Abstract
The amino acid glutamate is a major metabolic hub in many organisms and as such is involved in diverse processes in addition to its role in protein synthesis. Nitrogen assimilation, nucleoside, amino acid, and cofactor biosynthesis, as well as secondary natural product formation all utilize glutamate in some manner. Glutamate also plays a role in the catabolism of certain amines. Understanding glutamate's role in these various processes can aid in genome mining for novel metabolic pathways or the engineering of pathways for bioremediation or chemical production of valuable compounds.
Introduction
Organisms ranging from Escherichia coli to humans contain large intracellular pools of the amino acid glutamate [15,6,83]. In E. coli, glutamate is encoded only by roughly 6% of the codons in the genome [53], whereas the concentration of glutamate in its cytosol (96 mM) is over two orders of magnitude higher than that of the most commonly encoded amino acid, leucine [6]. Given the levels of glutamate present, it is perhaps not surprising that this metabolite plays a role in chemical transformations far beyond protein synthesis.
Amino acid biosynthesis and nitrogen metabolism
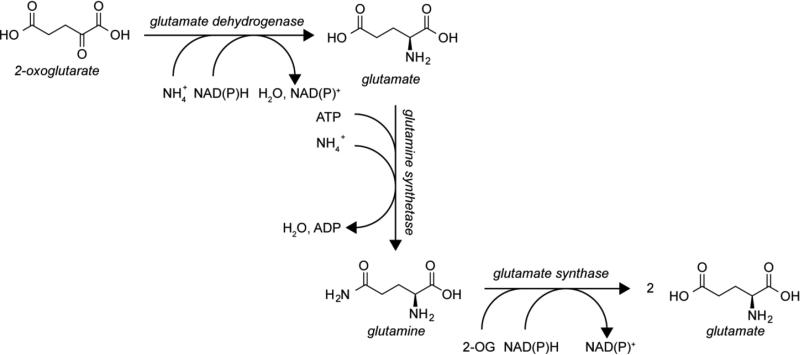
Glutamate is commonly produced through two pathways, both of which result in the overall conversion of 2-oxoglutarate, a citric acid cycle intermediate, to glutamate. One route is the reductive amination of 2-oxoglutarate with ammonium as the nitrogen donor via glutamate dehydrogenase (GDH) (Fig. 1) [4]. GDH belongs to a family of amino acid dehydrogenases, other members of which act on leucine, phenylalanine, or valine and physiologically tend to operate in the deamination direction [8,81]. The second route to glutamate is by way of the glutamate synthase (GltS)-catalyzed reductive amination of 2-oxoglutarate using glutamine as the nitrogen donor (Fig. 1) [4]. GltS is a flavin dependent iron sulfur cluster protein [86]. It is worth noting that the reverse of these reactions, producing 2-oxoglutarate, allows the carbon from glutamate to enter the citric acid cycle and many organisms are capable of using glutamate as their sole carbon source, possibly even preferring it over glucose [84].
Fig. 1.
Glutamate biosynthetic pathways. Glutamate is made from the citric acid cycle intermediate 2-oxoglutarate (2-OG) by reductive amination with either ammonium or glutamine as the nitrogen source
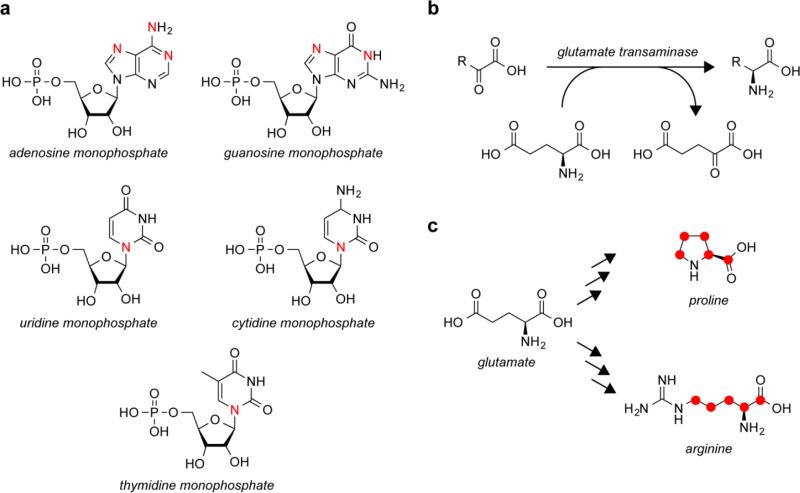
Glutamate biosynthesis is not only important for production of glutamate; it is also the major route for the assimilation of nitrogen [50]. Glutamine is produced by the action of the glutamine synthetase (GS), which catalyzes the ATP-dependent amidation of glutamate using ammonium as the nitrogen source [4]. GS is one of the most ancient functioning enzymes [39], and homologous enzymes are involved in other pathways of glutamate metabolism, as described in this review. While glutamine serves as the nitrogen donor for about half of the nitrogens in purines and pyrimidines, glutamate serves as the nitrogen donor for the remaining nitrogens in purines and pyrimidines (Fig. 2a) and the amino groups of all the amino acids, making the GS/GltS glutamate biosynthesis pathway, also known as the GS-GOGAT pathway, a major route for nitrogen assimilation [4]. Transaminases catalyze the transfer of the amine from glutamate to the 2-oxo precursors of serine, aspartate, alanine, valine, leucine, isoleucine, phenylalanine, and tyrosine (Fig. 2b) [82]. Additionally, all of the other amino acids are synthesized from these amino acids and therefore indirectly obtain their amine nitrogen from glutamate. Furthermore, all of the carbons in proline and the majority of carbons in arginine are also derived from glutamate in addition to their amino group (Fig. 2c).
Fig. 2.
Glutamate is the major nitrogen donor in metabolism. a Glutamate is the nitrogen donor, through glycine and aspartate, for about half of the nitrogens, indicated in red, in nucleotides. b Glutamate donates nitrogen for other amino acids through the action of glutamate transaminases. c Carbons in proline and arginine that come from glutamate are indicated by red circles
When organisms support both the GDH and GS-GOGAT glutamate biosynthetic routes, the choice of pathway depends on the nutrient availability. As the GS-GOGAT pathway consumes an ATP per ammonium assimilated, it is used under energy-rich conditions, while the GDH functions under energy limited conditions [28]. In fact it has been estimated that 15% of the ATP produced by E. coli under energy rich growing conditions is devoted to ammonium assimilation through the GS-GOGAT pathway [64]. In addition, the affinity of the GS for ammonium is higher than that of the GDH [51], such that the GS-GOGAT pathway is employed under ammonium limiting conditions [28].
