SUMMARY
Chromosomal rearrangements are a hallmark of acute lymphoblastic leukemia (ALL) and are important ALL initiating events. We describe four different rearrangements of the erythropoietin receptor gene EPOR in Philadelphia chromosome-like (Ph-like) ALL. All of these rearrangements result in truncation of the cytoplasmic tail of EPOR at residues similar to those mutated in primary familial congenital polycythemia, with preservation of the proximal tyrosine essential for receptor activation and loss of distal regulatory residues. This resulted in deregulated EPOR expression, hypersensitivity to erythropoietin stimulation, and heightened JAK-STAT activation. Expression of truncated EPOR in mouse B-cell progenitors induced ALL in vivo. Human leukemic cells with EPOR rearrangements were sensitive to JAK-STAT inhibition, suggesting a therapeutic option in high-risk ALL.
Graphical Abstract

INTRODUCTION
Acute lymphoblastic leukemia (ALL) remains a leading cause of cancer-related death in young people (Hunger and Mullighan, 2015a). ALL comprises a number of entities characterized by chromosomal rearrangements, aneuploidy, structural variants and sequence mutations that perturb cellular pathways including lymphoid development, tumor suppression, epigenetic regulation and kinase signaling (Hunger and Mullighan, 2015b). Chromosomal rearrangements are a hallmark of ALL and commonly deregulate hematopoietic transcription factors (e.g. ETV6-RUNX1, TCF3-PBX1), cytokine receptors (CRLF2, PDGFRB), and tyrosine kinases (BCR-ABL1).
BCR-ABL1-like, or Ph-like, B cell precursor ALL (B-ALL) is characterized by a leukemic cell transcriptional profile similar to BCR-ABL1-positive ALL, alterations of lymphoid transcription factor genes such as IKZF1, and a diverse range of alterations, most commonly chromosomal rearrangements that activate cytokine receptors and tyrosine kinases including ABL1, ABL2, CSF1R, JAK2 and PDGFRB (Den Boer et al., 2009; Mullighan et al., 2009b; Harvey et al., 2010; Roberts et al., 2012; Roberts et al., 2014). These findings are of clinical importance as proliferation of leukemic cells harboring these alterations is inhibited by tyrosine kinase inhibitors, and patients with Ph-like ALL refractory to conventional chemotherapy have exhibited profound and durable responses to respective tyrosine kinase inhibitors (Lengline et al., 2013; Weston et al., 2013; Roberts et al., 2014).
These studies identified several cases of Ph-like ALL with rearrangements of the erythropoietin receptor gene EPOR. EPOR forms a homodimeric receptor for erythropoietin, and EPOR signaling is required for normal erythroid development (Watowich, 2011). Normal B-progenitor cells do not express EPOR, but EPOR expression has been described in ETV6-RUNX1-positive B-ALL (Torrano et al., 2011) and in rare cases of B-progenitor ALL with chromosomal translocation t(14;19)(q32;p13) (Russell et al., 2009b; Russell et al., 2014). Together, these observations suggest that EPOR rearrangements are a recurring event that may be restricted to Ph-like ALL, and that deregulated EPOR signaling may drive leukemogenesis. In this study we sought to determine the prevalence and genomic structure of EPOR rearrangements, to define their role in lymphoid transformation and to examine the potential for inhibition of EPOR signaling in the treatment of EPOR-rearranged ALL.
RESULTS
Recurrent EPOR rearrangements in Ph-like ALL
To define the prevalence of EPOR rearrangements, we analyzed 3115 cases of childhood, adolescent and young adult B-ALL, 212 of which had a gene expression profile of Ph-like ALL. Analysis of genome and/or transcriptome sequencing data identified 19 cases with EPOR rearrangements, including 9 cases previously described (Roberts et al., 2012; Roberts et al., 2014), representing 8.9% of Ph-like ALL (Supplemental Experimental Procedures). Analysis of RNA-sequencing data generated by the Pediatric Cancer Genome Project showed that EPOR rearrangements were exclusively observed in Ph-like ALL. Genetic alterations of CDKN2A/CDKN2B, IKZF1 and PAX5 were common in EPOR-rearranged cases (Figure S1A and Table S1).
EPOR rearrangements result in expression of truncated receptors
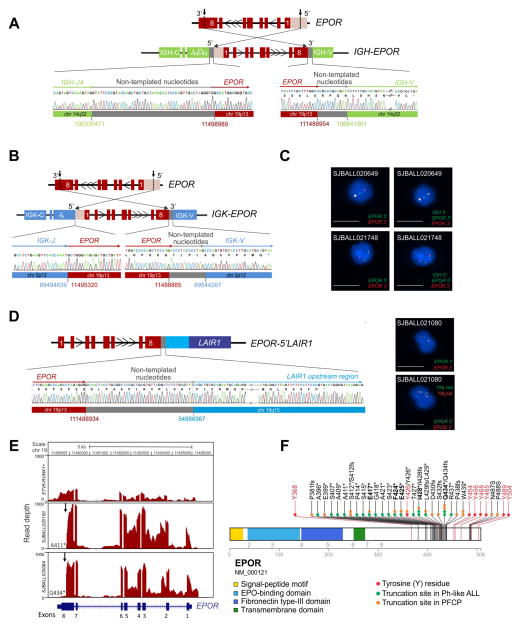
We identified four types of EPOR rearrangement: insertion of the EPOR locus distal to the immunoglobulin heavy chain (IGH) locus enhancer (14 cases; Figure 1A and Figure S1B); insertion of EPOR into the immunoglobulin kappa chain (IGK) locus (2 cases; Figure 1B)(Roberts et al., 2014); a reciprocal t(14;19)(q32;p13) rearrangement of IGH to EPOR (2 cases, Figure 1C and S1C) (Russell et al., 2009b; Roberts et al., 2014); and an intrachromosomal inversion of chromosome 19 that juxtaposes EPOR to the upstream region of LAIR1 (leukocyte-associated immunoglobulin-like receptor 1) at chromosome 19q13 (Table S1, Figure 1D and Figure S1D). All rearrangements were confirmed by tumor DNA and cDNA PCR and sequencing, and for reciprocal IGH-EPOR and EPOR-LAIR1 rearrangements, fluorescent in situ hybridization (FISH). FISH analysis showed the rearrangements to be clonal, consistent with the rearrangement being acquired early in leukemogenesis. Whole genome sequencing of matched non-tumor samples from two cases showed the alterations to be somatic.
Figure 1. EPOR rearrangements in Ph-like ALL.
(A, B) Schematic representation of cryptic insertion of EPOR within the immunoglobulin heavy chain (IGH) and kappa chain (IGK) loci in SJBALL020100 (A) and SJBALL020084 (B) and Sanger sequencing results. (C) FISH assay demonstrating rearrangement of EPOR in SJBALL020649 and SJBALL021748. Scale bars represent 10 μm. (D) Schematic representation of the fusion between EPOR exon 8 (red) and the LAIR1 upstream region (blue) in SJBALL021080 and Sanger sequencing electropherogram (left) and FISH assay of EPOR (right). The two right panels show the same cell with bottom panel having additional probes bracketing the 19q break point. Scale bars represent 10 μm. (E) Plot of read depth from RNA-seq showing EPOR expression in a representative case of ETV6-RUNX1-positive ALL and two cases of EPOR-rearranged ALL. Arrows indicate the EPOR breakpoints. The last translated amino acids, A411 and Q434 respectively, are shown following by the * sign. (F) Location of EPOR truncations occurring in Ph-like ALL and in PFCP (from http://www.erythrocytosis.org/scid/polycythemias_en/) (Kralovics and Prchal, 2001; Bento et al., 2014). Common truncation sites between Ph-like ALL and PFCP are in bold. Key tyrosine (Y) residues in EPOR are in red (NP_000112.1). See also Figure S1, Table S1 and Table S2.
