Abstract
Huntington’s disease (HD) is a progressive neurodegenerative disorder caused by a CAG-polyglutamine repeat expansion in the huntingtin (htt) gene. We found that peroxisome proliferator-activated receptor delta (PPARδ) interacts with htt and that mutant htt represses PPARδ-mediated transactivation. Increased PPARδ transactivation ameliorated mitochondrial dysfunction and improved cell survival of HD neurons. Expression of dominant-negative PPARδ in CNS was sufficient to induce motor dysfunction, neurodegeneration, mitochondrial abnormalities, and transcriptional alterations that recapitulated HD-like phenotypes. Expression of dominant-negative PPARδ specifically in the striatum of medium spiny neurons in mice yielded HD-like motor phenotypes, accompanied by striatal neuron loss. In mouse models of HD, pharmacologic activation of PPAR δ, using the agonist KD3010, improved motor function, reduced neurodegeneration, and increased survival. PPAR δ activation also reduced htt-induced neurotoxicity in vitro and in medium spiny-like neurons generated from human HD stem cells, indicating that PPAR δ activation may be beneficial in individuals with HD and related disorders.
The PPARs are ligand-activated transcription factors that belong to the nuclear hormone receptor superfamily. The three subtypes, termed PPARα, - δ, and -γ, serve as lipid sensors in response to increased energy requirements1. PPARs are activated by lipids and fatty acid derivatives, and perform essential functions in lipid homeostasis, glucose metabolism, energy production, and cellular differentiation.
The least-studied member of the PPAR nuclear receptor family is PPAR δ, which shows a fairly ubiquitous expression pattern, but is highly abundant in skeletal muscle and brain2,3. The function of PPAR δ has been studied in skeletal muscle, where PPAR δ regulates skeletal muscle energy metabolism and mitochondrial biogenesis4,5. Indeed, PPAR δ promotes mitochondrial biogenesis and regulates fiber type switching in muscle6,7. PPAR δ is at least two-fold more highly expressed in brain than in muscle8, making it the most abundant PPAR subtype in the CNS, but its functional relevance there remains unknown.
HD is a progressive autosomal dominant neurodegenerative disorder in which patients develop motor and cognitive impairment9. Although the mutant htt protein is widely expressed, neurodegeneration and atrophy occur principally in medium-sized spiny neurons (MSNs) of the striatum and cerebral cortex pyramidal neurons that project to the striatum10. In 1993, a CAG trinucleotide repeat expansion mutation in the coding region of the htt gene was identified as the cause of HD11. Htt glutamine tracts that exceed a certain length threshold (approximately 37 repeats in HD) adopt a pathogenic conformation, yielding conformers that are resistant to the normal processes of protein turnover, resulting in the accumulation of pathogenic mutant htt, cellular toxicity, and neurodegeneration12.
Neurons in the brain have high energy demands and require mitochondrial production of ATP. Chronic administration of a mitochondrial toxin, 3-nitropropionic acid, results in selective loss of MSNs in the striatum13. This finding has been corroborated in HD cell culture models, mice, and samples from HD individuals (reviewed in 14), and suggested that mitochondrial dysfunction may underlie HD pathogenesis and account for the cell-type specificity. At the same time, the necessity of htt nuclear localization for HD disease pathogenesis highlighted nuclear pathology as a likely early step in the neurotoxicity cascade15. N-terminal fragments of mutant htt protein interfere with gene transcription in the early stages of the HD disease process (reviewed in 16). We, and others linked the mitochondrial dysfunction and metabolic defects in HD to transcriptional dysregulation of peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α), a co-activator that controls a network of transcriptional programs that culminate in mitochondrial biogenesis and enhanced energy production17–19. The importance of PGC-1α for HD pathogenesis is underscored by the observation that PGC-1α over-expression in HD mice is sufficient to rescue motor phenotypes, reduce accumulation of misfolded htt protein in CNS, and ameliorate neurodegeneration20.
When an unbiased screen for htt-interacting proteins yielded PPARs as candidate interactors, we evaluated the different PPARs, and documented an interaction between PPARδ and htt. We investigated the role of PPARδ repression in HD, and determined that transcriptional dysregulation of PPARδ underlies HD mitochondrial dysfunction and neurodegeneration. By directing expression of dominant-negative PPARδ to the CNS or specifically to MSNs in striatum, we demonstrated that expression of dominant-negative PPARδ is sufficient to produce motor phenotypes, neurodegeneration, mitochondrial defects, and transcriptional abnormalities that parallel disease phenotypes in HD. In light of these results, we evaluated the PPARδ agonist KD3010 as a therapy for HD, and found that KD3010 could ameliorate HD phenotypes in transgenic mice and in striatal medium spiny-like neurons derived from induced pluripotent stem cells from HD individuals. Our findings offer novel insights into the pathogenesis of HD, provide evidence for an unexpectedly crucial role for PPARδ in maintaining neural health, and highlight a promising therapeutic strategy for HD and related neurodegenerative disorders.
Results
Huntingtin and PPARδ physically interact
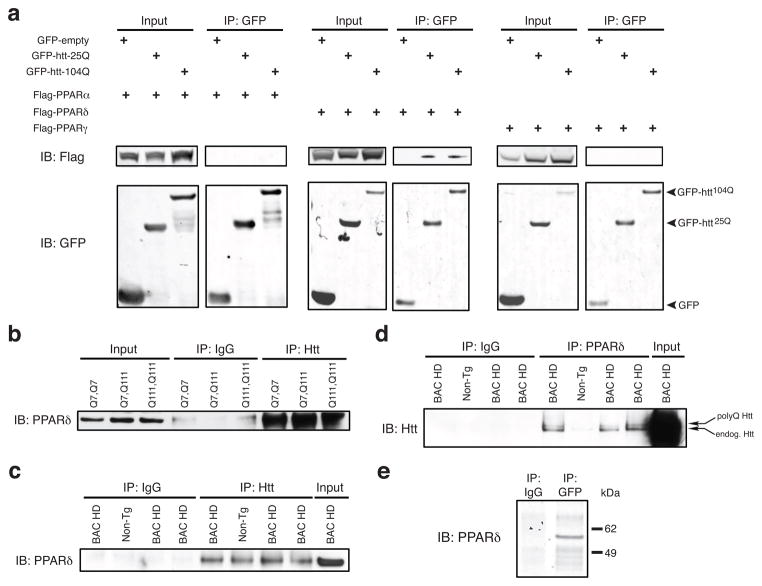
To identify transcription factors that interact with htt, we transfected HEK293 cells with GFP-tagged N-terminal htt protein, immunoprecipitated GFP-htt-25Q, and applied the htt-25Q immunoprecipitated material to a transcription factor binding site array. This unbiased screen yielded PPAR response element (PPRE) binding proteins as candidate interacting proteins (Supplementary Fig. 1a), and led us to examine whether htt interacts with one of the PPARs. To test this hypothesis, we performed transient transfections of HEK293 cells with GFP-htt-25Q, GFP-htt-104Q, or empty GFP vector, and with Flag-tagged PPARα, PPARδ, or PPARγ. After GFP immunoprecipitation, we immunoblotted with anti-Flag antibody and detected PPARδ in htt-25Q and htt-104Q immunoprecipitates, but found no evidence for an interaction between htt and PPARα, or htt and PPARγ (Fig. 1a). Quantification of PPARδ band intensity upon htt immunoprecipitation indicated that the polyQ tract expansion promotes greater interaction between PPARδ and htt (Supplementary Fig. 1b). PPARδ is the most abundantly expressed PPAR subtype in the CNS8; however, its functional relevance in CNS is ill-defined. To determine if endogenous PPARδ and htt interact, we performed co-immunoprecipitations in ST-Hdh striatal-like cells. After immunoprecipitation of htt with an antibody directed against its C-terminus (Supplementary Fig. 1c) and immunoblotting with a PPARδ-specific antibody, we found that PPARδ interacts with full-length htt in Q7/Q7, Q7/Q111, and Q111/Q111 ST-Hdh cells (Fig. 1b). To establish if this interaction occurs in the CNS under physiological conditions, we performed co-immunoprecipitation on protein lysates isolated from the cortex of BAC-HD97 mice21. When we immunoprecipitated htt and immunoblotted with a PPARδ-specific antibody, we detected PPARδ protein in BAC-HD and non-transgenic mice (Fig. 1c). We then immunoprecipitated PPARδ and upon immunoblotting with a htt-specific antibody, we observed both polyQ-htt and endogenous htt protein in PPARδ immunoprecipitates from BAC-HD cortex, and detected endogenous htt in PPARδ immunoprecipitates from non-transgenic cortex (Fig. 1d). Finally, to determine the nature of the interaction, we performed in vitro transcription and translation of PPARδ and GFP-htt-Q25 (aa 1-171), and then did a pull-down with a GFP or IgG-specific antibody. Immunoblotting of the pull-down material yielded PPARδ protein for the GFP immunoprecipitation, but not for the IgG immunoprecipitation (Fig. 1e). These findings indicate that PPARδ and htt physically interact in the CNS.
