Abstract
Antagonistic interactions between transcription factors contribute to cell fate decisions made by multipotent hematopoietic progenitor cells. Concentration of the transcription factor PU.1 affects myeloid/lymphoid development with high levels of PU.1 directing myeloid cell fate acquisition at the expense of B cell differentiation. High levels of PU.1 may be required for myelopoiesis in order to overcome inhibition of its activity by transcription factors that promote B cell development. The B cell transcription factors, E2A and EBF, are necessary for commitment of multipotential progenitors and lymphoid primed multipotential progenitors to lymphocytes. In this report we hypothesized that factors required for early B cell commitment would bind to PU.1 and antagonize its ability to induce myeloid differentiation. We investigated whether E2A and/or EBF associate with PU.1. We observed that the E2A component, E47, but not EBF, directly binds to PU.1. Additionally E47 represses PU.1-dependent transactivation of the MCSFR promoter through antagonizing PU.1's ability to bind to DNA. Exogenous E47 expression in hematopoietic cells inhibits myeloid differentiation. Our data suggest that E2A antagonism of PU.1 activity contributes to its ability to commit multipotential hematopoietic progenitors to the lymphoid lineages.
1. Introduction
E2A, EBF, and Pax5 (BSAP) are early acting transcription factors regulating B lymphopoiesis [1]. Mice lacking any of these factors do not generate B cells with E2A and EBF deficiency blocking B lymphopoiesis at a slightly earlier stage of development than Pax5 deficiency [2–4]. Early B cell progenitors cell lines can be grown out from the fetal liver or bone marrow isolated from mice deficient in each of these factors. These lines all have the striking phenotype that besides being unable to differentiate into mature B cells they can be induced to differentiate into other hematopoietic lineages in vitro and in vivo [5–7]. Interestingly E2A −/− , Ebf1 −/−, and Pax5 −/− pro-B cells all overexpress the myeloid cytokine receptor gene MCSFR (macrophage-colony stimulating factor receptor). Hematopoietic expression of MCSFR is dependent on the Ets family transcription factor PU.1 as demonstrated by the lack of detectable MCSFR mRNA in PU.1 −/− hematopoietic cells [8, 9].
Mice lacking PU.1 do not make lymphocytes, myeloid cells, or the progenitors for these cells (CMPs, GMPs, or CLPs) [10, 11]. PU.1 activity is also critical for determining cell fate acquisition of uncommitted progenitors. High expression of PU.1 in hematopoietic progenitors directs myeloid development, and low expression directs B cell development [12]. Levels of PU.1 also have been shown to be important in the development of erythroid and myeloid cells [13]. PU.1 physically associates with the essential erythroid transcription factor, GATA1 [14]. The two factors mutually antagonize each other's transactivation potential. The ratio of GATA1 to PU.1 is important in determining whether an uncommitted precursor will develop into an erythroid or myeloid cell, respectively [15–19]. If PU.1 protein levels rise in an uncommitted cell, PU.1 continues to inhibit GATA1, and free PU.1 is able to activate genes required for myeloid differentiation.
Similar to GATA1, Pax5 binds to PU.1 and inhibits its ability to activate transcription [20]. However, Pax5 has a very limited ability to commit multipotential progenitors (MPPs) to early lymphoid progenitors, suggesting that, unlike GATA1, Pax5 does not repress PU.1 to promote B cell fate. Instead it likely functions to maintain the repression of myeloid genes in cells already committed to the B cell fate. EBF and E2A are necessary for the development of MPPs and lymphoid primed multipotential progenitors (LMPPs) into B lymphocytes [7, 21]. In our current study we examined whether E2A and/or EBF bind to PU.1 and antagonize its activity. The E2A protein E47, but not EBF, was observed to bind to PU.1 and block PU.1's association with DNA. This interaction inhibits PU.1 induced transcription from the MCSFR promoter and its induction of macrophage differentiation from hematopoietic progenitor cells. Our study suggests that E2A association with PU.1 may contribute to E2A's promotion of B lymphoid cell fate from multipotent hematopoietic progenitors.
2. Methods
2.1. In Vitro Binding Assays
Glutathione-S-transferase (GST) fusion proteins were prepared as previously described [22]. The GST-E47bHLH expression plasmid was generated by PCR. GST-PU.1, GST-E47bHLH, or GST bound to glutathione agarose were incubated with 35S-methionine labeled in vitro translated protein in 500 μL of NETN buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, and 0.5% NP-40). In vitro translated products were prepared by the TNT reticulocyte lysate system (Promega). After 4 h incubation bound complexes were washed 4x in NETN buffer and eluted in sample buffer followed by separation by SDS-PAGE. Plasmids containing PU.1 deletion mutants and full-length Pax5 were obtained from Dr. M. Atchison (University of Pennsylvania) [20, 23]. Full-length human E47 was in vitro translated from the T7-E2/5 plasmid supplied by Dr. T. Kadesch (University of Pennsylvania). Truncations CT1 and CT2 were generated by linearizing T7-E2/5 plasmid with NotI and XhoI, respectively, before in vitro transcription/translation reaction. The amino terminal E47 truncations were generated by PCR. Human EBF protein was in vitro translated from pSport-EBF obtained from ATCC. ARNT protein was in vitro translated from pcDNA3-ARNT, which was provided by Dr. B. Keith (University of Pennsylvania).
2.2. Coimmunoprecipitations
Nuclear extracts were prepared from K562 cells. 2 mg of nuclear extract was incubated in binding buffer (20 mM Hepes, 25% glycerol, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 0.5 mM DTT) with 1 μg anti-PU.1 or 1 μg anti-GMCSFRα (Santa Cruz Biotechnology, sc-352 and sc-691) antibody prebound to protein A agarose beads overnight at 4°C. Agarose beads and captured protein complexes were washed 5X in binding buffer. Protein lysates were eluted in SDS-PAGE sample buffer.
2.3. Immunoblotting
Immunoblots were performed by resolving protein lysates via SDS-PAGE and transferring to nitrocellulose membrane (Gibco-BRL). The membranes were incubated with indicated antibodies and anti-mouse or anti-rabbit (Cell Signaling) horseradish peroxidase- (HRP-) conjugated secondary antibodies. Immunoreactive bands were detected using Supersignal (Pierce). Anti-E47 (Cat. #554077) and anti-PU.1 (Cat. #554268) were obtained from BD Biosciences.