All cases exhibited increased expression of all EPOR exons with the exception of the terminal portion of the last exon, exon 8 (Figure 1E and Figure S1E). Reverse transcription and genomic PCR of tumor RNA and DNA showed that distal EPOR rearrangement breakpoints were clustered in a 42 amino acid region of the intracytoplasmic tail of EPOR, with transcriptional read through into the distal immunoglobulin and LAIR loci appending additional amino acid residues to the truncated EPOR C-terminus (Table S2). Remarkably, these truncations were located at the same residues that are mutated in primary familial congenital polycythemia (PFCP), an inherited disorder in which germline EPOR frameshift and nonsense mutations truncate the C-terminal tail of the receptor (Figure 1F) (de la Chapelle et al., 1993; Bento et al., 2014; Gross et al., 2014).
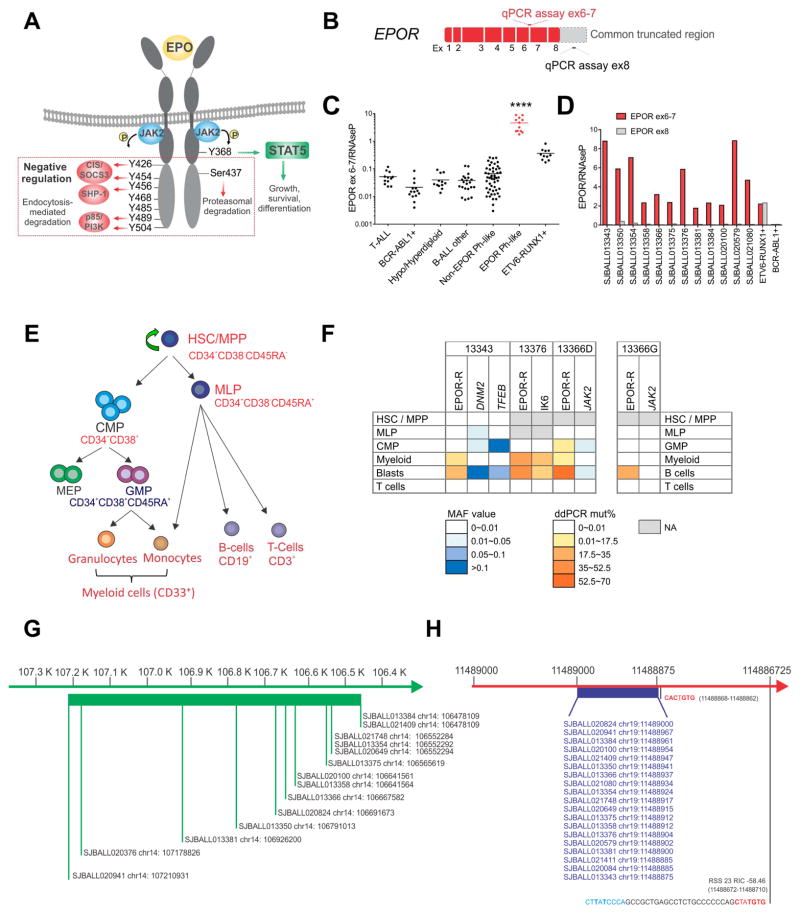
The intracytoplasmic tail of EPOR contains 8 tyrosine residues, phosphorylation of which mediates activation, negative regulation, and degradation of the receptor (Huang et al., 2010; Watowich, 2011). The most proximal residue, Y368, is required for docking of STAT5 and activation of JAK-STAT signaling upon binding of EPO (Figure 2A). Distal tyrosine residues, particularly Y426, Y454, Y456 and Y504, are required for negative regulation of receptor signaling mediated by suppressor of cytokine signaling 3 (SOCS3) and phosphatidylinositol-3-kinase (PI3K) p85-mediated ubiquitination, internalization and degradation of the receptor (Bulut et al., 2013). As in PFCP, all EPOR rearrangements preserved the most proximal tyrosine required for EPOR activation, but removed most or all of the distal tyrosine residues (Figure 2A). This was confirmed by quantitative real-time PCR assay that measured differential EPOR expression proximal and distal to the truncation (Figure 2B–D). EPOR-rearranged cases and ETV6-RUNX1 ALL cases with expression of full length EPOR exhibit elevated EPOR expression using an exon 6–7 junction assay proximal to the site of rearrangement (Figure 2C). However, EPOR-rearranged cases show negligible expression distal to the rearrangement using an exon 8 specific assay (Figure 2D). As many EPOR rearrangements are not detectable by conventional karyotyping or FISH, this provides a convenient screening test to identify EPOR rearrangements.
Figure 2. EPOR expression.
(A) A scematic representation of EPOR, its regulation, and its signaling. The red box indicates the cytoplasmic part of the receptor that is lost with EPOR-rearrangements. (B) A schematic representation of EPOR mRNA and regions for quantitative RT-PCR for its expression, the junction of exons 6 and 7 (red) in the common retained EPOR region and within exon 8 (grey) distal to the truncation. (C) EPOR expression was assessed in 134 cases from different ALL subtypes including T-lineage ALL (12), BCR-ABL1 ALL (14), hypodiploid B-ALL (7), hyperdiploid B-ALL (5), B-other ALL (22), non-EPOR Ph-like (51), Ph-like EPOR-rearranged (12) and ETV6-RUNX1 ALL cases (11).. The mean expression is shown by the horizontal line in the scatter dot plot. (D) Comparison of expression levels of EPOR exon 6–7 v. exon 8. An ETV6-RUNX1-positive ALL case is used as positive control for the expression of both exon 6–7 and exon 8 as it expresses full-length EPOR; a BCR-ABL1-positive ALL case is used as negative control. Results show the mean from two replicates. (E) Schematic representation of human hematopoietic hierarchy showing the hematopoietic stem cell (HSC) and the multipotent progenitor (MPP) fraction (CD34+CD38−CD45RA−), the multilymphoid progenitor (MLP) cell fraction (CD34+CD38−CD45RA+), the common myeloid progenitor (CMP) population (CD34+CD38+CD7−CD10−), which includes the megakaryocyte erythroid progenitor (MEP) and the granulocyte monocyte progenitor (GMP) (CD34+CD38+CD7−CD10−CD135+CD45RA+) and the mature cells: myeloid cells (CD33+) including granulocytes and monocytes, B-cells (CD45brightCD19+) and T-cells (CD3+). In the scheme all cell populations highlighted in red were sorted and analyzed for EPOR rearrangements and additional alterations. Additional population sorted only in the remission sample (GMP fraction) is shown in blue. Blasts were defined as CD19+CD45dim. (F) Summary of the presence of EPOR rearrangements and additional somatic alterations in isolated stem/progenitor, mature and blast cell populations from 3 EPOR-rearranged Ph-like ALL cases at diagnosis (left) and at remission from 1 case (right) as determined by MiSeq sequencing and ddPCR. All putative missense and silent mutations identified by analysis of RNA-seq data without matching germline data that were also identified by MiSeq in normal T-cells were considered germline and not included in this schema. (G) Schematic representation of the breakpoints occurring in EPOR-rearranged Ph-like ALL in the IGH locus. They span a region of 732.8 kb. (H) Schematic representation of the breakpoints occurring in EPOR-rearranged Ph-like ALL in EPOR exon eight. They are clustered in a hot spot region including 125 nucleotides with a semi-conserved heptamer immediately downstream to all breakpoints. Recombination signal sequences (RSS) heptamer sequences are shown in red and nonamer sequences are in blue. Abbreviations: mut, mutation; het, heterozygous, homo, homozygous; neg, negative; MAF, minor allele frequency; ddPCR, droplet digital PCR; ex: exon; ****, p<0.0001. See also Figure S2, Table S3 and Table S4.