Nitrogen availability can also play an important role in secondary metabolism [7,85]. The biosynthesis of many secondary natural products is reduced or stopped altogether when the producing microbe is cultured in a medium with a readily assimilable nitrogen source such as ammonium [65]. When Streptomyces clavuligerus, a producer of the β-lactam antibiotic cephamycin and the β-lactamase inhibitor clavulanic acid, is grown with different amino acids as the sole nitrogen source, levels of production of these metabolites vary widely and not necessarily in the same manner. It is possible that the responses are related to guanosine tetra or pentaphosphate (ppGpp) signaling. When the available pool of amino acids drops to the point at which tRNA amino acylation is unable to support protein synthesis, the ppGpp synthetase RelA begins to produce ppGpp, which induces large changes in processes such as transcription and growth [62]. When a strain of Streptomyces coelicolor with a disrupted relA gene was grown under nitrogen limiting conditions both ppGpp and secondary metabolite production were greatly reduced compared to a strain with an intact relA gene [9]. Likewise, heterologous expression of RelA or a constitutively active truncated version of RelA in S. coelicolor led to increased production of ppGpp and secondary metabolites under nitrogen sufficient conditions [29].
Glutamate as a building block in biosynthesis
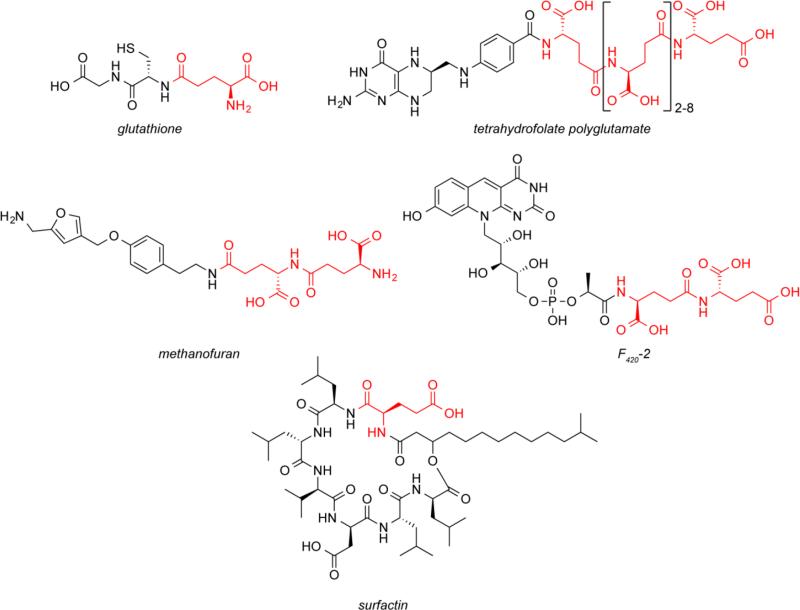
Nature has developed ways to use glutamate as a building block for the biosynthesis of molecules with complex structures beyond proteins. For example, glutathione is a low molecular weight thiol present in organisms ranging from bacteria to mammals [11]. Glutathione is a tripepide composed of cysteine, glycine and a γ-linked glutamate (Fig. 3). During the first step in glutathione biosynthesis γ-glutamylcysteine ligase, which is related to GS enzymes, catalyzes the ATP-dependent ligation of the γ-carboxylate of glutamate to the amine of cysteine. In the next step, glutathione synthetase ligates glycine to the cysteinyl carboxyl group to produce glutathione. Glutathione synthetase is a member of the ATP-grasp superfamily of proteins, which catalyze the ATP-dependent ligation of a carboxylic acid of one substrate to an amine, imine, alcohol or thiol of another molecule (or within the same molecule) [20] and are another class of enzymes that play a large role in the metabolism of glutamate. Glutathione is used to maintain redox homeostasis within the cell and to protect from oxidative damage [63,3]. It has been observed that the thiol group of glutathionine is oxidized less readily than that of cysteine, due to the protection of the free amine of cysteine by glutamylation [75,27].
Fig. 3.
Glutamate (shown in red) is used as a building block for the construction of more complex molecules
Several cofactors have been discovered that contain polyglutamate chains. Folate is an important cofactor in one-carbon metabolism of amino acids, nucleotides, and other metabolites [22]. In vivo folate exists as a pool of polyglutamylated derivatives with 2-8 γ-linked glutamate moieties (Fig. 3) [22]. The enzymes that produce these glutamylated derivatives are related to the amino acid ligases involved in the production of the bacterial cell wall precursor, UDP-N-acetylmuramoyl-pentapeptide [73]. These glutamate moieties are thought to be important for preventing the diffusion of folate out of cells and for enzyme recognition of the cofactor [67]. Other polyglutamylated cofactors include methanofuran, which is involved in the reduction of carbon dioxide to methane by methanogens [42]. Although different organisms produce different methanofurans [88], these cofactors share a common core structure that contains two γ-linked glutamate moieties. The enzyme from Methanocaldococcus fervens that installs one of these glutamates has been identified and is an ATP-grasp protein [87]. The redox cofactor F420 is also polyglutamylated and widely distributed in archaea [47]. During its biosynthesis, the intermediate F420-0 is glutamylated twice through γ-carboxylate linkages [45]. The enzyme that catalyzes the addition of these glutamates, CofE, phosphorylates F420-0 and F420-1 in a GTP-dependent manner to ligate the two glutamates [54]. Methanococcus jannaschii produces F420-3, which in addition contains an α-linked glutamate moiety [26].
Beyond primary metabolism, glutamate is also used as a building block by nonribosomal peptide synthetases (NRPS) [71]. The adenylation domains of these multimodular enzymes activate the α-carboxylate of their respective amino acids in an ATP-dependent manner, producing an amino acid adenylate. This activated amino acid is then transferred to the thiol of a phosphopantetheine prosthetic group of the peptidyl carrier protein domain of the NRPS forming a thioester. The condensation domain subsequently catalyzes the formation of an amide bond between the newly activated amino acid and the growing peptide chain residing on the peptidyl carrier protein domain of the previous module. Glutamate-specific adenylation domains can be identified through bioinformatic analysis of conserved residues in their active sites [10]. One example of a nonribosomal peptide containing a glutamate is the lipopeptide surfactin, produced by Bacillus subtilis (Fig. 3) [61].