Figure 1. Huntingtin and PPARδ physically interact.
(a) Immunoprecipitation with GFP antibody and immunoblot analysis of GFP-htt-25Q or GFP-htt-104Q and Flag-tagged PPARα, -δ, or -γ. GFP-empty-transfected HEK293 cells served as a negative control.
(b) Immunoprecipitation of ST-Hdh striatal-like neurons of the indicated genotypes was performed with Htt antibody, followed by immunoblot analysis of PPARδ.
(c) Immunoprecipitation of protein lysates from the cortex of 8 month-old BAC-HD mice and non-transgenic (Non-Tg) controls with Htt antibody, and immunoblot analysis of PPARδ.
(d) Immunoprecipitation of protein lysates from the cortex of 8 month-old BAC-HD mice and non-transgenic (Non-Tg) controls with PPARδ antibody, and immunoblot analysis of Htt. Note detection of both transgenic polyQ-Htt protein and endogenous mouse Htt protein in BAC-HD samples, and detection of endogenous mouse Htt protein in Non-Tg control.
(e) Immunoprecipitation of in vitro transcription-coupled translation of GFP-tagged htt-Q25 and PPARδ with GFP antibody, followed by immunoblot analysis of PPARδ. For (b) – (e), immunoprecipitation with IgG only served as a negative control.
Altered PPARδ transactivation contributes to HD mitochondrial dysfunction and neurotoxicity
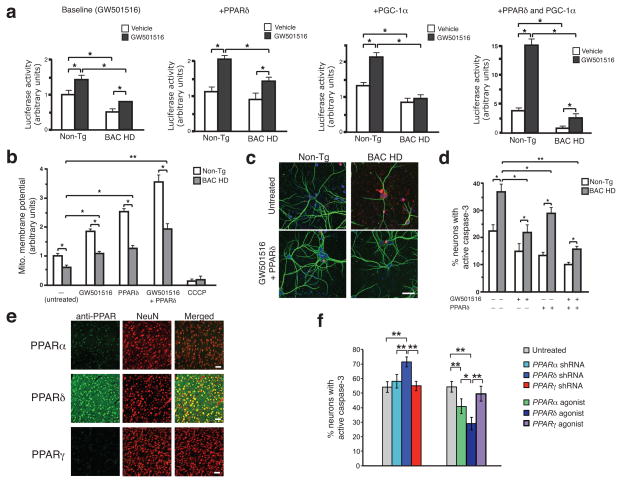
We evaluated the effect of polyQ-htt on PPARδ transactivation by co-transfecting N-terminal truncated htt (exon 1+2) and a 3x-PPAR response element (3x-PPRE) luciferase reporter into HEK293 cells, and noted polyQ length-dependent inhibition of PPARδ transactivation upon treatment with the PPARδ selective agonist GW501516 (Supplementary Fig. 2a). We then measured PPARδ transactivation in ST-Hdh striatal-like cells, and found that PPARδ transactivation was significantly repressed in a polyQ length-dependent fashion, but could be partially rescued by either PPARδ or PGC-1α, and fully rescued with PPARδ and PGC-1α together (Supplementary Fig. 2b). To examine the role of PPARδ transcription repression in HD neurons, we repeated the transactivation assays in primary cortical neurons from BAC-HD mice, and observed significantly impaired PPARδ transactivation at baseline, upon co-transfection with PPARδ or PGC-1α, and in response to co-transfection of PPARδ and PGC-1α together (Fig. 2a). When we measured mitochondrial membrane potential, we noted significantly reduced mitochondrial membrane potential in BAC-HD neurons (Fig. 2b). Importantly, over-expression of PPARδ, via infection with PPARδ lentivirus, or treatment with GW501516 rescued mitochondrial membrane polarization alterations in BAC-HD neurons, with combined PPARδ over-expression and GW501516 treatment resulting in the most robust effect (Fig. 2b). Caspase-3 activation was elevated and MAP2 immunoreactivity reduced in BAC-HD neurons, as compared to non-transgenic primary cortical neurons, indicating that HD neurons are more susceptible to cell death (Fig. 2c). We noted increased MAP2 staining and reduced caspase-3 activation in BAC-HD neurons transduced with PPARδ lentivirus in the presence of GW501516 (Fig. 2c). Increased PPARδ expression, GW501516 treatment, or PPARδ in combination with GW501516 could rescue cell death in BAC-HD neurons (Fig. 2d). Analysis of MAP2 immunoreactivity in BAC-HD neurons independently corroborated these results (Supplementary Fig. 2c).
Figure 2. PPARδ activation rescues transcriptional repression, mitochondrial membrane depolarization, and neurotoxicity in HD neurons.
(a) 3X-PPRE luciferase reporter activity in primary cortical neurons from BAC-HD or non-transgenic (Non-Tg) control mice, co-transfected with Renilla luciferase vector, and PPARδ and/or PGC-1α expression constructs, as indicated, and treated with GW501516 (100 nM) or vehicle. Results were normalized to Non-Tg neurons at baseline. *P < .05; t-test.
(b) Mitochondrial membrane potential of primary cortical neurons from BAC-HD and Non-Tg mice, treated as indicated, was determined from the ratio of mitochondrial to cytosolic JC-1 fluorescence. Results were normalized to Non-Tg neurons at baseline, and CCCP treatment served as a positive control for depolarization. *P < .05, **P < .01; t-test.
(c) Immunofluorescence of active caspase-3 (red) and microtubule-associated protein 2 (green) in primary cortical neurons from BAC-HD and Non-Tg mice, treated as indicated. Scale bar = 20 μm.
(d) Quantification of active caspase-3 staining shown in (e). Results were normalized to Non-Tg neurons at baseline. *P < .05, **P < .01; t-test.
(e) Cortex from Non-Tg mice immunostained with the indicated PPAR antibody (green) and NeuN antibody (red). Merged images reveal expression of indicated PPAR. Scale bar = 50 μm.
(f) Immunofluorescence of active caspase-3 in primary cortical neurons from BAC-HD mice, transfected with the indicated shRNA construct for 3 days, or treated with fenofibrate 100 nM (PPARα agonist), GW501516 100 nM (PPARδ agonist), or pioglitazone 20 nM (PPARγ agonist) for 24 h, prior to exposure to 25 μM H2O2. *P < .05, **P < .01; ANOVA with post-hoc Tukey. Error bars = s.e.m. All experiments were performed with 3 biological replicates and 9 technical replicates per condition.
