Abstract
Although environmental trace metals, such as copper (Cu), can disrupt normal olfactory function in fish, the underlying molecular mechanisms of metal-induced olfactory injury have not been elucidated. Current research has suggested the involvement of epigenetic modifications. To address this hypothesis, we analyzed microRNA (miRNA) profiles in the olfactory system of Cu-exposed zebrafish. Our data revealed 2, 10, and 28 differentially expressed miRNAs in a dose-response manner corresponding to three increasing Cu concentrations. Numerous deregulated miRNAs were involved in neurogenesis (e.g. let-7, miR-7a, miR-128 and miR-138), indicating a role for Cu-mediated toxicity via interference with neurogenesis processes. Putative gene targets of deregulated miRNAs were identified when interrogating our previously published microarray database, including those involved in cell growth and proliferation, cell death, and cell morphology. Moreover, several miRNAs (e.g. miR-203a, miR-199*, miR-16a, miR-16c, and miR-25) may contribute to decreased mRNA levels of their host genes involved in olfactory signal transduction pathways and other critical neurological processes via a post-transcriptional mechanism. Our findings provide novel insight into the epigenetic regulatory mechanisms of metal-induced neurotoxicity of the fish olfactory system, and identify novel miRNA biomarkers of metal exposures.
Introduction
Exposure to neurotoxic chemicals such as heavy metals has been linked to impaired chemosensory function and a loss of olfaction in vertebrates.1, 2 The olfactory system, a critically important component of the sensory system, transmits chemosensory signals from the peripherally located olfactory rosettes to the olfactory bulb and telencephalon.3 The fish peripheral olfactory system is in contact with the external environment leading to exposure to dissolved contaminants which can impair peripheral neurological function.4, 5 Metal-induced neurobehavioral injury in fish is especially problematic due to the loss of olfactory-driven behaviors critical to survival such as homing, predator avoidance, prey selection, and reproduction.6–8
Copper (Cu) is a ubiquitously distributed olfactory toxicant and a pervasive contaminant in urban runoff at concentrations varying from 3 to 64 ppb.9 Accordingly, vehicle emissions, pesticide formulations, and other industrial usages are major sources of Cu in runoff.10, 11 We previously reported that exposure to environmentally-relevant concentrations of Cu (6–40 ppb) impairs the transcription of genes involved in olfactory signal transduction in zebrafish.12 However, the complex molecular mechanisms by which Cu alters gene expression and leads to olfactory injury are still poorly understood. Thus, an intriguing research direction supporting epigenetic modifications as a mechanism of metal toxicity has emerged to the forefront of toxicological research.
As one of the major epigenetic markers, microRNAs (miRNAs) have received considerable attention.13 These molecules are short (~22 nucleotides) noncoding RNAs regulating mRNA translation and stability,13–15 in most cases resulting in negative regulation of target genes at the post-transcriptional level.13, 16 The mechanism of miRNA-mediated translational suppression requires binding of miRNA “seed sequence” to the 3′-UTR of the target mRNA.13 Ultimately, miRNAs can control diverse biological processes, such as metabolism, development, cell differentiation, apoptosis and proliferation.
Environmental chemicals such as heavy metals can interfere with the biogenesis and expression of miRNAs, leading to toxicological consequences. 17, 18 For example, cadmium and aluminum exposures have been linked to the alteration of miR-146a expression in mammalian cells.19 Additionally, extensive studies of miRNAs in non-mammalian models (e.g. zebrafish) have indicated a possible conserved function of miRNAs through evolution.20, 21 As vertebrates, fish are among the most diverse species, and are relevant models to study gene- and epigene-environmental interactions.22 However, little is known about the effect of environmental chemicals on the expression of miRNAs and their regulatory roles in fish olfactory molecular network and signaling pathways.
In the current study we hypothesized that the alteration of miRNA expression by Cu contributes to the differential expression of olfactory gene expression. We investigated miRNA profiles in Cu-exposed adult zebrafish olfactory system to identify individual miRNAs or miRNA families that may mediate metal toxicity. The mRNA targets, which are also differentially expressed following Cu exposure,12 encompass a wide range of biological processes, including chromatin structures, transcription factor activity, G-protein coupled receptor signaling, regulation of apoptosis and cell cycle, metal ion binding, and antioxidant activity. The results of our study provide novel insight into the epigenetic mechanisms of heavy metal-induced neurotoxicity in the fish olfactory system, and have yielded novel miRNA biomarkers in response to environmental toxicant exposure.
Materials and Methods
Animal Care and Maintenance
One-year-old adult AB strain zebrafish were housed in re-circulating aquaria maintained at 28 ± 0.5 °C in a 14 h light/10 h dark cycle. Fish received 2% of their body weight in flake food per day and were provided with supplemental artemia once daily. Source water was city municipal water passed through a Siemens treatment system containing ionic- and mixed-bed media, activated carbon, and 0.2 μm filtration. The resulting conductivity and chlorine-devoid water at pH 5.5 was stored within a holding reservoir and accordingly heated to 28 °C, reconstituted to 1000 ± 100 μS using Instant Ocean® salt, and adjusted to pH 7.2 using Na2HCO3. Total hardness was 115 mg/L. Critical water quality parameters (ammonia, nitrite, pH, and temperature) were recorded at least once per day.
