SUMMARY
Therapy development for adult diffuse glioma is hindered by incomplete knowledge of somatic glioma driving alterations and suboptimal disease classification. We defined the complete set of genes associated with 1,122 diffuse grade II-III-IV gliomas from The Cancer Genome Atlas and used molecular profiles to improve disease classification, identify molecular correlations, and provide insights into the progression from low- to high-grade disease. Whole genome sequencing data analysis determined that ATRX but not TERT promoter mutations are associated with increased telomere length. Recent advances in glioma classification based on IDH mutation and 1p/19q co-deletion status were recapitulated through analysis of DNA methylation profiles, which identified clinically relevant molecular subsets. A subtype of IDH-mutant glioma was associated with DNA demethylation and poor outcome; a group of IDH-wildtype diffuse glioma showed molecular similarity to pilocytic astrocytoma and relatively favorable survival. Understanding of cohesive disease groups may aid improved clinical outcomes.
INTRODUCTION
Diffuse gliomas represent 80% of malignant brain tumors (Schwartzbaum et al., 2006). Adult diffuse gliomas are classified and graded according to histological criteria (oligodendroglioma, oligoastrocytoma, astrocytoma and glioblastoma; grade II to IV). Although histopathologic classification is well established and is the basis of the WHO classification of CNS tumors (Louis et al., 2007), it suffers from high intra- and inter-observer variability, particularly amongst grade II-III tumors (van den Bent, 2010). Recent molecular characterization studies have benefited from the availability of the datasets generated by The Cancer Genome Atlas (TCGA) (Brennan et al., 2013; Eckel-Passow et al., 2015; Frattini et al., 2013; Kim et al., 2015; Suzuki et al., 2015; TCGA_Network et al., 2015), and have related genetic, gene expression and DNA methylation signatures with prognosis (Noushmehr et al., 2010; Sturm et al., 2012; Verhaak et al., 2010). For example, mutations in the isocitrate dehydrogenase genes 1 and 2 (IDH1/IDH2) define a distinct subset of glioblastoma (GBM) with a hypermethylation phenotype (G-CIMP) with favorable outcome (Noushmehr et al., 2010; Yan et al., 2009). Conversely, the absence of IDH mutations in LGG marks a distinct IDH-wildtype subgroup characterized by poor, GBM-like prognosis (Eckel-Passow et al., 2015; TCGA_Network et al., 2015). Recent work by us and others has proposed classification of glioma into IDH wildtype cases, IDH mutant samples additionally carrying codeletion of chromosome arm 1p and 19q (IDH mutant-codel) and samples with euploid 1p/19q (IDH-mutant-non-codel), regardless of grade and histology (Eckel-Passow et al., 2015; TCGA_Network et al., 2015). Mutation of the TERT promoter, which has been reported with high frequency across glioma, may be an additional defining feature. Current analyses have not yet clarified the relationships between LGGs and GBMs that share common genetic hallmarks like IDH mutation or TERT promoter mutation status. An improved understanding of these relationships will be necessary as we evolve toward an objective genome-based clinical classification.
To address the above issues, we assembled a dataset comprising all TCGA newly diagnosed diffuse glioma consisting of 1,122 patients, and comprehensively analyzed using sequencing and array based molecular profiling approaches. We have addressed crucial technical challenges in analyzing this comprehensive dataset, including the integration of multiple platforms and data sources (e.g. multiple methylation and gene expression platforms). We identified new diffuse glioma subgroups with distinct molecular and clinical features and shed light on the mechanisms driving progression of LGG (WHO grades II and III) into full-blown GBM (WHO grade IV).
RESULTS
Patient cohort characteristics
The TCGA LGG and GBM cohorts consist of 516 and 606 patients, respectively. Independent analysis of the GBM dataset was previously described, as was analysis of 290 LGG samples (Brennan et al., 2013; TCGA_Network et al., 2015). Two-hundred and twenty-six LGG samples were added to our current cohort (Table 1). Clinical data including age, tumor grade, tumor histology and survival was available for 93% (1046/1122) of cases (Table S1). The majority of samples were grade IV tumors (n=590, 56%), while 216 (21%) and 241 (23%) were grade II and III tumors, respectively. Similarly, 590 (56%) samples were classified as GBM, 174 (17%) as oligodendroglioma, 169 (16%) as astrocytoma and 114 (11%) as oligoastrocytoma.
Table 1.
Clinical characteristics of the sample set arranged by IDH and 1p/19q co-deletion status.
| Feature | IDH wt (n=520) | IDH mut - non-codel (n=283) | IDH mut - codel (n=171) | Unknown (n=148) |
|---|---|---|---|---|
| Clinical | ||||
| Histology (n) | ||||
| Astrocytoma | 52 (10.0%) | 112 (39.6%) | 4 (2.3%) | 1 (0.7%) |
| Glioblastoma | 419 (80.6%) | 32 (11.3%) | 2 (1.2%) | 137 (92.6%) |
| Oligoastrocytoma | 15 (2.9%) | 69 (24.4%) | 30 (17.5%) | 0 (0%) |
| Oligodendroglioma | 19 (3.7%) | 37 (13.1%) | 117 (68.4%) | 1 (0.7%) |
| Unknown | 15 (2.9%) | 33 (11.7%) | 18 (10.5%) | 9 (6.1%) |
| Grade (n) | ||||
| G2 | 19 (3.7%) | 114 (40.3%) | 81 (47.4%) | 2 (1.4%) |
| G3 | 67 (12.9%) | 104 (36.7%) | 70 (40.9%) | 0 (0%) |
| G4 | 419 (80.6%) | 32 (11.3%) | 2 (1.2%) | 137 (92.6%) |
| Unknown | 15 (2.9%) | 33 (11.7%) | 18 (10.5%) | 9 (6.1%) |
| Age | ||||
| Median (LQ-UQ) | 59 (51–68) | 38 (30–44) | 46 (35–54) | 55 (48–68) |
| Unknown (n) | 16 | 33 | 18 | 9 |
| Survival | ||||
| Median (CI) | 14.0 (12.6–15.3) | 75.1 (62.1–94.5) | 115.8 (90.5–Inf) | 12.6 (11.3–14.9) |
| Unknown (n) | 14 | 32 | 18 | 12 |
| KPS | ||||
| <70 | 85 (16.3%) | 8 (2.8%) | 5 (2.9%) | 21 (14.2%) |
| 70–80 | 196 (37.7%) | 41 (14.5%) | 18 (10.5%) | 60 (40.5%) |
| 90 | 29 (5.6%) | 60 (21.2%) | 32 (18.7%) | 2 (1.4%) |
| 100 | 51 (9.8%) | 44 (15.9%) | 30 (17.5%) | 14 (9.5%) |
| Unknown | 159 (30.6%) | 129 (45.6%) | 86 (50.3%) | 51 (34.5%) |
| Molecular | ||||
| MGMT promoter | ||||
| Methylated | 170 (32.7%) | 242 (85.5%) | 169 (98.8%) | 32 (21.6%) |
| Unmethylated | 248 (47.7%) | 36 (12.7%) | 1 (0.6%) | 34 (23.0%) |
| Unknown | 102 (19.6%) | 5 (1.8%) | 1 (0.6%) | 82 (55.4%) |
| TERT promoter | ||||
| Mutant | 67 (12.9%) | 8 (2.8%) | 86 (50.3%) | 1 (0.7%) |
| Wild-type | 19 (9.8%) | 146 (51.6%) | 2 (1.2%) | 0 (0%) |
| Unknown | 434 (83.5%) | 129 (45.6%) | 83 (48.5%) | 135 (99.3%) |
| TERT expression | ||||
| Expressed | 178 (34.2%) | 14 (4.9%) | 153 (89.5%) | 6 (4.1%) |
| Not expressed | 51 (9.8%) | 242 (85.5%) | 16 (9.4%) | 7 (4.7%) |
| Unknown | 291 (56.0%) | 27 (9.5%) | 2 (1.2%) | 135 (91.2%) |
Amongst the data sources considered in our analysis were gene expression (n = 1,045), DNA copy number (n = 1,084), DNA methylation (n = 932), exome sequencing (n = 820) and protein expression (n = 473). Multiple and overlapping characterization assays were employed (Table S1). All data files that were used in our analysis can be found at https://tcga-data.nci.nih.gov/docs/publications/lgggbm_2015/.
