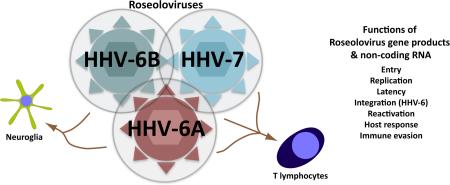
Abstract
Human herpesviruses 6A, −6B, and −7 (HHV-6A, −6B, and −7) are classified within the roseolovirus genus of the betaherpesvrus subfamily. Most humans likely harbor at least two of these large DNA viruses, and 1% of humans harbor germline chromosomally integrated HHV-6A or HHV-6B genomes. Differences at the genetic level manifest as distinct biologic properties during infection and disease. We provide a brief synopsis of roseolovirus replication and highlight the unique properties of their lifecycle and what is known about the viral gene products that mediate these functions. In the nearly 30 years since their discovery, we have only begun to unlock the molecular strategies these highly evolved pathogens employ to establish and maintain chronic infections in humans.
Graphical Abstract
The aims of this review are to provide an overview of roseolovirus molecular biology and highlight recent advances in our understanding of the molecular basis of the virus lifecycle, which in turn inform our understanding of pathogenesis, and illuminate paths to diagnosis, treatment, and prevention.
ROSEOLOVIRUSES: WHAT ARE THEY?
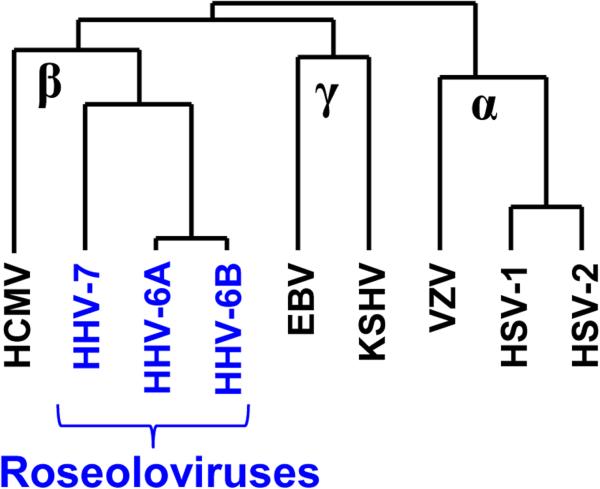
Human herpesviruses 6A, 6B, and 7 (HHV-6A, HHV-6B, and HHV-7) are the only formally recognized members of genus Roseolovirinae within order Herpesvirales, family Herpesviridae, and subfamily Betaherpesvirales (Fig. 1) (historical references are available in [1,2]). HHV-6A and HHV-6B were formerly described as variants, but are now formally classified as distinct virus species by the International Committee on Virus Taxonomy [3]. Roseoloviruses share numerous genetic and biologic properties with human cytomegalovirus (also a betaherpesvirus), yet have distinct genes and disease associations (Tables 1 and 2). The human roseoloviruses are contemporary representatives of an ancient lineage of herpesviruses that co-speciated with their hosts. Antibodies against HHV-6 have been detected in several species of Old and New World monkeys, suggesting the presence of viruses related to HHV-6 in these animals [4]. Consistent with this, relatives of HHV-6 and HHV-7 have been detected by PCR in chimpanzees, other great apes, and pig-tailed macaques [5-7]
Figure 1.
Dendrogram showing relationships among the human herpesviruses, based on sequences of the conserved protein, gB.
Table 1.
Genetic and biological properties of human roseoloviruses and HCMV.
| HHV-6A | HHV-6B | HHV-7 | HCMV | |
|---|---|---|---|---|
| Commonly used strains | U1102, GS | Z29, HST | JI, RK, SB, UCL-1 | AD169, Towne, Merlin, TB40E |
| Length of wild genomes | ? | ~170 kb | ? | 236 kb |
| Length of passaged genomes | 159 kb | 159-162 kb | 145 kb | ~230 kb |
| genes encoding unique proteins | ~102 | ~97 | ~86 | ~165 |
| miRNAs | 4 predicted | 4 | unknown | 16 |
| Replication | slow, extended | |||
| ballooning, refractile cytopathic effect | cytomegaly, nuclear and cytoplasmic inclusions | |||
| origin-binding protein for initiation of DNA replication | ||||
| Cell surface receptor | CD46 | CD134 | CD4 | EGFR, Integrins |
| Cell culture tropism | umbilical cord blood lymphocytes peripheral blood mononuclear cells | monocyte-macrophages | ||
| T cell lines: SupT-1, HSB2, J JAHN | T cell lines: Molt-3, Mt-4, SupT-1 | T cell lines: SupT-1 | CD34+ hematopoietic cells | |
| productive replication in astrocytes | low-level persistence in astrocytes | endothelial and epithelial cells, fibroblasts | ||
| Unique features | integration into host telomeres | |||
| Major disease associations | Hashimoto's thyroiditis | exanthem subitum | congenital birth defects | |
| febrile seizures/ status epilepticus | transplant complications | |||
| transplant complications | retinitis | |||
| post-transplant reactivation-associated encephalitis | hepatitis | |||
Table 2.