Cu exposure and tissue collection
All animal welfare and experimental procedures were carried out in strict accordance with the University of Washington Institutional Animal Care and Use Committee (IACUC) guidelines. Cu exposures were conducted as described previously.12 Briefly, adult zebrafish (n=15 per group, with a ratio of approximately 1:1 males/females) were exposed to the intended concentrations of 0, 6.3, 16 and 40 ppb Cu (as CuCl2, Alpha Aeser, Ward Hill, MA) for 24 h. These environmentally-relevant Cu concentrations are within the range of urban runoff concentrations,9 and which can inhibit the physiological responsiveness of olfactory receptor neurons in fish species.7, 23 All Cu exposures were spiked with 0.001% DMSO to equalize for carrier solvent effects.12 Exposures were conducted in 8 L of water in 9 L tanks maintained at 28 °C with individual heaters and air stones. The targeted nominal Cu concentrations closely tracked the measured waterborne Cu concentrations (Supplemental Table 1). At the conclusion of exposures, fish were euthanized by cervical dislocation. We combined the olfactory rosettes, olfactory bulbs and telencephalons as pooled tissue, referred henceforth as the olfactory system, consistent with our previously published microarray study.12 The pooled tissue from 3 animals was placed in Trizol® (Invitrogen, Carlsbad, CA) prior to freezing in liquid nitrogen and storage at −80 °C until RNA isolation. A total of 5 replicate pools of RNA were generated for each experimental group.
MiRNA microarray analysis
Total RNA was extracted using Nucleo®Spin miRNA kit (MACHEREY-NAGEL Inc., Bethlehem, PA) according to the manufacturer’s protocol. Integrity of RNA samples was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). 500 ng total RNA of each sample was processed for miRNA analysis. Processing of the RNA samples for the Affymetrix GeneChip miRNA 2.0 Array, which contains probesets designated for various species including 248 probes for zebrafish, was performed according to the standard protocol recommended by the manufacturer (www.affymetrix.com/). Raw GeneChip® miRNA array data was pre-processed with Affymetrix miRNA QCTool software (http://www.affymetrix.com/products_services/arrays/specific/mi_rna.affx#1_4). The pre-processing steps include: probe specific signal detection calls based on a Wilcoxon Rank-Sum test of the miRNA probe set signals compared to the distribution of signals from GC content matched anti-genomic probes, background estimation and correction, constant variance stabilization on probes, probe level quantile normalization, and finally probe summarization using median polish. MiRNA arrays included oligo spike-in controls for monitoring array quality. Arrays were required to pass manufacturer’s quality control recommendations before further analysis. MiRNAs with significant evidence for differential expression were identified using the Bioconductor limma package.24 P-values were calculated with a modified t-test with in conjunction with an empirical Bayes method to moderate the standard errors of the estimated Log-fold changes. P-values were adjusted for multiplicity with the Bioconductor package q value,25 which allows for selecting statistically significant miRNAs at a chosen estimated false discovery rate. Only miRNAs expressed at a fairly robust level (log2 average expression level > 5) were further investigated. The microRNA array data have been submitted to the Gene Expression Omnibus (GEO); accession number GSE46891. The corresponding mRNA array expression data is also publicly available and can be accessed via the NCBI GEO database under accession number GSE47039.
Target genes prediction of the differentially expressed miRNAs
The web resource MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) was used to determine the predicted mRNA targets of the differentially expressed miRNAs (>1.5-fold up/down-regulated, p<0.05). Given the number of biological replicates combined with the detection limit of the microRNA array platform used in this study, we chose a 1.5-fold change in expression cutoff as a reasonable choice to detect differential expression. We then identified a subset of the predicted targets that were also differentially expressed (>1.5-fold up/down-regulated, p<0.05) in our previously published mRNA array data set.12 This subset of the differentially expressed miRNA targets was uploaded into Ingenuity Pathway Analysis (IPA). The Core Analysis module of the IPA software (http://www.ingenuity.com/) was used to identify “Molecular and Cellular Function” and “Physiological System Development and Function” categories. IPA ranks each function using the right-tailed Fisher Exact Test and the top ranked (p<0.05) biofunctions for each Cu concentration was used for further analysis.
