Abstract
Potent and rapid-onset anticoagulation is required for several clinical settings, including cardiopulmonary bypass surgery. In addition, because anticoagulation is associated with increased bleeding following surgery, the ability to rapidly reverse such robust anticoagulation is also important. Previously, we observed that no single aptamer was as potent as heparin for anticoagulating blood. However, we discovered that combinations of two aptamers were as potent as heparin. Herein, we sought to combine two individual anticoagulant aptamers into a single bivalent RNA molecule in an effort to generate a single molecule that retained the potent anticoagulant activity of the combination of individual aptamers. We created four bivalent aptamers that can inhibit Factor X/Xa and prothrombin/thrombin and anticoagulate plasma, as well as the combination of individual aptamers. Detailed characterization of the shortest bivalent aptamer indicates that each aptamer retains full binding and functional activity when presented in the bivalent context. Finally, reversal of this bivalent aptamer with a single antidote was explored, and anticoagulant activity could be rapidly turned off in a dose-dependent manner. These studies demonstrate that bivalent anticoagulant aptamers represent a novel and potent approach to actively and reversibly control coagulation.
Introduction
Hemostasis is a highly regulated process involving the coordination of coagulation proteases and platelets that respond to tissue injury or prothrombotic stimuli to generate a fibrin clot and limit bleeding [1–3]. When inappropriately activated, this response can lead to thrombosis and result in stroke, myocardial infarction, deep vein thrombosis, and pulmonary embolism. Clinical use of anticoagulants in both chronic and acute settings can dampen the activation of the coagulation cascade to prevent fibrin clot formation. All anticoagulants carry an inherent bleeding risk, and few can be rapidly and reliably reversed in the event of hemorrhage.
Some clinical settings, such as cardiopulmonary bypass surgery, require robust anticoagulation that is achieved by inhibition of multiple coagulation enzymes. Unfractionated heparin (UFH) targets multiple coagulation enzymes; however, several drawbacks exist for heparin use, including the nonlinear relationship between dosing and anticoagulant effect and risk of bleeding [4]. Development of alternative therapeutics for clinical settings that require potent anticoagulation may necessitate combining agents that can simultaneously target multiple coagulation factors. Moreover, because such potent anticoagulants can increase bleeding risk, quick, easy, and reliable reversal of potent anticoagulant agents is essential following surgical procedures. The only directly neutralizable clinical anticoagulant is UFH, which can be reversed with protamine sulfate. However, in addition to heparin's side effects, reversal with protamine sulfate has many side effects, including hypotension, vasodilation, and cardiac dysfunction [5]. Thus, an unmet medical need exists for an anticoagulant that is as potent as heparin, yet easily reversible.
Aptamers are single-stranded oligonucleotides that bind with high affinity and specificity to their target, making them good candidates for therapeutics. Using a combinatorial chemistry technique known as systematic evolution of ligands by exponential enrichment (SELEX), we have generated several RNA aptamers targeting different coagulation cascade enzymes and their unactivated protease forms known as zymogens [6–11]. These aptamers derive their anticoagulant activity from their ability to inhibit protein–protein interactions [9,10,12]. By modulating critical protein–protein interactions important in the coagulation cascade, these aptamers are able to reduce thrombin generation, resulting in a delay in clot formation. Aptamer-mediated anticoagulant activity is easily modulated through two methods: sequence-specific oligonucleotide antidotes and universal antidotes [8,13]. Oligonucleotide antidotes are complementary to a portion of the aptamer sequence and form Watson–Crick base pairing duplexes that alter the conformation of the aptamer, thereby inhibiting aptamer function [8]. Alternatively, cationic universal antidotes can reverse aptamer activity by binding the backbone of the aptamer through electrostatic interaction and abolishing the aptamer's ability to bind its target [13]. Clinical studies have indicated that an aptamer–antidote pair targeting coagulation enzyme FIXa can effectively control clot formation during percutaneous coronary intervention and its antidote oligonucleotide can rapidly reverse the aptamer's activity to limit bleeding [14]. This aptamer–antidote pair was evaluated for this indication in Phase IIb and III clinical trials; the Phase III trial enrolled ∼3,200 patients, but was halted due to rare hypersensitivity reactions to the high molecular weight PEG carrier appended to the aptamer [14–17].
Our previous studies have indicated that individual anticoagulant aptamers are not able to achieve heparin-level anticoagulation in human blood [18]. However, we have observed that combinations of multiple aptamers targeting different individual coagulation enzymes can be as potent as heparin and can prevent fibrin clot formation during circulation of blood in extracorporeal circuits similar to those utilized during cardiopulmonary bypass surgery [18]. These observations suggest that a cocktail of two or more aptamers might be a promising novel approach to anticoagulation. The clinical development of a cocktail of novel agents poses a significant challenge from a development standpoint as preclinical IND enabling and Phase I studies need to be performed on each unapproved agent as well as the combination to ensure safety of each component [19]. Therefore, we sought to explore an alternative approach to this method using a single bivalent aptamer.
The creation of multimerized aptamers and bivalent aptamers often results in a greater overall effect compared to a single monovalent aptamer [20]. This process has been utilized because it can result in increased binding avidity and increased local concentration of the ligand once one of the ligands binds its target [20]. For example, we previously observed that monovalent aptamers to the T-cell receptors, CTLA-4, 4-1BB, and OX40 were unable to activate their cognate receptors alone, but multimerization of these aptamers on a scaffold was able to achieve activation of the target once additional aptamer copies were present [21–23]. In another example, Kim et al. were able to significantly increase anticoagulant activity when using a bivalent aptamer simultaneously targeting the two different surfaces on thrombin [24]. In addition, they found that linking two copies of the 15mer thrombin aptamer to an “anchor” thrombin aptamer resulted in even greater anticoagulation than the bivalent aptamer [25]. The same exosite targeting strategy was employed by Nimjee et al. with one DNA and one RNA aptamer [26]. This combination resulted in dramatically more potent anticoagulation when used in tandem than achieved with either aptamer alone [26]. Similarly, linkage of the thrombin aptamers into a weave tile conformation resulted in enhanced anticoagulant activity than when the aptamers were tested individually [27]. Altering the spatial arrangement of the aptamers within the tile also affected the activity in clotting assays, indicating that simply linking the aptamers is not sufficient for increasing anticoagulant activity [27]. All of these strategies utilize aptamers to a single target. While more copies generate increased activity, they still only recognize a single protein target.