We have previously shown that altered transcriptional activity of PGC-1α contributes to HD pathogenesis, with significant reductions in PGC-1α targets in HD mice and human individuals19. Although the expression of PGC-1α is reduced in HD, the magnitude of the decreases cannot fully account for the transcriptional alterations documented in HD, indicating that a PGC-1α-dependent transcription factor could be responsible for the transcriptional dysregulation. As PPARδ and htt physically interact in the CNS, and specific PPARδ target genes exhibit reduced expression in BAC-HD striatum, we postulated that impaired PPARδ function accounts for altered PGC-1α transcriptional activity in HD. Gene set analysis of striatal Affymetrix U133A/B microarray expression data from human HD individuals revealed significant alterations in 12 of 16 principal PPARδ target genes (P < 10−4, χ2) (Supplementary Fig. 3a), consistent with this hypothesis.
Physical interaction of polyQ-htt with PPARδ promotes HD neurotoxicity
To determine the physiological relevance of the PPARδ – htt interaction, we examined striatal sections from HD N171-82Q transgenic mice, and observed co-localization of polyQ-htt protein with PPARδ (Supplementary Fig. 3b). To test whether disruption of this interaction would prevent polyQ-htt neurotoxicity, we created PPARδ expression constructs with deletions of different PPARδ domains (Supplementary Fig. 4a). HEK293 cell co-transfections with GFP-tagged htt expression constructs and PPARδ domain deletion constructs, followed by co-immunoprecipitation, enabled mapping of the htt interaction site to the D domain (hinge region) of PPARδ (Supplementary Fig. 4b). To determine if physical interaction between polyQ-htt and PPARδ is necessary for neurotoxicity, we co-expressed different PPARδ deletion mutants in combination with full-length PPARδ in ST-Hdh Q111/Q111 striatal-like cells. Although co-expression of PPARδ lacking either the A – C domain or the E – F domain prevented polyQ-htt transcriptional repression, mitochondrial membrane depolarization, and cell death, co-expression of PPARδ lacking the D – F domain did not rescue any of these neurotoxicity phenotypes (Supplementary Fig. 5). These findings indicate that competition for mutant htt interaction with PPARδ deletion constructs specifically retaining the D domain is sufficient to blunt polyQ-htt neurotoxicity, underscoring the importance of the interaction for HD pathology.
Previous studies have implicated PPARγ transcriptional alterations in HD pathogenesis22–24. To examine the potential contribution of PPARα or PPARγ dysfunction to polyQ-htt neurotoxicity, we surveyed PPAR expression in CNS. Immunohistochemistry analysis of sections from the cortex of control mice revealed prominent neuronal expression of PPARδ, modest neuronal expression of PPARα, and barely detectable neuronal expression of PPARγ (Fig. 2e). In the cortex, striatum, and cerebellum of BAC-HD mice and non-transgenic littermates, prominent neuronal expression of PPARδ was observed, with limited expression of PPARα and PPARγ (Supplementary Fig. 6). These findings were corroborated upon immunoblotting of cortical lysates, cortical neurons, cerebellar lysates, and cerebellar granule neurons from BAC-HD and non-transgenic mice (Supplementary Fig. 7a–d). To directly determine the contribution of the different PPARs to polyQ-htt neurotoxicity, we cultured primary cortical neurons from BAC-HD mice and monitored cell death upon knock-down or agonist activation of each PPAR. Cell death was increased in BAC-HD neurons in which PPARδ was silenced by shRNA, while knock-down of PPARα or PPARγ did not affect cell death (Fig. 2f). Similarly, treatment with a PPARδ agonist reduced cell death in comparison to PPARα or PPARγ agonist-treated BAC-HD neurons (Fig. 2f).
To evaluate the contribution of PPARδ transcriptional alterations to HD pathogenesis in vivo, we measured the expression of 11 PPARδ target genes in the striatum of BAC-HD transgenic mice, and noted reductions for all tested PPARδ target genes in BAC-HD striatum (Supplementary Fig. 7e). RT-PCR analysis confirmed significant reductions in the expression levels of PPARδ targets in ST-Hdh Q111/Q111 striatal-like cells in comparison to Q7/Q7 cells (Fig. 3a). We then performed chromatin immunoprecipitation and found that PPARδ occupancy is reduced at the promoters of its target genes in Q111/Q111 striatal-like cells (Fig. 3a), suggesting that PPARδ binding to target promoters is prevented upon interacting with polyQ-htt.
Figure 3. PPARδ transcriptional interference yields neuron dysfunction, and induces neurological phenotypes and mitochondrial abnormalities in transgenic mice.
(a) RT-PCR analysis of PPARδ target gene expression (left) and ChIP analysis of PPARδ occupancy at the promoters of PPARδ target genes (right) in ST-Hdh wild-type (Q7/Q7) and homozygous HD mutant (Q111/Q111) striatal-like cells (n = 3 / genotype; 6 – 9 technical replicates). **P < .01, *P < .05; t-test.
(b) Nestin-Cre;PPARδ-E411P mice weigh less than littermate control mice. **P < .01; t-test.
(c) Nestin-Cre;PPARδ-E411P mice cannot easily dismount from the cage ledge . **P < .01; t-test.
(d) Nestin-Cre;PPARδ-E411P mice exhibit a prominent clasping phenotype. ***P < .001; t-test.
(e) Nestin-Cre;PPARδ-E411P mice display kyphosis. **P < .01; t-test.
(f) Nestin-Cre;PPARδ-E411P mice attained significantly worse ‘latency to fall’ times on the accelerating rotarod . **P < .01, *P < .05; ANOVA with post-hoc Tukey test.
(g) Decreases in mean stride length and reduced combined mesh and triangle grip strength were observed for Nestin-Cre;PPARδ-E411P mice. Grip strength is given in arbitrary units with littermate control performance set to 1. *P < .05; t-test. For (b) – (g), cohort age was 8 months, and group sizes were 8 – 9.
(h) Electron micrographs of mitochondria in striatum neurons of 8 month-old mice of indicated genotypes. Scale bar = 1 μm.
(i) Quantification of neuron mitochondrial size in the striatum and cortex, as assessed by mitochondrial area and perimeter (n = 4 mice / group). *P < .05; t-test.
(j) HPLC measurement of ATP concentrations in the striatum and cortex of 8 month-old mice of indicated genotypes (n = 3 mice / group). *P < .05; t-test. For all experiments, error bars = s.e.m.
Dominant-negative PPARδ expression in the CNS induces motor abnormalities, mitochondrial defects, and neurodegeneration
To determine if impaired PPARδ transactivation function is sufficient to elicit neurodegeneration in vivo, we derived a conditional transgenic PPARδ expression construct containing a single amino acid mutation, substituting proline for glutamate at residue 411 (Supplementary Fig. 8a). The E411P mutation inhibits PPARδ transactivation in a dominant-negative fashion25,26. After generating CAGGS-floxed STOP-PPARδ-E411P transgenic mice, we evaluated these mice for signs of systemic or neurological phenotypes, and did not detect any evidence of abnormality. We then crossed these mice with CMV-Cre driver mice to permit widespread expression of PPARδ-E411P, and akin to PPARδ knock-out mice27, we found that ubiquitous expression of PPARδ-E411P protein causes prenatal lethality. We next crossed the CAGGS-floxed STOP-PPARδ-E411P mice with Nestin-Cre mice to obtain bigenic Nestin-Cre;PPARδ-E411P mice, as the Nestin-Cre driver promotes expression of Cre recombinase throughout the neural lineage28. PPARδ-E411P protein was expressed at 1.5 to 2x endogenous PPARδ protein. Characterization of Nestin-Cre;PPARδ-E411P mice, in comparison to singly transgenic Nestin-Cre and CAGGS-floxed STOP-PPARδ-E411P mice, indicated that Nestin-Cre;PPARδ-E411P bigenic mice appear smaller by 4 – 6 months of age, such that by 8 months of age, they weigh less than littermate controls (Fig. 3b). Phenotype evaluation, using a neurological examination screening tool29, revealed that Nestin-Cre;PPARδ-E411P mice suffer prominent motor abnormalities by 8 months of age (Fig. 3c–e). Testing of Nestin-Cre;PPARδ-E411P mice on an accelerating rotarod confirmed motor impairment (Fig. 3f), while stride length analysis and grip strength testing provided further evidence for motor abnormalities (Fig. 3g). We also evaluated learning and memory in Nestin-Cre;PPARδ-E411P mice, and documented reduced novel object recognition and reduced novel environment exploration (Supplementary Fig. 8b–d).