2.4. Reporter Constructs and Expression Plasmids
The −74 to +67 141 bp human GCSFR promoter and −416 to +124 human MCSFR promoter were PCR amplified with previously described primer sets [24, 25] and cloned into pGL3 Basic (Promega). The pcDNA3-PU.1, MigR1-C/EBP alpha, MigR1-PU.1, (GAL4) 5-luciferase plasmid, and GAL4-PU.1 fusion plasmids have been previously described [22, 26]. The MigR1-E47 and (μE3)4-Luciferase plasmids were provided by Drs. W. Pear and T. Kadesch (University of Pennsylvania). (μE3)4-Luciferase contains 4 E box sites from the μ heavy chain locus upstream of the luciferase gene.
2.5. Transfections
U937 cells were electroporated as previously described [27] using 25 μg of luciferase reporter plasmid, 50 μg MigR1-E47 plasmid, and 5 μg of the thymidine kinase promoter renilla luciferase (pRL-tk) plasmid. 293 T cells were transfected using the BD CalPhos mammalian transfection kit (BD Biosciences). For MCSFR reporter assays, cells were cotransfected with 2 μg pGL3-MR, 4 μg MigR1-PU.1, 4 μg MigR1-E47, and 25 ng of pRL-tk. For E2A-reporter assays, cells were transfected with 2 μg (μE3)4-Luciferase, 4 μg MigR1-PU.1 and/or 4 μg MigR1-E47, and 25 ng of pRL-tk. Total amount of plasmid was kept constant with MigR1 plasmid. 48 h after transfection cell lysates were harvested using Promega cell lysis buffer. Firefly and renilla luciferase activity were measured using the Dual-Luciferase Assay System (Promega). Luciferase values were normalized to renilla luciferase values in order to adjust for potential differences in transfection efficiency.
2.6. Retroviral Transduction of PUER Cells
Retroviral vectors, MigR1 or MigR1-E47, were cotransfected into 293 T cells together with the retroviral packaging vector pCL-Eco (Imgenex) using calcium phosphate precipitation. 48 h after transfection retroviral supernatants were harvested. PUER progenitor cells were infected by resuspending in retroviral supernatant containing polybrene (8 μg/mL) and centrifuging at 2000 ×g for 2 h at 25°C. After the spin infection, the cells were recultured in fresh media containing rIL-3. Cell lines expressing high levels of GFP were obtained by limiting dilution cloning.
2.7. PUER Differentiation
The generation of PUER progenitor cells and their differentiation into macrophages is described [26, 28]. Cells were maintained in IMDM, 10% fetal calf serum (Hyclone), 1 U/mL penicillin/streptomycin, 2 mM L-glutamine, and 50 mM beta-mercaptoethanol. Media contained 5 ng/mL IL-3 (R&D systems). Cells were differentiated by adding 100 nM of OHT to the media. Immunohistochemistry was performed as previously described [29].
2.8. Primary Hematopoietic Cell Differentiation
The use of mice in these experiments was approved by the University of New Mexico LACUC (Protocol #07UNM027). In vitro culture of lineage negative bone marrow cells was performed as previously described [30]. Lineage negative bone marrow cells prepared with MACS lineage cell separation kit (Miltenyi Biotec). Cells were infected with MigR1 or MigR1-E47 retroviral supernatant. After infection cells were cocultured with OP9 cells in IMDM media containing 1 ng/mL IL-7 and 5 ng/mL Flt3L. After 12 d of coculture differentiation cells were stained with CD19-PE and CD11b-APC (Invitrogen) and analyzed with a FacsCalibur flow cytometer (Becton Dickinson).
2.9. Electrophoretic Mobility Shift Assay and Chromatin Immunoprecipitation
Nuclear extracts were prepared from GFP or E47 expressing PUER cell lines as described [31]. Nuclear extract were also prepared from 293 T cells transfected by calcium phosphate with PU.1 and/or E47 expression plasmids. 5 μg of nuclear extract was incubated with 10,000 counts per minute (cpm) of end-labeled oligonucleotide probe in binding buffer (7.5 mM Tris-HCl (pH 7.5), 0.74 mM EDTA, 3% ficoll, 56 mM KCl, and 0.75 mM DTT). It was indicated that 100 μg of unlabled wildtype or mutant competitor oligonucleotide was added to binding reactions to demonstrate specific DNA binding. Additionally it was indicated that 2 μl of rabbit anti-PU.1 antibody was added to binding reactions to determine if PU.1 was contained in DNA/protein complexes. After a 30-minute incubation at room temperature samples were separated on an 8% nondenaturing polyacrylamide gel. Oligonucleotides used for EMSA analysis were derived from the human MCSFR promoter, WT MCSFR: cctagctaaaaggggaagaagaggatcagc, MT MCSFR: cctagctaaaagggatcggtaccgatcagc. Underlined nucleotides indicate differences between wildtype and mutant oligonucleotides.
Chromatin immunoprecipitations were performed as described [32]. Chromatin preparation was incubated with normal rabbit serum, anti-PU.1 antibody (sc-352, Santa Cruz Biotechnology), prebound to protein A/G agarose. An aliquot of chromatin was saved for input. Immunoprecipitates were washed, crosslinks were reversed, and protein was digested with proteinase K. Isolated DNA was purified and DNA pellets were resuspended in sterile H2O. The MCSFR promoter was amplified by PCR.