Targeting of the prothrombinase complex and its substrate prothrombin with two individual anticoagulant aptamers is able to achieve heparin-level anticoagulation [18]. The prothrombinase complex contains activated Factor X (FXa) and activated Factor V (FVa), along with calcium ions, and requires a phospholipid surface for assembly [28,29]. This complex is responsible for converting prothrombin into the active enzyme thrombin, resulting in bioamplification of the coagulation cascade, activation of platelet receptors, and the conversion of fibrinogen to fibrin for clot formation [3]. Therefore, we explore whether a single bivalent aptamer can be generated to target and inhibit both prothrombin/thrombin and FXa. Our results demonstrate that this is not only possible but also results in a highly potent anticoagulant agent. By fusing the two aptamers, we also observe that a single oligonucleotide antidote can be created to control both aptamers simultaneously.
Materials and Methods
Bivalent aptamer constructs
A series of four bivalent aptamers (RNABA1-4 where BA stands for bivalent aptamer) were created by linking the stems of our FXa aptamer, 11F7t, and prothrombin aptamer, R9D-14t, together through three adenosine linkers (Table 1) [9,10]. Further truncation of the RNABA4 aptamer yielded only nonfunctional bivalent aptamers, so slight; additional modifications of RNABA4 focused on increasing in vitro transcription yields, resulting in the RNABA4M construct. The aptamers were in vitro transcribed as previously described [30]. All aptamers contain 2′-fluoro modifications on the pyrimidine nucleotides to increase stability in the presence of plasma nucleases. Mutant bivalent aptamers were created where single nucleotides previously shown to be important for each aptamer's activity were mutated. Mutant controls contained mutations in the FXa aptamer (FII functional BA), the prothrombin aptamer (FXa functional BA) (partial mutants), or both aptamers (nonfunctional BA) (double mutant). The oligonucleotide antidote sequence for RNABA4 comprised unmodified DNA 5′-GCT GGG GCT TTG TCG TCT GCC TCG-3′. The oligonucleotide antidote sequence for R9D-14t comprised unmodified DNA 5′-GTC TGC CTC GTA CAT TGG CT-3′. The oligonucleotide antidote sequence for 11F7t comprised unmodified DNA 5′-TAT TAT CTC GCT GGG-3′. Antidotes to R9D-14t and 11F7t were chosen by screening partially overlapping complementary oligonucleotides for their ability to return an activated partial thromboplastin time (aPTT) assay to baseline (see Coagulometer Assays section for antidote assay description). Oligonucleotide antidotes range from 12 to 20 nucleotides long to allow for sufficient interaction between the aptamer and antidote, without the antidote becoming long enough to form a structure. Two antidotes to RNABA4 were screened to return an aPTT to baseline levels; one antidote was 24 nucleotides and the other was 28 nucleotides in length (data not shown). There was no difference in their ability to return the aPTT to normal, therefore, only the 24mer was pursued as the antidote to minimize the chance of antidote structure formation. All oligonucleotide antidotes were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Table 1.
RNA Sequences of Bivalent Aptamer Constructs
| Name | Sequence 5′-3′ | Length (nt) |
|---|---|---|
| RNABA1 | GGGGCGGUCGAUCACACAGUUCAAACGUAAUAAGCCAAUGUACGAGGCAGACGACUCGCCCCAAAACGUAGAGAGCCCCAGCGAGAUAAUACUUGGCCCCGCUCUUUACGU | 111 |
| RNABA2 | GGCGGUCGAUCACACAGUUCAAACGUAAUAAGCCAAUGUACGAGGCAGACGACUCGCCAAAGAGAGCCCCAGCGAGAUAAUACUUGGCCCCGCUCUU | 97 |
| RNABA3 | GGGUCGAUCACACAGUUCAAACGUAAUAAGCCAAUGUACGAGGCAGACGACUCCAAAAGCCCCAGCGAGAUAAUACUUGGCCCCGCU | 87 |
| RNABA4 | GGUCGAUCACACAGUUCAAACGUAAUAAGCCAAUGUACGAGGCAGACGACAAAGCCCCAGCGAGAUAAUACUUGGCCCCGCU | 82 |
| RNABA4m | GGGGAUCACACAGUUCAAACGUAAUAAGCCAAUGUACGAGGCAGACCCCAAAGCCCCAGCGAGAUAAUACUUGGCCCCGCU | 81 |
Pyrimidines (C/U) are 2′-fluoro modified, and the purines (G/A) contain 2′OH.
Dephosphorylation and 5′-end radiolabeling
Bacterial alkaline phosphatase (Life Technologies, Carlsbad, CA) was used to dephosphorylate the in vitro transcribed RNAs. RNA was incubated at 65°C for 1 h followed by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) was used to add a 5′-[γ32P] ATP (PerkinElmer, Waltham, MA) to radiolabel the dephosphorylated RNAs. Counts per million readings for each radiolabeled RNA were determined using the Tri-Carb 2800TR Liquid Scintillation Analyzer (PerkinElmer).