EPOR rearrangements may arise in a progenitor with myeloid-lymphoid potential
The identification of a rearrangement of a gene encoding a cytokine receptor essential for erythroid development in the B lymphoid lineage, and the lack of erythroid features (e.g. glycophorin A expression, data not shown) of leukemic cells suggested acquisition of rearrangements in a lymphoid or multipotent progenitor. To directly address the lineage potential of EPOR rearranged cells, we isolated leukemic, stem cell, progenitor and mature cell populations from leukemic samples of three EPOR-rearranged ALL cases (Figures 2E and Figure S2A) and tested for EPOR rearrangements, IKZF1 deletions and mutations identified in each case (Table S3). EPOR rearrangements were identified in blast cells as expected, but also in myeloid (CD33+) cells in all cases, and myeloid progenitors in one case (SJBALL013366). Interestingly, analysis of sorted populations from a sample collected at the time of remission (SJBALL013366_G1) showed positivity for EPOR rearrangement in normal B-cells. Although sort purity was high for all samples, it was important to exclude leukemic cell contamination. Case SJBALL013343 harbored somatic DNM2 and TFEB mutations in blast cells that were absent in myeloid cells, suggesting identification of EPOR rearrangements in these cells was not due to contamination (Figure 2F and Figure S2B). Similarly, high percentages of mutant EPOR and IKZF1 exon 4–7 deletion (IK6) alleles in myeloid cells but not in other cell populations in case SJBALL13376 (Figure 2F) indicate these alterations were present in myeloid and lymphoid cells. A third case (SJBALL013366) with rearranged EPOR in myeloid progenitors lacked additional clonal genetic alterations in tumor cells to compare with progenitor populations and exclude contamination. Due to the high blast percentages we were not able to sort every possible progenitor population, including hematopoetic stem cell (HSC) and multipotent progenitor (MPP) cells in two samples. Thus, while the exact progenitor in which the rearrangement arises is unknown, our data indicate that EPOR rearrangements arise in a primitive progenitor with the capacity for myelo-lymphoid differentiation (Doulatov et al., 2010).
Recombination-activating gene (RAG) -mediated recombination drives the generation of many structural variations in ALL (Mullighan et al., 2008; Papaemmanuil et al., 2014). During lymphocyte development, the RAG1 and RAG2 endonucleases bind and cleave DNA at recombination signal sequences (RSS) motifs, consisting of conserved heptamer and nonamer sequences separated by a spacer region of 12 bp (12RSS) or 23 bp (23RSS) (Jung et al., 2006). Once cleaved the DNA ends are ligated by the RAG complex and non-templated nucleotides (NTN) are added by the terminal deoxynucleotidyl transferase DNTT (Teng et al., 2015). Although RAG activity is maximal at the proB/pre-B (Hardy stage B–D) stage (Figure S2C) (Hardy and Shinton, 2004; Holmfeldt et al., 2013), a subset of mouse common lymphoid progenitors (CLP) and human multilymphoid progenitors (MLP) expresses RAG (Figure S2D) (Mansson et al., 2010) (Zandi, Dick et al in preparation). RSS motifs and NTN were detected near the breakpoints in IGH/IGK and EPOR (Table S4 and Figure 2G–H). The expression of both EPOR and RAG in cells with myelo-lymphoid potential and the presence of myeloid cells harboring the EPOR rearrangements provide strong evidence for RAG-mediated rearrangement of EPOR in an early hematopoietic progenitor that then acquires secondary genetic alterations and follows a lymphoid developmental fate (Figure S2D).
EPOR truncation results in stabilization of receptor expression and hypersensitivity to EPO stimulation
Rearrangements or sequence mutations resulting in deregulated expression of cytokine receptor genes including CRLF2 and IL7R are recognized in ALL (Mullighan et al., 2009a; Russell et al., 2009a; Zenatti et al., 2011), however the functional consequences of receptor truncation are unknown. We hypothesized that these rearrangements may perturb receptor internalization and degradation, cell surface expression and thus augment JAK-STAT signaling, either constitutively or in response to ligand stimulation. To examine this, we generated constructs that expressed multiple truncated EPOR that terminated at the breakpoint of EPOR in exon 8 (EPOR*) or truncated EPOR with the amino acids translated from read through into IGH/IGK (EPOR-IGH*/-IGK*). Immunoblotting and RT-PCR of human leukemic cells and mouse Ba/F3 hematopoietic cells expressing these constructs confirmed that cells expressing truncated EPOR incorporate the additional amino acids from immunoglobulin read through (Figure S3A).
We expressed wild-type (WT) and truncated EPOR in mouse hematopoietic Ba/F3 and Arf−/− pre-B cells (Williams et al., 2006) that require exogenous cytokine (interleukin-3 (IL-3) and IL-7, respectively) to sustain proliferation. We confirmed cell surface expression of both wild-type and truncated EPOR (Figure S3B). In contrast to cells transduced with empty vector, cells expressing either wild-type or truncated EPOR proliferated in a dose-dependent fashion to exogenous EPO, with no significant difference between EPOR* and EPOR-IGH*/-IGK* (Figure 3A and Figure S3C). We observed significantly faster proliferation of Arf−/− pre-B cells expressing truncated EPOR than cells expressing WT EPOR when cells were cultured in EPO (Figure S3D). Notably, cell surface expression was significantly higher for truncated EPOR than WT EPOR, with reduced down-regulation in response to ligand stimulation (Figure 3B). Ba/F3 cells expressing truncated EPOR exhibited augmented JAK-STAT signaling compared to cells expressing WT EPOR (Figure 3C), with enhanced and prolonged STAT5 phosphorylation in response to increased exposure to EPO (Figures 3D). Activation of JAK-STAT signaling was abrogated by the JAK1/2 inhibitor ruxolitinib (Figure 3E).
Figure 3. Expression of EPOR in IL-3 dependent Ba/F3 mouse hematopoietic cell lines.
(A) Ba/F3 cells were transduced with empty vector (MIG) or vectors that express the wild-type EPOR, EPOR*, or EPOR-IGH*/-IGK*. Cells were grown with or without IL-3 and with increasing concentrations of rhEPO (0.01 U/mL to 10 U/ml) and cumulative cell numbers were measured. Data are means ± S.D. from triplicates from two independent experiments. (B) Ba/F3 cells expressing the empty vector, wild-type or indicated truncated EPOR were stimulated with rhEPO and cell surface EPOR and pSTAT5 were assessed by flow cytometric analysis at indicated time points since stimulation. Bars express the mean ± S.D. from three independent experiments. (C) Flow cytometric analysis of STAT5 phosphorylation after rhEPO stimulation (rhEPO 1 U/ml for 15 minutes) in cells expressing indicated EPOR. Dotted lines represent EPOR-IGH*/IGK*. (D) Ba/F3 cells expressing wild-type or truncated EPOR were stimulated over time with rhEPO and pSTAT5 was assessed by flow cytometry. (E) Ba/F3 cells were untreated (−) or treated (+) with ruxolitinib (1 μM) for 1 hour and levels of phosphorylated STAT5 were assessed by phosphoflow cytometric analysis. Abbreviations: NF, no factor; rhEPO, recombinant human erythropoietin; min, minutes; ***, 0.0001 < p < 0.001; ****, p <0.0001. See also Figure S3.
