Abstract
The rapidly growing area of “metabolomics,” in which a large number of metabolites from body fluids, cells or tissue are detected quantitatively, in a single step, promises immense potential for a number of disciplines including early disease diagnosis, therapy monitoring, systems biology, drug discovery and nutritional science. Because of its ability detect a large number of metabolites in intact biological samples reproducibly and quantitatively, nuclear magnetic resonance (NMR) spectroscopy has emerged as one of the most powerful analytical techniques in metabolomics. NMR spectroscopy of biological samples with isotope labeling of metabolites using nuclei such as 2H, 13C, 15N and 31P, either in vivo or ex vivo, has dramatically improved our ability to identify low concentrated metabolites and trace important metabolic pathways. Considering the somewhat limited sensitivity of NMR and high complexity of the spectra of biological samples, increased sensitivity and selectivity achieved through isotope labeling methods pave novel avenues to unravel biological complexity and understand cellular functions in health and various disease conditions. This chapter describes the current developments in isotope labeling of metabolites in vivo as well as ex vivo, and their potential metabolomics applications.
2. Metabolomics
Metabolomics, also sometimes referred to as metabonomics or metabolic profiling, is defined as “the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modifications” [1]. In metabolomics, a large number of small-molecule metabolites (typically less than 1000 Da) from body fluids, cells or tissue samples are detected quantitatively in a single step, and then analyzed typically with multivariate statistical methods to yield information that is essential for systems biology, drug discovery, toxicology, food and nutrition sciences and other studies. Metabolites in biological systems represent the end products of genes, transcripts and proteins functions, and therefore the study of concentrations and fluxes of such metabolites provides extremely important information for understanding the composition and function of biochemical networks. Hence, metabolomics may lead to solutions to many important questions related to human disease diagnosis, prognosis and therapeutic development [2-8].
The importance of metabolomics for biomarker discovery stems from the fact that a modest change in enzyme activity can cause substantial perturbations in the metabolic profile; identification of such perturbations represents a sensitive measure of the onset of many diseases. In addition, metabolomics enables the establishment of biological variation among individuals, and understanding such variations is important for achieving the goal of ‘personalized medicine’, which represents a new treatment procedure tailored to the individual's needs. In principle, this personalized approach could be based on the metabolic fingerprint of the patient [9]. A major advantage of the metabolomics approach over other related ‘omic’ areas such as genomics, transcriptomics and proteomics is its association with high throughput measurements, both quantitatively and reproducibly, using non-invasive or minimally invasive approaches, and its minimal requirements regarding sample preparation procedures. Several hundreds of metabolites can be characterized in parallel using modern analytical techniques such as high-resolution NMR spectroscopy, mass spectrometry (MS), or even Fourier transform infrared spectroscopy (FTIR) and electrochemical detection arrays, that provide efficient methods for monitoring altered biochemistry. However, because of the wealth of information they provide, the primary analytical methods that have been widely employed in the field of metabolomics are NMR, which we focus in this chapter, and MS. After a brief review of standard NMR methods used in metabolmics, we focus on isotope enhanced methods.
2.1 NMR spectroscopy in metabolomics
NMR spectroscopy is advantageous for metabolomics studies because it requires little or no sample preparation; is rapid, non-destructive, and non-invasive; and provides highly reproducible and easily quantifiable data. Moreover, NMR spectra can be reliably assigned to specific metabolic species, based on their chemical shifts and multiplet patterns; thus, NMR provides a wealth of information on the identity and quantity of a large number of metabolites in parallel from a single experiment [7, 10-13]. Independent of molecular identity, a particular type of nuclei (1H, 2H, 13C, 15N, 31P, etc.) are detected at the same sensitivity in one NMR experiment, and the absolute quantification of different metabolites can therefore be measured with a single internal or external standard. With advanced high-throughput NMR methodology, up to 200 samples can be measured within a day with the assistance of flow-injection probes and automated liquid handlers [7].
2.2 Biological samples used in metabolomics
To date, blood serum/plasma and urine are the most studied biofluids in the area of metabolomics. This is because they both contain hundreds to thousands of detectable metabolites and can be obtained minimally or non-invasively. Blood maintains a normal homeostasis in the human body by constant regulatory mechanisms. It perfuses all living cells and hence is presumed to carry vital information on virtually every cell in the human body. Hence metabolic profiling of blood serum or plasma enables global visualization of the metabolic status. The NMR spectrum of blood serum/plasma includes both narrow lines from small-molecule metabolites and broad lines from macromolecules such as proteins and lipids. A variety of spectral editing methods are often used to suppress the macromolecular components effectively and detect the small molecules selectively. Because of the relatively low concentrations of proteins and high concentrations of low molecular weight metabolites in urine, metabolomics study of urine is relatively simple in terms of the sample preparation and the NMR experiments. However, a major problem with urine results from its high salt concentration and varying pH across a sample set. In addition, variability in concentration due to dilution effects leads to spectra with significantly different integrated intensities (although normalization can compensate for this effect). Metabolic profiling of urine is particularly useful for studying organ toxicity and drug metabolism in animal models using a variety of drugs or toxins [14, 15]. Other fluids such as cerebrospinal fluid, bile, seminal fluid, amniotic fluid, synovial fluid, gut aspirate, and saliva have also been studied [16-20]. Studies have also involved intact tissues and its lipid and aqueous extracts [21]. Metabolic studies of cells including yeast [22], bacterium, tumor cells, and tissue spheroids have also been reported [23, 24]. Specifically, metabolic profiling of intact tissue has gained increased interest for understanding the molecular basis of diseases [25-27]. This is because, biomarkers due to pathophysiological stress are considered to be highly concentrated in the tissue due to their close association with the pathological source, such as tumors. The rich metabolic profile of tissue is thought to be particularly useful for guiding the detection of biomarkers in relatively easily accessible biofluids. The latest technological advancements in NMR have reduced the required sample quantity to a few mg such that even the biopsy tissue is sufficient to obtain good-quality NMR spectra with resolution that is often comparable to solution-state spectra. Detailed procedures to collect, store, and prepare biofluids or tissue samples for NMR analysis have been provided as guidelines for metabolomics applications [28, 29].
2.3 NMR experiments used in metabolomics
2.3.1 Common NMR experiments
A variety of NMR experiments are used to analyze complex samples such as biofluids, tissue and cells. 1D NOESY (nuclear Overhauser enhancement spectroscopy) and CPMG (Carr-Purcell-Meiboom-Gill) are the two most commonly used experiments. The 1D NOESY is particularly useful for samples with inherently narrow line shapes such as urine, while the CPMG experiment is often used for samples such as blood serum/plasma to suppress the broad background signals from macromolecules such as proteins and lipids. Since these signals are generally not of interest, the CPMG sequence [7, 30] attenuates them and improves the signal-to-noise ratio (SNR) of spectra of small-molecule metabolites. The number of spin echoes and the delay time between the pulses are critical for effective suppression of the broad signals without appreciably losing the sensitivity for the metabolites of interest. Alternatively, based on the large difference between the diffusion coefficients of small and macro molecules, their signals can be separated using diffusion ordered spectroscopy (DOSY) experiment. DOSY provides an alternative method for profiling small molecule metabolites by eliminating the signals of macromolecules [31-33].
Water signal suppression is common to most NMR experiments in metabolomics since most biological samples comprise water abundantly. A variety of water suppression sequences including the one pulse sequence with presaturation, presaturation utilizing relaxation gradients and echoes (PURGE), excitation sculpting and the combinations of gradient and weak rf pulses such as the WET sequence, have been developed and used [31, 34-36]. However, the most commonly used and simplest sequence uses presaturation that involves a continuous low-power irradiation at the water resonance position (∼ 4.8 ppm) during a relaxation delay of usually a few seconds [37]. Incorporation of the presaturation pulse into the 1D NOESY sequence is useful to obtain flat and undistorted baseline for the NMR spectrum.
2.3.2 Other NMR experiments
1D 1H NMR spectra obtained using common pulse sequences such as NOESY or CPMG with water suppression are generally complex with analyte signal intensities ranging over three orders of magnitude. In a typical 1H NMR spectrum, most signals are crowded into a region of ∼10 ppm and the resulting spectral overlap impedes identification and quantification of certain metabolites of interest. Alternative methods have been used to overcome such limitations.