Validation of microarray data by real-time qPCR
MiRNA quantification was carried out using NCode miRNA cDNA Synthesis Kit and EXPRESS SYBR GreenER miRNA qRT-PCR Kits (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Briefly, first strand cDNA was synthesized from 500 ng of total RNA using a universal RT primer and Superscript III® reverse transcriptase and diluted before use in real-time qPCR. PCR amplifications were performed in a Bio-Rad IQ5 thermocycler (Hercules, CA) with SYBR Green master mix (Finnzymes), 0.2 μM (final concentration) of universal RT primer, 0.2 μM miRNA-specific forward primers and 1 μL of diluted first strand cDNA (Supplemental Table 2). The PCR reaction began with template denaturation at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. U6 small nuclear RNA (RNU6) was used as the internal control for normalization26 as there were no significant differences in expression of this gene across exposure groups.. For quality control purposes, no template controls, as well as melt curve analyses, were completed for all reactions. Gene expression quantification was conducted as described previously.12
Results and Discussion
Identification of deregulated miRNAs by Cu exposure
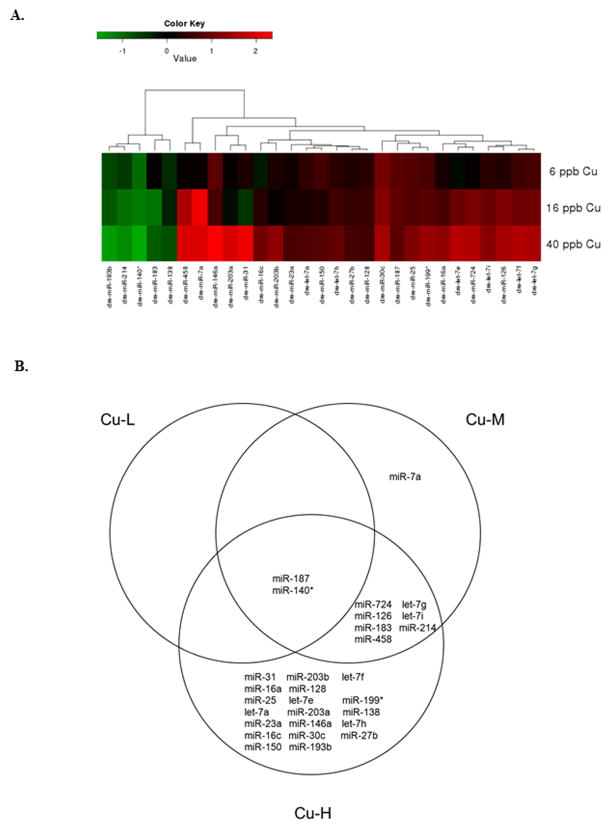
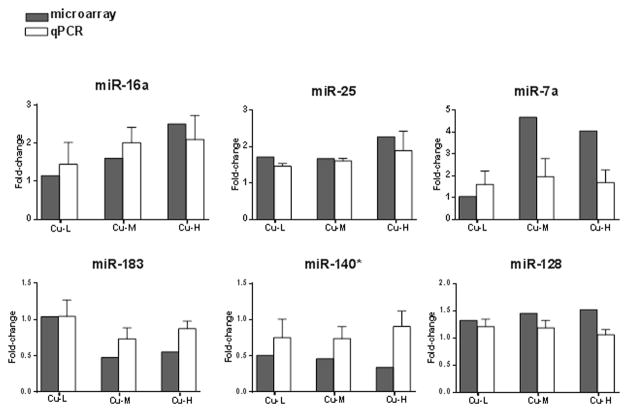
We observed 2, 10, and 28 differentially expressed miRNAs in response to 6.3 ppb (Cu-L), 16 ppb (Cu-M) and 40 ppb Cu (Cu-H), respectively (Figure 1A), that were > 1.5-fold up or down-regulated with a statistical significance of p<0.05. Among the altered miRNAs, 1, 7 and 23 miRNAs were up-regulated, and 1, 3, and 5 miRNAs were down-regulated in Cu-L, Cu-M, and Cu-H groups, respectively. Table 1 provides detailed data for the differentially expressed miRNAs, including levels of expression. Two miRNAs (miR-187 and miR-140*) were altered in all Cu exposure groups, and 7 miRNAs were differentially expressed in both Cu-M and Cu-H groups (Figure 1B). In addition, there were 1 and 19 significantly altered miRNAs unique to the Cu-M and Cu-H exposure groups, respectively. To validate the miRNA microarray data, we confirmed six differentially expressed miRNAs using real-time qPCR (Figure 2).
Figure 1.
(A) Hierarchical clustering of significantly altered miRNAs (>1.5-fold up/down-regulated, p<0.05) relative to vehicle controls; red indicates up-regulation and green indicates down-regulation. (B) Venn diagram analysis of the miRNA expression changes following Cu exposures. The numbers of transcripts meeting the cutoff (>1.5-fold up/down-regulated, p<0.05) for each Cu concentration are contained within each section of the labeled circle. The expression of 2 miRNAs was commonly altered in all three Cu concentrations, and 7 miRNAs were differentially expressed in both Cu-M and Cu-H groups. An additional 1 and 19 miRNAs were significantly and uniquely altered in the Cu-M and Cu-H groups, respectively.
Table 1.
Cu-induced differential expression of miRNAs ranked by p-value a.
| Cu exposure | miRNA name | FC | p-value | Log2 AE |
|---|---|---|---|---|
| Cu-L | miR-187 | 1.80 | 0.017 | 9.16 |
| miR-140* | 0.50 | 0.045 | 8.80 | |
| Cu-M | miR-183 | 0.48 | 0.004 | 8.73 |
| miR-187 | 1.87 | 0.013 | 9.16 | |
| miR-724 | 2.23 | 0.019 | 9.09 | |
| let-7g | 2.04 | 0.023 | 6.86 | |
| let-7i | 1.98 | 0.025 | 9.25 | |
| miR-140* | 0.45 | 0.025 | 8.80 | |
| miR-214 | 0.48 | 0.029 | 11.51 | |
| miR-458 | 3.01 | 0.040 | 5.75 | |
| miR-126 | 2.24 | 0.048 | 7.51 | |
| miR-7a | 4.68 | 0.049 | 6.39 | |
| Cu-H | miR-31 | 4.53 | 0.002 | 5.13 |
| miR-203b | 2.47 | 0.002 | 9.76 | |
| let-7g | 2.92 | 0.002 | 6.86 | |
| miR-724 | 3.06 | 0.003 | 9.09 | |
| miR-140* | 0.34 | 0.005 | 8.80 | |
| let-7f | 3.10 | 0.006 | 7.61 | |
| miR-16a | 2.50 | 0.007 | 5.98 | |
| let-7i | 2.36 | 0.008 | 9.25 | |
| miR-214 | 0.41 | 0.011 | 11.51 | |
| miR-183 | 0.55 | 0.013 | 8.73 | |
| miR-128 | 1.52 | 0.013 | 13.24 | |
| miR-458 | 4.08 | 0.013 | 5.75 | |
| miR-25 | 2.27 | 0.014 | 8.22 | |
| miR-126 | 2.85 | 0.015 | 7.51 | |
| miR-187 | 1.78 | 0.019 | 9.16 | |
| let-7e | 3.22 | 0.021 | 6.64 | |
| miR-199* | 2.63 | 0.022 | 8.84 | |
| let-7a | 1.64 | 0.023 | 12.99 | |
| miR-203a | 3.91 | 0.023 | 5.54 | |
| miR-138 | 0.60 | 0.024 | 12.93 | |
| miR-23a | 1.60 | 0.024 | 10.10 | |
| miR-146a | 5.14 | 0.029 | 5.33 | |
| let-7h | 1.90 | 0.030 | 7.30 | |
| miR-16c | 2.03 | 0.031 | 7.63 | |
| miR-30c | 2.88 | 0.035 | 5.34 | |
| miR-27b | 1.57 | 0.036 | 8.67 | |
| miR-150 | 1.72 | 0.045 | 9.02 | |
| miR-193b | 0.36 | 0.049 | 10.12 |
Data represents only those miRNAs >1.5-fold up-regulated (red), or down-regulated (green) with a statistical significance of p<0.05 and Log2 average expression >5. Down-regulation is indicated as percent of control. FC=fold-change; AE=average expression.
