Abstract
α-Synuclein (α-Syn) is an abundant cytosolic protein involved in the release of neurotransmitters in presynaptic terminal and its aberrant aggregation is found to be associated with Parkinson’s disease. Recent study suggests that the oligomers formed at the initial oligomerization stage may be the root cause of cytotoxicity. While characterizing this stage is challenging due to the inherent difficulties in studying heterogeneous and transient systems by conventional biochemical technology. Here we use solid-state nanopores to study the time-dependent kinetics of α-Syn oligomerization through a label-free and single molecule approach. A tween 20 coating method is developed to inhibit non-specific adsorption between α-Syn and nanopore surface to ensure successful and continuous detection of α-Syn translocation. We identify four types of oligomers formed in oligomerization stage and find an underlying consumption mechanism that the formation of large oligomers consumes small oligomers. Furthermore, the effect of lipid membrane on oligomerization of α-Syn is also investigated and the results show that 1,2-dioleoyl-sn-glycero-3-[phospho-L-serine] (DOPS) small unilamellar vesicles (SUVs) dramatically enhances the aggregation rate of α-Syn while do not alter the aggregation pathway.
α-Synuclein (α-Syn) is a small intrinsically disordered cytosolic protein of 140 amino acids (14.3 kDa) that is expressed abundantly in human brain tissue. α-Syn has been reported to function in the control of synaptic vesicle fusion and thus the release of neurotransmitters, subsequent vesicle recycling, and maintenance of synaptic integrity1,2,3,4. However, in pathophysiological conditions, adequate α-Syn monomers in solution undergo a nucleation-dependent mechanism growing into oligomers, which are able to elongate to form profibers through monomers addition and finally mature fibers that form the hallmark subcellular structure Lewy bodies5,6,7. It is well acknowledged that the abnormal aggregation process of α-Syn monomers is associated with pathophysiology of Parkinson’s disease (PD) and dementia7,8,9. Recent studies have shown that initial α-Syn oligomerization may be the root cause of cytotoxicity rather than the fibers themselves. Conway et al. distinguished sphere-like oligomers of different sizes in addition to chain-like and ring-like profibers during α-Syn aggregation by atomic force microscopy (AFM)10,11,12. Lashuel et al. identified pore-like oligomers in the aggregation of α-Syn mutations using negative stain transmission electron microscopy (TEM), which is suggested to be the cause of cell dysfunction and even cell death in PD13,14.
Natively, α-Syn localizes to synaptic vesicles and appears both in soluble and membrane-associated forms15,16. The α-Syn-lipid membrane interaction is supposed to be responsible for triggering the normal function of α-Syn5. However, considerable researches postulate that the lipid vesicles may both accelerate and inhibit α-Syn fibril formation depending on the varying experimental conditions6,17,18. Zhu et al. investigated the effects of lipid vesicles on the conformation and fibrillation kinetics of α-Syn through Thioflavin T (ThT) fluorescence assays, circular dichroism (CD) measurements, and AFM, suggesting that the lipid membrane of different composition modulates the α-Syn aggregation process in different ways19,20. Recently, Hellstrand et al. revealed that the lipid membrane could co-aggregate with α-Syn fibers which can influence the structure and function of lipid membrane and α-Syn-formed amyloid deposit by using phospholipid quantification, polarization transfer solid-state nuclear magnetic resonance and cryo-TEM21. In 2015, Galvagnion et al. demonstrated that the presence of anionic lipid 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS) small unilamellar vesicles (SUVs) dramatically accelerated the α-Syn primary nucleation process by ThT fluorescence assays, resulting in three orders or more of magnitude enhancement in overall aggregation rate22. These studies provide a deeper understanding of the effect of lipids on the kinetics and overall process of α-Syn aggregation.
However, identifying different oligomer types and tracking their oligomerization dynamics is difficult due to the inherent challenges of studying heterogeneous and transient system by using traditional biochemical techniques. Imaging techniques, such as TEM and AFM, are capable of characterizing morphologies of α-Syn aggregates, but the results will inevitably be affected by the biased adsorption of aggregates to sample substrates12,13,14,23,24,25. Circular dichroism (CD), Fourier transform infrared spectroscopy, fluorescence and Raman spectroscopy can reveal the ensemble structure information, while have difficulties in identifying oligomer species. Light-scattering techniques, which permit in situ measurements, are not suitable due to the poor performance for heterogeneous systems. Recently, Cremades et al. adapted improved single molecule two color coincidence detection (smTCCD) and intermolecular fluorescence resonance energy transfer (FRET) technique to investigate this heterogeneous and transient system, and successfully identified two type of oligomers, the normal form and the toxic form26. However, these experiments required fluorescence labeling paradigms.
A nanopore sensor is a device based on Coulter Counter27: a nanometer-scale pore imbedded in a thin membrane separating two electrolyte-filled reservoirs. Driven by electric force, a charged analyte suspended in solution translocates through the nanopore, producing an ionic current drop due to the displacement of electrolyte volume in the nanopore. The statistical analysis of the ionic current drops and their duration time can reveal the geometry and charge properties of the analyte28. Nanopore technology was proposed for the ultimate goal of high-throughput DNA sequencing nearly two decades ago29,30. And now it has been widely extended to detect and investigate protein molecules, such as mapping the structural dimension31,32,33,34, investigating the chemical bond35, describing DNA-complex interaction34,36, differentiating folded and unfolded protein state37,38,39,40, and distinguishing different microRNAs41. There are also a few reports on detection of α-Syn using protein nanopores (α-hemolysin)42,43 and it has been reported that the biological nanopore could offer the earliest steps of the α-synuclein aggregation pathway and provides the potential basis for the development of drugs that can prevent α-Syn aggregation at the initial stage. Solid-state nanopores, with a fabricated pore drilled in a thin insulating membrane, have advantages in controllable fabrication, tunable nanopore size and geometry, and long stability in aqueous solution44,45,46,47,48,49,50. As an in situ and label-free single molecule detection method with fairly high resolution, solid-state nanopores provide a viable and unique approach to characterize the oligomers formed during the process of aberrant protein aggregation, such as Amyloid-β protein associated with the Parkinson’s disease51 and lysozyme52.
In this study, we use solid-state nanopores to study the oligomerization of α-Syn protein in a label-free and single-molecule approach. A tween 20 surface coating method was developed to enable continuous and smooth α-Syn translocation through SiN nanopores. Through analysis of translocation events for α-Syn incubated over a range of times, we identified four types of oligomers formed during aggregation under controlled incubation condition. These four types of oligomers inferred as intermediates show a time-dependent quantity fraction change, suggesting that the formation of large oligomers is based on the consumption of small oligomers. Furthermore, we investigated the effect of lipid SUVs on α-Syn oligomerization process. The experimental results indicate that it is the presence of 20% 1,2-dioleoyl-sn-glycero-3-[phospho-L-serine] (DOPS) dramatically enhances the aggregation rate of α-Syn, but does not alter the aggregation pathway. Together, our data describes an effective label-free single-molecule methodology that permits the detailed investigation the oligomerization stage of α-Syn protein both in isolation and in the presence of synthetic vesicles that mimic in situ conditions.