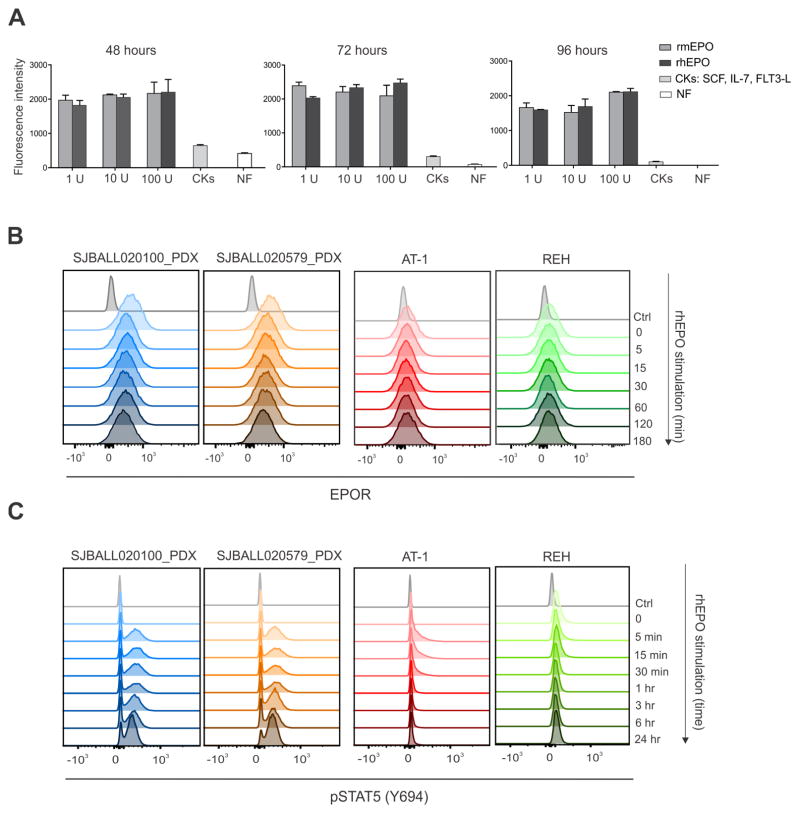
We next examined EPOR expression and signaling in human leukemic cells. We established two luciferase marked patient-derived xenografts (PDX) of EPOR-rearranged Ph-like ALL (Table S1, SJBALL020100 and SJBALL020579). Notably, proliferation of leukemic cells was sustained ex vivo by low concentrations of recombinant mouse or human EPO, but poorly by alternative cytokines (Figure 4A). We observed elevated receptor expression, reduced down regulation of receptor expression following ligand challenge, and augmented and sustained JAK-STAT signaling in EPOR-rearranged PDX cells (Figure 4B–C and Figure S4A). In contrast, the ETV6-RUNX1-positve ALL cell lines AT-1 and REH that express non-truncated EPOR transcripts (data not shown) showed low cell surface EPOR expression and minimal JAK-STAT activation following EPO stimulation (Figure 4B–C and Figure S4B), indicating that wild-type EPOR expression does not sustain leukemic cell proliferation in this subtype of B-ALL. To further explore signaling pathway activation upon EPO stimulation, we performed phosphosignaling analysis of the PI3K/mTOR and Ras-Raf-MEK-ERK signaling pathways. We observed augmented phosphorylation of the mTOR downstream target 4EBP1 and activation of the p44/42 MAPK (Erk1/2) signaling pathway upon EPO stimulation in EPOR-rearranged PDX cells but not in Ba/F3 cells expressing truncated EPOR (Figure S4C), indicating these pathways may also be inhibited to reduce leukemic cell proliferation.
Figure 4. Surface expression of EPOR and STAT5 phosphorylation in PDX cells and human B-ALL cell lines.
(A) EPOR-rearranged PDX cells were harvested and cultured ex vivo in media containing indicated concentrations of recombinant mouse erythropoietin (rmEPO) or recombinant human erythropoietin (rhEPO), a cytokine cocktail (IL-7, SCF, FLT3-L), or no exogenous cytokine (NF). Proliferation of PDX cells over time was measured. Data are means ± S.D. from triplicates from two independent experiments. (B) EPOR-rearranged PDX cells and human B-ALL cell lines known to overexpress EPOR were stimulated with rhEPO (10 U/ml) and cell surface EPOR was assessed by flow cytometric analysis at indicated time points after stimulation. (C) EPOR-rearranged PDX cells and human B-ALL cell lines known to overexpress EPOR were stimulated with rhEPO (10 U/ml) and phosphorylation of STAT5 was assessed by flow cytometric analysis at different time points since stimulation. Abbreviations: NF, no factor; CKs, cytokines; min, minutes; hr, hours. See also Figure S4.
Truncated EPOR induces B-progenitor ALL
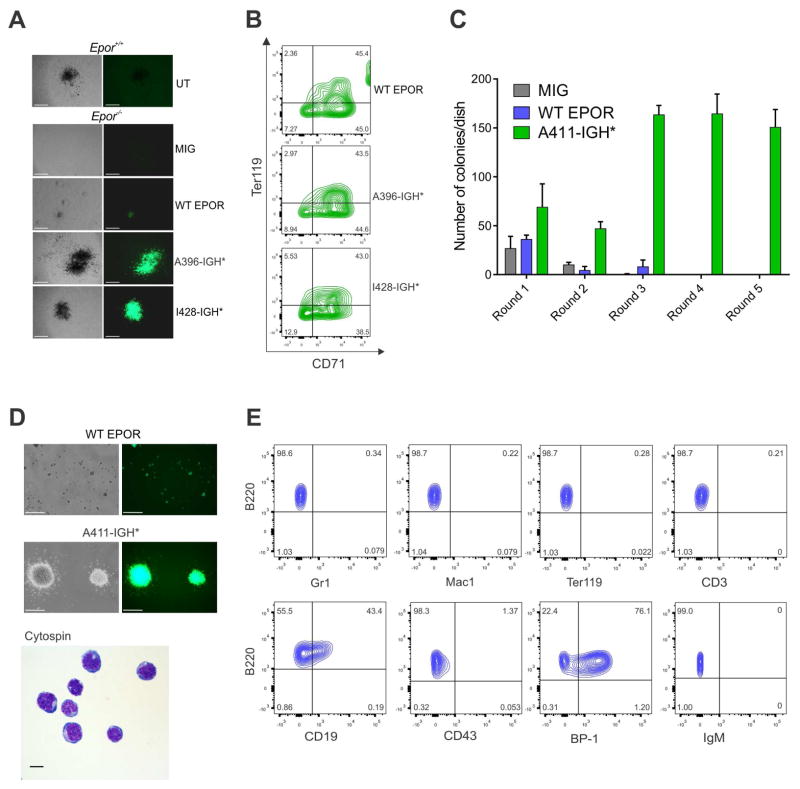
We examined the effect of expressing wild-type and truncated EPOR in mouse hematopoietic progenitors. As EPOR is required to sustain normal erythroid maturation, we expressed EPOR in Epor−/− fetal liver cells (Kieran et al., 1996). Expression of EPOR-IGH* resulted in larger size of burst-forming unit erythroid colonies in vitro than fetal liver cells expressing the WT EPOR (Figure 5A–B), consistent with the notion that the receptor truncation results in gain of function. To directly examine the role of truncated EPOR in leukemogenesis, we expressed empty vector, WT EPOR or EPOR A411-IGH* in C57Bl/6 lineage-negative bone marrow hematopoietic progenitors and assessed colony forming potential in vitro. Truncated EPOR resulted in increased and sustained growth of B-progenitor (B220+CD19+BP-1+) lymphoid colonies (Figure 5C–E).
Figure 5. Colony forming assay in Epor−/− fetal liver cells and clonogenic assay in lin- bone marrow cells.
(A) Embryonic day 12.5 Epor−/− fetal liver cells were isolated and transduced with empty vector (MIG) or vectors expressing WT EPOR or truncated EPOR (A396-IGH* or I428-IGH*) that are representative of the different locations of truncation in the receptor. Embryonic day 12.5 untransduced (UT) Epor+/+ fetal liver cells were isolated and used as positive control in the BFU-E assay. Colony morphology is shown. Scale bars represent 500 μm. (B) Erythroid colonies were harvested from culture dishes in (A), stained for CD71 and Ter119 antigens and analyzed by flow cytometry. (C) Clonogenic assays of lineage-negative mouse bone marrow cells expressing TR or WT EPOR or transduced with empty vector (MIG). Columns show means of three replicates ± S.D. (D) Representative colony morphology and cytospins from TR EPOR-expressing cells are shown. Scale bars represent 500 μm for colony morphology and 10 μm for cytospin. (E) Cells harvested from colony-forming assays after two or more replatings were subjected to flow cytometry and representative immunophenotype is showed.