Figure 2.
Validation of miRNA microarray data by quantitative real-time PCR. MiRNA fold-change from the controls (0 ppb Cu) detected in microarray (grey bars) and qPCR (white bars). The qPCR results were normalized using RNU6 as internal control (details see Materials and Methods). qPCR data represent the mean ± SEM of n= 5 individual pools.
Biological significance of differentially expressed miRNAs
Previous studies have reported the conserved seed sequence among CNS-specific vertebrate miRNAs,27 such as miR-128, miR-138, miR-25, miR-199*. However, the expression pattern of these miRNAs appears to be heterogeneously distributed in CNS tissue.27, 28 To better understand the biological significance of the deregulated miRNAs in zebrafish, we summarized their expression patterns and functional roles as reported in the literature (Supplemental Table 3). Our investigation revealed numerous deregulated miRNAs with known restricted expression in neuronal cells, implying important regulatory roles in neurons. For example, miR-724, miR-458, miR-16 (a, c), miR-30c, miR-193b, let-7 family members (a, f, i), miR-187, miR-128, miR-138, and miR-7a were found in brain; whereas miR-187, miR-128, miR-138, miR-183 and miR-7a have been shown to be specifically expressed in the olfactory system (Supplemental Table 3). In particular, miR-183 family members (miR-183, miR-182 and miR-96) are abundantly expressed in neurosensory organs of zebrafish, including olfactory sensory cells and hair cells in the lateral line.29 Knockdown of miR-183 family in zebrafish causes hair cell loss in the inner ear, suggesting an essential role in the development and function of sensory organs.30 In the current study, miR-183 showed a significant decrease in both Cu-M and Cu-H concentrations (52% and 45% decreases, respectively). Other studies have reported increased apoptosis and loss of sensory neurons in fish olfactory epithelium following low-dose Cu exposures.31, 32 Therefore, the decreased miR183 expression is potentially related to the loss of olfactory sensory neurons caused by Cu.
Predicted mRNA targets of differentially expressed miRNAs
Because the biological significance of altered miRNA expression by environmental stressors is intimately associated with their gene targets, we identified a subset of the predicted targets that were also differentially expressed in our previously published mRNA array data set derived from the olfactory system of zebrafish exposed to the same Cu concentrations (Supplemental Table 4).12 In addition, we listed putative target genes regulated by multiple miRNAs, as well as highlighted miRNA seed sequences, which illustrate the relationship between miRNAs and host mRNAs (Supplemental Table 5). The majority of the miRNAs under study and their target mRNAs originate from different regions of the genome, suggesting that they are derived from independent transcription units.16 MiRNAs from the same family (also known as homo-clusters), such as let-7a and f, miR-16a and c, usually target a similar set of genes.33 In contrast, miRNAs clustered from different families (also known as hetero-clusters), such as miR-199* and miR-214 (Supplemental Table 5), may share functional similarity even with different seed regions.
MiRNA regulatory network and Cu-induced cellular response
To gain a better understanding about the function of the target genes for each deregulated miRNA, we summarized the Top Molecular Function and Biological Process Gene Ontology annotations (GO Terms) based on the ZFIN database (http://zfin.org/) for each of the target genes (Supplemental Table 4). In addition, we used Ingenuity Pathway Analysis (IPA) software to analyze the significantly differentially expressed target genes and categorize them based upon functional similarity using the “Molecular and Cellular Function” (MCF) and “Physiological System Development and Function” (PSDF) categories (Supplemental Table 6). The results from the IPA analysis, which were based on mapping the zebrafish genes to their human orthologs, were similar to the results obtained with the aforementioned “GO Term” analysis which was based on the ZFIN database (see Supplemental Table 6, worksheet titled “gene list”). Supplemental Table 7 summarizes the deregulated miRNAs and their differentially expressed targets according to functional categories for each Cu dose.