Results
α-Syn protein translocation through Tween 20-coated nanopore
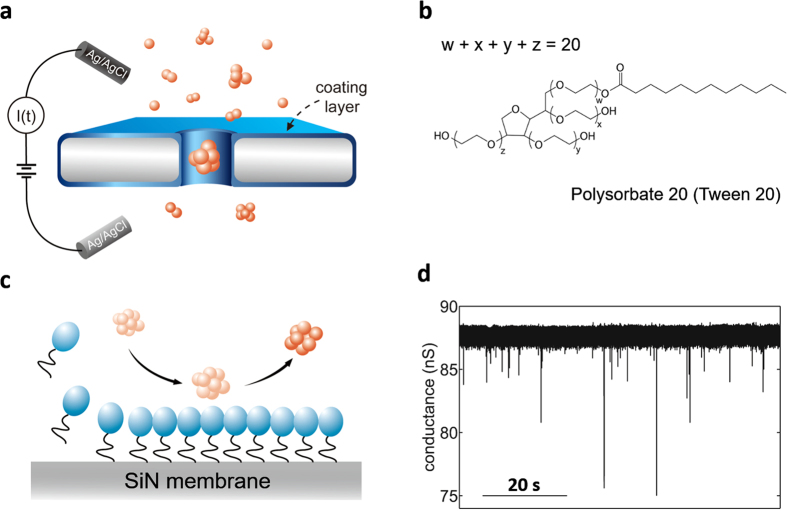
Figure 1a shows the schematic diagram of α-Syn protein detection experimental setup. A flow cell is separated by a silicon nitride membrane (Fig. 1a gray substrate) of around 40 nm in effective thickness, in which a 20 nm nanopore was drilled using focused electron beam in TEM. Two Ag/AgCl electrodes connected to patch-clamp amplifier were immersed in two reservoirs of the flow cell filled with electrolyte solution (1 M KCl, 10 mM HEPES, pH 9) to apply electric field across nanopore and to record the current signal through the nanopore. α-Syn protein samples (diluted to 1 μg/ml, 0.69 μM) after incubation under well-controlled condition were added into the cis side of the flow cell. An electric field was then applied across the nanopore to drive translocation of α-Syn proteins, leading to characteristic current blockade on the ionic current trace.
Figure 1. Detection of α-Syn using tween 20 coated solid-state nanopore.
(a) Schematic diagram of experimental setup. The flow cell is separated by silicon nitride membrane with a nanopore embedded on it. In order to provide a non-specific adsorption surface, the silicon nitride membrane is coated by a layer of tween 20 molecules (light blue layer). The α-Syn monomer is represented by particles just for clear reading in schematic diagram, which may not reveal its real morphology. (b) The chemical structure of tween 20. (c) Illustration of the assemble process of tween 20 on hydrophobic silicon nitride membrane and the compact coating layer reduce irreversible non-specific adsorption of α-Syn oligomers. Light blue ellipsoids and solid black lines represent hydrophilic ethylene glycol head groups and the hydrophobic alkyl tail of tween 20 molecule, respectively. (d) Current traces of nanopore experiment using tween 20 coated nanopore under 100 mV. The α-Syn sample had been incubated for 96 h at pH 9.
We found that bare nanopore is easily and irreversibly clogged by protein molecule adsorption on the nanopore walls by observing a dramatic current drop and a subsequent fluctuating current trace (Supplementary Fig. 1). It is believed that this phenomenon is caused by irreversible adsorption of α-Syn inside nanopore due to strong interaction between proteins and silicon nitride nanopore53,54. The non-specific adsorption is a common phenomenon51,52,55 and have been one of challenges impeding the successfully compliment of solid-state nanopore experiment. Several groups have made useful attempts to solve this problem, such as lipid coating51,56 and using glass nanopore52 to reduce the non-specific adsorption of aggregation-prone protein, PEG coating57,58,59 and chemically modified solid-state nanopore60 to inhibit the adsorption of DNA and other protein complex. Here by surface coating a layer of tween 20 molecules onto nanopore, we achieved smooth and continuous current trace with successful translocation of α-Syn samples (Fig. 1b–d). Tween 20 is a commercial used non-ionic surfactant (molecular weight: 1227.54 g/mol) consisting of hydrophilic ethylene glycol head groups and a hydrophobic alkyl tail (Fig. 1b). Here by self-assembling a compact layer of tween 20 on hydrophobic SiN surface (Supplementary Fig. 2), with its hydrophilic head groups exposed to solution and hydrophobic tail contacting with the hydrophobic SiN surface (Fig. 1c), irreversible non-specific adsorption of α-Syn proteins has been significantly reduced51,61,62,63,64, as evidenced by fluorescence microscopy experiments (Supplementary Fig. 3). The smooth current traces recorded from tween 20 coated nanopore (Fig. 1d) enables continuous translocation of α-Syn samples. Tween 20 exhibits an excellent ability against protein adsorption without affecting the protein’s original structure61,62,63,64, which is preferred over other lipid coating methods, since α-Syn-lipid membrane interaction have been implicated in affecting protein and membrane properties5,6,20,22,65.
Identifying four types of oligomers in α-Syn samples
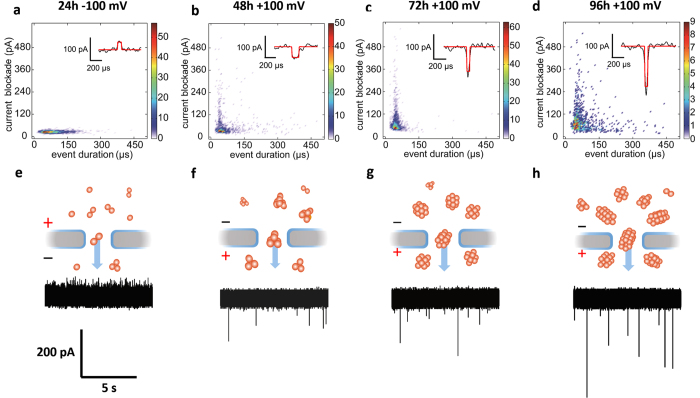
α-Syn samples were incubated for 24 h, 48 h, 72 h, and 96 h under well-controlled conditions prior to addition to the cis side of the flow cell. A temperature-controlled shaker (200 rpm, 37 oC) was used to accelerate the aggregation. Fig. 2a–d shows the density histograms for translocation events of α-Syn samples that had been incubated for 24 h, 48 h, 72 h, and 96 h, respectively. The insets are the corresponding typical current traces of single translocation events. Every translocation event is characterized by two significant parameters, current blockade ΔI and event duration Δt, corresponding to the exclude volume (i.e. size) and dwell time of proteins inside the nanopore. Fig. 2e–h depicts the scheme of the oligomerization of α-Syn and its corresponding original experimental current traces recorded for α-Syn samples incubated for different times. α-Syn monomers aggregate into oligomers and grow bigger along with longer incubation time, showing larger current blockades. Note that −100 mV instead of +100 mV is applied for 24 h sample to see successful translocation events due to the domination of electroosmosis as the driving force for α-Syn translocation in 24 h sample (Supplementary Fig. 4)66.