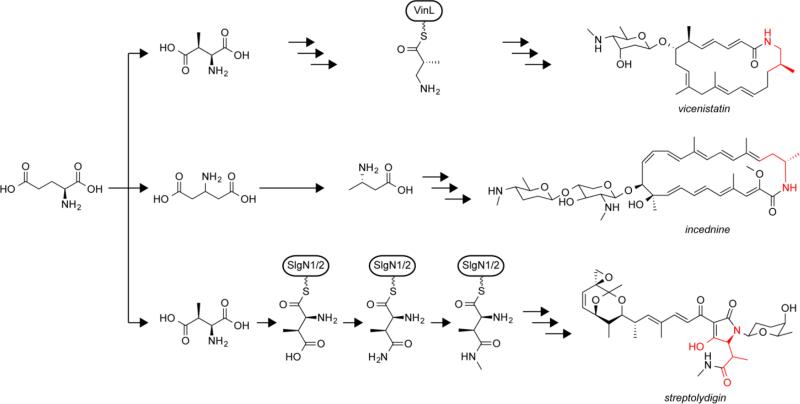
Glutamate can also play a role in the biosynthesis of polyketide natural products. The formation of macrolactam polyketides often commences with an amino acid starter unit [52]. Labeling studies suggested that the starter unit of the antibiotic macrolactam vicenistatin arises from a glutamate precursor [59]. When the biosynthetic cluster vicenistatin was identified it was found to contain genes encoding a glutamate mutase, which rearranges glutamate to 3-methylaspartate [55]. Decarboxylation of the 3-methylaspartate during the course of vicenistatin biosynthesis results in the 3-amino-2-methylpropionate derived moiety present in the final product (Fig. 4) [70]. The glutamate-derived starter unit of the cytotoxic macrolactam incednine (Fig. 4) is produced through a different route. The gene cluster was found to encode a glutamate 2,3-aminomutase and a decarboxylase [79]. The presence of these genes suggests glutamate is transformed to γ-glutamate by the aminomutase, which is then converted to 3-aminobutyrate by the decarboxylase, a route that was confirmed by labeling studies [78]. Genome mining for these glutamate-processing enzymes can be used to identify biosynthetic clusters for new macrolactams [72].
Fig. 4.
Glutamate derived building blocks (shown in red) can be used during biosynthesis of complex natural products
A more complex example of a glutamate-derived building block is found in the tetramic acid antibiotic streptolydigin, which contains a 5-membered lactam moiety that is partially derived from glutamate (Fig. 4). The biosynthetic cluster contains the genes for a glutamate mutase as well as a GltS [56]. These enzymes produce 3-methylaspartate, which is then loaded onto a peptidyl carrier protein of an NRPS module [30]. On the peptidyl carrier protein the 3-methylaspartate moiety is amidated to produce a 3-methylasparagine moiety, which is then N-methylated. This building block is subsequently incorporated during the final extension step in the pathway followed by cyclization to produce the lactam and release the product from the peptidyl carrier protein.
Glutamate as a protecting group and/or scaffold
Several pathways have been identified where glutamate is added to a compound, some transformations are performed on the glutamylated molecule, and then the glutamate is removed unchanged. In this context glutamate appears to be acting as a protecting group, preventing off-pathway reactions. Alternatively, instead of a protecting role, it is also possible that glutamate provides a scaffold for substrate recognition and that the enzymes acting on the glutamylated substrates were recruited from other pathways involving glutamate.
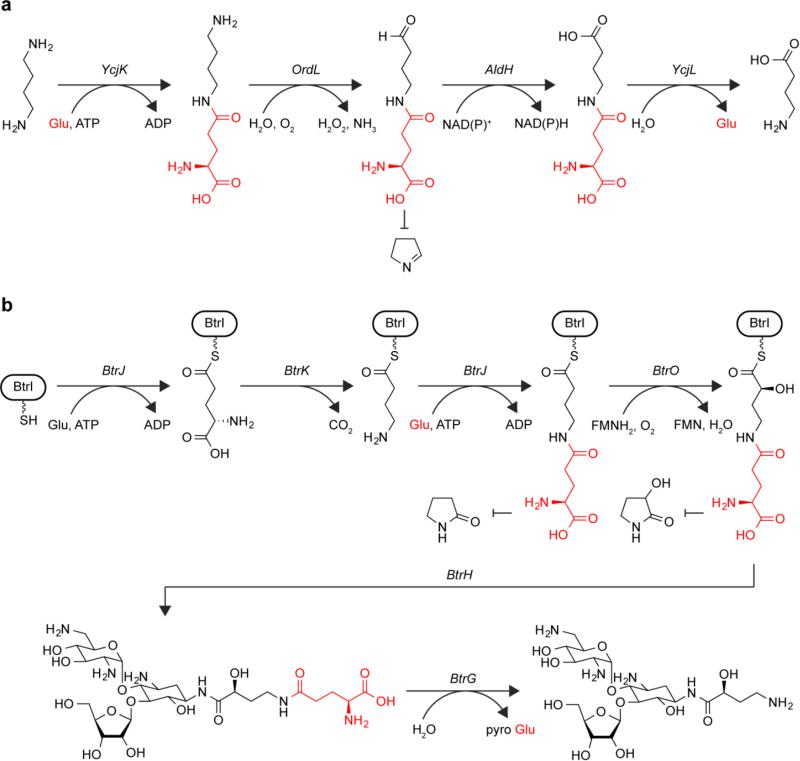
One type of off-pathway reaction that glutamylation can prevent is intramolecular cyclization. Putrescine is a diamine produced by a wide array of organisms that plays a role in a variety of biological processes [77]. Certain bacteria are capable of growing on putrescine as the sole nitrogen source [76]. A pathway for the catabolism of putrescine that proceeds through a γ-glutamylated species has been identified in E. coli and Pseudomonas aeruginosa wherein putrescine is oxidized to succinate, which can enter the citric acid cycle, releasing two equivalents of ammonium in the process [40,89]. The first enzyme in the E. coli pathway, PuuA, catalyzes the γ-glutamylation of putrescine in an ATP-dependent manner to produce γ-glutamylputrescine (Fig. 5a); it belongs to the GS family of enzymes [41]. In the next step γ-glutamylputrescine is oxidized to γ-glutamyl-γ-aminobutyraldehyde. This aldehyde is oxidized again to give γ-glutamyl-γ-aminobutyrate, at which point the glutamate is hydrolyzed by an enzyme belonging to the glutamine aminotransferase (GAT) superfamily to produce γ-aminobutyrate, which is then further catabolized. The glutamate in this pathway is installed before the production of the γ-aminobutyraldehyde species and removed as soon as that species is consumed, suggesting γ-aminobutyraldehyde is being protected. Indeed, E. coli also uses an alternate putrescine utilization pathway that proceeds through the same steps without glutamylation, and in that pathway γ-aminobutyraldehyde spontaneously cyclizes to form Δ1-pyrroline [68]. The relative roles of these two pathways in E. coli remain to be determined.
Fig. 5.
