Abstract
Objective
Previous investigations suggest that the embryonic origins of the calvarial tissues (neural crest or mesoderm) may account for the molecular mechanisms underlying sutural development. The aim of this study was to evaluate the differences in the gene expression of human cranial tissues and assess the presence of an expression signature reflecting their embryonic origins.
Methods
Using microarray technology, we investigated global gene expression of cells from the frontal and parietal bones and the metopic and sagittal intrasutural mesenchyme (ISM) of four human foetal calvaria. qRT-PCR of a selected group of genes was done to validate the microarray analysis. Paired comparison and correlation analyses were performed on microarray results.
Results
Of six paired comparisons, frontal and parietal compartments (distinct tissue types of calvaria, either bone or intrasutural mesenchyme) had the most different gene expression profiles despite being composed of the same tissue type (bone). Correlation analysis revealed two distinct gene expression profiles that separate frontal and metopic compartments from parietal and sagittal compartments. TFAP2A, TFAP2B, ICAM1, SULF1, TNC and FOXF2 were among differentially expressed genes.
Conclusion
Transcriptional profiles of two groups of tissues, frontal and metopic compartments vs. parietal and sagittal compartments, suggest differences in proliferation, differentiation and extracellular matrix production. Our data suggest that in the second trimester of human foetal development, a gene expression signature of neural crest origin still exists in frontal and metopic compartments while gene expression of parietal and sagittal compartments is more similar to mesoderm.
Keywords: Cranial suture, Differentiation, Extracellular matrix, Mesoderm, Neural crest, Proliferation
1. Introduction
Calvarial bones are formed by intramembranous ossification and are divided by mesenchymal tissues called sutures. The human calvaria has four major sutures (metopic, coronal, sagittal, and lambdoid). The presence of unossified sutures facilitates foetal movement through the birth canal and functions as a growth centre to allow for brain growth.1 Osteogenesis takes place at the osteogenic front, the leading edge of each bone.2
One important disorder of the cranial vault is craniosynostosis, the premature fusion of the sutures. Craniosynostosis occurs in 3–5 out of 10,000 live births and can cause malformations of the skull, increased intracranial pressure, and developmental delay.3 Many studies have characterized the genetic and environmental aetiology of craniosynostosis,4–6 but for the majority of cases the underlying molecular pathways are unclear. In order to understand these pathways, the basis of normal sutural development must be defined. The anatomic and developmental differences between the sutures suggest that distinct molecular mechanisms are controlling morphogenesis. These differences include a suture specific prevalence of synostosis2; predominance of coronal fusion in hereditary synostosis7,8; anatomic architecture (sutures with blunt vs. overlapping margins)2; the timing of physiologic fusion9,10; and distinct embryonic origins.11–13
The embryonic origins of the cranial compartments (bones of the calvaria and their intervening sutures), were not well understood until Jiang et al. used a Wnt1-Cre-recombinase LacZ reporter mouse model to identify craniofacial structures with neural crest (NC) origin. They demonstrated that in embryonic day 17.5 (E17.5) mice the frontal bone, the posterior frontal suture (equivalent to the metopic suture in humans), the sagittal suture, and the central portion of the interparietal bone were derived from NC cells while the parietal bone and coronal suture were of paraxial mesoderm origin.12 This was later substantiated by Yoshida et al. who used the same strategy in Wnt1-Cre and Msp1-Cre mice to map cells of neural crest and paraxial mesoderm origin.13 On the other hand, while supporting Jiang’s studies regarding the origins of frontal and parietal bones and posterior frontal suture, Gagan et al. found that the sagittal sutures of neonatal day 1 (N1) Wnt1-Cre-recombinase LacZ reporter mice were of mesodermal origin. Additionally, their data suggests that on N10 cells in dura mater, which originate entirely from NC, migrate into the sagittal intrasutural mechencyme (ISM).11 Deckelbaum et al. studied fate mapping using Wnt1, En1 and Gli1expression and demonstrated the complicated nature of cell mapping in the border of NC and mesoderm derived tissues due to cell mingling in some regions.14 Taken together, there is agreement on the embryonic origin of frontal and parietal bones and metopic suture in mice but the embryonic origin of the sagittal suture remains unclear.
Based on the targeted expression and cell-level studies in the biology and pathology of cranial sutures, it is clear that developmental origins play an important role in the interactions of the adjacent tissues.15–18 While these findings are enlightening, none of them investigate gene expression in human tissues. On the other hand, broad understanding of molecular pathways requires investigation of large array of genes. Recently, high throughput gene expression analysis (microarray analysis) has been used in investigating craniosynostosisaetiology19,20 but not in studying human calvarial development. Therefore, we aimed to investigate the global gene expression profile of human calvarial compartments.
2. Materials and methods
2.1. Ethics statement
Samples used in this study were obtained from the Department of Pathology and the Birth Defects Research Laboratory at the University of Washington. The study participants (mothers) signed an informed consent. All procedures were approved by the University of Washington and Washington State University institutional review boards.
2.2. Study design and samples
Tissues samples were obtained from foetal crania of four normal human foetuses. We received two females ages 94 and 103 days and two males ages 97 and 98 days. Tissues were transported and cultured in Dulbecco’s Modified Eagle Medium (DMEM) GlutaMAX (Life Technologies, Grand Island, NY) containing foetal calf serum (Life Technologies, Grand Island, NY) and antibiotic–antimycotic supplement (Life Technologies, Grand Island, NY) containing penicillin, streptomycin and amphotericin B.
2.3. Cell expansion
All dural and extracranial soft tissues were removed from the parietal and frontal bones. 2–3 mm tissue explants were excised from each compartment, frontal and parietal bones and metopic and sagittal ISM. To avoid contamination with osteoblasts, ISM tissue was dissected from the central portion of each suture. Similarly, bone was harvested at least 3 mm from the margin of the suture to avoid contamination with ISM. Media was changed every 3–4 days. After reaching 75–80% confluence, the cells were trypsinized with TrypLEExpress (Life Technologies, Denmark) and passaged. During the fourth and final passage, 180,000 cells were plated in triplicate in 6-well plates and cultured for 5 days followed by RNA extraction. Supplementary Table 1 shows summary of the information for the samples.
Supplementary Table 1 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
2.4. RNA isolation
RNA was isolated from the cells using Roche High Pure miRNA Isolation Kit (Indianapolis, IN) according to the manufacturer’s protocol. RNA was isolated from triplicate wells separately and then combined to have one RNA sample for microarray. The quality and quantity of the sample were assessed using Agilent 2100 Bioanalyzer and NanoDrop 1000 spectrophotometer.
2.5. Microarray analysis
We used Affymetrix HuGene 2.0 ST arrays (Affymetrix, Santa Clara, CA) containing DNA probes for 28,500 full-length transcripts. Preparation of labelled cDNA, hybridization of the arrays and the analysis were performed according to the manufacturer’s protocol.
2.6. Analyses
Raw data were normalized, background corrected, and summarized using a robust multi-array average (RMA) using the Bioconductor oligo package.21 A weighted analysis of variance (ANOVA) model was fit to the data that included factors for sample type and sex. All comparisons were made using empirical Bayes adjusted contrasts (Bioconductor limma package).22–24 We focused our analysis on genes that exhibited more than 1.5-fold change in expression (up or down) with p < 0.05. We used a combination of fold-change in expression and a p-value criteria based on recommendations by the MAQC consortium25 to minimize the false positive rate.