Figure 2. Translocation events of α-Syn samples incubated for different time.
Density histograms for translocation events of α-Syn only samples incubated for (a) 24 h, (b) 48 h, (c) 72 h and (d) 96 h. The insets are typical events for different α-Syn only samples. The black lines in these insets are filtered current trace and the red lines are the fitted square waves. Schematic graphs and characteristic current trace are also presented for (e) 24 h, (f) 48 h, (g) 72 h and (h) 96 h samples. Small orange spheres present α-Syn monomers. Under controlled incubation condition, α-Syn monomers begin to aggregate and grow bigger with longer incubation time, leading to deeper translocation events.
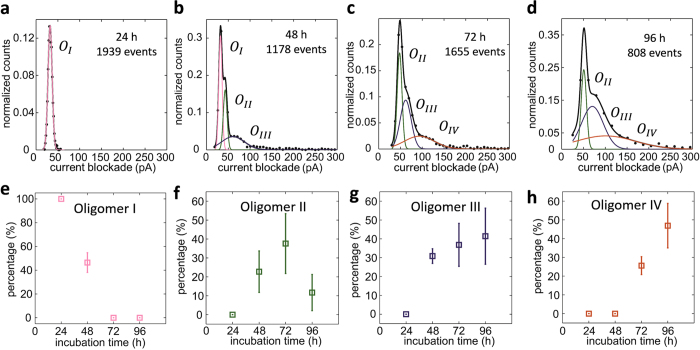
Figure 2a–d shows an increase in current blockade as a function of incubation time with increasing value of mean current blockade <ΔI>, from 32.1 pA (24 h) to 35.6 pA (48 h), to 53.0 pA (72 h), to 64.3 pA (96 h), indicating the aggregation nature of α-Syn8,9. Subsequently, multi peak Gaussian fitting is used to probe more details in current blockade distribution for 24 h, 48 h, 72 h, and 96 h samples and the fitting results are shown in Fig. 3a–d. Multi-Gaussian fitting was required to fit these distribution curves to give flat residuals except for 24 h sample (fitted by only one Gaussian component). Surprisingly, the peaks of Gaussian components from four samples can be well cataloged into four populations, OI to OIV based on the position of the peaks within a small error bar range. Detailed fitted peak positions for the four populations are summarized in Table 1. For instance, for the OIII peak, it positions at 61.0 pA, 60.7 pA and 65.4 pA in 48 h, 72 h, and 96 h samples, demonstrating a well-defined behavior. It is noteworthy to state that we did not observe translocation signal for non-incubated sample referred as 0 h sample which contains almost exclusively monomers and due to the small size of monomers (14.3 kDa) it is likely that their translocation events are below the resolution limit in our experiments. Therefore, we classify the observed populations in Fig. 3a–d into four types of oligomers: OI, OII, OIII, and OIV since mature fibers are much bigger than the nanopore size and they cannot be detected in our system.
Figure 3. Multi-Gaussian fitting results of current blockade histograms for α-Syn only samples.
Current blockade histograms and multi-Gaussian fitting results for (a) 24 h, (b) 48 h, (c) 72 h and (d) 96 h α-Syn only samples. The blockade histograms are normalized to the number of total events in each sample for clarity. The black solid lines are the Gaussian fitting results carried on the original data marked by black dot. The value of 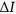 is cut off at 300 pA for statistical analysis. The individual Gaussian components are classified into four types, OI, OII, OIII, and OIV, based on its peak position. OI, OII, OIII, and OIV are marked by pink, green, purple and orange solid lines, respectively. The percentage changes of under-curve area for each Gauss contributions over incubation time for (e) oligomer I, (f) oligomer II, (g) oligomer III and (h) oligomer IV are depicted. The error arises from fitting error.
is cut off at 300 pA for statistical analysis. The individual Gaussian components are classified into four types, OI, OII, OIII, and OIV, based on its peak position. OI, OII, OIII, and OIV are marked by pink, green, purple and orange solid lines, respectively. The percentage changes of under-curve area for each Gauss contributions over incubation time for (e) oligomer I, (f) oligomer II, (g) oligomer III and (h) oligomer IV are depicted. The error arises from fitting error.
Table 1. The multi-Gaussian fitted peak position 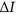 , calculated excluded volume
, calculated excluded volume 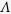 , and calculated oligomer diameter
, and calculated oligomer diameter 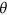 in 24 h, 48 h, 72 h, and 96 h α-Syn only samples.
in 24 h, 48 h, 72 h, and 96 h α-Syn only samples.
| Sample | 24 h |
48 h |
units | ||||
|---|---|---|---|---|---|---|---|
| cluster | OI | OII | OIII | OI | OII | OIII | – |
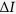 |
32.1 ± 2.2 | – | – | 34.2 ± 5.1 | 46.9 ± 7.7 | 61.0 ± 4.7 | pA |
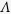 |
45.9 ± 3.1 | – | – | 48.8 ± 7.3 | 67.0 ± 11.0 | 87.1 ± 6.7 | nm3 |
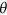 |
4.4 ± 0.1 | – | – | 4.53 ± 0.2 | 5.0 ± 0.3 | 5.5 ± 0.1 | nm |
| Sample | 72 h | 96 h | units | ||||
| cluster | OII | OIII | OIV | OII | OIII | OIV | – |
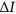 |
47.9 ± 2.4 | 60.7 ± 4.1 | 92.4 ± 20.3 | 43.2 ± 1.0 | 65.4 ± 4.7 | 98.3 ± 25.5 | pA |
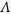 |
68.4 ± 3.4 | 86.7 ± 5.8 | 132.0 ± 29.0 | 61.7 ± 1.4 | 93.4 ± 6.7 | 140.4 ± 36.4 | nm3 |
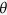 |
5.1 ± 0.1 | 5.5 ± 0.1 | 6.3 ± 0.5* | 4.9 ± 0.1 | 5.6 ± 0.1 | 6.4 ± 0.5* | nm |
The errors are fitting errors. The oligomers are regarded as spheres to calculate their diameter. (*the calculated diameter of OIV based on the sphere model may lead to an overestimation of the actual size, which are more likely have an asymmetric short rod-like structure due to the broad distribution of blockage of OIV56 and the TEM image in Supplementary Fig. 5d.)
Thanks to the advantages of label-free and single molecule detection based on nanopore experiments, we are able to probe distinct species within heterogeneous system based on the current blockade distribution of translocation events. The sample incubated under the aggregation promoting conditions for 24 h consists of almost only small oligomers (OI) of similar size based on its uniform and narrow distribution of current blockades (Fig. 3a), confirmed by negative stain TEM results (Supplementary Fig. 5). The peak position of Gaussian components listed in Table 1 reveal that distinct species of oligomers appeared after 48 h incubation (Fig. 3b–d), also supported by TEM analysis (Supplementary Fig. 5), presenting a heterogeneous system involving oligomers with different sizes in 48 h, 72 h, 96 h samples.
