The role of adenomatous polyposis coli (APC) tumor suppressor at mitochondria is unclear. We show that APC associates with the Miro/Milton/kinesin complex to stimulate anterograde transport of mitochondria. This identifies the first regulatory role of APC in organelle transport. APC cancer mutations block this activity.
Abstract
Mutations in adenomatous polyposis coli (APC) disrupt regulation of Wnt signaling, mitosis, and the cytoskeleton. We describe a new role for APC in the transport of mitochondria. Silencing of wild-type APC by small interfering RNA caused mitochondria to redistribute from the cell periphery to the perinuclear region. We identified novel APC interactions with the mitochondrial kinesin-motor complex Miro/Milton that were mediated by the APC C-terminus. Truncating mutations in APC abolished its ability to bind Miro/Milton and reduced formation of the Miro/Milton complex, correlating with disrupted mitochondrial distribution in colorectal cancer cells that could be recovered by reconstitution of wild-type APC. Using proximity ligation assays, we identified endogenous APC-Miro/Milton complexes at mitochondria, and live-cell imaging showed that loss of APC slowed the frequency of anterograde mitochondrial transport to the membrane. We propose that APC helps drive mitochondria to the membrane to supply energy for cellular processes such as directed cell migration, a process disrupted by cancer mutations.
INTRODUCTION
Adenomatous polyposis coli (APC) is a tumor-suppressor protein involved in many areas of normal cell growth and differentiation, including Wnt signaling, spindle formation, chromosome segregation, DNA damage response, and cell migration (Fearnhead et al., 2001; Hanson and Miller, 2005; Aoki and Taketo, 2007; Etienne-Manneville, 2009). Mutations in the APC gene occur in ∼80% of colorectal cancers (CRCs) and trigger progression of sporadic CRC. A gene mutation in one APC allele can cause the polyp syndrome familial adenomatous polyposis, whereas a “second hit” in the form of a somatic mutation in the remaining allele predisposes patients to cancer (Fodde et al., 2001; Polakis, 2007; Senda et al., 2007). Most APC mutations produce a premature stop codon, resulting in C-terminal–truncated APC proteins, and this leads to aberrant β-catenin signaling and, consequently, deregulated transcription of Wnt target proteins that can cause cell transformation (Fodde et al., 2001; Polakis, 2007). In addition, APC mutations give rise to alterations in chromosome stability, cell polarity, and cell migration (Nathke, 2006; Caldwell et al., 2007; Nelson et al., 2012). These processes are normally regulated by APC through its C-terminal interaction with the cytoskeleton, which is disrupted by truncating mutations (Askham et al., 2000; Wen et al., 2004).
APC localizes at the mitochondria (Brocardo et al., 2008), an organelle required for ATP synthesis, and regulation of apoptosis and calcium buffering (reviewed in McBride et al., 2006; Johannsen and Ravussin, 2009; Contreras et al., 2010; Tait and Green, 2012). The subcellular distribution of mitochondria is controlled through dynamic transport mechanisms critical for its ability to redistribute in response to cellular signals and provide ATP to cell regions with high bioenergetic demands (reviewed in Frederick and Shaw, 2007; da Silva et al., 2014). Mitochondrial transport has frequently been studied in the axons of neurons, and disruption of such transport can lead to neurological disorders (reviewed in Chaturvedi and Flint Beal, 2013). Recent studies in epithelial cells and T-cell lymphocytes show that altered mitochondrial transport can also impact a range of cell types and are linked to cancer processes such as cell migration and invasion (Campello et al., 2006; Desai et al., 2013; Zhao et al., 2013).
Mitochondrial transport is mediated by different routes, the best-characterized mechanism involves the Miro/Milton complex, which drives kinesin-dependent transport of mitochondria along microtubules and was identified by genetic screens in Drosophila (Stowers et al., 2002; Guo et al., 2005). The Miro and Milton proteins are evolutionarily conserved in mammals, and two orthologues have been identified for each protein: Miro-1/Miro-2 and Milton-1/Milton-2 (Fransson et al., 2003; Brickley et al., 2005). Milton acts as an adaptor between the mitochondrial transmembrane protein Miro and the kinesin-1/ kinesin superfamily protein 5 (KIF5) motor complex, which moves along microtubules in the anterograde direction toward the plasma membrane (Fransson et al., 2006; Glater et al., 2006; MacAskill et al., 2009a). This transport complex is regulated by metabolic fluctuations in calcium levels (Saotome et al., 2008; Macaskill et al., 2009b; Wang and Schwarz, 2009) and glucose availability (Pekkurnaz et al., 2014) that target the action of Miro and Milton, respectively. Interestingly, Wnt pathway proteins have been linked to mitochondrial processes, including transport, apoptosis, and oxidative phosphorylation (Brocardo et al., 2008; Yoon et al., 2010; Lopez-Domenech et al., 2012; Serrat et al., 2013; Pate et al., 2014). In this paper, we describe a new and unexpected role for APC in stimulating the initiation of mitochondrial transport.
RESULTS
Loss of wild-type APC causes mitochondria to redistribute from the membrane to the perinuclear zone
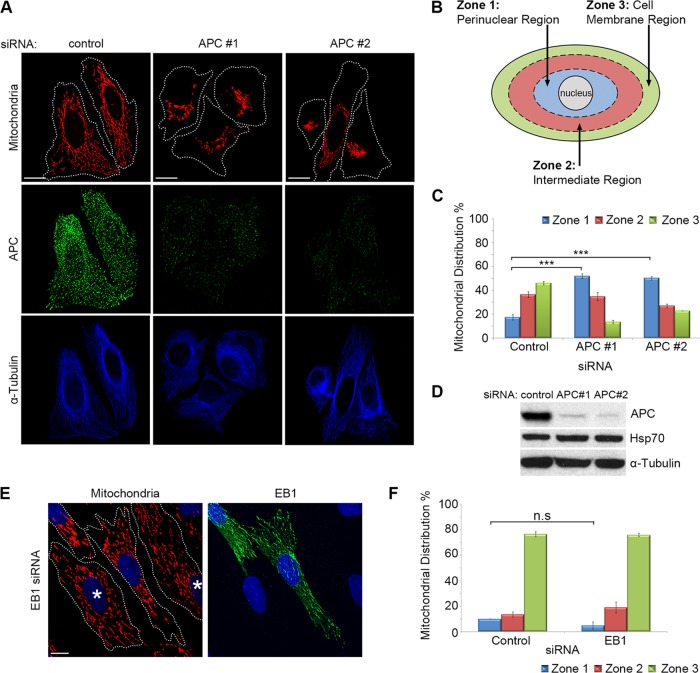
We previously detected APC at mitochondria (Brocardo et al., 2008). Here we silenced APC expression by small interfering RNA (siRNA) and assessed the status of mitochondria. U2OS osteosarcoma cells (express wild-type APC) were treated with control siRNA or APC-specific siRNAs; then cells were fixed and mitochondria visualized by immunofluorescence microscopy using the fluorescent MitoTracker dye CMX-Ros. We identified a dramatic shift in mitochondrial distribution following loss of APC. Mitochondria in control cells were uniformly distributed throughout the cell, whereas after loss of APC, most cells displayed distinct mitochondrial clustering around the nucleus (Figure 1A). To quantify this perinuclear shift, we scored cells for their relative mitochondrial distribution within three zones determined as a percentage (33, 66, and 100%) of the distance between the perinuclear region and the cell membrane (see schematic in Figure 1B and Materials and Methods for more detail). The loss of full-length APC elicited an ∼200% increase in cells that displayed mitochondrial clustering in the perinuclear region relative to control cells (p < 0.001; see Figure 1C). Conversely, the population of cells displaying spread-out mitochondria (extending to the cell membrane) significantly decreased following loss of APC (control = 46%, APC #1 siRNA = 13%, APC #2 siRNA = 23%; p < 0.001). The efficiency of APC knockdown was confirmed by both immunofluorescence microscopy and Western blot, with detection of mtHSP70 and α-tubulin as loading controls (Figure 1, A and D). A mitochondrial shift toward the perinuclear region was also observed when full-length APC was silenced in HDF1314 and NIH 3T3 fibroblasts (Supplemental Figure S1, A and B) and confirmed in U2OS cells with antibodies against mtHSP70 being used as an alternate mitochondrial marker (Supplemental Figure S1C).
FIGURE 1:
Loss of full-length APC induces perinuclear redistribution of mitochondria. (A) APC was silenced in U2OS cells by siRNA (APC #1 and #2), and mitochondrial distribution was analyzed by immunofluorescence microscopy after cells were stained for mitochondria (CMX-Ros) and APC. The microtubule network remained intact (α-tubulin). (B) The distribution of mitochondria in different “zones” was scored (C), revealing redistribution of mitochondria to the perinuclear region (zone 1) with APC siRNAs (***, p < 0.001). (D) Loss of APC in U2OS cells was confirmed by Western blot. (E) HDF1314 cells treated with EB1 siRNA were stained for mitochondrial distribution (CMX-Ros) and EB1. Cells displaying EB1 knockdown are indicated (*). (F) Scoring of mitochondrial distribution after EB1 silencing revealed no significant difference relative to control (n.s., not significant). Bar graph data are presented as mean (±SD), statistical analysis by unpaired two-tailed t test with Bonferroni correction (C and F). Scale bars: 10 μm.
The effect of APC silencing on mitochondrial redistribution is specific and not due to microtubule destabilization
Mitochondria primarily utilize the microtubule network for transport throughout the cytoplasm, and APC binds to and stabilizes microtubules (Zumbrunn et al., 2001). Therefore we ruled out microtubule destabilization as a cause of mitochondrial redistribution by immunostaining α-tubulin before and after treatment with APC siRNA. This revealed no obvious disruption in the microtubule network (Figure 1A) in comparison with the complete depolymerization of microtubules and mitochondrial redistribution seen upon treatment with nocodazole (Supplemental Figure S1, D and F). Moreover, the knockdown of the APC partner protein EB1, which also binds and stabilizes the plus ends of microtubules (Askham et al., 2000; Nakamura et al., 2001; Wen et al., 2004), did not alter mitochondrial spread (Figure 1, E and F). Furthermore, loss of APC is unlikely to affect mitochondria through changes in the actin cytoskeleton, as depolymerization of actin with latrunculin A did not shift mitochondria to the perinuclear zone (Supplemental Figure S1, E and F). The impact of APC silencing on other organelles known to be transported by microtubules (Hirokawa et al., 2009) was also analyzed. Markers detecting the endoplasmic reticulum, Golgi, lysosomes, and centrosome were examined before and after APC knockdown; however, no changes were observed (Supplemental Figure S1, G–I), implying that the microtubule network retains its organelle transport functionality despite loss of APC.
