Abstract
Constitutional heterozygous loss-of-function mutations in the SPRED1 gene cause a phenotype known as Legius syndrome, which consists of symptoms of multiple café-au-lait macules, axillary freckling, learning disabilities, and macrocephaly. Legius syndrome resembles a mild neurofibromatosis type 1 (NF1) phenotype. It has been demonstrated that SPRED1 functions as a negative regulator of the Ras-ERK pathway and interacts with neurofibromin, the NF1 gene product. However, the molecular details of this interaction and the effects of the mutations identified in Legius syndrome and NF1 on this interaction have not yet been investigated. In this study, using a yeast two-hybrid system and an immunoprecipitation assay in HEK293 cells, we found that the SPRED1 EVH1 domain interacts with the N-terminal 16 amino acids and the C-terminal 20 amino acids of the GTPase-activating protein (GAP)-related domain (GRD) of neurofibromin, which form two crossing α-helix coils outside the GAP domain. These regions have been shown to be dispensable for GAP activity and are not present in p120GAP. Several mutations in these N- and C-terminal regions of the GRD in NF1 patients and pathogenic missense mutations in the EVH1 domain of SPRED1 in Legius syndrome reduced the binding affinity between the EVH1 domain and the GRD. EVH1 domain mutations with reduced binding to the GRD also disrupted the ERK suppression activity of SPRED1. These data clearly demonstrate that SPRED1 inhibits the Ras-ERK pathway by recruiting neurofibromin to Ras through the EVH1-GRD interaction, and this study also provides molecular basis for the pathogenic mutations of NF1 and Legius syndrome.
Keywords: GTPase-activating protein (GAP), human genetics, mitogen-activated protein kinase (MAPK), protein domain, Ras protein, signal transduction, negative regulation
Introduction
Spred (Sprouty-related protein with an EVH1 domain) family proteins, initially discovered as c-Kit- and c-Fms-binding proteins, have been shown to suppress the Ras-ERK pathway. Spreds form a subfamily of the Sprouty/Spred family, which is characterized by the Sprouty-related C-terminal cysteine-rich (SPR)3 domain. In mammals, four Sprouty homologues and three members of the Spred family of proteins, Spred1, Spred2, and Spred3, have been discovered. Mammalian Spreds can negatively regulate the Ras-Raf-ERK pathway (1, 2); however, compared with Spred1 and Spred2, Spred3 has much weaker ERK suppression activity (3). The SPR domain has been shown to be palmitoylated, causing Sprouty and Spred to localize in the membrane fraction (4, 5). Spred1 and Spred2 are composed of an N-terminal enabled/vasodilator-stimulated protein homology 1 (EVH1) domain, a central c-Kit-binding domain, and a cysteine-rich C-terminal SPR domain, whereas Spred3 lacks a functional c-Kit-binding domain.
Germline loss-of-function mutations in the SPRED1 gene have been identified in patients fulfilling the clinical diagnostic criteria from the National Institutes of Health for neurofibromatosis type 1 (NF1) where no NF1 mutation could be identified. SPRED1 mutations account for at least 2% of the pathogenic mutations in patients clinically diagnosed with NF1 (6–11). The phenotype exhibited by such patients is known as NF1-like syndrome or Legius syndrome (OMIM 611431) and consists of multiple café-au-lait macules (CALM), axillary freckling, macrocephaly, and sometimes mild neurocognitive impairment, as well as a lack of certain features that are common in NF1, such as neurofibromas, iris Lisch nodules, and NF1-related malignancies (11). However, there might be an increased risk for leukemia in children with Legius syndrome (12). The similarities between NF1 and Legius syndrome, as well as that between the biochemical functions of neurofibromin and those of SPRED1, suggest that these two syndromes both result in part from hyperactive Ras-ERK signaling, as are Noonan syndrome, Noonan syndrome with lentigines (previous LEOPARD syndrome), cardio-facio-cutaneous syndrome, and Costello syndrome (13). Furthermore, Spreds are putative tumor suppressors. It has been reported that SPRED1 and SPRED2 expression is reduced in human hepatocellular carcinoma (14), and SPRED1 mutation and reduced expression are also found in acute myeloblastic leukemia (12). Overexpression of SPRD1 in tumor cells resulted in reduced tumorigenicity in nude mice (15). Moreover, the bi-allelic inactivation of SPRED1 has been demonstrated in melanocytes cultured from a café-au-lait macule in a patient with Legius syndrome (8).
It was revealed that the C-terminal deletion mutant of Spred1 functions as a dominant negative form against endogenous Spred1 and augments serum and nerve growth factor-induced ERK activation, suggesting that the EVH1 domain binds to a factor necessary for Ras inhibition (16, 17). However, despite lengthy and extensive screening, no binding partner of the EVH1 domain has been found so far. Recently, a molecular link between neurofibromin and SPRED1 was discovered. Stowe et al. (18) demonstrated that the SPRED1 protein binds to neurofibromin, the NF1 gene product, resulting in the plasma membrane localization of neurofibromin, which subsequently down-regulates Ras-GTP levels. This model explains why Legius syndrome resembles NF1 and how SPREDs suppress the Ras-ERK pathway. However, the molecular details of the Spred-neurofibromin interaction and the effects of mutations in SPRED1 and NF1 remain to be clarified. In this study, we found that the SPRED1 EVH1 domain interacts with the N- and C-terminal extended region of the GTPase-activating protein (GAP)-related domain (GRD) of neurofibromin. Some mutations in these N- and C-terminal regions of the GRD identified in NF1 patients reduced the binding of the GRD to the EVH1 domain. Furthermore, SPRED1 EVH1 mutations lost the ability to bind to the GRD, leading to reduced ERK suppression activity. Our data provide molecular details on the function of the EVH1 domain and the GRD, which contributes to the pathogenesis of NF1 and Legius syndrome features and the potential development of Ras pathway inhibitors for cancer therapy.
Experimental Procedures
Patient SPRED1 cDNA
SPRED1 mutation analysis was performed at the Department of Human Genetics, Catholic University of Leuven, Belgium, and in the UAB Medical Genomics Laboratory, University of Alabama at Birmingham, in individuals with a Legius syndrome phenotype (19). Some samples were sent by clinical geneticists from other centers. Individuals showed a phenotype compatible with Legius syndrome; specifically, the presence of CALM and/or freckling and the absence of neurofibromas, and comprehensive NF1 mutation analysis was performed in all patients with no NF1 mutations found. All patients carrying missense variants affecting the SPRED1 EVH1 domain, except one (p.Gly62Arg, described in the ARUP database), are summarized in the SPRED1 LOVD database, including references therein for those patients previously reported. Some patients were previously described (6, 7, 9, 19, 20). Mutation numbering was based on the cDNA sequence with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence (GenBankTM accession code NM_152594.2). For protein numbering, the initiation codon was codon 1 (NP_689807.1).