3. Results
3.1. PU.1 Interacts with the E2A Subunit E47 but Not with EBF
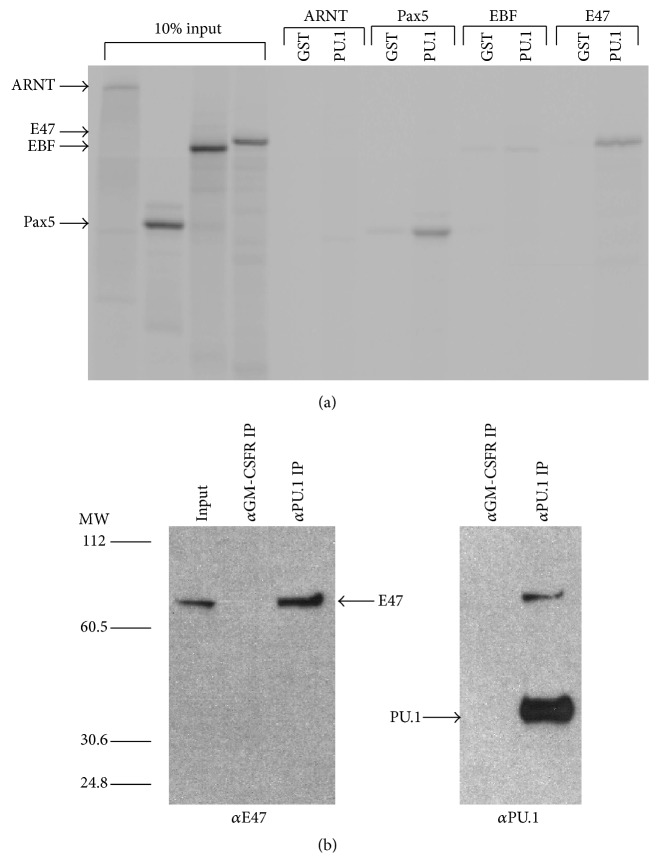
In vitro binding assays were performed to determine if E2A and/or EBF bind to PU.1. In vitro translated Pax5 (positive control), ARNT (negative control), EBF and the E2A subunit E47 were incubated with either purified GST or GST-PU.1 fusion protein. In agreement with previous data, Pax5 bound to GST-PU.1, but not GST (Figure 1(a)) [20]. EBF did not bind significantly to either protein; however, E47 specifically interacted with GST-PU.1 (Figure 1(a)). To demonstrate that this was not a nonspecific interaction between the basic domain of E47 and PU.1, the basic helix-loop-helix PAS domain protein ARNT was incubated with GST-PU.1 and no binding was detected [33].
Figure 1.
E47 but not EBF interacts with PU.1. (a) The transcription factors ARNT, PAX-5, EBF, and E47 were in vitro translated (IVT) and labeled with 35S-methionine. The labeled proteins were incubated with either GST or GST-PU.1 bacterially produced protein. 10% of IVT proteins and proteins bound to GST and GST-PU.1 were separated by SDS-PAGE and visualized by autoradiography. ARNT is a basic-helix-loop-helix transcription factor, which was used as a negative control (not expected to bind to PU.1). Pax-5 was used as a positive control. (b) E47 and PU.1 are coexpressed in the human hematopoietic cell line K562. Nuclear extracts were prepared from K562 cells and treated with anti-PU.1 antibody or anti-GM-CSFRα prebound to protein A agarose. Isolated complexes were separated on an SDS-PAGE gel and blotted. The presence of E47 in complexes was assayed using an anti-E47 antibody.
To determine if E47 and PU.1 interacted in vivo, nuclear extracts were prepared from the K562 hematopoietic cell line, which coexpresses both PU.1 and E47 endogenously. Extracts were incubated with anti-PU.1 or anti-GMSCFRα antibody prebound to protein A agarose beads. E47 was detected in PU.1 immunoprecipitates, but not in anti-GMCSFR immunoprecipitates (Figure 1(b)). These data demonstrate that PU.1 and E47 associate both in vitro and in vivo.
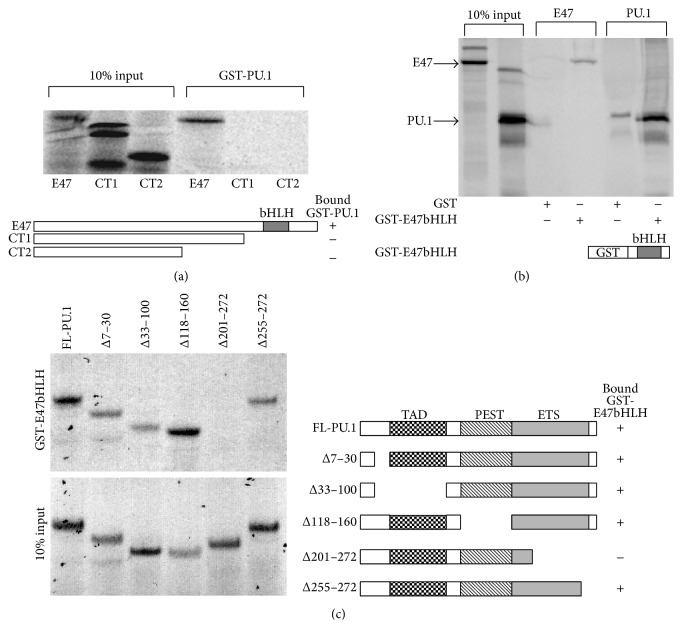
To delineate regions of both proteins that mediate the interaction, further in vitro binding assays were performed. In vitro translated full-length E47 and E47 truncation mutants were incubated with GST-PU.1. As shown previously, full-length E47 bound to the GST-PU.1 protein. However, neither of the two carboxyl terminal deletion mutants (CT1, CT2) lacking the basic-helix-loop-helix (bHLH) DNA binding domain could bind to PU.1 (Figure 2(a)). This suggested that the bHLH domain of E47 mediated the interaction with PU.1. To confirm this conclusion, a GST-E47bHLH fusion protein was prepared and incubated with either in vitro translated E47 or PU.1. As expected the E47 bHLH domain mediated homodimerization with E47. The bHLH domain also pulled down PU.1 demonstrating that it is sufficient for mediating the interaction with PU.1 (Figure 2(b)).
Figure 2.
Identification of domains required for mediating the interaction between PU.1 and E47. (a) In vitro translated 35S-methionine labeled full-length human E47 (FL, aa 1–654), E47 C-terminal truncation 1 (CT1, aa 1–492), and E47 C-terminal truncation 2 (CT2, aa 1–369) were incubated with bacterially produced GST-PU.1 fusion protein. 10% input proteins and proteins bound to GST-PU.1 were separated by SDS-PAGE and visualized by autoradiography. (b) 35S-methionine labeled in vitro translated PU.1 and E47 were incubated with GST-E47bHLH protein (E47, aa 521–623). In vitro translated E47 was used as a positive control since it should dimerize with the E47 bHLH domain. (c) 35S-methionine labeled in vitro translated PU.1 deletion mutants were incubated with GST-E47bHLH protein.