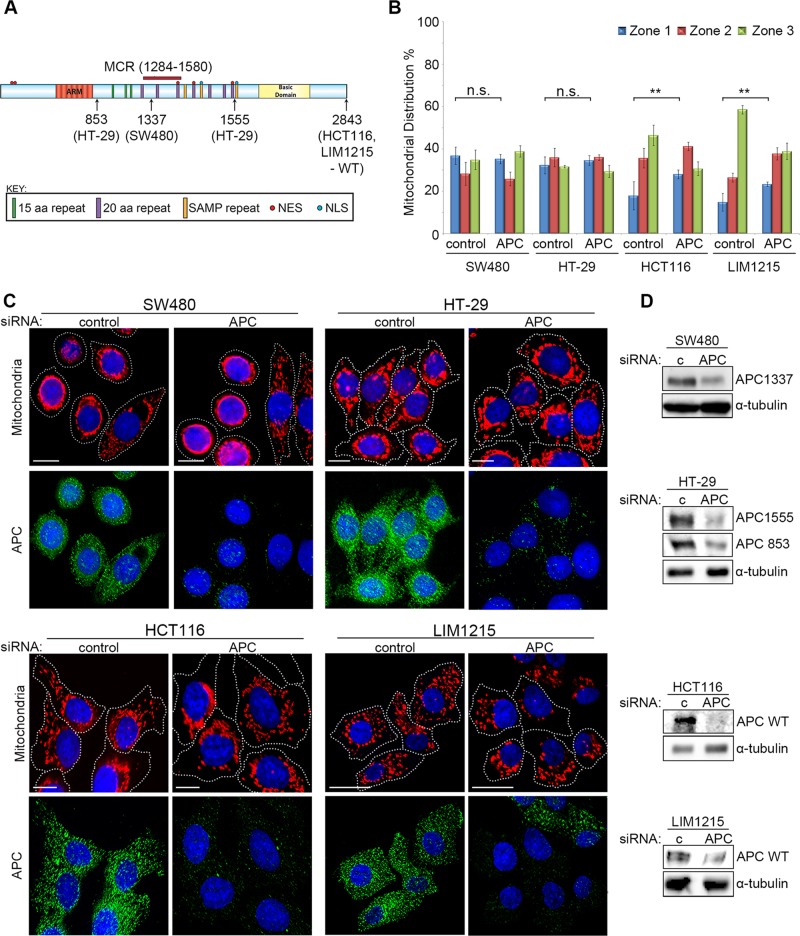
Cancer mutations disrupt the effect of APC on mitochondrial distribution
There is a high incidence of APC mutation in CRC, typically resulting in a truncated protein with altered function (Bienz, 2002; Brocardo and Henderson, 2008). To investigate whether mutation of APC alters mitochondrial distribution, we analyzed CRC cell lines that express either mutant truncated APC (SW480APC1337 and HT-29APC853/1555) or wild-type APC (HCT116 and LIM1215) (Figure 2A). Interestingly, the distribution of mitochondria in the mutant APC CRC cell lines SW480 and HT-29 was less frequently spread throughout the cytoplasm (Figure 2, B and C). This implies that truncation of APC may affect its ability to drive mitochondria to the membrane. Consistent with this idea, the knockdown of mutant APC by siRNA (Figure 2, B–D) had no significant impact (p > 0.05) on mitochondrial distribution in SW480 and HT-29 cells, while loss of full-length APC in HCT116 and LIM1215 caused a substantial shift (p < 0.01) toward the perinuclear region (see Figure 2, B and C). These results suggest that mutant truncated forms of APC, such as those commonly observed in colon cancer, are less able to facilitate transport of mitochondria to the cell periphery.
FIGURE 2:
Truncated mutant APC fails to regulate mitochondrial redistribution. (A) APC mutation status of CRC cell lines examined is indicated by schematic. (B and C) Cells treated with control or APC pooled siRNA (APC #1 and #2) were analyzed by immunofluorescence microscopy for mitochondrial distribution. (B) Mitochondrial localization patterns were scored and compared as previously described (Figure 1 legend). Graph indicates where loss of APC caused significant differences to perinuclear distribution relative to control (**, p < 0.01; n.s., not significant). Bar graph data presented as mean (±SD), statistical analysis by unpaired two-tailed t test. (C) Typical cell images after staining for mitochondria (CMX-Ros) and APC are shown. (D) Western blot confirms knockdown of full-length and mutant forms of APC in different CRC cell lines. α-Tubulin is the loading control. Scale bars: 10 μm.
Because the different cell types analyzed here for mitochondrial distribution varied in size and morphology, a subset of cells were selected at random from each cell line tested and were subjected to more rigorous quantitative analysis (described in detail in Supplemental Methods and Supplemental Figure S2). The results from this objective computer-based assessment were similar to those obtained in Figures 1C and 2B (see Supplemental Figure S2C).
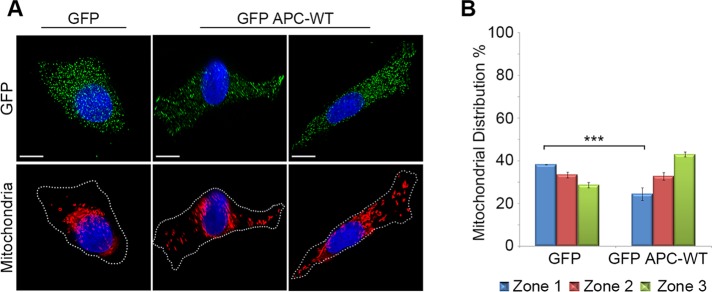
Mitochondrial transport toward the plasma membrane is recovered upon reconstitution of wild-type APC in mutant APC HT-29 cells
We next tested whether expression of wild-type APC could correct the defect in mitochondrial distribution observed in APC-mutant cells. HT-29APC853/1555 CRC cells were transfected with green fluorescent protein (GFP) alone or wild-type GFP-APC, and cells expressing only modest amounts of the GFP proteins were analyzed by immunofluorescence microscopy, revealing that reconstitution of wild-type APC significantly (p < 0.001) stimulated transport of mitochondria away from the perinuclear region (GFP = 38%, GFP-APC WT = 24%) and toward the cell membrane (GFP = 29%, GFP-APC WT = 43%) compared with GFP control cells (Figure 3). Rigorous computational analysis of a subset of these cells confirmed this finding (Supplemental Figure S2D). Thus reexpression of wild-type APC can at least partially correct the mitochondrial distribution defect we observed in the APC-mutant cells.
FIGURE 3:
Reconstitution of wild-type APC in APC-mutant HT-29 cells rescues mitochondrial transport. HT-29 cells transfected with plasmids that express GFP or GFP-APC-WT and stained for mitochondria (CMX-Ros) and GFP were analyzed for mitochondrial distribution by immunofluorescence microscopy. (A) Representative cell images are shown, and (B) scoring revealed that mitochondria were more often located in cytoplasm and near the membrane after low-level expression of GFP-APC. Significant differences relative to controls are indicated (***, p < 0.001). Bar graph data presented as mean (±SD), statistical analysis by unpaired two-tailed t test. Scale bars: 10 μm.
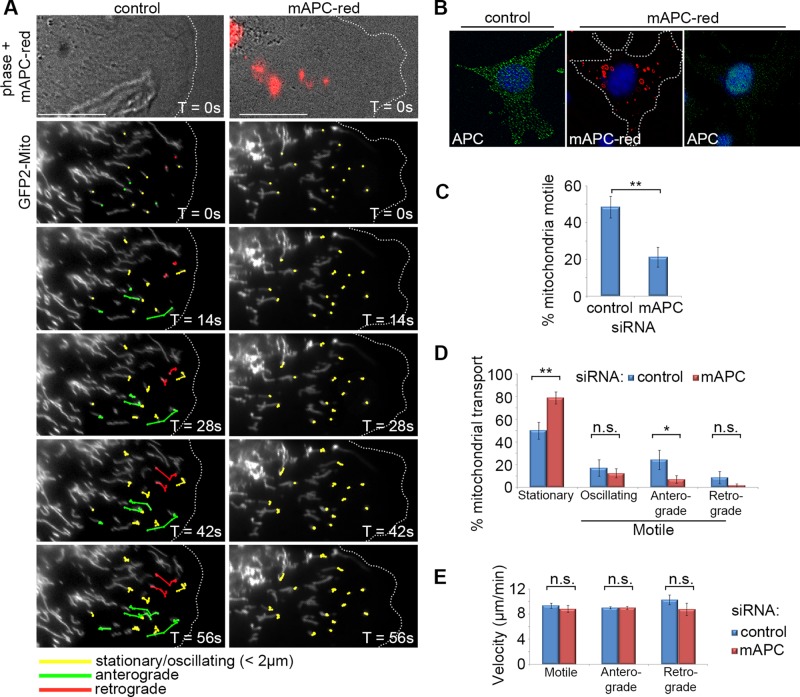
The loss of APC decreases initiation of mitochondrial transport
The above results in fixed cells suggest a role for APC in mitochondrial transport toward the cell periphery, which is lost upon C-terminal truncation and siRNA-mediated knockdown of APC. To investigate more directly whether APC stimulates the transport of mitochondria, we used live-cell imaging to determine what impact loss of APC had on real-time movement of mitochondria within the cell. Patil et al. (2013) reported a number of mitochondrial transport parameters in NIH 3T3 fibroblasts, and since robust APC staining is observed at microtubule-dependent protrusions of migrating NIH 3T3 fibroblasts (Sharma et al., 2006), we used this cell model to interrogate the role of APC in mitochondrial transport. NIH 3T3 fibroblasts were treated with control or red fluorescently tagged mouse APC siRNA (mAPC-red), transfected with a GFP-mitochondrial peptide marker to track mitochondria, and grown to confluence. Cells were then wounded and subjected 5 h later to time-lapse imaging by DeltaVision live-cell microscopy (Figure 4, A and B). Cells at the leading edge were analyzed for several mitochondrial transport parameters over 5-min intervals (more details are given in Materials and Methods).