According to previous studies in protein translocation through nanopores28,33,37,67, for the analyte that is much smaller than an idealized cylindrical nanopore, the instantaneous excluded volume 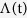 of the analyte is approximately proportional to
of the analyte is approximately proportional to 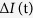 and the relationship is expressible as follows:
and the relationship is expressible as follows:
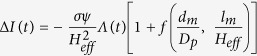 |
where 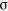 is the solution conductivity,
is the solution conductivity, 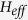 is the effective length of solid-state nanopore, and
is the effective length of solid-state nanopore, and 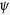 is the applied voltage. The correction factor
is the applied voltage. The correction factor 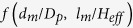 depends primarily on the geometry of nanopore and analyte. For an analyte much smaller than the length of nanopore, the correction factor
depends primarily on the geometry of nanopore and analyte. For an analyte much smaller than the length of nanopore, the correction factor 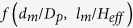 is approaching zero and the exclude volume of the analyte can be estimated from
is approaching zero and the exclude volume of the analyte can be estimated from 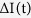 with
with 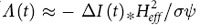 67. An effective length of 40 nm was used here to calculate the exclude volume and the diameter
67. An effective length of 40 nm was used here to calculate the exclude volume and the diameter of the defined four types of α-Syn oligomers. The detailed results are listed in Table 1. The calculated diameters for the four fitted clusters are 4.4 nm, 5.0 nm, 5.5 nm, and 6.5 nm. The general agreement between calculated diameter and that reported by other groups characterized by AFM demonstrate the rationality of this method5,12.
of the defined four types of α-Syn oligomers. The detailed results are listed in Table 1. The calculated diameters for the four fitted clusters are 4.4 nm, 5.0 nm, 5.5 nm, and 6.5 nm. The general agreement between calculated diameter and that reported by other groups characterized by AFM demonstrate the rationality of this method5,12.
The percentage of under-curve area for each Gaussian component is proportional to the number of captured molecules. Therefore, we are able to monitor the time-dependent quantity fraction of the four types of oligomers and give kinetic information of α-Syn oligomerization. The percentage of total under-curve area for each Gaussian components in 24 h, 48 h, 72 h, and 96 h samples have been calculated and the data is illustrated as a function of incubation time in Fig. 3e–h. The percentage for the smallest oligomer OI gradually decreased after 24 h (Fig. 3e), indicating its consumption during the aggregation of α-Syn. The percentage for OII increases from 48 h to 72 h and decreases after 96 h incubation (Fig. 3f). OIII oligomers appear at 48 h and grow steadily in percentage from 48 h to 96 h (Fig. 3g). OIV oligomers emerge at 72 h and exhibit a sharp increase in percentage at 96 h (Fig. 3h). From above analysis, it is reasonable to assure that at least four types of α-Syn oligomers have formed during the aggregation process under well-controlled condition and these four types of oligomers undergo a conversion between one another. In the 24 h sample, OI oligomers form and serve as aggregation seeds. As incubation time increases, oligomers OII, OIII, and OIV appear by the consumption of OI, resulting in reduction of quantity fraction of OI. The quantity fraction of OII reaches its maximum after 72 h incubation and then declines maybe because they are precursors for OIII and OIV. Both of the quantity fraction of oligomers OIII and OIV increase with longer incubation (up to 96 h), but they show different behavior indicating they are two distinct species. According to Smoluchowski rate equation, the rate of proteins captured by nanopore (J) can be expressed as: 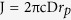 . In this equation, c is the bulk concentration of molecules; D is the bulk diffusion constant, and
. In this equation, c is the bulk concentration of molecules; D is the bulk diffusion constant, and 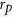 is the capture radius of nanopore, which is determined by the geometric parameters of nanopore and the applied voltage. Although the
is the capture radius of nanopore, which is determined by the geometric parameters of nanopore and the applied voltage. Although the 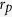 can be regarded as a constant value approximately in 48 h, 72 h, and 96 h samples for the same voltage applied and similar geometry of nanopore used in these experiment, the diffusion constant of different types of oligomer with dissimilar sizes are not the same, leading to an underestimation of percentage for larger oligomers with smaller diffusion constant in sample51.
can be regarded as a constant value approximately in 48 h, 72 h, and 96 h samples for the same voltage applied and similar geometry of nanopore used in these experiment, the diffusion constant of different types of oligomer with dissimilar sizes are not the same, leading to an underestimation of percentage for larger oligomers with smaller diffusion constant in sample51.
The current blockade histograms for various samples showed heterogeneous distribution, which were attributed to multiple oligomers formation during incubation. The event duration histograms for different samples (Supplementary Fig. 6) do not exhibit the same characteristic of heterogeneity. The first passage theory was used to fit the duration histograms and the extracted values of drift velocity, v. From four samples, we found that drift velocity increases along with incubation time up to 72 h, implying a faster translocation speed of samples with longer incubation time due to an increasing charge per aggregate from addition of monomers51.
Investigation of the effect of lipid SUVs on the kinetics of α-Syn’s oligomerization
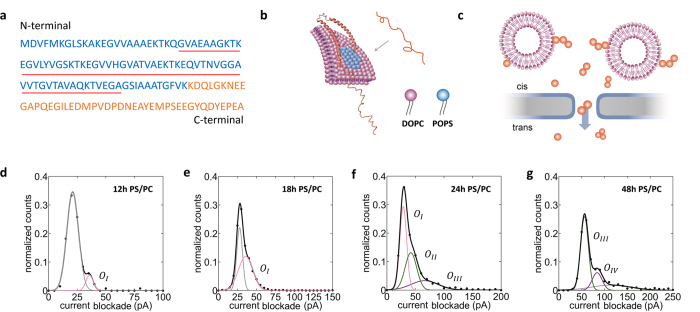
The presence of lipid membranes is believed to have a fundamental influence on the kinetics of α-Syn oligomerization, which depends on the relative proportion of the protein and lipids, as well as the lipid compositions used6. SUVs used here are composed of 20% 1,2-dioleoyl-sn- glycero-3- [phospho-L-serine] (DOPS) and 80% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) to imitate the lipid constitution of native vesicles. We investigated the effect of composite phospholipids SUVs consisting of 20% DOPS and 80% POPC (PS/PC) on the oligomerization of α-Syn. Samples were incubated under 37 °C in quiescence conditions to provide full interaction between PS/PC and α-Syn proteins. Figure 4a presents the amino acid sequence of α-Syn protein, which can be classified into three domains: a) the positive charged N-terminal that contains highly conserved and imperfect repeats KTKEGV, residues 1–60; b) the hydrophobic core at the central region, which is known as the non-amyloid- component (NAC), residues 61–90; c) an acidic C-terminal, residues 96–140. Although intrinsically unfolded in solution, the N-terminal of α-Syn will adopt an α-helix structure upon binding to lipid membrane, while the acidic C-terminal remains unfolded, as shown in Fig. 4b568,69.
component (NAC), residues 61–90; c) an acidic C-terminal, residues 96–140. Although intrinsically unfolded in solution, the N-terminal of α-Syn will adopt an α-helix structure upon binding to lipid membrane, while the acidic C-terminal remains unfolded, as shown in Fig. 4b568,69.