Identification of novel glioma-associated genomic alterations
To establish the set of genomic alterations that drive gliomagenesis, we called point mutations and indels on the exomes of 513 LGG and 307 GBM using the Mutect, Indelocator, Varscan2 and RADIA algorithms, and considered all mutations identified by at least two callers. Significantly mutated genes (SMGs) were determined using MutSigCV. This led to the identification of seventy five SMGs, of which ten had been previously reported in GBM (Brennan et al., 2013), twelve had been reported in LGG (TCGA_Network et al., 2015), and eight had been identified in both GBM and LGG studies. Forty five SMGs have not been previously associated with glioma and ranged in mutation frequency from 0.5% to 2.6% (Table S2A). We used GISTIC2 to analyze the DNA copy number profiles of 1,084 samples, including 513 LGG and 571 GBM, and identified 162 significantly altered DNA copy number segments (Table S2B). We employed PRADA and deFuse to detect 1,144 gene fusion events in the RNA-seq profiles available for 154 GBM and 513 LGG samples, of which 37 in-frame fusions involved receptor tyrosine kinases (Table S2C). Collectively, these analyses recovered all known glioma driving events, including in IDH1 (n = 457), TP53 (n = 328), ATRX (n = 220), EGFR (n = 314), PTEN (n = 168), CIC (n = 80), FUBP1 (n = 45). Notable newly predicted glioma drivers relative to the earlier TCGA analyses were genes associated with chromatin organization such as SETD2 (n = 24), ARID2 (n = 20), DNMT3A (n = 11), and the KRAS/NRAS oncogenes (n = 25 and n = 5, respectively).
We overlapped copy number, mutation (n = 793) and fusion transcript (n = 649) profiles and confirmed the convergence of genetic drivers of glioma into pathways including the Ras-Raf-MEK-ERK, p53/apoptosis, PI3K/AKT/mTOR, chromatin modification and cell cycle pathways. The Ras-Raf-MEK-ERK signaling cascade showed alterations in 106 of 119 members detected across 578 cases (73%), mostly occurring in IDH-wildtype samples (n = 327 of 357, 92%). Conversely, we found that a set of 36 genes involved in chromatin modification was targeted by genetic alterations in 423 tumors (54%, n = 36 genes), most of which belonged to the IDH-mutant-non-codel group (n = 230, 87%).
In order to identify new somatically altered glioma genes, we used MutComFocal to nominate candidates altered by mutation as well as copy number alteration. Prominent among these genes was NIPBL, a crucial adherin subunit that is essential for loading cohesins on chromatin (Table S2D)(Peters and Nishiyama, 2012). The cohesin complex is responsible for the adhesion of sister chromatids following DNA replication and is essential to prevent premature chromatid separation and faithful chromosome segregation during mitosis (Peters and Nishiyama, 2012). Alterations in the cohesin pathway have been reported in 12% of acute myeloid leukemias (Kon et al., 2013). Mutations of the cohesin complex gene STAG2 had been previously reported in GBM (Brennan et al., 2013). Taken together, 16% of the LGG/GBM showed mutations and/or CNAs in multiple genes involved in the cohesin complex, thus nominating this process as a prominent pathway involved in gliomagenesis.
Telomere length is positively correlated with ATRX but not TERT promoter mutations
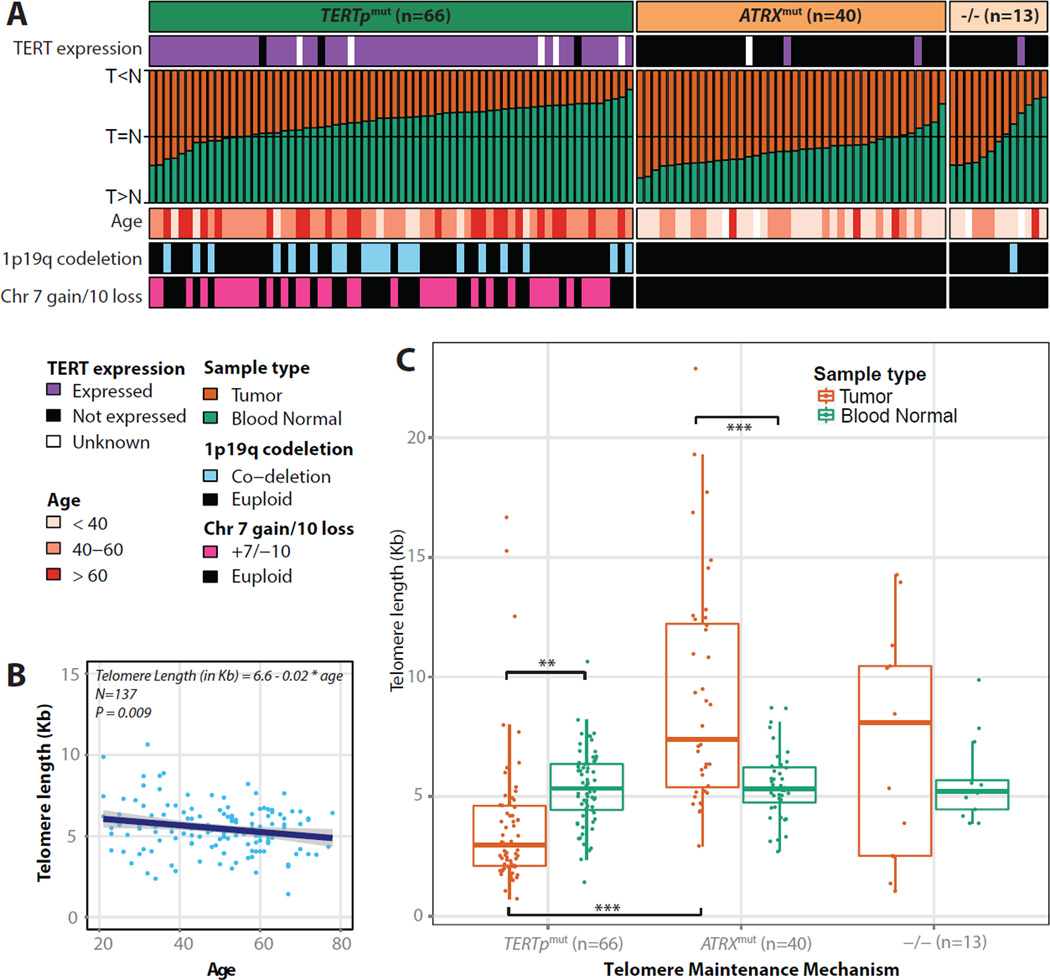
Mutations in the TERT promoter (TERTp) have been reported in 80% of GBM (Killela et al., 2013). We used TERTp mutation calls from targeted sequencing (n = 287) and complemented them with TERTp mutations inferred from whole genome sequencing (WGS) data (n = 42). TERTp mutations are nearly mutually exclusive with mutations in ATRX (Eckel-Passow et al., 2015), which was confirmed in our cohort. Overall, 85% of diffuse gliomas harbored mutations of TERTp (n=157, 48%) or ATRX (n=120, 37%). TERTp mutations activate TERT mRNA expression through the creation of a de novo ETS transcription factor-binding site (Horn et al., 2013) and we observed significant TERT upregulation in TERTp mutant cases (p-value < 0.0001, Figure S1A). TERT expression measured by RNA-seq was a highly sensitive (91%) and specific (95%) surrogate for the presence of TERTp mutation (Figure S1B). We correlated TERTp status with glioma driving alterations and observed that nearly all IDH-wildtype cases with chromosome 7 gain and chromosome 10 loss harbored TERTp mutations or upregulated TERT expression (n=52/53 and n = 134/147, respectively; Figure 1A). Conversely, only 45% of IDH-wildtype samples lacking chromosome 7/chromosome 10 events showed TERTp mutations or elevated TERT expression (n=15/33 and n = 43/82, respectively). Thus, TERTp mutations may precede the chr 7/chr 10 alterations which have been implicated in glioma initiation (Ozawa et al., 2014).
Figure 1. Telomere length associations in glioma.