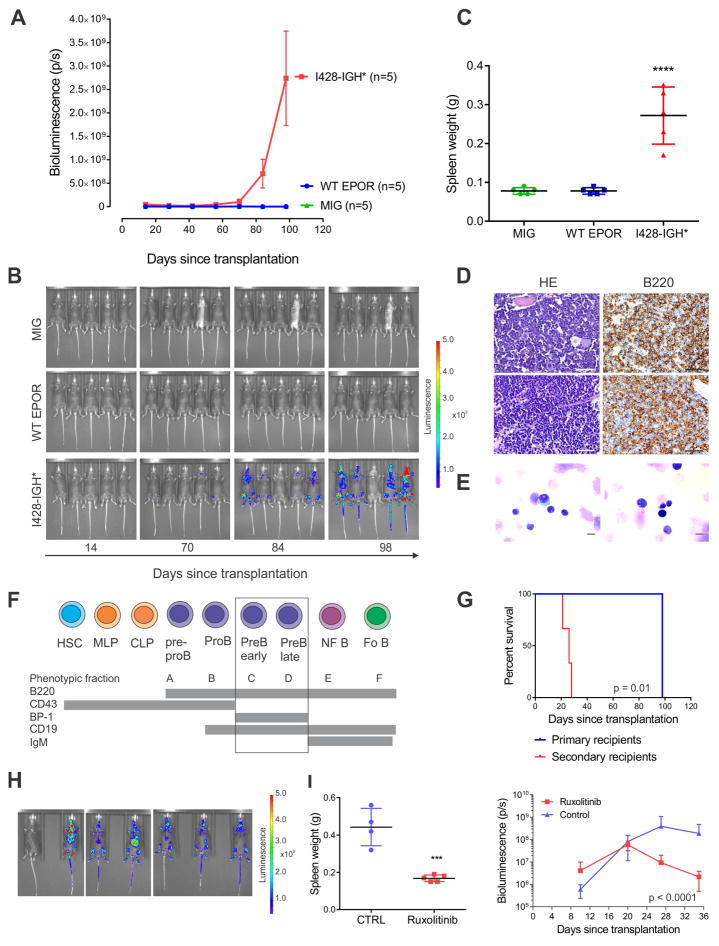
We next co-transduced Arf−/− pre-B cells with empty vector, WT EPOR, or EPOR I428-IGH*, together with a luciferase-expressing vector, and transplanted cells into sublethally irradiated C57Bl/6 mice. Recipient mice transplanted with EPOR I428-IGH* transduced cells, but not wild-type EPOR or empty-vector transduced cells developed B-ALL with a latency of approximately 3 months (Figure 6A–C). Leukemias were positive for B220 (Figure 6D–F) and negative for the erythroid markers (GATA1 and CD235a, data not shown). Leukemia was serially transplantable (Figure 6G–H, Figure S5A–B), demonstrating that the truncated EPOR are leukemogenic. Leukemia development was confirmed in sublethally irradiated C57Bl/6 mice transplanted with Arf−/− pre-B cells co-expressing EPOR I428-IGH* and the dominant negative Ikaros isoform IK6 with a latency of approximately two months (Figure S5C). Analysis of DNA copy number alterations in two tumors identified secondary genetic alterations involving an amplification of Jak2, confirming the central role of JAK-STAT signaling in leukemogenesis (Figure S5D). Leukemia cells from the tumor with amplification of Jak2 were harvested and transplanted in secondary sub lethally irradiated C57Bl/6 recipients for treatment with the JAK2 inhibitor ruxolitinib. Proliferation of leukemia cells in different tissues and organs were significantly attenuated in treated mice (Figure 6I and Figure S5E).
Figure 6. Leukemia development in mice transplanted with Arf−/− primary pre-B cells expressing truncated EPOR.
(A) Leukemia development in mice transplanted with indicated cells was assessed by bioluminescence analysis performed every two weeks for a total of 14 weeks. Results are means ± S.D. from five mice per group. (B) Bioluminescent images were captured after 14, 70, 84 and 98 days following transplantation from each group. One mouse in the truncated EPOR group did not show detectable engraftment. (C) Spleen weight at the end of study (112th day) in mice transplanted with indicated cells. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the S.D. ****, p < 0.0001. (D) Bone marrow sections from two representative C57Bl/6 mice (top and bottom row, respectively) transplanted with Arf−/− primary pre-B cells expressing truncated EPOR were stained with hematoxylin-eosin (HE) or labeled with an antibody specific for B220. Scale bars represent 50 μm. (E) Representative cytospins from two representative mice (right and left panels) with EPOR I428-IGH*. Scale bars represent 10 μm. (F) A schematic representation of Hardy stages (Hardy and Shinton, 2004) of cell surface markers expressed during B cell development. Abbreviations: HSC, hematopoietic stem cells; MLP, multilineage progenitor; CLP, common lymphoid progenitor; NF B, newly formed B cells; Fo B, mature follicular B cells. (G) Survival curves in primary and secondary recipients. (H) Bioluminescent images of secondary recipients 14 days following transplantation. (I) Spleen weight at the end of the treatment study (left panel) and the bioluminescence analysis results performed every week (right panel). The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the S.D. ***, 0.0001 < p < 0.001. See also Figure S5.
Synergism of JAK-STAT inhibition and chemotherapy in EPOR-rearranged ALL
To examine EPO sensitivity and the potential for therapeutic targeting of human EPOR-rearranged leukemic cells, EPOR-rearranged PDX cells were transduced with lentiviral vectors expressing yellow fluorescent protein and luciferase to enable in vivo imaging. Mice were randomized to receive the JAK1/2 inhibitor ruxolitinib and/or recombinant human EPO (darbepoietin alpha), or vehicle. Consistent with the in vitro data described above, we observed increased proliferation of leukemic cells in the presence of exogenous EPO. Ruxolitinib significantly attenuated proliferation of leukemic cells in vivo (Figure 7A–C), and multiple JAK inhibitors attenuated proliferation and STAT5 activation of xenografted EPOR-rearranged leukemic cells ex vivo (Figure 7D–E). Notably, these inhibitors exhibited synergy with dexamethasone, vincristine and daunorubicin, chemotherapeutic agents routinely used in ALL, including a xenograft that exhibited resistance to dexamethasone (Figures S6A–B). Significant synergy between dexamethasone and ruxolitinib was also observed in vivo (Figures 7F–H).
Figure 7. Sensitivity of EPOR-rearranged Ph-like ALL to JAK1/2 inhibition.