Ultrastructural analysis of Nestin-Cre;PPARδ-E411P mice revealed that mitochondria in the striatum appear smaller in size (Fig. 3h), in agreement with previous studies of HD30,31, as mitochondrial area and perimeter in the cortex and striatum was reduced (Fig. 3i). To evaluate metabolic function, we measured mitochondrial complex activities. Although mitochondrial complex I and II activities in Nestin-Cre;PPARδ-E411P and control mice were similar, mitochondrial complex IV and V activities were reduced in the striatum of Nestin-Cre;PPARδ-E411P mice (Supplementary Fig. 9a–d), and the overall ATP concentration was reduced in the striatum and cortex (Fig. 3j). Quantitative PCR analysis of mitochondrial and nuclear genomic DNA content corroborated abnormalities in mitochondrial bioenergetics in Nestin-Cre;PPARδ-E411P mice (Supplementary Fig. 9e).
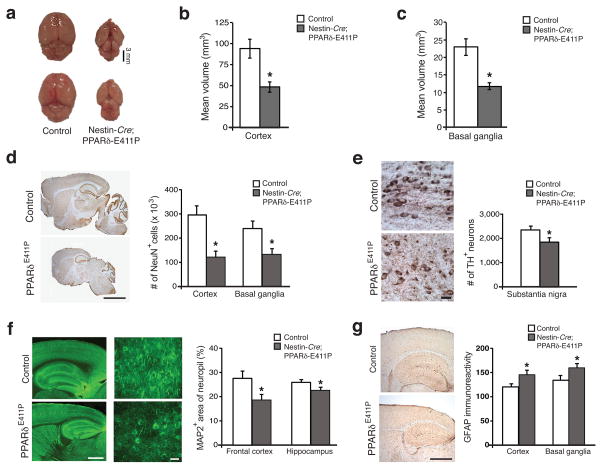
Brain size in adult Nestin-Cre;PPARδ-E411P mice was markedly reduced in comparison to littermate controls (Fig. 4a). Comparable reductions in the volume of the cortex and the basal ganglia were noted in Nestin-Cre;PPARδ-E411P mice (Fig. 4b,c). NeuN staining of Nestin-Cre;PPARδ-E411P brain sections revealed reduced neuron numbers (Fig. 4d). This was paralleled by a reduction in tyrosine hydroxylase (TH)-positive neurons in substantia nigra (Fig. 4e), decreased MAP2 immunostaining in the cortex and hippocampus (Fig. 4f), and reactive gliosis, as assessed by GFAP immunostaining, in the brains of Nestin-Cre;PPARδ-E411P mice (Fig. 4g). Hence, mice expressing a dominant-negative version of PPARδ in the CNS develop striking neurological abnormalities in the context of profound neurodegeneration.
Figure 4. Expression of dominant-negative PPARδ in neurons results in widespread neurodegeneration.
(a) Dissection of whole brain reveals dramatic global atrophy in Nestin-Cre;PPARδ-E411P mice. Cortex volume (b) and basal ganglia volume (c) are markedly reduced in Nestin-Cre;PPARδ-E411P mice. (d) Sections of whole brain stained for NeuN (left), and quantification of neuron number (right). Scale bar = 250 μm. (e) Substantia nigra stained for tyrosine hydroxylase (TH) (left), and quantification of TH+ neurons (right). Scale bar = 10 μm. (f) Frontal cortex and hippocampus stained for microtubule-associated protein 2 (MAP2) (left), and quantification of MAP2 immunoreactivity as a percentage of overall neuropil (right). Scale bar (low power) = 100 μm. Scale bar (high power) = 10 μm. (g) Cortex and basal ganglia stained for GFAP (left), and quantification of overall GFAP immunoreactivity (right). Scale bar = 100 μm.
For all experiments, *P < .05; t-test, cohort age was 10 months, group sizes were 4 – 6, and error bars = s.e.m.
Dominant-negative PPARδ expression in striatum induces Huntington’s disease-like neuronal and molecular pathology
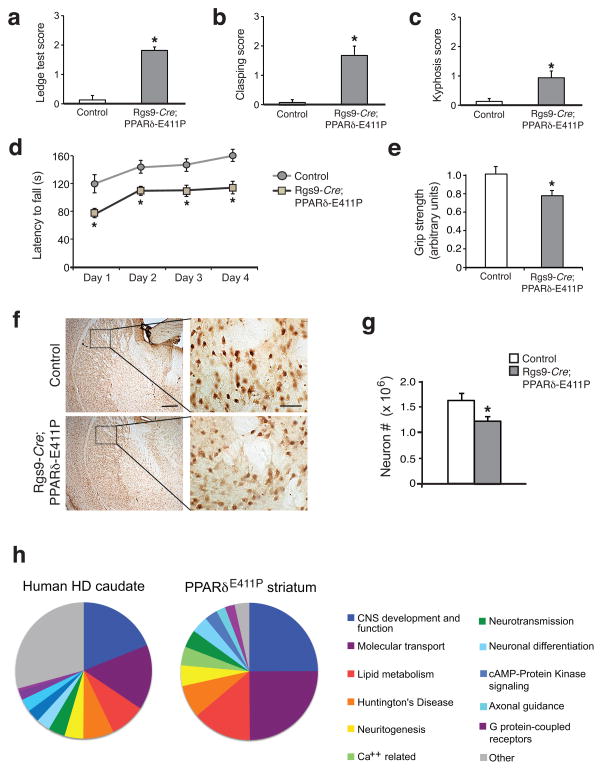
To assess the impact of PPARδ transcription interference in just the striatum, we obtained the Rgs9-Cre line, which restricts expression of Cre recombinase to striatal MSNs32, to derive bigenic Rgs9-Cre;PPARδ-E411P mice. Although Rgs9-Cre;PPARδ-E411P mice are of similar size and weight as gender-matched littermate controls, Rgs9-Cre;PPARδ-E411P mice develop motor abnormalities, as assessed by composite neurological examination29, rotarod analysis, and grip strength testing (Fig. 5a–e). Neuropathology examination of Rgs9-Cre;PPARδ-E411P mice revealed evidence for decreased striatal neuron number (Fig. 5f,g), due to a reduction in parvalbumin immunoreactive neurons, as acetylcholinesterase-positive neurons were spared (Supplementary Fig. 10). This pattern is consistent with striatal neuron loss observed in HD patients33,34. As CAGGS-floxed STOP-PPARδ-E411P transgenic crosses with the Nestin-Cre and Rgs9-Cre driver lines yielded distinct patterns of neurodegeneration that reflect Cre recombinase expression, observed phenotypes cannot be attributed to insertional mutagenesis.
Figure 5. Dominant-negative PPARδ expression in the striatum recapitulates HD-like motor dysfunction and transcriptional pathology.
(a) Rgs9-Cre;PPARδ-E411P mice cannot easily dismount from the cage ledge. *P < .05; t-test.
(b) Rgs9-Cre;PPARδ-E411P mice exhibit a prominent clasping phenotype. *P < .05; t-test.
(c) Rgs9-Cre;PPARδ-E411P mice display kyphosis. *P < .05; t-test.