FIGURE 4:
Loss of APC reduces initiation of mitochondrial transport but not velocity in NIH 3T3 cells. Cells were treated with either control or red fluorescently tagged mouse APC siRNA (mAPC-red), transfected with a mitochondrial marker (pGFP2-Mito), and then grown to confluence. Cells were then wounded and were analyzed 5 h later by live-cell time lapse, with images captured every 7 s for a total of 5 min. Post image acquisition, mitochondria were selected at random for tracking manually using the ImageJ plug-in MTrack. (A) Representative cell images are shown from ∼1 min after commencement of tracking. Mitochondria are marked for different types of movement: stationary/oscillating (yellow), anterograde/membrane-directed (green), and retrograde/nucleus-directed (red). The majority of directed movement of mitochondria was blocked when APC was silenced (successful APC silencing indicated by red fluorescence). Scale bars: 10 μm. (B) Confirmation of APC knockdown in fixed cells treated with mAPC-red siRNA (C) Analysis of mitochondrial motility revealed that loss of APC resulted in decreased overall mitochondrial transport (**, p < 0.01). (D) This was particularly evident in the anterograde direction (*, p < 0.05; n.s., not significant). (E) Analysis of mitochondrial velocity (μm/min) revealed that loss of APC had no impact on the speed of movement in either direction once transport was initiated (n.s., not significant). n over three independent experiments: control cells, n = 18, and mitochondria, n = 202; mAPC-red cells, n = 18, and mitochondria, n = 189. Bar graph data presented as mean (±SD), statistical analysis by two-tailed t test (C–E).
The most striking observation was that loss of APC elicited a dramatic decrease (∼2.3-fold reduction, p < 0.01) in the overall motility of mitochondria (Figure 4, A and C). Under control conditions, mitochondria were motile 48.4% of the time, and this presented as either directed (anterograde or retrograde) movement or oscillatory movement (no net displacement). In comparison, only 21.3% of cells transfected with mAPC siRNA were motile. A more detailed analysis revealed that, while oscillatory movements remained relatively constant following APC knockdown, the directed mitochondrial transport was severely diminished. In cells targeted by the APC siRNA, the frequency of mitochondria displaying anterograde transport dropped significantly from 24.3 to 7.0% (p < 0.05; Figure 4D). A decrease in mean retrograde transport from 8.7 to 1.7% was also observed; however, this was not found to be significant. Conversely, the proportion of stationary mitochondria increased after loss of APC. These results are reflected in the tiled cell images (Figure 4A) and the time-lapse movies (Supplemental Videos 1 and 2) that compare control and mAPC siRNA-treated NIH 3T3 cells. In line with these results, APC silencing decreased the average mitochondrial displacement and average distance traveled over the observed time period (Table 1). Our findings suggest that APC has a role in the initiation of long-range, directed mitochondrial transport in migrating cells.
TABLE 1:
Effect of loss of APC on parameters of mitochondrial transport in migrating NIH 3T3 cells.
| Control | mAPC-red | p Value | ||
|---|---|---|---|---|
| Overall mitochondrial movement | ||||
| % Motile mitochondria | 48.44 ± 5.92 | 21.25 ± 5.32 | ** | |
| % Directionality | Stationary | 49.91 ± 7.47 | 78.76 ± 5.32 | ** |
| Oscillating | 17.08 ± 7.29 | 12.50 ± 4.00 | n.s. | |
| Anterograde | 24.30 ± 8.43 | 7.03 ± 3.39 | * | |
| Retrograde | 8.74 ± 5.43 | 1.70 ± 1.49 | n.s. | |
| Average displacement (μm) (normalized to 1 min) | 1.19 ± 0.11 | 0.67 ± 0.16 | ** | |
| Average distance (μm) (normalized to 1 min) | 3.19 ± 0.14 | 2.10 ± 0.20 | ** | |
| Motile mitochondria | ||||
| Average velocity (μm/min) (does not include stationary time points) | Total | 9.27 ± 0.39 | 8.82 ± 0.57 | n.s. |
| Anterograde | 8.98 ± 0.19 | 8.99 ± 0.26 | n.s. | |
| Retrograde | 10.13 ± 0.76 | 8.75 ± 0.97 | n.s. | |
| Average distinct mitochondrial movements | Anterograde | 0.23 ± 0.03 | 0.13 ± 0.07 | ** |
| Retrograde | 0.15 ± 0.04 | 0.09 ± 0.03 | ** | |
| Average distance of mitochondrial movements (μm) | Anterograde | 1.35 ± 0.20 | 1.22 ± 0.16 | n.s. |
| Retrograde | 1.39 ± 0.18 | 1.16 ± 0.25 | n.s. | |
Mitochondria in migrating 3T3 cells treated with control or mAPC-red siRNA and transfected with pGFP2-mito were tracked using MTrackJ ImageJ plug-in as in Figure 4. Based on the raw tracking data, Microsoft Excel was used to calculate movement parameters of randomly selected mitochondria, including the pool of motile mitochondria. A distinct mitochondrial movement is classified as any directional motion without pause. *, p < 0.05; **, p < 0.01; n.s., not significant. Statistical analysis by two-tailed unpaired t test.
The loss of APC does not slow the rate of mitochondrial transport
Somewhat surprisingly, analysis of live-cell data from the same experiment revealed that loss of APC did not significantly slow the overall rate of mitochondrial transport once movement had been initiated (control = 9.27 ± 0.39 μms−1; mAPC siRNA = 8.82 ± 0.57 μms−1; p > 0.05). This lack of difference was also reflected in the average velocities of transport in both anterograde and retrograde directions (Figure 4E and Table 1). The average velocity of the control mitochondria is within a similar range to that observed elsewhere in NIH 3T3 cells (Patil et al., 2013). Furthermore, the average distance of these movements was not significantly altered following silencing of APC (Table 1). Collectively these results suggest a distinct and specific role for APC in stimulating the initiation of mitochondrial transport rather than its ongoing velocity.
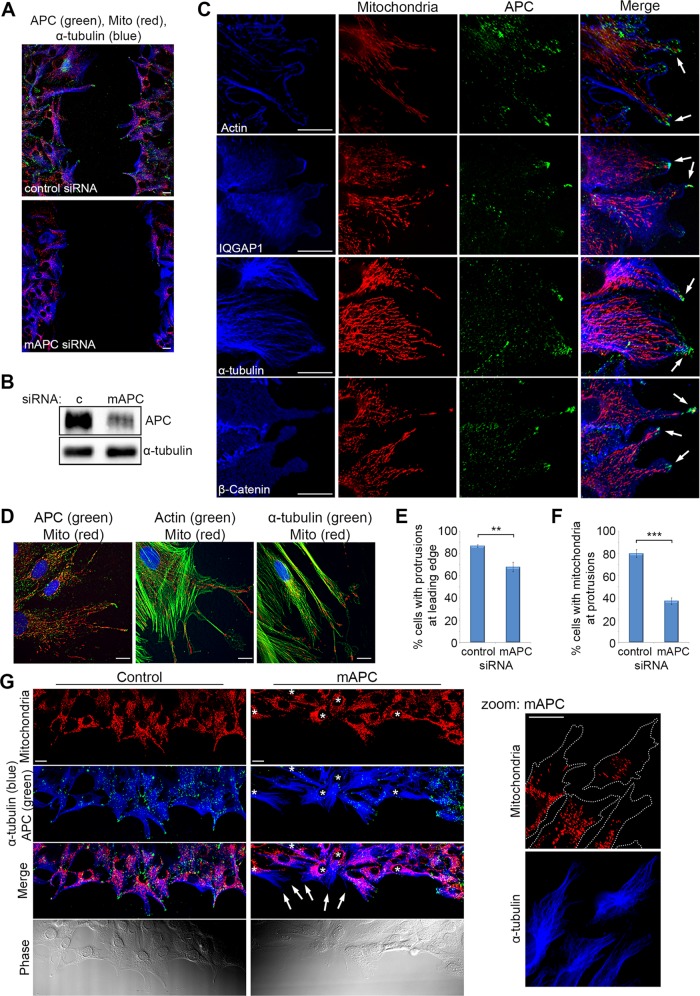
Mitochondria colocalize with APC at membrane protrusions of actively migrating cells
The accumulation of mitochondria at the membrane provides energy for different processes, including cell migration in epithelial breast and prostate cancer cells (Desai et al., 2013; Zhao et al., 2013). APC accumulates at microtubule-dependent membrane protrusions and is known to contribute to cell migration (Akiyama and Kawasaki, 2006; Nathke, 2006; Etienne-Manneville, 2009). The loss of APC can decrease the rate of cell migration (Kroboth et al., 2007). We confirmed this finding (Figure 5, A and B) and noted a reduction in mitochondria at the leading edge and cellular protrusions, prompting a more detailed analysis of mitochondrial localization at the membrane. Immunofluorescence staining of NIH 3T3 cells in wound-healing assays showed frequent colocalization of mitochondria with APC at cell migratory regions positive for actin, IQ-motif containing GTPase activating protein 1 (IQGAP1), microtubules (α-tubulin), and β-catenin (Figure 5C). Similar colocalization was seen in human HDF1314 cells (Figure 5D). To examine the impact of APC, we knocked down mouse APC in actively migrating 3T3 cells and observed an ∼20% reduction in the number of migrating cells with cellular protrusions at the leading edge (Figure 5E), in agreement with previous data (Sharma et al., 2006). In the remaining cell protrusions, mitochondrial localization at the ends of microtubules was found to decrease by >50% after APC knockdown (Figure 5, F and G), thus mirroring the prior results obtained in subconfluent cells. This APC-dependent alteration in mitochondrial localization could potentially decrease the supply of energy to the cell membrane required for cell migration, contributing to the observed slower rate of migration upon loss of APC (Kroboth et al., 2007; Figure 5A), or changes to the directed movement of cells (Nelson et al., 2012).
FIGURE 5:
Localization of mitochondria at microtubule-dependent cellular protrusions of actively migrating fibroblasts is lost upon APC silencing. Confluent cells were wounded; fixed 5 h later; counterstained for mitochondria (CMX-Ros), APC, and proteins involved in cell migration (actin, IQGAP1, α-tubulin, and β-catenin); and then analyzed by DeltaVision at the leading edge of the wound. (A) NIH 3T3 cells were treated with control or mAPC siRNA and counterstained for mitochondria (CMX-Ros), APC, and α-tubulin. Immunofluorescence analysis of cells at the leading edge of the wound revealed that cell migration was slowed upon loss of APC. (B) mAPC knockdown in NIH 3T3 cells was confirmed by Western blot. (C) Analysis of untreated NIH 3T3 cells at the wound edge indicated colocalization between mitochondria, APC, and cell migration factors at the membrane (arrows). (D) Colocalization of mitochondria and cell migration factors was confirmed by microscopy in human HDF1314 fibroblasts. (E) Formation of microtubule-dependent membrane protrusions was scored to reveal that treatment with mAPC siRNA reduced the number of protrusions detected relative to control cells (**, p < 0.01). (F) At the remaining protrusions, loss of mAPC decreased mitochondrial localization at protrusions (***, p < 0.001). Bar graph data presented as mean (±SD), statistical analysis by unpaired two-tailed t test. (G) Representative images of cells at the wound edge after transfection of control or APC-specific siRNAs (reduced green staining indicated by *). Note that mitochondria locate less frequently at microtubule-dependent protrusions after APC knockdown (indicated by arrows, closer view in zoomed panel). Scale bars: 10 μm.