The raw data were also used to analyze the correlation of gene expression. Singular value decomposition (SVD), a matrix factorization method, was used to analyze microarray data.19,26 An SVD of a matrix breaks expression variation into the following discrete components: patterns across genes (eigenarrays); pattern across samples (eigengenes); and the weights describing the relative importance of each eigenarray/eigengene (eigenweights). First, the eigenweights were inspected to determine the number of important variables and then the corresponding eigenarrays or eigengenes were used to determine the importance of genes, pathways or groups of samples. Two eigenweights were identified significantly larger than the remaining. Clustering the first two eigenarrays suggested the existence of three distinct groups of transcripts. Their corresponding patterns across the samples were used to define three distinct eigenpatterns or consensus patterns across samples.
2.7. qRT-PCR validation
Following statistical analysis of the expression array results, six genes of interest were chosen for qRT-PCR validation; TFAP2A, TFAP2B, ICAM1, SULF1, TNC and FOXF2. Prior to PCR, primers (Sigma–Aldrich, St Louis, MO) (Supplementary Table 2) were optimized. cDNA was prepared with Thermo Scientific RevertAid First Strand cDNA Synthesis Kit according to the manufacturer’s protocol. qRT-PCR was performed using SensiMix SYBR low-ROX kit (Bioline, Taunton, MA) and Applied Biosystems 7500 Fast Real Time PCR system. 18s rRNA was used as an internal control for normalization. The fold changes were calculated by the standard ΔΔCt method.27
Supplementary Table 2 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
3. Results
3.1. Paired comparisons
Six paired comparisons were made between the gene expression profiles of the four compartments (Table 1). Fold changes greater than ±1.5 with a p-value <0.05 were considered significant (Supplementary Tables 3–8). The frontal and parietal bone samples exhibited the largest number of differentially expressed genes while the frontal bone vs. metopic ISM and parietal bone vs. sagittal ISM had the lowest numbers of differentially expressed transcripts.
Table 1.
Number of differentially expressed genes in comparisons of compartments.
| Comparison | Count |
|---|---|
| Frontal vs. parietal | 104 |
| Frontal vs. sagittal | 100 |
| Parietal vs. metopic | 54 |
| Metopic vs. sagittal | 48 |
| Frontal vs. metopic | 22 |
| Parietal vs. sagittal | 13 |
Supplementary Table 3 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 4 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 5 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 6 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 7 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 8 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
3.2. Correlation analysis
Transcript correlation analysis revealed correlation of 795 genes represented in our array (Fig. 1). 264/795 genes (group 1) had higher expression in frontal bone and metopic ISM (Fig. 2; Supplementary Table 9) compared to parietal bone and sagittal ISM. In contrast, 394/795 genes (group 2), were expressed higher in parietal/sagittal compartments (Fig. 3; Supplementary Table 10). Therefore, the genes in groups 1 and 2 segregated the compartments into two groups of frontal/metopic and parietal/sagittal. 137/795 genes (group 3), were up-regulated in metopic and sagittal ISM (Fig. 4). Differential expression of these genes between compartments was not as large as in group 1 or 2.
Fig. 1.
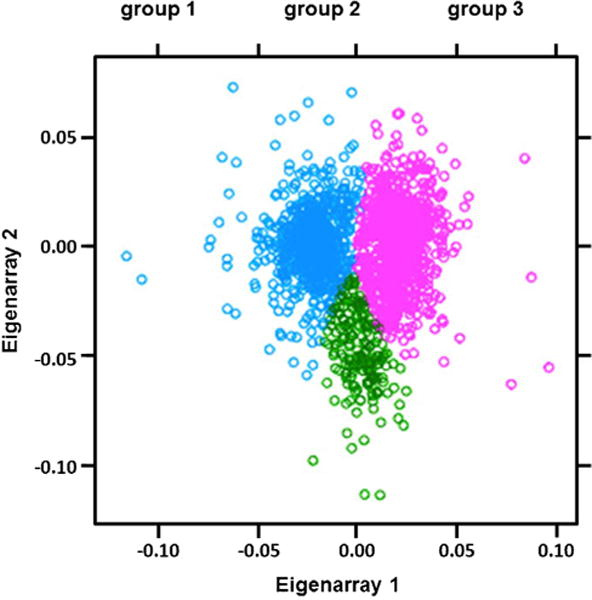
Correlation analysis. Clustering the first two eigenarrays suggested that three distinct gene groups (total of 795 genes), had correlated expression throughout all the microarrays.
Fig. 2.
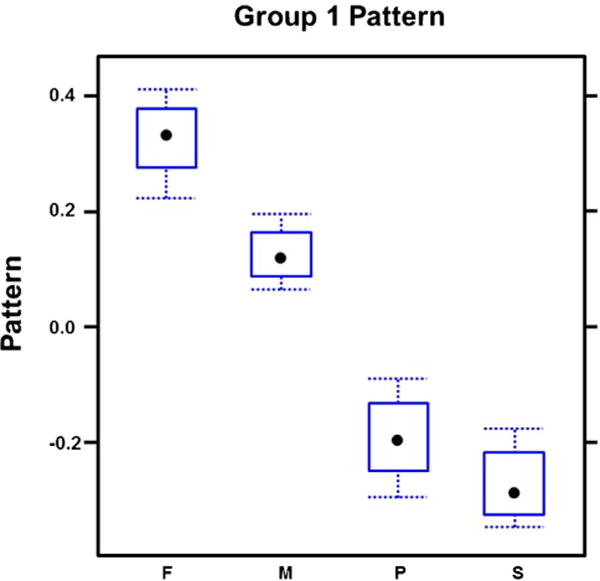
Expression of correlated genes in group1. This group was highly expressed in frontal and metopic compartments and segregated the compartments into two groups of frontal/metopic and parietal/sagittal.
Fig. 3.
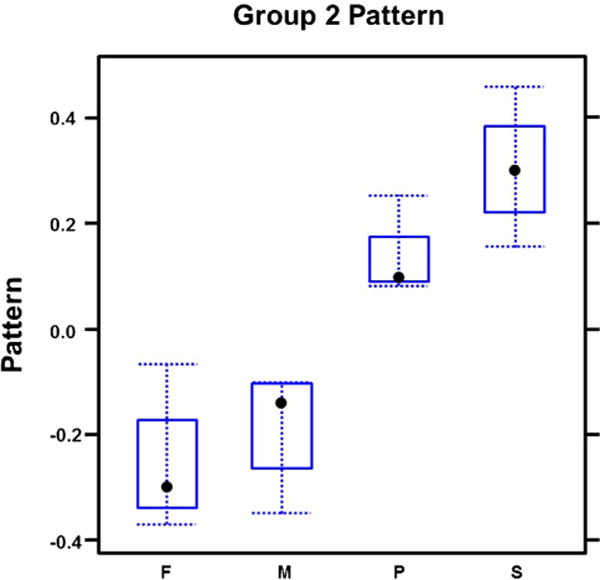
Expression of correlated genes in group 2. This group was highly expressed in parietal and sagittal compartments and segregated the compartments into two groups of frontal/metopic and parietal/sagittal.