To identify the domain of PU.1 that interacts with E47, in vitro translated deletion products of PU.1 were incubated with purified GST-E47bHLH protein. Deletion of the transactivation domain (Δ33–100) and the PEST region (Δ118–160) of PU.1 had no effect on binding to E47. However, a deletion construct lacking the Ets DNA binding domain (Δ201–272) was unable to bind to E47 (Figure 2(c)). These data demonstrate that PU.1 and E47 bind to each other through their DNA binding domains.
3.2. E47 Inhibits PU.1 Induced Transcription
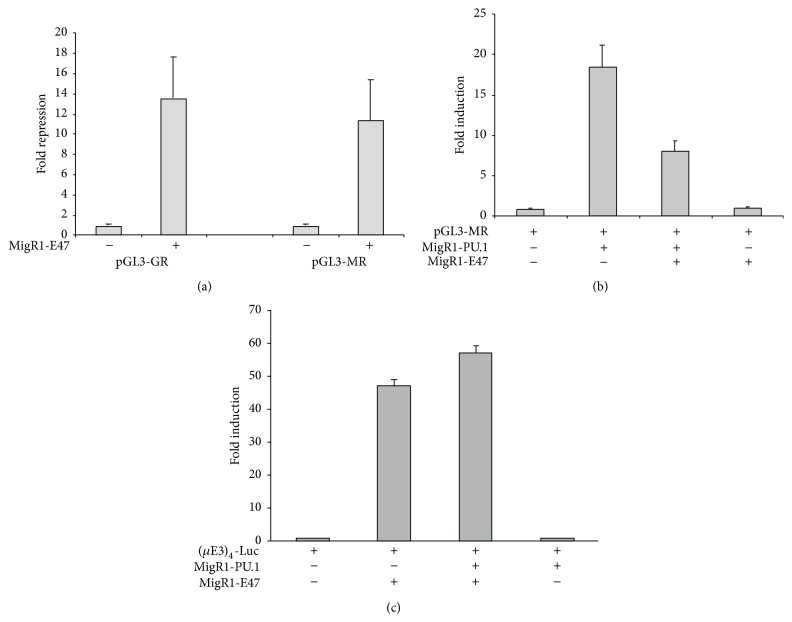
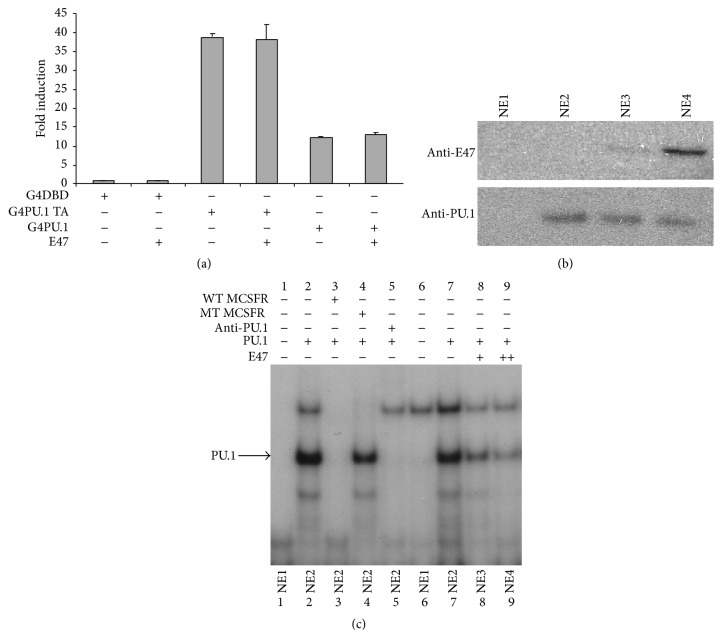
To investigate whether the interaction between PU.1 and E47 had any functional consequences, transient transfection reporter assays in U937 myeloid cells were performed. Two PU.1-dependent promoters were initially examined: the GCSFR and MCSFR promoters [24, 34]. E47 expression decreased expression from both the MCSFR and GCSFR promoter constructs over 10-fold (Figure 3(a)). Further experiments were carried out with the MCSFR promoter construct to determine if this repression was specifically targeting PU.1 activity. When the reporter was cotransfected into 293 T cells with PU.1 there was over 15-fold activation of reporter expression above that seen in the absence of PU.1 (Figure 3(b)). However, when E47 was expressed along with PU.1, MCSFR promoter activity was less than half that observed with PU.1 alone. E47 had no significant effect on the transcription of the MCSFR reporter plasmid in the absence of PU.1. We also determined whether PU.1 could repress E47 (E2A) induced transcriptional activity. Cells cotransfected with an E2A-reporter plasmid ((μE3)4-Luciferase) and E47 expression plasmid resulted in an almost 50-fold enhancement of luciferase activity over cells transfected with the E2A-reporter alone. Coexpression of PU.1 with E47 did not repress transcriptional activity from the reporter construct (Figure 3(c)).
Figure 3.
E47 inhibits promoter activity induced by PU.1. (a) Transient transfection of U937 cells with a MCSFR promoter and GCSFR promoter luciferase construct in the presence or absence of MigR1-E47 plasmid. (b) MCSFR promoter activity induced in 293 T cells with cotransfection of MigR1-PU.1 in the presence or absence of MigR1-E47 plasmid. MigR1-E47 by itself had no effect on MCSFR promoter activity. (c) (μE3)4-Luciferase activity induced by cotransfection with MigR1-E47 in the presence and absence of MigR1-PU.1. For (a) luciferase activity is reported as fold repression compared to activity in the absence of MigR1-E47. For (b) and (c) luciferase activity is reported as fold-induction above the activity seen in 293 T cells transfected with only the reporter plasmid and MigR1. Luciferase values are the mean ± standard deviation of three independent transfections.