Double filter nitrocellulose binding assay
Apparent binding affinity of the aptamers to each protein was determined using double filter nitrocellulose binding assays. Binding of the radiolabeled RNABA4 aptamer to proteins was assessed by incubating trace amounts of radiolabeled RNA with serial dilutions of the purified target protein, either human α-thrombin or human FXa over a range of 0–60 nM comparing the parent aptamers to RNABA4 or 0–500 nM when comparing RNABA4 to the partial and double mutant bivalent aptamers (Haematologic Technologies, Inc., Essex Junction, VT). The radiolabeled RNA was denatured and refolded in HEPES-buffered saline (HBS) with CaCl2 and 0.01% bovine serum albumin (BSA) (20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM CaCl2, 0.01% BSA) by incubating at 95°C for 3 min followed by cooling to room temperature for 3 min. The radiolabeled RNA alone was preincubated at 37°C for 5 min, followed by incubation with the protein in HBS with CaCl2 and 0.01% BSA at 37°C for 5 min. The RNA–protein complexes were partitioned from unbound RNA by passing the mixture through a 0.45-μm nitrocellulose filter (VWR, Radnor, PA), capturing unbound RNA on a nylon membrane (PerkinElmer) sandwich, and washing with 100 μL of HBS with CaCl2 without BSA. Bound and unbound RNA was quantitated using a Storm 825 phosphorImager (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The corrected fraction bound was determined for each protein concentration, and a nonlinear regression one site binding fit was applied in GraphPad Prism 6 to determine the apparent KD.
Coagulometer assays
The effects of the aptamers on clotting in aPTT and prothrombin time (PT) assays were determined using a STart 4 coagulometer (Diagnostica Stago, Asnières sur Seine Cedex, France) as described previously [26]. Aptamers were denatured and refolded in HBS with CaCl2 by incubation at 95°C for 3 min and then cooling to room temperature for 3 min. For experiments where combinations of aptamer were tested, the aptamers were folded separately and then mixed together at equal molar concentrations. For aPTT assays, pooled normal human platelet poor plasma (PPP) (50 μL) (George King Biomedical, Overland Parks, KS) was incubated with aptamer(s) for 5 min at 37°C, then TriniCLOT aPTT S (50 μL) (Trinity Biotech, Bray, Co Wicklow, Ireland) was added and incubated for another 5 min at 37°C. Clotting was initiated by 0.02 M CaCl2 (50 μL), and the time to clot formation was determined. For the PT assay, pooled normal human PPP (50 μL) was incubated with aptamer(s) for 5 min at 37°C before TriniCLOT PT Excel reagent (100 μL) was added to initiate clotting. Assays with more than one aptamer contained equal molar amounts of each aptamer and were graphed according to single aptamer concentrations (GraphPad Prism 6). When antidotes were tested in the aPTT, a third 5-min incubation with antidote was performed before initiation of clotting with CaCl2. The antidotes were treated the same as the aptamers; they were diluted in HBS with CaCl2 and denatured and refolded according to the same protocol.
Results
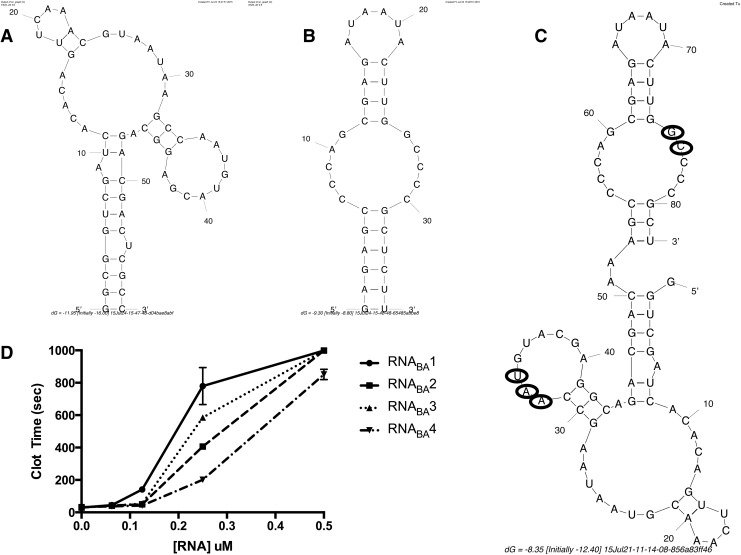
To generate a bivalent aptamer that can target both prothrombin/thrombin and FXa/Xa, we utilized the FXa aptamer, 11F7t, and prothrombin/thrombin aptamer, R9D-14t, which we previously developed and described (Fig. 1A, B) [9,10]. Four bivalent aptamer constructs (RNABA1–4) of different sizes were designed to try to identify one(s) that allow each aptamer to independently fold into its respective active structure and function as an anticoagulant. These variants were generated to determine the minimum bivalent aptamer length that still retains full function of both aptamers (Table 1). RNABA1 (111 nts) was generated by elongating and joining the two stems of the parent aptamers to create a bivalent structure with one aptamer on each end of the molecule (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/nat). In addition, three extra adenosine nucleotides were added as a linker between the two parent aptamers to create a flexible hinge and improve the possibility that the bivalent aptamer could interact with both targets simultaneously. RNABA2 (97 nts) is similar, but does not contain the stem elongation. RNABA3 (87 nts) has two base pairs from the prothrombin aptamer removed, and three base pairs of the FXa aptamer stem were eliminated. For RNABA4 (82 nts), two additional base pairs and a bulged uracil were removed from the prothrombin aptamer and one hinge adenosine was removed (Fig. 1C). The FXa aptamer remained unchanged from RNABA3 to RNABA4 (Supplementary Table S1). The functionality of all bivalent RNAs was tested in an aPTT assay and all were able to dose dependently increase clot time (Fig. 1D). RNABA4 was chosen for further study as it was the shortest construct that retained very potent anticoagulant activity, and additional shortening of RNABA4 resulted in a complete loss of anticoagulant activity. Optimization of the RNABA4 sequence to increase in vitro transcription yields produced RNABA4m, in which the first four nucleotides are all guanines that base pair with cytosine. In addition, mutant versions of RNABA4 were generated through point mutation of each aptamer at previously identified locations known to completely abrogate aptamer function (Fig. 1C) [9,10].
FIG. 1.
Fusion of FXa aptamer and FII aptamer to generate RNABA1-4. (A) FII aptamer, R9D-14t, mFold predicted structure, 58 nts in length. (B) FXa aptamer, 11F7t, mFold predicted structure, 36 nts in length. (C) RNABA4 mFold predicted structure, 82 nts in length. Circled nucleotides were mutated to generate the partial mutant bivalent aptamers and the double mutant bivalent aptamer. (D) Activated partial thromboplastin time (aPTT) assay comparing dose titrations of RNABA1–4.
