (scrambled siRNA); a.u.: arbitrary units; f.u.: fluorescence units. Scale bars represent 35 µm. See also Figures S6 and S7.
Both the model and the experimental results indicated that down-regulation of Tcf3 or Sox2 did not have any effects on the ME choice (Figure S7C and D). Similarly, Oct4 down-regulation had no effect on the NE choice (Figure S7F). Taken together, these results suggest an important central role for Nac1 in regulating ESC differentiation into both the ME and NE cell fates. While Nac1 in combination with Oct4 was required for the ME choice, it strongly opposed NE and its down regulation was required for the NE choice.
Nac1 differentially binds to the DNA regulatory regions in ES, ME and NE cells
How does Nac1 coordinate other key pluripotency TFs to regulate ESC differentiation? Fate-specific modulation of gene expression often results from direct binding of a TF to the regulatory regions of its target DNA (Davidson, 2006). In ESCs, pluripotency factors bind extensively to gene regions of each other to regulate their expression (Cole et al., 2008; Kim et al., 2008). Similarly, Oct4 is known to bind regulatory region of Sox2 during ME differentiation (Thomson et al., 2011). To know if Nac1 implemented similar mechanisms, we assessed it’s binding to genomic DNA in ES, ME and NE cells. We performed chromatin-immunoprecipitation (ChIP) to isolate Nac1 bound genomic DNA followed by next-generation sequencing (ChIP-seq) to identify the specific gene regions and their enrichment.
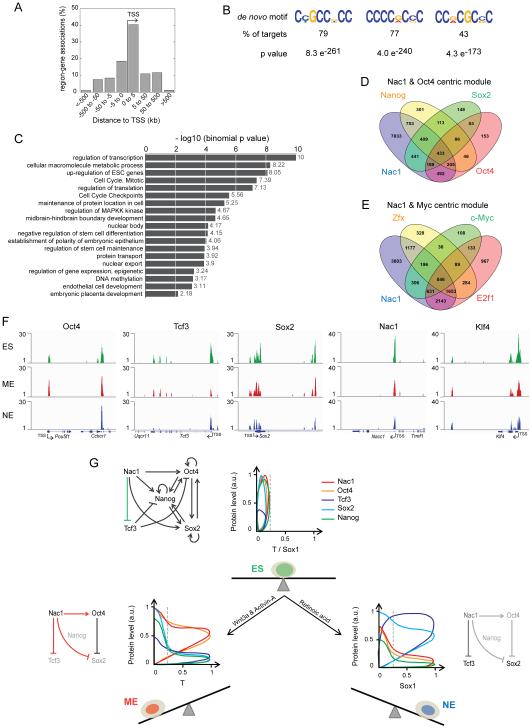
Since little is known about which target genes Nac1 regulates, we first assessed such regulation in ESCs. The majority of Nac1 bound regions were proximal to the transcription start site (TSS) of associated genes (within 5kb up or downstream of TSS), suggesting that it played a role in regulating the transcription of its target genes (Figure 7A). The de-novo Nac1 binding motifs identified are dominated by cytosine followed by guanine content and are found in majority of its target sequences (Figure 7B). Nac1 motifs shared similarity with motifs of TFs involved in regulation of ESCs, development and transcription - Klf4/5/7/1, Sp1, Smad3, etc. (Bouwman and Philipsen, 2002; Jiang et al., 2008; Mullen et al., 2011). In addition, we observed significant enrichment of Nac1 binding at gene regions associated with regulation of gene transcription, translation, ESC state, development, cell cycle and signaling (Figure 7C and Table S1).
Figure 7. Nac1 binds differentially to regulatory gene-regions in ES, ME and NE cells.