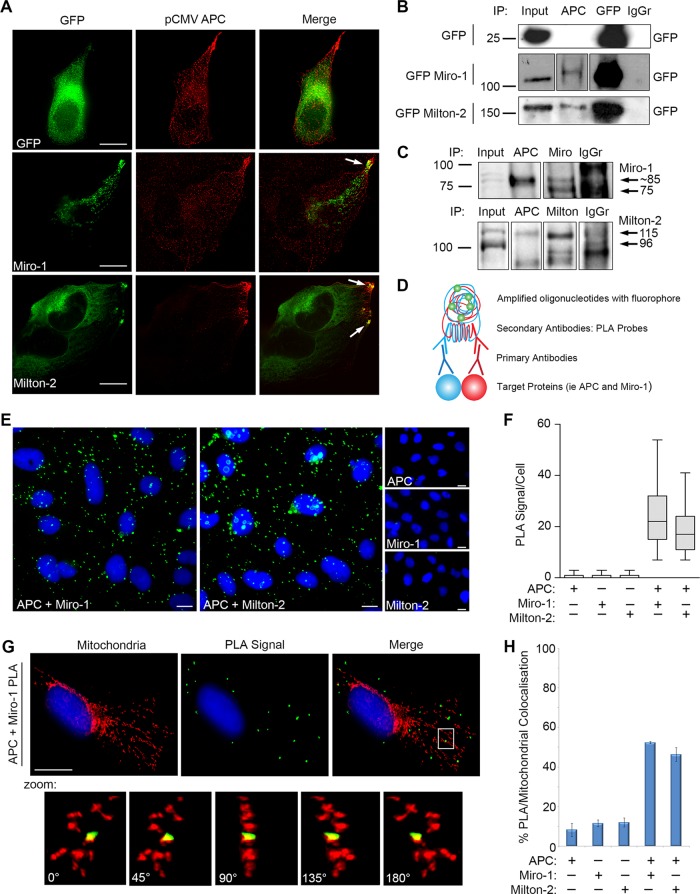
APC interacts with the Miro/Milton motor complex at mitochondria
We next investigated how APC stimulates mitochondrial movement. For determining whether APC associates with known mitochondrial transport complexes, pCMV-APC was transfected into U2OS cells, and the resultant untagged APC was analyzed for colocalization with GFP-tagged transport proteins. APC was found to colocalize with Miro-1 and Milton-2 at mitochondria and at cell membrane protrusions (Figure 6A). Miro and Milton form a bridging complex that connects mitochondria with kinesin motor proteins for anterograde transport along microtubules (Stowers et al., 2002; Guo et al., 2005; Glater et al., 2006), consistent with the actions observed by APC (Figures 1 and 4). Immunoprecipitation (IP) assays with U2OS cell lysates revealed that antibodies against endogenous APC could pull down both GFP-ectopic and endogenous forms of Miro-1 and Milton-2 (Figure 6, B and C), thus demonstrating binding of APC to Miro/Milton. APC/Miro-1 binding was confirmed in HEK293 cells with induced expression of wild-type APC, in which an antibody against APC successfully pulled down endogenous Miro-1 (Supplemental Figure S3A).
FIGURE 6:
Full-length APC interacts with the Miro/Milton complex at mitochondria in U2OS cells. (A) Cells were cotransfected with pCMV-APC and plasmids expressing GFP, GFP-Miro-1, or GFP-Milton-2; stained for APC and GFP; and analyzed by immunofluorescence microscopy. Points of colocalization are indicated by arrows. (B) Cells expressing GFP, GFP-Miro-1, or GFP-Milton-2 were subjected to IP. APC antibody pulled down GFP Miro-1 and GFP Milton-2 but not GFP control. Blots were spliced where indicated. (C) APC antibody captured endogenous Miro-1 and Milton-2 in cell lysates subjected to IP. Blots were spliced where indicated. (D) Duolink PLA, as described in the schematic, was used to visualize APC/Miro and APC/Milton interactions in situ using antibodies against APC and Miro-1 or Milton-2. (E) Representative cell images and (F) scoring indicate positive PLA signals between APC/Miro-1 and APC/Milton-2, indicated by green dots. PLA signals were absent when each antibody was tested alone. Box-and-whisker plot presented as median (line), upper/lower quartile (box), and min/max (error bars with 95% confidence interval [CI]). (G and H) Mitochondrial counterstaining (CMX-Ros) and scoring of immunofluorescence images show that APC/Miro-1 (3D projection pictured) and APC/Milton-2 PLA signals localize to the mitochondria. Bar graph data presented as mean (±SD). Scale bars: 10 μm.
We then sought to identify the subcellular location of APC-Miro/Milton complexes using the in situ Duolink proximity ligation assay (PLA; for details of the assay see Figure 6D and Materials and Methods). Antibodies targeting endogenous APC and either Miro-1 or Milton-2 were used to detect potential interactions. In line with the IP assays, APC/Miro-1 and APC/Milton-2 PLA experiments yielded a positive result, averaging ∼25 and ∼19 PLA signals per cell, respectively (Figure 6, E and F). Control samples using a single antibody had only low background signal (PLA signal/cell: APC = 0.7; Miro-1 = 0.7; Milton-2 = 0.8). Similar positive and specific Duolink PLA results between APC/Miro-1 and APC/Milton-2 were observed in Hela cells, with PLA signal/cell averaging 47 and 18, respectively (Supplemental Figure S3, B and C).
For determining whether the APC/Miro-1 complexes actually locate at mitochondria, PLA assays with U2OS cells were counterstained with the mitotracker dye CMX-Ros, revealing that ∼52% of the PLA-positive APC/Miro-1 complexes in each cell localized to the mitochondria (Figure 6, G and H). A similar striking colocalization (46%) of APC/Milton-2 PLA signals with mitochondria was observed. Precise verification of the PLA signal localization was obtained using three-dimensional (3D) image projections (see Figure 6G and Supplemental Figure S3D). Comparable 3D colocalization results were observed in Hela cells (Supplemental Figure S3, E and F).
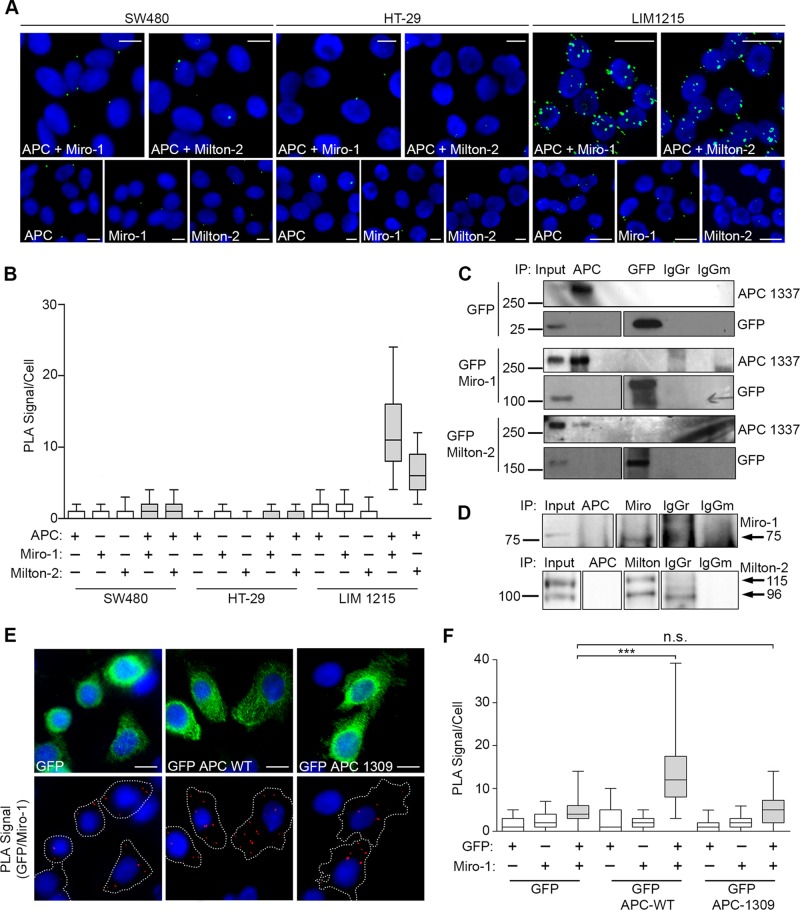
Truncating mutations disrupt APC binding to Miro/Milton
We showed that cancer-associated truncating mutations in APC reduce its influence on mitochondrial distribution (Figure 2). To determine whether they also altered APC binding to Miro/Milton, we used the sensitive PLA method to assess protein interactions in different CRC cell lines. We could not detect APC/Miro-1 or APC/Milton-2 protein complexes in mutant APC cell lines SW480 and HT-29; neither cell line elicited PLA signals above background (Figure 7, A and B). Conversely, under the same conditions, a consistent and positive result was observed for both APC/Miro-1 and APC/Milton-2 interactions in LIM1215 cells (which express wild-type APC), with on average ∼14 and ∼6 PLA signal/cell, respectively (Figure 7, A and B). Furthermore, in IP assays, the capture of endogenous mutant APC failed to identify binding of ectopic or endogenous Miro-1/Milton-2 in cell lysates (Figure 7, C and D), indicating that the ability of APC to bind Miro/Milton is disrupted by cancer mutations. Defective mutant APC/Miro-1 binding was also observed in HEK293 cells induced for mutant APC, in which antibodies against APC were unable to pull down endogenous Miro-1 (Supplemental Figure S3A).