A. Heatmap of relative tumor/normal telomere lengths of 119 gliomas, grouped by TERTp and ATRX mutation status. B. Telomere length decreases with increasing age (measured in years at diagnosis) in blood normal control samples (n=137). C. Quantitative telomere length estimates of tumors and blood normal, grouped by TERTp mutant (n=67, 56%), ATRX mutant (n=40, 33%) and double negative (n=13, 11%) status. *** = P<0.0001; ** = P<0.001.
To correlate TERTp mutations to telomere length, we used whole genome sequencing and low pass whole genome sequencing data to estimate telomere length in 141 pairs of matched tumor and normal samples. As expected, we observed an inverse correlation of telomere length with age at diagnosis in matching blood normal samples (Figure 1B) and tumor samples (Figure S1C). Glioma samples harboring ATRX mutations showed significantly longer telomeres compared to TERTp mutant samples (t-test p-value < 0.0001; Figure 1C). Among TERTp mutation gliomas, there was no difference in telomere length between samples with and without additional IDH1/IDH2 mutations, despite a difference in age. ATRX forms a complex with DAXX and H3.3, and the genes encoding these proteins are frequently mutated in pediatric gliomas (Sturm et al., 2012). Mutations in DAXX and H3F3A were identified in only two samples in our WGS dataset. The ATRX-DAXX-H3.3 complex is associated with the alternative lengthening of telomeres (ALT) and our observations confirm previously hypothesized fundamental differences between the telomere control exerted by telomerase and ALT (Sturm et al., 2014).
As demonstrated by the identification of TERTp mutations, somatic variants affecting regulatory regions may play a role in gliomagenesis. Using 67 matched whole-genome and RNA-seq expression pairs, we similarly sought to identify mutations located within 2kb upstream of transcription start sites and associated with a gene expression change. Using strict filtering methods we identified twelve promoter regions with mutations in at least six samples. Three of twelve regions related to a significant difference in the expression of the associated gene expression, suggesting possible functional consequences. Other than TERT (n=37), promoter mutations of the ubiquitin ligase TRIM28 (n=8) and the calcium channel gamma subunit CACNG6 (n=7) correlated with respectively upregulation and downregulation of these genes, respectively (Table S2E). TRIM28 has been reported to mediate the ubiquitin-dependent degradation of AMP-activated protein kinase (AMPK) leading to activation of mTOR signaling and hypersensitization to AMPK agonists, such as metformin (Pineda et al., 2015).
Unsupervised clustering of gliomas identifies six methylation groups and four RNA expression groups associated with IDH status
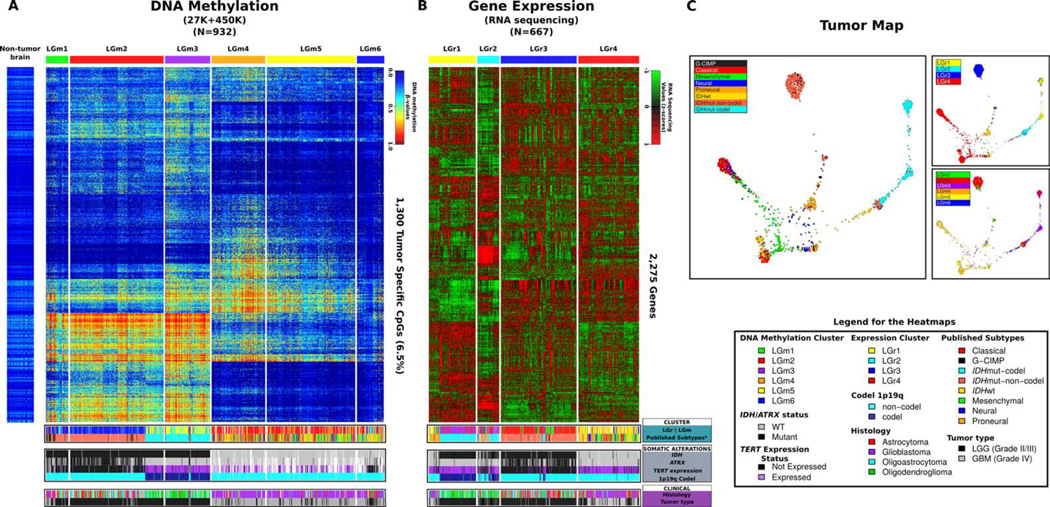
To segregate the DNA methylation subtypes across the pan-glioma dataset, we analyzed 932 glioma samples profiled on the HumanMethylation450 platform (516 LGG and 129 GBM) and the HumanMethylation27 platform (287 GBM). In order to incorporate the maximum number of samples, we merged datasets from both methylation platforms yielding a core set of 25,978 CpG probes. To reduce computational requirements to cluster this large dataset, we eliminated sites that were methylated (mean β-value ≥0.3) in non-tumor brain tissues and selected 1,300 tumor specific methylated probes (1,300/25,978, 5%) to perform unsupervised k-means consensus clustering. This identified six distinct clusters, labeled LGm1-6 (Figure 2A; Table S1; Table S3A). Next, we sought to determine pan-glioma expression subtypes through unsupervised clustering analysis of 667 RNA-seq profiles (513 LGG and 154 GBM) which resulted in four main clusters labeled LGr1-4 (Figure 2B; Table S1; Table S3A). An additional 378 GBM samples with Affymetrix HT-HG-U133A profiles (but lacking RNA-seq data) were classified into the four clusters using a k-nearest neighbor classification procedure. IDH mutation status was the primary driver of methylome and transcriptome clustering and separated the cohort into two macro-groups. The LGm1/LGm2/LGm3 DNA methylation macro-group carried IDH1 or IDH2 mutations (449 of 450, 99%) and was enriched for LGG (421/454, 93%) while LGm4/LGm5/LGm6 were IDH-wildtype (429/430, 99%) and enriched for GBM (383/478, 80%). LGm1-3 showed genome wide hypermethylation compared to LGm4-6 clusters (Figure S2A), documenting the association between IDH mutation and increased DNA methylation (Noushmehr et al., 2010; Turcan et al., 2012). Principal component analysis using 19,520 probes yielded similar results, thus emphasizing that our probe selection method did not introduce unwanted bias (Figure S2B). The gene expression clusters LGr1-3 harbored IDH1 or IDH2 mutations (438 of 553, 82%) and were enriched for LGG (436/563, 77%) while the LGr4 was exclusively IDH-wildtype (376 of 387, 97%) and enriched for GBM (399/476, 84%).
Figure 2. Pan-glioma DNA methylation and transcriptome subtypes.
A. Heatmap of DNA methylation data. Columns represent 932 TCGA glioma samples grouped according to unsupervised cluster analysis, rows represent DNA methylation probes sorted by hierarchical clustering. Non-neoplastic samples are represented on the left of the heatmap (n = 77) (Guintivano et al, 2013). B. Heatmap of RNA sequencing data. Unsupervised clustering analysis for 667 TCGA glioma samples profiled using RNA sequencing are plotted in the heatmap using 2,275 most variant genes. Previously published subtypes were derived from Brennan et al. Cell, 2013 and TCGA Research Network, NEJM, 2015. C. Tumor Map based on mRNA expression and DNA methylation data. Each data point is a TCGA sample colored coded according to their identified status. A live interactive version of this map is available at http://tumormap.ucsc.edu/?p=ynewton.gliomas-paper.
We extended our analysis using Tumor Map (Supplementary Methods) to perform integrated co-clustering analysis of, the combined gene expression (n = 1,196) and DNA methylation (n = 867) profiles. An interactive Tumor Map version is publicly available at http://tumormap.ucsc.edu/?p=ynewton.gliomas-paper. Tumor Map assigns samples to a hexagon in a grid so that nearby samples are likely to have similar genomic profiles and allows visualizing complex relationships between heterogeneous genomic data samples and their clinical or phenotypical associations. Thus, clusters in the map indicate groups of samples with high similarity of integrated gene expression and DNA methylation profiles (Figure 2C). The map confirms clustering by IDH status and additionally shows islands of samples that share previously reported GBM cluster memberships (Noushmehr et al., 2010; Verhaak et al., 2010). To assess clustering sensitivity to pre-processing we tried complementary methods and obtained similar results (Figure S2C).