(d) Rgs9-Cre;PPARδ-E411P mice exhibit impaired motor coordination on the accelerating rotarod. *P < .05; ANOVA with post-hoc Tukey test.
(e) Mesh grip strength analysis. Grip strength is given in arbitrary units with littermate control performance set to 1. *P < .05; t-test.
For (a) – (e), cohort age was 9 months, and group sizes were 7.
(f) Sections of striatum from 10 month-old Rgs9-Cre;PPARδ-E411P mice and littermate controls were immunostained for NeuN. Representative insets indicate reduced striatal neuron number in Rgs9-Cre;PPARδ-E411P mice. Scale bar (low power) = 200 μm; Scale bar (inset) = 20 μm.
(g) Quantification of neuron number from (f). *P < .05; t-test. n = 4 mice / group.
(h) RNA-Seq analysis on striatum from 10 month-old Nestin-Cre;PPARδ-E411P mice and littermate control mice (n = 3 / group). Pie charts indicate the percentage of genes for the highest ranked altered ‘Biofunctions’ pathways in microarray expression data from human HD caudate35 (left) and RNA-Seq analysis of PPARδ-E411P mouse striatum (right). Error bars = s.e.m.
Another defining feature of HD pathogenesis is transcriptional abnormalities16. Using RNA-Seq analysis to assess the striatal transcriptomes of Nestin-Cre;PPARδ-E411P mice, we found that 583 genes in Nestin-Cre;PPARδ-E411P striatum were altered in their expression in comparison to littermate controls. We performed Gene Ontology analysis on this 583-gene set, identified altered Biofunction pathways, and represented these Biofunction pathways in proportion to the percentage of total altered genes for each significantly perturbed pathway (Fig. 5h). To determine if the transcriptome changes found in the striatum of Nestin-Cre;PPARδ-E411P mice overlap with changes observed in HD brains, we performed Gene Ontology analysis on genes altered in their expression in the caudate of individuals with HD35, and represented the detected Biofunctions pathways in proportion to the percentage of total altered genes for each pathway (Fig. 5h). Ordering of altered Biofunctions pathways, for the human HD caudate transcriptome and Nestin-Cre;PPARδ-E411P striatum transcriptome (Supplementary Table 1), revealed nearly identical Biofunctions pathway rankings (P < 10−11). To validate gene expression alterations identified by RNA-Seq analysis, we performed qRT-PCR analysis on striatal RNAs from Nestin-Cre;PPARδ-E411P mice for 11 genes, and documented altered expression for 10 of the 11 tested genes, all in the predicted direction (Supplementary Fig. 11a). Finally, to assess the specificity of polyQ-htt repression of PPARδ, we performed 3x-PPRE reporter assays in HEK293 cells transfected with ataxin-7-10Q or ataxin-7-92Q, and found that polyQ-ataxin-7 did not repress PPARδ transactivation (Supplementary Fig. 11b).
The PPARδ agonist KD3010 rescues neurological phenotypes and neurodegeneration
PPARδ-selective compounds have been identified as potential therapies for type II diabetes and metabolic syndrome36. KD3010 ((S)-4-[cis-2,6-dimethyl-4-(4-trifluoromethoxy-phenyl)-piperazine-1-sulfonyl]-indan-2-carboxylic acid tosylate), is a highly selective and potent PPARδ agonist37. KD3010 in the low nanomolar range rescued PPARδ transcriptional repression and mitochondrial dysfunction in BAC-HD neurons, and could prevent cell death in primary cortical neurons transfected with a 586 amino acid N-terminal htt fragment containing 82 glutamines (Supplementary Fig. 12a–c). Intraperitoneal (i.p.) injection of 15 mg/kg of KD3010 into C57BL/6J mice resulted in ~10% brain/plasma ratios at 2 – 24 h post-injection (Supplementary Table 2). To test if KD3010 could promote PPARδ transactivation in brains of living mice, we injected 50 mg/kg /day of KD3010 and found that this treatment induced the expression of PPARδ target genes (Supplementary Fig. 12d). KD3010 did not cause weight loss or visible side effects, nor did we find any evidence of organ toxicity. Hence, KD3010 at 50 mg/kg /day is tolerated in mice and can promote PPARδ transactivation function in the mammalian CNS.
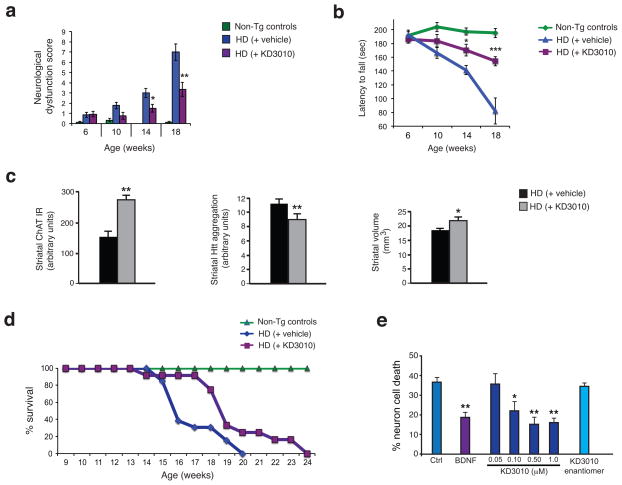
The HD N171-82Q mouse model recapitulates HD-like motor phenotypes and neurodegeneration within a time frame of 5 – 6 months38. Beginning at 6 weeks of age, HD mice were injected with either 50 mg/kg /day of KD3010 or vehicle, 5 times per week. Importantly, we adhered to recommended preclinical trial guidelines, intended to avoid spurious results39,40. We tracked the progression of disease phenotypes in vehicle-treated and agonist-treated HD mice by performing composite neurological examination29, rotarod analysis, and grip strength testing at 4 week intervals. KD3010 treatment attenuated neurological dysfunction and improved motor function in HD mice, as compared to vehicle-treated controls (Fig. 6a–b, Supplementary Fig. 12e–g). Striatal choline acetyltransferase (ChAT) levels, a measure of striatal interneuron dysfunction in HD33, were increased, aggregated htt protein was reduced, and striatal volume was maintained in KD3010-treated HD mice (Fig. 6c, Supplementary Fig. 13). In addition to rescuing neurological phenotypes and neurodegeneration, KD3010 yielded a 16% extension in mean lifespan in HD mice (Fig. 6d). Finally, we differentiated human HD induced pluripotent stem cells into striatal medium spiny-like neurons, and treated these human HD medium spiny-like neurons with KD3010. While the inactive enantiomer of KD3010 did not offer any protection from cell death, KD3010 rescued cell death in neurons derived from HD individuals (Fig. 6e).
Figure 6. Treatment with the PPARδ agonist KD3010 improves motor function, neurodegeneration, and survival in HD mice, and rescues neurotoxicity in human HD neurons.
Composite neurological examination (a) and accelerating rotarod analysis (b) of HD mice, receiving vehicle or KD3010, and non-transgenic (Non-Tg) littermate controls, at indicated ages. ***P < .001, **P < .01, *P < .05; ANOVA with post-hoc Tukey test. Cohort sizes were as follows: 9 Non-Tg controls, 13 HD mice on vehicle, and 12 HD mice on KD3010.
(c) Evaluation of neuropathology in striatum. Quantification of striatal choline acetyltransferase (ChAT) immunoreactivity (left), striatal htt aggregation (EM48) (center), and striatal volume by stereology (right) for 18 week-old HD mice receiving vehicle or KD3010 (n = 4 – 5 mice/group). **P < .01, *P < .05; t-test.
(d) Kaplan-Meier plot reveals extended lifespan in HD mice receiving KD3010. *P < .05, Log-rank test.