Fig. 4.
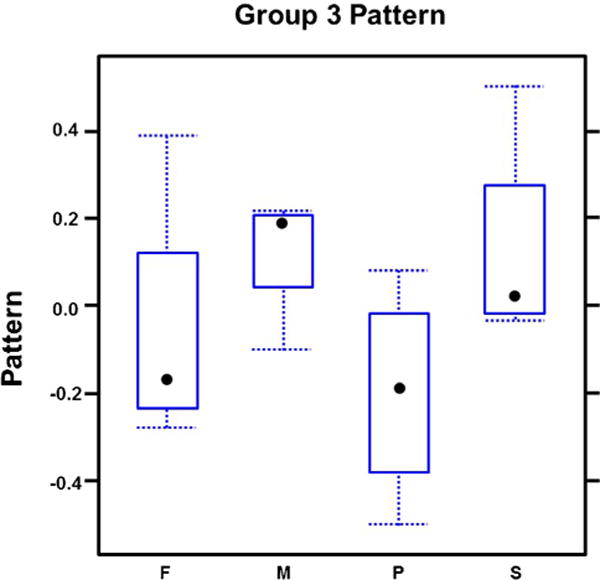
Expression of correlated genes in group 3. This group was highly expressed in metopic and sagittal compartments but the differential expression between the compartments was not as large as in groups 1 and 2.
Supplementary Table 9 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Supplementary Table 10 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
Since the correlation structures were defined independent of statistical significance and in order to enrich for genes with differential expression between compartments, we combined the results of two analyses. In order to be listed as highly correlated and significantly expressed genes, the expression of the genes in groups 1 and 2 had to be significantly different in four paired comparisons of frontal/metopic versus parietal/sagittal compartments and not different in frontal versus metopic or parietal versus sagittal comparisons (Tables 2 and 3). The same was performed for gene group 3. Our statistical analysis included transcripts with greater than ±1.2-fold change in expression level with a p-value of ≤0.05. None of the genes in group 3 met this requirement. Therefore, no further analysis was performed on this group.
Table 2.
Correlated genes, highly expressed in frontal and metopic compartments (table is sorted based on frontal–parietal fold change).
| Symbol | Gene name | Unigene | F–P foldΔ | p-Value | F–S foldΔ | p-Value | M–P foldΔ | p-Value | M–S foldΔ | p-Value | S–P foldΔ | p-Value | F–M foldΔ | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TFAP2B | Transcription factor AP-2 beta (activating enhancer binding protein 2 beta) | Hs.33102 | 3.78 | 0.0008 | 4.52 | 0.0002 | 5.85 | 0.0000 | 7 | 0.0000 | 0.84 | 0.6220 | 0.65 | 0.2349 |
| MAB21L1 | Mab-21-like 1 (C. elegans) | Hs.584776 | 3.68 | 0.0011 | 2.32 | 0.0278 | 3.87 | 0.0007 | 2.44 | 0.0202 | 1.58 | 0.2190 | 0.95 | 0.8905 |
| MTRNR2L2 | MT-RNR2-like 2 | Hs.666077 | 3.07 | 0.0001 | 3.07 | 0.0001 | 2.13 | 0.0047 | 2.13 | 0.0046 | 1 | 0.9937 | 1.44 | 0.1560 |
| DNAJC6 | DnaJ (Hsp40) homolog, subfamily C, member 6 | Hs.647643 | 2.6 | 0.0001 | 2.45 | 0.0001 | 2.5 | 0.0001 | 2.35 | 0.0002 | 1.06 | 0.7811 | 1.04 | 0.8474 |
| FAM105A | Family with sequence similarity 105, member A | Hs.477887 | 2.06 | 0.0028 | 1.69 | 0.0254 | 2.52 | 0.0002 | 2.07 | 0.0027 | 1.22 | 0.3877 | 0.82 | 0.3809 |
| DRAM1 | DNA-damage regulated autophagy modulator 1 | Hs.525634 | 1.92 | 0.0032 | 1.98 | 0.0022 | 1.79 | 0.0081 | 1.84 | 0.0057 | 0.97 | 0.8906 | 1.08 | 0.7263 |
| MRAP2 | Melanocortin 2 receptor accessory protein 2 | Hs.370055 | 1.88 | 0.0304 | 2.06 | 0.0141 | 2.21 | 0.0073 | 2.43 | 0.0031 | 0.91 | 0.7457 | 0.85 | 0.5579 |
| DNER | Delta/notch-like EGF repeat containing | Hs.234074 | 1.84 | 0.0153 | 1.78 | 0.0216 | 1.73 | 0.0272 | 1.67 | 0.0377 | 1.04 | 0.8850 | 1.06 | 0.8093 |
| IVNS1ABP | Influenza virus NS1A binding protein | Hs.497183 | 1.73 | 0.0011 | 1.55 | 0.0078 | 1.74 | 0.0011 | 1.55 | 0.0078 | 1.12 | 0.4724 | 1 | 0.9982 |
| PPAPDC1A | Phosphatidic acid phosphatase type 2 domain containing 1A | Hs.40479 | 1.72 | 0.0024 | 1.45 | 0.0311 | 1.9 | 0.0004 | 1.61 | 0.0070 | 1.18 | 0.3154 | 0.9 | 0.5439 |
| MT1F | Metallothionein 1F | Hs.513626 | 1.67 | 0.0033 | 1.54 | 0.0115 | 1.66 | 0.0035 | 1.54 | 0.0121 | 1.08 | 0.6331 | 1 | 0.9855 |
| ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | Hs.722260 | 1.66 | 0.0070 | 1.65 | 0.0079 | 1.5 | 0.0271 | 1.49 | 0.0304 | 1.01 | 0.9600 | 1.1 | 0.5818 |
| MT1X | Metallothionein 1X | Hs.374950 | 1.65 | 0.0006 | 1.49 | 0.0044 | 1.61 | 0.0009 | 1.46 | 0.0068 | 1.1 | 0.4627 | 1.02 | 0.8700 |
| GPR183 | G protein-coupled receptor 183 | Hs.784 | 1.61 | 0.0001 | 1.49 | 0.0004 | 1.44 | 0.0012 | 1.33 | 0.0084 | 1.08 | 0.4702 | 1.12 | 0.2910 |
| TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Hs.519880 | 1.53 | 0.0108 | 1.73 | 0.0013 | 1.74 | 0.0012 | 1.97 | 0.0001 | 0.88 | 0.4316 | 0.88 | 0.4189 |
| ACADM | Acyl-CoA dehydrogenase, C-4 to C-12 straight chain | Hs.445040 | 1.53 | 0.0220 | 1.5 | 0.0296 | 1.5 | 0.0306 | 1.46 | 0.0407 | 1.02 | 0.8997 | 1.03 | 0.8875 |
| ICAM1 | Intercellular adhesion molecule 1 | Hs.643447 | 1.5 | 0.0007 | 1.62 | 0.0001 | 1.26 | 0.0426 | 1.36 | 0.0076 | 0.92 | 0.4726 | 1.19 | 0.1179 |
| KRT7 | Keratin 7 | Hs.411501 | 1.49 | 0.0168 | 1.47 | 0.0213 | 1.47 | 0.0213 | 1.45 | 0.0269 | 1.02 | 0.9210 | 1.02 | 0.9211 |
