Abstract
Quality nutrition during the period of brain formation is a predictor of brain functional capacity and plasticity during adulthood; however it is not clear how this conferred plasticity imparts long-term neural resilience. Here we report that early exposure to dietary omega-3 fatty acids orchestrates key interactions between metabolic signals and Bdnf methylation creating a reservoir of neuroplasticity that can protect the brain against the deleterious effects of switching to a western diet (WD). We observed that the switch to a WD increased Bdnf methylation specific to exon IV, in proportion to anxiety-like behavior, in Sprague Dawley rats reared in low omega-3 fatty acid diet, and these effects were abolished by the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine. Blocking methylation also counteracted the reducing action of WD on the transcription regulator CTCF binding to Bdnf promoter IV. In vitro studies confirmed that CTCF binding to Bdnf promoter IV is essential for the action of DHA on BDNF regulation. Diet is also intrinsically associated to cell metabolism, and here we show that the switch to WD downregulated cell metabolism (NAD/NADH ratio and SIRT1). The fact that DNA methyltransferase inhibitor did not alter these parameters suggests they occur upstream to methylation. In turn, the methylation inhibitor counteracted the action of WD on PGC-1α, a mitochondrial transcription co-activator and BDNF regulator, suggesting that PGC-1α is an effector of Bdnf methylation. Results support a model in which diet can build an “epigenetic memory” during brain formation that confers resilience to metabolic perturbations occurring in adulthood.
Keywords: Epigenetics, Omega-3 fatty acid, Anxiety, BDNF, Metabolism
Introduction
An increasing body of evidence suggests that a large number of neuropsychiatric disorders are the result of complex interactions between genetic factors and the environment (Dauncey, 2012) and that epigenetic mechanisms may mediate these effects (Choi and Friso, 2010). Diet is one of the most crucial factors for species survival and adaptation, and here we examine the possibility that foods influence the brain by building an “epigenetic memory” that could be instrumental in resistance to neurological challenges. The strong dependence of the brain on energy implies that metabolic stimuli such as dietary factors have the intrinsic ability to influence brain plasticity (Agrawal et al., 2014) and epigenetic variability.
BDNF malfunction has been implicated in the pathology of neurological and psychiatric disorders (Greenberg et al., 2009; Zuccato and Cattaneo, 2009). Bdnf transcription and function is under regulation of DNA methylation (Martinowich et al., 2003), and abnormal epigenetic regulation of the Bdnf gene is emerging an underlying mechanism for various neurological disorders (Boulle et al., 2012). DNA methylation is the one of most stable forms of epigenetic variability involved in the control of transcription and function of selected genes (Godfrey et al., 2007; Moore et al., 2013) can repress Bdnf gene expression via transcriptional repressor methyl-CpG-binding protein (MeCP2) (Chen et al., 2003; Ma et al., 2009). Since BDNF can act on plasticity and metabolism it is termed a “metabotrophin” (Gomez-Pinilla, 2008; Gomez-Pinilla et al., 2008), and these inherent properties of BDNF may be crucial to translate the effects of foods on the brain. For example, one potential intermediary may be nicotinamide adenine dinucleotide (NAD+), a cofactor in oxidation/reduction (hydride transfer) reactions. The action of NAD on energy regulation is under the scope of the sirtuin family of proteins (Cantó and Auwerx, 2012). SIRT1 influences the activity of peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α), which is one of the most versatile metabolic transcriptional co-activators of genes involved in energy metabolism (Nemoto et al., 2005; Rodgers et al., 2008) and mitochondrial function (Finkel, 2006).
The omega-3 polyunsaturated fatty acid docosahexaenoic acid (DHA) is associated with reduced risk of Alzheimer's disease, schizophrenia and depression (Young and Conquer, 2005). On the other hand, dietary intakes of saturated fats do the opposite (Sánchez-Villegas et al., 2011; Tyagi et al., 2013). The current studies were designed to determine whether exposure to omega-3 fatty acids during the whole period of brain formation promotes epigenetic variability and protection against metabolic perturbations. Results introduce the idea that metabolism and epigenetics are closely associated events that contribute to program an epigenetic memory through long-term brain plasticity. These results provide fundamental information for the design of therapeutic applications and public health policy.
Materials and Methods
Animals and Experimental design
Female Sprague–Dawley rats (250-280 gm) were obtained from Charles River Laboratories (Wilmington, MA, USA) on 3rd day of pregnancy and were kept under standard housing condition (22–24 °C) with 12-h light/dark cycle in two dietary groups. Pregnant female rats were fed either a low omega-3 fatty acid (Low-n-3) or a high omega-3 fatty acid diet (High-n-3) through gestation and lactation. The two custom diets used were based on the composition of the American Institute of Nutrition diet (AIN-93G) and prepared commercially (Dyets Inc., PA, USA) as previously described (Greiner et al., 2003). Both diets had the same basal macronutrients, vitamins, minerals, and basal fats (hydrogenated coconut and safflower oils) (Table 1). The total fat content in both diets was ~10% (w/w) and the amount of omega-3 fatty acids in low and high omega-3 diets were 0.05% (w/w) and 1.8% (w/w) respectively. The amount of omega-3 fatty acid in high omega-3 diet was achieved by adding 0.48% (w/w) of flaxseed oil (source of α-linolenic acid, ALA), and 1.2% (w/w) of docosahexaenoic acid (DHA), 0.24% (w/w) of eicosapentaenoic acid (EPA) and 0.1% (w/w) of other omega-3 fatty acids (procured as ProDHA oil from Nordic Naturals, Inc., CA, USA) (Table 2). The study consists of a total 16 litters (8/diet group), and only male offspring were included such that each offspring within an experimental group was selected from different litter. At weaning on postnatal day 21, offspring were randomly divided into four experimental groups (n=8 in each group) based on their maternal (Low-n-3 or High-n-3) and transitioned diet [continued to either maternal diet (i.e. no transition to western diet, WD) or switched to the WD]. The male offspring from dams fed with low omega-3 diet were split into (1) low omega-3 diet offspring with no dietary transition i.e. continued to the same diet (Low-n-3/No WD) and (2) low omega-3 diet offspring switched to the WD (Low-n-3/WD) after 8 weeks of weaning (i.e. at 11 weeks age). The offspring from dams fed with high omega-3 diet were also split into (3) high omega-3 diet offspring with no dietary transition i.e. continued to the same diet (High-n-3/No WD) and (4) high omega-3 diet offspring switched to the WD (High-n-3/WD) after 8 weeks of weaning (i.e. at 11 weeks age). The duration of the dietary treatment period was chosen to cover the whole period of formation of the frontal cortex, including development of the synaptic machinery and establishment of the adult pattern of BDNF expression. It has been reported that the peak expression of BDNF mRNA in prefrontal cortex of human postmortem brains is reached at a period equivalent to the 7-10 week (P49-P70) age of laboratory rats (Sengupta, 2013; Webster et al., 2002). The highest expression of synaptic proteins: synapsin and synaptophysin in frontal cortex also occurs at 5-7 weeks (P37-P45) (Pinto et al., 2013) with continued pruning up to 14-21 weeks (P100- P150) (Kolb et al., 2012). Both low omega-3 and high omega-3 diets have same amount of sugar (10% w/w) and fat (10% w/w), however, WD (D12079B; Research Diets. Inc., NJ, USA) contains high sugar (34% w/w) and high fat (21% w/w). The diets were provided in powder in a bowl and animals had free access to food and water. There were no significant changes observed in body weight and weight gain of animals exposed to the experimental diet (Fig. S1). At 18 weeks of age (i.e. after 7 weeks of WD transition), the offspring were subjected to the elevated plus maze (EPM) test, to assess the anxiety-like behavior and were killed immediately by decapitation after the behavioral test. Fresh brain tissues were dissected out, frozen in dry ice and stored at −70 °C until use. All parameters were studied in the frontal cortex tissue from the same animals. All experiments were performed in accordance with the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the University of California at Los Angeles Chancellor's Animal Research Committee.