Our incorporation of both IPA and ZFIN analysis revealed conserved biochemical pathways impacted by Cu exposure among vertebrates. As the exposure levels of Cu increased, so did the number of altered zebrafish genes associated with cellular functions such as Cellular Growth and Proliferation, Cell Death, Cell Morphology, and Cellular Movement (Supplemental Table 6), suggesting that increased Cu levels exert a greater impact on cellular response. A closer examination indicated that a host of biological processes were significantly altered at every Cu concentration, such as chromatin/nucleosome assembly, cell cycle process, ion binding and transport (Supplemental Table 7). Furthermore, IPA analysis revealed the “Oxidative Stress” pathway as the top “Tox List” at all three Cu exposure concentrations (p=6.69×10−04 in Cu-L; p=6.67×10−03 in Cu-M; p=6.47×10−07 in Cu-H) (IPA data not shown). The deregulated miRNA target genes included those involved in protecting against oxidative stress (e.g. peroxiredoxin 1, NAD(P)H dehydrogenase, quinone 1), and other stress-induced factors such as NF-κB, Supplemental Table 4). Thus, cellular antioxidant pathways in the olfactory system appear to be an important defense mechanism against metal-induced olfactory injury at environmentally-relevant concentrations. This hypothesis is also supported by several recent studies from our laboratory.34–37
Effect of Cu on miRNAs in the context of olfactory signal transduction (OST)
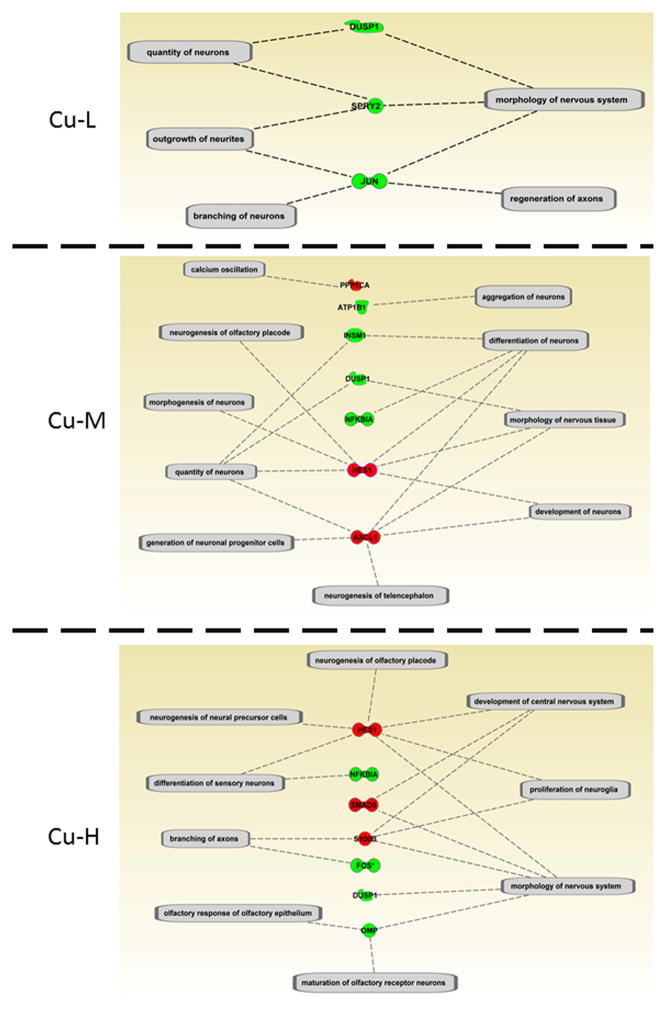
One of the significant PSDF categories IPA identified was “Nervous System Development and Function” and its sub-categories are shown in more detail in Supplemental Table 6 (worksheet: Nervous system development; Figure 3). Many of these transcripts are essential for odorant binding, as well as signaling cascades in the OST pathway, including olfactory marker protein, olfactory receptors, and S100 calcium binding proteins. These findings suggest a role of miRNAs in the molecular mechanism of metal-induced loss of olfactory signaling.
Figure 3.
IPA analysis of significantly differentially expressed miRNA target genes related to “Nervous system Development and Function” (details see Supplemental Table 6). The sub-function cluster that associated with olfactory function is presented here (red indicates up-regulation and green indicates down-regulation).
To address this hypothesis, we compared the target genes with gene clusters representing transcript trends in the zebrafish olfactory system in response to Cu exposures previously conducted under identical experimental conditions.12 In our previous study, we used principal components analysis to identify 12 gene clusters, encompassing altered transcripts in response to increasing Cu concentrations.12 The overwhelming majority of gene clusters (75%) showed a downward trend of mRNA expression with increasing Cu concentration, which was associated with deregulated miRNAs and their target genes. In order to conduct a valid comparison to our previous study, we selected six miRNA target genes using RNA samples from the current study, and compared with the microarray data12 (Supplemental Figure 1).
The gene regulatory process comprises multiple stages through a host of complex networks, including miRNAs, their target genes, and other classes of regulatory proteins, such as transcription factors.38 Previous computational analyses identified the coupling between transcription and post-transcriptional level regulation via forming regulatory feed-back and feed-forward loops.39 For example, an increase in miRNA expression often coincides with a decrease in their target gene transcripts.40, 41 This negative correlation between miRNA and their respective target genes was observed in our current study. For instance, several overall down-regulated gene clusters (Cluster 3 & 10, Cluster 4 & 11, see Table 2) contain genes putatively targeted by miRNAs that were inversely correlated in the present study. In particular, the up-regulated miR-203a, miR-199*, miR-16a, miR-16c, and miR-25 may contribute to decreased mRNA levels of their host genes in calcium signaling (e.g. parvalbumin 8, calbindin 2-like and s100 calcium binding protein z) via a post-transcriptional mechanism. Functionally, calcium is an intracellular ion that plays an essential role in regulating the sensitivity of ion channels during olfactory signal transduction.42, 43 Thus, the strong impact on ion homeostasis in neuronal cells by Cu may be an important mechanism of disruption in olfactory signaling pathways and olfactory-driven behaviors.
Table 2.