| ABI3BP | ABI family, member 3 (NESH) binding protein | Hs.477015 | 1.47 | 0.0465 | 1.54 | 0.0272 | 2.04 | 0.0005 | 2.13 | 0.0003 | 0.96 | 0.8114 | 0.72 | 0.0910 |
| CDH3 | Cadherin 3, type 1, P-cadherin (placental) | Hs.191842 | 1.47 | 0.0042 | 1.39 | 0.0132 | 1.64 | 0.0003 | 1.56 | 0.0012 | 1.06 | 0.6581 | 0.89 | 0.3630 |
| NGF | Nerve growth factor (beta polypeptide) | Hs.2561 | 1.46 | 0.0005 | 1.37 | 0.0035 | 1.32 | 0.0083 | 1.24 | 0.0411 | 1.07 | 0.5040 | 1.11 | 0.3221 |
| SULF1 | Sulfatase 1 | Hs.409602 | 1.45 | 0.0000 | 1.23 | 0.0034 | 1.46 | 0.0000 | 1.24 | 0.0029 | 1.18 | 0.0195 | 1 | 0.9466 |
| ASXL3 | Additional sex combs like 3 (Drosophila) | Hs.464876 | 1.42 | 0.0472 | 1.46 | 0.0334 | 1.73 | 0.0031 | 1.78 | 0.0020 | 0.97 | 0.8763 | 0.82 | 0.2709 |
| PLAT | Plasminogen activator, tissue | Hs.491582 | 1.42 | 0.0015 | 1.4 | 0.0019 | 1.36 | 0.0043 | 1.35 | 0.0053 | 1.01 | 0.9356 | 1.04 | 0.7049 |
| MYPN | Myopalladin | Hs.55205 | 1.42 | 0.0004 | 1.34 | 0.0021 | 1.53 | 0.0000 | 1.44 | 0.0002 | 1.06 | 0.5469 | 0.93 | 0.4051 |
| IL11 | Interleukin 11 | Hs.467304 | 1.37 | 0.0018 | 1.43 | 0.0005 | 1.25 | 0.0216 | 1.31 | 0.0069 | 0.96 | 0.6447 | 1.1 | 0.3360 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | Hs.510635 | 1.35 | 0.0324 | 1.56 | 0.0024 | 1.33 | 0.0446 | 1.53 | 0.0035 | 0.87 | 0.3032 | 1.02 | 0.8868 |
| LDOC1 | Leucine zipper, down-regulated in cancer 1 | Hs.45231 | 1.34 | 0.0027 | 1.48 | 0.0001 | 1.26 | 0.0169 | 1.39 | 0.0010 | 0.91 | 0.2976 | 1.07 | 0.4796 |
| TFAP2C | Transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) | Hs.473152 | 1.45 | 0.0309 | 1.42 | 0.0426 | 1.4 | 0.0489 | 1.37 | 0.0662 | 1.04 | 0.8367 | 1.02 | 0.8867 |
Table 3.
Correlated genes, highly expressed in parietal and sagittal compartments (table is sorted based on frontal-parietal fold change).
| Symbol | Gene name | Unigene | F–P foldΔ |
p-Value | F–S foldΔ |
p-Value | M–P foldΔ |
p-Value | M–S foldΔ |
p-Value | S–P foldΔ |
p-Value | F–M foldΔ |
p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST6GALNAC5 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 | Hs.303609 | −2.08 | 0.0008 | −2.28 | 0.0002 | −1.52 | 0.0434 | −1.67 | 0.0150 | 1.1 | 0.6476 | −1.37 | 0.1267 |
| C3orf62 | Chromosome 3 open reading frame 62 | Hs.403828 | −1.85 | 0.0080 | −1.58 | 0.0455 | −1.91 | 0.0054 | −1.63 | 0.0325 | −1.17 | 0.4676 | 1.03 | 0.8806 |
| PDE5A | Phosphodiesterase 5A, cGMP-specific | Hs.647971 | −1.78 | 0.0000 | −1.75 | 0.0000 | −1.4 | 0.0043 | −1.38 | 0.0058 | −1.01 | 0.9096 | −1.27 | 0.0360 |
| HSPB6 | Heat shock protein, alpha-crystallin-related, B6 | Hs.534538 | −1.74 | 0.0010 | −1.69 | 0.0017 | −1.53 | 0.0093 | −1.49 | 0.0147 | −1.03 | 0.8540 | −1.14 | 0.4193 |
| TNC | Tenascin C | Hs.143250 | −1.74 | 0.0002 | −1.55 | 0.0019 | −2.05 | 0.0000 | −1.83 | 0.0001 | −1.12 | 0.3864 | 1.18 | 0.2233 |
| FOXF2 | Forkhead box F2 | Hs.484423 | −1.71 | 0.0009 | −1.35 | 0.0489 | −1.72 | 0.0008 | −1.36 | 0.0436 | −1.26 | 0.1216 | 1.01 | 0.9581 |
| TNFRSF11B | Tumour necrosis factor receptor superfamily, member 11b | Hs.81791 | −1.69 | 0.0010 | −1.77 | 0.0004 | −1.64 | 0.0017 | −1.72 | 0.0007 | 1.05 | 0.7349 | −1.03 | 0.8473 |
| KLHL4 | Kelch-like family member 4 | Hs.49075 | −1.61 | 0.0006 | −1.95 | 0.0000 | −1.53 | 0.0021 | −1.84 | 0.0000 | 1.21 | 0.1465 | −1.06 | 0.6722 |
| KCNT2 | Potassium channel, subfamily T, member 2 | Hs.657046 | −1.61 | 0.0000 | −1.66 | 0.0000 | −1.24 | 0.0399 | −1.28 | 0.0175 | 1.04 | 0.7224 | −1.3 | 0.0120 |
| MFAP4 | Microfibrillar-associated protein 4 | Hs.296049 | −1.61 | 0.0032 | −1.62 | 0.0026 | −1.4 | 0.0296 | −1.42 | 0.0254 | 1.01 | 0.9469 | −1.14 | 0.3759 |
| KIAA1107 | KIAA1107 | Hs.21554 | −1.61 | 0.0001 | −1.48 | 0.0008 | −1.48 | 0.0007 | −1.36 | 0.0066 | −1.09 | 0.4255 | −1.09 | 0.4351 |
| ADH1B | Alcohol dehydrogenase 1B (class I), beta polypeptide | Hs.4 | −1.56 | 0.0173 | −1.6 | 0.0115 | −1.47 | 0.0349 | −1.52 | 0.0238 | 1.03 | 0.8683 | −1.06 | 0.7639 |
| PRSS12 | Protease, serine, 12 (neurotrypsin, motopsin) | Hs.445857 | −1.53 | 0.0009 | −1.57 | 0.0004 | −1.3 | 0.0316 | −1.34 | 0.0173 | 1.03 | 0.7968 | −1.18 | 0.1747 |
| GEM | GTP binding protein overexpressed in skeletal muscle | Hs.654463 | −1.51 | 0.0166 | −1.53 | 0.0130 | −1.53 | 0.0139 | −1.55 | 0.0108 | 1.02 | 0.9205 | 1.01 | 0.9418 |
| BTBD8 | BTB (POZ) domain containing 8 | Hs.676102 | −1.51 | 0.0005 | −1.48 | 0.0008 | −1.33 | 0.0110 | −1.31 | 0.0163 | −1.02 | 0.8726 | −1.13 | 0.2635 |