Table 1.
Composition of experimental diets
| Ingredient | Amount (g/100 g diet) | |
|---|---|---|
| High omega-3 diet | Low omega-3 diet | |
| Alacid 710, acid casein | 20 | 20 |
| Cornstarch | 15 | 15 |
| Sucrose | 10 | 10 |
| Dextrose | 19 | 19.9 |
| Maltose-dextrin | 15 | 15 |
| Cellulose | 5 | 5 |
| Salt-mineral mix | 3.5 | 3.5 |
| Vitamin mix | 1 | 1 |
| L-cystine | 0.3 | 0.3 |
| Choline bitartrate | 0.25 | 0.25 |
| TBHQ | 0.002 | 0.002 |
| Fat sources: | ||
| Hydrogenated coconut oil | 7.45 | 8.1 |
| Safflower oil | 1.77 | 1.9 |
| Flaxseed oil | 0.48 | -- |
| DHAa | 1.2 | -- |
| EPAa | 0.24 | -- |
| Other n-3sa | 0.1 | -- |
Note: Dashes indicate that component was not added.
Procured from Nordic Naturals, Inc., Watsonville, CA, USA as ProDHA capsule that contains 45% (w/w) DHA, 9% (w/w) EPA and 4% (w/w) other n-3s.
Table 2.
Fatty acid composition of the diets as percentage of total fatty acids
| Fatty acids | High omega-3 diet | Low omega-3 diet |
|---|---|---|
| Total saturated | 66.49 | 75.95 |
| Total monounsaturated | 3.87 | 2.18 |
| 18:2n-6 | 13.58 | 16.67 |
| 20:4n6 | 0.518 | ND |
| 18:3n-3 | 2.71 | 0.052 |
| 20:5n-3 | 2.82 | ND |
| 22:6n-3 | 9.07 | ND |
Note: ND indicates that component was not detected.
To examine the methylation specific changes in behavior, a separate cohort of animals transitioned from low omega-3 to the WD (n=8/group) were subjected to either intraperitoneal PBS vehicle (1 ml/kg body weight) or 5-aza-2'-deoxycytidine (5-AzaD; 0.4 mg/kg, ip), a DNA methyltransferase (DNMT) inhibitor, for 10 days daily till 1 h prior to the behavior. The 5-AzaD treatment was given only to low omega-3 groups as Bdnf hypermethylation was observed only in those animals. Animals were killed immediately by decapitation after the behavioral test and brain tissues were collected in dry ice and stored at -70 °C until use.
Elevated plus maze (EPM)
At 18 weeks of age (i.e. after 7 weeks of WD transition), EPM test was performed according to the (Walf and Frye, 2007). Briefly, the EPM apparatus made of laminated wood consisted of 2 opposing open arms (10 × 50 cm) and 2 opposing closed arms (10 × 50 cm with 30 cm high walls). The maze was placed 60 cm above the floor. White curtains surrounded the maze and behavior was recorded by an overhead video camera over a period of 5 min. Each rat was placed in the middle of the maze facing the open arm that faced away from the experimenter. The time spent in each arm, total distance traveled and total number of entries was analyzed using AnyMaze video tracking software (San Diego Instruments, CA, USA). The exploratory behavior of animals was depicted by the heat map occupancy plot generated by the AnyMaze video tracking software (Oaks et al., 2013). The total distance and total number of times rats entered in arms during the EPM test was calculated to account for differences in general motor activity in the maze.
Methylated DNA immunoprecipitation
Frontal cortical tissue from rats was collected and used to investigate methylation status of promoters for exons IV and VI via EpiQuik Tissue Methylated DNA immunoprecipitation Kit (Epigentek, NY, USA) according to the manufacturer's instruction. Briefly, tissue was lysed and genomic DNA was shearing by sonication. Samples were incubated with an antibody against 5-methylcytosine (1:1000, Millipore, MA, USA) or with an equivalent amount of normal IgG (anti-mouse). A portion of the sonicated DNA was left untreated to serve as input control. Immunoprecipitated DNA was subjected to quantitative real-time PCR using primers specific for 200-bp segments corresponding to CpG island sites within rat Bdnf exon IV and exon VI. For Bdnf exon IV the forward primer was 5′-AAGGTTTGGCTTCTGTGTG-3′ and the reverse primer was 5′-TGCACGAATTACCAGAATCAG-3′. For Bdnf exon VI forward primer was 5′-ACTCCCCGGCTTGGAGAAGGAAACC-3′ and the reverse primer was 5′-TCAGGGTCCACACAAAGCTCTCGGA-3′. The cumulative fluorescence for each amplicon was normalized to input amplification.
Chromatin immunoprecipitation assay
Frontal Cortical tissue from rats was collected and used for chromatin immunoprecipitation assay by Chromatin Immunoprecipitation (ChIP) assay kit (Millipore, MA, USA) according to the manufacturer's instruction. Briefly, tissue was disaggregated and incubated in 1% formaldehyde (cross-link). Chromatin was sheared by sonication. Samples were incubated with anti-CTCF antibody (1:1000, Millipore, Cat# 07-729, RRID: AB_441965) or with an equivalent amount of normal IgG (anti-mouse). A portion of the sonicated DNA was left untreated to serve as input control. Immunoprecipitated DNA was subjected to quantitative realtime PCR using primers specific to the rat Bdnf exon IV & VI. The cumulative fluorescence for each amplicon was normalized to input amplification.
NAD/NADH quantification assay
Intracellular NAD and NADH concentrations were determined by NAD/NADH Quantitation colorimetric Kit (BioVision Inc., CA, USA) as per manufacturer's instruction. Briefly, 20 mg of tissue homogenates were centrifuged in the NAD/NADH extraction buffer. The half of the lysate was used to determine total NAD concentration. The other half was heated to 60°C for 30 min, and used to determine NADH concentration. The reactions were prepared in 96-well plates, and read at 450 nm. Finally, the NAD concentration was determined by subtracting NADH concentration from total NAD (NADH and NAD) concentration.
Immunobloting
The frontal cortical tissues were homogenized in a lysis buffer containing 137 mM NaCl, 20 mM Tris–HCl pH 8.0, 1% NP40, 10% glycerol, 1 mM phenylmethylsulfonylfluoride (PMSF), 10 μg/ml aprotinin, 1 μg/ml leupeptin, 0.1 mM benzethonium chloride, 0.5 mM sodium vanadate. The homogenates were then centrifuged, the supernatants were collected and total protein concentration was determined according to MicroBCA procedure (Pierce, IL, USA), using bovine serum albumin (BSA) as standard. Briefly, protein samples were separated by electrophoresis on 10% polyacrylamide gel and electrotransferred to a PVDF membrane (Millipore, MA, USA). Non-specific binding sites were blocked in Tris-buffered saline (TBS), pH 7.6, containing 5% non-fat dry milk. Membranes were rinsed in buffer (0.05% Tween-20 in TBS) and then incubated with anti-BDNF (1:300; Santa Cruz Biotechnology, Cat# sc-546, RRID: AB_630940), anti-actin, (1:1000; Santa Cruz Biotechnology, Cat# sc-1616, RRID: AB_630836), anti-PGC-1α (Cat# ST1202, RRID: AB_2237237), anti-Sir2 (Cat# 07-131, RRID: AB_2188349), anti-CTCF (Cat# 07-729, RRID: AB_441965), anti-MeCP2 (Cat# 07-013, RRID: AB_2144004) (1:1000, Millipore) or anti-pMeCP2 (1:1000, Cell Signaling, Cat# AP3693a, RRID: AB_10612715) primary antibodies followed by anti-rabbit or anti goat or anti-mouse IgG horseradish peroxidase-conjugate (1:10,000; Santa Cruz Biotechnology) secondary antibodies. After rinsing with buffer, the immunocomplexes were visualized by chemiluminescence using the ECL kit (Amersham Pharmacia Biotech Inc., NJ, USA) according to the manufacturer's instructions. The signals were digitally scanned and then quantified using ImageJ software. Actin was used as an internal control such that data were standardized according to actin values.