Deregulated miRNAs and their target genes identified in Cluster 3 & 10 and Cluster 4 & 11 that were down-regulated at all Cu treatment groups (>1.5-fold up/down-regulated, p<0.05) a. These clusters are derived from our previous study.10
| Cluster 3 & 10 | |||
|---|---|---|---|
| Gene targets | Gene target FC (Cu dose) | miRNA | miRNA FC |
| parvalbumin 8 | 0.09 (high) | miR-203a | 3.91 |
| miR-199* | 2.63 | ||
| s100 calcium binding protein z (zgc:110464) | 0.11 (high) | miR-16a | 2.50 |
| miR-16c | 2.03 | ||
| similar to protein phosphatase 1, regulatory (inhibitor) subunit 3C like | 0.18 (high) | miR-16c | 2.03 |
| olfactory marker protein b | 0.18 (high) | miR-193b | 0.36 |
| calbindin 2, like | 0.21 (high) | miR-25 | 2.27 |
| similar genes: | |||
| ATPase, Na+/K+ transporting, beta 1a polypeptide | 0.60 (med) | miR-126 | 2.24 |
| 0.62 (high) | miR-126 | 2.85 | |
| miR-25 | 2.27 | ||
| Suppressor of cytokine signaling 1 (zgc:91868) b | 0.35; 0.31 (med) | miR-7a | 4.68 |
| 0.11; 0.08 (high) | miR-203a | 3.91 | |
| miR-30c | 2.88 | ||
| miR-203b | 2.47 | ||
| miR-16c | 2.03 | ||
| Cluster 4 & 11 | |||
| dopey family member 2 (zgc:63622) b | 0.29; 0.27; 0.18 (med) | miR-458 | 3.01 |
| 0.28; 0.25; 0.16 (high) | miR-458 | 4.08 | |
| zona pellucida glycoprotein 2.2 | 0.47 (high) | miR-16a | 2.50 |
| miR-128 | 1.52 | ||
| miR-193b | 0.36 | ||
| zona pellucida glycoprotein 3a.1 | 0.56 (med) | miR-214 | 0.48 |
| 0.53 (high) | miR-214 | 0.41 | |
| sulfotransferase family 2, cytosolic sulfotransferase 1 | 0.20 (high) | miR-27b | 1.57 |
| odorant receptor, family 5, member 1 | 0.26 (med) | miR-183 | 0.48 |
| 0.25 (high) | miR-183 | 0.55 | |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a | 0.62 (med) | let-7g | 2.04 |
| 0.47 (high) | let-7g | 2.92 | |
| let-7h | 1.90 | ||
| growth arrest and DNA-damage-inducible, beta b | 0.45;0.36 (high) | miR-16a | 2.50 |
| miR-25 | 2.27 | ||
| miR-16c | 2.03 | ||
| claudin d | 0.41 (med) | miR-214 | 0.48 |
| 0.40 (high) | miR-16a | 2.50 | |
| miR-16c | 2.03 | ||
| miR-214 | 0.41 | ||
| MpV17 transgene, murine homolog, glomerulosclerosis b | 0.62; 0.55; 0.40 (med) | let-7i | 1.98 |
| 0.56; 0.54; 0.40 (high) | miR-146a | 5.14 | |
| let-7f | 3.10 | ||
| let-7i | 2.36 | ||
| let-7h | 1.90 | ||
| dual specificity phosphatase 1 | 0.62 (low) | miR-140* | 0.50 |
| 0.47 (med) | miR-140* | 0.45 | |
| 0.44 (high) | miR-140* | 0.34 | |
| cyclin-dependent kinase inhibitor 3 | 0.41 (med) | miR-140* | 0.45 |
| 0.44 (high) | miR-16c | 2.03 | |
| miR-23a | 1.60 | ||
| miR-140* | 0.34 | ||
| fatty acid binding protein 10, liver basic | 0.39 (med) | miR-183 | 0.48 |
| 0.45 (high) | miR-183 | 0.55 | |
| miR-193b | 0.36 | ||
| odorant receptor, family 2, member 10 | 0.54 (med) | miR-214 | 0.48 |
| 0.47 (high) | miR-199* | 2.63 | |
| miR-214 | 0.41 | ||
Data represents only those miRNAs >1.5-fold up-regulated (red), or down-regulated (green) with a statistical significance of p<0.05 and Log2 average expression >5. Down-regulation is indicated as present of control. FC=fold-change.
More than one probe were used for the same gene target.
On the other hand, a few miRNAs (miR-193b, miR-214, miR-183 and miR-140*) showed a similar downward trend with their predicted target genes (e.g. olfactory marker protein b, odorant receptors, and dual specificity phosphatase 1). While expression levels of the majority of microRNAs are inversely correlated with their corresponding target mRNAs, exceptions have been reported.40, 41, 44, 45 MicroRNAs represent an important mechanism of regulating gene expression post-transcriptionally, but there are other epigenetic (e.g. methylation), as well as non-epigenetic transcriptional mechanisms mediated by transcription factors, that also regulate gene expression. All of these mechanisms can work in concert, forming a complex overall regulatory network.45 Other factors affecting net mRNA levels of microRNA targets include temporal aspects such as duration of chemical exposure, turnover of the target mRNAs, and stability of the microRNA-mRNA complex. Any of the aforementioned aspects may explain, at least in part, the parallel downward trend of the aforementioned microRNAs and their mRNA targets.
Role of miRNAs in regulating neurogenesis
Neurogenesis appears to be a universal phenomenon in the olfactory system of vertebrates and plays a critical role in modulating regeneration capacity in response to injury.46 Recent studies have shown that miRNAs function as key epigenetic regulators for maintenance and fate specification of neural stem/progenitor cells.47–50 Similarly, we observed that exposure to Cu deregulated the expression of certain miRNAs involved in regulating neural stem cells and neuronal fate, indicating a role for Cu-mediated toxicity via interference with neurogenesis. For example, our microarray profile indicated that 9 of 11 currently identified zebrafish let-7 isoforms51 were differentially expressed (all up-regulated). The let-7 family has been studied in various species and others have suggested a conserved function involving developmental regulation and neural cell differentiation in the CNS.52, 53 Recent studies in mammalian neural stem cells have revealed the mechanisms underlying the regulatory mechanism of let-7 in promoting neuronal differentiation.54–56 The conserved seed regions among multiple let-7 members suggests a similar regulatory function during adult neurogenesis.51
Likewise, another highly conserved miRNA, miR-7, appears to play an important role in neurite outgrowth and synapse formation during human neural stem cell-driven neurogenesis.57 A study in Drosophila melanogaster has proposed miR-7 is involved in photoreceptor neuron differentiation via the regulation of YAN expression in the progenitor cells.58 In zebrafish, miR-7a is highly enriched in vasotocinergic and RFamidergic neuron populations, both of which are components of the neurosecretory brain centers during early development.59 Our data suggests that up-regulation of miR-7a could stimulate neurogenesis by down-regulating its target gene, the suppressor of cytokine signaling 1 (socs1, Supplemental Table 4). A recent study has identified another suppressor of cytokine signaling family member, socs3, which shares extensive homology with socs160 and is a key regulator of hair cell regeneration in adult zebrafish.61
In addition to miR-7 and let-7 isoforms, we observed up-regulation of miR-128 and down-regulation of miR-138, two miRNAs highly expressed in the zebrafish olfactory bulb and other brain regions.62, 63 Others have demonstrated a role of miR-128 in promoting neuronal differentiation,64 and for miR-138 in inhibiting dendritic spine morphogenesis.65 When viewed collectively, our findings indicate that the observed Cu-induced modulation of the neurogenesis-related miRNAs and their putative targets may be associated with regeneration of olfactory sensory neurons under higher Cu concentrations (16 and 40 ppb exposure). Furthermore, our data suggest miR-724, miR-187, miR-126, miR-30c, miR-16c and miR-203 (a, b) may also be associated with neurogenesis processes. Therefore, in future studies, we will focus on in vivo validation of the target genes of these deregulated miRNAs and further explore their roles in determining neuronal identity under metal exposure.