The apparent binding affinity of RNABA4 toward purified FXa and α-thrombin proteins was compared to the parent 11F7t FXa and R9D-14t prothrombin aptamers to determine if the aptamers embedded in the bivalent structure retained full binding affinity (Table 2). In both cases, RNABA4 showed equivalent or slightly tighter binding to its targets relative to the parent monovalent aptamers; RNABA4 bound α-thrombin with a disassociation constant (KD) of 1.34 ± 0.24 nM versus 0.77 ± 0.22 nM for R9D-14t and to FXa with a KD of 1.72 ± 0.40 nM versus 2.59 ± 0.56 nM for 11F7t (Table 2). In addition, the partial mutant bivalent aptamers and the double mutant aptamer were evaluated and, as expected, retained binding to only one protein (partial mutants) or neither protein (double mutant), indicating that point mutations can inactivate one side of the molecule without disrupting construct folding (Table 3). The prothrombin partial mutant bivalent aptamer, which could not bind to α-thrombin due to mutation of three nucleotides in the prothrombin aptamer, was only able to bind to FXa (11.48 ± 8.15 nM versus 4.37 ± 0.91 nM for RNABA4), and the FXa partial mutant bivalent aptamer, which could not bind FXa due to mutation of three nucleotides in the FXa aptamer, was only able to bind α-thrombin (1.70 ± 0.04 nM versus 0.88 ± 0.12 nM for RNABA4) [9,10]. The double mutant bivalent aptamer was unable to bind to either FXa or α-thrombin because that bivalent aptamer contained point mutations in both aptamers (Table 3). These filter-binding assays indicated that the FXa and thrombin aptamers within the RNABA4 aptamer were still able to bind to their targets with an affinity similar to the parent aptamer, indicating that incorporation into a larger fused structure did not significantly impact aptamer folding or target binding.
Table 2.
Disassociation Constants (KD) for RNABA4, 11F7t, and R9D-14t as Determined in A Nitrocellulose-Based Binding Assay with Radiolabeled RNA
| Aptamer | KD to FXa (nM) ± SEM | KD to FIIa (nM) ± SEM |
|---|---|---|
| RNABA4 | 1.72 ± 0.40 | 1.34 ± 0.24 |
| 11F7t | 2.59 ± 0.56 | DNB |
| R9D-14t | DNB | 0.77 ± 0.22 |
DNB, does not bind.
Table 3.
Disassociation Constants (KD) for RNABA4, FXa Functional BA, FII Functional BA, and Nonfunctional BA as Determined in a Nitrocellulose-Based Binding Assay with Radiolabeled RNA
| Aptamer | KD to FXa (nM) ± SEM | KD to FIIa (nM) ± SEM |
|---|---|---|
| RNABA4 | 4.37 ± 0.91 | 0.88 ± 0.12 |
| FXa Functional BA | 11.48 ± 8.15 | DNB |
| FII Functional BA | DNB | 1.70 ± 0.04 |
| Nonfunctional BA | DNB | DNB |
DNB, does not bind.
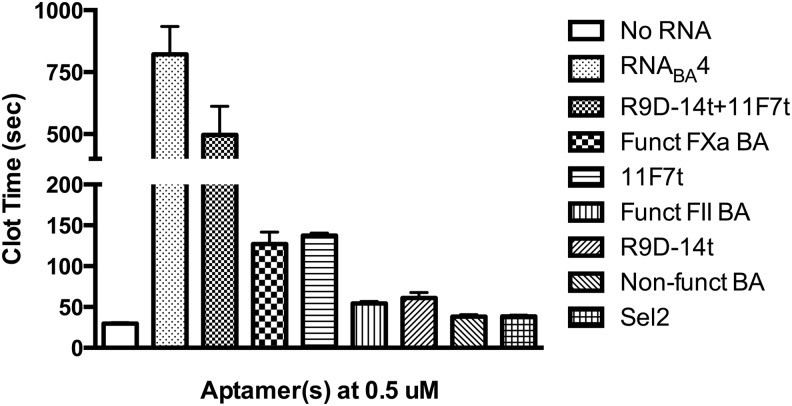
While the RNABA4 bivalent structure allowed the aptamers to bind their respective targets, we next wanted to study the anticoagulant ability of this bivalent aptamer and its derivatives in human plasma in more detail. As with the double filter binding assays, we tested if each portion of the bivalent aptamer still functioned as an anticoagulant in comparison to the single parent aptamer. The anticoagulant activity of RNABA4, its single or double mutant variants, and the parent aptamers was assayed using a clinical coagulation assay: the aPTT. This assay measured the time to fibrin clot formation by activating the intrinsic pathway. First, RNABA4 was compared to the partial mutant bivalent aptamer or double mutant bivalent aptamer to test if each individual aptamer could increase clotting time similar to the parent monovalent aptamers. The bivalent aptamer with FXa aptamer mutated anticoagulated human plasma to a similar extent as the prothrombin aptamer alone (Fig. 2). Similarly, the bivalent aptamer with a mutated prothrombin aptamer anticoagulated human plasma to a similar extent as the monovalent FXa aptamer (Fig. 2). Finally, the double mutant bivalent aptamer did not show any coagulant activity compared to RNA of similar size (Fig. 2). These results indicated that each aptamer within RNABA4 retains anticoagulant activity equivalent to the parent aptamers.
FIG. 2.
Anticoagulant activity of RNABA4, mutant derivatives, and individual parent aptamers (0.5 μM) in an aPTT clotting assay. R9D-14t is the aptamer targeting prothrombin/thrombin, and 11F7t is the aptamer targeting FX/FXa. BA stands for bivalent aptamer. Aptamers were tested in pooled normal human plasma. Each RNA was tested in triplicate and those data are represented as the mean clot time ± SEM.