Human SPRED1 cDNAs were cloned in pcDNA3 with a six-repeated Myc tag or pCMV2 with a FLAG tag at the N terminus. Mutant SPRED1 construction in a pMax vector (Amaxa Biosystems, Gaithersburg, MD) was performed by polymerase chain reaction (PCR)-directed mutagenesis as described previously (2).
Patient NF1 cDNA
NF1 mutation analysis was performed in the UAB Medical Genomics Laboratory, University of Alabama at Birmingham, using an RNA-based approach complemented by DNA-based dosage analyses, essentially as described previously (21, 22). NF1 mutations are described following recommendations of the Human Genome Variation Society using NM_001042492.2 as the reference sequence. Phenotypic data of all individuals carrying an NF1 constitutional missense mutation affecting amino acids 1202–1217 and 1511–1530, as provided by the referring physicians at the time of submission of the sample for clinical testing using a standardized phenotypic checklist, were summarized.
Ethics Information
This study was approved by the local institutional review board (IRB) of Catholic University of Leuven and University of Alabama at Birmingham.
Yeast Two-hybrid Assay
The yeast two-hybrid assay was performed as described (17, 23, 24). Briefly, DNA fragments encoding the human SPRED1 EVH1 domain (amino acid numbers 1–136) or the substitution mutants were subcloned into the pGBKT7 vector as fusions to the LexA DNA-binding domain. Human NF1 cDNAs or GRD cDNAs were subcloned into the pGADT7 vector as a fusion to GAL4 DNA-activating domain. We have used Y187 strain in which interaction of bait and prey induces both α-galactosidase and β-galactosidase. To detect interaction on the filter paper, yeasts were grown in Leu and Trp double dropout plates containing X-α-Gal for in situ staining, as described previously (23). Quantitative β-galactosidase assay for yeast two-hybrid systems was performed using β-galactopyranoside (Nacalai Tesque) as a substrate according to a previously described method (25).
Cell Culture and Transfection
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) in 10-cm dishes. Immunoprecipitation and immunoblotting were performed using anti-Myc (9E10), anti-HA (Clontech, rabbit polyclonal), and anti-FLAG (M2) antibodies as described (26). Briefly, the HEK293 cells were transfected with 1–2 μg of plasmid of SPRED1 or GRD expression vectors using the polyethyleneimine (PEI) method as described (27). Cells were lysed in 0.5 ml of TNE lysis buffer containing 150 mm Tris-HCl (pH 7.6), 50 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40 supplemented with protease inhibitor mixture (Nacalai Tesque) (28). After lysis, cellular debris was removed by centrifugation at 15,000 × g for 10 min. Protein from cell lysates was precipitated using 2 μg of antibody and 25 μl of TrueBlot® anti-mouse Ig immunoprecipitation beads or protein G-Sepharose (GE Healthcare) for 2 h at 4 °C. The immune complex was washed three times with a buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1% Nonidet P-40. For Western blotting, the immunoprecipitates or whole cell lysates were resolved using SDS-PAGE and transferred to Immobilon-P membranes (Millipore). The membranes were blotted with the indicated antibodies, and the bound antibodies were visualized using horseradish peroxidase-conjugated antibodies against goat, rabbit, or mouse IgG, and Chemi-Lumi One L Western blotting detection reagents (Nacalai Tesque).
ERK Reporter Assay and Luciferase Assay
The Elk-1 activation was measured by the GAL4 DNA-binding domain/Elk-1 fusion system according to the manufacturer's instructions (PathDetect in vivo signal transduction pathway trans-reporting system, Agilent Technologies) as described (3). HEK293 cells were transfected with 50 ng of Elk-1 consisting of GAL4 DNA-binding domain and Elk-1, 50 ng of pFR-Luc carrying the GAL4 UAS-fused luciferase gene, 50 ng of pCH110 encoding the β-galactosidase gene under the control of the SV40 promoter, and FLAG-tagged SPRED1 expression vectors. In some experiments, HA-tagged GRD expression vectors were included. After 24 h, cells were treated with 50 ng/ml epidermal growth factor (EGF) for 6 h and then collected and lysed with a PicaGene Reporter lysis buffer (TOYO Ink). The activity of luciferase and β-galactosidase was analyzed using beetle luciferin (Promega) and o-nitrophenyl β-galactopyranoside (Nacalai Tesque) as substrates. In all reporter assays, 2 × 105 HEK293 cells were plated on 12-well dishes and transfected according to the calcium-phosphate method.
Docking Model
The docking model of the SPREDl EVH1 domain and the NF1 GRD domain was calculated using ClusPro (29). Human SPREDl EVH1 domain (Protein Data Bank code 3SYX) and human NF1 GRD domain (Protein Data Bank code 1NF1) were used for the computation. Of the 30 clusters resulting from ClusPro, there are several models that can explain EVH1 missense mutations. A model came as cluster 10, which harbors 30 members, which can explain the result of our yeast two-hybrid assay (Fig. 5F).
FIGURE 5.
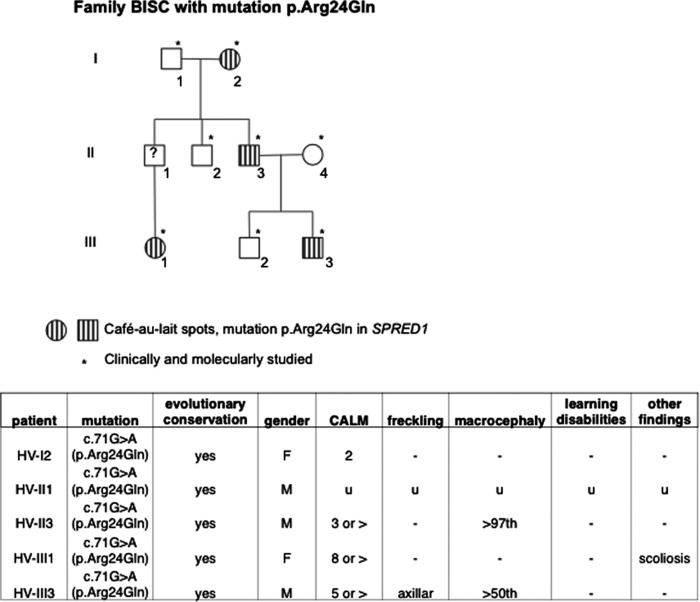
Information of the family carrying R24Q mutation. Upper panel, pedigree of the family. Open symbols indicate unaffected individuals; lined symbols indicate affected individuals. *, p < 0.05. Table, clinical phenotypes of individuals carrying R24Q mutation. CALM, café-au-lait spot; −, not present; u, unknown.