(A) Percentage of Nac1 bound regions associated to annotated genes of the mouse genome (build mm9) in ESCs. (B) De novo motifs identified from the Nac1 bound target region sequences in ESCs. Three motifs covering the highest percent of targets are shown. (C) Gene ontology and pathway terms enriched among the Nac1 target genes in ESCs. Data are shown graphically according to their p values (x axis) and the associated functional category (y axis). Comparison of Nac1 target genes with those of Oct4 centric module TFs – Oct4, Sox2 and Nanog (D), and the Myc centric module TFs – E2f1, c-Myc and Zfx (E). (F) Genome tracks showing Nac1 binding enrichment peaks detected at Oct4, Tcf3, Sox2, Nac1 and Klf4 regions, from ChIP-seq analysis in ES, ME and NE cells. Gene locus, TSS and the transcription orientation are indicated for each target. See also Figure S7 and Experimental Procedures. (G) The regulations among the key TFs – Nac1, Oct4, Sox2, Tcf3 and Nanog – may robustly maintain the mutually balanced ESC state (above). Single cell quantitative pattern of Nac1 (red), Oct4 (orange), Tcf3 (blue), Sox2 (cyan) and Nanog (green) protein levels with respect to T (ME fate marker) and Sox1 (NE fate marker) levels are shown, beside the sub-network schematics, for ES, ME and NE cells. The ES sub-network is updated to include Nac1 mediated Tcf3 inhibition. The balanced ES state (green cell) and its imbalance induced by indicated differentiation signals to favor either the ME (red cell) or the NE (blue cell) fate are shown. Dashed line indicates the threshold signal used to regard a cell as T or Sox1 positive. Arrows indicate activation and blunt ends indicate inhibition. Proteins and relationships shown in light gray are under repression. Residual levels of Nac1 and its inhibition of Tcf3 in NE cells are shown in dark gray (below right).
In ESCs, ChIP-seq profiling of several pluripotency TFs has led to identification of Oct4 and Myc centric modules (Chen et al., 2008; Ng and Surani, 2011). While Oct4 centric module includes the core factors such as Oct4, Sox2 and Nanog, among others, Myc centric module includes c-Myc, E2f1, Zfx, etc. In order to know how Nac1 targets compare with these TFs, we analyzed its target genes with respect to Oct4 and Myc module TF targets (Figure 7D and E). Nac1 shared a few hundred targets with each individual factor and all three factors of Oct4 module (433) (Figure 7D). Surprisingly, in comparison to Oct4 module, Nac1 shared ~ twice the number of targets with all three factors of Myc module (846) (Figure 7E). This cross-analysis suggests that Nac1 is an important member of Myc module rather than the Oct4 module. Moreover, majority of Nac1 binding regions are adjacent to the TSS (Figure 7A), which is a characteristic feature of Myc centric module (Chen et al., 2008; Ng and Surani, 2011). Together these cross-comparisons reveal important new insights into how Nac1 regulates its target genes.
We then analyzed differential binding patterns of Nac1 in ES, ME and NE cells by ChIP-seq. ESCs were grown in culture with LIF and BMP, and sorted T-GFP and Sox1-GFP positive cells at 72 hrs of differentiation were used as ME and NE cells respectively. Differential enrichment peaks were observed for all four differentiation promoting pluripotency factors (Figure 7F): Nac1 bound to regions of Oct4, Sox2, Tcf3, itself and Klf4 in ES and ME cells. In NE cells on the other hand, there were no peaks for Oct4 and the peaks at other TF regions were weakly enriched. To verify these observations further and to rule out the artifacts of sequencing, we performed quantitative polymerase chain reaction (qPCR) for the sites detected in ChIP-seq by using ChIP samples from ES, ME and NE cells. We validated ChIP-qPCR by testing known Nac1 binding regions for Oct4, Sox2 and Nanog in ESCs (Kim et al., 2008), and multiple non-target regions from ChIP-seq analysis. While there was no enrichment of non-target regions, known and new Nac1 binding regions were significantly enriched (Figure S7I) in ESCs. Nac1 in ESCs bound to its own regulatory regions and that of Tcf3 on a par with its binding to Sox2 and Nanog regions (Figures 7F and S7I). Nac1 bindings to these TFs was very similar in ME cells except for its slightly reduced binding to Oct4 and an increased binding to Tcf3 (Figure S7I). A striking difference was observed, however, in NE cells where Nac1 bound to none of the factors significantly (above 3 fold) except for Tcf3 and Sox2.
In assessing Nac1 ChIP-seq data, we also observed differential enrichment of gene-regions associated with various cell signaling pathways, mesoderm, neural tube patterning, and mouse phenotypes (Figure S7J and K). Particularly the ME related signaling pathways such as Wnt and Nodal were enriched in ME cells more than in NE cells (Gadue et al., 2006). Verification of selected gene-regions related to mesoderm development (Smad2/3/4) and Wnt/Activin/Nodal signaling genes by qPCR confirmed their high enrichment in ME cells, followed by ES cells, and lesser enrichment in NE cells (Figure S7I) (Fei et al., 2010; Gadue et al., 2006). Assessment of NE promoting retinoic acid signaling targets (Rhinn and Dolle, 2012) gave similar results – higher enrichment in ES and ME cells than in NE cells. These results are consistent with different Nac1 protein levels: higher levels in ES and ME cells allowed maximal binding to Nac1 targets, and its minimal level in NE cells resulted in loss of binding.