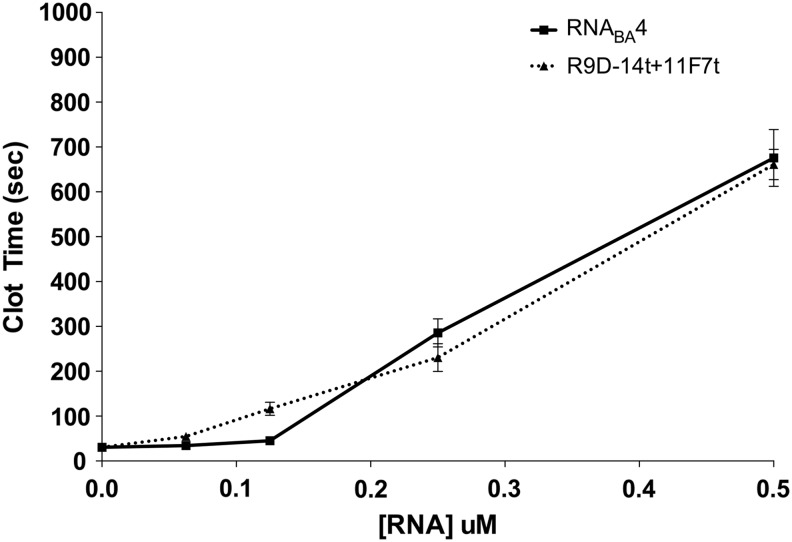
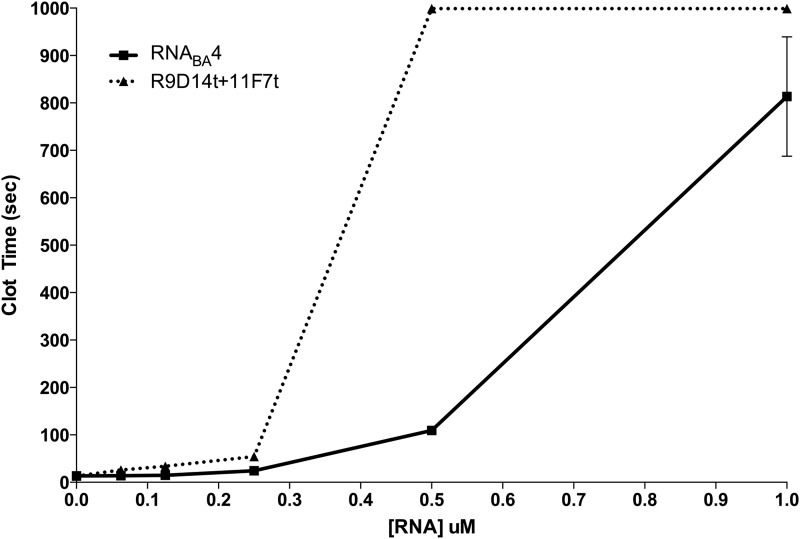
Because each half of the bivalent aptamer anticoagulated plasma to the same extent as the cognate parent aptamer, we next tested the anticoagulant ability of the fully functional RNABA4 aptamer relative to a combination of the individual FXa and prothrombin aptamers using both the aPTT and PT assays (Figs. 3 and 4). For both assays, the free aptamers were added at equal molar concentrations compared to the individual aptamers within the bivalent aptamer. For example, 1 μM of the RNABA4 aptamer was compared to 1 μM of the FXa aptamer plus 1 μM of the prothrombin aptamer. In the aPTT assay, at concentrations around 0.1 μM, the combination of individual parent aptamers was slightly more potent than RNABA4 (Fig. 3). However, at concentrations above 0.2 μM, RNABA4 was able to anticoagulate plasma at least as efficiently as the individual aptamers (Fig. 3). In a PT assay, at very low doses (0.0625–0.25 μM), the combination and RNABA4 were indistinguishable in their anticoagulant ability; however, at 0.5 μM, the combination was able to anticoagulate human plasma better than RNABA4 (Fig. 4). Once the combination and RNABA4 doses reached 1.0 μM, their ability to anticoagulate plasma once again became similar (Fig. 4).
FIG. 3.
Anticoagulant dose response of RNABA4 and the combination of individual parent aptamers in an aPTT clotting assay. RNABA4 is represented by the square (▪) connected with the solid line, and the individual parent aptamers are represented by the triangle (▴) connected with the dotted line. Titration was completed in pooled normal human plasma in triplicate, and each point is the mean clot time ± SEM.
FIG. 4.
Anticoagulant dose response of RNABA4 and the combination of the individual parent aptamers in a prothrombin time clotting assay. RNABA4 is represented by the square (▪) connected with the solid line, and the individual parent aptamers are represented by the triangle (▴) connected with the dotted line. Titration was completed in pooled normal human plasma in triplicate, and each point is the mean clot time ± SEM.