Results
Interaction between the EVH1 Domain and the GRD Detectedby a Yeast Two-hybrid System
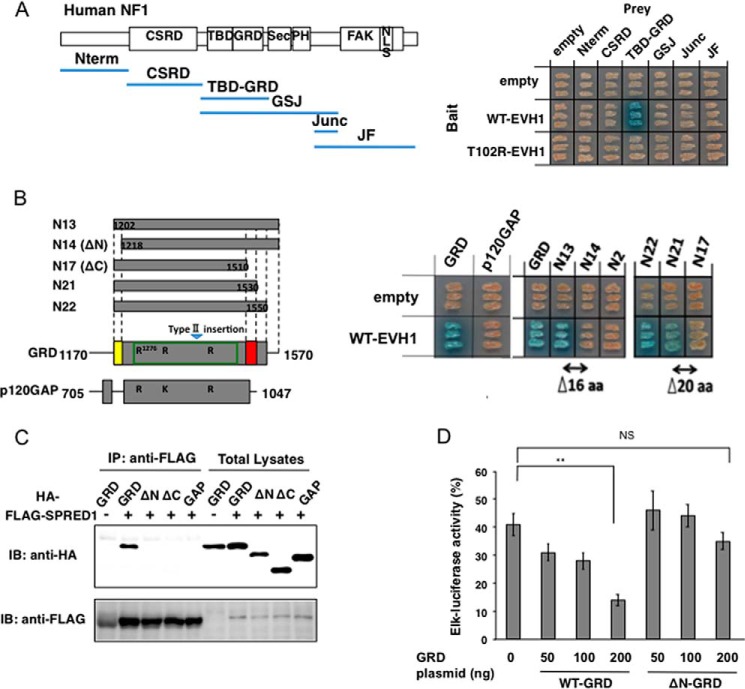
Neurofibromin is an ∼300-kDa large protein consisting of several domains, including the GRD (Fig. 1A). To identify the neurofibromin domain that interacts with the EVH1 domain of SPRED1, we performed a yeast two-hybrid assay. The human SPRED1 EVH1 domain was fused to the LexA-DNA binding domain (Bait), and six different segments of human neurofibromin were fused to the GAL4-transactivating domain (Prey). We found that only an internal region containing a tubulin-binding domain and a GRD can strongly interact with the EVH1 domain within yeasts. This interaction was not observed when EVH1 contained a T102R mutation, which disrupts the ERK suppression activity (Fig. 1A) (7). Further deletion mutation analysis revealed that the GRD region binds to the EVH1 domain. Fine deletion mutation analysis also revealed that the N-terminal 16 amino acids and C-terminal 20 amino acids outside the GAP domain are essential for the interaction between EVH1 and GRD (Fig. 1B). We noticed that EVH1 did not interact with p120RasGAP (Fig. 1B). The NF1 GRD possesses extensions of the N- and C-terminal peptides that are not present in p120RasGAP (Fig. 1B, yellow and red regions). These regions correspond exactly to the regions we have identified as the EVH1-binding sites. Previously, a 483-residue GRD of neurofibromin had been described as the Ras-GAP domain (30, 31). However, smaller fragments of p120GAP (GAP-273, residues Met714–His986) and neurofibromin (NF1–230, residues Asp1248–Phe1477) have also been shown to possess full Ras-GAP activity (32). Thus, extra N- and C-peptides unique to NF1 GRD are not necessary for the GAP function but are essential for the binding to the EVH1 domain.
FIGURE 1.
Yeast two-hybrid assay to detect EVH1 and neurofibromin interacting domains. A, yeast strains carrying pGBKT7-hEVH1(1–136) and the indicated neurofibromin domains cloned in pGADT7 were restreaked on a filter paper and stained for an in situ galactosidase assay. B, interaction between the EVH1 domain and deletion mutants of the GRD. Left, schematic view of the structural comparison between the NF-1 GRD and the GAP domain of p120GAP. The green box in the GRD indicates that it is minimum domain with full GTPase-activating activity (36). R and K in the GRD and GAP indicate conserved residues for Ras-GTP binding. Right, cDNAs of p120-RasGAP(705–1047) or neurofibromin GRD deletion mutants were subcloned into pACT2. N13 means full-length GRD(1170–1570). Yeasts were restreaked on a filter paper and stained for an in situ galactosidase assay. aa, amino acid. C, binding of EVH1 and GRD mutants in HEK293 cells. HEK293 cells grown in 10-cm dishes were transiently transfected with HA-tagged deletion mutants of GRD plasmids (1.5 μg/dish) together with FLAG-tagged SPRED1 expression plasmid (1 μg/dish). The immunoprecipitates (IP) with anti-FLAG antibody were immunoblotted (IB) with anti-HA and anti-FLAG antibodies. D, enhancement of SPRED1-dependent ERK suppression by GRD. HEK293 cells grown in 12-well plates were transfected with Elk-1 reporter plasmid, hSPRED1 expression vector (10 ng/well), together with indicated amounts of WT-GRD or ΔN-GRD expression vectors. After 24 h, cells were treated with 50 ng/ml EGF for 6 h, and luciferase activity was then measured. The Elk-luciferase activity of cells with EGF stimulation without transfecting SPRED1 and GRD plasmids is standardized as 100%. NS, not significant; **, p < 0.01.
We confirmed the binding of EVH1 and WT-GRD in HEK293 cells by means of an immunoprecipitation-Western blotting assay (Fig. 1C). GRD lacking these N- and C-terminal extensions could not bind to EVH1 (Fig. 1C, ΔN and ΔC). To examine whether these extensions were important for suppression of the Ras-ERK pathway, WT-GRD or ΔN-GRD, which lacked N-terminal 16 amino acids, were overexpressed together with a moderate amount of SPRED1, the concentration of which reduced EGF-induced ERK activity to ∼50% (Fig. 1D). We found that WT-GRD overexpression in HEK293 cells enhanced the SPRED1-mediated suppression of ERK activity, whereas ΔN-GRD did not (Fig. 1D). These data confirmed that SPRED1 recruits neurofibromin to suppress Ras activation through the interaction between EVH1 and GRD.