(A) In vivo response of an EPOR-rearranged xenograft to ruxolitinib, darbepoietin alpha, combination ruxolitinib and darbepoetin or vehicle (5 mice per group). Error bars represent means ± S.D. P values are from ANOVA test and show the results from the comparison of each treated group versus the control group. (B) In vivo luciferase imaging showing leukemia growth in mice treated with ruxolitinib compared to control mice. (C) Spleen weights of mice treated as indicated at the end of the study. P values are from ANOVA test and show the results from the comparison of each treated group versus the control group. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the S.D. (D) Ex vivo cytotoxicity assays of IGH-EPOR human leukemic cells (YFP+CD45+CD19+) harvested from xenografted mice and treated with imatinib and the JAK inhibitors ruxolitinib, fedratinib, momelotinib or pacritinib. Error bars represent means ± S.D. (E) Ex vivo phospho-STAT5 signaling analysis in engrafted leukemia cells (YFP+CD45+CD19+) treated with JAK1/2 inhibitors (1 μM for 1 hour). (F) Ex vivo combinatorial studies of JAK1/2 inhibitor and chemotherapeutic agents in two EPOR-rearranged PDX models. Plot of combination index values are from Calcusyn analysis. (G) In vivo response of two EPOR-rearranged xenografts to ruxolitinib, dexamethasone, combination of ruxolitinib and dexamethasone or vehicle (5 mice per group). Error bars represent means ± S.D. P values are from ANOVA (Tukey’s multiple comparisons test). (H) Spleen weights in mice treated as indicated at the end of the study. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the S.D. Abbreviations: EPO, erythropoietin; CTRL, vehicle treated control mice; Dauno, Daunorubicin; VNC, Vincristine; Dex, Dexamethasone. EPO, erythropoietin; CTRL, vehicle treated control mice; ruxo, ruxolitinib; **, 0.001 < p < 0.01; ***, 0.0001 < p < 0.001; ****, p < 0.0001. See also Figure S6.
These results suggest that JAK-STAT inhibition is a logical therapeutic approach in EPOR-rearranged ALL. In support of this, we report a 25 year-old male with refractory EPOR-rearranged Ph-like ALL (SJBALL021748). Following multiple salvage regimens, he received ruxolitinib 200 mg twice per day as single agent therapy in a Phase I/II clinical trial aimed to determine safety of ruxolitinib in patients with relapsed/refractory acute myeloid leukemia or ALL (Pemmaraju et al., 2015). The circulating leukemic cell count fell from 85% to a nadir of 15.9% at one week, followed by subsequent progression (Figure S6C).
DISCUSSION
These results demonstrate the high frequency of EPOR rearrangements in Ph-like precursor B-ALL and identify a mechanism of cytokine receptor deregulation in leukemia. A notable finding was the similarity in the position of EPOR truncations arising from somatic rearrangement to the inherited truncating mutations in PFCP. Although the presence of the rearrangement in cell populations in the non-leukemia samples obtained at remission in one patient raised the possibility that the EPOR rearrangements in ALL may also be germline events, there was no evidence of the rearrangement in non-leukemic T cells or whole genome sequencing data of remission material. This suggests that the rearrangement was present in a residual preleukemic progenitor in this case, rather than the germline. While the EPOR rearrangements were different from the inherited EPOR mutations observed in PFCP, both result in similar receptor truncations and activation. The rearrangements and receptor truncation facilitate EPOR signaling and leukemogenesis by multiple mechanisms: deregulation of EPOR gene expression, stabilization and impaired down-regulation of cell surface EPOR in response to ligand stimulation, and heightened JAK-STAT signaling. The dependence of B-progenitor leukemic cell proliferation on EPOR signaling extends prior studies implicating “hijacking” of cytokine receptor signaling in the pathogenesis of B-cell leukemia including CRLF2 (Mullighan et al., 2009a; Russell et al., 2009a), CSF1R (Roberts et al., 2014), IL7R (Shochat et al., 2011; Zenatti et al., 2011; Shochat et al., 2014) and PDGFRB (Roberts et al., 2012; Roberts et al., 2014). However, these previously reported examples involve deregulated expression of a non-truncated receptor (e.g. CRLF2), the formation of a chimeric fusion that deregulates the kinase domain of the receptor (e.g. CSF1R and PDGFRB) or mutations that stabilize and activate the receptor (e.g. IL7R). In contrast, the EPOR rearrangements couple deregulated expression with impaired negative regulation, rather than constitutive activation of a receptor tyrosine kinase domain. This mechanism of cytokine receptor truncation and deregulation has not been identified on analysis of other pediatric tumor transcriptomes, and appears restricted to EPOR-rearranged ALL. However, truncating rearrangements of oncogenes have been identified in other tumors, such as MYB in low grade glioma (Zhang et al., 2013). Moreover, the complex rearrangement of EPOR into repetitive genomic regions at immunoglobulin loci is not detected by any of RNA-sequencing analysis algorithms, suggesting that careful analysis of data from other hematologic and solid tumors is warranted. Truncated EPOR showed minimal constitutive signaling in cell lines but markedly augmented JAK-STAT signaling and cell proliferation in mouse and human EPOR-rearranged cells in response to exogenous erythropoietin. These findings suggest that tonic stimulation by EPO, rather than ligand-autonomous signaling, sustains leukemic cell proliferation, and raises the possibility that the anemia resulting from bone marrow replacement by lymphoblasts in ALL may result in further EPO-driven augmentation of tumor cell growth.
Several observations indicate that the EPOR rearrangements are driver events that are acquired early during leukemogenesis and are logical targets for therapeutic targeting. The alterations were clonal, and retained during disease progression (SJBALL013376, SJBALL021748). This is in contrast to other classes of genetic alterations, particularly sequence mutations of the Janus kinase genes, that are commonly secondary sub- or multi-clonal events and that may raise and fall during disease evolution (Ma et al., 2015). Secondly, using mutation-specific assays of purified cell populations at diagnosis and remission, we have shown that the EPOR alterations, but not sequence mutations, must have arisen in a progenitor with myeloid-lymphoid fate due to the presence of EPOR rearrangements in both leukemic blast and non-leukemic myeloid cells. The frequency of such cells may be low and precise identification would require the analysis of much greater amounts of primary patient material than was obtainable. The cell of origin of B-ALL has previously been considered to be a lymphoid-restricted cell but has more recently been postulated to be a multipotent progenitor, as suggested for ETV6-RUNX1-positive ALL from studies of monozygotic twins (Alpar et al., 2015), our study supports this conclusion. Sequencing of the genomic breakpoints of the EPOR rearrangements provided compelling evidence that the alterations arise from RAG-mediated recombination, and gene expression profiling demonstrates RAG expression in mouse and human progenitor cell populations, supporting acquisition of the EPOR rearrangement in a hematopoietic progenitor that then acquires additional alterations such as deletion of the CDKN2A/B tumor suppressor loci that contribute to the establishment of the leukemic clone.
Importantly, we provide direct evidence that these rearrangements result in leukemogenic gain of function alleles. Expression of truncated EPOR resulted in enhanced colony formation in Epor-null fetal liver cells, enhanced proliferation of Arf−/− mouse pre-B cells, and sustained replating of lymphoid colonies in vitro. Definitive evidence of leukemogenicity was provided by establishment of a serially transplantable lymphoid leukemia that recapitulated features of human pre-B ALL.
Our findings provide the rationale for clinical identification of EPOR rearrangements and therapeutic targeting with agents directed against the activated signaling pathways, particularly with JAK inhibitors, but potentially with agents that also target parallel signaling pathways. Identification of EPOR rearrangements is challenging due to the diversity of rearrangement partner genes and breakpoints, and the cryptic nature of the focal (~7 kb) insertions of EPOR on karyotyping and FISH. We have shown that flow cytometry demonstrates increased expression of EPOR on leukemic cells, and developed quantitative gene expression assays proximal and distal to the site of EPOR rearrangement that sensitively and specifically identifies EPOR-rearranged cases, providing a reliable diagnostic test to easily screen leukemia cases for EPOR rearrangements.
Finally, although we previously reported the sensitivity of PDX cells with EPOR-rearrangement to ruxolitinib ex vivo (Roberts et al., 2014), here we extended the analysis of the efficacy of JAK-STAT inhibition in model cell lines, engineered mouse leukemias and human xenografts (ex vivo and in vivo). Importantly, we demonstrate potent synergism with conventional chemotherapeutic agents in human leukemic cells in vitro and in vivo. Ph-like ALL, including EPOR-rearranged ALL, is commonly refractory or poorly responsive to conventional chemotherapy. We have shown that dexamethasone-resistant EPOR-rearranged ALL cells are exquisitely sensitive to combinatorial therapy, and moreover, partial responses to ruxolitinib are potentiated with chemotherapy. The importance of combinatorial therapy is highlighted by the partial response of a case of relapsed ALL treated empirically with ruxolitinib that was subsequently found to have an EPOR rearrangement.