(e) Quantification of neuron cell death for medium spiny-like neurons differentiated from iPSC lines derived from a HD individual with a 60Q allele, and treated with KD3010 at the indicated concentrations, BDNF at 20 ng/ml (positive control), or the inactive enantiomer of KD3010 at 1 μM (negative control). Untreated neurons served as another negative control (Ctrl). **P < .01, *P < .05; ANOVA with post-hoc Bonferroni test. Error bars = s.e.m.
Discussion
Here we document PPARδ transcription interference as a cause of the bioenergetics defects and mitochondrial abnormalities in HD. In addition to demonstrating that htt represses PPARδ transactivation in an interaction-dependent fashion to yield mitochondrial defects and neurotoxicity in a variety of model systems, we generated transgenic mice expressing dominant-negative PPARδ to recapitulate HD-like behavioral, metabolic, and transcriptional phenotypes. As interfering with PPARδ function in mice can phenocopy HD, our findings implicate PPARδ dysregulation as a key node in the HD pathogenic cascade.
Of the three PPARs, the least-studied member of this family is PPARδ. We found that treatment with a PPARδ-selective agonist evoked a robust response in transactivation assays, and demonstrated that PPARδ could elicit neuroprotection in HD primary neurons. To clarify the role of PPARδ vis-à-vis PPARα and PPARγ, we modulated each PPAR individually by shRNA knock-down or agonist treatment, and found that only PPARδ modulation yielded highly significant effects on HD neurotoxicity. We documented that PPARδ is much more highly expressed than PPARα or PPARγ in neurons of the cortex and striatum, in agreement with previous work41. While studies have shown that PPARγ agonist treatment can ameliorate disease phenotypes in HD22,42,43, the beneficial effects of PPARγ agonist therapy likely stem from improved function in peripheral tissues, in hypothalamic neurons, and in non-neural CNS cells41. As our studies suggested an important role for PPARδ in neural function, we generated lines of conditional transgenic PPARδ mice, and directed expression of dominant-negative PPARδ to neurons throughout the CNS or just to striatal MSNs. These experiments revealed that PPARδ is necessary for normal neural function, as profound neurological and neurodegenerative phenotypes emerged in mice expressing dominant-negative PPARδ in neural lineages. Our findings thus identify neurons as a cell type where PPARδ is essential for homeostasis.
If impaired PPARδ function is contributing to HD pathogenesis, we theorized that an attractive treatment option would be to agonize PPARδ. Of various possible PPARδ agonists, we opted for KD3010, as this PPARδ agonist is potent and specific, crosses the blood-brain barrier, and was approved for use in humans in a Phase 1b metabolic disease safety trial, where no incidences of side effects were reported. We performed a preclinical trial of KD3010 in HD N171-82Q transgenic mice, and observed compelling evidence for therapeutic efficacy. To further evaluate KD3010 as a treatment for HD, we tested KD3010 in human HD striatal medium spiny-like neurons derived from induced pluripotent stem cells, and documented robust neuroprotection in the nanomolar range. Our findings strongly suggest that repurposing PPARδ agonists capable of crossing the blood brain barrier is a viable treatment strategy for HD, and indicate that KD3010 should be pursued as a lead. As mitochondrial dysfunction and transcription interference with PGC-1α, the co-activator for PPARδ are recognized features of Parkinson’s disease (PD) pathogenesis44,45, and we observed marked loss of TH+ neurons in the substantia nigra of dominant-negative PPARδ conditional transgenic mice, PPARδ agonist therapy should also be considered as a treatment for PD.
Another facet of PPARδ biology with relevance to therapy development for neurodegenerative disease is that PPARδ forms a heterodimer with RXR, and RXR agonists are capable of promoting PPARδ transactivation46. Bexarotene is a synthetic drug, approved for use in humans, which is structurally similar to retinoic acid compounds that are known endogenous RXR ligands. One provocative study reported that administration of bexarotene to Alzheimer’s disease (AD) mice yielded dramatic improvements in cognitive, social, and olfactory deficits, accompanied by improved neural circuit function and enhanced clearance of soluble Aβ oligomers47. As PPARδ is highly expressed in neurons of the CNS8, and bexarotene would potently activate PPARδ, our results indicate that enhanced PPARδ activation could be contributing to bexarotene neuroprotection in AD47,48.
As the PPARδ – PGC-1α pathway promotes the expression of genes that drive high-level energy production and insure protein and organelle quality control, the neuroprotective effects of agonizing this pathway likely stem from boosting neuronal bioenergetics and proteostasis. HD, PD, and AD are all late-onset disorders, implying that a decline in the function of crucial homeostatic pathways with age is a central factor in the development of clinical disease. In HD, this view is all the more relevant, as affected patients express the mutant disease protein from the time of conception, yet can go more than five or even six decades without obvious symptoms. It seems reasonable to propose that activation of pathways promoting energy production (which is always under high demand in neurons and CNS cells) and supporting protein – organelle quality control (which constitutes a major challenge for neurons and other CNS cell types) might stave off cellular decompensation in the face of proteotoxic stress during this long prodromal phase. Thus, therapies directed at boosting PPARδ function could have broad applicability for a wide range of neurodegenerative diseases.
Methods
Real-time RT-PCR analysis
RNA samples were isolated using Trizol (Life Technologies). Genomic DNA was removed using RNAse free DNAse (Ambion). mRNA quantification was performed using the 7500 Real Time PCR System (ABI) with ABI Assays-on-Demand primers and TaqMan® based probes. ABI TaqMan primer and probe set designations are available upon request. 18S RNA was used as internal control. Relative expression levels were calculated via the ΔΔCt method.
Cell culture and primary neuron studies
ST-Hdh cells were cultured as previously described20. Primary cortical neurons were prepared as previously described50. HEK293T cells were cultured as previously described19,20. HEK293T cells tested negative for mycoplasma contamination, and were used for the purposes of preliminary testing prior to confirmation of results in multiple more physiologically relevant model systems, due to their robust protein expression levels of transfected proteins. Co-transfection with the indicated constructs (previously described19) was done with Lipofectamine 2000, as per manufacturer’s protocol (Invitrogen). Addgene provided constructs for PPARα (22751) and PPARγ (8895). PPARδ-Flag (Origene, MR207001) truncation constructs [ΔA–C (Δ1-141), ΔA–D (Δ1-214), ΔD–F (Δ143-440), and ΔE–F (Δ215-440)] were made by standard techniques and verified by sequencing. Lentiviral transduction was used to induce gene expression or knockdown in primary neurons, with infection achieved by adding 1 x 107 titer units of lentivirus to the culture media. For co-transfection assays, cells were drug-treated 24 h after reporter transfection, harvested 24 h later, and subjected to analysis using the Dual-Luciferase® Reporter Assay system (Promega). Mitochondrial membrane potential in primary cortical neurons was measured via live cell loading with a potential-sensitive dye, JC-1, using the Tecan M200Pro Reader. Analysis of cell death with immunofluorescence to activated caspase-3 and evaluation of MAP2 immunoreactivity to gauge neuron health was performed as described51. In all experiments, the investigator was blinded to culture conditions and cell treatments. Compound concentrations: fenofibrate (Sigma, 100 nM), GW501516 (Santa Cruz, 100 nM), pioglitazone (Sigma, 20 nM), and H2O2 (Sigma, 25 μM).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described20. Briefly, St-Hdh cells were cross-linked with 1.0% formaldehyde. After sonication, lysates were incubated with PPARδ antibody (sc-7197, Santa Cruz Biotechnology, 1:100) or control rabbit IgG (Santa Crux Biotechnology, 1:100). Quantitative PCR analysis was performed with the SYBR-Green PCR master mix (ABI) on a 7500 Real Time PCR System (ABI).