DISCUSSION
The data presented here reveal differentiation-promoting functions of Nac1, confirm known roles of Oct4, Sox2 and Tcf3 (Atlasi et al., 2013; Kishi et al., 2000; Thomson et al., 2011), and extend the repertoire of pluripotency TFs also utilized to regulate early stages of ESC differentiation. Through integrative and quantitative approaches, we have shown that these four pluripotency factors regulate the ME vs NE choice of ESCs in distinct ways. Nac1 in particular appears to play important roles in fine-tuning the extent of ME and NE fate selection, as well as the ESC state. Although we cannot rule out direct or indirect participation of other TFs in regulating the ME and NE fates, the TFs we have identified play significant roles in fate selection.
Although Nac1 had been shown to be an important TF for the pluripotency (Kim et al., 2008; Wang et al., 2006), its functions in ESCs as well as in other cell types remain surprisingly poorly understood. Nac1 is known to promote tumor proliferation and gain-of-resistance to chemotherapy, including in cell types that originate from ME (Jinawath et al., 2009; Nakayama et al., 2006). It is conceivable that Nac1 may play a role in the putative link between stem cells and cancer with diverse functions in regulating both development and disease.
Nac1-knockout mice exhibit a lower survival rate for embryos or newborns, with surviving mice showing gross skeletal abnormalities (Yap et al., 2013). We observed significant enrichment of Nac1 binding at regions associated with mouse phenotypes such as abnormal rib development (Figure S7K) but the effect of Nac1-knockout on early embryogenesis, especially in mice that do not survive, has not been studied and would complement the results obtained here.
In a similar way to our results, certain TFs are reused during cell fate specification from progenitors during hematopoietic development (Mercer et al., 2011; Rothenberg et al., 2010). For instance, during early hematopoiesis PU.1 regulates myeloid, B-cell and T-cell fate choices from their common multipotent progenitor cells (Mercer et al., 2011). PU.1 along with CEBPα also regulates the macrophage vs. neutrophil fate choice of granulocyte-macrophage progenitor cells (Laslo et al., 2006). In these cases, it is still not clear how the causal networks are reorganized from the progenitor cells to differentiated cell types. However, taken together with our findings in ESCs, these results suggest a common strategy of reorganizing the progenitor cell networks around a subset of shared factors. This may be an efficient way to specify successive and alternative cell types during development.
The data presented here suggest that the “ES state” may exist as a composite balance between several of the differentiation-promoting TFs (Figure 7G). Regulation of the quantitative pattern of Nac1, Oct4, Sox2, Tcf3 and the central factor Nanog may dictate the balance of the pluripotent ES state vs. the ME and NE states. In ESCs, Nac1, Oct4, Sox2 and Nanog spanned all three possible ranges (low, medium and high) of protein levels and are expressed in strong correlation with each other (Figure S3A - G). This suggests that, irrespective of large cell-to-cell variations, the self-reinforcing regulations among these TFs may promote a mutually-balanced stable ES state that resisted differentiation. A strict stoichiometry of protein levels between these TFs may reinforce the ES state, and Nac1 might play a key role in it. The differentiation signals, however, effectively compromised the ES state by eliminating Nanog-dependent positive regulations (Cole and Young, 2008; MacArthur et al., 2012; Niwa, 2007; Silva and Smith, 2008). They promoted a distinct fate choice by restraining the levels of specific TFs within quantitative windows and repressing those required for the alternative fate. However, the roles of extrinsic signals for ME and NE lineage commitment are not completely resolved in our data. While we show that the signals reconfigure the TFs into forming new lineage-specific networks, it is not clear how the reconfigured TFs integrate with signaling effectors such as Smad 2/3, RARs, etc. The connections between the transcriptional and signaling networks are currently not well understood.
In this paper, we have investigated the differentiation process through which pluripotent ESCs decide between the ME and NE fates. We used computational analysis of population-averaged data (Figure 2) to suggest which pluripotency TFs might play a key role in these cell fate transitions and then exploited quantitative single-cell analysis and siRNA perturbation to identify the causal interactions which underlie differentiation (Figures 3, 4 & 5). This allowed us to make predictions at the molecular level, such as the role of Nac1, which emerged as a central regulator of differentiation. Our integrative methodology fully exploits protein-level data, in contrast to previous work on pluripotency-network reconstruction in ESCs which relies on a Boolean (on/off) approximation (Dunn et al., 2014; Xu et al., 2014). We believe our approach can be extended to analyze complex molecular networks underlying cell fate choice in other developmental and disease contexts, especially when augmented with new methods for measuring multiple proteins in single cells (Bendall et al., 2011).