Together, these findings provide a comprehensive description of the genomic and biological basis of a subtype of B-progenitor ALL characterized by a unique form of cytokine receptor deregulation. This is emphasized by the parallel between the somatic EPOR truncating rearrangements in B-ALL and the germline truncating mutations in PFCP. This further highlights the remarkable interplay of germline and somatic events in hematological disorders, and suggests that ongoing sequencing efforts in ALL are likely to yield additional key insights and therapeutic vulnerabilities in this disease.
EXPERIMENTAL PROCEDURES
Patients and genome sequencing
Analysis of 3115 B-ALL cases, including genome and/or RNA-sequencing of 212 Ph-like ALL cases was performed. Their detailed molecular and clinical information are reported in the Supplemental Experimental Procedures. Library construction was performed using Truseq exome capture baits and RNA-sequencing library preparation kits (Illumina), and sequencing was performed on the HiSeq 2500. RNA-seq data were mapped using StrongArm and rearrangements were identified using CICERO (Roberts et al., 2014). The study was approved by the St Jude Institutional Review Board, and all patients and/or their parents/guardians provided informed consent and/or assent.
Hematopoietic progenitor cell assays
Hematopoietic cell populations from three EPOR-rearranged Ph-like cases (SJBALL013376, SJBALL013366, and SJBALL013343) were isolated by fluorescent activated cell sorting, including the HSC-MPP fraction (CD34+CD38−CD45RA−), the MLP (CD34+CD38−CD45RA+), the common myeloid progenitor (CMP) populations (CD34+CD38+CD7−CD10−) including the granulocyte monocyte progenitor (GMP) (CD34+CD38+CD7−CD10−CD135+CD45RA+) and the mature fractions: myeloid cells (CD33+) including granulocytes and monocytes, B-cells (CD45brightCD19+) and T-cells (CD3+). Genomic PCR and digital droplet PCR (ddPCR; RainDrop Digital PCR System, RainDance Technologies) were performed in each population using primers specific for the EPOR rearrangement and for additional genetic alterations, including IKZF1 deletions and missense mutations identified in each case from analysis of transcriptome sequencing data.
In vitro functional studies
Wild-type and EPOR-rearranged cDNAs were cloned in the mouse stem cell virus- internal ribosome entry site - green fluorescent protein (MSCV-IRES-GFP) retroviral vector and expressed in IL-3-dependent mouse hematopoietic Ba/F3 cells and IL-7-dependent pre-B cells harboring alterations of Arf with or without the dominant negative IKZF1 allele IK6. Cell proliferation and downstream signaling were examined in the absence or presence of recombinant human erythropoietin. Surface expression of the WT or truncated EPOR was analyzed in Ba/F3 cells, Arf−/− pre-B cells, B-ALL leukemia cell lines (REH and AT-1) and EPOR-rearranged PDX cells by staining non-permeabilized cells with allophycocyanin (APC)-conjugated mouse monoclonal anti-human EPOR antibody (FAB3072A, R&D Systems). For intracellular signaling, cells (0.6×106 per well) were starved for 16 hours in RPMI-1640 supplemented with 0.5% BSA, stimulated with EPO (1–10 U/ml) for 5 minutes to 24 hours, fixed, permeabilized and stained with anti-phosphot-STAT5 (pY694)-Ax647 (BD Biosciences), pS6 Ribosomal (2211L), pAkt (Thr 308; 9275L), pAkt (Ser473; 4060L), p4E-BP1(T37/46; 2855L), or p44-42 MAPK (T202/Y204; 9101L) from Cell Signaling. Cellular fluorescence data were collected on an LSR II flow cytometer (BD Biosciences) using DIVA software (BD Biosciences), and analyzed with FlowJo v.10.0 (Tree Star).
Mouse modeling of EPOR-rearranged leukemia
Sublethally irradiated 8 week-old wild-type C57Bl/6 female mice were transplanted by tail vein injection of IL-7 dependent Arf−/− pre-B cells transduced with retroviral supernatants expressing MSCV-EPOR-IGH*-IRES-GFP, MSCV-EPOR-WT-IRES-GFP or empty MSCV-IRES-GFP, and MSCV-luciferase-IRES-mCherry. Leukemia development was monitored over time by biweekly measurement of bioluminescence at Xenogen IVIS-200 (PerkinElmer). Region of interest (ROI) measurements and total fluxes (photons/second, p/s) were recorded and analyzed by the Living Imaging v.4.4 software (Caliper Life Sciences). Animals that became moribund were euthanized, and blood, bone marrow, and spleen samples were analyzed for evidence of leukemia, using automated complete blood count analyses, morphology, flow cytometry, and histopathologic analysis.
Two luciferase marked PDX models were established in NSG mice by tail vein injection of human primary EPOR-rearranged leukemia cells transduced with a lentiviral vector (vCL20SF2-Luc2a-YFP) expressing luciferase and yellow fluorescent protein. EPOR expression, EPOR signaling, EPO-dependent proliferation and sensitivity to the JAK1/2 inhibitors were assessed ex vivo and in vivo. For in vivo drug treatment studies the ruxolitinib rodent chow was manufactured from LabDiet #5002 Rodent Chow and INCB18424 Phosphate (ruxolitinib) in the ratio of 2 g of ruxolitinib to 1 kg of chow. The study was approved by the St Jude Children’s Research Hospital Animal Care and Use Committee. Mice were housed in an American Association of Laboratory Animal Care (AALAC)-accredited facility and were treated on Institutional Animal Care and Use Committee (IACUC)-approved protocols in accordance with NIH guidelines.
Statistical analysis
Data analyses were performed using Prism Version 6.0 (GraphPad). Data are presented as mean ± S.D. Significance was determined using student’s t test and ANOVA (with Tukey and Dunnet tests as appropriate). Kaplan-Meier analysis and the Mantel-Cox log rank test were used for survival data. A p value less than 0.05 was used to define statistical significance.
Other Procedures
All methods are described in detail in the Supplemental Experimental Procedures.
Supplementary Material
SIGNIFICANCE.
Truncations of the erythropoietin receptor represent a mechanism of cytokine receptor deregulation in leukemia. Multiple, frequently cryptic rearrangements of EPOR are present in high-risk ALL that define a specific subgroup of Ph-like ALL. Each rearrangement juxtaposes EPOR to enhancer regions in immunoglobulin and immunoglobulin-like genes and result in deregulated EPOR expression, hypersensitivity to EPO, enhanced JAK-STAT signaling and sensitivity to JAK-STAT inhibition. The clinical importance of these findings is further emphasized by the significant synergy between JAK-STAT inhibition and chemotherapeutic agents in inhibiting proliferation of these leukemic cells. The potential for JAK-STAT inhibition represents an attractive therapeutic prospect for patients with high-risk B-ALL who are often refractory to conventional therapy.
HIGHLIGHTS.