Mutations of the N- and C-terminal Extended Regions of the GRD Observed in NF1 Patients with Reduced EVH1 Binding Affinity
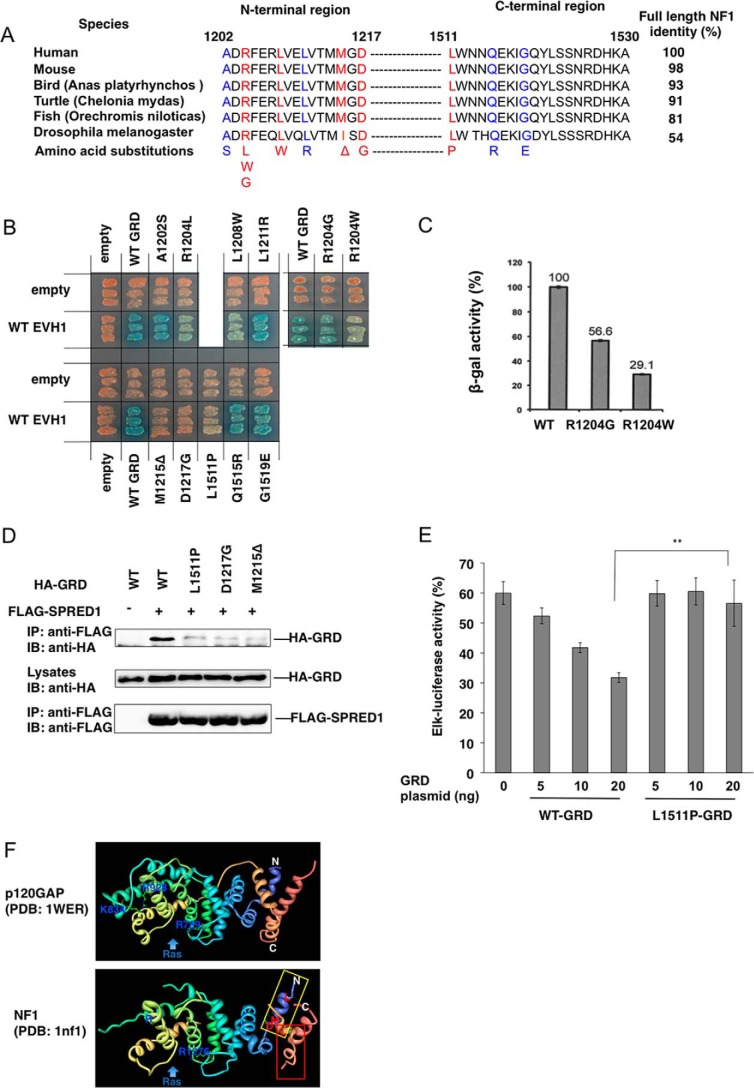
The extended N- and C-terminal peptides of the GRD of neurofibromin are highly conserved among species (Fig. 2A). Because we have used the type II NF1 gene that contains a 21-amino acid insertion within the GRD (33), the C-terminal region corresponds to Leu1511- Ala1530. Among the mutations reported in NF1 patients, we noticed that two previously reported point mutations, p.Arg1204Gly (R1204G) and p.Arg1204Trp (R1204W), were localized in the N-terminal EVH1 interaction region (34). We also analyzed eight additional mutations in these regions that were detected in NF1 patients (Fig. 2A). (Patient information is shown in Supplemental Table 1.) All of these amino acids were highly conserved among various species (Fig. 2A). All NF1 missense mutations were classified for pathogenicity according to the criteria of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (35).
FIGURE 2.
GRD mutations in the N-terminal extended region reduce EVH1 binding affinity. A, amino acid sequences of the N- and C-terminal EVH1 binding region of GRDs from various species. Red letters indicate amino acids whose mutations severely affected binding to the EVH1 domain. Blue letters show amino acids whose mutations had little effect on the binding. B, in situ yeast two-hybrid assay of the interaction between EVH1 and NF1 GRD mutants. Yeast strains carrying pGBKT7-hEVH1 and the indicated GRD mutants in pGADT7 were restreaked on a filter paper and stained for an in situ galactosidase assay. C, quantitative β-gal assay of yeast strains carrying pGBKT7-hEVH1 and pGADT7-GRD with indicated mutations. D, binding of EVH1 and GRD mutants in HEK293 cells. HEK293 cells were transiently transfected with HA-tagged GRD mutants and FLAG-tagged SPRED1. The immunoprecipitates (IP) with anti-FLAG antibody were immunoblotted (IB) with anti-HA and anti-FLAG antibodies. E, GRDL1511P showed a weaker effect on SPRED1-mediated ERK suppression than WT-GRD. HEK293 cells were transfected with Elk-1 reporter plasmid and hSPRED1 expression vector, together with indicated amounts of WT-GRD or GRDL1511P expression vectors. After 24 h, cells were treated with 50 ng/ml EGF for 6 h and luciferase activity was then measured. The Elk-luciferase activity of cells with EGF stimulation without transfecting SPRED1 and GRD plasmids is standardized as 100%. F, ribbon drawing structures of GRDs of p120GAP and NF1. Blue, N-terminal region. **, p < 0.01. Beige color, C-terminal region. The positions of Leu1511, Leu1208, Met1212, and Asp1217 are shown.
As shown in Fig. 2B and summarized in Fig. 2A, the two-hybrid assay revealed that p.Arg1204Trp (R1204W), p.Arg1204Leu (R1204L), p.Leu1208Trp (L1208W), p.Met1215del(M1215Δ), p.Asp1217Gly (D1217G), and p.Leu1511Pro (L1511P) severely reduced the GRD-EVH1 interaction, whereas p.Ala1202Ser (A1202S), p.Arg1204Gly (R1204G), p.Leu1211Arg (L1211R), p.Gln1515Arg (Q1515R), and p.Gly1519Glu (G1519E) did not significantly affect the GRD binding activity to the EVH1 domain (Fig. 2B). Three (p.Met1215del, p.Asp1217Gly, and p.Leu1511Pro) from the five likely pathogenic NF1 missense mutations (p.Arg1204Gly, p.Met1212del, p.Asp1217Gly, p.Leu1511Pro, and p.Gln1515Arg)profoundly reduced binding of GRD with the SPRED1 EVH1 domain. We observed 40% reduction of the interaction between EVH1 and GRD mutant carrying p.Arg1204Gly (R1204G), which is likely a pathogenic NF1 mutation by using quantitative β-galactosidase assay (Fig. 2C), indicating that the pathogenic mechanism of these mutations may also related to an impaired binding of the EVH1 domain of SPRED1. Three (p.Arg1204Leu, p.Arg1204Trp, and p.Leu1208Trp) of the five variants of uncertain significance also showed a severely reduced GRD-EVH1 interaction.
We confirmed reduced binding between EVH1 and GRDL1511P, GRDD1217G, and GRDM1215Δ by means of IP-Western blotting in HEK293 cells (Fig. 2D). The suppression activity of GRDL1511P for EGF-induced ERK activation was much lower than that of WT in HEK293 cells (Fig. 2E). These data suggest that pathogenicity of the mutations outside the GAP domain can be explained by the disruption of GRD-EVH1 interaction, thereby reduced Ras-ERK suppression activity.
We also introduced mutations into putative phosphorylation sites, Tyr1521, Ser1523, and Ser1524; however, there were no apparent effects on the binding (data not shown). Because Glu1206 is a conserved residue with a negative charge, we substituted this residue to Ala; however, we did not see any effects on the interaction between EVH1 and GRD (data not shown). As shown through crystal structural analysis (NF1–333; residues 1198–1530, which does not contain a 21-amino acid insertion) (36), these two extensions form two α-helix tubes that crossover each other, and severely affected mutations were mostly present in the surface of the N-terminal α-helix (Fig. 2F). Leu1511 is located on the same side of Leu1208, Met1215, and Asp1217; thus, we suspect that these residues form the binding site for the EVH1 domain (Fig. 2F).