2.3.2.1 Selective TOCSY
The 1D selective TOCSY (total correlation spectroscopy) methodology was explored for the analysis of metabolites in biofluids and was shown to detect metabolites quantitatively, even at concentrations much lower than those of the major components [38-40]. In a 1D selective TOCSY experiment, protons signals of interest are selectively excited by a shaped soft pulse; the magnetization is then transferred to all sequentially coupled spin system through J-couplings. This targeted approach suppresses unwanted and intense signals from other molecules significantly, and thus alleviates any dynamic-range problem and enables detection of molecules at low concentrations.
2.3.2.2 2D NMR
2D NMR experiments improve spectral resolution and information content compared to 1D NMR, which can be very helpful for identifying unknown metabolites. Extensive use of such experiments has been limited, however, due to their longer acquisition times, larger data sizes and increased complexity in data analysis. Nevertheless, due to increased interest in unraveling the complexity of biological samples, the use of two-dimensional NMR methods is on the rise [11, 31, 41-48]. Commonly used 2D NMR experiments include 2D J-resolved spectroscopy, correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), heteronuclear single quantum coherence spectroscopy (HSQC), and heteronuclear multiple bond correlation spectroscopy (HMQC). 2D J-resolved spectroscopy separates the chemical shifts and spin-spin couplings (J-coupling) along two frequency axes [46, 49]. The projection of this experiment is a simplified 1D spectrum; the simplification is achieved by removing the multiplicity associated with J-couplings and retaining only the chemical shift information.
2.3.2.3 Chromatography resolved NMR spectroscopy
There is a growing interest to couple NMR with high pressure liquid chromatography (HPLC). This combination offers access to low-concentration metabolites and hence promises numerous possibilities for metabolomics applications. One area that has highly benefited from this approach is pharmaceutical research and development [50]. Reverse phase columns commonly used in HPLC suffer from poor chromatographic resolution and long elution times (≥30 min each run). Recent developments in hydrophilic interaction liquid chromatography (HILIC) columns promise significant improvement in the separation of polar metabolites [51]. Moreover, developments in ultrahigh pressure liquid chromatography (UPLC) have enabled significant improvements in chromatographic resolution, reductions in separation time and enhancement in detection limits (by 3-5 folds) [52].
2.4 Advanced NMR methods
The inherently low sensitivity of NMR poses a major challenge for identifying low concentration metabolites in complex biological samples. Numerous efforts have been focused on improving the resolution and sensitivity to alleviate the limitations of NMR. Strong magnets and cryogenic probes have significantly lowered the NMR detection limit [53]. The use of cryogenic probes can significantly reduce the level of thermal noise resulting in an increase in the signal-to-noise ratio, typically by fourfold. In addition, advancements in micro-coil probe technology has shown that small-diameter detection coils have better signal-to-noise ratios and are particularly useful for mass limiting samples [54-56]. While saddle coils are widely used in conventional NMR, metabolomics applications are shown to benefit from solenoidal micro-coils because solenoidal coil enhances sensitivity by factor of 2-3 fold [55, 56]. Using isolated molecules, such coils have been demonstrated to significantly lower the sample volume as well as the mass detection limit. More recently, a variety of micro-coil probes with different sample volumes and multiple coils have been designed and constructed, which are potentially useful for profiling metabolites in biofluids with limited sample quantities [57-60].
3. Isotope labeling
While the standard and advanced methods discussed above have benefited the field of metabolomics, stable isotope labeling methods provide numerous capabilities and potential to take NMR-based metabolomics to a new level. Both NMR and MS based methods use isotope labeling for applications that include metabolites flux analysis, identification and quantitation of low concentration metabolites in complex biological samples. The unique advantage of using isotope enhanced NMR for metabolomics is that it enables tracing of biochemical pathways and the derivation of metabolic fluxes accurately in animal and human subjects. It allows unraveling interconnectivity of pathways and provides vital clues to disease mechanisms. Moreover, the isotope labeling approach, ex vivo, enables detection of a large number of (nearly 200) metabolites with improved resolution and sensitivity, which is unprecedented from NMR point of view. The use of mixtures of metabolites containing light and heavy isotopes is a popular method currently for relative and absolute quantitation of metabolites using MS methods [61-64], while the addition of isotope labeled internal standards has been used for MS quantitation for decades. Broadly, isotope labeling methods in NMR-based metabolomics are used in two major applications. One area focuses on metabolic pathway and flux analysis, and the other provides enhancement of resolution and sensitivity for complex mixture analysis.
3.1 In vivo isotope labeling for metabolic pathway and flux analysis
Metabolic pathway and flux analysis have significantly evolved as a consequence of the combination of stable isotopes and sensitive detection using advanced analytical methods. Flux analysis allows a detailed understanding of metabolic pathways, enables quantification of all intracellular metabolite levels and determination of the rates at which metabolites are produced or consumed. Although both NMR and MS methods are used in metabolic flux analysis, NMR is especially powerful as it allows both identification and quantification of metabolites in intact biological samples, and determination of positional isotopomer distributions derived from precursors enriched with the stable isotopes [65-69] (Figure 1).
Figure 1.
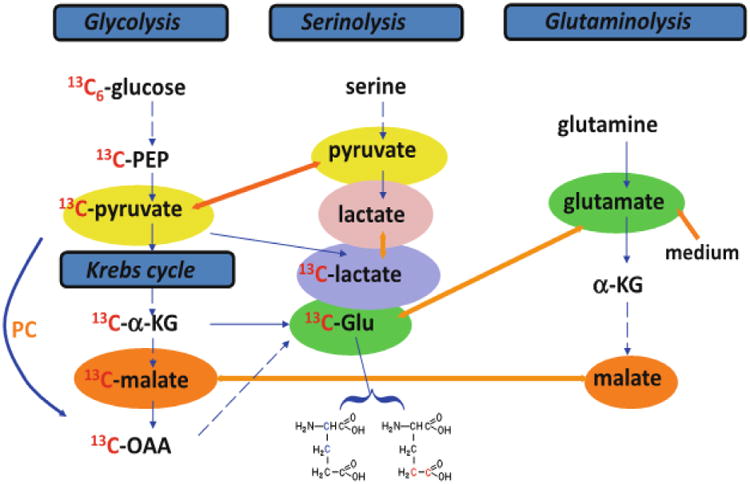
Tracers are necessary for delineating metabolic pathways. In the scheme depicted, pyruvate, lactate, α-ketoglutarate (α-KG), malate and glutamic acid (Glu) participate in multiple pathways including glycolysis, the Krebs cycle, serinolysis, and glutaminolysis. Without labeled tracers, it is impractical to resolve the specific pathway(s) involved in their production. With the use of 13C6-glucose, the synthesis of pyruvate, lactate, α-KG, malate, and Glu via glycolysis and the Krebs cycle can be distinguished from that via serinolysis or glutaminolysis by the 13C labeling pattern of these metabolites. Further, the 13C positional isotopomers of Glu (i.e. 13C-2,3-Glu and 13C-4,5-Glu) can be used to delineate respectively the Krebs cycle with or without pyruvate carboxylation (PC) input. [Reproduced with permission from reference 69].
Metabolite concentrations within a cell are controlled by regulation through homeostasis and a large number of metabolites are associated with many metabolic pathways. Hence global analysis of such metabolites does not enable a clear understanding of the biosynthesis of metabolites. For example, pyruvate can be obtained from several precursors and it is itself a precursor of many other metabolites [70-74]. Changes in pyruvate's concentration can therefore be due to any or all perturbations in such pathways. Similarly, lactate is secreted at a very high rate in cancer cells and accounts for significant fraction of the glucose consumed by these cells [73]. However, the total lactate concentration represents both the amount of glycolysis as well as amino acid oxidation. Therefore, it is not possible to quantify the contributions of different pathways based on the total concentrations of pyruvate or lactate.
The cells supplied with 13C-labeled glucose enable measurements of the labeled glucose consumption as well as the concentrations of the fractions of the metabolites produced from labeled glucose. Thus, for lactate, two forms - one containing 12C at the methyl position and the other containing 13C derived solely from the glycolysis of 13C labeled glucose- can be clearly seen in the NMR spectra [75, 76]. In the same way, a number of additional labeled metabolites can be used to quantitatively measure their rate of consumption as well as the fractions of the labeled metabolites produced. A number of stable isotopes such as 2H, 13C and 15N can be incorporated as tracers and the formed metabolites can be detected through their isotopically enriched nuclei. Stable isotopes used in these studies are biocompatible and are easily distinguished from highly abundant natural isotopes using NMR spectroscopy; thus they facilitate distinguishing the same metabolite derived from multiple pathways, and identification and measurement of the contributions of specific pathways involved in the biosynthesis of specific metabolites.