Genes unique to roseoloviruses.
| Functiona | Roseolovirus ORF | %S with HHV-6Ab | % I with HHV-6Ac | %S with HHV-7 | %I with HHV-7 |
|---|---|---|---|---|---|
| Roseolovirus specific genes | |||||
| U13 | 93.4 | 92.5 | 44.9 | 35.7 | |
| U15EX1 | 91.4 | 86.7 | 76.4 | 67.9 | |
| U15EX2 | 100 | 95.8 | 83.3 | 75 | |
| U15EX3 | 96.7 | 91.7 | 82 | 75.4 | |
| Glycoprotein | U20 | 95.6 | 95.6 | 31.8 | 22.2 |
| Down-regulation of MHC class I | U21 | 91 | 89.8 | 42.8 | 31.6 |
| Glycoprotein | U23 | 94.6 | 94.1 | 26.9 | 20.9 |
| U24 | 88.3 | 82.7 | 46.2 | 31.2 | |
| U24A | 94.7 | 91.2 | 40.3 | 28.1 | |
| U26 | 93.8 | 92.9 | 60.5 | 47.4 | |
| OX-2 homology, glycoprotein | U85 | 93.1 | 91.7 | 46.9 | 36.8 |
| IE-A (IE1), transactivator | U90EX1 | 73.7 | 68.4 | 42.8 | 35.7 |
| U90EX2 | 70.3 | 67.2 | 67.1 | 57.1 | |
| U90EX3 | 76.7 | 71.5 | 32.9 | 25.2 | |
| IE-A | U91EX1 | 67.8 | 57.1 | 33.3 | 25 |
| U91EX2 | 69.2 | 67.9 | 45.6 | 40 | |
| Spliced envelope glycoprotein; HHV-6 pg82-gp105, HHV-7gp65 | U100EX1 | 78.1 | 73.4 | 27.2 | 19.7 |
| U100EX2 | 84.9 | 81.7 | 53.8 | 38.7 | |
| U100EX3 | 82.9 | 79.3 | 40.9 | 32.7 | |
| U100EX4 | 96 | 88 | 44 | 40 | |
| U100EX5 | 88.6 | 80 | 34.3 | 28.6 | |
| U100EX6 | 91.9 | 91.9 | 48.6 | 37.8 | |
| U100EX7 | 88.7 | 83 | 35.3 | 27.4 | |
| U100EX8 | 100 | 100 | |||
| U100EX9 | 92.8 | 90.5 | 35.7 | 23.8 | |
| U100EX10 | 83.9 | 76.5 | 24 | 13.3 | |
| HHV-6 specific genes | |||||
| DR3 | 87 | 86.4 | |||
| U6 | 97.1 | 97.1 | |||
| U9 | 94.2 | 94.2 | |||
| Glycoprotein | U22 | 91.2 | 89.6 | ||
| Intercrine cytokine | U83 | 87.6 | 85.6 | ||
| Parvovirus rep homolog | U94 | 98.4 | 97.6 | ||
| HHV-6A gene | |||||
| U78 | |||||
| HHV-6B genes | |||||
| B3,B4,B5,B6,B7,B8 | |||||
Implied functions of homologous genes or experimental validation.
Percentage of amino acid similarity between homologs.
Percentage of amino acid identity between homologs.
ROSEOLOVIRUSES AND HUMAN HEALTH
HHV-6B is the most common cause of roseola infantum (exanthem subitum) and related febrile rash illnesses that often accompany primary infection in early childhood [8]; this can also be caused by HHV-7. HHV-6B and HHV-7 have also been associated with febrile seizures in young children. Immune suppressed hemopoietic stem cell transplant recipients can experience limbic encephalitis and other mental disorders during HHV-6B reactivations [9]. HHV-6A has been associated with Hashimoto's thyroiditis [10] and neurological disorders, including multiple sclerosis, but proof of causality is incomplete [11].
A striking feature of roseoloviruses is the presence of mammalian telomeric sequences at the ends of the virus genome [12-14]. Approximately 1% of the human population world-wide harbors inherited chromosomally integrated (ci) HHV-6A and HHV-6B. Germline integration may be a byproduct of the use of integration as a hypothesized mechanism for establishing latency in somatic cells, with virus infection of spermatocytes leading to occasional germline transmission. The health effects of ciHHV-6 have not been elucidated.
ROSEOLOVIRUS GENOMES AND GENES
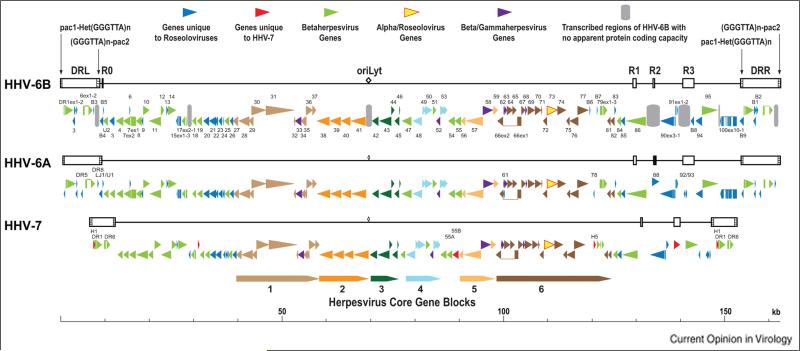
Roseoloviruses genomes consist of a long unique region (U) bracketed by a pair of direct repeats (DR) (Fig. 2). Roseolovirus genomes have heterogeneous and perfect arrays of mammalian telomeric repeats at the left and right ends of the DR elements, respectively, and consequently at the left and right genomic termini. At least for HHV-6B, genomes of wild viruses can be several kb longer than those of laboratory-adapted strains, due to repetitive sequences in the DR that are lost upon passage in cultured cells. Roseolovirus genomes are approximately 65 to 90 kb shorter than the 235 kb HCMV genome. The origins of lytic genome replication (oriLyt) are located between U41 and U42, and are structurally similar to oriLyts of alphaherpesviruses.