To identify genes whose copy number changes are associated with concordant changes in gene expression, we combined expression and copy number profiles from 659 samples to define a signature of 57 genes with strong functional copy number (fCN) change (Table S3B). The fCN signature clustered gliomas into three macro-clusters, LGfc1-3, strongly associated with IDH and 1p/19q status (Figure S2D). The fCN analysis revealed the functional activation of a cluster of HOXA genes in the IDH-wildtype LGfc2 cluster, which were previously associated with glioma stem cell maintenance (Kurscheid et al., 2015).
Finally, we clustered reverse phase protein array profiles, consisting of 196 antibodies on 473 samples. Two macro clusters were observed and in contrast to the transcriptome/methylome/fCNV clustering, the primary discriminator was based on glioma grade (LGG vs GBM) rather than IDH status (Figure S2E). Compared to the LGG-like cluster, the GBM-like cluster had elevated expression of IGFBP2, fibronectin, PAI1, HSP70, EGFR, phosphoEGFR, phosphoAKT, Cyclin B1, Caveolin, Collagen VI, Annexin1 and ASNS, whereas the LGG class showed increased activity of PKC (alpha, beta and delta), PTEN, BRAF, and phosphoP70S6K.
The above results confirm IDH status as the major determinant of the molecular footprints of diffuse glioma. To further elucidate the subtypes of diffuse glioma, we performed unsupervised clustering within each of the two IDH-driven macroclusters. We used 1,308 tumor specific CpG probes defined among the IDH mutation cohort (n = 450) and identified three IDH-mutant specific DNA methylation clusters (Figure S3A). Using 914 tumor specific CpG probes in the IDH-wildtype cohort (n = 430), we uncovered three IDH-wildtype specific clusters (Figure S4A). The sets of CpG probes used to cluster each of the two IDH-driven datasets overlapped significantly with the 1,300 probes that defined the pan-glioma DNA methylation clustering (1162/1,300, 89% and 853/1,300, 66%, for IDH-mutant and IDH-wildtype, respectively). The clusters identified by separating IDH-mutant and IDH-wildtype gliomas showed strong overall concordance with pan-glioma DNA methylation subtypes (Table S3A). Similarly, unsupervised clustering of 426 IDH-mutant RNA-seq profiles resulted in three subtypes (Figure S3A) and analysis of the 234 IDH-wildtype samples led to four mixed LGG/GBM clusters that showed enrichment for previously identified GBM expression subtypes (Figure S4C)(Verhaak et al., 2010).
An epigenetic signature associated with activation of cell cycle genes segregates a subgroup of IDH-mutant LGG and GBM with unfavorable clinical outcome
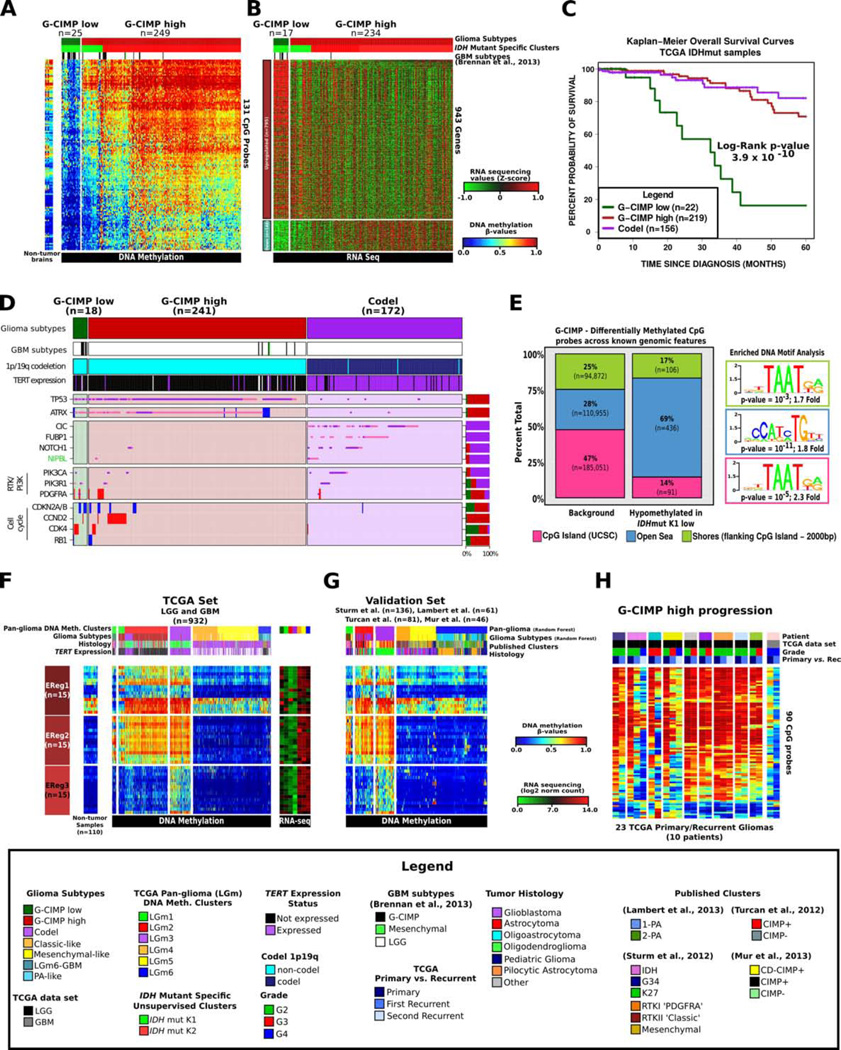
The three epigenetic subtypes defined by clustering IDH-mutant glioma separated samples harboring the 1p/19q co-deletion into a single cluster and non-codel glioma into two clusters (Figure S3A). Conversely, non-codel glioma grouped nearly exclusively into a single expression cluster and codels were split in two separated expression clusters (Figure S3A). A distinct subgroup of samples within the IDH-mutant-non-codel DNA methylation clusters manifested relatively reduced DNA methylation (Figure S3B). The unsupervised clustering of IDH-mutant glioma was unable to segregate the lower methylated non-codel subgroup as the 1,308 probes selected for unsupervised clustering included only 19 of the 131 differentially methylated probes characteristic for this subgroup (FDR < 10−15, difference in mean methylation beta-value > 0.27). The low-methylation subgroup consisted of both G-CIMP GBM (13/25) and LGGs (12/25) and was confirmed using a non-TCGA dataset (Figure S3C). The tumors with higher methylation in the split cluster were very similar to those grouped in the second non-codel cluster and a supervised comparison identified only 12 probes as differentially DNA methylated (Figure 3A; Figure 3B). We concluded that IDH-mutant glioma is composed of three coherent subgroups: 1. The Codel group, consisting of IDH-mutant-codel LGGs; 2. The G-CIMP-low group, including IDH-mutant-non-codel glioma (LGG and GBM) manifesting relatively low genome wide DNA methylation; and 3. The G-CIMP-high group including IDH-mutant-non-codel glioma (LGG and GBM) with higher global levels of DNA methylation. The newly identified G-CIMP-low group of glioma was associated with significantly worse survival as compared to the G-CIMP-high and Codel groups (Figure S3D). The clinical outcome of the tumors classified as G-CIMP-high was as favorable as that of Codel tumors, the subgroup generally thought to have the best prognosis among glioma patients (Figure 3C; Figure S3D). We compared the frequencies of glioma driver gene alterations between the three types of IDH-mutant glioma and found that 15 of 18 G-CIMP-low cases carried abnormalities in cell cycle pathway genes such as CDK4 and CDKN2A, relative to 36/241 and 2/172 for G-CIMP-high and Codels, respectively (Figure 3D). Supervised analysis between gene expression of G-CIMP-low and G-CIMP-high resulted in 943 differentially expressed genes. We mapped the 943 deregulated genes to 767 nearest CpG probes (max distance 1kb) and found the majority of the CpG probes (486/767, 63%) to show a significant methylation difference (FDR<0.05, difference in mean methylation beta-value > 0.01) between G-CIMP-low and G-CIMP-high, suggesting a mechanistic relation between loss of methylation and increased transcript levels.
Figure 3. Identification of a distinct G-CIMP subtype defined by epigenomics.