3.3. PU.1 Induced Myeloid Differentiation Is Blocked by Exogenous E47 Expression
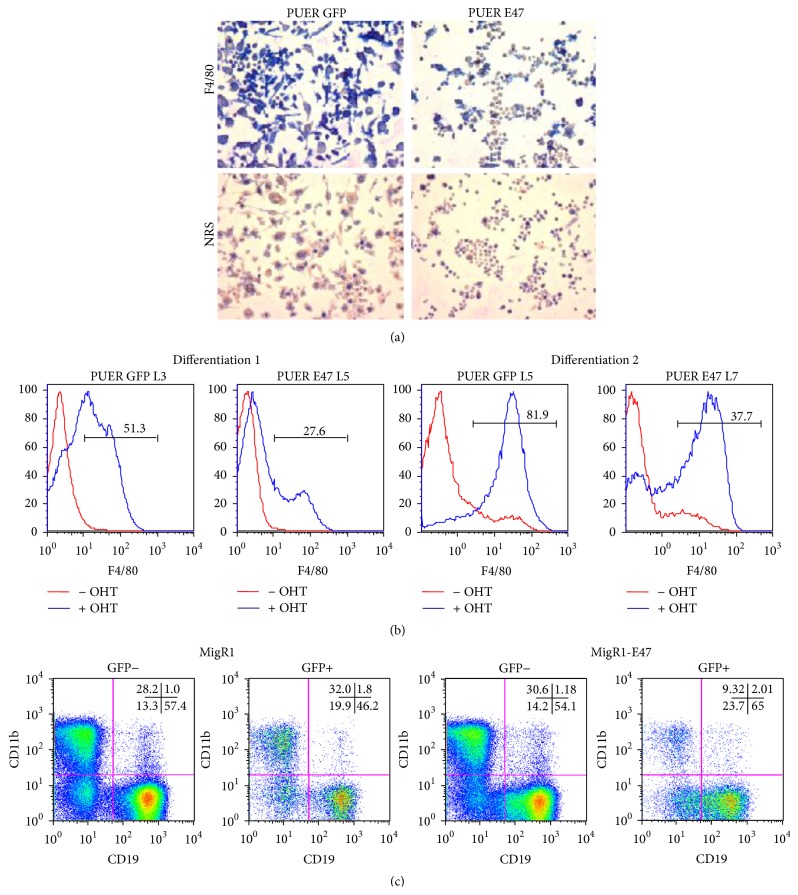
We tested the ability of E47 to affect PU.1's biological activity by assaying the differentiation of the PUER myeloid progenitor cell line [28]. These cells are derived from the fetal liver of PU.1 −/− mice and express a fusion protein between PU.1 and the estrogen receptor ligand-binding domain. In the absence of the ligand hydroxytamoxifen (OHT), these cells remain undifferentiated. In the presence of 100 nM OHT, PUER cells differentiate into macrophages. These cells were superinfected with either an E47-IRES-GFP virus or a GFP only virus. Cell lines expressing high levels of GFP were generated and their ability to differentiate into macrophages was assayed. After 8 d of OHT treatment, PUER GFP cultures became adherent and expressed the macrophage specific marker F4/80 (Figure 4(a)). In contrast the majority of PUER E47 cells remained nonadherent and were predominantly negative for F4/80. Additionally we analyzed surface expression of F4/80 by flow cytometry. Two separate differentiations were examined with 2 independent PUER GFP and PUER E47 cell lines before and after 8 d of culture with OHT to induce PU.1 activity. In both differentiations we observed that E47 decreased the expression of F4/80 (Figure 4(b)) similar to what was observed with the immunohistochemistry results.
Figure 4.
E47 inhibits myeloid differentiation of hematopoietic cells. (a) PUER cells were superinfected with MigR1 or MigR1-E47 virus and pure GFP expressing clones generated. Cell lines were either undifferentiated (no OHT) or differentiated (100 nM OHT) for 8 d. Immunohistochemistry of adherent PUER cell lines induced to differentiate with 100 nM OHT for 8 d. Cells were stained either with the macrophage differentiation marker F4/80 or with normal rat serum (NRS). Magnification is 400x. (b) F4/80 surface expression as determined by flow cytometry of independent PUER GFP and PUER E47 lines with no addition of OHT and after 8 d of OHT treatment. (c) Flow cytometry analysis of lineage minus hematopoietic progenitors infected with control MigR1 virus or MigR1-E47 virus. Cells were cultured for 12 d on OP9 stromal cell layer in the presence of 1 ng/mL IL-7 and 5 ng/mL Flt3L. Differentiation was analyzed by staining cells with anti-CD11b (myeloid) and anti-CD19 (B cell). Percentages of total cells positive for CD11b and CD19 expression are shown in the upper right corner of FACs plots.
To determine if E47 inhibited myeloid differentiation of primary cells, lineage negative hematopoietic progenitors were isolated and infected with control GFP or E47-IRES-GFP retroviruses. To assay myeloid versus lymphoid differentiation, the infected progenitors were cultured on an OP9 stromal cell layer in the presence of IL-7 and Flt3L cytokines. These culture conditions strongly promote B cell development but are also permissive for myeloid development. After 12 d of culture, differentiation was assayed by cell surface expression of the myeloid specific protein CD11b and B cell specific protein CD19 (Figure 4(c)). In the control (GFP only virus) cultures both the uninfected and the infected (GFP+) populations contained approximately 30% CD11b+ cells. In the E47 retrovirus treated cultures the uninfected GFP− population also contained approximately 30% CD11b+ cells, but in the infected GFP+ population only 10% of the cells were CD11b+, which is similar to the inhibition of myeloid development seen with the PUER cell line (Figures 4(a) and 4(b)). The E47 infected population also had an increased percentage of CD19+ B cells.