In summary, our studies have led to a better understanding of the epigenetic mechanisms of metal-induced olfactory injury. To our knowledge, this is the first study, or one of the select few to investigate miRNA regulation as an underlying mechanism of metal-induced olfactory impairment. Our results provide mechanistic insight to our previous report of Cu-mediated transcriptional depression of key gene elements comprising the zebrafish olfactory signal transduction pathway. Of note is that environmentally-relevant Cu concentrations alter the expression of a diverse number of miRNAs in the olfactory system of zebrafish with predicted gene targets preferentially involved in olfactory signal transduction and other critical neurological processes. This observation provides a basis for extending the use of miRNAs as biomarkers to environmental applications involving metal exposures. Collectively, our results support Cu modulation of miRNA biogenesis, expression, and key miRNA-regulated genes in the olfactory signaling pathways.
Supplementary Material
Acknowledgments
This work was sponsored in part by the University of Washington Superfund Research Program [NIEHS P42ES004696], NIEHS University of Washington Center for Ecogenetics & Environmental Health [P30ES07033], and the University of Washington Sea Grant Program, pursuant to National Oceanic and Atmospheric Administration Project R/OCEH-8 [NA10OAR4170057]. The authors gratefully acknowledge the technical assistance of Herbert M. Espinoza.
Footnotes
Table S1 shows the nominal (and measured) Cu concentrations from waterborne exposures; Table S2 provides the PCR primers of selected miRNAs; Table S3 summarizes the expression patterns and functional roles of deregulated miRNAs based on previous studies in zebrafish; Table S4 lists differentially expressed miRNAs and their significantly altered putative gene targets; Table S5 lists host genes that are regulated by multiple miRNAs and includes comparisons of the seed sequences and chromosome locations; Table S6 shows all the genes from the IPA “Molecular and Cellular Function” and “Physiological System Development and Function” analysis; Table S7 summarizes target gene functions and regulated miRNAs; Supplemental Figure 1 shows quantitative real-time PCR comparison of the current study with previously reported microarray response.
References
- 1.Baldwin DH, Tatara CP, Scholz NL. Copper-induced olfactory toxicity in salmon and steelhead: extrapolation across species and rearing environments. Aquat Toxicol. 2011;101(1):295–7. doi: 10.1016/j.aquatox.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Bondier JR, Michel G, Propper A, Badot PM. Harmful effects of cadmium on olfactory system in mice. Inhal Toxicol. 2008;20(13):1169–77. doi: 10.1080/08958370802207292. [DOI] [PubMed] [Google Scholar]
- 3.Hamdani el H, Doving KB. The functional organization of the fish olfactory system. Prog Neurobiol. 2007;82(2):80–6. doi: 10.1016/j.pneurobio.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Brain Res Rev. 2001;36(1):46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- 5.Tierney KB, Baldwin DH, Hara TJ, Ross PS, Scholz NL, Kennedy CJ. Olfactory toxicity in fishes. Aquat Toxicol. 2010;96(1):2–26. doi: 10.1016/j.aquatox.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin DH, Sandahl JF, Labenia JS, Scholz NL. Sublethal effects of copper on coho salmon: impacts on nonoverlapping receptor pathways in the peripheral olfactory nervous system. Environ Toxicol Chem. 2003;22(10):2266–74. doi: 10.1897/02-428. [DOI] [PubMed] [Google Scholar]
- 7.Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. A sensory system at the interface between urban stormwater runoff and salmon survival. Environ Sci Technol. 2007;41(8):2998–3004. doi: 10.1021/es062287r. [DOI] [PubMed] [Google Scholar]
- 8.Scott GR, Sloman KA, Rouleau C, Wood CM. Cadmium disrupts behavioural and physiological responses to alarm substance in juvenile rainbow trout (Oncorhynchus mykiss) J Exp Biol. 2003;206(Pt 11):1779–90. doi: 10.1242/jeb.00353. [DOI] [PubMed] [Google Scholar]
- 9.Soller J, Stephenson J, Olivieri K, Downing J, Olivieri AW. Evaluation of seasonal scale first flush pollutant loading and implications for urban runoff management. Journal of environmental management. 2005;76(4):309–18. doi: 10.1016/j.jenvman.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Davis AP, Shokouhian M, Ni S. Loading estimates of lead, copper, cadmium, and zinc in urban runoff from specific sources. Chemosphere. 2001;44(5):997–1009. doi: 10.1016/s0045-6535(00)00561-0. [DOI] [PubMed] [Google Scholar]
- 11.Good JC. Roof runoff as a diffuse source of metals and aquatic toxicity in storm water. Water Science & Technology. 1993;28:317–321. [Google Scholar]
- 12.Tilton F, Tilton SC, Bammler TK, Beyer R, Farin F, Stapleton PL, Gallagher EP. Transcriptional biomarkers and mechanisms of copper-induced olfactory injury in zebrafish. Environ Sci Technol. 2008;42(24):9404–11. doi: 10.1021/es801636v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhuri S. Small noncoding RNAs: biogenesis, function, and emerging significance in toxicology. J Biochem Mol Toxicol. 2010;24(3):195–216. doi: 10.1002/jbt.20325. [DOI] [PubMed] [Google Scholar]
- 14.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 15.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–51. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutat Res. 2011;714(1–2):105–12. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36(Pt 6):1211–5. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 20.Sifuentes-Romero I, Milton SL, Garcia-Gasca A. Post-transcriptional gene silencing by RNA interference in non-mammalian vertebrate systems: where do we stand? Mutat Res. 2011;728(3):158–71. doi: 10.1016/j.mrrev.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. 2007;17(2):145–50. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Cossins AR, Crawford DL. Fish as models for environmental genomics. Nat Rev Genet. 2005;6(4):324–33. doi: 10.1038/nrg1590. [DOI] [PubMed] [Google Scholar]