Glutamylation (shown in red) serves as a protecting group to prevent cyclization. a During putrescine catabolism, intermediates on the pathway are glutamylated to potentially prevent the spontaneous cyclization of the γ-aminobutyraldehyde to Δ1-pyrroline. b During butirosin biosynthesis, glutamylation may block cyclization of aminobutyryl thioester intermediates to 5-membered lactams
Another example of glutamylation as potential protection against undesired cyclization comes from the biosynthesis of butirosin (Fig. 5b). Butirosin is an aminoglycoside antibiotic produced by Bacillus circulans [16]. It contains an unusual (2S)-4-amino-2-hydroxybutyryl moiety that protects it from certain aminoglycoside modifying enzymes [37]. During the synthesis of this moiety, an ATP-grasp enzyme, BtrJ, activates the γ-carboxylate of glutamate by ATP-dependent phosphorylation and ligates it to the phosphopantetheine prosthetic group of an acyl carrier protein (ACP), BtrI [46]. In the next step, a PLP-dependent decarboxylase, BtrK, decarboxylates the γ-glutamyl-ACP to produce γ-aminobutyryl-ACP. Interestingly, when BrtK and BtrJ were co-incubated in an in vitro reaction, a second glutamate was appended to the γ-aminobutyryl-ACP, generating γ-glutamyl-γ-amidobutyryl-ACP. The next enzyme in the pathway, the flavin dependent monooxygenase BtrO, then hydroxylates the 2 position of the γ-glutamyl-γ-amidobutyryl-ACP. BrtO is not capable of carrying out this reaction on the γ-aminobutyryl-acyl carrier protein, suggesting that the second glutamylation is on pathway for the production of the (2S)-4-amino-2-hydroxybutyryl moiety. The γ-glutamyl-γ-amidobutyryl group is then transferred to the aminoglycoside core of butirosin by the N-acetyltransferase, BtrH, followed by removal of the glutamate by a cyclotransferase to yield pyroglutamate and mature butirosin [48]. In the absence of a glutamate protecting group the γ-aminobutyryl thioester produced in the second step of the pathway could cyclize to form a 5-membered lactam, cleaving the group from the acyl carrier protein and aborting the synthesis of the moiety. However, the amide linkage of the γ-aminobutyryl group in the final product should be stable to such cyclization events.
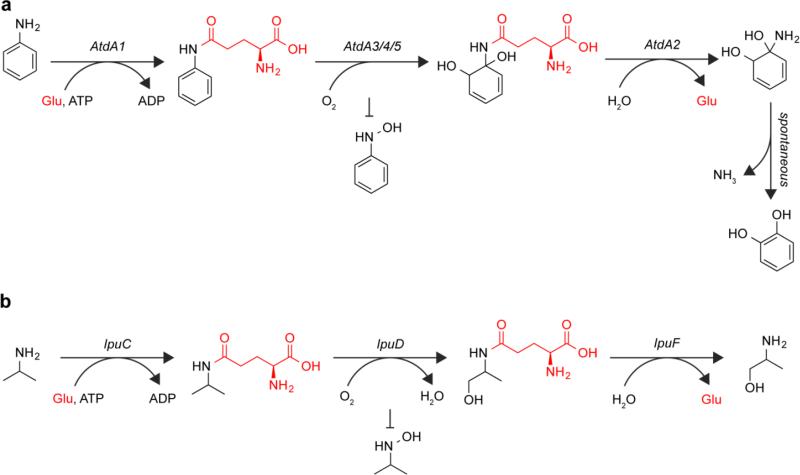
Glutamate might also have a role as a protecting group preventing off-pathway N-oxidation. Aniline and its derivatives are involved in many industrial and agricultural processes and are often found in the environment [1]. These compounds can be toxic, mutagenic, or carcinogenic [5]. However, they can be removed from the environment through degradation by microorganisms. The bacterium Acinetobacter sp. strain YAA is capable of degrading aniline and 2-methylaniline to their respective catechols [23]. The gene cluster responsible for aniline degradation contains a gene encoding an enzyme named AtdA1 that is homologous to GS enzymes. This enzyme is capable of catalyzing the production of γ-glutamylaniline in an ATP-dependent manner (Fig. 6a) [80]. AtdA1 was also found to act on aniline derivatives such as 2-, 3-, and 4-chloroaniline and 2-, 3-, and 4-methylaniline. Other studies have shown that AtdA1 also accepts 2-ethylaniline and 2,4-dimethylaniline [2], suggesting this pathway could be involved in the degradation of a range of aniline derivatives. After glutamylation by AtdA1, a Rieske non-heme dioxygenase produces catechol from γ-glutamylaniline. Rieske non-heme dioxygenases that perform a similar reaction on nitrotoluene, producing nitrite and methylcatechol, have been shown to produce nitrobenzyl alcohol as an off-pathway product [60,43]. The nitro moiety cannot be further oxidized and therefore does not need not to be protected, however variability in site of oxidation by these enzymes could suggest that if a free amine were present in the aniline degradation pathway, it might be oxidized to some extent. Finally, a GAT superfamily enzyme releases glutamate to produce a 1-amino-1,2-diol-3,5-cyclohexadiene intermediate that spontaneously eliminates ammonia to produce catechol (Fig. 6a).
Fig. 6.
Glutamylation (shown in red) may block off-pathway N-oxidation. Intermediates during aniline (a) and isopropylamine (b) catabolism are glutamylated, which might prevent N-oxidation of the amine
As with anilines, isopropylamine enters the environment through industrial and agricultural processes, particularly as part of the active ingredient of the herbicide Roundup. The bacterium Pseudomonas sp. Strain KIE171 is capable of degrading isopropylamine to L-alinol [14]. The gene cluster encoding this pathway contains an enzyme homologous to GS enzymes (IpuC), a cytochrome P-450 monooxygenase (IpuD), and a GAT family protein (IpuF). IpuC was shown to produce γ-glutamylisopropylamide in an ATP-dependent manner (Fig 6b). This enzyme was also capable of glutamylating linear amines, branched amines including t-butylamine, and amino alcohols such as ethanolamine, amino-2-propanol, and 2-amino-1-butanol all with activities similar to or higher than that observed for isopropylamine. The next step in the pathway is the oxidation of γ-glutamylisopropylamide to γ-glutamylalinol, presumably by the cytochrome P-450 monooxygenase in the cluster. IpuF then cleaves off glutamate, releasing alinol. The role of glutamylation in this pathway could be to protect the primary amine of isopropylamine from off-pathway oxidation by the monooxygenase. This degradation pathway has been identified in Arthrobacter aurescens TC1, which is capable of growing on the herbicide atrazine as the sole carbon and nitrogen source [69]. The degradation of atrazine releases both ethylamine and isopropylamine, which can then be assimilated through the glutamate-dependent pathway.
Interestingly, the use of glutamate as a protecting group and/or scaffold may be much more widespread than currently realized. Analysis of genomes revealed a very common occurrence of a pair of GS and amide hydrolase or cyclotransferase genes in gene clusters [32]. One intriguing example is in a set of five genes that appear to be conserved in clusters of natural products containing N-N bonds such as the hydrazine-containing fosfazinomycin [24] and the diazo-containing lomaiviticin [35]. Thus, it is possible that glutamate may be in some way involved in the process of N-N bond formation.
Use of Glutamyl-tRNA in biosynthesis
While glutamate is the predominant metabolite in many organisms, in E. coli tRNAGlu levels are more consistent with the amino acid content of its proteome [19]. Despite the lack of a large pool of glutamylated tRNAGlu, pathways have evolved that use it as a substrate.