| HECW2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | Hs.654742 | −1.5 | 0.0090 | −1.57 | 0.0044 | −1.51 | 0.0080 | −1.58 | 0.0039 | 1.04 | 0.7799 | 1.01 | 0.9635 |
| FAM129A | Family with sequence similarity 129, member A | Hs.518662 | −1.41 | 0.0154 | −1.37 | 0.0231 | −1.43 | 0.0110 | −1.4 | 0.0167 | −1.02 | 0.8665 | 1.02 | 0.8925 |
| FXYD1 | FXYD domain containing ion transport regulator 1 | Hs.442498 | −1.39 | 0.0122 | −1.45 | 0.0055 | −1.31 | 0.0400 | −1.36 | 0.0196 | 1.04 | 0.7567 | −1.07 | 0.6158 |
| CCND2 | Cyclin D2 | Hs.376071 | −1.38 | 0.0171 | −1.41 | 0.0109 | −1.39 | 0.0144 | −1.43 | 0.0091 | 1.02 | 0.8538 | 1.01 | 0.9426 |
| MYLIP | Myosin regulatory light chain interacting protein | Hs.484738 | −1.35 | 0.0094 | −1.42 | 0.0031 | −1.28 | 0.0319 | −1.34 | 0.0116 | 1.05 | 0.6717 | −1.06 | 0.6119 |
| ARHGEF35 | Rho guanine nucleotide exchange factor (GEF) 35 | Hs.534621 | −1.28 | 0.0407 | −1.39 | 0.0067 | −1.31 | 0.0240 | −1.43 | 0.0037 | 1.09 | 0.4580 | 1.03 | 0.8167 |
Genes such as TFAP2 (A–C), FOXF2, ICAM1, SULF1 and TNC (Tables 2 and 3) are among the genes that segregate the two compartment groups (frontal/metopic vs. parietal/sagittal) and are expressed significantly different between them. Biological function of these genes28–36 confer functional differences between the two compartment groups regarding proliferation, differentiation, cell migration and production of extracellular matrix. These 2 groups are also different in the expression of several NC regulatory genes.36–38 There was no significant difference in the expression of other NC regulatory genes37,38 in our study (Supplementary Table 11).
Supplementary Table 11 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.archoralbio.2015.06.008.
3.3. qRT-PCR validation
qRT-PCR was performed on six of the genes in Tables 2 and 3, TFAP2A, TFAP2B, FOXF2, ICAM1, SULF1 and TNC. These genes were selected due to their importance in neural crest development, cell proliferation, differentiation and extracellular matrix.28–36 Six paired comparisons were made in the same manner as for microarray results. The results (Table 4) were mostly in concert with microarray results. Similar to the microarray results, the expression of TFAP2A, TFAP2B, FOXF2 and TNC was significantly different in four paired comparisons of frontal/metopic versus parietal/sagittal compartments. The difference was not significant in frontal versus metopic or parietal versus sagittal comparisons. The expression of ICAM1 and SULF1 was significantly higher in frontal versus parietal and frontal versus sagittal comparisons. However unlike the microarray results the difference was not significant comparing metopic versus parietal and sagittal compartments by qRT-PCR.
Table 4.
qRT-PCR validation result.
| Symbol | F–P foldΔ | p-Value | F–S foldΔ | p-Value | M–P foldΔ | p-Value | M–S foldΔ | p-Value | F–M foldΔ | p-Value | P–S foldΔ | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TFAP2B | 19.65 | 0.0240 | 34.76 | 0.0017 | 30.93 | 0.0200 | 54.71 | 0.0024 | −1.57 | 0.4700 | 1.77 | 0.6300 |
| TFAP2A | 6.48 | 0.0059 | 5.4 | 0.0070 | 6.54 | 0.0016 | 5.45 | 0.0016 | −1.01 | 0.9800 | −1.2 | 0.5600 |
| ICAM1 | 1.85 | 0.0074 | 2.12 | 0.0097 | 1.22 | 0.5412 | 1.4 | 0.2884 | −1.14 | 0.4662 | 1.52 | 0.1919 |
| SULF1 | 2.11 | 0.0314 | 1.71 | 0.0527 | 1.8 | 0.0631 | 1.46 | 0.1642 | 1.23 | 0.0274 | 1.18 | 0.2748 |
| TNC | −2 | 0.0083 | −1.85 | 0.0458 | −3.19 | 0.0016 | −2.95 | 0.0429 | −1.08 | 0.7427 | 1.59 | 0.0724 |
| FOXF2 | −2.8 | 0.0001 | −1.91 | 0.0006 | −2.23 | 0.0067 | −1.52 | 0.0750 | −1.25 | 0.2600 | 1.46 | 0.0230 |
4. Discussion
The aim of this paper was to analyze gene expression of human cell lines derived from foetal cranial bone and ISM to investigate the existence of gene expression patterns indicating their embryonic origins.
In this study, the pairwise comparisons and correlation analyses determined that among the four calvarial compartments, those with unlike tissues (frontal bone/metopic suture vs. parietal bone/sagittal suture), have more similar gene expression profiles. We found specific biological themes in these patterns and suggest that these expression profiles reveal developmental and functional differences related to neural crest regulation and extracellular matrix production between the two compartment groups (frontal/metopic vs. parietal/sagittal) (Table 5).
Table 5.
Genes of interest in comparison of frontal/metopic vs. parietal/sagittal compartments.