RNA isolation and real-time quantitative polymerase chain reaction
An RNA STAT 60 kit (Tel-Test Inc., Friendswood, TX, USA) was used for total RNA isolation and the procedure was followed by the manufacturer's protocol. RNA concentration was assessed using a NanoPhotometer (Implen, Germany). The ratio of the absorbance at 260 and 280 nm (A260/A280) was within the ratio 1.9 and 2.1 for all samples. Real-time quantitative polymerase chain reaction (RT-PCR) was performed to measure the mRNA levels of Bdnf, Glyceraldehyde-3-phosphate dehydrogenase served as an internal control for sample normalization. Total RNA (100 ng) was used for cDNA synthesis (iScript cDNA Synthesis kit, Bio-Rad Laboratories Inc., Hercules, CA, USA). The synthesized cDNA was the template for the real-time polymerase chain reaction (PCR) amplification carried out by the CFX96 real-time PCR detection system (Bio-Rad Laboratories Inc.) using a SsoFast EvaGreen Supermix kit (Bio-Rad Laboratories Inc.) and forward/reverse primers. The sequences of primers were as follows: Bdnf exon IV primer pair (Lubin et al., 2008), forward: 5′-TGCGAGTATTACCTCCGCCAT-3′, reverse: 5′-TCACGTGCTCAAAAGTGTCAG-3′; and glyceraldehyde- 3-phosphate dehydrogenase primer pair, forward: 5′-TGCCACTCAGAAGACTGTGG-3′, reverse: 5′-TTCAGCTCTGGGATGACCTT-3′. Glyceraldehyde-3-phosphate dehydrogenase primers were generated with primer3 software. The reverse transcription reaction steps for each cycle consisted of an initial 30 s incubation step at 95 C for enzyme activation and were followed by 5 s at 95 C for denaturation and then 5 s at 60 C for annealing/extension. Three negative (no template) controls were performed to verify the sample genomic DNA contamination. Quantification of the RT-PCR results was performed using the CFX manager software version 1.6 (Bio-Rad Laboratories Inc.).
In vitro experiments
CTCF blocking by siRNA in N2a cells
Neuro-2a cells (mouse neuroblastoma cell line from American Type Culture Collection; ATCC, VA, USA) were maintained as monolayer cultures in Dulbecco's modified Eagle medium (DMEM) (Gibco by Life Technologies Inc., NY, USA) supplemented with 10% (v/v) fetal bovine serum (Gibco by Life Technologies Inc., NY, USA), penicillin (100 units/ml) and streptomycin (100 μg/ml) at 37 °C, in a humidified atmosphere containing 95% air and 5% CO2. Commercially available siRNA-CTCF duplexes and control siRNA-A were purchased from Santa Cruz Biotechnology. siRNA-CTCF is a pool of three target-specific 19- to 25-nucleotide siRNAs designed to knock down CTCF gene expression. Negative control siRNA (siRNA-con) consisted of a scrambled sequence that does not lead to the specific degradation of any known cellular mRNA. The N2a cells were transfected using siRNA duplexes according to the manufacturer's protocol. After 24 h, the transfected cells were treated with DHA (10-40 μM; Nu-Chek Prep, Inc., MN, USA) for 24 h and the treatment cells were harvested for protein expression studies. All cells were grown in DMEM medium containing α-tocopherol (2 μM). Cell lysates were prepared in 1x RIPA lysis buffer containing protease inhibitors for immunobloting. Total protein concentration was determined according to MicroBCA procedure (Pierce, IL, USA), using bovine serum albumin (BSA) as standard and immnobloting was performed as described above.
Immunocytochemistry
N2a cells were plated onto 8-chamber slides for immunocytochemical analysis and were cultured for 1 day. The transfection and treatment for cells was followed as described above. After 24 h of treatment, cells were fixed with 4% para-formaldehyde for 10 min, washed with PBS and incubated for 1 h at room temperature with TBS solution containing 3% BSA and 0.3% Triton X-100 to inhibit non-specific binding. Anti-BDNF (1:300; Santa Cruz Biotechnology, CA, USA) and anti-CTCF (1:300, Millipore, MA, USA) antibodies, diluted in 1% bovine serum albumin (BSA)/TBST were added to the cells and incubated overnight at 4°C. After thorough washing with PBS, the sections cells were incubated with appropriate dilutions of fluorescent sheep or mouse secondary antibodies (FITC; 1:1000, Cy3; 1:4000; Jackson ImmunoResearch Laboratories Inc., PA, USA) for 1 h at room temperature. Slides were washed in PBS and mounted using aquamount. Negative controls were performed by omission of the primary antibody. The results of immunocytochemistry controls were negative as no staining was observed in cell structures. The staining was visualized under Zeiss microscope (Zeiss Imager.Z1) using the Axiovision 4.6 software.
Statistical Analysis
The results are represented as mean ± standard error of the mean (SEM). Data for in vivo experiments were analyzed by two-way ANOVA [(Maternal diet: Low-n-3 vs. High-n-3) and (Transitioned diet: No WD vs. WD)]. Post hoc analyses were conducted using Tukey's multiple comparison tests to determine the significance of difference among various groups. The data with treatment of DNMT inhibitor, 5-AzaD were analyzed by Student (unpaired) ‘t’ test. Data for CTCF binding at promoter region of Bdnf exon IV and in vitro experiments were analyzed by oneway ANOVA followed by Tukey's multiple comparison Post hoc tests. A level of 5% probability was considered as statistically significant. Pearson correlation analysis was performed on individual samples to evaluate the association between variables.
Results
Dietary interventions influences anxiety-like behavior
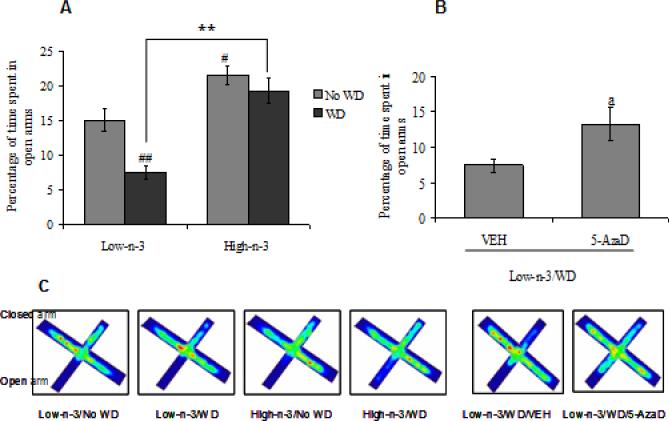
We assessed the influence of dietary manipulations on anxiety-like behavior by utilizing elevated plus maze test (EPM). Two-way ANOVA analysis revealed a significant effect of maternal diet (F1,28=39.38, p<0.01) and transitioned diet (F1,28=11.47, p<0.01) for percentage of time spent in open arms. We found that transition from low omega-3 to WD decreased percentage of time spent in open arms as an indication of elevated anxiety-like behavior (Low-n-3/WD Vs Low-n-3/No WD, p<0.01). Animals that had been reared in a diet enriched in omega-3 did not show behavioral alterations after switching to WD (High-n-3/WD Vs High-n-3/No WD, p>0.05). Results suggest that prior exposure of high omgea-3 diet conferred protection against WD induced anxiety like behavior (High-n-3/WD Vs Low-n-3/WD, p<0.01). Animals reared on high omega-3 diet until sacrifice, spent more time on the open arms compared to the low omega-3 diet group (High-n-3/No WD Vs Low-n-3/No WD, p<0.05; Fig 1A). The heat map occupancy plots for EPM also show the increased exploration of the open arms in animals pre-exposed to the high omega-3 (Fig. 1C). The total distance in meter (Low-n-3/No WD: 3.5±0.6; Low-n-3/WD: 4.7±1.0; High-n-3/No WD: 4.2±0.8; High-n-3/WD: 4.9±0.7; there was no statistical difference (P>0.05) between any of the groups) and total number of entries (data not shown) was similar among all the groups during the EPM test suggesting that the changes in anxiety-like behavior were not due to changes in motor activity.