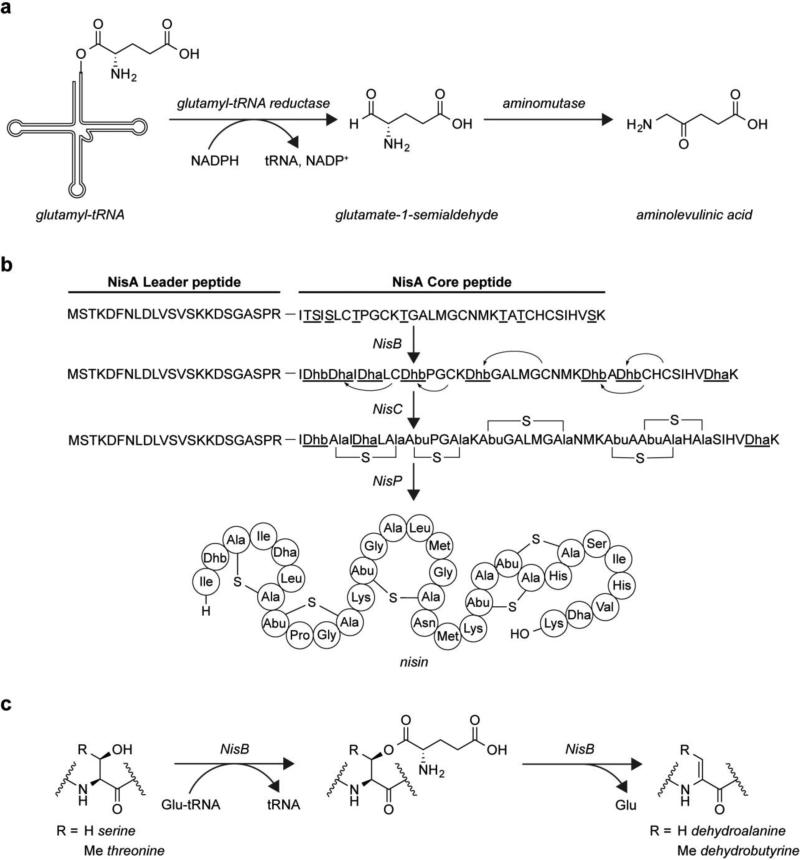
Tetrapyrroles, such as heme and chlorophyll, are important prosthetic groups in biology. In certain organisms, including plants, bacteria, and archaea aminolevulinic acid is synthesized from glutamate through a Glu-tRNAGlu intermediate as part of tetrapyrrole biosynthesis [34]. The glutamyl-tRNAGlu is produced by the glutamate tRNA synthetase in the same manner as for protein synthesis. Then the Glu-tRNAGlu is reduced to glutamate-1-semialdehyde by glutamyltRNA reductase, thereby freeing it from the tRNA [66]. In the next step an aminomutase converts the glutamate-1-semialdehyde to aminolevulinic acid (Fig. 7a) [74]. Competition between protein and tetrapyrrole synthesis can be an issue in organisms that produce large amounts of tetrapyrroles. Indeed, the bacterium Acidithiobacillus ferrooxidans, which is able to use iron as the terminal electron acceptor during respiration requiring a large number of heme proteins to do so, has evolved a mechanism for separating heme and protein synthesis [90]. This organism contains two glutamyl tRNA synthetases and three tRNAGlu [44]. While all the tRNAGlu isoforms bind to elongation factor Tu, one of them is not a substrate for glutamyl-tRNA reductase, allowing it to be used solely for protein biosynthesis. Additionally, one of the glutamyl-tRNA synthetases does not efficiently activate this tRNA, further allowing specific tRNAs to support either tetrapyrrole or protein biosynthesis. We note that a similar mechanism of using different glutamylated tRNA molecules for distinct purposes is found in some bacteria, archaea and organelles that lack an aminoacyl-tRNA synthetase that can produce glutaminyltRNAGln [31]. Instead, the genomes of these cells encode an aminoacyl-tRNA synthetase capable of loading glutamate onto both tRNAGlu and tRNAGln. An amidotransferase then catalyzes an ATP-dependent transamidation reaction specifically on glutamyl-tRNAGln using glutamine as the nitrogen donor, producing Gln-tRNAGln and glutamate [12].
Fig. 7.
Glutamylated-tRNA is involved in biosynthetic pathways. a The biosynthesis of the tetrapyrrole precursor aminolevulinic acid proceeds through a Glu-tRNAGlu intermediate. b Biosynthesis of the antimicrobial lanthipeptide nisin. c During biosynthesis of nisin, dehydrated amino acids are produced in a Glu-tRNAGlu dependent manner
The antimicrobial peptide nisin [49] belongs to a class of ribosomally synthesized and post translationally modified peptides known as lanthipeptides that are cyclized through thioether crosslinks and has been used as a food preservative for over 40 years without the appearance of significant bacterial resistance [38]. Two enzymes install the thioether crosslinks on the nisin precursor peptide, NisA (Fig. 7b). The enzyme NisB dehydrates eight serine and threonine residues, producing dehydroalanine (Dha) and dehydrobutyrine (Dhb) residues, respectively. The second enzyme, named NisC, catalyzes the Michael-type addition of cysteine thiols onto the dehydrated amino acids, producing the thioether crosslinks. During studies of the activity of NisB mutants on NisA in E. coli, adducts corresponding to glutamylated NisA were observed [25]. Incubation of these glutamylated species with wild type NisB yielded the dehydrated NisA, suggesting these glutamylated species are intermediates in the dehydration reaction. Fractionation of the cell extract led to the observation that nucleic acids supported dehydration, and glutamylated tRNAGlu was identified as being sufficient for NisB to carry out the reaction on NisA (Fig. 7c) [58]. Structural characterization of NisB revealed a basic cavity that is likely involved in tRNA binding, and docking the structure of a bacterial tRNA onto NisB places the site on the tRNA of the activated glutamate near residues in NisB known to be important for glutamylation activity.
These studies have laid the groundwork for genome mining for novel lanthipeptides by heterologous expression in E. coli. Indeed, analysis of 830 actinobacterial genomes has revealed a vast reservoir of previously uncharacterized lanthipeptides [91]. This finding may be particularly important given that several such peptides from actinobacteria such as microbisporicin (NAI-107) [33,17,18], duramycin [57,36], a semisynthetic derivative of actagardine [13], and labyrinthopeptins [21] are all in clinical development. In addition, the tRNA-dependent dehydratases are also found in the biosynthetic gene clusters of nonlanthipeptide natural products including the thiopeptides [58], which also are widespread in actinobacterial genomes. Hence, the discovery of glutamylation as a cryptic intermediate in dehydration may also assist genome mining for this class of natural products.
Conclusion
Glutamate plays a critical role in the central metabolism of many organisms, including nitrogen assimilation, amino acid biosynthesis, and cofactor production. It is also involved in the production of secondary metabolites such as antibiotics. Whether the large intracellular pool of glutamate present in many organisms evolved to support a wide array of processes, or whether these processes evolved to make use of the large pool of glutamate is unclear. It is possible that the utilization of glutamate in these various processes is adaptation of ancient non-enzymatic processes that relied on the unique capability of glutamate and aspartate to form cyclic anhydrides. Regardless of the evolutionary history, increased understanding of the roles of glutamate could allow for the engineering of processes such as bioremediation of industrial contaminates and the production of important compounds in heterologous hosts.