Figure 2.
Genomic and genetic architectures of the human roseoloviruses. Based on information from [15,63-67].
HHV-6A and HHV-6B are ~90% identical across their genomes, with ~95% identity across the herpesvirus core genes. Regions in the vicinity of the genomic termini are less conserved, with as little as 50% identity in the region that encodes the major immediate early transactivators [15]. While its overall organization and gene content are similar to those of HHV-6A and HHV-6B, the HHV-7 genome is shorter and more compactly arranged across its length, with many genes being 5 to 10% shorter than their HHV-6 counterparts. In intrastrain comparisons, roseolovirus genomes are typically ~99.9% identical, except for pockets of elevated heterogeneity.
The core herpesvirus genes (43 genes conserved among members of the Herpesviridae) are clustered across the central portion of the genomes in an arrangement colinear with the core genes in HCMV and other betaherpesviruses. In contrast to HCMV and most other betaherpesviruses, the roseoloviruses, along with elephant endotheliotropic herpesviruses, encode homologs (roseolovirus gene U73) of the origin of DNA replication binding protein (OBP) encoded by all alphaherpesviruses. Most of the genes shared only among betaherpesviruses or unique to one or more roseoloviruses lie in or near the DR, or between conserved gene blocks (Table 2 and Fig. 2).
HHV-6B expresses several small RNAs of unknown function, including some that map to oriLyt and microRNAs that map to the DR3/B1 and B2 immediate early gene locus in DR. These miRNAs are conserved in HHV-6A, and one is an ortholog of human miRNA miR-582-5p [16].
Major functions of many roseolovirus genes are known only by inference from known functions of their homologs in HCMV or other herpesviruses. Most virion proteins are likely to have significant biological roles that go beyond structural, such as tegument proteins that modify host cell activities before de novo viral gene expression begins. Only a handful of genes unique to roseoloviruses have been studied functionally. These include transactivators encoded by DR6 and U3, the U94 parvovirus rep gene homolog, immunoevasins encoded by U21, a non-essential Golgi-localizing non-structural glycoprotein encoded by U23 [17], and the gQ1 and gQ2 glycoproteins.
Major research priorities include assessment of genome sequences and genetic variation of wild viruses, and identification of the functions of genes unique to roseoloviruses. A bacterial artificial chromosome (BAC) system has enabled targeted genetic analysis for HHV-6A [18]; analogous systems are needed for HHV-6B and HHV-7.
PRODUCTIVE REPLICATION
Roseolovirus tropism: beyond T cells
The human roseoloviruses were discovered on the basis of their lytic replication activity in cultured PBMCs. Some strains have adapted to growth in specific T cell lines and are commonly used for laboratory studies. Other cell types such as monocytes, dendritic cells, astrocytes, and glial cells are permissive for infection. HHV6A and HHV-6B can bind to the sperm acrosome, providing a possible route to germline integration [19]. The ability of HHV-6A and HHV-6B to infect olfactory-ensheathing glial cells that are present in the nasal cavity may provide a route to the central nervous system [20].
Mechanisms of attachment and entry are important determinants of cell tropism and latency reservoirs in the host. Each roseolovirus has a distinct entry receptor: CD46 for HHV-6A [21], CD134 for HHV-6B [22], and CD4 for HHV-7 [23]. Receptors are targets for neutralization [24] and can be used to create receptor-transgenic animal models that support infection [25]. The essential components for membrane fusion by HHV-6A and HHV-6B are gB and the gH/gL/gQ1/gQ2 complex [26-28]. gQ2 and gM are essential for virus production of HHV-6A since virus stocks could not be generated from BACs with disruptions in these ORFs [28,29]. The degree of functional homology between roseolovirus genes can be examined in transcomplementation assays and by gene substitutions in the HHV-6A BAC. For instance, the HHV-6B gH gene can functionally replace HHV-6A gH for replication [30].
De novo gene expression and productive replication
Roseolovirus lytic gene expression follows the general herpesvirus paradigm: immediate early genes are transcribed in the absence of new protein synthesis, expression of early genes is dependent on prior synthesis or immediate early proteins, and late genes are expressed at high levels upon viral DNA replication. Approximately 10 genes have spliced transcripts (some have multiple spliced isoforms), and some transcripts are kinetically regulated. Roseolovirus major IE genes are spliced and have promoters that can be highly active in T cells.