Fig. 1.
(A) Percentage of time spent in open arms of elevated plus maze in low omega-3 (Low-n-3) or high omega-3 diet (High-n-3) fed animals, which were either continued to the same diet i.e. no transition to western diet (Low-n-3/No WD and High-n-3/No WD) or switched to the WD (Low-n-3/WD and High-n-3/WD). (B) Percentage of time spent in open arms in animals transitioned from low omega-3 to the WD and treated with either vehicle (Low-n-3/WD/VEH) or 5-aza-2'-deoxycytidine (Low-n-3/WD/5-AzaD; 0.4 mg/kg, ip). (C) Average heat map occupancy plots for EPM in each group. Data are expressed as mean ± SEM. #P<0.05, ##P<0.01 Vs Low-n-3/No WD, **P<0.01 Vs Low-n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test. aP<0.05 Vs Low-n-3/WD/VEH; Student (unpaired) ‘t’ test.
The treatment with 5-AzaD, a DNMT inhibitor, normalized the anxiety-like behavior in WD animals pre-exposed to the low omega-3 diet (Low-n-3/WD), as reflected by an elevated levels of percentage of time spent in open arms in 5-AzaD treated group as compared to the vehicle treated group (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p<0.05; Fig 1B). The increased exploration of the open arms is also manifested by heat map occupancy plots in 5-AzaD treated animals (Fig. 1C). There was no significant difference observed in total distance (meter) in vehicle and drug treated (Low-n-3/WD/VEH: 4.6±0.4; Low-n-3/WD/5-AzaD= 5.4±1.1) groups. These results emphasize the involvement of methylation on diet mediated behavioral responses.
Dietary manipulations influences Bdnf methylation
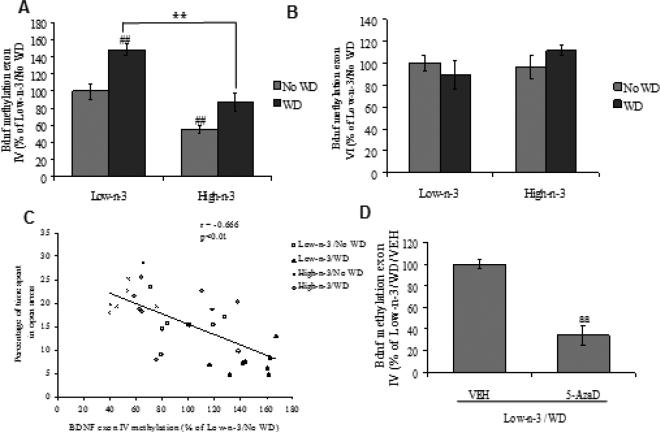
We examined the possibility that Bdnf methylation could mediate the effects of the dietary interventions on behavior. Thus, we analyzed the methylation pattern at promoter region of Bdnf exon IV and VI. Two-way ANOVA analysis showed a significant effect of maternal diet (F1,28=44.031, p<0.01) and transitioned diet (F1,28=24.395, p<0.01) for Bdnf exon IV promoter methylation levels. There were no significant changes were observed in methylation levels of promoter of Bdnf exon VI. Post hoc analysis showed a significant increase in methylation at Bdnf exon IV promoter after WD exposure (Low-n-3/WD Vs Low-n-3/No WD, p<0.01), only in animals raised on low omega-3 diet. Animals exposed to high omega-3 diet showed less methylation at Bdnf exon IV promoter as compared to animal exposed to the low omega-3 diet (High-n-3/No WD Vs Low-n-3/No WD, p<0.01; Fig 2A), both diets fed until sacrifice. No significant change was observed in Bdnf exon VI promoter with any of the dietary intervention (Fig 2B). There was a negative correlation between Bdnf exon IV promoter methylation and percentage of time spent in open arms (r = - 0.666, p<0.01; Fig 2C), which appears to indicate an association between methylation of Bdnf exon IV promoter and the anxiety-like behavior. We found that the 5-AzaD treatment normalized the WD induced Bdnf methylation at promoter IV region (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p<0.01; Fig 2D).
Fig. 2.
DNA methylation at Bdnf promoter (A) exon IV, (B) exon VI and (C) correlation analysis of methylation status of Bdnf exon IV with percentage of time spent in open arms in low omega-3 (Low-n-3) or high omega-3 diet (High-n-3) fed animals, which were either continued to the same diet i.e. no transition to WD (Low-n-3/No WD and High-n-3/No WD) or switched to the western diet (Low-n-3/WD and High-n-3/WD). (D) DNA methylation at Bdnf exon IV in animals transitioned from low omega-3 to the WD and treated with either vehicle (Low-n-3/WD/VEH) or 5-aza-2'-deoxycytidine (Low-n-3/WD/5-AzaD; 0.4 mg/kg, ip). Data are expressed as percentage of Low-n-3/No WD or Low-n-3/WD/VEH (mean ± SEM). ##P<0.01 Vs Low-n-3/No WD, **P<0.01 Vs Low-n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test. aaP<0.01 Vs Low-n-3/WD/VEH; Student (unpaired) ‘t’ test.