Mouse studies
All animal experimentation adhered to NIH guidelines and was approved by and performed in accordance with the University of Washington Institutional Animal Care and Use Committee and the UCSD Institutional Animal Care and Use Committee. The pCAGGS-loxP-STOP-loxP murine wild-type PPARδ transgenic construct52 was altered with the E411P mutation25, and then prepared for pronuclear injection. Resulting transgenic mice were crossed with Cre hemizygous mice to derive floxed-STOP PPARδ E411P – Cre driver bigenic mice. All HD transgenic, PPARδ transgenic, and Cre driver lines were backcrossed onto the C57BL/6J strain background for >9 generations, before genetic crosses were performed for purposes of experimentation. Blinded observers visually inspected mice for obvious neurological signs and examined mice with a composite neurological evaluation tool [Ledge test, clasping, kyphosis, and gait were scored on a scale of 0 (normal) to 3 (severely impaired)], as described previously53. Blinded investigators examined motor phenotypes, by performing rotarod testing, as well as grip strength and stride length analysis, as previously described20. Cohort sizes were designated based upon power analysis for threshold effects of at least 25% difference. Novel object recognition and Y maze spatial memory testing were performed by blinded investigators as previously described54.
For neuropathology experiments, brains were harvested as described20. Histopathology, volume measurements, and stereology analysis were performed, as previously described20. For PPAR expression analysis, brain sections were incubated with: NeuN (MAB377, Millipore; 1:100), and either PPARδ (PA1-823A, Pierce; 1:200), PPARα (PA1-822A, Pierce; 1:200), or PPARγ (PA3-821A, Pierce; 1:200), and imaged with a Zeiss LSM 780 inverted microscope. Validation of PPARγ immunostaining of brain sections was achieved by detection of immunoreactivity in neurons of the hypothalamus, in agreement with previous work41. For electron microscopy analysis, brains were fixed, embedded, and sectioned, and imaged with a Zeiss OM 10 electron microscope. Images were analyzed with the NIH Image J program. In all cases, the scorer was blinded to the genotype status of the mice.
KD3010 pharmacology
KD3010 was injected intraperitoneally (15 mg/kg) as a suspension in corn oil (3 mg/ml) to fasted male C57BL/6J mice. Three cohorts of 3 mice were dosed, and plasma and brains were taken at 2, 8, and 24 h, and analyzed by LC-MS/MS (AB Sciex API4000). For plasma samples, an aliquot of 10 μl sample was protein precipitated with 100 μl IS, the mixture was vortex-mixed for 1 min, and centrifuged at 13000 rpm for 15 min, 20–25 °C. 80 μl supernatant was then mixed with 160 μl water/ACN (v:v, 95:5) with 0.1% FA, vortex-mixed 10 min and centrifuged for 10 min at 4000 rpm, 4 °C. 10 μl sample was injected for LC-MS/MS analysis. Brain homogenates were prepared by homogenizing brain tissue for 2 min with 4 volumes (w:v) of homogenizing solution (deionized water). An aliquot of 60 μl sample was protein precipitated with 600 μl IS, the mixture was vortex-mixed for 1 min and centrifuged at 13000 rpm for 15 min, 20–25 °C. 80 μl supernatant was then mixed with 160 μl water/ACN (v:v, 95:5) with 0.1% FA, vortex-mixed 10 min and centrifuged for 15 min at 4000 rpm, 4 °C. 10 μl sample was injected for LC-MS/MS analysis. A standard curve of KD3010 was determined with a LLOQ of 1 ng/ml for plasma and 5 ng/g in brain homogenate.
Treatment of mice with KD3010
After genotyping mice, we performed motor baseline assessment prior to group assignments, and littermates were randomly assigned to experimental groups while balancing genders, in accordance with guidelines intended to avoid spurious results39,40. Exclusion criteria determined prior to the start of the trial were an adverse response to the injections, or death prior to start of treatment. These conditions did not occur. After group assignment, 6 week-old mice were injected initiated daily, five times a week (from Monday to Friday) intraperitoneally with 50mg/kg/ day of KD3010 suspended in corn oil (3 mg/ml). Blinded observers visually inspected mice for obvious neurological signs and examined mice with a composite neurological evaluation tool, as described previously29. Blinded investigators examined motor phenotypes, by performing rotarod testing, as previously described20, as well as grip strength and stride length analysis55. Cohort sizes were designated based upon power analysis for threshold effects of at least 25% difference. For neuropathology experiments, brains were harvested as previously described56,57. Histopathology, volume measurements, and stereology analysis were performed, as previously20. In all cases, the scorer was blinded to the genotype status of the mice.
Bioenergetics assays
Mitochondrial complex activity assays were performed as described previously and adapted to a 96-well format58. For quantification of ATP, HPLC of adenine nucleotides isolated from regions of the brain was performed, as done previously20. In all cases, the investigator was blinded to the genotype status of the mice.
Transcriptome analysis
RNA-Sequencing (RNA-Seq) was performed by isolating RNA from the striatum of individual mice. The Illumina TruSeq™ RNA Sample Preparation Kit was used to prepare libraries for sequencing on the Illumina HiSeq 2000 system. RNA-Seq data was mapped with the alignment software GSNAP to the mouse reference genome, obtained from NCBI (mm9, Build 37 assembly). The expression of genes was quantified by assigning reads to gene locations, determined by the Ensembl gene annotations for mouse, and subsequently normalizing to annotated gene length (exons only) and depth of sequencing. Differentially expressed genes were called using the local z-score method on pair-wise sample comparisons, and were ranked by consistency of differential expression between similar comparisons of conditions. Order of magnitude threshold of 20%, a P value cut-off of 0.01, and False Discovery Rate of 0.005 were used to identify significantly altered genes upon comparison of transgenic samples to non-transgenic controls. A representative subset of identified gene expression alterations was validated by qRT-PCR, as above. For gene ontology (GO) classification, differentially expressed genes in E411P mice and human HD35 were associated with the GO categories of cellular component and biological process. The GO enriched biofunctional categories were compared using Ingenuity Pathway Analysis, and only GO categories with at least 10 altered genes were reported, and then ranked based upon the percentage of altered genes in a particular GO category.
Protein biochemistry analysis
The unbiased screen for htt protein interactors was performed using a Panomics transcription factor (TF) immunoprecipitation array, according to the manufacturer’s instructions: http://www.veritastk.co.jp/attached/2209/TFInteractionArrayKitcombined.pdf. Htt IP for the Panomics TF IP array utilized anti-htt antibody EM48. Protein lysates were prepared as previously described55. For co-immunoprecipitations, homogenized protein was incubated with indicated co-immunoprecipitation antibody (GFP: 3E6 A11120, Life Technologies (1:100); Htt: MCA2050, Abcam (1:100); PPARδ: SC-7197, Santa Cruz; (1:100)). Proteins were run on 10% Bis-Tris gels (Invitrogen) and transferred to PVDF membranes (Millipore) prior to blocking in Odyssey Blocking Buffer (LI-COR biosciences). Membranes were incubated with antibodies as indicated: Flag (F7425, Sigma; 1:1000), Htt (MAB2166, Millipore; 1:1000), GFP (A11121, Life Technologies, 1:1000), PPARδ (PA1-823A, Pierce; 1:1000), PPARα (PA1-822A, Pierce; 1:1000), or PPARγ (PA3-821A, Pierce; 1:1000), or β-actin (ab8226, Abcam; 1:10,000), and imaged on the Odyssey instrument (Licor). To obtain proteins in vitro for the direct interaction co-immunoprecipitation, the Promega TnT T7 Quick Coupled Transcription/ Translation System (Promega, #L1170) was used prior to co-immunoprecipitation.