To date, stable isotope based metabolomics methods are used for a number of applications including studies of cancer using cells, animal models and even human subjects [65-68, 77, 78]. These methods have also been applied to other higher organisms and plants [44, 79]. Cell studies are particularly important as they provide the means for understanding metabolic pathways under controlled conditions. On the other hand, studies using animal models enable understanding pathogenesis in more realistic tissues that are still under controlled conditions. Mouse models, for example, can provide reasonable (though not perfect) surrogates for human diseases. The knowledge gained from the studies of cells and animal models can then be translated to understanding diseases directly using humans [65, 67, 80, 81].
Mammalian cells depend on the catabolism of glucose and glutamine for their viability and growth [82, 83]. While it is well known that many cancer cell lines depend on high rates of glucose uptake and metabolism to maintain their viability, it has only recently been observed that high rates glutamine metabolism are also exhibited by some cancer cells. In 2007, it was shown that the high rates at which the glioma cells uptake and metabolize glutamine exceed the cells' use of glutamine for protein and nucleotide biosynthesis [74]. Subsequently, based on the identification of the downstream products of 13C isotope labeled glutamine using NMR, oncogenes known to contribute to the malignant transformation of the cells were tested for the ability to induce glutaminolysis [84]. Results of this study show that the transcriptional regulatory oncogene Myc is involved in the catabolism of glutamine leading to glutamine addiction of the tumor cells to provide needed energy via the TCA cycle. It is thought that Myc binds to promoters and induces expression of many key regulatory genes that are associated with the glutamine catabolism. These studies promise a number of enzymatic targets through which the growth of Myc-transformed tumor cells can be prevented.
Numerous investigations are focused on improving isotope labeled approaches for metabolomics applications. For example, many studies have used a combination of isotopes such as 2H and 13C to trace pathways and analyze fluxes in humans and animal models [85-89]. The advantage of such an approach is that it allows the use of multiple isotope tracers, which enable investigations of multiple pathways such as glycolysis gluconeogensis and tricarboxylic acid cycle, simultaneously. Investigations of pathways using non-invasive approaches, using urine samples for example, are shown to be useful in metabolic studies of populations where sampling of blood and tissue is limited [90]. Dynamic nuclear polarization of isotope labeled nuclei is shown to provide an additional advantage of improved sensitivity of nearly 4 orders of magnitude and enable real-time assessment of metabolic pathways [91, 92].
For flux analysis using cancer or control cells, the cells are typically grown in media containing isotope labeled substrates such as [U-13C]-glucose or [U-13C]-glutamine that serve as source of tracers. For studies using mice, isotope labeled substrates are often injected through a tail vein; and for studies using humans, stable isotope labeled tracers such as [U-13C]-glucose, 2H2O or [U-13C]-propionate, that are fully compatible with human experimentation are injected intravenously into recruited patients prior to surgical resection or ingested. Isotope enriched metabolites in the resulting cells or animal/human tissue are then extracted and subjected to quantitative analysis using NMR methods.
Numerous isotope edited 2D NMR experiments have been used to identify and quantify specific isotopomers. These include 1H-13C HSQC, HSQC-TOCSY, HCCH-TOCSY, HACACO, 1H-31P HSQC-TOCSY, 1H-15N HSQC-TOCSY and 1H-1H TOCSY [69, 74]. Of these, the 2D HSQC experiment is the most often used and is a highly valuable method for identifying essentially all of the metabolites containing isotope tracers with good resolution and sensitivity. To aid identification of metabolites, a database of over 1000 1H and 13C chemical shifts corresponding to nearly 150 metabolites has been developed under identical conditions [44]. A large number of metabolites peaks in cell extracts have been identified using the combination of 2D NMR and metabolite databases (Figure 2). More recently, a new pulse sequence, isotope edited total correlation spectroscopy (ITOCSY), that filters two-dimensional 1H-1H NMR spectra from 12C-and 13C-containing molecules into separate and quantitatively equivalent spectra has been proposed for metabolic flux analysis [93]. The ITOCSY spectra of labeled and unlabeled molecules are directly comparable, and it effectively separates signals from labeled and unlabeled molecules (Figure 3).
Figure 2.
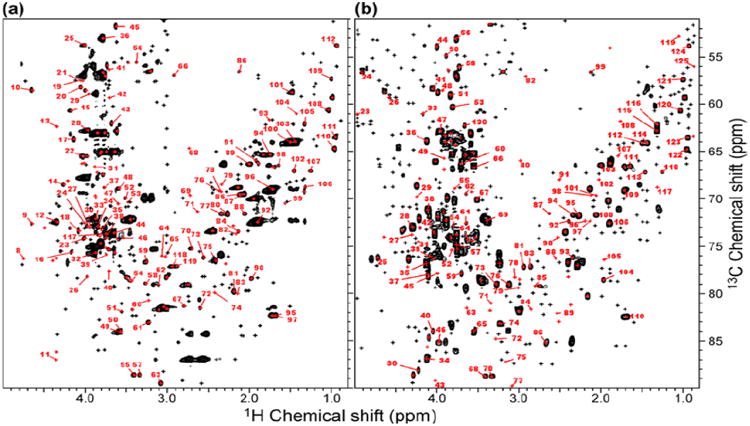
Metabolite identification using chemical shift database. (a) A total of 453 peaks were detected in the B. mori HSQC spectrum, of which 174 had candidate matches in the database (red) and 119 were uniquely identified, and 279 had no candidate matches (black). (b) A total of 544 peaks were detected in extracts from T87 cultured cells, of which 192 had candidate matches in the database (red) and 124 were uniquely identified, and 353 had no candidate matches (black). [Reproduced from reference 44].
Figure 3.
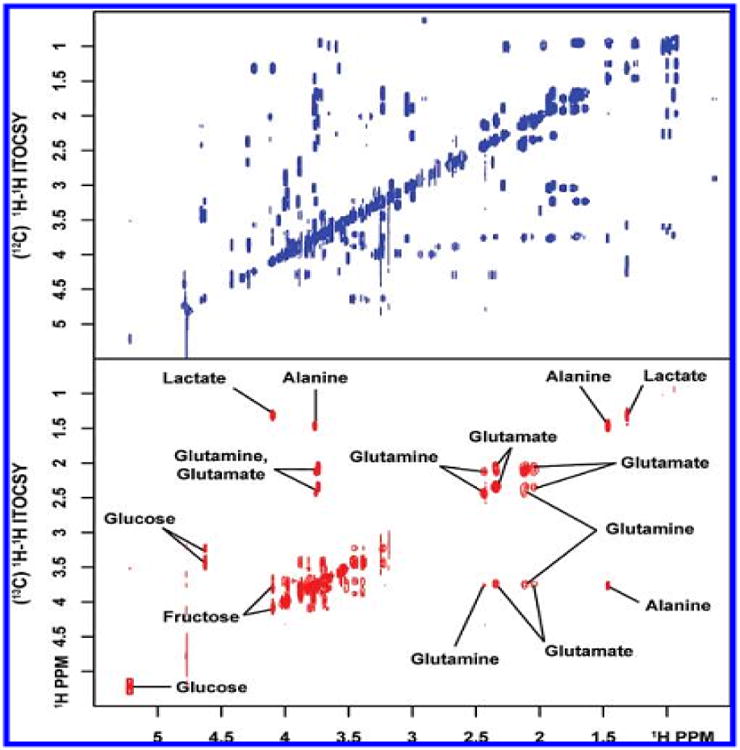
Isotope-filtered (12C) and difference edited (13C) 2D 1H-1H isotope edited total correlation spectroscopy (ITOCSY) spectra of a synthetic mixture containing 30 unlabeled and six 13C-labeled metabolites. Resonance assignments for 13C-enriched compounds are shown [Reproduced with permission from reference 93].
Ex vivo isotope labeling for metabolites analysis
The complexity and spectral overlap apparent in NMR spectra of biofluids remain significant obstacles to the identification and quantification of a large number of metabolites. Eukaryotic organisms possess more than 3000 metabolites with very diversified molecular structures and physical/chemical properties [94]. The quantitative determination of this magnitude of compounds in a single analysis remains out of reach for current technologies. Only a small fraction of the metabolites can currently be accurately and precisely detected. The information derived from this small sampling of metabolites is insufficient to reveal enough biochemical detail. To circumvent this problem, targeted profiling has shown to be a powerful approach for better understanding the metabolome in complex biological systems. As every metabolite contains at least one chemical functional group, the use of chemoselective tags can reduce the molecular complexity of the samples and potentially improve the detection of low-concentration metabolites by reducing the contribution of less interesting chemical signals.