EXPERIMENTAL PROCEDURES
ESC culture and differentiation
Mouse ESCs - R1 (E14-Tg2A), T-GFP and Sox1-GFP, were cultured by standard methods in Knock-out DMEM medium supplemented with non-essential amino acids, sodium pyruvate, L-Glutamine, β-mercaptoethanol, 15% fetal calf serum (Hyclone) and Leukemia Inhibitory Factor (LIF – 1000u/ml) (Millipore # ESG1106). CHIR99021 (Stemgent # 04-0004-02) and Activin-A (R&D Systems # 338-AC-010), and Retinoic acid (Sigma) were used at the indicated concentrations for ME and NE differentiation respectively.
Quantitative Immuno-fluorescence
Immunofluorescence was performed as described previously (Munoz Descalzo et al., 2012). The primary antibodies used are listed in the supplemental information. A Nikon Ti inverted confocal microscope fitted with perfect focus, 20x Plan-Apochromatic objective (NA .75) and Hamamatsu ORCA-AG cooled CCD camera was used for imaging.
Image analysis and normalization
Quantification of fluorescence intensities were performed by semi-automated image analysis using Cell Profiler (Broad Institute) (Carpenter et al., 2006). Cells were segmented using the DAPI signals. Protein normalizations were performed across multiple conditions using the maximum signal of a protein as described.
Perturbations
Target specific ON-TARGET plus SMART siRNA pools, consisting of four constructs, from Dharmacon (Thermo scientific) were used to down-regulate TFs. Cells were transfected with 25nM of either scrambled siRNA (negative control) or target specific siRNA pools using DharmaFECT reagent 12 hrs prior to addition of differentiation signals. Rescue of TFs during differentiation was achieved through their ectopic over-expression from pmCherry-N1 vector under CMV promoter.
Chromatin Immunoprecipitation
ChIP was performed using the Magna ChIP reagents (Upstate # 17-409) according to manufacturer guidelines. ESCs grown in LIF and BMP4 medium and FACS sorted T-GFP (ME) and Sox1-GFP (NE) positive cells were used to perform ChIP with 3µg of Nac1 antibody (Abcam ab29047).
ChIP-seq analysis
Nac1 bound DNA was sequenced by Next-generation sequencing using Illumina HiSeq2500. Sequences were aligned to mm9 mouse genome build using Bowtie, peaks were called by MACS, and genes were assigned to peaks using GREAT and ChIP-Enrich. While DREME and TOMTOM were used for motif discovery, gene ontology and pathway enrichment were performed using GREAT and ChIP-Enrich.
ChIP-qPCR
Quantitative PCR (qPCR) on ChIPed DNA from ES, ME and NE cells was performed in triplicates on CFX96 RT-System (Bio-Rad) using the RT2 SYBR Green Fluor (Qiagen #330510). Ct values were first normalized to the input and then to the normal mouse IgG (negative control) to calculate fold enrichment (2ΔΔCt). Relative fold enrichment was calculated over Igx1a. The primers used for qPCR analysis are listed in the Supplemental Information.
Principal component analysis and dynamic Bayesian network
PCA was performed with custom code written in R by using singular value decomposition of the centered matrix of median values. DBN on ME and NE data was performed by implementing a custom modified version of the EDISON package (Dondelinger, 2013) in R. Modifications introduced to the EDISON package are described in the Supplemental Information. The dynamic Bayesian network code is available for download at https://github.com/meghapadi/StemCellDBN.
Mathematical modelling
Simulation and analysis of mathematical model was performed using open source tools for Python (SciPy). Description of the model, its assumptions and parameters are provided in the Supplemental Information.
Full descriptions of the experimental and computational methods are provided in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anna-Katerina Hadjantonakis and Gordon Keller for the ES cell lines used in this study; Rebecca Ward, Marc Kirschner, Stephen Michnick, Tathagata Dasgupta, Satabhisa Mukhopadhyay, Victor Li and members of the J.G. and A.M.A labs for comments and discussions; Panos Xenopoulos and Silvia Munoz-Descalzo for help with ESC culture and immuno-fluorescence; Tathagata Dasgupta for help with the Bayesian analysis; Nikon Imaging Center at Harvard Medical School for help with microscopy; and Flow Cytometry facility at the Department of Systems Biology.