Dietary Regulation of BDNF expression
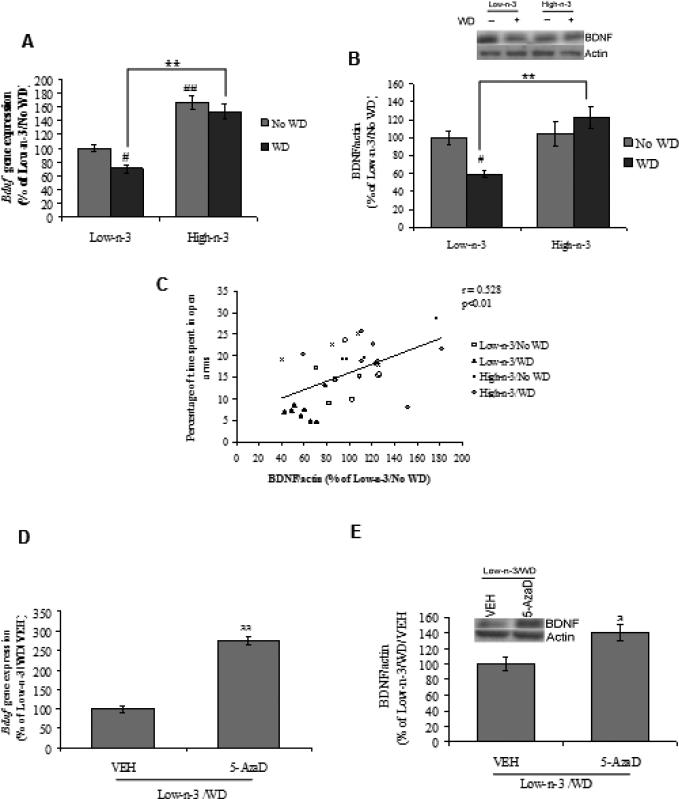
We evaluated the possibility that changes in methylation pattern related to dietary interventions could translate into changes of Bdnf gene as well as BDNF protein. Two-way ANOVA analysis showed a significant effect of maternal (F1,28=81.706, p<0.01) and transitioned diet (F1,28=7.167, p<0.05) for Bdnf gene expression. With regards to BDNF protein level there was a significant effect of maternal (F1,28=9.806, p<0.01) and its interaction with transitioned diet (F1,28=6.995, p<0.05). Post-hoc analysis revealed a significant down-regulation in BDNF protein as well as gene expression pattern after WD exposure in low omega-3 fed animals (Low-n-3/WD Vs Low-n-3/No WD, p<0.05). The treatment with high omega-3 diet elevated BDNF levels (both protein and gene) with WD transition in comparison to low omega-3 fed animals (High-n-3/WD Vs Low-n-3/WD, p<0.01; Fig 3A,B). We observed a positive correlation between BDNF protein levels and percentage of time spent in open arms (r = 0.528, p<0.01; Fig 3C), which suggests that increase in BDNF may contribute to decrease anxiety-like behavior. The 5-AzaD treatments also elevated BDNF at the mRNA (p<0.01) and protein (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p<0.05; Fig 3D,E) levels, which corroborates the repressive action of Bdnf methylation on BDNF.
Fig. 3.
(A) Bdnf gene (B) protein expression and (C) correlation analysis of BDNF protein level with percentage of time spent in open arms in low omega-3 (Low-n-3) or high omega-3 diet (High-n-3) fed animals, which were either continued to the same diet i.e. no transition to western diet (Low-n-3/No WD and High-n-3/No WD) or switched to the WD (Low-n-3/WD and High-n-3/WD). (D) Bdnf gene and (E) protein expression in vehicle (VEH) and 5-aza-2'-deoxycytidine (5-AzaD; 0.4 mg/kg, ip) treated animals, which were reared on low omega-3 diet and switched to the western diet (Low-n-3/WD/VEH and Low-n-3/WD/5-AzaD). Data are expressed as percentage of Low-n-3/No WD or Low-n-3/WD/VEH (mean ± SEM). #P<0.05, ##P<0.01 Vs Low-n-3/No WD, **P<0.01 Vs Low-n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test. aP<0.05, aaP<0.01 Vs Low-n-3/WD/VEH; Student (unpaired) ‘t’ test.
Transcriptional Regulation of BDNF expression
Binding of CTCF at Bdnf exon IV promoter
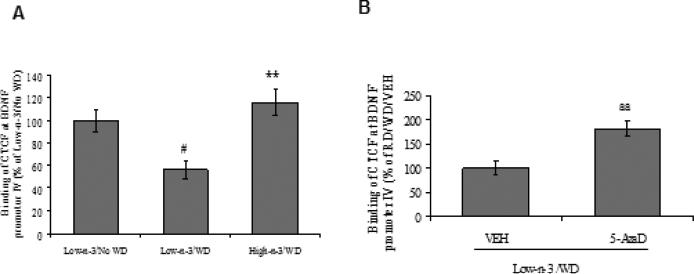
CCCTC binding factor (CTCF), a nuclear protein, is a uniquely versatile transcription regulator linked to epigenetics and genetic diseases (Ohlsson et al., 2001). We explored whether the binding of CTCF at promoter region of Bdnf exon IV could be sensitive to dietary interventions and could play role in BDNF-related behavioral regulation. The WD transition reduced the binding of CTCF at Bdnf promoter region IV only for animals reared on low omega-3 diet (Low-n-3/WD Vs Low-n-3/No WD, p<0.05). This also implies that preconditioning with high omega-3 diet preserved the binding of CTCF at Bdnf promoter IV (High-n-3/WD Vs Low-n-3/WD, p<0.01; Fig 4.1 A). We used 5-AzaD treatment to confirm the action of methylation on CTCF binding, and found that this treatment increased CTCF binding in the WD group transitioned from low omega-3 diet (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p<0.01; Fig 4.1 B).
Fig. 4.1.
(A) CTCF Binding at promoter region of Bdnf exon IV in low omega-3 (Low-n-3) or high omega-3 diet (High-n-3) fed animals, which were either continued to the same diet i.e. no transition to western diet (Low-n-3/No WD and High-n-3/No WD) or switched to the WD (Low-n-3/WD and High-n-3/WD). (B) CTCF Binding at promoter region of Bdnf exon IV in vehicle (VEH) and 5-aza-2'-deoxycytidine (5-AzaD; 0.4 mg/kg, ip) treated animals, which were raised on low omega-3 diet and switched to the western diet (Low-n-3/WD/VEH and Low-n-3/WD/5-AzaD). Data are expressed as percentage of Low-n-3/No WD or Low-n-3/WD/VEH (mean ± SEM). #P<0.05 Vs Low-n-3/No WD, **P<0.01 Vs Low-n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test. aaP<0.01 Vs Low-n-3/WD/VEH; Student (unpaired)
CTCF binding leads to the BDNF expression
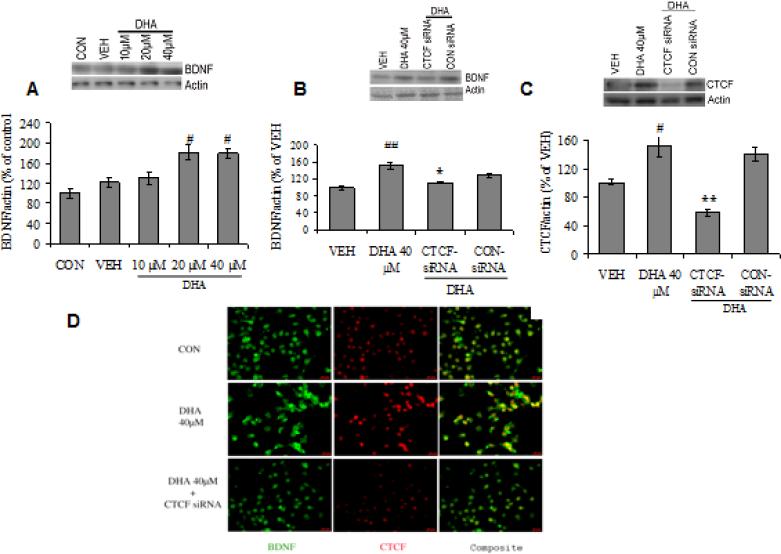
To confirm that the CTCF binding is pivotal for the effects of omega-3 on BDNF expression, we employed RNA interference technology to selectively reduce levels of CTCF in Neuro-2a (N2a)-neuroblastoma cells. Treatment with DHA for 24 h in N2a cells resulted in a significant increase in BDNF protein levels in a dose dependent manner (20-40 μM; P<0.05; Fig. 4.2 A). Blocking of CTCF using siRNA followed by DHA resulted in a significant downregulation of BDNF (P<0.05; Fig. 4.2 B) and CTCF protein (P<0.01; Fig. 4.2 C) levels, determined by immunoblot analysis in cell lysate. Furthermore, we assessed the phenotypic expression of CTCF and BDNF by immunocytochemical analysis and observed a qualitative increase in the staining intensity of CTCF/BDNF co-localization, after 24 h of DHA treatment (40 μM). However, the treatment of DHA (40 μM, 24 h) in CTCF-siRNA treated N2a cells, showed reduced staining intensity of CTCF/BDNF co-localization, suggesting that CTCF binding is crucial for omega-3 fatty acid induced changes in BDNF (Fig. 4.2 D).