Roseoloviruses diverge from most betaherpesviruses in their mechanism of initiating viral DNA replication. Their homologs of the alphaherpesvirus origin binding protein bind to, and presumably facilitate unwinding of the origin of lytic replication to initiate viral DNA synthesis [31]. The OBPs of HHV-6B and HHV-7 have slight differences in preferential binding sites that may explain a lack of complete reciprocity between HHV-6B and HHV-7 in transient oriLyt replication assays.
Information about HHV-6 virion assembly and egress is sparse. An interesting feature of HHV-6A virion envelopes is the presence of ganglioside GM1, a component of lipid rafts [32]. Along with other evidence, this suggests that virions may assemble via lipid rafts. Envelopment and egress are via a cellular CD63-associated exosomal pathway [33].
LATENCY and REACTIVATION
Gene expression during latency
Roseolovirus latency is poorly defined in molecular terms. CD34-positive hematopoietic cells are a site of HHV-6 latency, and circulating lymphocytes positive for HHV-7 DNA but not for lytic gene transcripts have been detected. Latency associated transcripts have been identified in two loci: antisense to the major IE locus, with splicing patterns reminiscent of an HCMV latency transcript [34], and from the U94 gene [35]. No laboratory has reported detection of both of these transcripts.
Integration
One of the most unique and biologically intriguing aspects of HHV-6A and HHV-6B is their integration into the germline of some humans (~1%), which can result in inherited transmission among families [14]. All three human roseoloviruses contain mammalian telomeric sequences at their genomic termini, and telomeres are the site of integration of HHV-6A and HHV-6B in patients with chromosomally integrated HHV-6 [36,37]. Telomeric integration occurs in infected cultured Jjhan and HEK-293 cells, establishing a system for mapping and characterizing the mechanistic processes of integration. The efficiency of integration in cultured cells has led to the hypothesis that chromosomal integration is a normal part of HHV-6 latency.
The U94 gene of HHV-6A and HHV-6B is a homolog of the parvovirus Rep gene, an integrase with single-stranded and double-stranded DNA binding properties. Cytomegaloviruses of rats [38] and bats [39] encode U94 homologs, indicating that the gene may have been acquired prior to the divergence of roseoloviruses and cytomegaloviruses. HHV-6 U94 binds ssDNA [40] and its ectopic expression inhibits betaherpesvirus replication [41] and impairs lymphatic endothelial cell angiogenesis [42]. Given its homology with the parvovirus integrase, U94 is hypothesized to promote integration and excision of HHV-6A and HHV-6B, either by host-mediated base excision repair or by exonuclease strand invasion [14]. The transcriptome of ciHHV-6 cells has not been reported, but spliced U90 transcripts have been detected in B cells harboring integrated HHV-6 [43]. Genome-wide analyses of viral and cellular gene expression are needed in individuals with ciHHV-6 and in ciHHV-6 cell culture systems.
Reactivation
Uncontrolled or aberrant primary infection and HHV-6 reactivation are associated with neurological syndromes and transplant failure. Very little is known about the molecular basis of reactivation. Mitogen stimulation of PBMCs leads to reactivation and enables infection of T cell lines. Lytic replication can also be stimulated by apoptosis [44]. If integration is a mechanism of latency, a functional virus genome must be excised from telomeres in order to reactivate full lytic infection. HEK293 cells with integrated HHV-6A can produce viral genome concatamers upon treatment with the histone deacetylase inhibitor trichostatin A [36]. Huang et al. [43] noted that the telomeres attached to integrated HHV-6 genomes are frequently shortened and associated with detection of circular viral genomes. Such short, unstable telomeres are thought to facilitate excision of viral genomes via telomere-loops within the viral genome [43]. Interestingly, Chlamydia trachomatis drives reactivation of ciHHV-6 and transient shortening of telomere ends [45]; the signaling pathways and mechanism of excision remain to be defined.
VIRUS-HOST INTERACTIONS
All herpesviruses manipulate host cell processes to promote replication. Roseoloviruses push the cell cycle into G2/M, presumably to ramp up cellular processes that promote DNA replication [46]. Virally-induced degradation of Rb and activation of E2F1 further benefits HHV-6A and HHV-6B by enhancing the expression of some lytic genes [47]. Many roseolovirus gene products inhibit both innate immune responses (U20, IE1) and adaptive immune responses (U21), and interfere with cell death (U19, U20, DR6) and T cell signaling (U21, U54) [48]. Functions should not be assumed to be conserved among all roseolovirus homologs. Virus-specific differences in gene function such as U54 modulation of IL-2 signaling, the chemotactic properties of the roseolovirus U83 chemokines, and IE1 inhibition of interferon stimulated genes have been noted [49-51]. BAC-based recombinant viruses will facilitate examination of gene function in the context of infection.