Figure 4. Detection of α-Syn and PS/PC SUVs co-incubated samples.
(a) Protein sequence of wild type α-Syn. The positive charged N-terminal (residues 1–106) of α-Syn is colored blue. The C-terminal is labeled by orange color and the NAC region is denoted by red underline. (b) Illustrating graph that shows the transition of an unstructured α-Syn molecule into an α-helices structure upon binding to lipid membrane. The N-terminal adopts the α-helices structure and acts as an anchor when α-Syn binds to phospholipid and the C-terminal is usually unstructured. (c) Schematic diagram of nanopore experiment for α-Syn and lipid SUVs co-incubated sample. Current blockade histograms for PS/PC co-incubated samples for (d) 12 h, (e) 18 h, (f) 24 h and (g) 48 h are shown. The original data is marked by black dot; multi-Gaussian fitted curves are labeled by black solid line. Individual Gaussian components are classified depending on oligomeric species defined in α-Syn only samples and marked by corresponding color. The error is from fitted error.
Figure 4c shows the schematic diagram of nanopore experiment for PS/PC and α-Syn co-incubated samples. The size of PS/PC SUVs is around 60 nm (Supplementary Fig. 7a), which is much larger than the 20 nm nanopore used here. Prior to addition of sample, ultrasonic treatment is adopted to break the weak binding between α-Syn and lipid membrane. This ultrasonic treatment is found to have little effect on α-Syn aggregates (see Supplementary Fig. 8 for details). It is believed that the translocation events belong to the α-Syn oligomers other than the α-Syn-SUVs complex for its similar behavior of blockade histograms as samples without SUVs.
Current blockade histograms for lipid co-incubated α-Syn samples incubated for 12 h, 18 h, 24 h, and 48 h under quiescence condition are presented in Fig. 4d–g. Multiple Gaussian fitting is used to fit the multi-peak histograms. The range that the current blockade values cover becomes wider and multiple peaks emerge as the increase of incubation time. According to the fitted peak values (Supplementary Table 1), the peaks can also be categorized into four types of oligomers as we previously defined in α-Syn translocation experiment. The OI Gaussian component appeared in 12 h, 18 h and 24 h PS/PC samples and shows an increase in percentage of under-curve area along with longer incubation time, indicating more OI oligomers has formed as a function of incubation time. As time increases, the defined larger oligomers such as OII, OIII and OIV have emerged indicating the aggregation nature of lipid co-incubated α-Syn samples. The analogous evolution behavior of defined oligomeric types in lipid co-incubation samples compared with α-Syn only samples suggests that the formation of larger oligomers is also based on the consumption of small oligomers formed earlier.
In order to give an intuitive comparison of the oligomeric species formed in α-Syn only sample and PS/PC SUVs co-incubated α-Syn sample, the multi-Gaussian fitted peak values are plotted as the function of incubation time in Fig. 5a. It can be seen that the peak values of current blockade for PS/PC SUVs co-incubated α-Syn sample fall into an acceptable region of the corresponding four oligomers defined in α-Syn only samples although the kinetics is accelerated. For α-Syn only sample, 24 hours incubation only results in small oligomer OI; after 48 hours incubation, larger oligomer OII and oligomer OIII appear. For samples incubated for more than 72 hours, small oligomer OI disappears and large oligomer OIV emerges. For PS/PC SUVs co-incubated α-Syn sample, the aggregation pathway is similar to that of α-Syn only sample, which also goes through the conversion of four types of oligomer, while the time need for the formation of large oligomer is reduced from 96 h to 48 h, leading to an 100% enhancing in aggregation rate. Note that the incubation condition for these two samples is not exactly the same: we used an aggregation-promoting condition for the α-Syn only sample to speed up the experiment schedule and a quiescence condition for SUVs co-incubated sample to guarantee full interaction between SUVs and α-Syn, therefore the aggregation rate enhancement is underestimated here. In addition, two small peaks (<30 pA) have been observed lipid SUVs co-incubated α-Syn samples under 12 h and 18 h incubation, which may be due to folded state of α-Syn when binding to lipid membrane.
Figure 5. Comparison between α-Syn only and PS/PC SUVs co-incubated α-Syn samples.
(a) Multi-Gaussian fitted peak values of α-Syn only sample (marked by red square) and PS/PC co-incubation α-Syn sample (marked by blue square) in current blockade distributions as a function of incubation time. The pink, green, purple, and orange bands indicate four types of oligomers we discussed above. The band width is determined by error bar. (b) The scheme depicts the possible mechanism of α-Syn aggregation process in the absence or presence of PS/PC SUV. Note only part of the lipid SUVs is shown in the scheme to show the binding area more clearly.
It is acknowledged that the anionic lipid DOPS SUVs are responsible for this enhancement of aggregation rate. The possible mechanism is that the negative charged DOPS attracts positive charged N-terminal of α-Syn, leading to a locally high concentration of α-Syn monomers, which accelerates the primary nucleation process22,65. We also did the control experiment with 100% POPC SUVs co-incubated α-Syn samples incubated for 6 h, 12 h, 24 h, and 48 h (Supplementary Fig. 9). According to multi-Gaussian fitting results, no peak larger than 60 pA can be seen in current blockade histograms after 48 hours incubation, indicating a very slow aggregation rate, which agrees with results reported by other groups65. Control experiments suggest that POPC has no notable effect in α-Syn aggregation, and the enhanced aggregation rate is mainly caused by DOPS in lipid SUVs.
Figure 5b illustrates a possible mechanism of α-Syn aggregation process in the absence or presence of lipid SUVs. The aggregation process of α-Syn protein exhibits a sigmoidal growth profile, which has an initial lag phase due to the high barrier of nuclei formation26. In the absence of PS/PC SUVs, the α-Syn monomers form nuclei through a high barrier homogenous nucleation process, leading to a relatively slow aggregation rate (Fig. 5b). While in the presence of PS/PC SUVs, the α-Syn monomers form nuclei through a low barrier heterogeneous nucleation process68. In addition, the weak attractive force between the negatively charged DOPS hydrophobic head and the positive α-Syn N-terminal attracts more monomers to gather around DOPS molecules, leading to a local relatively higher concentration of α-Syn monomers, which contributes to the growth of nuclei to large oligomers65. Both of these two factors count for the enhancement of aggregation rate for the PS/PC SUVs co-incubated α-Syn sample. Our experimental results indicate the presence of PS/PC SUVs only accelerates the aggregation rate of α-Syn while does not alter the aggregation pathway.