EVH1-GRD Interaction Is Essential for the Suppression of ERK by Neurofibromin
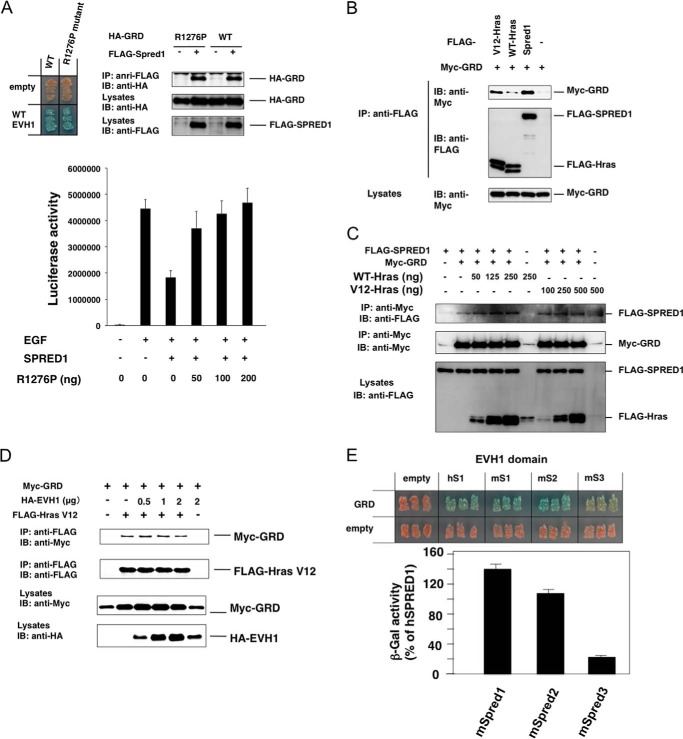
To show the functional importance of the EVH1-GRD interaction in the Ras-ERK inactivation in mammalian cells, we overexpressed a GRD mutant (R1276P) that lacked GAP activity (37). Because this mutant GRDR1276P still possessed full EVH1 binding activity (Fig. 3A), it should function as a dominant negative form against the endogenous full-length neurofibromin. As expected, we found that the SPRED1-dependent suppression of EGF-induced ERK activation was reverted by the GRDR1276P mutant (Fig. 3A, bottom panel). This strongly supports the idea that the binding of the EVH1 of SPRED1 to GRD is critical for the suppressive function of neurofibromin.
FIGURE 3.
GRD-EVH1 interaction is independent of the GRD-Ras interaction. A, effect of the GAP-null mutant of GRD on SPRED1 suppression activity. Upper panels, in situ yeast two-hybrid assay and IP-Western blotting assay for the interaction between WT EVH1 and R1276P mutant GRD. Lower panel, reversal of the effect of hSPRED1 on R1276P GRD. HEK293 cells were transfected with or without 10 ng of hSPRED1 cDNA in the presence of increased concentrations of R1276P GRD cDNA. EGF-induced Elk-1 reporter activity was measured 1 day after transfection. IP, immunoprecipitation; IB, immunoblot. B, comparison of the binding of GRD between SPRED1 and Ras. HEK293 cells were transfected with FLAG-tagged SPRED1 (2 μg), WT (0.5 μg), or V12-H-Ras (0.25 μg) and Myc-GRD (2 μg) expression plasmids. Cell extracts were immunoprecipitated with anti-FLAG antibody and then blotted with anti-FLAG or anti-Myc antibodies. C, effect of the overexpression of Ras on the SPRED1-GRD interaction. HEK293 cells were transfected with FLAG-SPRED1 cDNA (3 μg) and Myc-GRD cDNA (0.1 μg) together with increased concentrations of FLAG-tagged WT or V12 H-Ras cDNA plasmids. One day after transfection, cell extracts were immunoprecipitated with anti-Myc antibody and then blotted with anti-Myc or anti-FLAG antibodies. D, effect of the overexpression of the SPRED1-EVH1 domain on the V12-HRas-GRD interaction. HEK293 cells were transfected with 500 ng of FLAG-Ras cDNA and 1.5 μg of Myc-GRD cDNA together with increased concentrations of HA-tagged SPRED1 (EVH1) cDNA plasmids. One day after transfection, cell extracts were immunoprecipitated with anti-FLAG antibody and then blotted with anti-Myc or anti-FLAG antibodies. E, yeast strains carrying pGBKT7-EVH1s from mSpred1–3 (mS1, mS2, and mS3) and pGADT7-GRD were restreaked on a filter paper and stained for an in situ galactosidase assay (upper panels). A quantitative β-galactosidase (β-gal) assay was performed and normalized against the GRD-hSPRED1 EVH1 interaction.
Because it has been shown that GRD binds to the Ras-GTP form through a Ras-binding groove (36), we investigated whether EVH1 binding modulates the GRD-Ras interaction or whether Ras modulates the EVH1-GRD interaction. As shown in Fig. 3B, V12-Ras (oncogenic Ras), but not WT-Ras, bound to GRD; however, the V12-Ras-GRD interaction was weaker than the EVH1-GRD interaction. When WT or V12-Ras was co-expressed, the EVH1-GRD interaction was not affected (Fig. 3C). We also found that the overexpression of EVH1 did not affect the GRD-V12-Ras interaction as observed by the IP-Western assay (Fig. 3D). These data suggest that the EVH1-GRD interaction is independent of the GRD-Ras interaction. This is reasonable because the extended N- and C-terminal EVH1-binding peptides of the GRD distantly locate from the Ras-binding groove of the GRD (see Fig. 4F).
FIGURE 4.
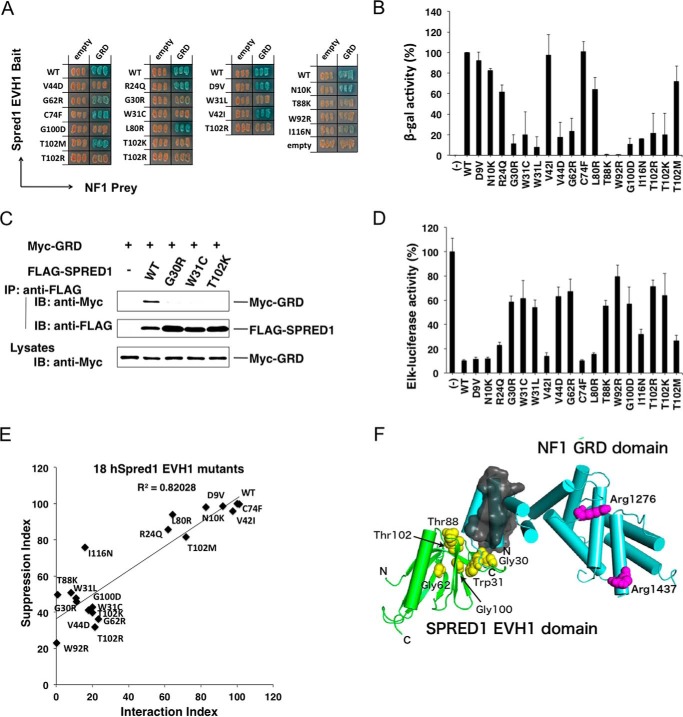
Effects of EVH1 mutations on the EVH1-GRD interaction and ERK suppression activity. A and B, yeast strains carrying pGBKT7-hEVH1 with indicated mutations and pGADT7-GRD were restreaked on a filter paper and stained for an in situ galactosidase assay (A) and a quantitative β-gal assay (B). Error bars denote the mean ± S.D. of triplicate experiments. C, lack of interaction of the GRD with mutant EVH1 domains in 293 cells. Cells were transfected with Myc-tagged GRD and indicated FLAG-tagged SPRED1 mutants. Immunoprecipitates (IP) with anti-FLAG antibody were blotted with anti-Myc antibody. IB, immunoblot. D, ERK suppression activity by mutant SPREDs. HEK293 cells were transfected with the Elk-1 reporter system and 30 ng of mutant SPRED1 cDNAs. One day after transfection, HEK293 cells were stimulated with 50 ng/ml EGF for 6 h, and luciferase activity was then measured. E, correlation between ERK suppression activity and GRD binding activity of mutant EVH1 domains. Plotting of the EVH1-GRD interaction index and ERK suppression activity index is shown. The EVH1-GRD interaction index is calculated from B. WT EVH1 β-gal activity is standardized as 100%. ERK suppression index (calculated from D) = ((Elk-1 reporter with mutant Spred1) − (Elk-1 reporter with WT Spred1))/(Elk-1 reporter without Spred1). F, in silico modeling of the EVH1 domain (green) and GRD (light blue). The side chains of the arginine residues in charge of binding to Ras (Arg1276 and Arg1416) in GRD are shown in magenta. The N- and C-terminal helices in the GRD, which are important for EVH1 binding, are covered with a gray surface. Plausible residues in EVH1 for interaction with GRD suggested from the results of yeast two-hybrid assay and ERK suppression assay (Gly30, Trp31, Gly62, Thr88, Gly100, and Thr102) are shown in yellow.