A. Heatmap of probes differentially methylated between the two IDH-mutant-non-codel DNA methylation clusters allowed the identification of a low-methylation subgroup named G-CIMP-low. Non-tumor brain samples (n=12) are represented on the left of the heatmap. B. Heatmap of genes differentially expressed between the two IDH-mutant-non-codel DNA methylation clusters. C. Kaplan-Meier survival curves of IDH-mutant methylation subtypes. Ticks represent censored values. D. Distribution of genomic alterations in genes frequently altered in IDH-mutant glioma. E. Genomic distribution of 633 CpG probes differentially demethylated between co-clustered G-CIMP-low and G-CIMP-high. CpG probes are grouped by UCSC genome browser defined CpG Islands, shores flanking CpG island +/− 2kb and open seas (regions not in CpG islands or shores). F. DNA methylation heatmap of TCGA glioma samples ordered per Figure 2A, and the epigenetically regulated (EReg) gene signatures defined for G-CIMP-low, G-CIMP-high and Codel subtypes. NThe mean RNA sequencing counts for each gene matched to the promoter of the identified cgID across each cluster are plotted to the right. G. Heatmap of the validation set classified using the random forest method1,300 probes defined in Figure 2A. H. Heatmap of probes differentially methylated between G-CIMP-low and G-CIMP-high in longitudinally matched tumor samples.
Recent analysis of epigenetic profiles derived from colon cancers showed that transcription factors may bind to regions of demethylated DNA (Berman et al., 2012). Therefore, we asked whether transcription factors may be recruited to the DNA regions differentially methylated between G-CIMP-low samples and G-CIMP-high samples from the same methylation cluster, using 450K methylation profiles (n=39). Globally, we detected 643 differentially methylated probes between 27 G-CIMP-low and 12 G-CIMP-high samples (absolute diff-mean difference >= 0.25, FDR <= 5%). Most of these probes (69%) were located outside of any known CpG island but positioned within intergenic regions known as open seas (Figure 3E). This represents a 2.5-fold open sea enrichment compared to the expected genome-wide distribution of 450K CpG probes (Chi-Square p-value < 2.2*10−16). We also observed a 3.4-fold depletion within CpG islands (Chi-Square p-value < 2.2*10−16).
Using this set of intergenic CpG probes, we asked whether a DNA motif signature associated with distal regulatory elements. Such a pattern would point to candidate transcription factors involved in tumorigenesis of the G-CIMP-low group. A de novo motif scan and known motif scan identified a distinct motif signature –TGTT- (geometric test p-value = 10−11, fold enrichment = 1.8), known to be associated with the OLIG2 and SOX transcription factor families (Figure 3E)(Lodato et al., 2013). This observation was corroborated by the higher expression levels of SOX2 as well as 17 out of 20 other known SOX family members in G-CIMP-low compared to G-CIMP-high (fold difference > 2). The primary function of SOX2 in the nervous system is to promote self-renewal of neural stem cells and, within brain tumors, the glioma stem cell state (Graham et al., 2003). Interestingly, SOX2 and OLIG2 have been described as neurodevelopmental transcription factors being essential for GBM propagation (Suva et al., 2014). Supervised gene expression pathway analysis of the genes activated in the G-CIMP-low group as opposed to G-CIMP-high group revealed activation of genes involved in cell cycle and cell division consistent with the role of SOX in promoting cell proliferation (Figure S3E). The enrichment in cell cycle gene expression provides additional support to the notion that development of the G-CIMP-low subtype is associated with activation of cell cycle progression and may be mediated by a loss of CpG methylation and binding of SOX factors to candidate genomic enhancer elements.
To validate the G-CIMP-low, G-CIMP-high and Codel IDH-mutant subtypes, we compiled a validation cohort from published studies including 324 adult and pediatric gliomas (Lambert et al., 2013; Mur et al., 2013; Sturm et al., 2012; Turcan et al., 2012). The CpG probe methylation signatures used to classify the validation set are provided on the publication portal accompanying this publication (https://tcga-data.nci.nih.gov/docs/publications/lgggbm_2015/). Among them, 103 were identified as IDH-mutant on the basis of their genome wide DNA methylation profile. We classified samples in the validation set using the probes that defined the IDH-mutant specific DNA methylation cluster analysis integrated in a supervised random forest method. The analysis recapitulated the clusters generated from the TCGA collection (Figure S3C). In order to determine epigenetically regulated (EReg) genes that may be characteristic of the biology of the IDH-mutant diffuse glioma subtypes, we compared 450k methylation DNA methylation profiles and gene expression levels between 636 IDH-mutant and IDH-wildtype gliomas and 110 non-tumor samples from eleven different tissue types (Guintivano et al., 2013). From the list of epigenetically regulated genes we extracted 263 genes that were grouped into EReg gene signatures which showed differential signals amongst the three IDH-mutant subtypes (Figure 3F). These trends were confirmed in the validation set (Figure 3G).
We investigated the possibility that the G-CIMP-high group is a predecessor to the G-CIMP-low group by comparing the DNA methylation profiles from ten IDH-mutant-non-codel LGG and GBM primary-recurrent cases with the TCGA cohort. We evaluated the DNA methylation status of probes identified as differentially methylated (n = 90) between G-CIMP-low and G-CIMP-high (FDR < 10−13, difference in mean methylation beta-value > 0.3 and < −0.4). Four out of ten IDHmut-non-codel cases showed a demethylation pattern after disease recurrence while partial demethylation was demonstrated in the remaining six recurrences, supporting the notion of a progression from G-CIMP-high to G-CIMP-low phenotype (Figure 3H).
An IDH-wildtype subgroup of histologically-defined diffuse glioma is associated with favorable survival and shares epigenomic and genomic features with pilocytic astrocytoma
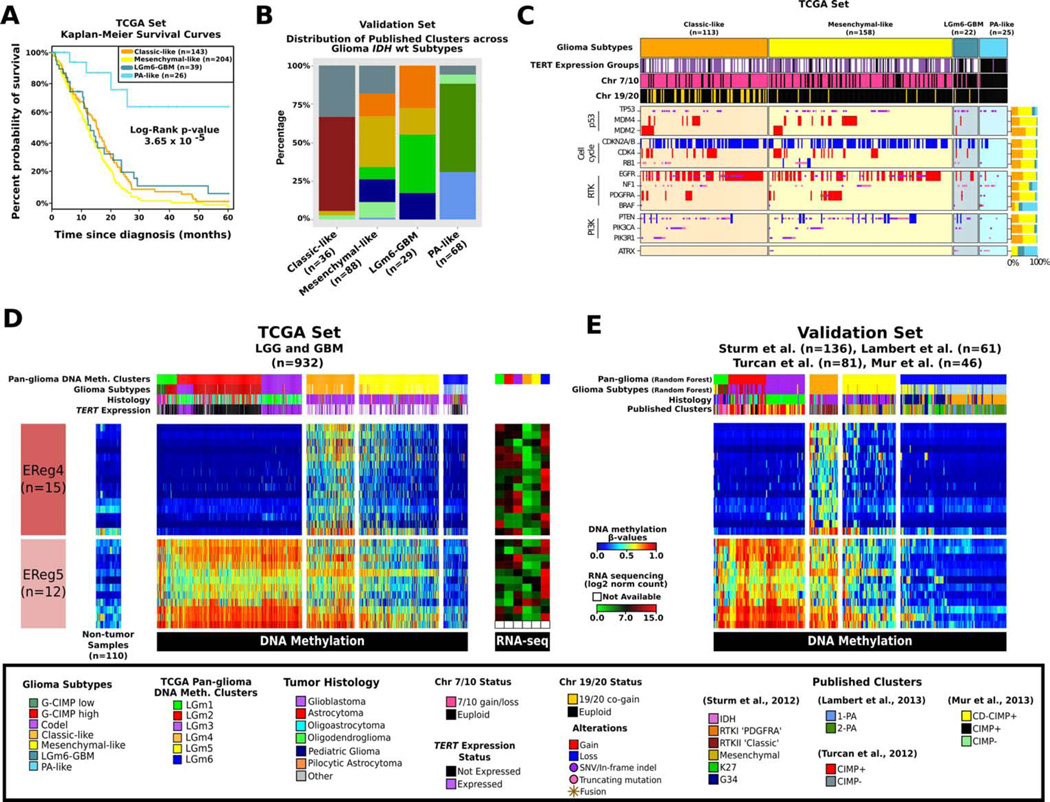
IDH-wildtype gliomas segregated into three DNA methylation clusters (Figure S4A). The first is enriched with tumors belonging to the classical gene expression signature and was labeled Classic-like, whereas the second group, enriched with mesenchymal subype tumors, was labeled Mesenchymal-like (Table S1)(Verhaak et al., 2010). The third cluster contained a larger fraction of LGG in comparison to the other IDH-wild type clusters. We observed that the IDH-wildtype LGGs but not the IDH-wildtype GBM in this cluster displayed markedly longer survival (log-rank p-value =3.6 × 10−5; Figure 4A) and occurred in younger patients (mean 37.6yr vs 50.8yr, t-test p-value = 0.002). Supervised analysis of differential methylation between LGG and GBM in the third DNA methylation cluster did not reveal any significant probes despite significant differences in stromal content (p-value < 0.005; Figure S4D), suggesting that this group cannot be further separated using CpG methylation markers.