Discussion
In this study, we have demonstrated that solid-state nanopore sensors can investigate the time-dependent intrinsic and membrane-facilitated oligomerization of α-Syn in a single-molecule and label-free approach. Tween 20 coating is found to effectively inhibit non-specific adsorption of α-Syn on silicon nitride nanopore membrane. Four types of oligomers were identified as intermediates formed during α-Syn aggregation by multi-Gaussian fitting of current blockade distributions. The quantity fraction of these oligomers is found to change over time, suggesting a consumption mechanism of small oligomers for the formation of large oligomers. In addition, we found that the negative charged DOPS dramatically accelerate the aggregation rate of α-Syn and the PS/PC SUVs co-incubated samples share similar α-Syn aggregation pathway with the α-Syn only sample. As a label-free single molecule detection method, solid-state nanopores are suitable for characterizing heterogeneous and vital oligomerization of α-Syn in situ, giving important insights into the formation and conversion of oligomeric species, which not only represent significant steps of aggregation process but also regarded as key role in the pathogenesis of PD.
Method
Nanopore Fabrication
2 μm-thick silicon oxide film and a layer of 200 nm thick silicon nitride were deposited on both sides of the 400 μm-thick silicon wafers. Then photolithography and reactive ion etching (RIE) were used to form a square of 20 μm × 20 μm freestanding membrane of two layers (SiN/SiO2). A focused ion beam (FIB, DB235) was adopted to remove about 1.5 μm thick silicon oxide of 1 μm × 1 μm in the center of the freestanding membrane, and the remaining 500 nm thick silicon oxide was removed by a subsequent timed buffered oxide etch (BOE), resulting in a 2 μm × 2 μm freestanding mini membrane of silicon nitride. The outside silicon nitride layer was estimated to be 120 nm by ellipsometry after the timed hot KOH thinning process. The chips were cleaned with NH4OH, H2O2 and water (1:1:6 v/v) right after RCA. Then the chips are dried by nitride-flow before nanopore drilling. The nanopores were drilled in the center of mini membranes by a 300 kV focused electron beam from transmission electron microscope (TEM, Tecnai F30). Note the effective thickness of SiN membrane is ~40 nm due to the truncated double-cone geometry of the drilled pore70,71. We store the drilled chips in air in a constant temperature and humidity drying oven for further use.
Tween 20 Coating Buffer Preparation
0.1% w/v Tween 20 was added in 1 M KCl with 10 mM HEPES buffer at pH 9. The mixture solution was stored at room temperature for further use.
Contact Angle Experiment
20 chips are separated to two groups as bare SiN membrane and tween 20 coated ones to compare the difference of hydrophilicity. Tween 20 coated chips are bathed in 0.1% w/v tween 20 solution for 1 h at room temperature and blew-dry by nitrogen gas gun. Control uncoated chip samples are bathed in pure water for 1 h at room temperature. Contact angle experiments are conducted by dripping a drop of pure water onto the membrane surface, after which the profile is captured by the optical system of a contact angle goniometer (Datephysics OCA20). The contact angle between liquid/solid interface and liquid/vapor interface is then analyzed automatically by software.
Fluorescent Experiment
Cover slip (Fisher Brand, Microscope cover glass: 12-545-E 22*60-1) is cleaned by solution (NH4OH : H2O2 : water = 1:1:6) at 85 °C for 20 min, then rinsed by ultrapure water and blew-dry by Nitrogen gas gun. After the cleaning process, PDMS micro flow channels of 40 μm in width and 10 μm in height is mounted on the cover glass. The dyed α-Syn sample diluted by the buffer with and without 0.1% w/v Tween 20 are loaded separately through the tiny hole connecting to the micro fluidic channel. The fluorescent images are recorded after 10 min. Green fluorescence of YOYO-1 stained α-Syn oligomers anchored onto the glass surface is observed with Leica TCS SPE Confocal Microscope (Leica, German) after excitation with a 488 nm laser. Images are captured in 512 pixels × 512 pixels.
Lipid SUVs Preparation
Two populations of small unilamellar vesicles (SUVs) containing different ratio of (mol/mol) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dioleoyl-sn- glycero-3- [phospho-L-serine] (DOPS) (Avanti Polar Lipids, Birmingham, AL) were mixed in a molar ratio of 100:0 and 80:20, respectively. The mixtures were dried in a vacuum to form films. The membrane films were re-suspended in 10 mM HEPES buffer at the pH 7.45 and stirred at 45 °C for 1 hour. The sonication (Ultra Sonics, 760, 3 × 5 min, 30% maximum power) was used to prepare SUVs of the lipids.
Dynamic Light Scattering Measurements
Dynamic light scattering measurements were carried out using a spectrometer of standard design (ALV-5000/E/WIN Multiple Tau Digital Correlator) with a Spectra-Physics 2017 200 mW Ar laser (wavelength: 514.5 nm). The scattering angle was 90 and CONTIN method was used to analyze the distribution of the radii of micelles. Unweighted data were recorded for all experiments. The scattering intensities were recorded and normalized with respect to the total concentration.
α-Syn only Oligomers Preparation
The wild-type α-Syn proteins before aggregation was purchased from AnaSpec, Inc. in the form of powder. Then the powder was dissolved with 10 mM HEPES buffer at pH 7.45 to a concentration of 100 μg/ml (6.9 μm). For the sake of experiment, equimolecular concentrations of protein samples (6.9 μm) were sealed in 1 mL siliconized plastic microcentrifuge tube. For α-Syn only experiments, a tube containing 6.9 μm α-Syn at pH 7.45 was incubated under constant agitation at 200 rpm, 37 °C in the dark for 1–4 days to accelerate aggregation process. At each time point, 20 μl aliquot was diluted with Tween 20 buffer (10 mM HEPES, pH 9, 1 M KCl, 0.1% w/v Tween 20) into an experimental concentration of 0.69 μM α-Syn for nanopore translocation measurement.
Lipid SUVs Doped α-Syn Oligomers Preparation
Wild-type α-Syn was prepared and diluted as above. Then 69 μm 80% PS/20% PC or 100% PC in the control experiment was added into the sample tube that means the molar ratio of α-Syn and lipid SUVs are 1:10, which will accelerate the aggregation process of α-Syn according to the work by Galvagnion et al.22. In order not to disrupt the interaction between α-Syn proteins and lipid SUVs surface, the samples are incubated at quiescence condition at the controlled temperature 37oC for 12 h, 18 h, 24 h, and 48 h in 10 mM HEPES buffer at pH 7.45. At each time point, 20 μl aliquot was diluted with Tween 20 buffer (10 mM HEPES, pH 9, 1 M KCl, 0.1% w/v Tween 20) into a concentration of 0.69 μM α-Syn and 6.9 μM lipid SUVs for nanopore translocation measurement.