| Symbol | Unigene | F–P foldΔ | p-Value | F–S foldΔ | p-Value | M–P foldΔ | p-Value | M–S foldΔ | p-Value | S–P foldΔ | p-Value | F–M foldΔ | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TFAP2B | Hs.33102 | 3.78 | 0.0008 | 4.52 | 0.0002 | 5.85 | 0.0000 | 7 | 0.0000 | 0.84 | 0.6220 | 0.65 | 0.2349 |
| TFAP2A | Hs.519880 | 1.53 | 0.0108 | 1.73 | 0.0013 | 1.74 | 0.0012 | 1.97 | 0.0001 | 0.88 | 0.4316 | 0.88 | 0.4189 |
| ICAM1 | Hs.643447 | 1.5 | 0.0007 | 1.62 | 0.0001 | 1.26 | 0.0426 | 1.36 | 0.0076 | 0.92 | 0.4726 | 1.19 | 0.1179 |
| SULF1 | Hs.409602 | 1.45 | 0.0000 | 1.23 | 0.0034 | 1.46 | 0.0000 | 1.24 | 0.0029 | 1.18 | 0.0195 | 1 | 0.9466 |
| TFAP2C | Hs.473152 | 1.45 | 0.0309 | 1.42 | 0.0426 | 1.4 | 0.0489 | 1.37 | 0.0662 | 1.04 | 0.8367 | 1.02 | 0.8867 |
| TNC | Hs.143250 | −1.74 | 0.0002 | −1.55 | 0.0019 | −2.05 | 0.0000 | −1.83 | 0.0001 | −1.12 | 0.3864 | 1.18 | 0.2233 |
| FOXF2 | Hs.484423 | −1.71 | 0.0009 | −1.35 | 0.0489 | −1.72 | 0.0008 | −1.36 | 0.0436 | −1.26 | 0.1216 | 1.01 | 0.9581 |
Neural crest regulatory genes which are also important in proliferation and differentiation are differently expressed between frontal/metopic vs. parietal/sagittal compartments. During embryonic development there are NC regulatory genes that have a tissue specific expression pattern in either NC or the surrounding mesodermal tissues.38 We identified regulatory gene expression profiles similar to NC in frontal/metopic compartments and profiles similar to mesoderm in parietal/sagittal compartments. We found high levels of expression of TFAP2 gene family in frontal/metopic compartments and high levels of expression of FOXF2 in parietal/sagittal compartments. These genes are among the regulatory genes of NC development.36,38 They have distinct roles in proliferation and differentiation as well.39–45
Transcription factor TFAP2 family consists of five members, A, B, C, D and E. Among the orthologs TFAP2A, B and C are structurally similar. These genes are expressed in the early development of NC and considered to be NC specifiers.29,33,34,38,46,47 Mutations of these genes in humans and animal models cause deficiencies in craniofacial tissues derived from NC.48–50 In our study, orthologs important in NC development (TFAP2 (A–C)) are the only members that are differentially expressed. These transcripts are upregulated in frontal and metopic compartments. Members of the FOX family are among NC regulatory genes as well.36 Fox genes such as FOXF2 are primarily expressed in the mesodermal layers of the embryo including the paraxial, axial, lateral and lateral splenic mesoderm.51 However, their expression in mesoderm affects development of adjacent neural crest derived tissues.52 They are important in NC patterning and craniofacial organization.53 We found higher expression of FOXF2 in parietal/sagittal cells compared to frontal/metopic cells.
In this study, differential gene expression of TFAP2 (A–C) and FOXF2 in frontal/metopic compartments compared to parietal/sagittal compartments supports previous studies demonstrating the distinct embryonic origins of these compartments. High expression of TFAP2 (A–C) in frontal and metopic compartments suggests that they are originated from NC. On the other hand, high expression of FOXF2 and low expression of TFAP2 (A–C) in parietal and sagittal compartments suggest a different developmental origin for these compartments. These findings are in agreement with the previous studies on the origin of frontal, parietal bones and metopic suture but not sagittal suture. In previous studies, the findings regarding the sagittal suture were inconsistent and not conclusive.11–13 Since mingling and migration of NC and mesoderm cells happen during development specifically in sagittal suture,11,14 the disagreement on the origin of sagittal suture can be due to different time points that were studied.
FOXF2 and the TFAP2 family are also important in cell proliferation and differentiation. Animal studies on different cell types show that reduction of expression of TFAP2 reduces cell proliferation and induces early differentiation while reduction of FOXF2 leads to higher proliferation and less production of ECM.45,51 Similar results in the investigations of cancerous cells indicate association of high cell proliferation with high expression of TFAP2 and low expression of FOXF2.39,40,44,54 Based on the function of TFAP2 and FOXF239–41,43,45 and their expression profile presented in this study, we suggest that the frontal and metopic compartments have an increased potential for proliferation and reduced differentiation while the converse is true for the parietal and sagittal compartments. High proliferation potential in frontal and metopic compartments can be a contributory factor in early physiologic fusion of metopic suture.
While this hypothesis requires more study on human cells the studies on the cellular phenotype of rodent frontal and parietal osteoblasts evaluated proliferation and differentiation potential using BrdU proliferation assay and alkaline phosphatase (ALP) assay, respectively.17,18,55 These studies support our hypothesis on higher proliferation capacity of the frontal bone. However, the results on the early differentiation of the cells are not consistent. This discrepancy may be due to different experimental methods.
ZIC family, MSX1/2, SOX9/10, ANAI2, PAX3/7 and MYC are other NC specifiers in NC gene regulatory network.37,38 In the present study the expression of these genes is not significantly different between the frontal and metopic group compared to parietal and sagittal group. However, a tendency of higher expression of PAX3 and lower expression of ZIC family is seen in frontal and metopic compartments. These transcripts can be future targets for a study with a larger samples size.
Based on gene expression, the extracellular matrix (ECM) in frontal/metopic compartments has a different composition compared to the ECM in parietal/sagittal compartments. We have demonstrated that SULF1, ICAM1 and TNC, important genes in the composition of ECM, are differentially expressed in the two compartment groups. SULF1 is an extracellular protein involved in remodelling proteoglycans on cell membranes so that NC cells are recognized by other NC cells during development. SULF1 modulates the path through which NC cells migrate31 and therefore is important in the regulation of NC development.38,56 Protein encoded by ICAM1 is a surface glycoprotein that regulates cell–cell contact and cell movement through ECM.30,32 It is highly expressed in the tissues with high proliferation and high potential for migration like prostate, breast, lung and bone cancer.30,32 Furthermore, ICAM1 is regulated by TFAP-2 which is one of the genes of interest in this study.57 The protein encoded by TNC (Tenascin) is a major extracellular matrix protein important in cell adhesion, proliferation and differentiation.58 Although it is known to play an important role in development, there are controversies surrounding its exact roles. While some studies suggest that the protein is secreted by NC cells playing a role in delamination and migration, others believe that tenascin expression has an inhibitory effect on NC migration.28,35,58 Regardless of the exact effect of tenascin on NC migration, these studies demonstrate its importance on ECM and NC development.
SULF1, ICAM1 are highly expressed in frontal/metopic compartments and TNC is high in parietal/sagittal compartments. These differences suggest that the two groups have ECM with distinct characteristics. The ECM composition of frontal/metopic compartments is more similar to NC suggesting facilitation of cell migration. The similarity of the expression of ECM genes in frontal/metopic compartments in this study and the expression of these genes in NC further supports previous studies on the NC origin of frontal and metopic tissues.11–13
Considering the function of these differentially expressed genes and the fact that metopic suture is the only suture that fuses during the first year of life in humans,10 we propose that the distinct expression of these genes plays an important role in suture fate and the development of cranium. Since ICAM1 facilitates adhesion of the cells to heterotypic cells, it can facilitate adhesion of frontal osteoblasts and metopic mesenchymal cells and increase their motility and migration. SULF1 can modulate the responsiveness of frontal and metopic cells to signalling pathways important in cell migration and osteogenesis and finally, the TFAP2 enhances proliferation. We suggest that the expression of these genes in frontal/metopic compartments makes the ECM appropriate for mingling of high proliferative frontal osteoblasts and metopic mesenchymal cells and results in the early physiologic closure of the suture. However, this is not the case in the sagittal suture complex in which the calvaria and ISM are not NC derived.
5. Conclusion
In order to elucidate the aetiology of craniosynostosis more studies on molecular mechanisms of suture fusion are required. This study sheds light on the physiological differences between cranial compartments. Our data demonstrates that during the early second trimester of humans, foetal cranial compartments have differential gene expression profiles that persist in cell culture. Gene expression profiles are more similar in the compartments with unlike tissues (frontal bone/metopic suture vs. parietal bone/sagittal suture) reflecting the NC and paraxial mesoderm origins of the compartments. Due to the difference in embryonic origin of distinct parts of occipital bone in mammalians59,60 investigating this finding in occipital bone is of great interest. However, since early fusion of lambdoid suture is the rarest cranyosynistosis61 this study is focused on frontal, parietal, metopic and sagittal compartments.