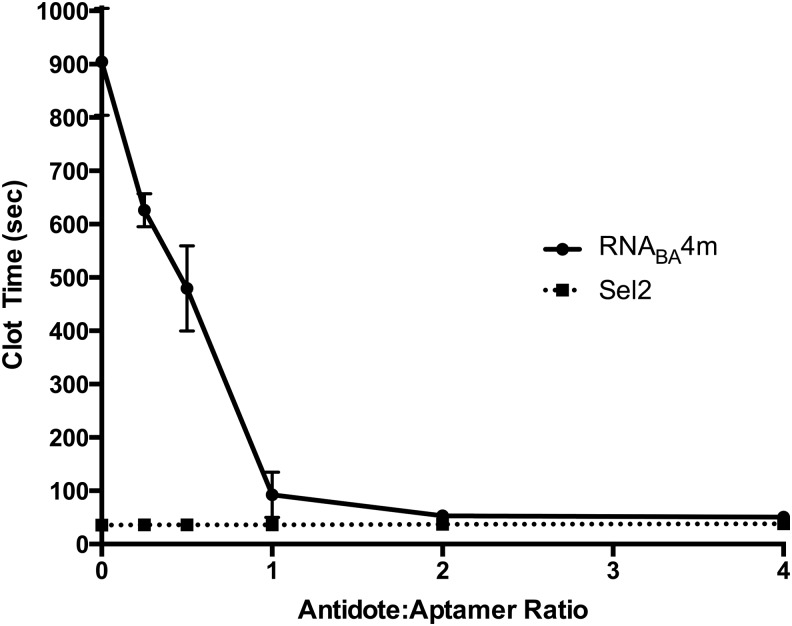
Using a modified aPTT, we assayed the ability of each aptamer within RNABA4 and fully functional RNABA4 to be reversed by the individual FXa aptamer, prothrombin aptamer-specific antidotes, or an antidote that can simultaneously target both aptamers within RNABA4 (Fig. 5A). The bivalent aptamer oligonucleotide antidote was created based on the RNABA4 sequence to attempt to reverse both aptamers simultaneously. All antidotes took advantage of the RNA sequence of the aptamers they targeted. The antidotes were designed to be complementary to the aptamer and are thought to form a double-stranded duplex upon interaction [8]. Initially, we tested the ability of the antidotes to reverse the activities of each individual aptamer embedded in RNABA4. When the FXa aptamer antidote (11F7t AO) was added to RNABA4, the anticoagulant ability of RNABA4 was reduced and became equivalent to that of the prothrombin parent aptamer alone. The reciprocal was also true, when the prothrombin aptamer antidote (R9D-14t) was added, RNABA4 anticoagulated plasma to a similar degree as to the FXa parent aptamer alone (Fig. 5B). To avoid using two oligonucleotide antidotes to reverse each functional half of the construct, we designed and tested a new oligonucleotide antidote that spans the junction of the bivalent aptamer and has the ability to bind and disrupt both aptamers simultaneously (Fig. 5B). The RNABA4 antidote oligonucleotide was able to completely reverse anticoagulant activity of RNABA4 at 1:2 stoichiometry (Figs. 5B and 6). Furthermore, as the antidote to RNABA4 stoichiometry was decreased, the anticoagulant activity of RNABA4 scaled accordingly, allowing for tunable anticoagulation to be achieved with the RNABA4 aptamer–antidote pair (Fig. 6).
FIG. 5.
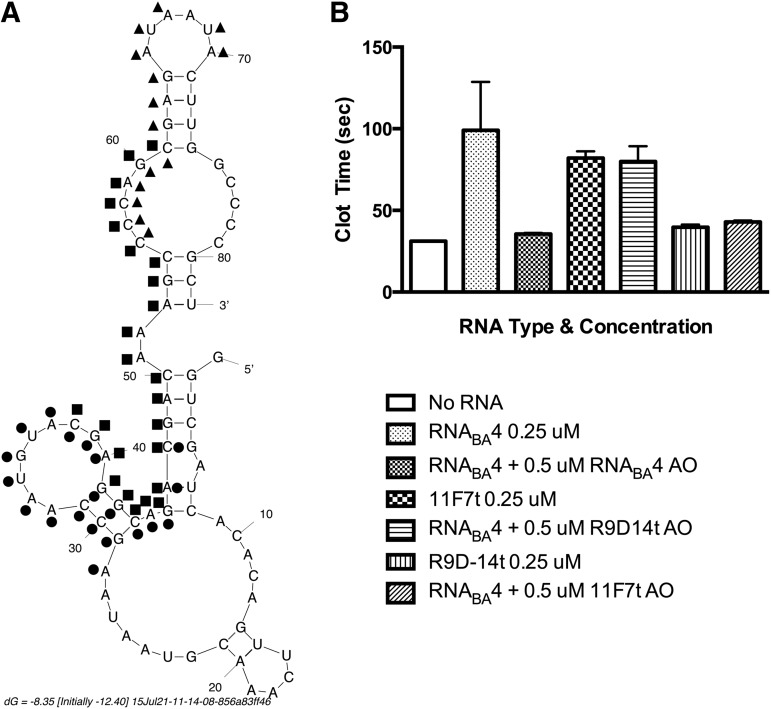
Antidote-mediated reversal of bivalent anticoagulant aptamer RNABA4. (A) Predicted mFold structure of RNABA4 with antidote oligonucleotide binding mapped onto the structure. RNABA4 AO is a 24mer represented by the squares (▪). The R9D-14t AO is a 20mer represented by the circles (●). The 11F7t AO is a 15mer represented by the triangles (▴). Nucleotides between the R9D-14t antidote or the 11F7t antidotes and the RNABA4 antidote are represented by being dotted on the exterior of the structure (RNABA4) or the interior of the structure (R9D-14t and 11F7t). (B) Normal human plasma was anticoagulated with RNABA4 (0.25 μM), and the ability of various antidotes to rapidly reverse this effect (5 min) was determined. Antidotes tested are bivalent aptamer-specific antidote (RNABA4 AO), an antidote targeting the FII aptamer (R9D-14t AO), or an antidote targeting the FXa aptamer (11F7t AO). The individual parent aptamers were dosed at 0.25 μM for comparison to the partially reversed RNABA4 aptamer. Data are represented as mean clot time ± SEM and were completed in triplicate in pooled normal human plasma.
FIG. 6.
Antidote dose response. Reversal of RNABA4m with a sequence-specific antidote targeting both aptamers simultaneously. RNABA4m was dosed at 0.5 μM, and the bivalent aptamer-specific antidote was dosed at varying molar ratios (0:1–4:1). Sel2 is a control RNA of similar size with no inherent anticoagulant activity. Sel2 was dosed at 0.5 μM, and the bivalent aptamer-specific antidote was dosed at the same molar ratios. Data are represented as mean clot time ± SEM in triplicate in pooled normal human plasma.