α-Syn Oligomers Translocation Experiments
The chips containing nanopore were mounted in a sealed fluid cell and were separated into two electrically isolated reservoirs of electrolytes. First the 0.1% w/v Tween 20 buffer was added into cis side and after 10 min the protein sample diluted with the 0.1% w/v Tween 20 was introduced into the cis side of the fluid cell. The voltage was applied through two Ag/AgCl electrodes coupled to the two opposite electrolyte reservoirs by an Axon 200B patch clamp amplifier (Molecular Devices, Sunnyvale, CA). Attached with an 8 pole, 40 kHz, low pass Bessel filter operating in resistive feedback mode, the patch clamp was also used for ionic currents measurement. The output of patch clamp was digitized at 250 kHz and continuously recorded by an Axon Digidata 1440A digitizer and pClamp 10.3 software. Then the data recorded was analyzed through custom MATLAB code (The MathWorks, Natick, MA). All the events showed in figures have been filtered by 10 kHz low-pass Bessel filter through MATLAB code for clarity.
Samples for Transmission Electron Microscopy Preparation
TEM images were obtained by negative-staining method. First, 7 μl aliquots of 0.1 mg/mL α-Syn in 10 mM HEPES buffer at pH 7.4 were transferred to plasma processed, fresh glow-discharged, carbon-coated copper grids. After 1 minute, the sample solution was wicked off with a piece of filter paper and the grid was rinsed with fresh deionized water, then a 7 μl droplet of 2% (w/v) phosphotungstic acid staining solution was placed on the grid. After 1 minute, the excess fluid on the grid was wicked off and the grid was allowed to dry. The samples were viewed by a transmission electron microscope at 300 kV (TEM, Tecnai F30).
Additional Information
How to cite this article: Hu, R. et al. Intrinsic and membrane-facilitated a-synuclein oligomerization revealed by label-free detection through solid-state nanopores. Sci. Rep. 6, 20776; doi: 10.1038/srep20776 (2016).
Supplementary Material
Acknowledgments
This work was supported by National 973 projects (2013CB932602, 2011CB707601, 2015CB856304, MOST) from Ministry of Science and Technology, China, National Natural Science Foundation of China (NSFC51272007, 61571015, 11234001, 91433102, 11327902). Q.Z acknowledges Beijing Nova Program (XX2013003) and the Program for New Century Excellent Talents in University of China.
Footnotes
Author Contributions J.-J.D. and Q.Z. conceived the idea and designed the experiment. R.H. performed the experiment and analyzed the data. J.L. and Z.-P.T. fabricated the nanopore sensor chips and provided technical help. R.H. and X.-Q.L. prepared lipid SUVs and protein samples. J.-G.L., J.-K.L., J.-J.D., Q.Z. and D.-P.Y. supervised the project during the whole process. R.H., J.-J.D., J.L. and Q.Z. co-wrote the manuscript. All authors read and comment on the manuscript.
References
- Bonini N. M. & Glasson B. I. Snaring the function of alpha-synuclein. Cell 123, 359–361 (2005). [DOI] [PubMed] [Google Scholar]
- Lotharius J. & Brundin P. Pathogenesis of Parkinson’s disease: Dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci 3, 932–942 (2002). [DOI] [PubMed] [Google Scholar]
- Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2, 492–501 (2001). [DOI] [PubMed] [Google Scholar]
- Uversky V. N. & Eliezer D. Biophysics of Parkinson’s Disease: Structure and Aggregation of alpha-Synuclein. Curr Protein Pept Sc 10, 483–499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink A. L. The aggregation and fibrillation of alpha-synuclein. Accounts Chem Res 39, 628–634 (2006). [DOI] [PubMed] [Google Scholar]
- Auluck P. K., Caraveo G. & Lindquist S. alpha-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu rev cell dev bi 26, 211–233 (2010). [DOI] [PubMed] [Google Scholar]
- Tuomas P. J. K., Michele V. & Christopher M. D. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Bio 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- Chiti F. & Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M. & Goedert M. alpha-synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. P Natl Acade Sci USA 95, 6469–6473 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K. A., Harper J. D. & Lansbury P. T. Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat med 4, 1318–1320 (1998). [DOI] [PubMed] [Google Scholar]
- Conway K. A., Harper J. D. & Lansbury P. T. Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochem 39, 2552–2563 (2000). [DOI] [PubMed] [Google Scholar]
- Conway K. A. et al. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. P Natl Acade Sci USA 97, 571–576 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel H. A., Hartley D., Petre B. M., Walz T. & Lansbury P. T. Neurodegenerative disease - Amyloid pores from pathogenic mutations. Nature 418, 291–291 (2002). [DOI] [PubMed] [Google Scholar]
- Lashuel H. A. et al. α-Synuclein, Especially the Parkinson’s Disease-associated Mutants, Forms Pore-like Annular and Tubular Protofibrils. J Mol Biol 322, 1089–1102 (2002). [DOI] [PubMed] [Google Scholar]
- Dikiy I. & Eliezer D. Folding and misfolding of alpha-synuclein on membranes. BBA-Biomembranes 1818, 1013–1018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruipérez V., Darios F. & Davletov B. Alpha-synuclein, lipids and Parkinson’s disease. Prog Lipid Res 49, 420–428 (2010). [DOI] [PubMed] [Google Scholar]
- Chaudhary H., Stefanovic A. N. D., Subramaniam V. & Claessens M. M. A. E. Membrane interactions and fibrillization of alpha-synuclein play an essential role in membrane disruption. FEBS Lett 588, 4457–4463 (2014). [DOI] [PubMed] [Google Scholar]
- Iyer A., Petersen N. O., Claessens M. & Subramaniam V. Amyloids of Alpha-Synuclein Affect the Structure and Dynamics of Supported Lipid Bilayers. Biophys J 106, 2585–2594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. & Fink A. L. Lipid binding inhibits alpha-synuclein fibril formation. J Biol Chem 278, 16873–16877 (2003). [DOI] [PubMed] [Google Scholar]
- Zhu M., Li J. & Fink A. L. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J Biol Chem 278, 40186–40197 (2003). [DOI] [PubMed] [Google Scholar]
- Hellstrand E., Nowacka A., Topgaard D., Linse S. & Sparr E. Membrane Lipid Co-Aggregation with alpha-Synuclein Fibrils. PloS one 8, 77235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagnion C. et al. Lipid vesicles trigger alpha-synuclein aggregation by stimulating primary nucleation. Nat chem biol 11, 229–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner B. et al. In vivo demonstration that α-synuclein oligomers are toxic. P Natl Acade Sci USA 108, 4194–4199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W., Cherny D., Subramaniam V. & Jovin T. M. Rapid Self-assembly of α-Synuclein Observed by In Situ Atomic Force Microscopy. J Mol Miol 340, 127–139 (2004). [DOI] [PubMed] [Google Scholar]
- Bucciantini M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002). [DOI] [PubMed] [Google Scholar]
- Cremades N. et al. Direct Observation of the Interconversion of Normal and Toxic Forms of α-Synuclein. Cell 149, 1048–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg W. R., Liedholz G. A. & Coulter W. H. Test on particles suspended in fluid - comprises method and arrangement for analysis of such particles Patent DE2438401-A; FR2241069-A; US3944917-A; GB1476209-A; GB1476210-A; CA1013432-A.