Future studies will focus on correlations between gene expression and cellular phenotype including proliferation and differentiation and ECM production in each compartment. Furthermore, the gene expression at different time points through the course of development in both cases of normal and early sutural fusion requires more investigation.
Supplementary Material
Acknowledgments
Funding: Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research under Award Number R01DE018227 (MLC), the Jean Renny Endowment for Craniofacial Research (MLC), the (MLC), the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number P30ES007033 (FMF, TKB and JWB) and the National Institute of Child Health and Human Development under Award Number P30 HD02274 (FMF, TKB and JWB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest
None.
Competing interests
None declared.
Ethical approval
Not applicable.
References
- 1.Moore KL. The developing human clinically oriented embryology. 6th. Philadelphia: Saunders; 1998. [Google Scholar]
- 2.Cohen MM., Jr . Craniosynostosis. 2nd. New York: Oxford University Press; 2000. [Google Scholar]
- 3.Kimonis V, Gold JA, Hoffman TL, Panchal J, Boyadjiev SA. Genetics of craniosynostosis. Semin Pediatr Neurol. 2007;14(3):150–61. doi: 10.1016/j.spen.2007.08.008. Epub 2007/11/06. [DOI] [PubMed] [Google Scholar]
- 4.Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37(3):275–81. doi: 10.1038/ng1511. Epub 2005/02/26. [DOI] [PubMed] [Google Scholar]
- 5.Rawlins J, Opperman L. Craniofacial sutures development disease and treatment. Basel: Karger; 2008. p. 18. [Google Scholar]
- 6.Ting MC, Wu NL, Roybal PG, Sun J, Liu L, Yen Y, et al. EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development. 2009;136(5):855–64. doi: 10.1242/dev.028605. Epub 2009/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passos-Bueno MR, Serti Eacute AE, Jehee FS, Fanganiello R, Yeh E. Genetics of craniosynostosis: genes, syndromes, mutations and genotype–phenotype correlations. Front Oral Biol. 2008;12:107–43. doi: 10.1159/000115035. Epub 2008/04/09. [DOI] [PubMed] [Google Scholar]
- 8.Wilkie AO. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6(10):1647–56. doi: 10.1093/hmg/6.10.1647. Epub 1997/01/01. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham ML, Heike CL. Evaluation of the infant with an abnormal skull shape. Curr Opin Pediatr. 2007;19(6):645–51. doi: 10.1097/MOP.0b013e3282f1581a. Epub 2007/11/21. [DOI] [PubMed] [Google Scholar]
- 10.Vu HL, Panchal J, Parker EE, Levine NS, Francel P. The timing of physiologic closure of the metopic suture: a review of 159 patients using reconstructed 3D CT scans of the craniofacial region. J Craniofac Surg. 2001;12(6):527–32. doi: 10.1097/00001665-200111000-00005. Epub 2001/11/17. [DOI] [PubMed] [Google Scholar]
- 11.Gagan JR, Tholpady SS, Ogle RC. Cellular dynamics and tissue interactions of the dura mater during head development. Birth Defects Res C Embryo Today. 2007;81(4):297–304. doi: 10.1002/bdrc.20104. Epub 2008/01/30. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241(1):106–16. doi: 10.1006/dbio.2001.0487. Epub 2002/01/11. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125(9–10):797–808. doi: 10.1016/j.mod.2008.06.007. Epub 2008/07/12. [DOI] [PubMed] [Google Scholar]
- 14.Deckelbaum RA, Holmes G, Zhao Z, Tong C, Basilico C, Loomis CA. Regulation of cranial morphogenesis and cell fate at the neural crest–mesoderm boundary by engrailed 1. Development. 2012;139(7):1346–58. doi: 10.1242/dev.076729. Epub 2012/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morriss-Kay GM, Wilkie AO. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. J Anat. 2005;207(5):637–53. doi: 10.1111/j.1469-7580.2005.00475.x. Epub 2005/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Meyer NP, Quarto N, Longaker MT. Integration of multiple signaling regulates through apoptosis the differential osteogenic potential of neural crest-derived and mesoderm-derived osteoblasts. PLOS ONE. 2013;8(3):e58610. doi: 10.1371/journal.pone.0058610. Epub 2013/03/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Quarto N, Longaker MT. Activation of FGF signaling mediates proliferative and osteogenic differences between neural crest derived frontal and mesoderm parietal derived bone. PLoS ONE. 2010;5(11):e14033. doi: 10.1371/journal.pone.0014033. Epub 2010/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quarto N, Wan DC, Kwan MD, Panetta NJ, Li S, Longaker MT. Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res. 2010;25(7):1680–94. doi: 10.1359/jbmr.091116. Epub 2009/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamper BD, Mecham B, Park SS, Wilkerson H, Farin FM, Beyer RP, et al. Transcriptome correlation analysis identifies two unique craniosynostosis subtypes associated with IRS1 activation. Physiol Genomics. 2012;44(23):1154–63. doi: 10.1152/physiolgenomics.00085.2012. Epub 2012/10/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamper BD, Park SS, Beyer RP, Bammler TK, Cunningham ML. Unique sex-based approach identifies transcriptomic biomarkers associated with non-syndromic craniosynostosis. Gene Regul Syst Biol. 2012;6:81–92. doi: 10.4137/GRSB.S9693. Epub 2012/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–7. doi: 10.1093/bioinformatics/btq431. Epub 2010/08/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. Epub 2003/08/20. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinform. 2006;7:261. doi: 10.1186/1471-2105-7-261. Epub 2006/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3, Epub 2006/05/02. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–61. doi: 10.1038/nbt1239. Epub 2006/09/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97(18):10101–6. doi: 10.1073/pnas.97.18.10101. Epub 2000/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. Epub 2002/02/16. [DOI] [PubMed] [Google Scholar]
- 28.Breau MA, Dahmani A, Broders-Bondon F, Thiery JP, Dufour S. Beta1 integrins are required for the invasion of the caecum and proximal hindgut by enteric neural crest cells. Development. 2009;136(16):2791–801. doi: 10.1242/dev.031419. Epub 2009/07/28. [DOI] [PubMed] [Google Scholar]
- 29.Chazaud C, Oulad-Abdelghani M, Bouillet P, Decimo D, Chambon P Dolle. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mech Dev. 1996;54(1):83–94. doi: 10.1016/0925-4773(95)00463-7. Epub 1996/01/01. [DOI] [PubMed] [Google Scholar]
- 30.Chen PC, Lin TH, Cheng HC, Tang CH. CCN3 increases cell motility and ICAM-1 expression in prostate cancer cells. Carcinogenesis. 2012;33(4):937–45. doi: 10.1093/carcin/bgs108. Epub 2012/02/22. [DOI] [PubMed] [Google Scholar]