Discussion
In this study, we have taken two anticoagulant aptamers and combined them into a single bivalent molecule that retains the activities of both aptamers to generate a potent anticoagulant agent targeting multiple coagulation enzymes [18]. The shortest construct that retained activity, RNABA4, was able to achieve the same level of anticoagulation as that of the two free parent aptamers when dosed equivalently. This observation suggests that the FXa and prothrombin aptamers are still able to synergistically anticoagulate human plasma, despite being fused into a single molecule as we previously observed evaluating the monovalent versions of these aptamers [18]. The availability of a single bivalent molecule is important from a safety and drug development standpoint. The most feared complication when administering anticoagulants is hemorrhage that cannot be controlled. By combining the aptamers into a single bivalent molecule, we have shown that the anticoagulant activity of the RNABA4 construct can be controlled with a single oligonucleotide antidote at a 1:2 molar ratio. This control allows for rapid reversal of anticoagulant activity to limit bleeding risk. Moreover, the cost of drug development for two independent aptamers and two antidotes is likely to be prohibitive, whereas the development of a single bivalent molecule and its antidote should be easier to achieve even though the synthesis of long RNAs is currently challenging.
Currently, the cost of oligonucleotide synthesis for a bivalent 2′-fluoro-modified aptamer of this size on a large scale remains high. In this study, we used in vitro transcription to generate the amounts of the construct required for in vitro testing. However, solid-phase oligonucleotide synthesis has significantly improved and is becoming a reasonable alternative production method to generate enough material for in vivo and ex vivo testing. Thus, future advances in technology and oligonucleotide manufacturing/production may render the use of a bivalent anticoagulant aptamer as a feasible alternative to heparin anticoagulation just as bivalent antibodies have recently proven to be clinically useful [31].
Bivalent aptamers have already been reported to be useful in preclinical cancer studies. For example, we recently reported that a tumor-targeting aptamer can be used to deliver a second therapeutic aptamer into cancer cells [32]. Kotula et al. used a combination of the nucleolin aptamer to bring an aptamer targeting β-arrestin 2 into blast crisis chronic myelogenous leukemia cells [32]. Using the nucleolin aptamer, we were able to deliver the bivalent aptamer selectively to cancerous cells versus normal lymphoblastoid cells because the cancerous cells expressed 30 times more nucleolin than normal cells [32]. Bispecific aptamers have also proven useful as antitumor immunotherapeutic agents compared to antibodies. Agonistic stimulation of 4-1BB has been explored as a way to boost cancer patients' immunity toward their tumors. However, antibodies targeting 4-1BB have been associated with cytokine storm issues resulting in dysfunction of multiple organs. Schrand et al. conjugated a 4-1BB aptamer to aptamers that recognize either vascular endothelial growth factor or osteopontin, proteins commonly found in the tumor stroma [33]. This resulted in preferential accumulation of the bivalent aptamers in the tumor stroma and localized stimulation of the immune system. Superior antitumor immunity was seen in several cancer types.
The bivalent aptamer, RNABA4, joins this growing list of potentially useful bivalent aptamers and adds a new facet to this emerging approach in that it targets two different proteins in the same pathway resulting in potent pathway inhibition. Most of the bivalent aptamers employ one aptamer to deliver a “payload” aptamer into the proper location to execute its function. The RNABA4 aptamer results in the disruption of a critical enzymatic complex with its substrate in the coagulation cascade: the prothrombinase complex and its substrate prothrombin. The FXa aptamer functions by blocking the FVa-FXa interaction, which prevents the complex from assembling, resulting in greatly reduced thrombin generation [9]. The prothrombin aptamer then can inhibit even these low levels of thrombin generated [10].
While a powerful level of anticoagulation is necessary if heparin is going to be replaced, control of this effect is of paramount importance. Aptamers are currently the only class of anticoagulant that can theoretically be used in synergistic combinations, yet directly controlled with antidotes. We have shown that a single oligonucleotide antidote can be used to turn off both aptamers in a bivalent construct simultaneously; this could only be achieved by fusing the aptamers into a bivalent molecule. While dosing the aptamers individually and controlling them individually is possible, making a single bivalent molecule allows for simpler dosing and reversal. In addition, with the individual aptamer antidotes and the mutants, we have shown that each portion of the aptamer is still able to function as an anticoagulant on its own.
Anticoagulants that can match the ability of heparin in cardiopulmonary bypass and percutaneous coronary intervention have been sought for quite some time; however, many promising candidates have fallen short, primarily because they have been unable to achieve sufficient anticoagulation. They likely have missed the mark because they are unable to target as many proteins within the coagulation cascade as heparin. Our laboratory has shown that anticoagulant synergy can be achieved through specific combinations of anticoagulant aptamers [18]. We have taken that a step further by combining two such synergistic anticoagulant aptamers into a single bivalent molecule with anticoagulant ability that is equivalent to dosing of the individual aptamers. This suggests that the fusion of anticoagulant aptamers into a single molecule may be a way to achieve heparin-like anticoagulation with a lower side effect profile.
Supplementary Material
Acknowledgments
The authors would like to thank Juliana Layzer for technical assistance with the generation of the FXa aptamer antidote. This study was supported by National Institutes of Health grants to B.A.S. (R01HL65222) and (U54HL112307).
Author Disclosure Statement
B.A.S. is a scientific founder of Regado Biosciences Inc. All other authors have no disclosures.