Previously, we reported that mSpred3 has much weaker ERK suppression activity compared with mSpred1 and mSpred2 (3). Thus, we compared the binding activity among the Spred1/2/3 EVH1 domains to the GRD. As shown in Fig. 3E, the binding of the EVH1 domain of mSpred2 to the GRD was similar to that of mSpred1, whereas the mSpred3 EVH1 domain interacted with the GRD with much weaker affinity than mSpred1 or mSpred2 (Fig. 3E). Thus, the lower binding affinity of the Spred3 EVH1 domain may explain its lower Ras-ERK suppression activity compared with that of Spred1 and Spred2.
EVH1 Missense Mutations Found in Legius Syndrome Reduce the EVH1-GRD Interaction
At least 45 different SPRED1 missense mutations in humans have been reported so far (SPRED1 LOVD and one selected from the ARUP database); however, the pathogenicity is not always predictable. Of these, we selected 18 mutations in the EVH1 domain (unpublished patient information is shown in Supplemental Table 2). Pathogenicity of the missense mutations was also classified according to the criteria of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (35). Mutations p.Asp9Val (D9V), p.Asn10Lys (N10K), and p.Val42Ile (V42I) were classified as benign. Mutations p.Cys74Phe (C74F), p.Leu80Arg (L80R), p.Thr102Met (T102M), and p.Ile116Asn (I116N) fell in the category of variants of uncertain significance. However, p.Arg24Gln (R24Q), p.Gly30Arg (G30R), p.Trp31Leu (W31L), p.Gly44Asp (G44D), p.Gly62Arg (G62R), p.Thr88Lys (T88K), p.Trp92Arg(W92R), p.Gly100Asp (G100D), p.Thr102Arg (T102R), and p.Thr102Lys (T102K) were identified as probably pathogenic, and p.Trp31Cys (W31C) was confidently classified as pathogenic.
The EVH1-GRD interaction was assessed by in situ staining of yeast two-hybrid assay (Fig. 4A), and the results were quantified by a β-gal enzyme activity assay (Fig. 4B). Disruption of the EVH1-GRD interaction by such mutations was confirmed by IP-Western blotting assay in 293 cells (Fig. 4C). All except one pathogenic and likely pathogenic mutations, such as G30R, W31C, W31L, V44D, G62R, T88K, W92R, G100D, T102R, and T102K, severely reduced the EVH1-GRD binding (Fig. 4, A and B). The three benign mutations and one variant of uncertain significance (D9V, N10K, V42I, and C74F) did not affect the interaction and behaved as wild type EVH1 (Fig. 4, A and B).
ERK suppression activity was then measured by the Elk reporter assay in HEK293 cells (Fig. 4D), and the ERK suppression activity versus the strength of the EVH1-GRD interaction was plotted (Fig. 4E). The ERK suppression activity was well correlated to the EVH1-GRD binding activity. We noticed that the mutations fell into three classes as follows: those with almost completely diminished GRD binding; those with partially reduced binding (R24Q, L80R, V42T, and T102M); and those with unaltered binding (D9V, N10K, V42I, V21I, and C74F). W31C, T102R, and V44D have been shown previously to be (likely) pathogenic missense mutations, whereas C74F was a sequence variant that had previously been shown to have no effect on SPRED1 function (11), suggesting a correlation between the EVH1-GRD interaction and pathogenicity. Therefore, overall, mutations that disrupt the EVH1-GRD interaction are pathogenic and less suppressive for ERK. However, there are a couple of exceptions. The likely pathogenic mutation R24Q showed only a mildly reduced EVH1-GRD binding (the pedigree and clinical phenotypes of a family carrying R24Q mutation are shown in Fig. 5), and a variant of uncertain significance I116N showed a severely reduced EVH1-GRD binding, but retained relatively higher ERK suppression activity (Fig. 4E).
The SPRED1 EVH-l domain has been suggested to bind to a ligand in a groove formed by β-strands βl, β2, β6, and β7 (38). Missense mutations with a severe effect on GRD binding such as G30R, W31C/W31L, V44D, G62R, G100D, and T102K/T102R are probably present in the groove for the binding to the GRD or important for the structure of the binding groove. As V42I and T102M are substitutions of similar amino acids, they may not affect the entire structure of the EVH1 domain. Cys74 and Leu80 are located in an outer strand of the EVH1 β-sandwich (β4), and accordingly, the effects of these mutations on the EVH1-GRD interaction and ERK suppression are less drastic compared with those of other mutations. We used computer simulation to provide a model for the interaction between EVH1 and GRD (Fig. 4F). These mutations are very likely to disrupt the EVH1 binding pocket for interaction with the GRD-N- and C-terminal regions. Several amino acids that severely affected binding, Gly30, Trp31, Val44, Gly62, Gly100, and Thr102, are located in this pocket.
Discussion
In this study, we demonstrate that the SPRED EVH1 domain interacts with the N- and C-terminal extended regions of the GRD of neurofibromin, which is essential for the function of SPREDs as well as neurofibromin. Most pathogenic and likely pathogenic missense mutations in EVH1 found in Legius syndrome and some NF1 GRD pathogenic missense mutations from NF1 patients severely disrupt this interaction and reduce suppression activity against the Ras-ERK pathway. Using the data on the severely reduced EVH1-GRD binding in this work, we could potentially “upgrade” the classification of three NF1 missense mutations of uncertain significance to likely pathogenic mutations (p.Arg1204Leu, p.Arg1204Trp, and p.Leu1208Trp) and of three likely pathogenic mutations to pathogenic mutations (p.Met1215del, p.Asp1217Gly, and p.Leu1511Pro).