Stable isotope tagging of metabolites in biofluids ex vivo using MS methods has found widespread applications for the identification as well as quantitation of metabolites [61-64]. The approach used for ex vivo isotope labeling for the NMR analysis of metabolites is shown in Figure 4. To date, stable isotopes such as 13C and 15N and abundant heteronuclei such as 31P have been used to tag metabolites with specific functional groups and thereby significantly enhance the resolution and sensitivity of the NMR experiments [95-99].
Figure 4.
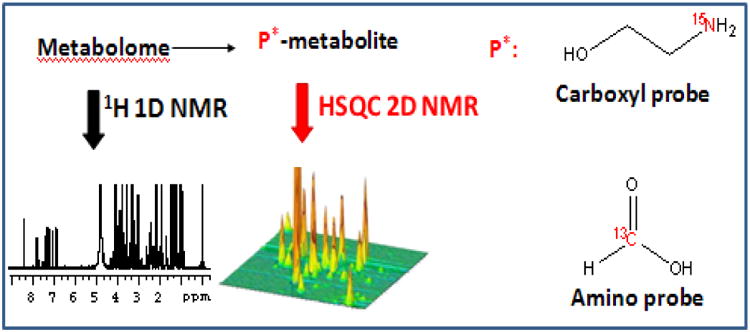
Isotope tagging of metabolites in biological mixtures and their detection by 2D NMR enables access to low concentrated metabolites that are inaccessible to 1D NMR [95-97].
3.2.1 13C isotope labeling
13C NMR can potentially serve as a useful alternative to 1H NMR for identifying and quantifying metabolites [100]. The combination of the larger chemical shift range of 13C and NMR experiments such as 2D 1H-13C HSQC significantly reduces spectral complexity by spreading the peaks in a two-dimensional plane. Recently constant time experiments have been developed to enable improved quantitation from HSQC type experiments [101, 102]. However, the low natural abundance of 13C results in unacceptably long data acquisition times to compensate for the poor sensitivity. Hence, despite the improved resolution offered by 13C NMR, it is impractical to use experiments involving 13C at natural abundance for routine metabolomics applications unless concentrations are very high [11].
In contrast, isotope tagging of metabolites with the amine functional group using a 13C isotope labeled tagging molecule significantly improves both the resolution and sensitivity of NMR methods for metabolite analysis [95, 96]. Tagging a specific functional group using 13C enables selection of only a particular class of molecules, and using 1H-13C HSQC contributes to a significant simplification of the 2D spectrum. 13C labeling using an acetylation reaction provides a useful approach for the analysis of the amine class of metabolites in complex mixtures [95]. The acetylation reaction is performed using 1,1′-13C2 acetic anhydride followed by 1D 13C or 1H-13C HSQC experiments to obtain a spectrum enhanced in both resolution and sensitivity. Typically, the pH of aqueous solutions of the biofluids samples are set to 8.0, to which 1, 1′-13C2 acetic anhydride is added while stirring at room temperature. The pH of the reaction medium is maintained at 8.0 by addition of a 1 M NaOH solution at regular intervals. The resulting mixture containing 13C labeled metabolites is then used for NMR analysis. Important aspects of this labeling approach are that it is facile, quantitative, and can be carried out directly in aqueous solution at ambient temperature. This is especially attractive for the analysis of complex mixtures such as urine, serum or other bio-fluids on a large set of samples.
More recently the performance of 13C isotope enhanced amino metabolites profiling was improved by using 13C-formic acid as the isotope tag [97, 99]. In the 13C-formylated metabolite, the short, one-bond distance between the labeled 13C and its closest 1H produces a large J-coupling, which facilitates efficient transfer of polarization between 1H and 13C in HSQC experiments. In particular, the J-coupling of 200 Hz allows for a short 2.5 ms INEPT transfer delay compared to the 83 ms delay, due to the small J-coupling of 6 Hz, which was used for acetic anhydride tagging. For the 13C-formic acid tagging, 13C-formic acid and N-hydroxysuccinimide are dissolved in tetrahydrofuran; N, N-dicyclohexylcarbodiimide in tetrahydrofuran is then added to the mixture. After the reaction, the supernatant containing 13C-N-formyloxysuccinimide is separated and added to the biofluid sample along with an aqueous solution of NaHCO3. The mixture is then stirred at room temperature, dried under vacuum and dispersed in D2O. This solution is then used for NMR analysis after adjusting the pH to 7.0. Alternatively, 13C-N-formyloxysuccinimide can be purified by recrystallization in ethanol and used for the tagging reaction instead of in situ generation.
Amino group containing metabolites are an important class of molecules associated with biological processes. Amino acids, for example, are not only the building blocks for proteins but also precursors for nucleotides [103] and energy sources through transamination, urea cycle, citric acid cycle and gluconeogenesis [104-106]. Other common amino metabolites include derivatives of amino acids, taurine, dimethylamine, methylamine, and many neurotransmitters such as dopamine, serotonin and histamine. Drugs such as amphetamine, procain, rimantadine and their metabolites also belong to this group of compounds. Selection of this class of metabolites using 13C isotope enhanced NMR promises a number of metabolomics applications.
3.2.2 15N isotope labeling
Carboxyl functional group containing metabolites, which are associated with almost all the metabolic pathways, represent another major class of molecules in biological systems. Efforts focused on the detection of this class of metabolites using NMR spectroscopy have lead to the development of a 15N isotope enhanced strategy, in which metabolites with carboxyl groups are chemically tagged with 15N-ethanolamine and detected using a 2D heteronuclear correlation NMR experiment [96, 99]. This approach, which significantly improves the sensitivity and resolution of the NMR method, is capable of detecting metabolites at concentrations as low as a few micromolar in biological samples, both quantitatively and reproducibly. By this approach nearly 200 well-resolved signals corresponding to well over 100 carboxyl-containing metabolites can be routinely detected in serum and urine samples, which is unprecedented from an NMR point of view (Figures 5 and 6). The resolution improvement, in this approach, is imparted from the dispersion of the chemical shifts of the 15N tag, and the high sensitivity is derived from the combination of factors such as isotope labeling and the strong, 90 Hz J-coupling between the observed nuclei (15N and 1H). A single peak, devoid of multiplicity, for each tagged metabolite and effective filtering of nontagged metabolites significantly add to the sensitivity and background suppression. Further, the 15N tag shows excellent reproducibility for complex biological samples. These characteristics are important for advanced metabolic profiling as well as for identifying unknown potential metabolite biomarkers.
Figure 5.
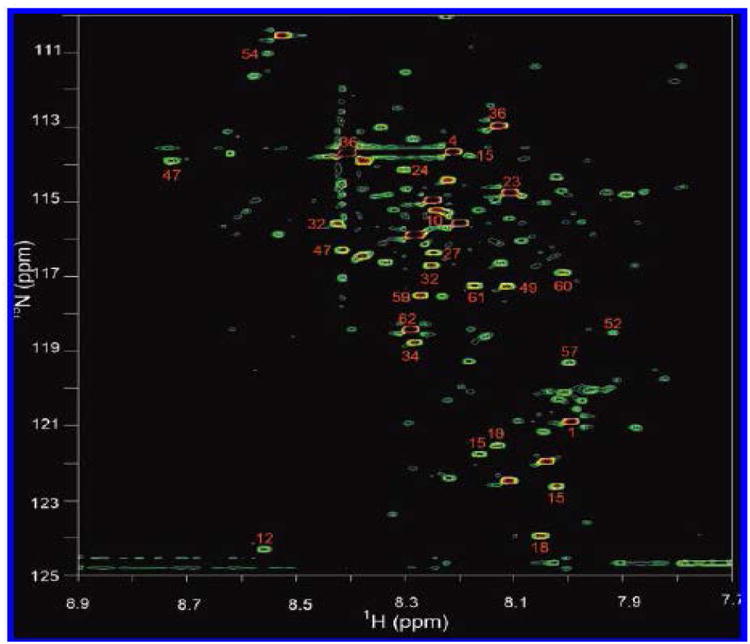
A 2D 1H-15N HSQC spectrum of human serum obtained after tagging carboxyl-containing metabolites with 15N-ethonalime. Nearly 180 metabolite peaks are detected, and the annotated peaks indicate identified metabolites [Reproduced with permission from reference 96].
Figure 6.