- Ledden B., Fologea D., Talaga D. S. & Li J. Sensing Single Protein Molecules with Solid-State Nanopores. 129–150 (2011). [Google Scholar]
- Kasianowicz J. J., Brandin E., Branton D. & Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. P Natl Acade Sci USA 93, 13770–13773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat Nanotech. 2, 209–215 (2007). [DOI] [PubMed] [Google Scholar]
- Han A. et al. Sensing protein molecules using nanofabricated pores. Appl Phys Lett 88, 093901 (2006). [Google Scholar]
- Han A. et al. Label-free detection of single protein molecules and protein-protein interactions using synthetic nanopores. Anal Chem 80, 4651–4658 (2008). [DOI] [PubMed] [Google Scholar]
- Fologea D., Ledden B., McNabb D. S. & Li J. Electrical characterization of protein molecules by a solid-state nanopore. Appl Phys Lett 91, 539011–539013 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shasha C. et al. Nanopore-Based Conformational Analysis of a Viral RNA Drug Target. ACS Nano 8, 6425–6430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabard-Cossa V. et al. Single-Molecule Bonds Characterized by Solid-State Nanopore Force Spectroscopy. ACS Nano 3, 3009–3014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivankin A., Carson S., Kinney S. R. & Wanunu M. Fast, label-free force spectroscopy of histone-DNA interactions in individual nucleosomes using nanopores. J Am Chem Soc 135, 15350–15352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaga D. S. & Li J. Single-Molecule Protein Unfolding in Solid State Nanopores. J Am Chem Soc 131, 9287–9297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madampage C., Tavassoly O., Christensen C., Kumari M. & Lee J. S. Nanopore analysis: An emerging technique for studying the folding and misfolding of proteins. Prion 6, 116–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Larrea D. & Bayley H. Multistep protein unfolding during nanopore translocation. Nat Nanotech 8, 288–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman K. J., Haq S. R., Edel J. B., Jemth P. & Kim M. J. Single molecule unfolding and stretching of protein domains inside a solid-state nanopore by electric field. Sci Rep 3, 1638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotech 5, 807–814 (2010). [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Ying Y. L., Li Y., Kraatz H. B. & Long Y. T. Nanopore Analysis of β-Amyloid Peptide Aggregation Transition Induced by Small Molecules. Anal Chem 83, 1746–1752 (2011). [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Gu Z., Cao C., Wang J. & Long Y. T. Analysis of a Single alpha-Synuclein Fibrillation by the Interaction with a Protein Nanopore. Anal Chem 85, 8254–8261 (2013). [DOI] [PubMed] [Google Scholar]
- Storm A. J., Chen J. H., Ling X. S., Zandbergen H. W. & Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater 2, 537–540 (2003). [DOI] [PubMed] [Google Scholar]
- Storm A., Chen J., Zandbergen H. & Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E 71, 051903 (2005). [DOI] [PubMed] [Google Scholar]
- Krapf D. et al. Fabrication and Characterization of Nanopore-Based Electrodes with Radii down to 2 nm. Nano Lett 6, 105–109 (2005). [DOI] [PubMed] [Google Scholar]
- Merchant C. A. et al. DNA translocation through graphene nanopores. Nano Lett 10, 2915–2921 (2010). [DOI] [PubMed] [Google Scholar]
- Larkin J. et al. Slow DNA Transport through Nanopores in Hafnium Oxide Membranes. ACS Nano 7, 10121–10128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. Boron Nitride Nanopores: Highly Sensitive DNA Single-Molecule Detectors. Adv Mate 25, 4549–4554 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J., You L., Ye H. & Yu D. Fabrication of ultrafine nanostructures with single-nanometre precision in a high-resolution transmission electron microscope. Nanotech 18, 155303 (2007). [Google Scholar]
- Yusko E. C. et al. Single-Particle Characterization of Aβ Oligomers in Solution. ACS Nano 6, 5909–5919 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyushenko N., Bell N. A., Lamboll R. D. & Keyser U. F. Nanopore analysis of amyloid fibrils formed by lysozyme aggregation. Analyst 140, 4882–4886 (2015). [DOI] [PubMed] [Google Scholar]
- Roach P., Farrar D. & Perry C. C. Interpretation of Protein Adsorption: Surface-Induced Conformational Changes. J Am Chem Soc 127, 8168–8173 (2005). [DOI] [PubMed] [Google Scholar]
- Rabe M., Verdes D. & Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv Colloid Interface Sci 162, 87–106 (2011). [DOI] [PubMed] [Google Scholar]
- Niedzwiecki D. J., Grazul J. & Movileanu L. Single-Molecule Observation of Protein Adsorption onto an Inorganic Surface. J Am hem Soc 132, 10816–10822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko E. C. et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotech 6, 253–260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton L. T. et al. Resistive-pulse studies of proteins and protein/antibody complexes using a conical nanotube sensor. J Am Chem Soc 129, 13144–13152 (2007). [DOI] [PubMed] [Google Scholar]
- Sexton L. T. et al. An Adsorption-Based Model for Pulse Duration in Resistive-Pulse Protein Sensing. J Am Chem Soc 132, 6755–6763 (2010). [DOI] [PubMed] [Google Scholar]
- Tang Z. et al. Surface Modification of Solid-State Nanopores for Sticky-Free Translocation of Single-Stranded DNA. Small 10, 4332–4339 (2014). [DOI] [PubMed] [Google Scholar]
- Wanunu M. & Meller A. Chemically Modified Solid-State Nanopores. Nano Lett 7, 1580–1585 (2007). [DOI] [PubMed] [Google Scholar]
- Shen L., Guo A. & Zhu X. Tween surfactants: Adsorption, self-organization, and protein resistance. Surf Sci 605, 494–499 (2011). [Google Scholar]
- Towns J. K. & Regnier F. E. Capillary electrophoretic separations of proteins using nonionic surfactant coatings. Anal Chem 63, 1126–1132 (1991). [DOI] [PubMed] [Google Scholar]
- Chen R. J. et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. P Natl Acade Sci USA 100, 4984–4989 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua B. et al. An improved surface passivation method for single-molecule studies. Nat Methods 11, 1233–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E. R. & Rhoades E. Effects of curvature and composition on alpha-synuclein binding to lipid vesicles. Biophys J 99, 2279–2288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnkes M., Pedone D., Knezevic J., Döblinger M. & Rant U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett 10, 2162–2167 (2010). [DOI] [PubMed] [Google Scholar]
- DeBlois R. W. Counting and Sizing of Submicron Particles by the Resistive Pulse Technique. Rev Sci Instrum, 41, 909–916 (1970). [Google Scholar]
- Eliezer D., Kutluay E., Bussell R. Jr & Browne G. Conformational properties of α-synuclein in its free and lipid-associated states1. J mol Biol 307, 1061–1073 (2001). [DOI] [PubMed] [Google Scholar]
- Fusco G. et al. Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat commun 5, 3827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Wanunu M., Bell D. C. & Meller A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv Mater 18, 3149–3153 (2006). [Google Scholar]
- Kim M. J., McNally B., Murata K. & Meller A. Characteristics of solid-state nanometre pores fabricated using a transmission electron microscope. Nanotech 18, 205302 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.