Overall, there is a strong correlation among pathogenic mutations, disruption of the EVH1-GRD interaction, and ERK suppression activity. The likely pathogenic SPRED1 missense mutations G30R, W31L, G62R, T88K, W92R, G100D, T102R, and T102K in the EVH1 domain showed a severely reduced binding affinity to the GRD, whereas the three benign mutations and one mutation of uncertain significance (D9V, N10K, V42I, and C74F) did not affect binding to the GRD. Thus, the effect of the EVH1 missense mutations on the EVH1-GRD interaction explains the pathogenetic mechanism. However, some SPRED1 mutations (R24Q, L80R, and T102M), especially R24Q, have only a modest effect on the GRD interaction, but they clearly are associated with a Legius syndrome phenotype with café-au-lait macules in three generations. In addition, we cannot explain at the moment why variant of uncertain significance I116N has a low EVH1-GRD interaction index but a relatively high ERK suppression index. Further study is necessary to define the pathogenetic mechanism of these mutations.
The overlapping phenotypes between NF1 and Legius syndrome are explained by the model proposed by Stowe et al. (18), i.e. SPREDs recruit neurofibromin to the Ras-enriched cell membrane. Because there are three functionally overlapping SPREDs in humans, the relatively benign phenotype of Legius syndrome can be explained by SPRED2/3 activity compensating for SPRED1 mutations in most cell types. It is notable that the EVH1 domains of mSpred1 and mSpred2 have similar GRD binding affinity and similar ERK suppression activity. Physiological redundancy of Spred1 and Spred2 was confirmed by gene-disruption studies in which Spred1/2 double KO mice were embryonically lethal, and Spred1 and Spred2 single KO mice were viable (15).
In conclusion, pathogenic and likely pathogenic mutations in the EVH1 domain of SPRED1 cause loss of binding affinity to neurofibromin GRD, and our yeast two-hybrid assay correlates excellently with the Ras-ERK suppression activity of SPRED1. The information obtained in this study will be helpful for the classification of missense mutations in Legius syndrome and NF1. This unique EVH1-GRD interaction may also be useful in the development of agents that could induce Ras inactivation in cancer.
Author Contributions
H. B., F. M., L. M., E. L., and A. Y. conceived and designed the experiments. Y. H., M. K., M. O., and T. O. performed the experiments. H. B., M. S., M. K., M. O., and R. M. analyzed the data. H. B., I. L. R., F. M., L. M., and E. L. contributed reagents/materials/analysis tools. H. B., L. M., E. L., and A. Y. wrote the paper.
Supplementary Material
Acknowledgments
We thank R. Komine, S. Tsuruta, N. Shiino, Y. Noguchi, and M. Asakawa for technical assistance; Dr. F. McCormick (University of California at San Francisco) for neurofibromin cDNA and discussion; and Y. Ushijima and R. Mizoguchi for manuscript preparation. We thank M. J. Martínez-González (Neuropediatric Unit, Pediatric Department, Cruces University Hospital, Biocruces Health Research Institute) and Mao Rong (ARUP Laboratories, University of Utah, Salt Lake City) for providing patient information.
This work was supported by special Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan 25221305, Advanced Research and Development Programs for Medical Innovation (AMED-CREST), the Takeda Science Foundation, the Uehara Memorial Foundation, and “Opening the Future” grant from KU Leuven (to E. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Tables S1 and S2.
- SPR
- Sprouty-related C-terminal cysteine-rich domain
- GAP
- GTPase-activating protein
- GRD
- GAP-related domain
- CALM
- café-au-lait macule.
References
- 1. Yoshimura A. (2009) Regulation of cytokine signaling by the SOCS and Spred family proteins. Keio J. Med. 58, 73–83 [DOI] [PubMed] [Google Scholar]
- 2. Bundschu K., Walter U., and Schuh K. (2007) Getting a first clue about SPRED functions. BioEssays 29, 897–907 [DOI] [PubMed] [Google Scholar]
- 3. Kato R., Nonami A., Taketomi T., Wakioka T., Kuroiwa A., Matsuda Y., and Yoshimura A. (2003) Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem. Biophys. Res. Commun. 302, 767–772 [DOI] [PubMed] [Google Scholar]
- 4. Nonami A., Taketomi T., Kimura A., Saeki K., Takaki H., Sanada T., Taniguchi K., Harada M., Kato R., and Yoshimura A. (2005) The Sprouty-related protein, Spred-1, localizes in a lipid raft/caveola and inhibits ERK activation in collaboration with caveolin-1. Genes Cells 10, 887–895 [DOI] [PubMed] [Google Scholar]
- 5. Impagnatiello M. A., Weitzer S., Gannon G., Compagni A., Cotten M., and Christofori G. (2001) Mammalian sprouty-1 and -2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J. Cell Biol. 152, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., De Schepper S., Fryns J. P., Cools J., Marynen P., Thomas G., Yoshimura A., and Legius E. (2007) Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 39, 1120–1126 [DOI] [PubMed] [Google Scholar]
- 7. Messiaen L., Yao S., Brems H., Callens T., Sathienkijkanchai A., Denayer E., Spencer E., Arn P., Babovic-Vuksanovic D., Bay C., Bobele G., Cohen B. H., Escobar L., Eunpu D., Grebe T., et al. (2009) Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 302, 2111–2118 [DOI] [PubMed] [Google Scholar]
- 8. Pasmant E., Sabbagh A., Hanna N., Masliah-Planchon J., Jolly E., Goussard P., Ballerini P., Cartault F., Barbarot S., Landman-Parker J., Soufir N., Parfait B., Vidaud M., Wolkenstein P., Vidaud D., and France R. N. (2009) SPRED1 germline mutations caused a neurofibromatosis type 1 overlapping phenotype. J. Med. Genet. 46, 425–430 [DOI] [PubMed] [Google Scholar]
- 9. Spurlock G., Bennett E., Chuzhanova N., Thomas N., Jim H. P., Side L., Davies S., Haan E., Kerr B., Huson S. M., and Upadhyaya M. (2009) SPRED1 mutations (Legius syndrome): another clinically useful genotype for dissecting the neurofibromatosis type 1 phenotype. J. Med. Genet. 46, 431–437 [DOI] [PubMed] [Google Scholar]
- 10. Muram-Zborovski T. M., Stevenson D. A., Viskochil D. H., Dries D. C., Wilson A. R., and Rong M. (2010) SPRED 1 mutations in a neurofibromatosis clinic. J. Child Neurol. 25, 1203–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brems H., and Legius E. (2013) Legius syndrome, an update. Molecular pathology of mutations in SPRED1. Keio J. Med. 62, 107–112 [DOI] [PubMed] [Google Scholar]