Multiple EPOR rearrangements are recurrent in high risk acute lymphoblastic leukemia
EPOR rearrangements truncate the C-terminus of EPOR and stabilize the receptor
Truncated EPOR is hypersensitive to EPO stimulation
Truncated EPOR are leukemogenic and sensitive to JAK-STAT inhibition
Acknowledgments
This work was supported in part by the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital; by a Stand Up to Cancer Innovative Research Grant and St Baldrick’s Foundation Scholar Award (to Dr Mullighan); by a St. Baldrick’s Consortium Award (to Dr. Hunger), by a Leukemia and Lymphoma Society Specialized Center of Research grant (to Drs. Hunger and Mullighan), by a Lady Tata Memorial Trust Award (to Dr Iacobucci), by a Leukemia and Lymphoma Society Special Fellow Award and Alex’s Lemonade Stand Foundation Young Investigator Awards (to Dr Roberts), and by National Cancer Institute Grants CA21765 (St Jude Cancer Center Support Grant), U01 CA157937 (to Drs Willman and Hunger), U24 CA114737, U10 CA180820 (ECOG-ACRIN Operations) and CA180827 (to Dr Paietta); U10 CA101140 (The Alliance Leukemia Correlative Science); CA145707 (to Drs. Willman and Mullighan); and grants to the Children’s Oncology Group: U10 CA98543 (Chair’s grant and supplement to support the COG ALL TARGET project), U10 CA98413 (Statistical Center), and U24 CA114766 (Specimen Banking).
We thank the staff of the St Jude Biorepository, the Hartwell Centre for Bioinformatics and Biotechnology, the Flow Cytometry and Cell Sorting Core Facility, the Cell and Tissue Imaging Facility, the Animal Resources Center and the Small Animal Imaging Center of St. Jude Children’s Research Hospital. We thank Incyte Corporation for providing ruxolitinib, Drs E. Parganas and Dr J. Ihle for providing Epor−/+ Balb/c mice, and Amgen Inc. for providing the rabbit monoclonal antibody specific to human EpoR A82.
Footnotes
Accession Numbers
Affymetrix single nucleotide polymorphism microarray (SNP6) and RNA-sequencing data are deposited at the European Genome Phenome archive, accession EGAS00001000654. Data from array-based comparative genomic hybridization of mouse tumors are deposited in Gene Expression Omnibus (GEO) repository under accession code GSE76061. Mouse Affymetrix gene expression data from Hardy B cell progenitor fractions are deposited in GEO under accession code GSE38463.
AUTHOR CONTRIBUTIONS
I.I., S.M.D., and C.G.M. designed experiments. I.I., K.G.R., S.M.D., J.C.K., D.P.T., R.C. H., M.V., M.C.C., D.Y., S.Z., L.J.J., Y. S., and C.G.M. performed experiments. I.M.C, M.R., S.M.K., M.K., E.J., E.M.P., J.M.R., C.H.P., J.G.F., S.R., M.L., J.R., M.S.T., P.H.W., M.R.L., J.D.M., C.L.W., J.R.W., and S.P.H. provided clinical samples and data. I.I., Y.L., S.M.D., J.C.K., D. Y., S.Z., Z.G., L.J., J.E.D., J.Z., and C.G.M. analyzed data. I.I. and C.G.M. wrote the manuscript.
Co-first authors: I.I. designed and performed experiments; analyzed data and contributed to the manuscript writing. Y.L. analyzed genomic and transcriptome data and prepared genomic figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alpar D, Wren D, Ermini L, Mansur MB, van Delft FW, Bateman CM, Titley I, Kearney L, Szczepanski T, Gonzalez D, et al. Clonal origins of ETV6-RUNX1(+) acute lymphoblastic leukemia: studies in monozygotic twins. Leukemia. 2015;29:839–846. doi: 10.1038/leu.2014.322. [DOI] [PubMed] [Google Scholar]
- Bento C, Percy MJ, Gardie B, Maia TM, van Wijk R, Perrotta S, Della Ragione F, Almeida H, Rossi C, Girodon F, et al. Genetic basis of congenital erythrocytosis: mutation update and online databases. Hum Mutat. 2014;35:15–26. doi: 10.1002/humu.22448. [DOI] [PubMed] [Google Scholar]
- Bulut GB, Sulahian R, Yao H, Huang LJ. Cbl ubiquitination of p85 is essential for Epo-induced EpoR endocytosis. Blood. 2013;122:3964–3972. doi: 10.1182/blood-2013-05-506212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A, Traskelin AL, Juvonen E. Truncated erythropoietin receptor causes dominantly inherited benign human erythrocytosis. Proc Natl Acad Sci U S A. 1993;90:4495–4499. doi: 10.1073/pnas.90.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Gross M, Ben-Califa N, McMullin MF, Percy MJ, Bento C, Cario H, Minkov M, Neumann D. Polycythaemia-inducing mutations in the erythropoietin receptor (EPOR): mechanism and function as elucidated by epidermal growth factor receptor-EPOR chimeras. Br J Haematol. 2014;165:519–528. doi: 10.1111/bjh.12782. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Shinton SA. Characterization of B lymphopoiesis in mouse bone marrow and spleen. Methods Mol Biol. 2004;271:1–24. doi: 10.1385/1-59259-796-3:001. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LJ, Shen YM, Bulut GB. Advances in understanding the pathogenesis of primary familial and congenital polycythaemia. Br J Haematol. 2010;148:844–852. doi: 10.1111/j.1365-2141.2009.08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med. 2015a;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015b;125:3977–3987. doi: 10.1182/blood-2015-02-580043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kieran MW, Perkins AC, Orkin SH, Zon LI. Thrombopoietin rescues in vitro erythroid colony formation from mouse embryos lacking the erythropoietin receptor. Proc Natl Acad Sci U S A. 1996;93:9126–9131. doi: 10.1073/pnas.93.17.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovics R, Prchal JT. Genetic heterogeneity of primary familial and congenital polycythemia. Am J Hematol. 2001;68:115–121. doi: 10.1002/ajh.1162. [DOI] [PubMed] [Google Scholar]
- Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98:e146–148. doi: 10.3324/haematol.2013.095372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009a;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009b;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemmaraju N, Kantarjian H, Kadia T, Cortes J, Borthakur G, Newberry K, Garcia-Manero G, Ravandi F, Jabbour E, Dellasala S, et al. A phase I/II study of the Janus kinase (JAK)1 and 2 inhibitor ruxolitinib in patients with relapsed or refractory acute myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2015;15:171–176. doi: 10.1016/j.clml.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009a;114:2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- Russell LJ, De Castro DG, Griffiths M, Telford N, Bernard O, Panzer-Grumayer R, Heidenreich O, Moorman AV, Harrison CJ. A novel translocation, t(14;19)(q32;p13), involving IGH@ and the cytokine receptor for erythropoietin. Leukemia. 2009b;23:614–617. doi: 10.1038/leu.2008.250. [DOI] [PubMed] [Google Scholar]
- Russell LJ, Enshaei A, Jones L, Erhorn A, Masic D, Bentley H, Laczko KS, Fielding AK, Goldstone AH, Goulden N, et al. IGH@ translocations are prevalent in teenagers and young adults with acute lymphoblastic leukemia and are associated with a poor outcome. J Clin Oncol. 2014;32:1453–1462. doi: 10.1200/JCO.2013.51.3242. [DOI] [PubMed] [Google Scholar]
- Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, Cario G, Cazzaniga G, Kulozik AE, Stanulla M, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208:901–908. doi: 10.1084/jem.20110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Gryshkova V, Birger Y, Bandapalli OR, Cazzaniga G, Gershman N, Kulozik AE, Biondi A, Mansour MR, et al. Novel activating mutations lacking cysteine in type I cytokine receptors in acute lymphoblastic leukemia. Blood. 2014;124:106–110. doi: 10.1182/blood-2013-10-529685. [DOI] [PubMed] [Google Scholar]
- Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood. 2011;118:4910–4918. doi: 10.1182/blood-2011-05-354266. [DOI] [PubMed] [Google Scholar]
- Watowich SS. The erythropoietin receptor: molecular structure and hematopoietic signaling pathways. J Investig Med. 2011;59:1067–1072. doi: 10.231/JIM.0b013e31820fb28c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston BW, Hayden MA, Roberts KG, Bowyer S, Hsu J, Fedoriw G, Rao KW, Mullighan CG. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31:e413–416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, Tritapoe J, Hixon JA, Silveira AB, Cardoso BA, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43:932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.