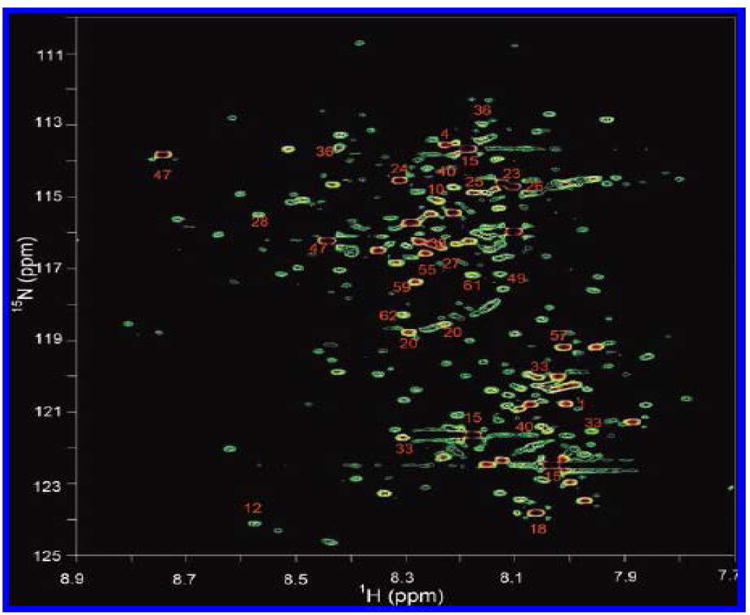
A 2D 1H-15N HSQC spectrum of human urine obtained after tagging carboxyl-containing metabolites with 15N-ethanolamine. Nearly 200 metabolites are detected, and the annotated peaks indicate identified metabolites [Reproduced with permission from reference 96].
Chemical derivatization reaction to tag 15N isotope to carboxyl metabolites is performed by adding 15N-ethanolamine and a catalyst, DMT-MM (4-(4,6-dimethoxy[1.3.5]triazin-2-yl)-4-methylmorpholinium chloride) to the biological mixture. In order to maintain the 15N amide protonation in the tagged metabolites, the pH is adjusted to 5.0 by adding HCl or NaOH, and then the solution volume is adjusted to 600 μL by adding water prior to detection of isotope tagged carboxyl metabolites using 2D HSQC NMR experiment. While the samples such as urine are used with no pretreatment, samples such as blood serum/plasma are deproteinized before chemical derivatization, by precipitating out the proteins by mixing serum/plasma samples with methanol in a 1:2 (v / v) ratio.
An important aspect of the isotope labeling using 15N-ethanolamine is that it easily and selectively combines with carboxyl-containing metabolites under aqueous conditions, which is suited for the study of biological mixtures without subjecting the samples to complex preprocessing. The use of the 15N labeled tag ensures that other nitrogen containing metabolites are invisible in the spectrum because of the low natural abundance of 15N (0.37%). The tagged metabolite retains the solubility since it contains at least one polar group (the hydroxyl in ethanolamine).
3.2.3 31P labeling
The analysis of lipids in biological mixtures is a vital part of metabolomics studies due to the important role of lipids in human diseases and health. Using 1H NMR, aliphatic fatty acid chain resonances can be assigned to sub-groups such as methyl, methylene, olefin, and allylic features but it is not possible to identify individual lipophilic components. Extending the scope of metabolite class selection using isotope labels, a method to detect members of the lipid class of metabolites with hydroxyl, carboxyl or aldehyde groups in complex biological fluids such as serum was recently developed [98]. This method employs the 31P containing reagent, 2-chloro-4,4,5,5-tetramethyldioxaphospholane (CTMDP), to derivatize the lipid metabolites. Derivatized metabolites are then detected with enhanced resolution using one-dimensional 31P NMR.
Typically, serum samples are lyophilized to dryness, reconstituted in stock solution (pyridine/chloroform), and subjected to derivatization with 31P containing the reagent, CTMDP. In the 31P NMR spectra, newly formed P-O groups derived from hydroxyl groups appear in the 144.5 - 150 ppm region (Figure 7), while the P-O groups derived from carboxylic acids appear at 134.5 - 135.5 ppm. 31P labeled unesterified cholesterol appears at 144.93 ppm. Mono and diglycerides can also be distinguished by this methodology, whereas derivatized 1,2 diglycerides in serum appear at 148.26 ppm, while for mono glycerides, derivatized primary and secondary hydroxyl groups appear at 147.6 and 146.54 ppm, respectively. Regardless of the length of the carbon chain, polar head group or unsaturation, lyso lipids produce two resonances at 146.68 ppm and 146.74 ppm in serum. Free fatty acids appeared at 134.78 ppm.
Figure 7.
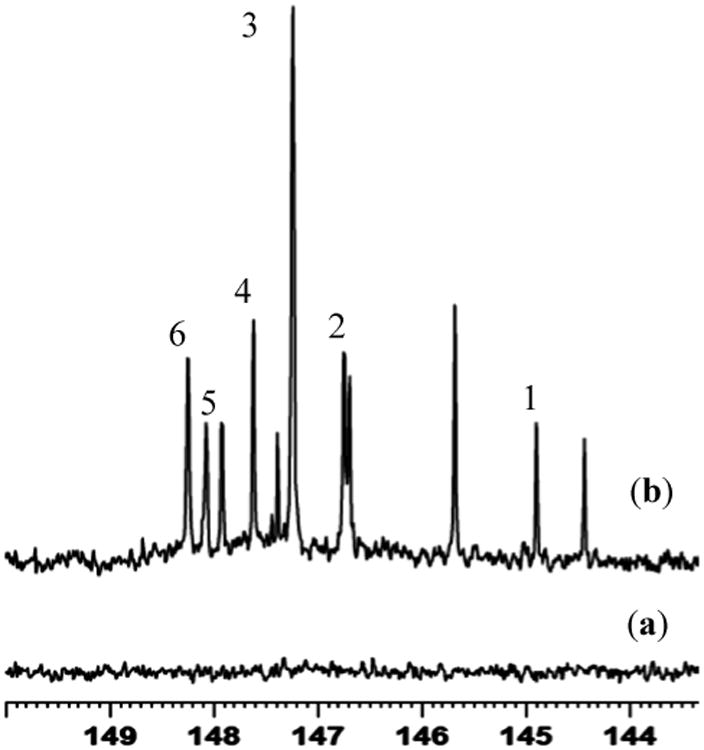
A portion of the 31P NMR spectrum of serum from a healthy individual: (a) Non-derivatized and (b) 31P-labeled. 31P labeled compounds include: cholesterol 1; lyso lipids 2; possible fatty aldehydes 3; phosphotidyl mono glycerides (primary) 4; free n-alkanol 5; 1,2-diacylglycerol 6.
Overall, this 31P labeling strategy offers the possibility of identification of a number of lipophilic compounds that contain hydroxyl and carboxylic acid functionalities in different chemical environments. The method provides sufficient sensitivity and spectral resolution, and derivatized species have unique and well-resolved resonances in the 31P NMR spectrum. This approach is consistent with the requirements for a fast screening method for lipid pathologies in human serum making possible the efficient use of 31P NMR spectroscopy alongside 1H NMR spectroscopy in metabolomics studies.
3.2.4 Applications of ex vivo isotope labeling
The reduced complexity of the spectra due to the absence of less interesting chemical signals is particularly important for the analysis of low-concentration metabolites. Chemoselective isotope tags produce a single peak for a metabolite with a single functional group. The resulting spectra from the isotope labeling strategies produce altogether new NMR chemical shifts for several hundreds of metabolites. Identification of these metabolites requires the knowledge of chemical shifts of isotope tags for authentic compounds. In view of this, from the spectra of isotope labeled standard compounds, a chemical shifts library has been developed for nearly 150 metabolites. By matching with this library of chemical shifts, a number of metabolites in the 13C, 15N and 31P isotope labeled spectra have so far been identified [96-98]. Subsequently, using a combination of isotope labeled methods, metabolites in human plasma procured from the National Institute of Standards and Technology (NIST, Gaithersburg, MD), were quantified after validating the experimental protocols using a mixture of synthetic compounds [99] (Figure 8). It was demonstrated that the combination of isotope tagging approaches, along with conventional 1D and 2D NMR methods enables the quantitative analysis of a large number of metabolites in human biofluids on a routine basis. For example, the 13C isotope labeling approach was used for the analysis of urine from patients with pre-diagnosed metabolic disorders such as tyrosinemia type II, argininosuccinic aciduria, homocystinuria and phenylketonuria [95].