FIGURE 7:
Truncated APC does not interact with the Miro/Milton complex. (A and B) Test for interaction between APC and Miro/Milton in colon cancer cell lines SW480 (APC1377), HT-29 (APC853/1555), and LIM1215 (APC WT) in situ using Duolink PLA. (A) Representative cell images (green dots are positive signals) were quantified in the graph (B). The interaction between APC and Miro/Milton is lost when APC is truncated (SW480 and HT-29 CRC cells) but remains intact when APC is full length (LIM1215 CRC cells). (C) SW480 cells expressing GFP, GFP-Miro-1, or GFP-Milton-2 were subjected to IP assays using anti-APC to pull down the respective GFP-tagged protein and anti-GFP to pull down APC. No bands were detected. Blots were spliced where indicated. (D) SW480 cell lysates were used for IP of endogenous protein; however, antibodies against APC were unable to pull down endogenous Miro-1 or Milton-2. Blots were spliced where indicated. (E) SW480 cells transiently expressing GFP-APC-WT and GFP-APC-1309 were subject to Duolink PLA where interactions between GFP-tagged APC sequences and Miro-1 were observed in situ by immunofluorescence microscopy. (F) Quantification of PLA signals revealed a significant positive signal above GFP background for GFP-APC-WT that was not observed for the GFP-APC mutant (***, p < 0.001; n.s., not significant). n > 125 per sample over two independent experiments. Box-and-whisker plot presented as a median (line), upper/lower quartile (box), and min/max (error bars with 95% CI). Statistical analysis by Mann-Whitney U-test with Bonferroni correction. Scale bars: 10 μm.
To ensure that the inability of mutant APC 1337 to interact with Miro/Milton in SW480 cells was due to APC truncation alone, rather than to additional mutations in Miro, Milton, or potential intermediate proteins, ectopic GFP-tagged forms of APC were expressed in SW480 cells, and their ability to bind endogenous Miro-1 was determined by PLA. In standard PLA reactions using antibodies against GFP and/or Miro-1, we could readily detect the interaction between wild-type GFP-APC and Miro-1 in transfected SW480 cells (2.8-fold higher than GFP alone); however, no complex formation (above GFP background) was observed between mutant GFP-APC-1309 and Miro-1 (Figure 7, E and F).
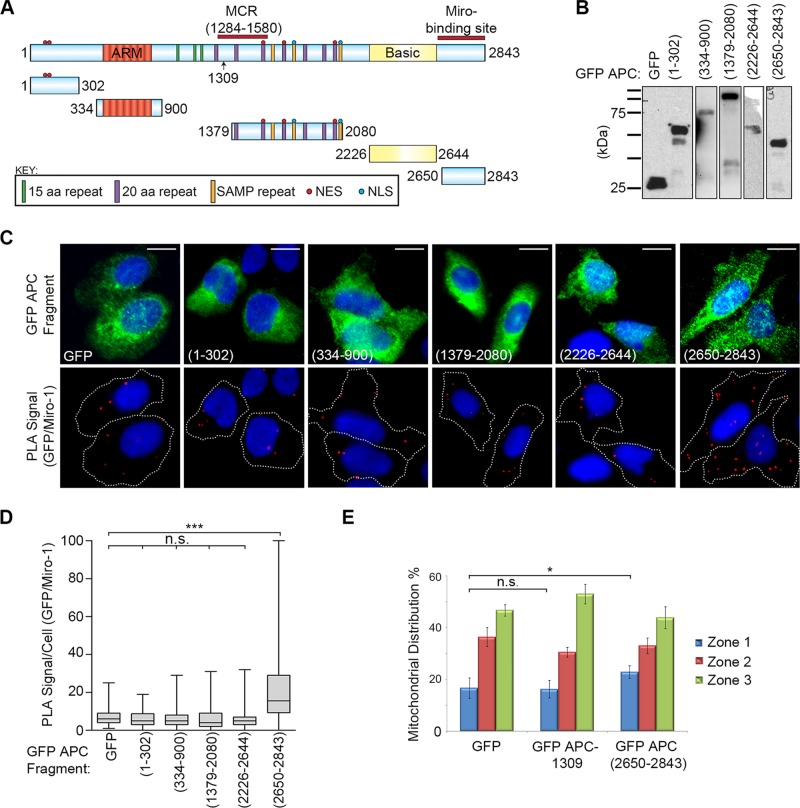
Mapping the Miro-binding region to the C-terminal amino acids 2650–2843 of APC
The above findings indicated that APC might associate with Miro through its C-terminal sequences. To identify the Miro-binding site, we performed detailed mapping in U2OS cells using Duolink PLA and antibodies against GFP and Miro-1 in cells transfected with plasmids expressing a range of GFP-tagged APC fragments (Figure 8, A and B). Of five sequences tested, only GFP-APC(2650–2843) gave a positive signal above GFP baseline (p < 0.001; Figure 8, C and D and Supplemental Figure S4, A and B). This sequence corresponds to the far C-terminal end of APC and would be lost in the majority of known truncating mutations. We validated the interaction by IP and found that antibodies against GFP captured both GFP-APC(2650–2843) and Miro-1 in U2OS cell lysates, whereas Miro was not observed in pull downs from cells transfected with GFP alone, GFP-APC(1–302) or GFP-APC(1379–2080) (Supplemental Figure S4C). Overexpression of the GFP-APC(2650–2843) fragment also caused a small but significant (p < 0.05) shift of mitochondria toward the perinuclear region compared with GFP control cells (Figure 8E). No perinuclear shift was observed following overexpression of the GFP APC-1309 fragment (Figure 8E). These data suggest that competition with the APC/Miro-1 interaction by overexpression of the APC C-terminus can alter mitochondrial distribution, although not to the same extent as observed for APC knockdown.
FIGURE 8:
Miro-1 interacts with the C-terminal fragment (2650–2843) of APC. (A) Schematic of GFP-tagged APC fragments overexpressed in U2OS cells and used to map the Miro-binding domain. (B) Western blot to confirm expression and integrity of the GFP-APC fragments. (C) GFP-APC fragments and Miro-1 were analyzed for their ability to form protein complexes in situ using Duolink PLA. Representative cell images are shown, with PLA GFP-Miro complexes indicated by red dots. Scale bars: 10 μm. (D) The PLA signals per cell were scored to reveal that only one GFP-APC fragment (2650–2843) associated with Miro (***, p < 0.001) above the GFP background, indicative of a positive interaction. n > 97 cells for each sample over two independent experiments. Box-and-whisker plot presents a median (line), upper/lower quartile (box), and min/max (error bars with 95% CI). Statistical analysis for samples above background by Mann-Whitney U-test with Bonferroni correction. (E) U2OS cells expressing GFP or GFP-APC fragments were scored for mitochondrial distribution. Overexpression of the APC(2650–2843) fragment caused a statistically significant increase in perinuclear mitochondrial distribution relative to GFP control. Bar graph data presented as mean (±SD), statistical analysis by unpaired two tailed t test (*, p < 0.05; n.s., not significant).
The C-terminal fragment of APC also associated with Milton-2 (Supplemental Figure S4, D–F). The mapping of the Miro/Milton binding site to the C-terminus of APC is intriguing, given that a different kinesin complex (that of KAP3A) was previously found to bind to the Armadillo repeat region of APC (Jimbo et al., 2002). The new findings provide an explanation for why truncated APC is not active in transporting mitochondria in CRC cells (Figure 2).
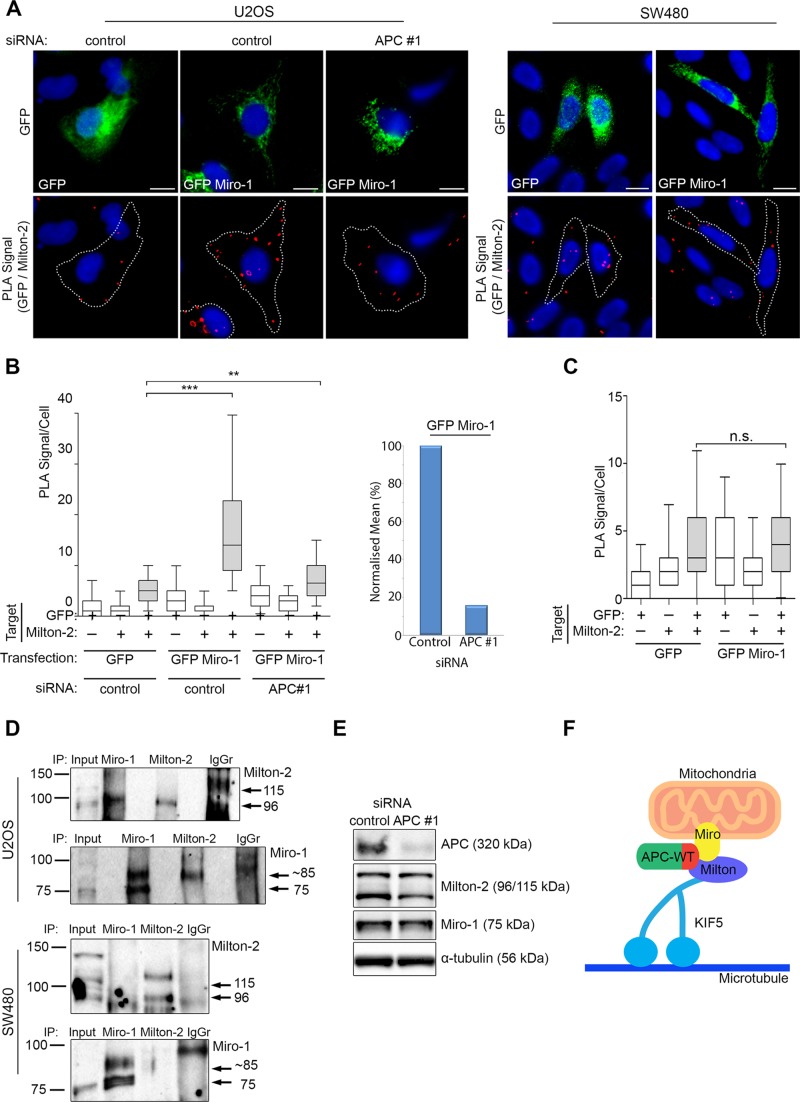
Wild-type APC stabilizes the interaction between Miro-1 and Milton-2
APC can act as a scaffold in a number of protein complexes (Lui et al., 2012), in particular the β-catenin destruction complex (Stamos and Weis, 2013). For investigating whether APC has a similar role in the formation of the Miro/Milton complex, GFP-tagged Miro-1 was analyzed for its ability to bind endogenous Milton-2 by Duolink PLA in U2OS cells treated with control or APC #1 siRNAs (Figure 9, A and B). In control cells, a strong PLA signal indicative of GFP-Miro/Milton-2 complexes was detected, compared with cells expressing GFP alone. In contrast, when APC was silenced, the mean number of GFP-Miro/Milton-2 PLA complexes detected per cell was greatly diminished relative to GFP control (Figure 9, A and B). When values were normalized by subtraction of the GFP background, it was revealed that loss of APC reduced Miro/Milton complex formation by ∼85% compared with the control (Figure 9B). These results suggest that APC is required for the interaction between Miro-1 and Milton-2. When a similar assay was applied to SW480APC1337 cells (Figure 9, A and C), we detected no GFP-Miro/Milton-2 PLA signal above background in transfected cells, a finding supported by IP assays in which antibodies successfully captured Miro/Milton complexes in U2OS cells but not in extract from SW480 cells (Figure 9D). Thus APC truncations, as found in CRC cells, correlate with reduced formation of Miro/Milton complexes. This result is quite specific, as APC truncations did not alter the binding of Milton to the kinesin motor KIF5 (Supplemental Figure S5, A and B).