Fig. 4.2.
BDNF protein expression in Neuro-2a (N2a)-neuroblastoma cells treated with (A) 10, 20 and 40 μM of DHA for 24 h and (B) CTCF-siRNA followed by 40 μM of DHA treatment. (C) CTCF protein expression in N2a cells treated with CTCF-siRNA followed by 40 μM of DHA. (D) BDNF (FITC) and CTCF (Cy3) in N2a cells treated with 40μ M DHA and CTCF-siRNA. Fluorescent immunocytochemical images reveal the co-localization of BDNF and CTCF in N2a cells (green for BDNF, red for CTCF and yellow for co-stained cells, scale bar: 20 μm). Data are expressed as percentage of control or vehicle group (mean ± SEM) of four independent experiments. #P<0.05, ##P<0.01 Vs VEH, *P<0.05, **P<0.01 Vs DHA 40 μM group; ANOVA (one-way) followed by Tukey's post-hoc test.
Regulation of MeCP2 expression is important for BDNF transcription
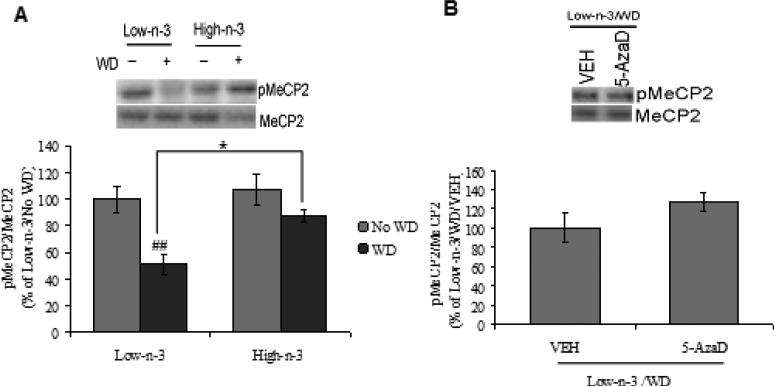
It has been shown that MeCP2 can modulate Bdnf gene expression and function by interacting with methylated chromatin and suppressing promoter IV transcription (Martinowich et al., 2003). Two-way ANOVA analysis revealed a significant effect of maternal (F1,28=6.413, p<0.05) and transitioned (F1,28=15.556, p<0.01) diet for MeCP2 phosphorylation.. The WD paradigm, showed a significant reduction of MeCP2 phosphorylation only in animals reared with low omega-3 diet (Low-n-3/WD Vs Low-n-3/No WD, p<0.01). The prior exposure of high omega-3 diet prevented the effect of WD on MeCP2 phosphorylation in comparison to low omega-3 diet pre-exposure (High-n-3/WD Vs Low-n-3/WD, p<0.05; Fig. 4.3A). The treatment with 5-AzaD did not show any effect on WD induced MeCP2 phosphorylation in low omega-3 diet fed animals (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p>0.05; Fig. 4.3B).
Fig. 4.3.
(A) Phosphorylation of MeCP2 in low omega-3 (Low-n-3) or high omega-3 diet (High-n-3) fed animals, which were either continued to the same diet i.e. no transition to western diet (Low-n-3/No WD and High-n-3/No WD) or switched to the WD (Low-n-3/WD and High-n-3/WD). (B) Phosphorylation of MeCP2 in vehicle (VEH) and 5-aza-2'-deoxycytidine (5-AzaD; 0.4 mg/kg, ip) treated animals, which were reared on low omega-3 diet and switched to the western diet (Low-n-3/WD/VEH and Low-n-3/WD/5-AzaD). Data are expressed as percentage of Low-n-3/No WD or Low-n-3/WD/VEH (mean ± SEM). #P<0.05 Vs Low-n-3/No WD, *P<0.05 Vs Low n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test.
Dietary interventions influence metabolic sensors
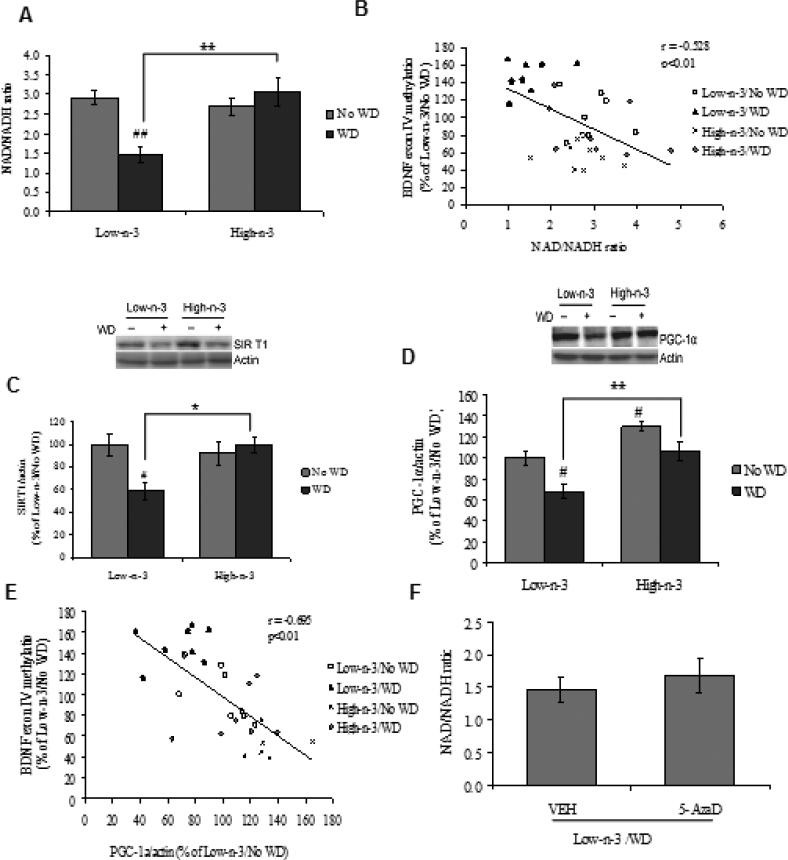
We have examined a possible involvement of metabolic sensors in the interaction between dietary interventions and epigenetic regulation of plasticity and behavior. We focused on the NAD/NADH ratio and the expression pattern of protein molecules such as SIRT1 and PGC1α that controls the balance and transduction of cellular energy. A two-way ANOVA analysis (maternal vs. transitioned diet) showed a significant effect of maternal diet (F1,28=7.557, p<0.05), transitioned diet (F1,28=4.885, p<0.05) and an interaction between maternal vs. transitioned diet (F1,28=13.074, p<0.01) for the levels of NAD/NADH ratio. The Tukey's post-hoc test for multiple comparisons showed that the exposure to WD resulted in a significant reduction in NAD/NADH ratio in low omega-3 diet fed animals (Low-n-3/WD Vs Low-n-3/No WD, p<0.01), while prior exposure to high omega-3 diet prevented these effects (High-n-3/WD Vs Low-n-3/WD, p<0.01). Animals reared on high omega-3 diet until sacrifice did not exhibit changes in the levels of NAD/NADH ratio (High-n-3/No WD Vs Low-n-3/No WD, p>0.05; Fig. 5A). The NAD/NADH ratio was negatively correlated with Bdnf exon IV methylation (r = -0.528, p<0.01; Fig. 5B), suggesting that cell energy regulation could be a factor for the methylation of Bdnf exon IV. Twoway ANOVA analysis showed a significant interaction between maternal vs. transitioned diet (F1,28=7.901, p<0.01) on SIRT1 levels. With regards to the levels of PGC-1α, there was a significant effect of maternal diet (F1,28=21.465, p<0.01) and transitioned diet (F1,28=14.640, p<0.01). Post hoc analysis revealed a significant decrease in both SIRT1 (Low-n-3/WD Vs Low-n-3/No WD, p<0.05; Fig. 5C) and PGC-1α (Low-n-3/WD Vs Low-n-3/No WD, p<0.05; Fig. 5D) levels after WD exposure in animals fed on low omega-3 diet. However, high omega-3 diet preconditioning (High-n-3/WD Vs Low-n-3/WD, p<0.05-0.01) appeared to prevent the effects of WD. We observed that PGC-1α and Bdnf exon IV methylation are inversely correlated (r = -0.695, p<0.01; Fig. 5E), thus suggesting an association between Bdnf exon IV methylation and PGC-1α expression.