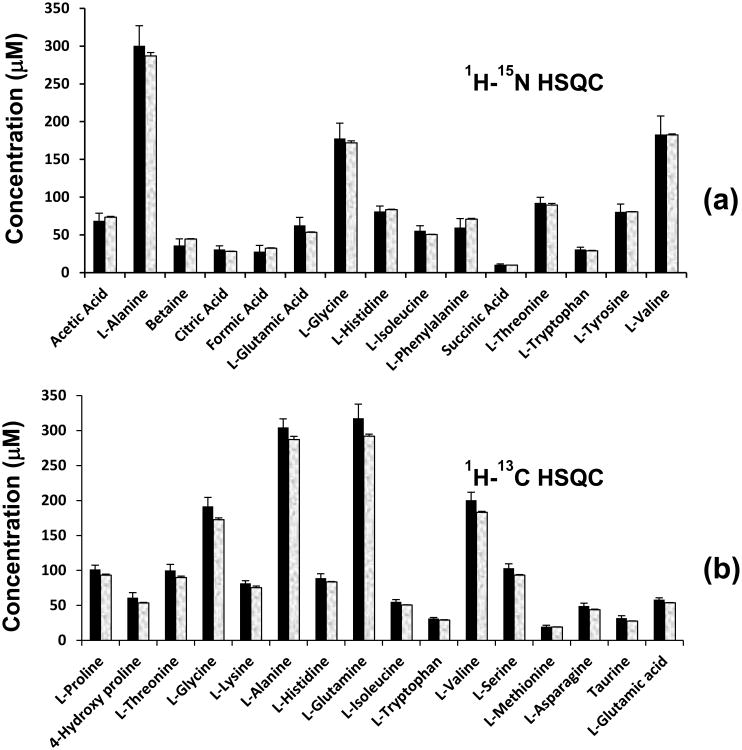
Figure 8.
Concentration of metabolites obtained with 15N or 13C tagging: (a) obtained from 1H-15N HSQC NMR after 15N tagging; (b) obtained from 1H-13C HSQC NMR after 13C tagging. The shaded bar on the right in each pair represents the actual concentration of the metabolite.
The combination of improved sensitivity and resolution, and less time required when compared to natural abundance heteronuclear NMR methods, is attractive for the routine and accurate analysis of metabolites in complex biological samples. Although, the isotope tagging methods often use 2D NMR experiments, each 2D experiment requires 30 min or less, and hence the approach can be useful for high throughput analysis of human plasma as well as other biological fluids.
4. Conclusions
While mass spectrometry is highly sensitive for metabolomics applications, the performance of NMR spectroscopy is significantly better in both reproducibility and quantitative ability and hence it promises a number of unique applications in the area of metabolomics. Despite vast advancements in the area, however, challenges remain for NMR for the detection, identification and quantification of a large number of metabolites in complex biological samples. Aimed at alleviating these limitations and gaining insights into in vivo biochemistry in cells, animal models and humans, the number of NMR investigations utilizing stable isotope labeling methods in vivo and ex vivo is growing rapidly. In in vivo isotope labeled studies, the targeted metabolites are always products of the metabolism of isotope labeled substrates and their detection, selectively, prevents the contributions from many confounding factors. This is extremely important for metabolomics studies since confounding factors arising from unrelated pathways or source are still a major challenge to the growth of metabolomics research. Development of ex vivo isotope labeling methods has enabled the detection of hundreds of metabolites with improved sensitivity and resolution, from a single experiment, which is unprecedented for NMR of complex biological samples. Further, a combination of the isotope tagging approach with the latest advancements in NMR technology, such as detection using micro-coil probes and cryoprobes, for example, can significantly minimize the volume of biofluid samples and time required for routine analysis. Future advancements in this area promise far reaching implications for systems biology, early detection of diseases and therapy monitoring, and personalized medicine.
References
- 1.Nicholson JK, Lindon JC, Holmes E. Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Fiehn O. Metabolomics-the link between genotype and phenotype. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 3.Saghatelian A, Cravatt BF. Global strategies to integrate the proteome and metabolome. Curr Opin Chem Biol. 2005;9:62–68. doi: 10.1016/j.cbpa.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Assfalg M, Bertini I, Colangiuli D, et al. Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci USA. 2008;105:1420–1424. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 6.van der Greef J, Smilde AK. Symbiosis of chemometrics and metabolomics: past, present, and future. J Chemomet. 2005;19:376–386. [Google Scholar]
- 7.Nagana Gowda GA, Zhang S, Gu H, et al. Metabolomics Based Methods for Early Disease Diagnostics: A Review. Exp Rev Mol Diagn. 2008;8:627–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagana Gowda GA, Ijare OB, Shanaiah N, et al. Combining NMR Spectroscopy and Mass Spectrometry in Biomarker Discovery. Biomarkers Med. 2009;3:307–322. doi: 10.2217/bmm.09.22. [DOI] [PubMed] [Google Scholar]
- 9.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 10.Weljie AM, Newton J, Mercier P, et al. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 11.Lewis IA, Schommer SC, Hodis B, et al. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional 1H-13C NMR spectra. Anal Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinette SL, Zhang F, Brüschweiler-Li L, et al. Web server based complex mixture analysis by NMR. Anal Chem. 2008;80:3606–3611. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 13.Chikayama E, Sekiyama Y, Okamoto M, et al. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal Chem. 2010;82:1653–1658. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 14.Keun HC. Metabonomic modeling of drug toxicity. Pharmacol Therapeut. 2006;109:92–106. doi: 10.1016/j.pharmthera.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Coen M, Holmes E, Lindon JC, et al. NMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chem Res Toxicol. 2008;2:9–27. doi: 10.1021/tx700335d. [DOI] [PubMed] [Google Scholar]
- 16.Bollard ME, Stanley EG, Lindon JC, et al. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18:143–162. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- 17.Nagana Gowda GA, Ijare OB, Somashekar BS, et al. Single-step analysis of individual conjugated bile acids in human bile using 1H NMR spectroscopy. Lipids. 2006;41:591–603. doi: 10.1007/s11745-006-5008-7. [DOI] [PubMed] [Google Scholar]
- 18.Nagana Gowda GA. Human Bile as a Rich Source of Biomarkers for Hepatopancreatobiliary Cancers. Biomark Med. 2010;4:299–314. doi: 10.2217/bmm.10.6. [DOI] [PubMed] [Google Scholar]
- 19.Nagana Gowda GA. NMR spectroscopy for discovery and quantitation of biomarkers of disease in human bile. Bioanalysis. 2011;3:1877–1890. doi: 10.4155/bio.11.152. [DOI] [PubMed] [Google Scholar]
- 20.Bala L, Ghoshal UC, Ghoshal U, et al. Malabsorption syndrome with and without small intestinal bacterial overgrowth: A study on upper-gut aspirate using 1H NMR spectroscopy. Magn Reson Med. 2006;56:738–744. doi: 10.1002/mrm.21041. [DOI] [PubMed] [Google Scholar]
- 21.Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J Proteome Res. 2007;6:498–505. doi: 10.1021/pr060464h. [DOI] [PubMed] [Google Scholar]
- 22.Villas-Boas SG, Hojer-Pedersen J, Akesson M, et al. Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast. 2005;22:1155–1169. doi: 10.1002/yea.1308. [DOI] [PubMed] [Google Scholar]
- 23.Bollard ME, Xu JS, Purcell W, et al. Metabolic profiling of the effects of D-galactosamine in liver spheroids using 1H NMR and MAS-NMR spectroscopy. Chem Res Toxicol. 2002;15:1351–1359. doi: 10.1021/tx025571e. [DOI] [PubMed] [Google Scholar]
- 24.Boroujerdi AF, Vizcaino MI, Meyers A, et al. NMR-based microbial metabolomics and the temperature-dependent coral pathogen Vibrio coralliilyticus. Environ Sci Technol. 2009;43:7658–7664. doi: 10.1021/es901675w. [DOI] [PubMed] [Google Scholar]
- 25.Lyng H, Sitter B, Bathen TF, et al. Metabolic mapping by use of high-resolution magic angle spinning 1H MR spectroscopy for assessment of apoptosis in cervical carcinomas. BMC Cancer. 2007;7:11. doi: 10.1186/1471-2407-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitter B, Bathen T, Hagen B, et al. Cervical cancer tissue characterized by high-resolution magic angle spinning MR spectroscopy. Magn Reson Mat Phys Biol Med. 2004;16:174–181. doi: 10.1007/s10334-003-0025-5. [DOI] [PubMed] [Google Scholar]
- 27.Schenetti L, Mucci A, Parenti F, et al. HR-MAS NMR spectroscopy in the characterization of human tissues: Application to healthy gastric mucosa. Conc Magn Reson Part A. 2006;28A:430–443. [Google Scholar]
- 28.Beckonert O, Keun HC, Ebbels TMD, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 29.Beckonert O, Coen M, Keun HC, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Prot. 2010;5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson JK, Foxall PJD, Spraul M, et al. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood-plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 31.Balayssac S, Delsuc MA, Gilard V, et al. Two-dimensional DOSY experiment with Excitation Sculpting water suppression for the analysis of natural and biological media. J Magn Reson. 2009;196:78–83. doi: 10.1016/j.jmr.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 32.Van Lokeren L, Kerssebaum R, Willem R, et al. ERETIC implemented in diffusion-ordered NMR as a diffusion reference. Magn Reson Chem. 2008;46:S63–S71. doi: 10.1002/mrc.2315. [DOI] [PubMed] [Google Scholar]