References
- 1.Davie EW. and Ratnoff OD. (1964). Waterfall sequence for intrinsic blood clotting. Science 145:1310–1312 [DOI] [PubMed] [Google Scholar]
- 2.Macfarlane RG. (1964). An enzyme cascade in the blood clotting mechanism, and its function as a biochemical amplifier. Nature 202:498–499 [DOI] [PubMed] [Google Scholar]
- 3.Hoffman M. and Monroe DM 3rd., (2001). A cell-based model of hemostasis. Thromb Haemost 85:958–965 [PubMed] [Google Scholar]
- 4.Bussey H. and Francis JL. (2004). Heparin overview and issues. Pharmacotherapy 24(8 Pt 2):103s–107s [DOI] [PubMed] [Google Scholar]
- 5.Hirsh J, Warkentin TE, Raschke R, Granger C, Ohman EM. and Dalen JE. (1998). Heparin and low-molecular-weight heparin: mechanisms of action, pharmacokinetics, dosing considerations, monitoring, efficacy, and safety. Chest 114(5 Suppl):489s–510s [DOI] [PubMed] [Google Scholar]
- 6.Ellington AD. and Szostak JW. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822 [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C. and Gold L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510 [DOI] [PubMed] [Google Scholar]
- 8.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D. and Sullenger BA. (2002). RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419:90–94 [DOI] [PubMed] [Google Scholar]
- 9.Buddai SK, Layzer JM, Lu G, Rusconi CP, Sullenger BA, Monroe DM. and Krishnaswamy S. (2010). An anticoagulant RNA aptamer that inhibits proteinase-cofactor interactions within prothrombinase. J Biol Chem 285:5212–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bompiani KM, Monroe DM, Church FC. and Sullenger BA. (2012). A high affinity, antidote-controllable prothrombin and thrombin-binding RNA aptamer inhibits thrombin generation and thrombin activity. J Thromb Haemost 10:870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Layzer JM. and Sullenger BA. (2007). Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides 17:1–11 [DOI] [PubMed] [Google Scholar]
- 12.Sullenger B, Woodruff R. and Monroe DM. (2012). Potent anticoagulant aptamer directed against factor IXa blocks macromolecular substrate interaction. J Biol Chem 287:12779–12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oney S, Lam RT, Bompiani KM, Blake CM, Quick G, Heidel JD, Liu JY, Mack BC, Davis ME, Leong KW. and Sullenger BA. (2009). Development of universal antidotes to control aptamer activity. Nat Med 15:1224–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Povsic TJ, Vavalle JP, Alexander JH, Aberle LH, Zelenkofske SL, Becker RC, Buller CE, Cohen MG, Cornel JH, et al. (2014). Use of the REG1 anticoagulation system in patients with acute coronary syndromes undergoing percutaneous coronary intervention: results from the phase II RADAR-PCI study. EuroIntervention 10:431–438 [DOI] [PubMed] [Google Scholar]
- 15.Cohen MG, Purdy DA, Rossi JS, Grinfeld LR, Myles SK, Aberle LH, Greenbaum AB, Fry E, Chan MY, et al. (2010). First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation 122:614–622 [DOI] [PubMed] [Google Scholar]
- 16.Povsic TJ, Wargin WA, Alexander JH, Krasnow J, Krolick M, Cohen MG, Mehran R, Buller CE, Bode C, et al. (2011). Pegnivacogin results in near complete FIX inhibition in acute coronary syndrome patients: RADAR pharmacokinetic and pharmacodynamic substudy. Eur Heart J 32:2412–2419 [DOI] [PubMed] [Google Scholar]
- 17.Povsic TJ, Vavalle JP, Aberle LH, Kasprzak JD, Cohen MG, Mehran R, Bode C, Buller CE, Montalescot G, et al. (2013). A phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur Heart J 34:2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bompiani KM, Lohrmann JL, Pitoc GA, Frederiksen JW, Mackensen GB. and Sullenger BA. (2014). Probing the coagulation pathway with aptamers identifies combinations that synergistically inhibit blood clot formation. Chem Biol 21:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyke CK, Steinhubl SR, Kleiman NS, Cannon RO, Aberle LG, Lin M, Myles SK, Melloni C, Harrington RA, et al. (2006). First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation 114:2490–2497 [DOI] [PubMed] [Google Scholar]
- 20.Musumeci D. and Montesarchio D. (2012). Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol Ther 136:202–215 [DOI] [PubMed] [Google Scholar]
- 21.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E. and Sullenger BA. (2008). Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol 15:675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B. and Gilboa E. (2008). Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest 118:376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilboa E, McNamara J 2nd, and Pastor F. (2013). Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin Cancer Res 19:1054–1062 [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Cao Z. and Tan W. (2008). Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci U S A 105:5664–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y, Dennis DM, Morey T, Yang L. and Tan W. (2010). Engineering dendritic aptamer assemblies as superior inhibitors of protein function. Chem Asian J 5:56–59 [DOI] [PubMed] [Google Scholar]
- 26.Nimjee SM, Oney S, Volovyk Z, Bompiani KM, Long SB, Hoffman M. and Sullenger BA. (2009). Synergistic effect of aptamers that inhibit exosites 1 and 2 on thrombin. RNA 15:2105–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangnekar A, Zhang AM, Li SS, Bompiani KM, Hansen MN, Gothelf KV, Sullenger BA. and LaBean TH. (2012). Increased anticoagulant activity of thrombin-binding DNA aptamers by nanoscale organization on DNA nanostructures. Nanomedicine 8:673–681 [DOI] [PubMed] [Google Scholar]
- 28.Subbaiah PV, Bajwa SS, Smith CM. and Hanahan DJ. (1976). Interactions of the components of the prothrombinase complex. Biochim Biophys Acta 444:131–146 [DOI] [PubMed] [Google Scholar]
- 29.Mann KG, Nesheim ME, Tracy PB, Hibbard LS. and Bloom JW. (1982). Assembly of the prothrombinase complex. Biophys J 37:106–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M. and Sullenger B. (2001). Generation of species cross-reactive aptamers using “Toggle” SELEX. Mol Ther 4:567–573 [DOI] [PubMed] [Google Scholar]
- 31.Laszlo GS, Gudgeon CJ, Harrington KH. and Walter RB. (2015). T-cell ligands modulate the cytolytic activity of the CD33/CD3 BiTE antibody construct, AMG 330. Blood Cancer J 5:e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotula JW, Sun J, Li M, Pratico ED, Fereshteh MP, Ahrens DP, Sullenger BA. and Kovacs JJ. (2014). Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS One 9:e93441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrand B, Berezhnoy A, Brenneman R, Williams A, Levay A. and Gilboa E. (2015). Reducing toxicity of 4-1BB costimulation: targeting 4-1BB ligands to the tumor stroma with bi-specific aptamer conjugates. Oncoimmunology 4:e970918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.