FIGURE 9:
Full-length APC promotes assembly of the Miro-1/Milton-2 complex. (A) U2OS cells (treated with control siRNA or APC #1 siRNA) and SW480 cells expressing GFP alone or GFP-Miro-1 were subjected to Duolink PLA and analyzed for their ability to form protein complexes with endogenous Milton-2. Representative images from immunofluorescence microscopy are shown with PLA-positive protein complexes indicated by red dots. Scale bars: 10 μm. (B) The PLA signals per cell were scored in U2OS to reveal that the number of complexes formed between GFP Miro-1 and endogenous Milton-2 was significantly reduced following loss of APC (left panel). This was also presented as mean PLA signal/cell after subtraction of GFP background (right panel). (C) In SW480 cells, no significant interaction above background level was detected. Statistical significances are indicated on the graphs (***, p < 0.001; **, p <0.05; n.s., not significant). Box-and-whisker plots are presented as a median (line), upper/lower quartile (box), and min/max (error bars with 95% CI). Statistical analysis by Mann-Whitney U-test, with Bonferroni correction applied to (B). (D) U2OS and SW480 cells were subjected to IP assays using antibodies to pull down Miro and Milton. In U2OS cells, Milton was successfully detected in Miro pull downs and vice versa. In SW480 cells, however, Milton/Miro complexes were not detected by IP. (E) Total-cell extracts from U2OS cells treated with control siRNA or APC #1 siRNA were analyzed by Western blot to detect APC, Milton-2, Miro-1, and α-tubulin (loading control). No change in protein expression level was detected between the two samples. n > 100 cells for each sample from more than two independent experiments. (F) A proposed summary model is shown to indicate that APC has a role in initiation of anterograde mitochondrial transport through a C-terminal (indicated in red) interaction with the Miro/Milton complex. Note that the illustration is conceptual and that specific details of protein–protein interactions and the role of potential additional binding partners are not yet known.
Miro and Milton have been reported to be ubiquitinated and consequently degraded via the PINK/Parkin pathway (Liu et al., 2012; Birsa et al., 2014). To confirm that APC is not regulating assembly of Miro/Milton complexes by causing a change in Miro or Milton protein levels, we treated U2OS cells with control or APC siRNAs and analyzed cell lysates by Western blot (Figure 9E). The knockdown of APC elicited no changes in Miro-1 and Milton-2 protein expression. The mitochondrial localization pattern of Miro-1 also remained unchanged after APC knockdown (Supplemental Figure S5, C and D), indicating that disruption of the Miro/Milton complex by silencing of APC was not simply due to relocalization of Miro-1 away from mitochondria. We therefore conclude that wild-type APC promotes the interaction between Miro-1 and Milton-2 to facilitate mitochondrial transport (Figure 9F).
DISCUSSION
In 2008, we discovered that wild-type and truncated mutant forms of APC locate at mitochondria (Brocardo et al., 2008). In this study, we identify a new role for APC in the transport of mitochondria through its interaction with Miro/Milton. These new findings indicate that APC interacts via its C-terminus with the Miro/Milton complex and that this interaction is abolished by APC-truncating mutations commonly associated with CRC.
Our live-cell imaging data suggest that APC specifically influences the initiation of mitochondrial movement, rather than the ongoing velocity or rate of kinesin-dependent translocation. This is supported by our observation that silencing of APC caused the pool of motile mitochondria to decrease from 48 to 21% (Figure 4, A–D), while the velocity of motile mitochondria remained unchanged (Figure 4E). Remarkably, similar observations were reported previously following the silencing of Milton-1 in axons (Brickley and Stephenson, 2011) of a newly discovered regulator of the Miro/Milton complex, Alex3 (Lopez-Domenech et al., 2012), and of Miro itself (Wang and Schwarz, 2009; Wang et al., 2011). Collectively these studies suggest that loss of Milton, Miro, Alex3, or APC each causes a decrease in anterograde, and sometimes retrograde, transport of mitochondria and support the view that APC is a positive regulator of the Miro/Milton/KIF5 complex.
In fixed-cell experiments, APC depletion caused mitochondria to change from being uniformly spread throughout the cell to being more concentrated in the perinuclear region (Figure 1). The anterograde (outward membrane–directed) transport of mitochondria normally occurs in a series of pulsed ratchet-type movements (da Silva et al., 2014; Sheng, 2014) along microtubules, and this occurred less frequently in the absence of APC (Figure 4), explaining the net shift in mitochondria away from the membrane toward the nucleus. Evidence from Duolink PLA and IP assays indicate that both APC knockdown and truncation disrupt Miro/Milton-complex formation (Figure 9), suggesting that APC C-terminal sequences may contribute to the interaction between Miro and Milton. While the Miro/Milton interaction has been conserved from Drosophila to humans and the binding sites mapped to the N-termini (Glater et al., 2006; MacAskill et al., 2009a), it is less clear from the literature whether these two proteins interact without assistance of other cofactors. We propose that APC may act as a scaffold to assist in the assembly of the mitochondrial transport complex, an idea consistent with the known scaffolding roles of APC in other cellular pathways (Lui et al., 2012). Another possibility is that APC may regulate a posttranslational modification required for the Miro/Milton association, especially given the reported susceptibility of the mitochondrial transport complex to regulation by phosphorylation, ubiquitination, and O-GlcNAcylation (Wang et al., 2011; Liu et al., 2012; Pekkurnaz et al., 2014). In this regard, it is interesting that both APC (Figure 6C and Supplemental Figure S3A) and Milton (Figure 9D) appeared to preferentially associate with an ∼85-kDa form of Miro in IP assays; however, the nature of this form is yet to be defined. Further investigation will be required to more fully elucidate the mechanism and determine whether APC acts on Miro/Milton directly or indirectly by recruiting an additional bridging molecule(s).
Mitochondrial transport has often been studied in the context of neurodegenerative diseases in neuronal cells (reviewed in Chaturvedi and Flint Beal, 2013). However, more recent studies in different cell types, including epithelial and lymphocytic cells (Campello et al., 2006; Desai et al., 2013; Zhao et al., 2013), have shed light on the importance of regulated mitochondrial targeting to other facets of cell function related to cancer (Desai et al., 2013; Zhao et al., 2013). For example, Desai and collegues (2013) reported that siRNA silencing of Miro-1 reduced mitochondria at the membrane in breast cancer epithelial cells, correlating with decreased speed and loss of directional persistence. The cytoskeletal remodeling that occurs in cell migration is bioenergetically demanding. The localized concentration of mitochondria could provide swift ATP turnaround by recycling ADP back into the mitochondria for oxidative phosphorylation. Consistent with this idea, Zhao and colleagues (2013) showed that blocking mitochondrial distribution to the leading edge reduced F-actin polymerization, lamellipodia formation, and cell migration in breast cancer cells (Zhao et al., 2013). By analogy, the ATP supplied indirectly by the APC-mediated localization of mitochondria at membrane protrusions (Figure 5) may provide energy for tubulin polymerization and thus an additional mechanism by which APC influences cell polarity or directionality of migration (Etienne-Manneville, 2009; Okada et al., 2010).
The Miro/Milton-binding region was mapped to the C-terminal sequence 2650–2843 of APC, and loss of this sequence in CRC-associated APC mutants prevented binding to the Miro/Milton complex. This suggests that APC mutants, which can efficiently associate with mitochondria and contribute to cell survival (Brocardo et al., 2008), may be compromised in their ability to drive mitochondria to the membrane. Moreover, preliminary observations in SW480APC1337 cells indicate that APC truncation correlates with a reduced association of Miro and Milton, which are key to the attachment of mitochondria to the KIF5 kinesin motor (Figure 9; Glater et al., 2006; MacAskill et al., 2009a). Indeed, we observed diminished mitochondrial transport to the cell periphery in SW480APC1337 and HT29APC853/1555 cell lines. Importantly, this defect was partly rescued by reconstitution of full-length APC, demonstrating that not only does APC influence mitochondrial spread but that this function can in some instances of CRC be disrupted by mutation of APC (Figure 3). We speculate that the disruption of ATP delivery might contribute to the recently defined loss of directed cell migration attributed to peptides of truncated APC, especially in epithelial cells along the crypt/villus axis (Nelson et al., 2012).
Tumor cells, including CRC cells, display increased rates of glycolysis in the cytosol (Warburg effect) in comparison with normal cells, which primarily obtain their energy from mitochondrial oxidative-phosphorylation pathways (Koppenol et al., 2011). Furthermore, pyruvate dehydrogenase kinase, an enzyme that promotes aerobic glycolysis through inhibition of the PHD complex and, in turn, oxidative phosphorylation, has recently been identified as a Wnt target gene (Pate et al., 2014). Despite this, it is important for bioenergetic structures, such as dynamic membrane regions, to achieve a high level of site-specific ATP synthesis through targeting of mitochondria. Thus APC-stimulated movement of mitochondria to the membrane can provide a means for consistent and localized ATP output through fast ADP recycling. It is also metabolically ∼18-fold more efficient to produce ATP by oxidative phosphorylation than by glycolysis (Zheng, 2012).