- 33.Smith LM, Maher AD, Cloarec O, et al. Statistical correlation and projection methods for improved information recovery from diffusion-edited NMR spectra of biological samples. Anal Chem. 2007;79:5682–5689. doi: 10.1021/ac0703754. [DOI] [PubMed] [Google Scholar]
- 34.Simpson AJ, Brown SA. Purge NMR: Effective and easy solvent suppression. J Magn Reson. 2005;175:340–346. doi: 10.1016/j.jmr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Ogg RJ, Kingsley PB, Taylor JS. Wet, a T-1-Insensitive and B-1-Insensitive Water-Suppression Method for in-Vivo Localized 1H NMR Spectroscopy. J Magn Reson Series B. 1994;104:1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 36.Mo H, Raftery D. Improved residual water suppression: WET180. J Biomol NMR. 2008;41:105–111. doi: 10.1007/s10858-008-9246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoult DI. Solvent Peak Saturation with Single-Phase and Quadrature Fourier Transformation. J Magn Reson. 1976;21:337–347. [Google Scholar]
- 38.Sandusky P, Raftery D. Use of selective TOCSY NMR experiments for quantifying minor components in complex mixtures: Application to the metabonomics of amino acids in honey. Anal Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 39.Sandusky P, Raftery D. Use of semiselective TOCSY and the pearson correlation for the metabonomic analysis of biofluid mixtures: Application to urine. Anal Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 40.Sandusky P, Appiah-Amponsah E, Raftery D. Use of optimized 1D TOCSY NMR for improved quantitation and metabolomic analysis of biofluids. J Biomol NMR. 2011;49:281–290. doi: 10.1007/s10858-011-9483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumas ME, Canlet C, André F, et al. Metabonomic assessment of physiological disruptions using 1H-13C HMBC-NMR spectroscopy combined with pattern recognition procedures performed on filtered variables. Anal Chem. 2002;74:2261–2273. doi: 10.1021/ac0156870. [DOI] [PubMed] [Google Scholar]
- 42.Tang HR, Wang Y, Nicholson JK, et al. Use of relaxation-edited one-dimensional and two dimensional nuclear magnetic resonance spectroscopy to improve detection of small metabolites in blood plasma. Anal Biochem. 2004;325:260–272. doi: 10.1016/j.ab.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 43.Xi Y, de Ropp JS, Viant MR, et al. Automated screening for metabolites in complex mixtures using 2D COSY NMR spectroscopy. Metabolomics. 2006;2:221–233. [Google Scholar]
- 44.Chikayama E, Suto M, Nishihara T, et al. Systematic NMR analysis of stable isotope labeled metabolite mixtures in plant and animal systems: coarse grained views of metabolic pathways. PLoS One. 2008;3:e3805. doi: 10.1371/journal.pone.0003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan TWM, Bandura LL, Higashi RM, et al. Metabolomics-edited transcriptomics analysis of Se anticancer action in human lung cancer cells. Metabolomics. 2005;1:325–339. [Google Scholar]
- 46.Ludwig C, Viant MR. Two-dimensional J-resolved NMR Spectroscopy: Review of a Key Methodology in the Metabolomics Toolbox. Phytochem Anal. 2010;21:22–32. doi: 10.1002/pca.1186. [DOI] [PubMed] [Google Scholar]
- 47.Parsons HM, Ludwig C, Gunther UL, et al. Improved classification accuracy in 1-and 2-dimensional NMR metabolomics data using the variance stabilising generalised logarithm transformation. BMC Bioinformatic. 2007;8:234. doi: 10.1186/1471-2105-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hyberts SG, Heffron GJ, Tarragona NG, et al. Ultrahigh-resolution 1H-13C HSQC spectra of metabolite mixtures using nonlinear sampling and forward maximum entropy reconstruction. J Am Chem Soc. 2007;129:5108–5116. doi: 10.1021/ja068541x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Commun. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 50.Lindon JC, Holmes E, Nicholson JK. Metabonomics techniques and applications to pharmaceutical research & development. Pharm Res. 2006;23:1075–1088. doi: 10.1007/s11095-006-0025-z. [DOI] [PubMed] [Google Scholar]
- 51.Kind T, Tolstikov V, Fiehn O, et al. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 52.Wilson ID, Nicholson JK, Castro-Perez J, et al. High resolution “ultra performance” liquid chromatography coupled to a-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 53.Spraul M, Freund AS, Nast RE, et al. Advancing NMR sensitivity for LC-NMR-MS using a cryo-flow probe: application to the analysis of acetaminophen metabolites in urine. Anal Chem. 2003;75:1536–1541. doi: 10.1021/ac026203i. [DOI] [PubMed] [Google Scholar]
- 54.Lacey ME, Subramanian R, Olson DL, et al. High-Resolution NMR Spectroscopy of Sample Volumes from 1 nL to 10 μL. Chem Rev. 1999;99:3133–3152. doi: 10.1021/cr980140f. [DOI] [PubMed] [Google Scholar]
- 55.Olson DL, Peck TL, Webb AG, et al. High-Resolution Microcoil 1H-NMR for Mass-Limited, Nanoliter-Volume Samples. Science. 1995;270:1967–1970. [Google Scholar]
- 56.Webb AG. Microcoil nuclear magnetic resonance spectroscopy. J Pharm Biomed Anal. 2005;38:892–903. doi: 10.1016/j.jpba.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 57.Kc R, Henry ID, Park GHJ, et al. Design and construction of a versatile dual volume heteronuclear double resonance microcoil NMR probe. J Magn Reson. 2009;197:186–192. doi: 10.1016/j.jmr.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henry D, Park GHJ, Kc R, et al. Design and construction of a microcoil NMR probe for the routine analysis of 20-mu L samples. Conc Magn Reson Part B-Magn Reson Engineering. 2008;33B:1–8. [Google Scholar]
- 59.Bergeron SJ, Henry ID, Santini RE, et al. Saturation transfer double-difference NMR spectroscopy using a dual solenoid microcoil difference probe. Magn Reson Chem. 2008;46:925–929. doi: 10.1002/mrc.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kc R, Gowda YN, Djukovic D, et al. Susceptibility-matched plugs for microcoil NMR probes. J Mag Reson. 2010;205:63–68. doi: 10.1016/j.jmr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo K, Bamforth F, Li L. Qualitative metabolome analysis of human cerebrospinal fluid by 13C-/12C-isotope dansylation labeling combined with liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2011;22:339–347. doi: 10.1007/s13361-010-0033-4. [DOI] [PubMed] [Google Scholar]
- 62.Huang X, Regnier FE. Differential metabolomics using stable isotope labeling and two-dimensional gas chromatography with time-of-flight mass spectrometry. Anal Chem. 2008;80:107–114. doi: 10.1021/ac071263f. [DOI] [PubMed] [Google Scholar]
- 63.Yang WC, Adamec J, Regnier FE. Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding. Anal Chem. 2007;79:5150–5157. doi: 10.1021/ac070311t. [DOI] [PubMed] [Google Scholar]
- 64.Yang WC, Regnier FE, Sliva D, et al. Stable isotope-coded quaternization for comparative quantification of estrogen metabolites by high-performance liquid chromatography-electrospray ionization mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;870:233–240. doi: 10.1016/j.jchromb.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lane AN, Fan TWM, Bousamra M, II, et al. Stable Isotope-Resolved Metabolomics (SIRM) in Cancer Research with Clinical Application to NonSmall Cell Lung Cancer. OMICS A Journal of Integrative Biology. 2011;15:173–182. doi: 10.1089/omi.2010.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan TW, Lane AN, Higashi RM, et al. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–269. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan TW, Lane AN, Higashi RM, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan TW, Lane AN. NMR-based stable isotope resolved metabolomics in systems biochemistry. J Biomol NMR. 2011;49:267–280. doi: 10.1007/s10858-011-9484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petch D, Butler M. Profile of energy metabolism in a murine hybridoma: glucose and glutamine utilization. J Cell Physiol. 1994;161:71–76. doi: 10.1002/jcp.1041610110. [DOI] [PubMed] [Google Scholar]