3.4. E47 Abrogates PU.1 DNA Binding
Since E47 binds to the DNA binding domain of PU.1, we determined if E47 represses PU.1 activity if DNA binding was not dependent on the Ets domain. Full-length PU.1 and the PU.1 transactivation domain alone were fused to the GAL4 DNA binding domain and transactivation activity of the hybrid proteins was assayed by transient transfection of 293 T cells with a GAL4 reporter plasmid. Both fusion proteins activated luciferase expression. The full-length PU.1 fused to GAL4 did not activate transcription as robustly as the PU.1 transactivation domain alone, GAL4 fusion, potentially due to the Ets domain in the full-length allowing the fusion to be competed from the GAL4 reporter plasmid by Ets sites in the genome. Importantly though, neither fusion construct was inhibited by E47 (Figure 5(a)). This result suggested that E47 represses PU.1 activity by interfering with PU.1's ability to bind to DNA.
Figure 5.
E47 inhibits PU.1 binding to DNA. (a) Transient transfections of 293 T cells with a GAL4 responsive luciferase reporter construct. Cells were cotransfected with either GAL4 full-length PU.1 expression plasmid or GAL4-PU.1 activation domain in the presence or absence of MigR1-E47 plasmid. Luciferase activity is the mean ± standard deviation of three independent transfections and luciferase activity is reported as fold-induction above the activity seen in 293 T cells transfected with the GAL4 DNA binding domain expression plasmid. (b) Immunoblot of nuclear extracts prepared from 293 T cells transfected with empty expression plasmids (NE1), PU.1 expression plasmid (NE2), and PU.1 and E47 expression plasmids (NE3, NE4). NE4 was prepared from cells transfected with an increased amount of E47 expression plasmid (++). Immunoblots were probed with anti-PU.1 or anti-E47 antibody as indicated. (c) EMSA performed with nuclear extracts from (b) and a 32P-labeled MCSFR probe. The PU.1 containing complex was identified by performing competitions with unlabeled wildtype and mutant MCSFR probes as indicated. Additionally the PU.1 containing complex could be ablated by inclusion of anti-PU.1 antibody in the binding mix.
To test this hypothesis we assayed PU.1 DNA binding in nuclear extracts prepared from 293 T cells transfected with PU.1 and/or E47 expression plasmids. Nuclear extracts were incubated with a labeled MCSFR promoter fragment (Figures 5(b) and 5(c)). We identified protein complex that was competed away by unlabeled wildtype MCSFR probe (lane 3) but not a similar probe containing a mutated PU.1 binding site (lane 4) from the extracts expressing only PU.1. This complex was ablated by addition of a PU.1 antibody (lane 5). In nuclear extracts coexpressing PU.1 and increasing amounts of E47 we observed a decrease in the PU.1/MCSFR complex (lanes 8, 9). Expression of PU.1 and E47 in the 293 T extracts is shown by immunoblot (Figure 5(b)).
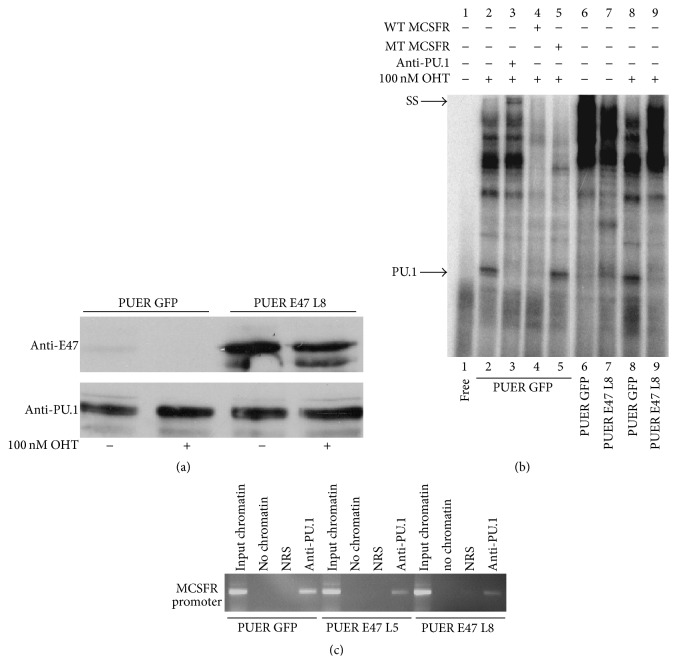
To determine if E47 abrogates PU.1 DNA binding in hematopoietic cells we performed EMSAs with GFP− and E47− expressing PUER cell nuclear extracts. As shown by immunoblot E47-infected cells expressed substantially more E47 in the presence or absence of OHT, than GFP-infected PUER (Figure 6(a)). As done previously a PU.1 containing DNA binding complex was identified in the OHT-treated PUER GFP cells using unlabeled wildtype and mutant MCSFR probes. This complex was supershifted (indicated by SS) by a PU.1 antibody (lane 3). Additionally this complex was induced by OHT treatment as it was not present in extracts from untreated PUER GFP cells (lane 6). Significantly the PU.1 containing complex was not detectable in PUER E47 OHT-treated cells (Lane 9).
Figure 6.
E47 blocks PU.1 from binding to DNA in hematopoietic cells. (a) Immunoblot of PUER and E47 protein expression in nuclear extracts prepared from PUER GFP and PUER E47 cell lines which were grown in the absence or presence (+) of 100 nM OHT. (b) Electrophoretic mobility shift assay (EMSA) with a 32P-labeled DNA probe from the MCSFR promoter. Nuclear extracts were prepared from PUER GFP and PUER E47 cell lines that were grown for 2 d in the absence or presence (+) of 100 nM OHT. SS indicates supershifted complex. (c) Chromatin immunoprecipitation (ChIP) of the MCSFR promoter from PUER GFP and 2 independent PUER E47 cell lines using anti-PU.1 antibody. PUER cell lines were treated for 7 d with OHT. Chromatin also precipitated with normal rabbit serum (NRS) to demonstrate specificity of the anti-PU.1 immunoprecipitations.
Using PUER GFP cells and two independent lines of PUER E47 cells, chromatin immunoprecipitations (ChIP) with anti-PU.1 were performed to determine if E47 would antagonize PU.1 DNA binding in cells. Anti-PU.1 efficiently precipitated the MCSFR promoter, whereas the promoter was not detected in normal rabbit serum immunoprecipitates. Consistent with our findings with the EMSA assays, less MCSFR promoter was precipitated with anti-PU.1 from chromatin isolated from both PUER lines expressing E47 (Figure 6(c)). The results of EMSAs and ChIP assays support the conclusion that E47 inhibits the ability of PU.1 to bind to DNA.