Preliminary experiments indicate that stimulation of the canonical Wnt signaling pathway does not affect mitochondrial distribution in U2OS cells or alter the ability of APC to drive mitochondrial transport (Supplemental Figure S5E). However, there are additional Wnt-independent implications of altered mitochondrial distribution in APC-mutant cancer cells to be considered, such as a potential impact on rates of mitochondrial fission/fusion, calcium response, or cell survival. This last point is of interest, given that APC truncation mutants associate strongly at mitochondria to promote cell survival (Brocardo et al., 2008) and can influence CRC cell resistance to drugs (Martino-Echarri et al., 2014). Currently, certain anticancer drugs are being developed that target mitochondria, such as the CT20 peptide that promotes mitochondrial aggregation and membrane hyperpolarization leading to death of metastatic breast cancer cells (Lee et al., 2014). In addition, impairment of mitochondrial respiration by graphene was shown to inhibit the migration and invasion of cancer cells (Zhou et al., 2014). Further research into reagents that target mitochondria might prove useful when used in combination with current CRC drugs.
MATERIALS AND METHODS
Cell culture and transfection
U2OSAPC WT osteosarcoma and SW480APC 1309, HT-29APC 853/1555, HCT116APC WT, and LIM1215APC WT CRC cell lines were cultured under standard conditions in DMEM supplemented with 10% fetal bovine serum. NIH 3T3APC WT mouse fibroblasts and HDF1314APC WT human fibroblasts were cultured under standard conditions in DMEM supplemented with 10% calf serum. All cell lines were maintained at 37°C and confirmed to be mycoplasma free. For most transient transfections, cells seeded for at least 12 h were transfected with 500 ng–2 μg/ml DNA in Optimol medium using Fugene HD (Promega Corporation, Madison, WI) according to the manufacturer’s instructions. For transfections in wound-healing experiments 500 ng–2 μg/ml DNA in Optimol medium was transfected using the K2 transfection system (Biontex, Munich, Germany) according to the manufacturer’s instructions. Cells were processed 48 h posttransfection.
RNA interference
Double-stranded 21mer RNA oligonucleotides homologous to sequences in APC were purchased as purified duplexes (Qiagen, Hilden, Germany) and used to silence endogenous APC in cells seeded for at least 12 h. The APC DNA target sequences used were APC #1 (human): 5′-AGGGGCAGCAACTGATGAAAA-3′, APC #2 (human): 5′AACGAGCACAGCGAAGAATAG-3′ and mAPC/mAPC-red (mouse), 5′-AAGGACTGGTATTATGCTCAA-3′. The control sequence used was 5′-AACGAGCAGTCGCTTCAATAG-3′. EB1 siRNA and control siRNA were purchased from Santa Cruz Biotechnology (Dallas, TX). Cells were transfected with 3 μg/ml RNA duplexes in DMEM using Lipofectamine (Invitrogen, Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Cells were processed 72 h posttransfection.
Plasmids
The construction of pGFP-tagged forms of APC(1–302), APC(334–900), APC(2226–2644), and APC(2650–2843) was previously described (Sharma et al., 2006). pGFP-APC(1–1309) was kindly supplied by Myth Mok (Chinese University of Hong Kong), pGFP-APC(1379–2080) was provided by Mariann Bienz (Rosin-Arbesfeld et al., 2000), pCMV-APC by Bert Vogelstein (Morin et al., 1997), and pGFP-APCWT was provided by Angela Barth (Barth et al., 2002). pGFP-Miro-1 and pGFP-Milton-2 were provided by Mike Ryan (Koutsopoulos et al., 2010). pTagGFP2-mito is commercially available and was purchased from Evrogen (Moscow, Russia). pEGFP-C1 (Clontech, Mountainview, CA) was used as a control.
Immunofluorescence staining and deconvolution microscopy
Cells were grown for at least 24 h on glass coverslips in six-well plates before fixation with methanol-acetone. Cells were incubated for 1 h in 3% bovine serum albumin/phosphate-buffered saline (PBS), 1 h with the appropriate primary antibodies, and then an additional 1 h with secondary antibodies at concentrations described below. Each step was followed by three washes with PBS. The nucleus was detected with Hoechst 33258 (Invitrogen, Thermo Fisher Scientific). Coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA), and cells were analyzed using an Olympus IX71 microscope with a DeltaVision deconvolution system (GE Healthcare, Buckinghamshire, UK) equipped with a CoolSNAP HQ2 camera for image capture. When required, sequential transverse optical sections of the cell (z-stack images) were acquired in incremental steps of 0.2 μm, which could be rendered into a 3D projection using Softworx (version 3.7.1, Applied Precision, Seattle, WA). Images collected using the DeltaVision system were further resolved using Softworx deconvolution software. Data from scoring experiments were collected from at least 300 cells acquired from at least three independent experiments.
Scoring mitochondrial distribution
The distribution of mitochondria in distinct cell zones (see Figure 1B) within the cytoplasm was scored by microscopy after cells were stained with the MitoTracker Red CMX-Ros dye (Molecular Probes, Thermo Fisher Scientific). These zones radiated stepwise from the nucleus and constituted 33% (zone 1), 66% (zone 2), and 100% (zone 3) of the area and distance toward the cell membrane. Cells were selected at random and scored according to the zone in which mitochondria were located. SW480 is a biclonal cell line expressing two morphologically distinct cell types that appear as round or elongated, and hence additional parameters were observed. Elongated cells typically present a higher number of mitochondria in zone 3 compared with round cells and appeared more sensitive to toxicity associated with siRNA treatment. To avoid selecting out a single population while scoring, it was ensured that equal numbers of round and elongated cells were scored before and after APC knockdown, and results were pooled. For validation of the key experimental results, additional rigorous computational analysis was performed on a subset of randomly selected cells in each sample to take into account the varying morphological features of the cell lines tested (see Supplemental Methods).
Wound healing
NIH 3T3 cells were grown to confluence following the appropriate treatment, wounded with a 0.8-mm needle (PrecisionGlide, Becton Dickinson, Franklin Lakes, NJ), and washed twice with PBS before the addition of new medium. Subsequent fixation/immunostaining or live-cell imaging was performed 5 h after wounding.
IP
Cells seeded in flasks were trypsinized, washed with PBS, and lysed using Hunt’s buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5% NP-40) for 30 min on ice. For IP analysis, 1 mg lysate was continuously mixed with 2 μg antibody overnight at 4°C. Antibodies used were GFP pAb (Invitrogen, Thermo Fisher Scientific), APC Ab5 mAb (Merck Millipore, Darmstadt, Germany), APC C20 pAb (Santa Cruz), Miro1 pAb (Sigma-Aldrich, St. Louis, MO), immunoglobulin G (IgG) mouse (Sigma-Aldrich), and IgG rabbit (Sigma-Aldrich). Lysates were continuously mixed for an additional 2 h at 4°C with 30 μl protein A-Sepharose beads (Amersham Biosciences, GE Healthcare) to pull down immunocomplexes. Immunocomplexes attached to the beads were pelleted by centrifugation and washed three times with TBS. Precipitates were boiled in Laemmli buffer and subjected to SDS–PAGE and Western blotting as described below.
Western blotting
Cells seeded in flasks were trypsinized, washed with PBS, and lysed using RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40) for 30 min on ice. Cell debris in the lysates were pelleted by centrifugation, and the supernatant was denatured in Laemmli buffer. Samples (immunoprecipitates described above or ∼30 μg total-cell lysate) were separated by 5 or 7.5% SDS–PAGE and transferred onto a nitrocellulose membrane. The membrane was blocked in 5% skim milk/PBS for 1 h, and the protein of interest was detected using the appropriate primary antibody (concentrations listed below, in 5% skim milk) for 1 h and a secondary α-mouse/rabbit-HRP antibody (Sigma-Aldrich) for 1 h (1:5000 in 5% skim milk). Immunoblotting was visualized using the ChemiDoc MP imaging system (Bio-Rad, Hercules, CA) or on Amersham Hyperfilm developed using a CP1000 X-Ray film processor (AGFA). Experiments were replicated a minimum of three times.
Duolink PLA
Duolink PLA (Olink Biosciences, Uppsala, Sweden) uses specifically designed “PLA” probes, species-specific secondary antibodies joined to a short oligonucleotide sequence, which attach to the primary antibodies of target proteins. If the target proteins are in proximity (<40 nm), PLA probes are able to ligate and, through rolling circle amplification, which is followed by attachment of a complementary fluorophore, generate a quantifiable signal for visualization of protein–protein interactions in the cell (see Figure 6C). Cells were seeded in eight-well Lab-Tek chamber slides (Nunc) coated with poly-l-lysine (Sigma-Aldrich) for at least 24 h and fixed with methanol-acetone as previously described. Duolink PLA, including any subsequent counterstaining, was performed according to the manufacturer’s instructions. Primary antibody concentrations are listed below. Immunofluorescence analysis was performed using the DeltaVision system, and images were acquired and deconvolved as described above. Interactions were scored manually with a differential interference contrast (DIC) channel overlay used to define the cell periphery. If a construct was transfected, only cells expressing modest levels of the protein were scored. Data for endogenous assays were obtained from at least 250 cells per condition, while data for assays with transfected proteins are described in the figure legends or corresponding Supplemental Material. Each experiment was repeated at least twice.
Antibodies
Antibodies and concentrations used for immunofluorescence microscopy (IF), Western blotting (WB), and Duolink PLA (D) are as follows. APC antibodies used were Ab1 mAb (1:100 WB; Merck, OP44), Ab7 mAb (1:300 D; Merck, OP80) and H-290 pAb (1:500 IF; Santa Cruz, sc7930). GFP antibodies used were GFP mAb (1:1000 IF and WB, 1:10000 D; Roche, 11814460001) and GFP pAb (1:1000 IF; Invitrogen, Thermo Fisher Scientific, ab27043). Other primary antibodies used were α-tubulin mAb (1:1500 IF and WB; Sigma-Aldrich, T9026), EB1 mAb (1:200 IF; BD, 610535), mtHSP70 mAb (1:500 IF and WB; Thermo Fisher Scientific, MA3-028), Miro-1 pAb (1:100 IF, 1:500 WB, 1:300 D; Sigma-Aldrich, HPA010687), Milton-2 pAb (1:100 IF, 1:500 WB, 1:300 D; Sigma-Aldrich, HPA015827), actin mAb (1:500 IF; Sigma-Aldrich, A2228), IQGAP1 mAb (1:500 IF; BD, 610612), and β-catenin mAb (1:100 IF; BD, 610154). If required, CMX-Ros (1:10000; Molecular Probes, Life Technologies) was added directly to seeded cells before fixation for 15 min at 37°C. Secondary antibodies used in immunofluorescence microscopy studies were Alexa Fluor 405 mAb (1:300; Invitrogen, Thermo Fisher Scientific), Alexa Fluor 488 mAb and pAb (1:500; Invitrogen, Thermo Fisher Scientific), and Alexa Fluor 594 mAb and pAb (1:2000; Invitrogen, Thermo Fisher Scientific).