Roseoloviruses impact cytokine profiles of cultured cells [52,53]. Cytokine dysregulation also occurs in patients undergoing acute illness associated with primary infection [54-56] and reactivation [57], and in animal models of HHV-6 infection [25,58]. The viral gene products that induce these changes in host signaling are not known. Inactivated virions induce an interferon-lambda 1 (IL-29) response in dendritic cells that might skew T cell responses to infection [59]. The host immune response may play a large role in the immune pathology of reactivation-associated diseases and facilitate roseolovirus transit across the blood brain barrier [60]. In addition, HHV-6B reactivation might be triggered in response to pro-inflammatory cytokines such as TNF-alpha and immunosuppression with corticosteroids. Such a mechanism might contribute to the frequent detection of HHV-6B reactivation in patients diagnosed with Drug Reaction with Eosinophilia and Systemic Symptom (DRESS), a potential fatal syndrome initiated by adverse drug reactions [61,62].
Understanding the functional changes described above will be enhanced by deep analysis of the effects of roseolovirus infection on host cell transcription, translation, and export of gene products.
RESEARCH PRIORITIES
Understanding of the molecular virology of roseoloviruses lags behind that for all other human herpesviruses. Understanding the genetic content of roseoloviruses has not been extended far beyond basic sequence analysis of laboratory-adapted strains. Modern methods of DNA sequencing need to be applied to understanding the sequence composition of wild, uncultured roseolovirus genomes, as well as inter- and intrahost sequence variation at the genome level in immune competent and immune compromised individuals. Among other things, such genetic analyses are necessary to ensure that animal studies and other experiments are done with viruses that appropriately represent wild viruses. Functional analysis of the the genes unique to roseoloviruses and betaherpesviruses will provide information as to how these viruses have adapted to their specific and specialized niches. Genetic approaches using BAC-based recombination strategies are critical to identify the viral factors and cis-determinants of replication, integration, and reactivation. Even in the absence of well-established genetically tractable systems for HHV-6B and HHV-7, transcript and proteomic profiles can rapidly confirm putative genes and identify novel ORFs, novel transcript forms, and non-coding RNAs. Vaccine development typically involves attenuation, but intelligently designed attenuation will not be possible for the roseoloviruses without fundamental knowledge of replication and host interaction determinants.
SUMMARY
Roseolovirues have unique cellular tropisms and biological properties, and encode ORFs distinct from the other human betaherpesvirus, HCMV. Each HHV-7 gene has a homolog in HHV-6A and HHV-6B. However, HHV-6A and HHV-6B have several genes not found in HHV-7, including a homolog of the parvovirus rep protein, U94. Roseolovirus gene products mediate cell entry and viral replication, modulate the host cell's growth, survival, signaling, and immune responses, and regulate latency. RNA analyses and proteomics coupled with new genetic tools and advances in systems biology are needed to advance the identification and function of known, as well as uncharacterized and novel ORFs, and transcripts such as miRNAs. Advancements in understanding roseolovirus gene function will reveal novel virus-host interactions and better define the mechanism of integration and excision of the virus genome into and from host chromosomes, a potential form of latency that would be unique among the human herpesviruses. Investments in understanding the fundamental molecular processes of roseolovirus infections will inform our understanding of the dynamic process of persistence and disease in humans and identify targets for therapeutic intervention.
Highlights.
HHV-6A, HHV-6B, and HHV-7 are distinct members of the Roseolovirus genus.
HHV-6A and HHV-6B can integrate into the telomeres of the host chromosome.
Lytic infection involves regulated expression of viral proteins and non-coding RNAs.
Infection alters cellular processes, and innate and adaptive immune responses.
Many aspects of their molecular biology remain to be defined.
ACKNOWLEDGEMENTS
L.T.K. was supported by an American Cancer Society Research Scholar Grant RSG-11-160-01-MPC and NIH AI111129-01 and AI097875-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Braun DK, Dominguez G, Pellett PE. Human herpesvirus 6. Clinical Microbiology Reviews. 1997:521–567. doi: 10.1128/cmr.10.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanishi K, Mori Y, Pellett PE. Human herpesviruses 6 and 7. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B, editors. Fields Virology. edn 6. Vol. 2. Lippincott Williams & Wilkins; 2013. pp. 2058–2079. Chapt. 64. [Google Scholar]
- 3.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol. 2014;159:863–870. doi: 10.1007/s00705-013-1902-5. 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higashi K, Asada H, Kurata T, Ishikawa K, Hayami M, Spriatna Y, Sutarman, Yamanishi K. Presence of antibody to human herpesvirus 6 in monkeys. J Gen Virol. 1989;70(Pt 12):3171–3176. doi: 10.1099/0022-1317-70-12-3171. [DOI] [PubMed] [Google Scholar]