4. Discussion
We have demonstrated that the E2A protein E47 binds to PU.1 and blocks its ability to bind to DNA. This inhibition of DNA binding resulted in repression of MCSFR promoter activity and inhibited myeloid differentiation. The E47 bHLH domain mediates the interaction with PU.1, so potentially other bHLH proteins may also be able to bind to PU.1 and affect its activity. Preliminary results demonstrated that another E2A related molecule HEB binds to PU.1 (data not shown). However, there is specificity to the binding of PU.1 to bHLH proteins as the bHLH PAS domain protein, ARNT, cannot bind to PU.1 (Figure 1(a)). The Ets family member, Ets1, has been shown to bind to the bHLH factor USF1 through interactions between their respective DNA binding domains similar to what we describe for PU.1 and E2A [35]. However, in the case of Ets1 and USF1 the interaction enhances the pairs DNA binding and transactivation of target genes. Interestingly PU.1 was shown not to interact with USF1, demonstrating that there is specificity in the binding of bHLH and Ets family members. Since PU.1 binds to the DNA binding domain of E47 it is somewhat surprising that PU.1 is unable to block E47 binding to DNA. However, it has been shown that PU.1 binds to the DNA binding domain of GATA1, without affecting GATA1's association with DNA [19], and similarly c-Jun, GATA1, C/EBP alpha, and Pax5 bind to the Ets domain of PU.1 without disrupting PU.1's association with DNA [17, 18, 20, 36].
E47 inhibited macrophage differentiation of PUER cells and myeloid differentiation of primary hematopoietic cells suggesting that E47 may play a role in cell fate decisions. This may explain why high levels of PU.1 are required for inducing myeloid differentiation [12]. In an uncommitted progenitor cell there may need to be sufficient PU.1 protein to bind to E47 and other inhibitors yet still leave enough free PU.1 protein available for binding to the regulatory regions of myeloid target genes. By stochastic mechanisms or through cytokine signaling one factor may increase in concentration in relation to the other and commit cells to either myeloid (PU.1) or B cell (E2A) differentiation. Unlike in the cases of GATA1 we did not observe a coordinate ability of PU.1 to repress E2A driven transcription (Figure 3(c)). However, overexpression of PU.1 in early CD4−CD8− double negative thymocytes and induction of PU.1 in PUER cells have demonstrated that PU.1 upregulates the expression of the bHLH inhibitor Id-2 [37, 38]. A similar upregulation of Id-2 by PU.1 in early hematopoietic progenitors may be an indirect mechanism by which PU.1 represses E2A activity. There is evidence that E2A specifies hematopoietic cell fate of multipotent cells. 70Z/3 pre-B lymphocytes can be converted to macrophages and this is associated with the loss of E2A and EBF expression. If the E2A protein E12 is expressed in 70Z/3 macrophages, they are converted to B cells [39]. An analysis of LMPPs isolated from E2A −/− mice showed that the lack of E2A results in increased development of monocytes and granulocytes from these progenitors [21]. Similarly ectopic expression of the E2A inhibitor, Id1, in LMPPs promotes myeloid differentiation [40].
Consistent with our observations Bhalla et al. previously observed that E2A component E12 could induce B cell differentiation of fetal liver MPPs [41]. E2A activity can be heterodimers or homodimers of E12 and/or E47. The two proteins are products of alternatively spliced transcripts generated from the same gene (Tcf3) that differ by the use of two exons that encode alternative bHLH domains. The distinct bHLH domains of E12 and E47, however, share >80% amino acid identity and interact with the same sets of proteins [42]. Interestingly the Bhalla report showed that E2A inhibition of macrophage development required the transcription factor's transactivation domain. Since we observe that the E2A DNA binding domain mediates the interaction between PU.1 and E2A this might suggest that other non-PU.1 functions may be needed to inhibit macrophage development. Alternatively the activation domain may not be required for direct binding to PU.1 but may still be needed to inhibit PU.1's DNA binding and/or stabilize the PU.1/ E2A interaction.
5. Conclusions
Our data demonstrates that E2A and PU.1 directly bind to each other, which results in antagonism of PU.1's DNA binding activity. Previous inhibitory binding partners of PU.1 such as Pax5, GATA1, and C/EBP alpha have either recruited in transcriptional corepressors or displaced coactivators without affecting association with DNA. The observations support the hypothesis that during hematopoiesis E47 can antagonize PU.1 activity to direct B cell development. If PU.1 levels are high though, PU.1 overcomes this antagonism and directs myeloid differentiation.
Acknowledgments
The authors would like to thank the members of the UNM Animal facility for expert animal care. They are grateful to Karen Cowden Dahl for commenting on the paper. This work was supported by The American Cancer Society (RSG-06-170-01-LIB, Richard Dahl), American Society of Hematology (Junior Faculty Scholar Grant, Richard Dahl), the Abramson Family Cancer Research Institute (M. Celeste Simon), and the Howard Hughes Medical Institute (M. Celeste Simon).
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Jason H. Rogers and Kristin S. Owens contributed equally to this paper.