Live-cell imaging
NIH 3T3 cells were grown in two-well Lab-Tek chamber slides (Nunc), transfected with mAPC-red siRNA and pTagGFP2-mito, grown to confluence, and wounded as described above. Live-cell imaging was performed using the Olympus IX71 DeltaVision core deconvolution microscope equipped with a CoolSNAP HQ2 camera for image capture at 40×. Cells were maintained at 37°C and 5% CO2 and were imaged every 7 s for 5 min. Three z-stack images were acquired in fluorescein isothiocyanate (FITC) and DIC channels for each time point. The MTrackJ ImageJ plug-in (Meijering et al., 2012) was used to track randomly selected mitochondria in cells at the leading edge over three independent experiments, and the raw data were processed using Microsoft Excel. Cells chosen for imaging were healthy, positive for APC siRNA, expressed modest GFP2-mito, and did not show significant movement over the 5-min time period (visualized in DIC channel).
Supplementary Material
Acknowledgments
We are grateful to Mike Ryan for plasmids and antibody reagents and to members of our team for helpful discussions. This work was supported by grants to B.R.H. from the Cancer Institute, New South Wales, Australia (Career Development Fellowship 12/CDF/2-12), the Cancer Council New South Wales (project grant RG 11-04), and the National Health and Medical Research Council of Australia (project grant APP512372). K.M.M. was supported by a University of Sydney postgraduate award, and B.R.H. and M.G.B. were supported by bridging grants from the Westmead Institute for Medical Research.
Abbreviations used:
- 3D
three-dimensional
- APC
adenomatous polyposis coli
- CRC
colorectal cancer
- GFP
green fluorescent protein
- IF
immunofluorescence microscopy
- IgG
immunoglobulin G
- IP
immunoprecipitation
- IQGAP1
IQ-motif containing GTPase-activating protein 1
- KIF5
kinesin superfamily protein 5
- mAPC-red
red fluorescently tagged mouse APC siRNA
- mtHSP70
mitochondrial heat shock protein 70
- PBS
phosphate-buffered saline
- PLA
proximity ligation assay
- siRNA
small interfering RNA
- WB
Western blotting
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0632) on December 10, 2015.
REFERENCES
- Akiyama T, Kawasaki Y. Wnt signalling and the actin cytoskeleton. Oncogene. 2006;25:7538–7544. doi: 10.1038/sj.onc.1210063. [DOI] [PubMed] [Google Scholar]
- Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- Askham JM, Moncur P, Markham AF, Morrison EE. Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene. 2000;19:1950–1958. doi: 10.1038/sj.onc.1203498. [DOI] [PubMed] [Google Scholar]
- Barth AI, Siemers KA, Nelson WJ. Dissecting interactions between EB1, microtubules and APC in cortical clusters at the plasma membrane. J Cell Sci. 2002;115:1583–1590. doi: 10.1242/jcs.115.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. The subcellular destinations of APC proteins. Nat Rev Mol Cell Biol. 2002;3:328–338. doi: 10.1038/nrm806. [DOI] [PubMed] [Google Scholar]
- Birsa N, Norkett R, Wauer T, Mevissen TE, Wu HC, Foltynie T, Bhatia K, Hirst WD, Komander D, Plun-Favreau H, Kittler JT. Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley K, Smith MJ, Beck M, Stephenson FA. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J Biol Chem. 2005;280:14723–14732. doi: 10.1074/jbc.M409095200. [DOI] [PubMed] [Google Scholar]
- Brickley K, Stephenson FA. Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J Biol Chem. 2011;286:18079–18092. doi: 10.1074/jbc.M111.236018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocardo M, Henderson BR. APC shuttling to the membrane, nucleus and beyond. Trends Cell Biol. 2008;18:587–596. doi: 10.1016/j.tcb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Brocardo M, Lei Y, Tighe A, Taylor SS, Mok MT, Henderson BR. Mitochondrial targeting of adenomatous polyposis coli protein is stimulated by truncating cancer mutations: regulation of Bcl-2 and implications for cell survival. J Biol Chem. 2008;283:5950–5959. doi: 10.1074/jbc.M708775200. [DOI] [PubMed] [Google Scholar]
- Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi RK, Flint Beal M. Mitochondrial diseases of the brain. Free Radical Biol Med. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Contreras L, Drago I, Zampese E, Pozzan T. Mitochondria: the calcium connection. Biochim Biophys Acta. 2010;1797:607–618. doi: 10.1016/j.bbabio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- da Silva AF, Mariotti FR, Maximo V, Campello S. Mitochondria dynamism: of shape, transport and cell migration. Cell Mol Life Sci. 2014;71:2313–2324. doi: 10.1007/s00018-014-1557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. APC in cell migration. Adv Exp Med Biol. 2009;656:30–40. doi: 10.1007/978-1-4419-1145-2_3. [DOI] [PubMed] [Google Scholar]
- Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- Fransson A, Ruusala A, Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Hanson CA, Miller JR. Non-traditional roles for the adenomatous polyposis coli (APC) tumor suppressor protein. Gene. 2005;361:1–12. doi: 10.1016/j.gene.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, Akiyama T. Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol. 2002;4:323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- Johannsen DL, Ravussin E. The role of mitochondria in health and disease. Curr Opin Pharmacol. 2009;9:780–786. doi: 10.1016/j.coph.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim Biophys Acta. 2010;1803:564–574. doi: 10.1016/j.bbamcr.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Kroboth K, Newton IP, Kita K, Dikovskaya D, Zumbrunn J, Waterman-Storer CM, Nathke IS. Lack of adenomatous polyposis coli protein correlates with a decrease in cell migration and overall changes in microtubule stability. Mol Biol Cell. 2007;18:910–918. doi: 10.1091/mbc.E06-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Bassiouni R, Sparrow NA, Iketani A, Boohaker RJ, Moskowitz C, Vishnubhotla P, Khaled AS, Oyer J, Copik A, et al. The CT20 peptide causes detachment and death of metastatic breast cancer cells by promoting mitochondrial aggregation and cytoskeletal disruption. Cell Death Dis. 2014;5:e1249. doi: 10.1038/cddis.2014.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Domenech G, Serrat R, Mirra S, D’Aniello S, Somorjai I, Abad A, Vitureira N, Garcia-Arumi E, Alonso MT, Rodriguez-Prados M, et al. The Eutherian Armcx genes regulate mitochondrial trafficking in neurons and interact with Miro and Trak2. Nat Commun. 2012;3:814. doi: 10.1038/ncomms1829. [DOI] [PubMed] [Google Scholar]
- Lui C, Mills K, Brocardo MG, Sharma M, Henderson BR. APC as a mobile scaffold: regulation and function at the nucleus, centrosomes, and mitochondria. IUBMB Life. 2012;64:209–214. doi: 10.1002/iub.599. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Brickley K, Stephenson FA, Kittler JT. GTPase dependent recruitment of Grif-1 by Miro1 regulates mitochondrial trafficking in hippocampal neurons. Mol Cell Neurosci. 2009a;40:301–312. doi: 10.1016/j.mcn.2008.10.016. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009b;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino-Echarri E, Henderson BR, Brocardo MG. Targeting the DNA replication checkpoint by pharmacologic inhibition of Chk1 kinase: a strategy to sensitize APC mutant colon cancer cells to 5-fluorouracil chemotherapy. Oncotarget. 2014;5:9889–9900. doi: 10.18632/oncotarget.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Zhou XZ, Lu KP. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr Biol. 2001;11:1062–1067. doi: 10.1016/s0960-9822(01)00297-4. [DOI] [PubMed] [Google Scholar]
- Nathke I. Cytoskeleton out of the cupboard: colon cancer and cytoskeletal changes induced by loss of APC. Nat Rev Cancer. 2006;6:967–974. doi: 10.1038/nrc2010. [DOI] [PubMed] [Google Scholar]
- Nelson SA, Li Z, Newton IP, Fraser D, Milne RE, Martin DM, Schiffmann D, Yang X, Dormann D, Weijer CJ, et al. Tumorigenic fragments of APC cause dominant defects in directional cell migration in multiple model systems. Dis Model Mech. 2012;5:940–947. doi: 10.1242/dmm.008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, McQuade MM, Garner C, Digman MA, Teitell MA, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33:1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil H, Cho KI, Lee J, Yang Y, Orry A, Ferreira PA. Kinesin-1 and mitochondrial motility control by discrimination of structurally equivalent but distinct subdomains in Ran-GTP-binding domains of Ran-binding protein 2. Open Biol. 2013;3:120183. doi: 10.1098/rsob.120183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz G, Trinidad JC, Wang X, Kong D, Schwarz TL. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 2014;158:54–68. doi: 10.1016/j.cell.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci USA. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda T, Iizuka-Kogo A, Onouchi T, Shimomura A. Adenomatous polyposis coli (APC) plays multiple roles in the intestinal and colorectal epithelia. Med Mol Morphol. 2007;40:68–81. doi: 10.1007/s00795-006-0352-5. [DOI] [PubMed] [Google Scholar]
- Serrat R, Lopez-Domenech G, Mirra S, Quevedo M, Garcia-Fernandez J, Ulloa F, Burgaya F, Soriano E. The non-canonical Wnt/PKC pathway regulates mitochondrial dynamics through degradation of the arm-like domain-containing protein Alex3. PLoS One. 2013;8:e67773. doi: 10.1371/journal.pone.0067773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Leung L, Brocardo M, Henderson J, Flegg C, Henderson BR. Membrane localization of adenomatous polyposis coli protein at cellular protrusions: targeting sequences and regulation by beta-catenin. J Biol Chem. 2006;281:17140–17149. doi: 10.1074/jbc.M513027200. [DOI] [PubMed] [Google Scholar]
- Sheng ZH. Mitochondrial trafficking and anchoring in neurons: new insight and implications. J Cell Biol. 2014;204:1087–1098. doi: 10.1083/jcb.201312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on Milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell signalling. J Cell Sci. 2012;125:807–815. doi: 10.1242/jcs.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, Lavoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4:1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhang B, Zheng J, Yu M, Zhou T, Zhao K, Jia Y, Gao X, Chen C, Wei T. The inhibition of migration and invasion of cancer cells by graphene via the impairment of mitochondrial respiration. Biomaterials. 2014;35:1597–1607. doi: 10.1016/j.biomaterials.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Zumbrunn J, Kinoshita K, Hyman AA, Nathke IS. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.