- 5.Lacoste V, Verschoor EJ, Nerrienet E, Gessain A. A novel homologue of Human herpesvirus 6 in chimpanzees. J Gen Virol. 2005;86:2135–2140. doi: 10.1099/vir.0.81034-0. 10.1099/vir.0.81034-0. [DOI] [PubMed] [Google Scholar]
- 6.Lavergne A, Donato D, Gessain A, Niphuis H, Nerrienet E, Verschoor EJ, Lacoste V. African great apes are naturally infected with roseoloviruses closely related to Human herpesvirus 7. J Virol. 2014 doi: 10.1128/JVI.01490-14. 10.1128/JVI.01490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staheli JP, Dyen MR, Lewis P, Barcy S. Discovery and biological characterization of two novel pig-tailed macaque homologs of HHV-6 and HHV-7. Virology. 2014 doi: 10.1016/j.virol.2014.10.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tesini B, Epstein LG, Caserta MT. Clinical impact of primary infection with roseoloviruses. Curr Opin Virol. 2014;9C doi: 10.1016/j.coviro.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill J, Zerr DM. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol. 2014;9C doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caselli E, Zatelli MC, Rizzo R, Benedetti S, Martorelli D, Trasforini G, Cassai E, degli Uberti EC, Di Luca D, Dolcetti R. Virologic and immunologic evidence supporting an association between HHV-6 and Hashimoto's thyroiditis. PLoS Pathog. 2012;8:e1002951. doi: 10.1371/journal.ppat.1002951. 10.1371/journal.ppat.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leibovitch E, Jacobson S. An update on the evidence linking HHV-6 with multiple sclerosis. Curr Opin Virol. 2014;9C doi: 10.1016/j.coviro.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 13**.Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, Flamand L, Gautheret-Dejean A, Hall CB, Kamble RT, et al. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 2012;22:144–155. doi: 10.1002/rmv.715. 10.1002/rmv.715. [An international collaboration of many experts in the field that addresses the consequences of HHV-6A and HHV-6B integration into the host chromosome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufer BB, Flamand L. Chromosomally integrated HHV-6: impacts on virus, cell and organismal biology. Curr Opin Virol. 2014;9C doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol. 2012;86:1638–1649. doi: 10.1128/JVI.05911-11. 10.1128/JVI.05911-11. [Identification of HHV-6B miRNAs and other noncoding RNAs expressed during lytic infection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi M, Yoshida K, Tang H, Sadaoka T, Kawabata A, Jasirwan C, Mori Y. Characterization of the human herpesvirus 6A U23 gene. Virology. 2014;450-451:98–105. doi: 10.1016/j.virol.2013.12.004. 10.1016/j.virol.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Tang H, Kawabata A, Yoshida M, Oyaizu H, Maeki T, Yamanishi K, Mori Y. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology. 2010;407:360–367. doi: 10.1016/j.virol.2010.08.018. 10.1016/j.virol.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Kaspersen MD, Larsen PB, Kofod-Olsen E, Fedder J, Bonde J, Höllsberg P. Human herpesvirus-6A/B binds to spermatozoa acrosome and is the most prevalent herpesvirus in semen from sperm donors. PLoS One. 2012;7:e48810. doi: 10.1371/journal.pone.0048810. 10.1371/journal.pone.0048810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harberts E, Yao K, Wohler JE, Maric D, Ohayon J, Henkin R, Jacobson S. Human herpesvirus-6 entry into the central nervous system through the olfactory pathway. Proc Natl Acad Sci U S A. 2011;108:13734–13739. doi: 10.1073/pnas.1105143108. 10.1073/pnas.1105143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 22*.Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, Yamanishi K, Mori Y. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc Natl Acad Sci U S A. 2013;110:9096–9099. doi: 10.1073/pnas.1305187110. 10.1073/pnas.1305187110. [Discovery of the HHV-6B cellular entry receptor.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusso P, Secchiero P, Crowley RW, Garzino-Demo A, Berneman ZN, Gallo RC. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci U S A. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawabata A, Oyaizu H, Maeki T, Tang H, Yamanishi K, Mori Y. Analysis of a neutralizing antibody for human herpesvirus 6B reveals a role for glycoprotein Q1 in viral entry. J Virol. 2011;85:12962–12971. doi: 10.1128/JVI.05622-11. 10.1128/JVI.05622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Reynaud JM, Jégou J-F, Welsch JC, Horvat B. Human herpesvirus 6A infection in CD46 transgenic mice: viral persistence in the brain and increased production of proinflammatory chemokines via Toll-like receptor 9. J Virol. 2014;88:5421–5436. doi: 10.1128/JVI.03763-13. 10.1128/JVI.03763-13. [Development of an animal model based on mice that express human CD46, the HHV-6A receptor, with evidence of HHV-6A in the brain.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeki T, Hayashi M, Kawabata A, Tang H, Yamanishi K, Mori Y. Identification of the human herpesvirus 6A gQ1 domain essential for its functional conformation. J Virol. 2013;87:7054–7063. doi: 10.1128/JVI.00611-13. 