- 31.Guiral EC, Faas L, Pownall ME. Neural crest migration requires the activity of the extracellular sulphatases XtSulf1 and XtSulf2. Dev Biol. 2010;341(2):375–88. doi: 10.1016/j.ydbio.2010.02.034. Epub 2010/03/09. [DOI] [PubMed] [Google Scholar]
- 32.Lin YM, Chang ZL, Liao YY, Chou MC, Tang CH. IL-6 promotes ICAM-1 expression and cell motility in human osteosarcoma. Cancer Lett. 2013;328(1):135–43. doi: 10.1016/j.canlet.2012.08.029. Epub 2012/09/04. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5(1):105–19. doi: 10.1101/gad.5.1.105. Epub 1991/01/01. [DOI] [PubMed] [Google Scholar]
- 34.Moser M, Ruschoff J, Buettner R. Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Dev Dyn. 1997;208(1):115–24. doi: 10.1002/(SICI)1097-0177(199701)208:1<115::AID-AJA11>3.0.CO;2-5. Epub 1997/01/01. [DOI] [PubMed] [Google Scholar]
- 35.Tucker RP, McKay SE. The expression of tenascin by neural crest cells and glia. Development. 1991;112(4):1031–9. doi: 10.1242/dev.112.4.1031. Epub 1991/08/01. [DOI] [PubMed] [Google Scholar]
- 36.Nelms BL, Labosky PA. Transcriptional control of neural crest development, San Rafael, CA. (1st) 2010 [PubMed] [Google Scholar]
- 37.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7(3):291–9. doi: 10.1016/j.devcel.2004.08.007. Epub 2004/09/15. [DOI] [PubMed] [Google Scholar]
- 38.Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69(22):3715–37. doi: 10.1007/s00018-012-0991-8. Epub 2012/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beger M, Butz K, Denk C, Williams T, Hurst HC, Hoppe-Seyler F. Expression pattern of AP-2 transcription factors in cervical cancer cells and analysis of their influence on human papillomavirus oncogene transcription. J Mol Med (Berl) 2001;79(5–6):314–20. doi: 10.1007/s001090100211. Epub 2001/08/04. [DOI] [PubMed] [Google Scholar]
- 40.Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene. 1996;13(8):1701–7. Epub 1996/10/17. [PubMed] [Google Scholar]
- 41.Ding X, Yang Z, Zhou F, Wang F, Li X, Chen C, et al. Transcription factor AP-2alpha regulates acute myeloid leukemia cell proliferation by influencing Hoxa gene expression. Int J Biochem Cell Biol. 2013;45(8):1647–56. doi: 10.1016/j.biocel.2013.04.024. Epub 2013/05/11. [DOI] [PubMed] [Google Scholar]
- 42.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, et al. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2 RECK and MTSS1 genes in human prostate cancer. PLOS ONE. 2013;8(1):e55502. doi: 10.1371/journal.pone.0055502. Epub 2013/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoei-Hansen CE, Nielsen JE, Almstrup K, Sonne SB, Graem N, Skakkebaek NE, et al. Transcription factor AP-2gamma is a developmentally regulated marker of testicular carcinoma in situ and germ cell tumors. Clin Cancer Res. 2004;10(24):8521–30. doi: 10.1158/1078-0432.CCR-04-1285. Epub 2004/12/30. [DOI] [PubMed] [Google Scholar]
- 44.Nik AM, Reyahi A, Ponten F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144(5):1001–11. doi: 10.1053/j.gastro.2013.01.045. Epub 2013/02/05. [DOI] [PubMed] [Google Scholar]
- 45.Pfisterer P, Ehlermann J, Hegen M, Schorle H. A subtractive gene expression screen suggests a role of transcription factor AP-2 alpha in control of proliferation and differentiation. J Biol Chem. 2002;277(8):6637–44. doi: 10.1074/jbc.M108578200. Epub 2001/12/14. [DOI] [PubMed] [Google Scholar]
- 46.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6(13):246. doi: 10.1186/gb-2005-6-13-246. Epub 2006/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Luo T, Sargent TD. Expression of TFAP2beta and TFAP2gamma genes in Xenopus laevis. Gene Expr Patterns. 2006;6(6):589–95. doi: 10.1016/j.modgep.2005.11.011. Epub 2006/01/18. [DOI] [PubMed] [Google Scholar]
- 48.Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82(5):1171–7. doi: 10.1016/j.ajhg.2008.03.005. Epub 2008/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao F, Weismann CG, Satoda M, Pierpont ME, Sweeney E, Thompson EM, et al. Novel TFAP2B mutations that cause Char syndrome provide a genotype–phenotype correlation. Am J Hum Genet. 2001;69(4):695–703. doi: 10.1086/323410. Epub 2001/08/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381(6579):238–41. doi: 10.1038/381238a0. Epub 1996/05/16. [DOI] [PubMed] [Google Scholar]
- 51.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, et al. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133(5):833–43. doi: 10.1242/dev.02252. Epub 2006/01/28. [DOI] [PubMed] [Google Scholar]
- 52.Aitola M, Carlsson P, Mahlapuu M, Enerback S, Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epitheliomesenchymal interactions. Dev Dyn. 2000;218(1):136–49. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. Epub 2000/05/24. [DOI] [PubMed] [Google Scholar]
- 53.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18(8):937–51. doi: 10.1101/gad.1190304. Epub 2004/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLOS ONE. 2013;8(4):e61591. doi: 10.1371/journal.pone.0061591. Epub 2013/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Malladi P, Zhou D, Longaker MT. Molecular and cellular characterization of mouse derived from neural crest and paraxial mesoderm. Plast Reconstr Surg. 2007;120(7):1783–95. doi: 10.1097/01.prs.0000279491.48283.51. Epub 2007/12/20. [DOI] [PubMed] [Google Scholar]
- 56.Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27(2):678–88. doi: 10.1128/MCB.01279-06. Epub 2006/11/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grether-Beck S, Olaizola-Horn S, Schmitt H, Grewe M, Jahnke A, Johnson JP, et al. Activation of transcription factor AP-2 mediates UVA radiation- and singlet oxygen-induced expression of the human intercellular adhesion molecule 1 gene. Proc Natl Acad Sci U S A. 1996;93(25):14586–91. doi: 10.1073/pnas.93.25.14586. Epub 1996/12/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tucker RP. Abnormal neural crest cell migration after the in vivo knockdown of tenascin-C expression with morpholino antisense oligonucleotides. Dev Dyn. 2001;222(1):115–9. doi: 10.1002/dvdy.1171. Epub 2001/08/17. [DOI] [PubMed] [Google Scholar]
- 59.Gross JB, Hanken J. Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Dev Biol. 2008;317(2):389–400. doi: 10.1016/j.ydbio.2008.02.046. Epub 2008/04/12. [DOI] [PubMed] [Google Scholar]
- 60.Koyabu D, Maier W, Sanchez-Villagra MR. Paleontological and developmental evidence resolve the homology and dual embryonic origin of a mammalian skull bone, the interparietal. Proc Natl Acad Sci U S A. 2012;109(35):14075–80. doi: 10.1073/pnas.1208693109. Epub 2012/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellenbogen RG, Gruss JS, Cunningham ML. Update on craniofacial surgery: the differential diagnosis of lambdoid synostosis/posterior plagiocephaly. Clin Neurosurg. 2000;47:303–18. Epub 2001/02/24. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