References
- 1.Adrian P, Andreux F, Viswanathan R, Freitag D, Scheunert I. Fate of anilines and related compounds in the environment. A review. Toxicol Environ Chem. 1989;20-21:109–120. doi:10.1080/02772248909357366. [Google Scholar]
- 2.Ang EL, Obbard JP, Zhao H. Probing the molecular determinants of aniline dioxygenase substrate specificity by saturation mutagenesis. FEBS J. 2007;274:928–939. doi: 10.1111/j.1742-4658.2007.05638.x. doi:10.1111/j.1742-4658.2007.05638.x. [DOI] [PubMed] [Google Scholar]
- 3.Åslund F, Beckwith J. Bridge over troubled waters: Sensing stress by disulfide bond formation. Cell. 1999;96:751–753. doi: 10.1016/s0092-8674(00)80584-x. doi:10.1016/S0092-8674(00)80584-X. [DOI] [PubMed] [Google Scholar]
- 4.Bender DA. Amino acid metabolism. John Wiley & Sons, Ltd; 2012. Nitrogen metabolism. pp. 1–65. doi:10.1002/9781118357514.ch1. [Google Scholar]
- 5.Benigni R, Passerini L. Carcinogenicity of the aromatic amines: from structure–activity relationships to mechanisms of action and risk assessment. Mutat Res. 2002;511:191–206. doi: 10.1016/s1383-5742(02)00008-x. doi:10.1016/S1383-5742(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 6.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. doi:10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. doi:10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Britton KL, Baker PJ, Engel PC, Rice DW, Stillman TJ. Evolution of substrate diversity in the superfamily of amino acid dehydrogenases: Prospects for rational chiral synthesis. J Mol Biol. 1993;234:938–945. doi: 10.1006/jmbi.1993.1647. doi:10.1006/jmbi.1993.1647. [DOI] [PubMed] [Google Scholar]
- 9.Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. doi:10.1016/S1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 11.Copley S, Dhillon J. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 2002;3:1–16. doi: 10.1186/gb-2002-3-5-research0025. doi:10.1186/gb-2002-3-5-research0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curnow AW, Hong K-w, Yuan R, Kim S-i, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson MJ. Lantibiotics as antimicrobial agents. Expert Opin Ther Pat. 2007;17:365–369. doi:Doi 10.1517/13543776.17.4.365. [Google Scholar]
- 14.de Azevedo Wäsch SI, van der Ploeg JR, Maire T, Lebreton A, Kiener A, Leisinger T. Transformation of isopropylamine to L-alaninol by Pseudomonas sp. Strain KIE171 involves N-glutamylated intermediates. Appl Environ Microbiol. 2002;68:2368–2375. doi: 10.1128/AEM.68.5.2368-2375.2002. doi:10.1128/aem.68.5.2368-2375.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. doi:10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dion HW, Woo PWK, Willmer NE, Kern DL, Onaga J, Fusari SA. Butirosin, a new aminoglycosidic antibiotic complex: Isolation and characterization. Antimicrob Agents Chemother. 1972;2:84–88. doi: 10.1128/aac.2.2.84. doi:10.1128/aac.2.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donadio S, Maffioli S, Monciardini P, Sosio M, Jabés D. Antibiotic discovery in the twenty-first century: Current trends and future perspectives. J Antibiot. 2010;63:423–430. doi: 10.1038/ja.2010.62. doi:10.1038/ja.2010.62. [DOI] [PubMed] [Google Scholar]
- 18.Donadio S, Maffioli S, Monciardini P, Sosio M, Jabés D. Sources of novel antibiotics-aside the common roads. Appl Microbiol Biotechnol. 2010;88:1261–1267. doi: 10.1007/s00253-010-2877-8. doi:10.1007/s00253-010-2877-8. [DOI] [PubMed] [Google Scholar]
- 19.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. doi:10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 20.Fawaz MV, Topper ME, Firestine SM. The ATP-grasp enzymes. Bioorg Chem. 2011;39:185–191. doi: 10.1016/j.bioorg.2011.08.004. doi:10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Férir G, Petrova MI, Andrei G, Huskens D, Hoorelbeke B, Snoeck R, Vanderleyden J, Balzarini J, Bartoschek S, Brönstrup M, Süssmuth RD, Schols D. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. Plos One. 2013;8 doi: 10.1371/journal.pone.0064010. doi:10.1371/journal.pone.0064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. In: Gerald L, editor. Vitamins & Hormones. Vol. 79. Academic Press; 2008. pp. 1–44. doi:10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 23.Fujii T, Takeo M, Maeda Y. Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology (Reading, UK) 1997;143:93–99. doi: 10.1099/00221287-143-1-93. doi:10.1099/00221287-143-1-93. [DOI] [PubMed] [Google Scholar]
- 24.Gao JT, Ju KS, Yu XM, Velasquez JE, Mukherjee S, Lee J, Zhao CM, Evans BS, Doroghazi JR, Metcalf WW, van der Donk WA. Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster. Angew Chem Int Edit. 2014;53:1334–1337. doi: 10.1002/anie.201308363. doi:10.1002/anie.201308363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg N, Salazar-Ocampo LMA, van der Donk WA. In vitro activity of the nisin dehydratase NisB. Proc Natl Acad Sci USA. 2013;110:7258–7263. doi: 10.1073/pnas.1222488110. doi:10.1073/pnas.1222488110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graupner M, White RH. Methanococcus jannaschii coenzyme F420 analogs contain a terminal α-linked glutamate. J Bacteriol. 2003;185:4662–4665. doi: 10.1128/JB.185.15.4662-4665.2003. doi:10.1128/jb.185.15.4662-46652003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Held KD, Biaglow JE. Mechanisms for the oxygen radical-mediated toxicity of various thiol-containing compounds in cultured mammalian cells. Radiat Res. 1994;139:15–23. doi:10.2307/3578727. [PubMed] [Google Scholar]
- 28.Helling RB. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hesketh A, Sun J, Bibb M. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol Microbiol. 2001;39:136–144. doi: 10.1046/j.1365-2958.2001.02221.x. doi:10.1046/j.1365-29582001.02221.x. [DOI] [PubMed] [Google Scholar]
- 30.Horna DH, Gómez C, Olano C, Palomino-Schätzlein M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA. Biosynthesis of the RNA polymerase inhibitor streptolydigin in Streptomyces lydicus: Tailoring modification of 3-methyl-aspartate. J Bacteriol. 2011;193:2647–2651. doi: 10.1128/JB.00108-11. doi:10.1128/jb.00108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibba M, Curnow AW, Söll D. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem Sci. 1997;22:39–42. doi: 10.1016/s0968-0004(96)20033-7. doi:10.1016/S0968-0004(96)20033-7. [DOI] [PubMed] [Google Scholar]