- 71.Portais JC, Voisin P, Merle M, et al. Glucose and glutamine metabolism in C6 glioma cells studied by carbon-13. NMR Biochimie. 1996;78:155–164. doi: 10.1016/0300-9084(96)89500-9. [DOI] [PubMed] [Google Scholar]
- 72.Mazurek S, Grimm H, Oehmke M, et al. Tumor M2-PK and glutaminolytic enzymes in the metabolic shift of tumor cells. Anticancer Res. 2000;20(6D):5151–5154. [PubMed] [Google Scholar]
- 73.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lane AN, Fan TW, Higashi RM, et al. Prospects for clinical cancer metabolomics using stable isotope tracers. Exp Mol Pathol. 2009;86:165–73. doi: 10.1016/j.yexmp.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lloyd SG, Zeng H, Wang P, et al. Lactate isotopomer analysis by 1H NMR spectroscopy: consideration of long-range nuclear spin-spin interactions. Magn Reson Med. 2004;51:1279–1282. doi: 10.1002/mrm.20075. [DOI] [PubMed] [Google Scholar]
- 76.Lane AN, Fan TW. Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics. 2007;3:79–86. [Google Scholar]
- 77.Burgess SC, Babcock EE, Jeffrey FM, et al. NMR indirect detection of glutamate to measure citric acid cycle flux in the isolated perfused mouse heart. FEBS Lett. 2001;505:163–167. doi: 10.1016/s0014-5793(01)02799-5. [DOI] [PubMed] [Google Scholar]
- 78.Perdigoto R, Furtado AL, Porto A, et al. Sources of glucose production in cirrhosis by 2H2O ingestion and 2H NMR analysis of plasma glucose. Biochim Biophys Acta. 2003;1637:156–163. doi: 10.1016/s0925-4439(03)00018-8. [DOI] [PubMed] [Google Scholar]
- 79.Kikuchi J, Shinozaki K, Hirayama T. Stable isotope labeling of Arabidopsis thaliana for an NMR-based metabolomics approach. Plant Cell Physiol. 2004;45:1099–104. doi: 10.1093/pcp/pch117. [DOI] [PubMed] [Google Scholar]
- 80.Lane AN, Fan TW, Higashi RM. Isotopomer based metabolic analysis by NMR and mass spectrometry. Biophys Tools Biol. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- 81.Lane AN, Fan TW, Xie Z, et al. Isotopomer analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal Chim Acta. 2009;651:201–208. doi: 10.1016/j.aca.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coles NW, Johnstone RM. Glutamine metabolism in Ehrlich ascites-carcinoma cells. Biochem J. 1962;83:284–291. doi: 10.1042/bj0830284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 84.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weis BC, Margolis D, Burgess SC, et al. Glucose production pathways by 2H and 13C NMR in patients with HIV-associated lipoatrophy. Magn Reson Med. 2004;51:649–654. doi: 10.1002/mrm.20057. [DOI] [PubMed] [Google Scholar]
- 86.Jones JG, Solomon MA, Cole SM, et al. An integrated (2)H and (13)C NMR study of gluconeogenesis and TCA cycle flux in humans. Am J Physiol Endocrinol Metab. 2001;281:E848–56. doi: 10.1152/ajpendo.2001.281.4.E848. [DOI] [PubMed] [Google Scholar]
- 87.Hausler N, Browning J, Merritt M, et al. Effects of insulin and cytosolic redox state on glucose production pathways in the isolated perfused mouse liver measured by integrated 2H and 13C NMR. Biochem J. 2006;394(Pt 2):465–473. doi: 10.1042/BJ20051174. Erratum in: Biochem J 2006; 395:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perdigoto R, Rodrigues TB, Furtado AL, et al. Integration of [U-13C]glucose and 2H2O for quantification of hepatic glucose production and gluconeogenesis. NMR Biomed. 2003;16:189–198. doi: 10.1002/nbm.826. [DOI] [PubMed] [Google Scholar]
- 89.Jin ES, Jones JG, Merritt M, et al. Glucose production, gluconeogenesis, and hepatic tricarboxylic acid cycle fluxes measured by nuclear magnetic resonance analysis of a single glucose derivative. Anal Biochem. 2004;327:149–155. doi: 10.1016/j.ab.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 90.Burgess SC, Weis B, Jones JG, et al. Noninvasive evaluation of liver metabolism by 2H and 13C NMR isotopomer analysis of human urine. Anal Biochem. 2003;312:228–234. doi: 10.1016/s0003-2697(02)00465-7. [DOI] [PubMed] [Google Scholar]
- 91.Merritt ME, Harrison C, Sherry AD, et al. Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc Natl Acad Sci USA. 2011;108:19084–19089. doi: 10.1073/pnas.1111247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schroeder MA, Atherton HJ, Ball DR, et al. Real-time assessment of Krebs cycle metabolism using hyperpolarized 13C magnetic resonance spectroscopy. FASEB J. 2009;23:2529–2538. doi: 10.1096/fj.09-129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis IA, Karsten RH, Norton ME, et al. NMR Method for Measuring Carbon-13 Isotopic Enrichment of Metabolites in Complex Solutions. Anal Chem. 2010;82:4558–4563. doi: 10.1021/ac100565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernie AR, Trethewey RN, Krotzky AJ, et al. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 95.Shanaiah N, Desilva A, Nagana Gowda GA, et al. Metabolite class selection of amino acids in biofluids using chemical derivatization and their enhanced 13C NMR. Proc Natl Acad Sci USA. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ye T, Mo H, Shanaiah N, et al. Chemoselective 15N Tag for Sensitive and High-Resolution Nuclear Magnetic Resonance Profiling of the Carboxyl-Containing Metabolome. Anal Chem. 2009;81:4882–4888. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ye T, Zhang S, Mo H, et al. 13C-Formylation for Improved NMR Profiling of Amino Metabolites in Biofluids. Anal Chem. 2010;82:2303–2309. doi: 10.1021/ac9024818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DeSilva MA, Shanaiah N, Nagana Gowda GA, et al. Application of 31P NMR spectroscopy and chemical derivatization for metabolite profiling of lipophilic compounds in human serum. Magn Reson Chem. 2009;47:S74–80. doi: 10.1002/mrc.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nagana Gowda GA, Tayyari F, Ye T, et al. Quantitative Analysis of Blood Plasma Metabolites Using Isotope Enhanced NMR Methods. Anal Chem. 2010;82:8983–8990. doi: 10.1021/ac101938w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan TW. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog NMR Spectrosc. 1996;28:161–219. [Google Scholar]
- 101.Hu K, Ellinger JJ, Chylla RA, et al. Measurement of Absolute Concentrations of Individual Compounds in Metabolite Mixtures by Gradient-Selective Time-Zero (1)H-(13)C HSQC with Two Concentration References and Fast Maximum Likelihood Reconstruction Analysis. Anal Chem. 2011;83:9352–9360. doi: 10.1021/ac201948f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu K, Wyche TP, Bugni TS, et al. Selective Quantification by 2D HSQC(0) Spectroscopy of Thiocoraline in an Extract from a Sponge-Derived Verrucosispora sp. J Nat Prod. 2011;74:2295–2298. doi: 10.1021/np200503c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W. H. Freeman & Co; New York: 2002. [Google Scholar]
- 104.Sakami W, Harrington H. Amino acid metabolism. Annu Rev Biochem. 1963;32:355–398. doi: 10.1146/annurev.bi.32.070163.002035. [DOI] [PubMed] [Google Scholar]
- 105.Brosnan JT. Glutamate, at the interface between amino acid and carbohydrate metabolism. J Nutri. 2000;130:988S–990S. doi: 10.1093/jn/130.4.988S. [DOI] [PubMed] [Google Scholar]
- 106.Young VR, Ajami AM. Glutamine: the emperor or his clothes? J Nutr. 2001;131:2449S–2459S. doi: 10.1093/jn/131.9.2449S. [DOI] [PubMed] [Google Scholar]