Figure 4. A distinct subgroup of IDH-wildtype diffuse glioma with molecular features of pilocytic astrocytoma.
A. Kaplan-Meier survival curves for the IDH-wildtype glioma subtypes. Ticks represent censorship. B. Distribution of previous published DNA methylation subtypes in the validation set, across the TCGA IDH-wildtype specific DNA methylation clusters. C. Distribution of genomic alterations in genes frequently altered in IDH-wildtype glioma. D. Heatmap of TCGA glioma samples ordered according to Figure 2A and two EReg gene signatures defined for the IDH-wildtype DNA methylation clusters. Mean RNA sequencing counts for each gene matched to the promoter of the identified cgID across each cluster are plotted to the right. E. Heatmap of the validation set classified using the random forest method using the 1,300 probes defined in Figure 2A.
Next, we sought to validate the methylation-based classification of IDH-wildtype glioma in an independent cohort of 221 predicted IDH-wildtype glioma samples, including 61 grade I pilocytic astrocytomas (PA). Towards this aim, we used a supervised random forest model built with the probes that defined the IDH-wildtype clusters. Samples classified as Mesenchymal-like showed enrichment for the Sturm et al. Mesenchymal subtype (29/88) and gliomas predicted as Classic-like were all RTK II ‘Classic’ (22/22), per the Sturm et al. classification (Figure 4B; Figure S4B)(Sturm et al., 2012). We observed that PA tumors were unanimously classified as the third, LGG-enriched group (Figure S4B). Based on the molecular similarity with PA we labeled the LGGs in the third methylation cluster of IDH-wild type tumors as PA-like. The GBMs in this group were best described as LGm6-GBM, for their original pan-glioma methylation cluster assignment and tumor grade.
Pilocytic astrocytomas are characterized by frequent alterations in the MAPK pathway, such as FGFR1 mutations, KIAA1549-BRAF and NTRK2 fusions (Jones et al., 2013). The frequency of mutations, fusions and amplifications in eight PA-associated genes (BRAF, NF1, NTRK1, NTRK2, FGFR1, and FGFR2) rated from 11% (n=12/113) of Classic-like, 13% (n=21/158) of Mesenchymal-like IDH-wildtype tumors to 32% (n=7/22) of LGm6-GBM and 52% (n=13/25) of PA-like LGG (Fisher Exact Test (FET) p-value < 0.0001; Figure 4C). Conversely, only two of 25 (8%) PA-like LGG tumors showed TERT expression, compared to five of 12 LGm6-GBM (43%), 60 of 65 Classic-like (92%) and 82 of 98 Mesenchymal-like (84%, FET p-value < 0.0001). The PA-like group was characterized by relatively low frequency of typical GBM alterations, in genes such as EGFR, CDKN2A/B, and PTEN and displayed euploid DNA copy number profiles (Figure S4E). To ascertain that the histologies of the PA-like subgroup had been appropriately classified, we conducted an independent re-review. This analysis confirmed the presence of the histologic features of diffuse glioma (grade II or grade III) in 23 of the 26 cases in the cluster. The remaining three cases were re-named as PA (grade I). An independent review of the magnetic resonance diagnostic images from thirteen cases showed a similar pattern, with the majority of tumors showing behavior consistent with grade II or grade III glioma. Taken together, the epigenetic analysis of the IDH-wildtype group of adult glioma revealed the existence of a novel subgroup sharing genetic and DNA methylation features with pediatric PA, and favorable clinical outcome compared to diffuse IDH-wildtype glioma. This group may include but extends beyond BRAF-mutated grade II oligodendroglioma that were previously recognized as a unique clinical entity (Chi et al., 2013)
Through comparison of the methylation profiles of 636 glioma and 110 non-neoplastic normal samples from different tissue types, we defined EReg signatures consisting of 27 genes that showed differential signals amongst IDH-wildtype subtypes in the TCGA (Figure 4D) and the validation set (Figure 4E). EReg4 comprised a group of 15 genes hypermethylated and downregulated in particularly Classic-like. EReg5 was defined as a group of 12 genes associated with hypomethylation in LGm6/PA-like compared to all other LGm clusters. These ERegs aided in characterizing the biological importance of IDH-wildtype subtypes and were subsequently used to evaluate the prognostic importance of the IDH-wildtype clusters.
The epigenetic classification of glioma provides prognostic value independent of age and grade
In order to assess whether the DNA methylation-based subtypes we identified carry prognostically relevant information independent of known overall survival predictors, we constructed a series of survival regression models. To find the optimal model for survival prediction we studied covariates individually and in combination with other covariates. Age at diagnosis, histology, IDH/codel subtype, TERT expression and epigenetic subtype all contribute to survival in single-predictor analysis (log-rank p-value < 0.05, Table S4). As expected, age was a highly significant predictor (P<0.0001, C-Index 0.78) and was included in all subsequent multi-predictor models. We found that histology and grade are highly correlated. Histology provided only marginal improvement to a model that includes grade (likelihood ratio test (LRT) p-value = 0.08) and was therefore not included in further analyses. Conversely, grade markedly impacted a histology-based predictor model (LRT p-value = 0.0005, Table S4) and was retained in the subsequent models. In contrast to previous reports (Eckel-Passow et al., 2015), we failed to observe a statistically significant and independent survival association with TERT expression (LRT p-value = 0.82, Table S4) or TERTp mutations, after accounting for age and grade (LRT p-value = 0.85, not shown). Thus, the optimal survival prediction model includes age, grade and epigenetic subtype (LRT p-value< 0.0001, C-Index 0.836; Table 2).
Table 2.
Epigenetics subtypes are prognostically relevant in multivariable analysis and in external validation data. Survival regression analysis indicates that an optimal model of prognosis includes age, grade and methylation subtype. These predictors are statistically significant in both our discovery dataset and an external validation dataset.
| Discovery (N=809) C-Index: 0.835 ± 0.019 |
Validation (N=183) C-Index: 0.745 ± 0.032 |
||||||
|---|---|---|---|---|---|---|---|
| Predictor | Levels | N | HR (95% CI) | Signif. | N | HR (95% CI) | Signif. |
| Age at diagnosis | per year | 809 | 1.05 (1.03–1.06) | *** | 183 | 1.02 (1–1.04) | * |
| WHO Grade | II | 214 | 1.0 (ref) | 41 | 1.0 (ref) | ||
| III | 241 | 1.96 (1.15–3.33) | * | 51 | 1.24 (0.55–2.76) | ||
| IV | 354 | 2.38 (1.3–4.34) | * | 91 | 2.6 (1.08–6.3) | * | |
| Subgroup | IDHmut-codel | 156 | 1.0 (ref) | 57 | 1.0 (ref) | ||
| G-CIMP-low | 22 | 5.6 (2.49–12.62) | *** | 2 | 0 (0-Inf) | ||
| G-CIMP-high | 219 | 1.92 (1.05–3.51) | * | 15 | 1.25 (0.43–3.66) | ||
| Classic-like | 143 | 5.4 (2.79–10.44) | *** | 22 | 4.55 (1.8–11.49) | * | |
| Mesenchymal-like I | 204 | 8.71 (4.59–16.53) | *** | 61 | 5.55 (2.52–12.21) | *** | |
| LGm6-GBM | 39 | 5.79 (2.78–12.1) | *** | 22 | 6.8 (2.58–17.91) | ** | |
| PA-like | 26 | 2.02 (0.71–5.71) | 4 | 3.64 (0.79–16.78) | . | ||
Significance codes: 0
0.001
0.05
0.1.