- 23.Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. Odor-evoked field potentials as indicators of sublethal neurotoxicity in juvenile coho salmon (Oncorhynchus kisutch) exposed to copper, chlorpyrifos, or esfenvalerate. Can J Fish Aquat. 2004;61:404–413. [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Li YY, Zeng HC, Wei J, Wan YJ, Chen J, Xu SQ. MicroRNA expression changes during zebrafish development induced by perfluorooctane sulfonate. J Appl Toxicol. 2011;31(3):210–22. doi: 10.1002/jat.1583. [DOI] [PubMed] [Google Scholar]
- 27.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–44. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103(39):14385–9. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240(4):808–19. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30(9):3254–63. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen JA, Rose JD, Jenkins RA, Gerow KG, Bergman HL. Chinook salmon (Oncorhynchus tshawytscha) and rainbow trout (Oncorhynchus mykiss) exposed to copper: Neurophysiological and histological effects on the olfactory system. Environmental Toxicology and Chemistry. 1999;18(9):1979–1991. [Google Scholar]
- 32.Julliard AK, Saucier D, Astic L. Time-course of apoptosis in the olfactory epithelium of rainbow trout exposed to a low copper level. Tissue & cell. 1996;28(3):367–77. doi: 10.1016/s0040-8166(96)80023-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Haubrock M, Cao KM, Hua X, Zhang CY, Wingender E, Li J. Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network. BMC systems biology. 2011;5:199. doi: 10.1186/1752-0509-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Gallagher EP. Role of Nrf2 antioxidant defense in mitigating cadmium-induced oxidative stress in the olfactory system of zebrafish. Toxicol Appl Pharmacol. 2013;266(2):177–86. doi: 10.1016/j.taap.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Espinoza HM, Williams CR, Gallagher EP. Effect of cadmium on glutathione S-transferase and metallothionein gene expression in coho salmon liver, gill and olfactory tissues. Aquat Toxicol. 2012;110–111:37–44. doi: 10.1016/j.aquatox.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Harris SM, Espinoza HM, McClain V, Gallagher EP. Characterization of phospholipid hydroperoxide glutathione metabolizing peroxidase (gpx4) isoforms in Coho salmon olfactory and liver tissues and their modulation by cadmium. Aquat Toxicol. 2012;114–115:134–41. doi: 10.1016/j.aquatox.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espinoza HM, Shireman LM, McClain V, Atkins W, Gallagher EP. Cloning, expression and analysis of the olfactory glutathione S-transferases in coho salmon. Biochemical pharmacology. 2013;85(6):839–48. doi: 10.1016/j.bcp.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahan O, Gingold H, Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 2011;27(8):316–22. doi: 10.1016/j.tig.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26(5):753–67. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 41.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103(8):2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menini A. Calcium signalling and regulation in olfactory neurons. Curr Opin Neurobiol. 1999;9(4):419–26. doi: 10.1016/S0959-4388(99)80063-4. [DOI] [PubMed] [Google Scholar]
- 43.Klimmeck D, Mayer U, Ungerer N, Warnken U, Schnolzer M, Frings S, Mohrlen F. Calcium-signaling networks in olfactory receptor neurons. Neuroscience. 2008;151(3):901–12. doi: 10.1016/j.neuroscience.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 44.Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. Journal of immunology. 2010;184(11):6053–9. doi: 10.4049/jimmunol.0902308. [DOI] [PubMed] [Google Scholar]
- 45.Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41(5):2817–31. doi: 10.1093/nar/gks1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermingham-McDonogh O, Reh TA. Regulated reprogramming in the regeneration of sensory receptor cells. Neuron. 2011;71(3):389–405. doi: 10.1016/j.neuron.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawahara H, Imai T, Okano H. MicroRNAs in Neural Stem Cells and Neurogenesis. Frontiers in neuroscience. 2012;6:30. doi: 10.3389/fnins.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30(45):14931–6. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell. 2012;22(5):1052–64. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cellular and molecular life sciences : CMLS. 2012;69(1):89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32(21):6284–91. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21(2):415–26. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 54.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136(5):913–25. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135(2):227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107(5):1876–81. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Githinji J, McLaughlin B, Wilczek K, Nolta J. Role of miRNAs in Neuronal Differentiation from Human Embryonic Stem Cell-Derived Neural Stem Cells. Stem cell reviews. 2012 doi: 10.1007/s12015-012-9411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123(7):1267–77. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 59.Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129(7):1389–400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 60.Wang T, Gorgoglione B, Maehr T, Holland JW, Vecino JL, Wadsworth S, Secombes CJ. Fish Suppressors of Cytokine Signaling (SOCS): Gene Discovery, Modulation of Expression and Function. Journal of signal transduction. 2011;2011:905813. doi: 10.1155/2011/905813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang J, Wang D, Renaud G, Wolfsberg TG, Wilson AF, Burgess SM. The stat3/socs3a pathway is a key regulator of hair cell regeneration in zebrafish stat3/socs3a pathway: regulator of hair cell regeneration. J Neurosci. 2012;32(31):10662–73. doi: 10.1523/JNEUROSCI.5785-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 64.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42(4):500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11(6):705–16. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.