Fig. 5.
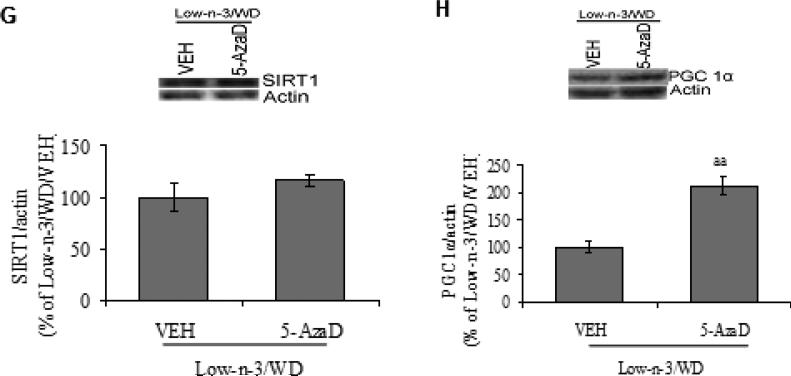
(A) NAD/NADH ratio, (B) correlation analysis of NAD/NADH ratio with Bdnf promoter exon IV, (C) levels of SIRT1, (D) PGC-1α and (E) correlation analysis of PGC-1α with Bdnf promoter exon IV, in animals fed with either low omega-3 (Low-n-3) or high omega-3 diet (High-n-3), which were continued to the same diet i.e. no transition to western diet (Low-n-3/No WD and High-n-3/No WD) or switched to the WD (Low-n-3/WD and High-n-3/WD). (F) NAD/NADH ratio, (G) levels of SIRT1 and (H) PGC-1α in vehicle (VEH) and 5-aza-2'-deoxycytidine (5-AzaD; 0.4 mg/kg, ip) treated animals, which were reared on low omega-3 diet and switched to the western diet (Low-n-3/WD/VEH and Low-n-3/WD/5-AzaD). Data are expressed as percentage of Low-n-3/No WD or Low-n-3/WD/VEH (mean ± SEM). #P<0.05, ##P<0.01 Vs Low-n-3/No WD, *P<0.05, **P<0.01 Vs Low-n-3/WD; ANOVA (two-way) followed by Tukey's post-hoc test. aaP<0.01 Vs Low-n-3/WD/VEH; Student (unpaired) ‘t’ test.
Further the analysis of these metabolic markers after 5-AzaD treatment showed a significant increase in the protein expression of PGC-1α (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p<0.01; Fig. 5H). The treatment with 5-AzaD was not able to show any effect on WD induced changes in NAD/NADH ratio (p>0.05) and SIRT1 (Low-n-3/WD/5-AzaD Vs Low-n-3/WD/VEH, p>0.05; Fig. 5E,F).
Discussion
We report a potential mechanism by which early nutrition can influence long-term plasticity by acting on epigenetic regulation of the Bdnf gene. For example, this epigenetic memory could provide an underlying mechanism for the process by which the brain builds up cognitive reserve to resist periods of hardship. Early feeding with a diet rich in n-3 fatty acids protected against cognitive decay resulting from subsequent exposure to a WD. The WD increased Bdnf methylation specific to exon IV in animals previously reared with a diet low in n-3 fatty acids. This also implies that n-3 fatty acid feeding has a priming effect by conferring resistance to future environmental challenges. A DNA methytransferase inhibitor counteracted the Bdnf hypermethylation and cognitive decay. The WD diet exposure reduced the binding of the insulator protein CTCF to the Bdnf promoter of exon IV and increased the levels of transcription repressor MeCP2. Studies in vitro confirmed that CTCF binding to Bdnf promoter is a necessary step for the action of n-3 fatty acid diet on BDNF protein production. We also found that the metabolic sensor NAD/NADH and SIRT1 levels appear to act as an intermediate step between the WD and Bdnf methylation. The fact that the DNMT inhibitor did not affect the WD-induced changes on NAD/NADH suggests that NAD-NADH engagement is upstream to methylation. In turn, the fact that the methyltransferase inhibitor blocks the WD-reducing effects on the mitochondrial transcription regulator PGC-1α suggests that PGC-1α action is downstream to the Bdnf methylation. Results show the existence of an “epigenetic memory” in which the long-term effects of the omega-3 diet are conserved in the epigenome through metabolic signals, and may confer protection against future metabolic challenges.
Bdnf methylation regulated by dietary intervention
Our results show that WD promoted hypermethylation of the Bdnf gene at promoter region IV. DNA methylation plays a crucial role in the remodeling of chromatin and can repress the transcription and function of Bdnf (Martinowich et al., 2003). Therefore, our results suggest that DNA methylation is a crucial mediator between diet and the regulation of Bdnf expression. Further, the blocking of DNA methyltransferase with 5AzaD reduced the expression of Bdnf mRNA and protein, and reduced anxiety-like behavior in animals administered AzaD compared to WD fed animals. Our results are in accordance with studies suggesting that elevated DNA methylation of Bdnf exon IV is a mechanism to down-regulate BDNF in adverse environmental circumstances (Roth et al., 2009). For example, traumatic stress experience, such as continuous psychosocial stress, has been shown to induce hypermethylation of the Bdnf exon IV and to reduce the Bdnf expression in the dorsal CA1 area of the hippocampus in a rat model of PTSD (Roth et al., 2011). In addition, increased DNA methylation at the promoter region of Bdnf exon IV in the hippocampus of adult mice has been correlated with depression-like behavior (Onishchenko et al., 2008). Interestingly, we found that preconditioning with n-3 diet was able to protect against the hypermethylation of the Bdnf exon IV promoter in WD fed animals. Bdnf methylation levels varied in proportion with the time spent in the open arms of the EPM, suggesting that regulation of BDNF transcription may have behavioral consequences.
Bdnf methylation is a pivotal point for the regulation of brain plasticity and behavior
An increasing body of research indicates that numerous disorders of the nervous system are related to abnormal DNA methylation (Ikegame et al., 2013). DNA methylation may remain stable over long periods and likely provides a niche for the impact of the environment on the pathology of neurological and psychiatric disorders. DNMT inhibitors such as the nucleoside analogues 5-aza-cytidine and 5-aza-2-deoxycytidine have been shown to mimic the behavioral effects of the antidepressant imipramine in adult rodents by inhibiting Bdnf methylation in the hippocampus (Sales et al., 2011). Moreover, the DNMT inhibitor zebularine has been shown to induce exon-specific Bdnf expression through DNA demethylation mechanisms in the rat hippocampus (Lubin et al., 2008).