To confirm that the epigenetic subtypes provide independent prognostic information we tested the survival model on the validation dataset. Epigenetic subtypes in these samples were determined as described above. The distinction between LGm6-GBM and PA-like gliomas was made on the basis of tumor grade and not by DNA methylation signature. Using a subset of 183 samples in the validation cohort with known survival, age and grade we found that epigenetic subtypes are significant independent predictors of survival in the multivariate analysis (LRT p-value < 0.0001, C-Index 0.746, Table 2). This generalization of our model supports the epigenetic subtypes as a means to improve the prognostication of glioma.
Activation of cell cycle/proliferation and invasion/microenvironmental changes marks progression of LGG to GBM
We observed that in spite of morphological differences between LGG and GBM, such as high cell density and microvascular proliferation, clustering of gene expression profiles frequently grouped LGG and GBM together within the same subtype. Gene Set Enrichment Analysis of the genes activated in G-CIMP GBM as opposed to the IDH-mutant-non-codel within LGr3 (Figure 2B) revealed four major groups, including cell cycle and hyperproliferation, DNA metabolic processes, response to stress, and angiogenesis (Figure S5A, Table S5). These biological functions are consistent with the criteria based on mitotic index used by pathologists to discriminate lower and high-grade glioma and the significance of activated microglia for tumor aggressiveness (Roggendorf et al., 1996). Conversely, compared with the G-CIMP GBM, IDH-mutant-non-codel LGG in LGr3 were characterized by enrichment of genes associated with neuro-glial functions such as ion transport and synaptic transmission, possibly suggesting a more differentiated nature. The comparison of co-clustered GBM and LGG in LGr3 by the PARADIGM algorithm that integrates DNA copy number and gene expression to infer pathway activity, confirmed that GBMs express genes associated with cell cycle, proliferation and aggressive phenotype, through activation of a number of cell cycle, cell replication and NOTCH signaling pathways whereas LGGs exhibit an enrichment of neuronal differentiation specific categories including synaptic pathways (Figure S5C, Table S5).
The analysis of the genes activated in GBM versus the LGG component of LGr4, which grouped IDH-wildtype tumors, identified an inflammation and immunologic response signature characterized by the activation of several chemokines (CCL18, CXCL13, CXCL2, CXCL3) and interleukins (IL8, CXCR2) enriching sets involved in inflammatory and immune response, negative regulation of apoptosis, cell cycle and proliferation, and the IKB/NFKB kinase cascade Map (Figure S5B, Table S5). These characteristics suggest differences in the relative amount of microglia. We used the ESTIMATE method to estimate the relative presence of stromal cells which revealed significantly lower (p-value 10−6) stromal scores of LGG IDH-wildtype versus GBM IDH-wildtype (Figure S5F)(Yoshihara et al., 2013). Resembling the functional enrichment for LGG within LGr3, functional enrichment of LGG IDH-wildtype in comparison to GBM within LGr4 showed activation in LGG of special glial-neuronal functions involved in ion transport, synaptic transmission and nervous system development.
Finally, we aimed to identify transcription factors that may exert control over prominent gene expression programs, known as master regulators. Master regulator analysis comparing the IDH-wildtype group to the IDH-mutant group revealed transcription factors that were upregulated in IDH-wildtype gliomas and showed an increase in expression of target genes, including NKX2-5, FOSL1, ETV4, ETV7, RUNX1, CEBPD, NFE2L3, ELF4, RUNX3, NR2F2, PAX8 and IRF1 (Table S5). No TFs were found to be upregulated in IDH-mutant gliomas relative to IDH-wildtype gliomas (at a log fold change >1).
DISCUSSION
This study represents the largest multi-platform genomic analysis performed to date of adult diffuse glioma (WHO grades II, III and IV). A simplified graphical summary of the identified groups and their main clinical and biological characteristics is reported in Figure 5. The clustering of all diffuse glioma classes and grades within similarly shaped methylation-based and expression-based groups has allowed us to pinpoint specific molecular signatures with clinical relevance. The DNA methylation classification proposed should be considered as a basis and it is likely that future studies involving significantly larger cohorts and more refined profiling methods will be able to further reduce intra-subtype heterogeneity. The dissection of the IDH-mutant non-codel G-CIMP LGG and GBM into two separate subgroups (G-CIMP-low and G-CIMP-high) based on the extent of genome-wide DNA methylation has crucial biological and clinical relevance. In particular, the identification of the G-CIMP-low subset, characterized by activation of cell cycle genes mediated by SOX binding at hypomethylated functional genomic elements and unfavorable clinical outcome is an important finding that will guide more accurate segregation and therapeutic assessment in a group of patients in which correlations of conventional grading with outcome are modest (Olar et al., 2015; Reuss et al., 2015). The finding that G-CIMP-high tumors can emerge as G-CIMP-low glioma at recurrence identify variations in DNA methylation as crucial determinants for glioma progression and provides a clue to the mechanisms driving evolution of glioma. Our results unify previous observations that linked the cell cycle pathway to malignant progression of low grade glioma (Mazor et al., 2015). Future updates of the TCGA glioma clinical annotation and independent validation of our findings may be able to consider additionally important clinical confounders such as extent of resection and performance status, to further optimize the weights of the currently known prognostic variables and their association to the molecular subtypes we identified.
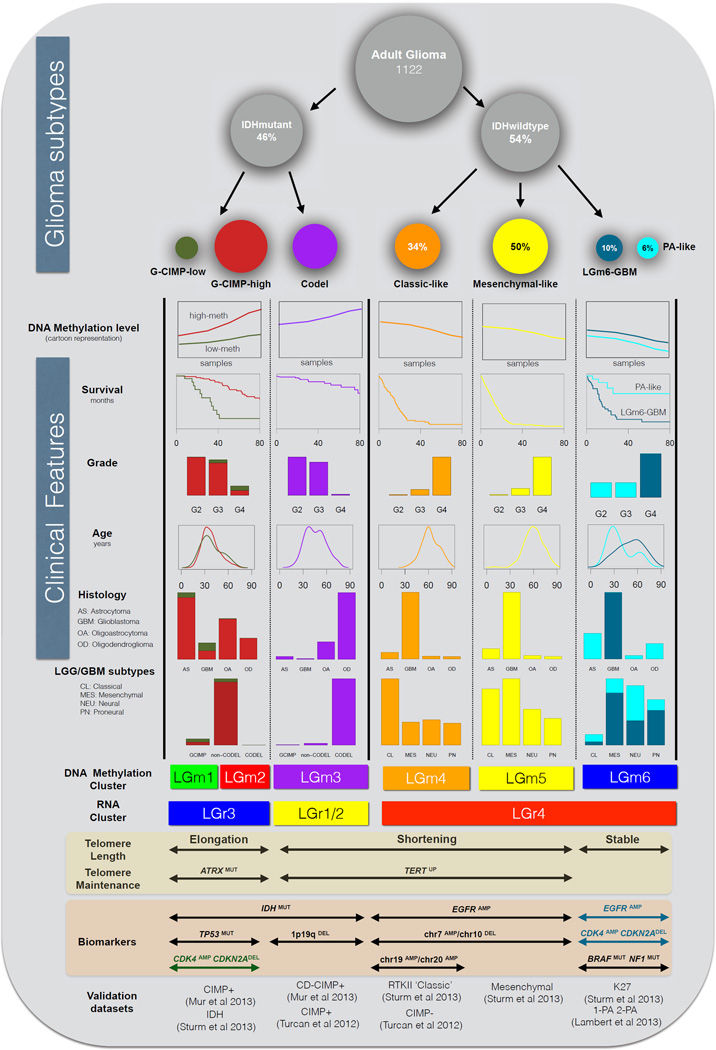
Figure 5. Overview of major subtypes of adult diffuse glioma.
Integrative analysis of 1,122 adult glioma resulted in seven different subtypes with distinct biological and clinical characteristics. The groups extend across six DNA methylation subtypes of which the LGm6 cluster was further separated by tumor grade, into PA-like and LGm6-GBM. The size of the circles is proportional to the percentages of samples within each group. DNA methylation plot is a cartoon representation of overall genome-wide epigenetic pattern within glioma subtypes. Survival information is represented as a set of Kaplan-Meier curves, counts of grade, histology and LGG/GBM subtypes within the groups are represented as bar-plots, whereas age is represented as density. Labeling of telomere length and maintenance status is based on the enrichment of samples within each column, similarly for the biomarkers and the validation datasets.