- 32.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: Synthesis of novel metabolites and peptide modifications of proteins. Mol Biosyst. 2009;5:1636–1660. doi: 10.1039/b917682a. doi:10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jabés D, Brunati C, Candiani G, Riva S, Romanó G, Donadio S. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother. 2011;55:1671–1676. doi: 10.1128/AAC.01288-10. doi:10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahn D, Verkamp E, Söll D. Glutamyl-transfer RNA: A precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci. 1992;17:215–218. doi: 10.1016/0968-0004(92)90380-r. doi:10.1016/0968-0004(92)90380-R. [DOI] [PubMed] [Google Scholar]
- 35.Janso JE, Haltli BA, Eustáquio AS, Kulowski K, Waldman AJ, Zha L, Nakamura H, Bernan VS, He H, Carter GT, Koehn FE, Balskus EP. Discovery of the lomaiviticin biosynthetic gene cluster in Salinispora pacifica. Tetrahedron. 2014;70:4156–4164. doi: 10.1016/j.tet.2014.03.009. doi:10.1016/j.tet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones AM, Helm JM. Emerging treatments in cystic fibrosis. Drugs. 2009;69:1903–1910. doi: 10.2165/11318500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi H, Naito T, Nakagawa S, Fujisawa KI. BB-K 8, a new semisynthetic aminoglycoside antibiotic. J Antibiot (Tokyo) 1972;25:695–708. doi: 10.7164/antibiotics.25.695. [DOI] [PubMed] [Google Scholar]
- 38.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. doi:10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 39.Kumada Y, Benson DR, Hillemann D, Hosted TJ, Rochefort DA, Thompson CJ, Wohlleben W, Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA. 1993;90:3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, Suzuki H. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J Biol Chem. 2005;280:4602–4608. doi: 10.1074/jbc.M411114200. doi:10.1074/jbc.M411114200. [DOI] [PubMed] [Google Scholar]
- 41.Kurihara S, Oda S, Tsuboi Y, Kim HG, Oshida M, Kumagai H, Suzuki H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J Biol Chem. 2008;283:19981–19990. doi: 10.1074/jbc.M800133200. doi:10.1074/jbc.M800133200. [DOI] [PubMed] [Google Scholar]
- 42.Leigh JA, Rinehart KL, Wolfe RS. Methanofuran (carbon dioxide reduction factor), a formyl carrier in methane production from carbon dioxide in Methanobacterium. Biochemistry. 1985;24:995–999. doi: 10.1021/bi00325a028. doi:10.1021/bi00325a028. [DOI] [PubMed] [Google Scholar]
- 43.Lessner DJ, Johnson GR, Parales RE, Spain JC, Gibson DT. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl Environ Microbiol. 2002;68:634–641. doi: 10.1128/AEM.68.2.634-641.2002. doi:10.1128/aem.68.2.634-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levicán G, Katz A, Valenzuela P, Söll D, Orellana O. A tRNAGlu that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett. 2005;579:6383–6387. doi: 10.1016/j.febslet.2005.09.100. doi:10.1016/j.febslet.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Graupner M, Xu H, White RH. CofE catalyzes the addition of two glutamates to F420-0 in F420 coenzyme biosynthesis in Methanococcus jannaschii. Biochemistry. 2003;42:9771–9778. doi: 10.1021/bi034779b. doi:10.1021/bi034779b. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Llewellyn NM, Giri R, Huang F, Spencer JB. Biosynthesis of the unique amino acid side chain of butirosin: Possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem Biol. 2005;12:665–675. doi: 10.1016/j.chembiol.2005.04.010. doi:10.1016/j.chembiol.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Lin XL, White RH. Occurrence of coenzyme F420 and its gamma-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol. 1986;168:444–448. doi: 10.1128/jb.168.1.444-448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llewellyn NM, Li Y, Spencer JB. Biosynthesis of butirosin: Transfer and deprotection of the unique amino acid side chain. Chem Biol. 2007;14:379–386. doi: 10.1016/j.chembiol.2007.02.005. doi:10.1016/j.chembiol.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. doi:10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller RE, Stadtman ER. Glutamate synthase from Escherichia coli: An iron-sulfide flavoprotein. J Biol Chem. 1972;247:7407–7419. [PubMed] [Google Scholar]
- 52.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep. 2002;19:70–99. doi: 10.1039/b003939j. doi:10.1039/B003939J. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. doi:10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nocek B, Evdokimova E, Proudfoot M, Kudritska M, Grochowski LL, White RH, Savchenko A, Yakunin AF, Edwards A, Joachimiak A. Structure of an amide bond forming F420:γγ-glutamyl ligase from Archaeoglobus fulgidus - A member of a new family of non-ribosomal peptide synthases. J Mol Biol. 2007;372:456–469. doi: 10.1016/j.jmb.2007.06.063. doi:10.1016/j.jmb.2007.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogasawara Y, Katayama K, Minami A, Otsuka M, Eguchi T, Kakinuma K. Cloning, sequencing, and functional analysis of the biosynthetic gene cluster of macrolactam antibiotic vicenistatin in Streptomyces halstedii. Chem Biol. 2004;11:79–86. doi: 10.1016/j.chembiol.2003.12.010. doi:10.1016/j.chembiol.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Olano C, Gómez C, Pérez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA. Deciphering biosynthesis of the RNA polymerase inhibitor streptolydigin and generation of glycosylated derivatives. Chem Biol. 2009;16:1031–1044. doi: 10.1016/j.chembiol.2009.09.015. doi:10.1016/j.chembiol.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 57.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. Apmis. 2010;118:982–990. doi: 10.1111/j.1600-0463.2010.02680.x. doi:10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 58.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517:509–512. doi: 10.1038/nature13888. doi:10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otsuka M, Eguchi T, Shindo K, Kakinuma K. Non-fatty acyl polyketide starter in the biosynthesis of vicenistatin, an antitumor macrolactam antibiotic. Tetrahedron Lett. 1998;39:3185–3188. doi:10.1016/S0040-4039(98)00455-9. [Google Scholar]
- 60.Parales JV, Parales RE, Resnick SM, Gibson DT. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the c-terminal region of the α subunit of the oxygenase component. J Bacteriol. 1998;180:1194–1199. doi: 10.1128/jb.180.5.1194-1199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peypoux F, Bonmatin JM, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. doi:10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 62.Potrykus K, Cashel M. (p)ppGpp: Still magical?. Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. doi:10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 63.Prinz WA, Åslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. doi:10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 64.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. doi:10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez S, Demain AL. Metabolic regulation of fermentation processes. Enzyme Microb Technol. 2002;31:895–906. doi:10.1016/S0141-0229(02)00172-2. [Google Scholar]