10.1128/JVI.00611-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeki T, Mori Y. Features of human herpesvirus-6A and -6B entry. Adv Virol. 2012;2012:384069. doi: 10.1155/2012/384069. 10.1155/2012/384069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Tang H, Hayashi M, Maeki T, Yamanishi K, Mori Y. Human herpesvirus 6 glycoprotein complex formation is required for folding and trafficking of the gH/gL/gQ1/gQ2 complex and its cellular receptor binding. J Virol. 2011;85:11121–11130. doi: 10.1128/JVI.05251-11. 10.1128/JVI.05251-11. [Development and application of an HHV-6A BAC suitable for genetic analysis of viral gene functions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawabata A, Jasirwan C, Yamanishi K, Mori Y. Human herpesvirus 6 glycoprotein M is essential for virus growth and requires glycoprotein N for its maturation. Virology. 2012;429:21–28. doi: 10.1016/j.virol.2012.03.027. 10.1016/j.virol.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Oyaizu H, Tang H, Ota M, Takenaka N, Ozono K, Yamanishi K, Mori Y. Complementation of the function of glycoprotein H of human herpesvirus 6 variant A by glycoprotein H of variant B in the virus life cycle. J Virol. 2012;86:8492–8498. doi: 10.1128/JVI.00504-12. 10.1128/JVI.00504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krug LT, Inoue N, Pellett PE. Sequence requirements for interaction of human herpesvirus 7 origin binding protein with the origin of lytic replication. J Virol. 2001;75:3925–3936. doi: 10.1128/JVI.75.8.3925-3936.2001. 10.1128/JVI.75.8.3925-3936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawabata A, Tang H, Huang H, Yamanishi K, Mori Y. Human herpesvirus 6 envelope components enriched in lipid rafts: evidence for virion-associated lipid rafts. Virol J. 2009;6:127. doi: 10.1186/1743-422X-6-127. 10.1186/1743-422X-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic. 2008;9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo K, Shimada K, Sashihara J, Tanaka-Taya K, Yamanishi K. Identification of human herpesvirus 6 latency-associated transcripts. J Virol. 2002;76:4145–4151. doi: 10.1128/JVI.76.8.4145-4151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci U S A. 1998;95:13911–13916. doi: 10.1073/pnas.95.23.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arbuckle JH, Pantry SN, Medveczky MM, Prichett J, Loomis KS, Ablashi D, Medveczky PG. Mapping the telomere integrated genome of human herpesvirus 6A and 6B. Virology. 2013;442:3–11. doi: 10.1016/j.virol.2013.03.030. 10.1016/j.virol.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. 10.1073/pnas.0913586107. [This landmark paper described the telomeric targets of HHV-6A chromosomal integration in human specimens and in a cell culture model.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Cleef KWR, Scaf WMA, Maes K, Kaptein SJF, Beuken E, Beisser PS, Stassen FRM, Grauls GELM, Bruggeman CA, Vink C. The rat cytomegalovirus homologue of parvoviral rep genes, r127, encodes a nuclear protein with single- and double-stranded DNA-binding activity that is dispensable for virus replication. J Gen Virol. 2004;85:2001–2013. doi: 10.1099/vir.0.79864-0. 10.1099/vir.0.79864-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Todd S, Tachedjian M, Barr JA, Luo M, Yu M, Marsh GA, Crameri G, Wang L-F. A novel bat herpesvirus encodes homologues of major histocompatibility complex classes I and II, C-type lectin, and a unique family of immune-related genes. J Virol. 2012;86:8014–8030. doi: 10.1128/JVI.00723-12. 10.1128/JVI.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhepakson P, Mori Y, Jiang YB, Huang HL, Akkapaiboon P, Okuno T, Yamanishi K. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J Gen Virol. 2002;83:847–854. doi: 10.1099/0022-1317-83-4-847. [DOI] [PubMed] [Google Scholar]
- 41.Caselli E, Bracci A, Galvan M, Boni M, Rotola A, Bergamini C, Cermelli C, Dal Monte P, Gompels UA, Cassai E, et al. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology. 2006;346:402–414. doi: 10.1016/j.virol.2005.11.018. 10.1016/j.virol.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Caruso A, Caselli E, Fiorentini S, Rotola A, Prandini A, Garrafa E, Saba E, Alessandri G, Cassai E, Di Luca D. U94 of human herpesvirus 6 inhibits in vitro angiogenesis and lymphangiogenesis. Proc Natl Acad Sci U S A. 2009;106:20446–20451. doi: 10.1073/pnas.0905535106. 10.1073/pnas.0905535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Huang Y, Hidalgo-Bravo A, Zhang E, Cotton VE, Mendez-Bermudez A, Wig G, Medina-Calzada Z, Neumann R, Jeffreys AJ, Winney B, et al. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. 2014;42:315–327. doi: 10.1093/nar/gkt840. 10.1093/nar/gkt840. [Discovery that somatic cell chromosomes with integrated HHV-6 have short telomeres, and evidence that telomeric loop formation is involved in excision of ciHHV-6 genomes during viral reactivation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad A, Remick J, Zeichner SL. Activation of human herpesvirus replication by apoptosis. J Virol. 2013;87:10641–10650. doi: 10.1128/JVI.01178-13. 10.1128/JVI.01178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prusty BK, Siegl C, Hauck P, Hain J, Korhonen SJ, Hiltunen-Back E, Puolakkainen M, Rudel T. Chlamydia trachomatis infection induces replication of latent HHV-6. PLoS One. 2013;8:e61400. doi: 10.1371/journal.pone.0061400. 10.1371/journal.pone.0061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shamir E, Zeigerman H, Frenkel N. Roseoloviruses: viral host interactions and cell cycle arrest. Curr Opin Virol. 2014;9C doi: 10.1016/j.coviro.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Sharon E, Volchek L, Frenkel N. Human herpesvirus 6 (HHV-6) alters E2F1/Rb pathways and utilizes the E2F1 transcription factor to express viral genes. Proc Natl Acad Sci U S A. 2014;111:451–456. doi: 10.1073/pnas.1308854110. 10.1073/pnas.1308854110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudson A. Roseoloviruses: a treasure trove of immune modulators. Curr Opin Virol. 2014;9C [Google Scholar]