- 12. Pasmant E., Gilbert-Dussardier B., Petit A., de Laval B., Luscan A., Gruber A., Lapillonne H., Deswarte C., Goussard P., Laurendeau I., Uzan B., Pflumio F., Brizard F., Vabres P., Naguibvena I., et al. (2015) SPRED1, a RAS MAPK pathway inhibitor that causes Legius syndrome, is a tumour suppressor downregulated in paediatric acute myeloblastic leukaemia. Oncogene 34, 631–638 [DOI] [PubMed] [Google Scholar]
- 13. Rauen K. A. (2013) The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida T., Hisamoto T., Akiba J., Koga H., Nakamura K., Tokunaga Y., Hanada S., Kumemura H., Maeyama M., Harada M., Ogata H., Yano H., Kojiro M., Ueno T., Yoshimura A., and Sata M. (2006) Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene 25, 6056–6066 [DOI] [PubMed] [Google Scholar]
- 15. Miyoshi K., Wakioka T., Nishinakamura H., Kamio M., Yang L., Inoue M., Hasegawa M., Yonemitsu Y., Komiya S., and Yoshimura A. (2004) The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene 23, 5567–5576 [DOI] [PubMed] [Google Scholar]
- 16. Taniguchi K., Kohno R., Ayada T., Kato R., Ichiyama K., Morisada T., Oike Y., Yonemitsu Y., Maehara Y., and Yoshimura A. (2007) Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol. Cell. Biol. 27, 4541–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., and Yoshimura A. (2001) Spred is a Sprouty-related suppressor of Ras signalling. Nature 412, 647–651 [DOI] [PubMed] [Google Scholar]
- 18. Stowe I. B., Mercado E. L., Stowe T. R., Bell E. L., Oses-Prieto J. A., Hernández H., Burlingame A. L., and McCormick F. (2012) A shared molecular mechanism underlies the human rasopathies Legius syndrome and neurofibromatosis-1. Genes Dev. 26, 1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Denayer E., Chmara M., Brems H., Kievit A. M., van Bever Y., Van den Ouweland A. M., Van Minkelen R., de Goede-Bolder A., Oostenbrink R., Lakeman P., Beert E., Ishizaki T., Mori T., Keymolen K., Van den Ende J., et al. (2011) Legius syndrome in fourteen families. Hum. Mutat. 32, E1985–E1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brems H., Pasmant E., Van Minkelen R., Wimmer K., Upadhyaya M., Legius E., and Messiaen L. (2012) Review and update of SPRED1 mutations causing Legius syndrome. Hum. Mutat. 33, 1538–1546 [DOI] [PubMed] [Google Scholar]
- 21. Messiaen L. M., Callens T., Mortier G., Beysen D., Vandenbroucke I., Van Roy N., Speleman F., and Paepe A. D. (2000) Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 15, 541–555 [DOI] [PubMed] [Google Scholar]
- 22. Messiaen L., and Wimmer K. (2008) NF1 Mutational Spectrum. Monogr. Hum. Genet. 16, 63–77 [Google Scholar]
- 23. Sasaki A., Yasukawa H., Suzuki A., Kamizono S., Syoda T., Kinjyo I., Sasaki M., Johnston J. A., and Yoshimura A. (1999) Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4, 339–351 [DOI] [PubMed] [Google Scholar]
- 24. Endo T., Sasaki A., Minoguchi M., Joo A., and Yoshimura A. (2003) CIS1 interacts with the Y532 of the prolactin receptor and suppresses prolactin-dependent STAT5 activation. J. Biochem. 133, 109–113 [DOI] [PubMed] [Google Scholar]
- 25. Schneider S., Buchert M., and Hovens C. M. (1996) An in vitro assay of β-galactosidase from yeast. BioTechniques 20, 960–962 [DOI] [PubMed] [Google Scholar]
- 26. Sasaki A., Taketomi T., Wakioka T., Kato R., and Yoshimura A. (2001) Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor–but not epidermal growth factor-induced ERK activation. J. Biol. Chem. 276, 36804–36808 [DOI] [PubMed] [Google Scholar]
- 27. Taniguchi K., Ishizaki T., Ayada T., Sugiyama Y., Wakabayashi Y., Sekiya T., Nakagawa R., and Yoshimura A. (2009) Sprouty4 deficiency potentiates Ras-independent angiogenic signals and tumor growth. Cancer Sci. 100, 1648–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorenz S., Lissewski C., Simsek-Kiper P. O., Alanay Y., Boduroglu K., Zenker M., and Rosenberger G. (2013) Functional analysis of a duplication (p.E63_D69dup) in the switch II region of HRAS: new aspects of the molecular pathogenesis underlying Costello syndrome. Hum. Mol. Genet. 22, 1643–1653 [DOI] [PubMed] [Google Scholar]
- 29. Kozakov D., Beglov D., Bohnuud T., Mottarella S. E., Xia B., Hall D. R., and Vajda S. (2013) How good is automated protein docking? Proteins 81, 2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., and Cawthon R. M. (1990) The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63, 843–849 [DOI] [PubMed] [Google Scholar]
- 31. Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., and Tamanoi F. (1990) The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63, 835–841 [DOI] [PubMed] [Google Scholar]
- 32. Ahmadian M. R., Wiesmüller L., Lautwein A., Bischoff F. R., and Wittinghofer A. (1996) Structural differences in the minimal catalytic domains of the GTPase-activating proteins p120GAP and neurofibromin. J. Biol. Chem. 271, 16409–16415 [DOI] [PubMed] [Google Scholar]
- 33. Andersen L. B., Ballester R., Marchuk D. A., Chang E., Gutmann D. H., Saulino A. M., Camonis J., Wigler M., and Collins F. S. (1993) A conserved alternative splice in the von Recklinghausen neurofibromatosis (NF1) gene produces two neurofibromin isoforms, both of which have GTPase-activating protein activity. Mol. Cell. Biol. 13, 487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas L., Richards M., Mort M., Dunlop E., Cooper D. N., and Upadhyaya M. (2012) Assessment of the potential pathogenicity of missense mutations identified in the GTPase-activating protein (GAP)-related domain of the neurofibromatosis type-1 (NF1) gene. Hum. Mutat. 33, 1687–1696 [DOI] [PubMed] [Google Scholar]
- 35. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W. W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H. L., and ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheffzek K., Ahmadian M. R., Wiesmüller L., Kabsch W., Stege P., Schmitz F., and Wittinghofer A. (1998) Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 17, 4313–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klose A., Ahmadian M. R., Schuelke M., Scheffzek K., Hoffmeyer S., Gewies A., Schmitz F., Kaufmann D., Peters H., Wittinghofer A., and Nürnberg P. (1998) Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum. Mol. Genet. 7, 1261–1268 [DOI] [PubMed] [Google Scholar]
- 38. Harmer N. J., Sivak J. M., Amaya E., and Blundell T. L. (2005) 1.15 A crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family. FEBS Lett. 579, 1161–1166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.