- 66.Schauer S, Chaturvedi S, Randau L, Moser J, Kitabatake M, Lorenz S, Verkamp E, Schubert W-D, Nakayashiki T, Murai M, Wall K, Thomann H-U, Heinz DW, Inokuchi H, Söll D, Jahn D. Escherichia coli glutamyl-tRNA reductase: Trapping the thioester intermediate. J Biol Chem. 2002;277:48657–48663. doi: 10.1074/jbc.M206924200. doi:10.1074/jbc.M206924200. [DOI] [PubMed] [Google Scholar]
- 67.Schirch V, Strong WB. Interaction of folylpolyglutamates with enzymes in one-carbon metabolism. Arch Biochem Biophys. 1989;269:371–380. doi: 10.1016/0003-9861(89)90120-3. doi:10.1016/0003-9861(89)90120-3. [DOI] [PubMed] [Google Scholar]
- 68.Shaibe E, Metzer E, Halpern YS. Metabolic pathway for the utilization of L-arginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol. 1985;163:933–937. doi: 10.1128/jb.163.3.933-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shapir N, Mongodin EF, Sadowsky MJ, Daugherty SC, Nelson KE, Wackett LP. Evolution of catabolic pathways: Genomic insights into microbial s-triazine metabolism. J Bacteriol. 2007;189:674–682. doi: 10.1128/JB.01257-06. doi:10.1128/jb.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shinohara Y, Kudo F, Eguchi T. A natural protecting group strategy to carry an amino acid starter unit in the biosynthesis of macrolactam polyketide antibiotics. J Am Chem Soc. 2011;133:18134–18137. doi: 10.1021/ja208927r. doi:10.1021/ja208927r. [DOI] [PubMed] [Google Scholar]
- 71.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. doi:10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 72.Skellam EJ, Stewart AK, Strangman WK, Wright JLC. Identification of micromonolactam, a new polyene macrocyclic lactam from two marine Micromonospora strains using chemical and molecular methods: Clarification of the biosynthetic pathway from a glutamate starter unit. J Antibiot. 2013;66:431–441. doi: 10.1038/ja.2013.34. doi:10.1038/ja.2013.34. [DOI] [PubMed] [Google Scholar]
- 73.Smith CA. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol. 2006;362:640–655. doi: 10.1016/j.jmb.2006.07.066. doi:10.1016/j.jmb.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 74.Smith MA, Kannangara CG, Grimm B. Glutamate 1-semialdehyde aminotransferase: Anomalous enantiomeric reaction and enzyme mechanism. Biochemistry. 1992;31:11249–11254. doi: 10.1021/bi00160a041. doi:10.1021/bi00160a041. [DOI] [PubMed] [Google Scholar]
- 75.Sundquist AR, Fahey RC. The function of gamma-glutamylcysteine and bis-gamma-glutamylcystine reductase in Halobacterium halobium. J Biol Chem. 1989;264:719–725. [PubMed] [Google Scholar]
- 76.Suzuki H, Kurihara S. Kusano T, Suzuki H, editors. Polyamine catabolism in prokaryotes. Polyamines. Springer Japan. 2015:47–59. doi:10.1007/978-4-431-55212-3_4. [Google Scholar]
- 77.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. doi:10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 78.Takaishi M, Kudo F, Eguchi T. A unique pathway for the 3-aminobutyrate starter unit from L-glutamate through β-glutamate during biosynthesis of the 24-membered macrolactam antibiotic, incednine. Org Lett. 2012;14:4591–4593. doi: 10.1021/ol302052c. doi:10.1021/ol302052c. [DOI] [PubMed] [Google Scholar]
- 79.Takaishi M, Kudo F, Eguchi T. Identification of the incednine biosynthetic gene cluster: characterization of novel β-glutamate-β-decarboxylase IdnL3. J Antibiot. 2013;66:691–699. doi: 10.1038/ja.2013.76. doi:10.1038/ja.2013.76. [DOI] [PubMed] [Google Scholar]
- 80.Takeo M, Ohara A, Sakae S, Okamoto Y, Kitamura C, Kato D-i, Negoro S. Function of a glutamine synthetase-like protein in bacterial aniline oxidation via γ-glutamylanilide. J Bacteriol. 2013;195:4406–4414. doi: 10.1128/JB.00397-13. doi:10.1128/jb.00397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang L, Hutchinson CR. Sequence, transcriptional, and functional analyses of the valine (branched-chain amino acid) dehydrogenase gene of Streptomyces coelicolor. J Bacteriol. 1993;175:4176–4185. doi: 10.1128/jb.175.13.4176-4185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Umbarger HE. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. doi:10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 83.van Eunen K, Bouwman J, Daran-Lapujade P, Postmus J, Canelas AB, Mensonides FIC, Orij R, Tuzun I, van den Brink J, Smits GJ, van Gulik WM, Brul S, Heijnen JJ, de Winde JH, Teixeira de Mattos MJ, Kettner C, Nielsen J, Westerhoff HV, Bakker BM. Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 2010;277:749–760. doi: 10.1111/j.1742-4658.2009.07524.x. doi:10.1111/j.1742-4658.2009.07524.x. [DOI] [PubMed] [Google Scholar]
- 84.van Wezel GP, Krabben P, Traag BA, Keijser BJF, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol. 2006;72:5283–5288. doi: 10.1128/AEM.00808-06. doi:10.1128/aem.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011;28:1311–1333. doi: 10.1039/c1np00003a. doi:10.1039/C1NP00003A. [DOI] [PubMed] [Google Scholar]
- 86.Vanoni MA, Curti B. Glutamate synthase: A complex iron-sulfur flavoprotein. Cell Mol Life Sci. 1999;55:617–638. doi: 10.1007/s000180050319. doi:10.1007/s000180050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Xu H, Harich KC, White RH. Identification and characterization of a tyramine–glutamate ligase (MfnD) involved in methanofuran biosynthesis. Biochemistry. 2014;53:6220–6230. doi: 10.1021/bi500879h. doi:10.1021/bi500879h. [DOI] [PubMed] [Google Scholar]
- 88.White RH. Structural diversity among methanofurans from different methanogenic bacteria. J Bacteriol. 1988;170:4594–4597. doi: 10.1128/jb.170.10.4594-4597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yao X, He W, Lu C-D. Functional characterization of seven γ-glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and β-alanine utilization in Pseudomonas aeruginosa PAO1. J Bacteriol. 2011;193:3923–3930. doi: 10.1128/JB.05105-11. doi:10.1128/jb.05105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yarzábal A, Brasseur G, Bonnefoy V. Cytochromes c of Acidithiobacillus ferrooxidans, vol 209. FEMS Microbiol Lett. 2002;2 doi: 10.1111/j.1574-6968.2002.tb11130.x. doi:10.1111/j.1574-69682002.tb11130.x. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Q, Doroghazi JR, Zhao X, Walker MC, van der Donk WA. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in Actinobacteria. Appl Environ Microbiol. 2015 doi: 10.1128/AEM.00635-15. doi:10.1128/AEM.00635-15. [DOI] [PMC free article] [PubMed] [Google Scholar]