Analysis of IDH-wildtype glioma revealed the PA-like LGG subset that harbors a silent genomic landscape, confers favorable prognosis relative to other IDH-wildtype diffuse glioma and displays a molecular profile with high similarity to PA. Re-review by neuropathologists and neuroradiologists confirmed that the majority were correctly diagnosed as diffuse glioma, emphasizing the need for integration of molecular signatures intro clinical classification (Chi et al., 2013) for this subgroup of patients that may be spared potentially unnecessary intensive treatments.
The large number of exomes in our dataset allowed identification of novel glioma-associated somatic alterations, including in the KRAS and NRAS genes which were frequently used to genetically engineered glioma mouse models (Holland et al., 2000). Our analysis further nominates glial tumors to join an increasing number of tumor types characterized by a deactivated cohesin pathway (Kon et al., 2013; Solomon et al., 2011). Cohesin mutant tumors may infer increased sensitivity to DNA damage agents and PARP inhibitors (Bailey et al., 2014) suggesting that gliomas with genetic alterations of key cohesin regulatory factors may represent biomarkers and therapeutic opportunities.
Overexpression of TERT mRNA was found to be associated with increased telomere length in urothelial cancer (Borah et al., 2015). Our results revealed that in gliomas, increased telomere length is associated with ATRX mutations, suggesting an ALT mechanism. ALT has been associated with sensitivity to inhibition of the protein kinase ATR (Flynn et al., 2015).
In summary, our pan-glioma analysis has expanded our knowledge of the glioma somatic alteration landscape, emphasized the relevance of DNA methylation profiles as a modality for clinical classification, and quantitatively linked somatic TERT pathway alterations to telomere maintenance. Combined, these findings are an important step forward in our understanding of glioma as discrete disease subsets, and the mechanisms driving gliomagenesis.
EXPERIMENTAL PROCEDURES
Patient and Sample Characteristics
Specimens were obtained from patients, with appropriate consent from institutional review boards. Details of sample preparation are described in the Extended Experimental Procedures.
Data Generation
In total, tumors from 1,132 patients were assayed on at least one molecular profiling platform, which platforms included: (1) whole genome sequencing, including high coverage and low pass whole genome sequencing, (2) exome sequencing (3) RNA sequencing (4) DNA copy-number and single-nucleotide polymorphism arrays, including Agilent CGH 244K, Affymetrix SNP6.0 and Illumina 550K Infinium HumanHap550 SNP Chip microarrays (5) gene expression arrays, including, Agilent 244K Custom Gene Expression, Affymetrix HT-HGU133A and Affymetrix Human Exon 1.0 ST arrays (5) DNA methylation arrays, including Illumina GoldenGate Methylation, Illumina Infinium HumanMethylation27, and Illumina Infinium HumanMethylation450 BeadChips (7) reverse phase protein arrays, (8) miRNA sequencing and (9) miRNA Agilent 8 × 15K Human miRNA-specific microarrays. Details of data generation have been previously reported (Brennan et al., 2013; TCGA_Network et al., 2015). To ensure across-platform comparability, features from all array platforms were compared to a reference genome.
Data Analysis
The data and analysis results can be explored through the Broad Institute FireBrowse portal (http://firebrowse.org/?cohort=GBMLGG), the cBioPortal for Cancer Genomics (http://www.cbioportal.org/study.do?cancer_study_id=lgggbm_tcga_pub), in a Tumor Map (http://tumormap.ucsc.edu/?p=ynewton.gliomas-paper), the TCGA transcript fusion portal (http://www.tumorfusions.org), TCGA Batch Effects (http://bioinformatics.mdanderson.org/tcgambatch/), Regulome Explorer (http://explorer.cancerregulome.org/), Next-Generation Clustered Heat Maps (http://bioinformatics.mdanderson.org/TCGA/NGCHMPortal/). See also Supplemental Information and the TCGA publication page (https://tcga-data.nci.nih.gov/docs/publications/lgggbm_2015/).
Supplementary Material
Acknowledgments
This study was supported by NIH grants: U24CA143883, U24CA143858, U24CA143840, U24CA143799, U24CA143835, U24CA143845, U24CA143882, U24CA143867, U24CA143866, U24CA143848, U24CA144025, U54HG003067, U54HG003079, U54HG003273, U24CA126543, U24CA126544, U24CA126546, U24CA126551, U24CA126554, U24CA126561, U24CA126563, U24CA143731, U24CA143843, P30CA016672, P50 CA127001, R01 CA190121, P01 CA085878, U54CA193313, R01CA179044, R01CA185486; Cancer Prevention & Research Institute of Texas (CPRIT) R140606; São Paulo Research Foundation (FAPESP) 2014/02245-3, 2015/07925-5, 2015/02844-7 and 2015/08321-3.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization and project administration: R.G.W.V., A.I., and H.N.; Supervision: S.R.S., K.D.A., P.W.L., M.G., D.H., D.J. B., D.H.G., R.R., C.C.L., J.B.S., C.G.C.J., D.P.C.T., W.K.A.Y., J.H., L.C., M.M., and T.M.; Formal Analysis: R.G.W.V., A.I., H.N., M.C., F.P.B., T.M.M., T.S.S., O.M., Y.N., S.M.P., P.Z., L.P., Amie R., G.R., R.A., J.W. G.M., S.L., S.A., Arvind R., B.A.M., A.D.C., and H.Z.; Investigation: D.J.B., L.C., and L.P.; Data Curation: D.J.B., L.P., and F.P.B; Writing - Original Draft: R.G.W.V., A.I., H.N., M.C., F.P.B., T.M.M., and T.S.S.; Manuscript review: DJB, KAD, SRS, MW, NL, DHG.
REFERENCES
- Bailey ML, O'Neil NJ, van Pel DM, Solomon DA, Waldman T, Hieter P. Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition. Molecular cancer therapeutics. 2014;13:724–732. doi: 10.1158/1535-7163.MCT-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Batchelor TT, Yang D, Dias-Santagata D, Borger DR, Ellisen LW, Iafrate AJ, Louis DN. BRAF V600E mutation identifies a subset of low-grade diffusely infiltrating gliomas in adults. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:e233–e236. doi: 10.1200/JCO.2012.46.0220. [DOI] [PubMed] [Google Scholar]
- Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. The New England journal of medicine. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nature genetics. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome research. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nature genetics. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, Criekinge WV, Daniel RT, van den Bent MJ, Marosi C, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. doi: 10.1186/s13059-015-0583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SR, Witt H, Hovestadt V, Zucknick M, Kool M, Pearson DM, Korshunov A, Ryzhova M, Ichimura K, Jabado N, et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol. 2013;126:291–301. doi: 10.1007/s00401-013-1124-7. [DOI] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS genetics. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJ, Smirnov IV, Reis GF, Phillips JJ, Barnes MJ, et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur P, Mollejo M, Ruano Y, de Lope AR, Fiano C, Garcia JF, Castresana JS, Hernandez-Lain A, Rey JA, Melendez B. Codeletion of 1p and 19q determines distinct gene methylation and expression profiles in IDH-mutated oligodendroglial tumors. Acta Neuropathol. 2013;126:277–289. doi: 10.1007/s00401-013-1130-9. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, Armstrong TS, Sulman EP, Cahill DP, Vera-Bolanos E, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta neuropathologica. 2015;129:585–596. doi: 10.1007/s00401-015-1398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, Holland EC. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, White MA, Potts PR. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–728. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss DE, Mamatjan Y, Schrimpf D, Capper D, Hovestadt V, Kratz A, Sahm F, Koelsche C, Korshunov A, Olar A, et al. IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta neuropathologica. 2015;129:867–873. doi: 10.1007/s00401-015-1438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggendorf W, Strupp S, Paulus W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta neuropathologica. 1996;92:288–293. doi: 10.1007/s004010050520. [DOI] [PubMed] [Google Scholar]
- Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. quiz 491 p following 516. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- TCGA_Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathol. 2010;120:297–304. doi: 10.1007/s00401-010-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.