This work was supported by the following funding sources: MM, AMA and JG received funding from the Human Frontier Science Program (http://www.hfsp.org/) grant (RGP0029/2010); MM from the Canadian Institutes of Health Research (http://www.cihr-irsc.gc.ca/) award (201411MFE-339075-254265); MM and JG from National Institutes of Health (http://www.nih.gov/) grant (R01GM105375); PR and AMA from the European Research Council Advanced Investigator award (http://erc.europa.eu/); MP from the National Human Genome Research Institute (http://www.genome.gov/) grant (K25HG006031); and JQ from the National Institutes of Health (http://www.nih.gov/) grant (1R01 HL111759).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The European Nucleotide Archive (ENA) accession numbers for the ChIP-Seq data reported in this paper are ERS651665, ERS651666 and ERS651667.
AUTHOR CONTRIBUTIONS
M.M. and J.G. conceived the study. M.M. designed and performed research. M.M., M.P., and P.R. planned and M.P. and P.R. performed computational data analysis and mathematical modeling, respectively. M.M., M.P., P.R., J.Q., A.M.-A., and J.G. analyzed the results. M.M. and J.G. wrote the paper with input from the other authors.
The authors declare no conflict of financial and non-financial interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven supplemental figures, one supplemental table, and supplemental experimental procedures which can be found with this article online.
REFERENCES
- Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ, et al. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS genetics. 2013;9:e1003424. doi: 10.1371/journal.pgen.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amirel AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Molecular and cellular endocrinology. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & development. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Young RA. Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harbor symposia on quantitative biology. 2008;73:183–193. doi: 10.1101/sqb.2008.73.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Elsevier; Burlington, MA, USA: 2006. [Google Scholar]
- Dondelinger F, Lebre S, Husmeier D. Non-homogeneous dynamic Bayesian networks with Bayesian regularization for inferring gene regulatory networks with gradually time-varying structure. Machine Learning. 2013;90:191–230. [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell stem cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fei T, Zhu S, Xia K, Zhang J, Li Z, Han JD, Chen YG. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell research. 2010;20:1306–1318. doi: 10.1038/cr.2010.158. [DOI] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. Journal of computational biology : a journal of computational molecular cell biology. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Nostro MC, Kattman S, Keller GM. Germ layer induction from embryonic stem cells. Experimental hematology. 2005;33:955–964. doi: 10.1016/j.exphem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nature cell biology. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Jinawath N, Vasoontara C, Yap KL, Thiaville MM, Nakayama K, Wang TL, Shih IM. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–1948. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT. Principal Component Analysis. Springer; New York: 2002. [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma'ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur BD, Sevilla A, Lenz M, Muller FJ, Schuldt BM, Schuppert AA, Ridden SJ, Stumpf PS, Fidalgo M, Ma'ayan A, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nature cell biology. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackler SA, Korutla L, Cha XY, Koebbe MJ, Fournier KM, Bowers MS, Kalivas PW. NAC-1 is a brain POZ/BTB protein that can prevent cocaine-induced sensitization in the rat. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:6210–6217. doi: 10.1523/JNEUROSCI.20-16-06210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer EM, Lin YC, Murre C. Factors and networks that underpin early hematopoiesis. Seminars in immunology. 2011;23:317–325. doi: 10.1016/j.smim.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Descalzo S, Rue P, Garcia-Ojalvo J, Martinez Arias A. Correlations between the levels of Oct4 and Nanog as a signature for naive pluripotency in mouse embryonic stem cells. Stem cells. 2012;30:2683–2691. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nakayama N, Davidson B, Sheu JJ, Jinawath N, Santillan A, Salani R, Bristow RE, Morin PJ, Kurman RJ, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18739–18744. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, Park IH, Kim KS, Daley GQ, Kornblum HI, et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell stem cell. 2010;7:671–681. doi: 10.1016/j.stem.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nature cell biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Jakt LM, Era T. Embryonic stem-cell culture as a tool for developmental cell biology. Nature reviews Molecular cell biology. 2007;8:502–507. doi: 10.1038/nrm2189. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Pe'er D. Bayesian network analysis of signaling networks: a primer. Science's STKE : signal transduction knowledge environment. 2005;2005:pl4. doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Molecular and cellular biology. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunological reviews. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The Strategy of the Gene. George Allen and Unwin; London: 1957. [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nature cell biology. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ang YS, Sevilla A, Lemischka IR, Ma'ayan A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput Biol. 2014;10:e1003777. doi: 10.1371/journal.pcbi.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Sysa-Shah P, Bolon B, Wu RC, Gao M, Herlinger AL, Wang F, Faiola F, Huso D, Gabrielson K, et al. Loss of NAC1 expression is associated with defective bony patterning in the murine vertebral axis. PloS one. 2013;8:e69099. doi: 10.1371/journal.pone.0069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nature biotechnology. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.