References
- 1.Hagman J., Lukin K. Transcription factors drive B cell development. Current Opinion in Immunology. 2006;18(2):127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Bain G., Maandag E. C. R., Izon D. J., et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79(5):885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 3.Urbánek P., Wang Z.-Q., Fetka I., Wagner E. F., Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79(5):901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhuang Y., Soriano P., Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79(5):875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 5.Ikawa T., Kawamoto H., Wright L. Y. T., Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20(3):349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- 6.Nutt S. L., Heavey B., Rolink A. G., Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401(6753):556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 7.Kapur R., Everett E. T., Uffman J., et al. Overexpression of human stem cell factor impairs melanocyte, mast cell, and thymocyte development: a role for receptor tyrosine kinase-mediated mitogen activated protein kinase activation in cell differentiation. Blood. 1997;90(8):3018–3026. [PubMed] [Google Scholar]
- 8.Anderson K. L., Smith K. A., Perkin H., et al. PU.1 and the granulocyte- and macrophage colony-stimulating factor receptors play distinct roles in late-stage myeloid cell differentiation. Blood. 1999;94(7):2310–2318. [PubMed] [Google Scholar]
- 9.DeKoter R. P., Walsh J. C., Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. The EMBO Journal. 1998;17(15):4456–4468. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKercher S. R., Torbett B. E., Anderson K. L., et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. The EMBO Journal. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 11.Scott E. W., Simon M. C., Anastasi J., Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 12.DeKoter R. P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288(5470):1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 13.Cantor A. B., Orkin S. H. Hematopoietic development: a balancing act. Current Opinion in Genetics and Development. 2001;11(5):513–519. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- 14.Pevny L., Simon M. C., Robertson E., et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349(6306):257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 15.Nerlov C., Querfurth E., Kulessa H., Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95(8):2543–2551. [PubMed] [Google Scholar]
- 16.Zhang P., Zhang X., Iwama A., et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96(8):2641–2648. [PubMed] [Google Scholar]
- 17.Zhang P., Behre G., Pan J., et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rekhtman N., Radparvar F., Evans T., Skoultchi A. I. Direct interaction of hematopoietic transcription factors PU.1 and GATA- 1: functional antagonism in erythroid cells. Genes and Development. 1999;13(11):1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rekhtman N., Choe K. S., Matushansky I., Murray S., Stopka T., Skoultchi A. I. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Molecular and Cellular Biology. 2003;23(21):7460–7474. doi: 10.1128/mcb.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maitra S., Atchison M. BSAP can repress enhancer activity by targeting PU.1 function. Molecular and Cellular Biology. 2000;20(6):1911–1922. doi: 10.1128/mcb.20.6.1911-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan B. L., Hong L., Munugalavadla V., Kapur R. Functional and biochemical consequences of abrogating the activation of multiple diverse early signaling pathways in Kit. Role for Src kinase pathway in Kit-induced cooperation with erythropoietin receptor. The Journal of Biological Chemistry. 2003;278(13):11686–11695. doi: 10.1074/jbc.m207068200. [DOI] [PubMed] [Google Scholar]
- 22.Rao S., Matsumura A., Yoon J., Simon M. C. SPI-B activates transcription via a unique proline, serine, and threonine domain and exhibits DNA binding affinity differences from PU.1. The Journal of Biological Chemistry. 1999;274(16):11115–11124. doi: 10.1074/jbc.274.16.11115. [DOI] [PubMed] [Google Scholar]
- 23.Perkel J. M., Atchison M. L. A two-step mechanism for recruitment of Pip by PU.1. Journal of Immunology. 1998;160(1):241–252. [PubMed] [Google Scholar]
- 24.Smith L. T., Hohaus S., Gonzalez D. A., Dziennis S. E., Tenen D. G. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88(4):1234–1247. [PubMed] [Google Scholar]
- 25.Zhang D.-E., Hetherington C. J., Chen H.-M., Tenen D. G. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Molecular & Cellular Biology. 1994;14(1):373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl R., Walsh J. C., Lancki D., et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nature Immunology. 2003;4(10):1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 27.Heydemann A., Juang G., Hennessy K., Parmacek M. S., Simon M. C. The myeloid-cell-specific c-fes promoter is regulated by Sp1, PU.1, and a novel transcription factor. Molecular & Cellular Biology. 1996;16(4):1676–1686. doi: 10.1128/mcb.16.4.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh J. C., DeKoter R. P., Lee H.-J., et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17(5):665–676. doi: 10.1016/S1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 29.Fisher R. C., Olson M. C., Pongubala J. M. R., et al. Normal myeloid development requires both the glutaminerich transactivation domain and the PEST region of transcription factor PU.1 but not the potent acidic transactivation domain. Molecular and Cellular Biology. 1998;18(7):4347–4357. doi: 10.1128/MCB.18.7.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong K. Y., Owens K. S., Rogers J. H., et al. MIR-23A microRNA cluster inhibits B-cell development. Experimental Hematology. 2010;38(8):629.e1–640.e1. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Research. 1991;19(9):p. 2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson K. D., Bresnick E. H. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods. 2002;26(1):27–36. doi: 10.1016/S1046-2023(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 33.Reisz-Porszasz S., Probst M. R., Fukunaga B. N., Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT) Molecular and Cellular Biology. 1994;14(9):6075–6086. doi: 10.1128/MCB.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D.-E., Fujioka K.-I., Hetherington C. J., et al. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1) Molecular and Cellular Biology. 1994;14(12):8085–8095. doi: 10.1128/MCB.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieweke M. H., Tekotte H., Jarosch U., Graf T. Cooperative interaction of ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. The EMBO Journal. 1998;17(6):1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy V. A., Iwama A., Iotzova G., et al. Granulocyte inducer C/EBPα inactivates the myeloid master regulator PU.1: possible role in lineage commitment decisions. Blood. 2002;100(2):483–490. doi: 10.1182/blood.v100.2.483. [DOI] [PubMed] [Google Scholar]
- 37.Anderson M. K., Weiss A. H., Hernandez-Hoyos G., Dionne C. J., Rothenberg E. V. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16(2):285–296. doi: 10.1016/S1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 38.Laslo P., Spooner C. J., Warmflash A., et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 39.Kee B. L., Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. Journal of Experimental Medicine. 1998;188(4):699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akin C., Brockow K., D'Ambrosio C., et al. Effects of tyrosine kinase inhibitor STI571 on human mast cells bearing wild-type or mutated c-kit. Experimental Hematology. 2003;31(8):686–692. doi: 10.1016/s0301-472x(03)00112-7. [DOI] [PubMed] [Google Scholar]
- 41.Bhalla S., Spaulding C., Brumbaugh R. L., et al. Differential roles for the E2A activation domains in B lymphocytes and macrophages. The Journal of Immunology. 2008;180(3):1694–1703. doi: 10.4049/jimmunol.180.3.1694. [DOI] [PubMed] [Google Scholar]
- 42.Murre C. Helix-loop-helix proteins and lymphocyte development. Nature Immunology. 2005;6(11):1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]