Stem cell modeling
60i4 HD induced pluripotent stem cells (iPSCs) were differentiated into medium spiny neurons for 56 days using a modified protocol59, as described previously60. On day 42 of differentiation, cell aggregates were plated into 24-well plates coated with Matrigel (BD). Two weeks later, the cells were transferred to Neural Induction Medium (NIM). KD3010 or its enantiomer was added at the time of the medium change to NIM. After 48 h, cells were fixed with 4% paraformaldehyde in PBS for 30 min. After 3 washes with PBS, cells were stained with 0.8 μg/ml of Hoechst 33342 (Sigma) to visualize cell nuclei. A nuclear condensation assay to measure the extent of cell death was performed on 3 different clonal lines, as previously described60.
Statistical analysis
Statistical analysis was done using Microsoft Excel, Prism 4.0 (Graph Pad), or the VassarStats website <http://faculty.vassar.edu/lowry/VassarStats.html>. For ANOVA (one-way), if statistical significance (P < 0.05) was achieved, we performed post-hoc analysis corresponding to the experiment, as specified in the figure legend, to account for multiple comparisons. Hypergeometric distribution analysis was performed using the Stat Trek calculator <http://stattrek.com/online-calculator/hypergeometric.aspx>. All t-tests were two-tailed unless otherwise indicated, and the level of significance (alpha) was always set at 0.05.
Supplementary Material
Acknowledgments
We are grateful to S. Luquet for the gift of the CAGGS-floxed STOP PPARδ expression construct. ST-Hdh cells were a kind gift from M. MacDonald20,49. HD N171-82Q mice38 were originally obtained from C.A. Ross, and BAC-HD mice21 were originally obtained from X.W. Yang. This work was supported by funding from the Hereditary Disease Foundation, the Cure Huntington’s Disease Initiative, and grants from the National Institute of Health: R01 NS065874 (A.R.L.), R01 AG033082 (A.R.L.), NRSA F32 NS081964 (A.S.D.), and P01 HL110873 (E.R.L.).
Footnotes
Accession codes
NCBI GEO accession code: GSE74583
Author Contributions
A.R.L. provided the conceptual framework for the study. A.S.D., V.V.P., T.T., B.L.S., E.R.L., E.Y., C.A.R., G.M., A.P., E.M., and A.R.L. designed the experiments, which were performed by A.S.D., V.V.P., P.P.L., H.C.M., S.K.G.-H., N.L., K.R.S., A.B., M.-J.M.T., A.L.F., M.A., N.A., S.A., T.G., B.L.S., E.R.L., E.Y., E.M., G.M., A.P., and A.R.L. A.S.D. and A.R.L. wrote the manuscript.
References
- 1.Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 2.Auboeuf D, et al. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: no alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46:1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 3.Kliewer SA, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luquet S, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 5.Schuler M, et al. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang YX, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narkar VA, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girroir EE, et al. Quantitative expression patterns of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) protein in mice. Biochem Biophys Res Commun. 2008;371:456–461. doi: 10.1016/j.bbrc.2008.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nance MA. Genetic testing of children at risk for Huntington’s disease. US Huntington Disease Genetic Testing Group. Neurology. 1997;49:1048–1053. doi: 10.1212/wnl.49.4.1048. [DOI] [PubMed] [Google Scholar]
- 10.Ross CA, et al. Huntington’s disease and dentatorubral-pallidoluysian atrophy: proteins, pathogenesis and pathology. Brain Pathol. 1997;7:1003–1016. doi: 10.1111/j.1750-3639.1997.tb00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.HD Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 12.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beal MF, et al. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 15.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 16.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Tsunemi T, et al. PGC-1alpha Rescues Huntington’s Disease Proteotoxicity by Preventing Oxidative Stress and Promoting TFEB Function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang MC, et al. Modulation of energy deficiency in Huntington’s disease via activation of the peroxisome proliferator-activated receptor gamma. Hum Mol Genet. 2010;19:4043–4058. doi: 10.1093/hmg/ddq322. [DOI] [PubMed] [Google Scholar]
- 23.Jin YN, Hwang WY, Jo C, Johnson GV. Metabolic state determines sensitivity to cellular stress in Huntington disease: normalization by activation of PPARgamma. PLoS One. 2012;7:e30406. doi: 10.1371/journal.pone.0030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastie C, Luquet S, Holst D, Jehl-Pietri C, Grimaldi PA. Alterations of peroxisome proliferator-activated receptor delta activity affect fatty acid-controlled adipose differentiation. J Biol Chem. 2000;275:38768–38773. doi: 10.1074/jbc.M006450200. [DOI] [PubMed] [Google Scholar]
- 26.Holst D, Luquet S, Kristiansen K, Grimaldi PA. Roles of peroxisome proliferator-activated receptors delta and gamma in myoblast transdifferentiation. Exp Cell Res. 2003;288:168–176. doi: 10.1016/s0014-4827(03)00179-4. [DOI] [PubMed] [Google Scholar]
- 27.Peters JM, et al. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 29.Guyenet SJ, et al. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp. 2010 doi: 10.3791/1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirendeb UP, et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang MT, et al. Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc Natl Acad Sci U S A. 2006;103:15254–15259. doi: 10.1073/pnas.0601758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrante RJ, Beal MF, Kowall NW, Richardson EP, Jr, Martin JB. Sparing of acetylcholinesterase-containing striatal neurons in Huntington’s disease. Brain Res. 1987;411:162–166. doi: 10.1016/0006-8993(87)90694-9. [DOI] [PubMed] [Google Scholar]
- 34.Reiner A, et al. Striatal parvalbuminergic neurons are lost in Huntington’s disease: implications for dystonia. Mov Disord. 2013;28:1691–1699. doi: 10.1002/mds.25624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 36.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 37.Iwaisako K, et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor delta agonist. Proc Natl Acad Sci U S A. 2012;109:E1369–1376. doi: 10.1073/pnas.1202464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schilling G, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 39.Landis SC, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490:187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrin S. Preclinical research: Make mouse studies work. Nature. 2014;507:423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 41.Lu M, et al. Brain PPAR-gamma promotes obesity and is required for the insulin-sensitizing effect of thiazolidinediones. Nat Med. 2011;17:618–622. doi: 10.1038/nm.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang MC, Chern Y, Huang RN. PPARgamma rescue of the mitochondrial dysfunction in Huntington’s disease. Neurobiol Dis. 2012;45:322–328. doi: 10.1016/j.nbd.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Jin J, et al. Neuroprotective effects of PPAR-gamma agonist rosiglitazone in N171–82Q mouse model of Huntington’s disease. J Neurochem. 2013;125:410–419. doi: 10.1111/jnc.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng B, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin JH, et al. PARIS (ZNF746) Repression of PGC-1alpha Contributes to Neurodegeneration in Parkinson’s Disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cramer PE, et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitz NF, Cronican AA, Lefterov I, Koldamova R. Comment on “ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models”. Science. 2013;340:924-c. doi: 10.1126/science.1235809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 50.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. The Journal of biological chemistry. 2009;284:2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young JE, et al. Polyglutamine-expanded androgen receptor truncation fragments activate a Bax-dependent apoptotic cascade mediated by DP5/Hrk. J Neurosci. 2009;29:1987–1997. doi: 10.1523/JNEUROSCI.4072-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luquet S, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 53.Guyenet SJ, et al. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp. 2010 doi: 10.3791/1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nithianantharajah J, Barkus C, Murphy M, Hannan AJ. Gene-environment interactions modulating cognitive function and molecular correlates of synaptic plasticity in Huntington’s disease transgenic mice. Neurobiol Dis. 2008;29:490–504. doi: 10.1016/j.nbd.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Sopher BL, et al. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41:687–699. doi: 10.1016/s0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 56.Garden GA, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.La Spada AR, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 58.Janssen AJ, et al. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem. 2007;53:729–734. doi: 10.1373/clinchem.2006.078873. [DOI] [PubMed] [Google Scholar]
- 59.Aubry L, et al. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.