- 49.Iampietro M, Morissette G, Gravel A, Flamand L. Inhibition of interleukin-2 gene expression by human herpesvirus 6B U54 tegument protein. J Virol. 2014;88:12452–12463. doi: 10.1128/JVI.02030-14. 10.1128/JVI.02030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark DJ, Catusse J, Stacey A, Borrow P, Gompels UA. Activation of CCR2+ human proinflammatory monocytes by human herpesvirus-6B chemokine N-terminal peptide. J Gen Virol. 2013;94:1624–1635. doi: 10.1099/vir.0.050153-0. 10.1099/vir.0.050153-0. [DOI] [PubMed] [Google Scholar]
- 51.Jaworska J, Gravel A, Flamand L. Divergent susceptibilities of human herpesvirus 6 variants to type I interferons. Proc Natl Acad Sci U S A. 2010;107:8369–8374. doi: 10.1073/pnas.0909951107. 10.1073/pnas.0909951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi J, Wang F, Li L, Feng D, Qin J, Xie F, Zhou F, Chen Y, Wang J, Yao K. The role of MAPK in CD4(+) T cells toll-like receptor 9-mediated signaling following HHV-6 infection. Virology. 2012;422:92–98. doi: 10.1016/j.virol.2011.09.026. 10.1016/j.virol.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 53.Murakami Y, Tanimoto K, Fujiwara H, An J, Suemori K, Ochi T, Hasegawa H, Yasukawa M. Human herpesvirus 6 infection impairs Toll-like receptor signaling. Virol J. 2010;b:91. doi: 10.1186/1743-422X-7-91. 10.1186/1743-422X-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci U S A. 2013;b:12792–12797. doi: 10.1073/pnas.1302968110. 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawamura Y, Yamazaki Y, Ohashi M, Ihira M, Yoshikawa T. Cytokine and chemokine responses in the blood and cerebrospinal fluid of patients with human herpesvirus 6B-associated acute encephalopathy with biphasic seizures and late reduced diffusion. J Med Virol. 2014;86:512–518. doi: 10.1002/jmv.23788. 10.1002/jmv.23788. [DOI] [PubMed] [Google Scholar]
- 56.Kawamura Y, Nakai H, Sugata K, Asano Y, Yoshikawa T. Serum biomarker kinetics with three different courses of HHV-6B encephalitis. Brain Dev. 2013;35:590–595. doi: 10.1016/j.braindev.2012.08.005. 10.1016/j.braindev.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 57.Kawamura Y, Sugata K, Ihira M, Mihara T, Mutoh T, Asano Y, Yoshikawa T. Different characteristics of human herpesvirus 6 encephalitis between primary infection and viral reactivation. J Clin Virol. 2011;51:12–19. doi: 10.1016/j.jcv.2011.02.002. 10.1016/j.jcv.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Tanner A, Carlson SA, Nukui M, Murphy EA, Berges BK. Human herpesvirus 6A infection and immunopathogenesis in humanized Rag2−/− γc−/− mice. J Virol. 2013;87:12020–12028. doi: 10.1128/JVI.01556-13. 10.1128/JVI.01556-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nordström I, Eriksson K. HHV-6B induces IFN-lambda1 responses in cord plasmacytoid dendritic cells through TLR9. PLoS One. 2012;7:e38683. doi: 10.1371/journal.pone.0038683. 10.1371/journal.pone.0038683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kittaka S, Hasegawa S, Ito Y, Ohbuchi N, Suzuki E, Kawano S, Aoki Y, Nakatsuka K, Kudo K, Wakiguchi H, et al. Serum levels of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinases-1 in human herpesvirus-6-infected infants with or without febrile seizures. J Infect Chemother. 2014 doi: 10.1016/j.jiac.2014.07.017. 10.1016/j.jiac.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 61.Uno H, Kabashima K, Tohyama M, Watanabe T, Hashimoto K, Iijima M, Sueki H, Watanabe H. J Dermatol Sci. Vol. 74. Netherlands: 2014. TNF-α as a useful predictor of human herpesvirus-6 reactivation and indicator of the disease process in drug-induced hypersensitivity syndrome (DIHS)/drug reaction with eosinophilia and systemic symptoms (DRESS). pp. 177–179. [DOI] [PubMed] [Google Scholar]
- 62.Ishida T, Kano Y, Mizukawa Y, Shiohara T. The dynamics of herpesvirus reactivations during and after severe drug eruptions: their relation to the clinical phenotype and therapeutic outcome. Allergy. 2014;69:798–805. doi: 10.1111/all.12410. 10.1111/all.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, et al. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 65.Megaw AG, Rapaport D, Avidor B, Frenkel N, Davison AJ. The DNA sequence of the RK strain of human herpesvirus 7. Virology. 1998;244:119–132. doi: 10.1006/viro.1998.9105. 10.1006/viro.1998.9105. [DOI] [PubMed] [Google Scholar]
- 66.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Øster B, Höllsberg P. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J Virol. 2002;76:7578–7586. doi: 10.1128/JVI.76.15.7578-7586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]