In the present set of experiments, we have utilized the DNMT inhibitor 5-AzaD to examine the action of Bdnf hypermethylation on BDNF regulation and behavior. Although 5-AzaD was able to reverse the effects of WD on anxiety, Bdnf hypermethylation, and BDNF transcription and translation, we cannot discard that demethylation of other molecules may be involved with the action of 5-AzaD. It is significant that the 5-AzaD treatment had a positive effect on behavior (reduce anxiety) which is consistent with the idea that reducing BDNF methylation is the main target for the action of 5-Azad. Overall, the results of this study provide mechanistic evidence for the role of Bdnf methylation on brain plasticity and behavior under the action of dietary factors. In vitro experiments revealed the capacity of DHA to induce BDNF protein expression. Using siRNA to block the binding of CTCF to the Bdnf promoter region further corroborated this possibility. CTCF is a DNA binding factor that plays important roles in development, and regulates important gene transcriptions by interacting with CCCTC motifs in the DNA-regulatory element (Awad et al., 1999; Burcin et al., 1997; Filippova et al., 1996). CTCF deficiency has been shown to disturb mouse development (Fedoriw et al., 2004) and cause abnormal behavior (Hirayama et al., 2012). Our results indicate that CTCF mediated transcription regulation is an important step for the dietary influence on BDNF expression. Reduced immunofluroscence staining of BDNF after blocking of CTCF binding also supports these results.
Studies have suggested that MeCP2 acts as a global chromatin regulator (Skene et al., 2010), and selectively regulates BDNF in response to neuronal activity (Chen et al., 2003; Zhou et al., 2006). It has been recently shown that the loss of activity-dependent phosphorylation of MeCP2 not only increased Bdnf exon IV expression but also improved synaptogenesis, long-term potentiation and spatial memory in mice (Li et al., 2011). The effects of WD on increasing phosphorylation of MeCP2 and the counteracting effects of the n-3 diet are in accordance with the described role of MeCP2 in the regulation of BDNF expression.
Metabolic sensors mediate the effects of diet on Bdnf methylation
Management of cellular energy is crucial for cell survival and function, such that a disruption of energy metabolism is getting recognition for being an integral aspect in the pathology of various neurological (Farooqui et al., 2011) and psychiatric (Khaitovich et al., 2008) disorders. Based on the current results, we pose that interaction between energy metabolism and epigenetic is essential for gating the effects of the environment on brain function and plasticity. Environmental stimuli may alter the levels of DNA methylation and consequently gene expression by using metabolic signals, and these events are operational for building long-lasting neural resilience. We found that WD altered the levels of important energy regulators and preconditioning with n-3 diet preserved these effects. Results indicate that WD decreased levels of metabolic sensors such as NAD/NADH, SIRT1 and PGC 1α, which have been implicated in regulation of BDNF signaling (Chang et al., 2010; Cheng et al., 2012). Low NAD levels have been shown to increase DNA methylation at the Bdnf exon IV promoter and can trigger the dissociation of the insulator protein CTCF in neuronal cultures, while increased NAD can restore Bdnf level (Chang et al., 2010). The binding of the DNA methylation–sensitive factor CTCF has been shown to play a critical role in the NAD-dependent regulation of Bdnf expression through chromatin remodeling (Chang et al., 2010). We found that WD decreased the NAD/NADH ratio and levels of SIRT1 and PGC1α in the frontal cortex. Preconditioning with an omega-3 diet negated these effects, suggesting the importance of metabolic signals in the maintenance of brain plasticity. The fact that methyltransferase inhibitor was not able to block changes in NAD/NADH ratio and SIRT1 expression, suggests that this regulation is upstream to Bdnf methylation. The action of these metabolic sensors on BDNF regulation is further emphasized by the positive correlation observed between the NAD/NADH ratio and Bdnf methylation. The methylation inhibitor did not block the effects of WD on the mitochondrial transcription regulator PGC-1α suggesting that PGC1α action is downstream to Bdnf methylation. The overall results indicate that metabolic signals are integral aspects in the epigenetic regulation of Bdnf gene by dietary factors. In addition, results seem to point out to the existence of an “epigenetic memory” in which the long-term effects of the n-3 diet are saved in the epigenome using metabolic signals. These results are further significant based on information that DHA is crucial for the brain, and that consumption of DHA is below recommended levels in the western society (Simopoulos, 2006). The adequate intake of n-3; however, is likely influenced by baseline consumption and varies according to the age and gender, as suggested by the Institute of Medicine (Report 2002/2005).
Conclusion
These studies show that early life exposure to omega-3 fatty acids can promote epigenetic variability, which confers resilience to metabolic perturbations incurred in later life. Further detailed studies are required to determine the action of particular periods of brain development on plasticity of the BDNF system during adulthood. The results also portray cell energy metabolism as an important factor by which the environment impacts the epigenome. In turn, results are consistent a central role of Bdnf methylation in the mechanisms that preserve the effects of diet on plasticity and behavior (Fig. 6). The overall study introduces the likelihood that metabolism and epigenetics are closely associated events, and together contributes to build an “epigenetic memory” serving long-term brain plasticity. These results provide fundamental information to understand the influence of environmental factors on brain plasticity and function, and its potential to redirect the course of pathology.
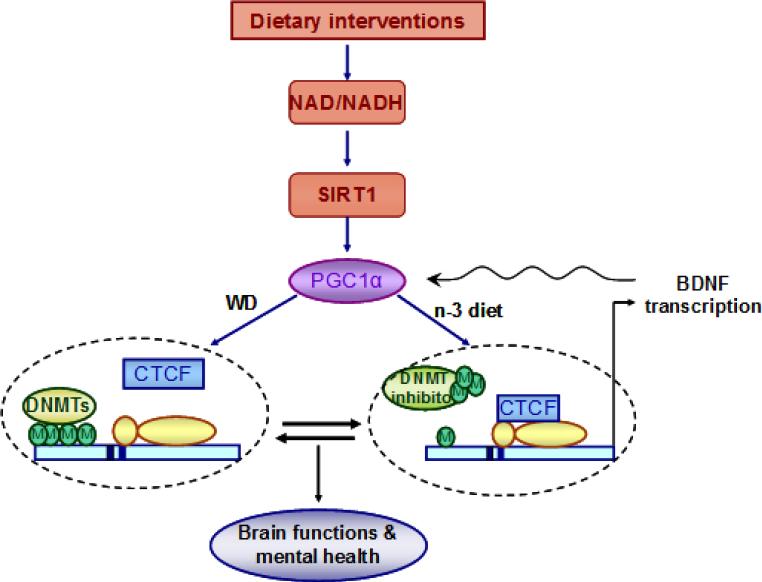
Fig. 6.
Proposed mechanism by which dietary interventions impact neurological function and mental health by engaging an interplay between epigenetic and energy management system. The pre-exposure of dietary n−3 fatty acid and transition to the western diet (WD) alter neuronal metabolic status by affecting NAD/NADH and SIRT1 levels, thereby influencing the co transcriptional regulator PGC-1α. The transition to WD promotes DNA hypermethylation in the Bdnf exon IV promoter that may result in dissociation of CTCF binding and leading to the suppression of the gene transcription. The prior exposure of n-3 diet preserves the effect of WD transition by promoting DNA demethylation in Bdnf promoter IV, which is associated with CTCF binding that leads to Bdnf gene transcription. The effects of dietary interventions on BDNF regulation may also involve the action of DNA methyltransferase (DNMT) inhibitor as the inhibition of DNA methylation activates Bdnf gene transcription by stimulating the binding of CTCF to the Bdnf promoter IV. The changes in Bdnf gene transcription are associated with altered BDNF protein expression that may influence neuronal function and mental health by involving PGC-1α. Overall, the pre-exposure of dietary n-3 fatty acid (n-3 diet) during early life may act as a determining factor for the outcome of WD exposure.
Supplementary Material
Highlights.
Western diet promotes methylation of the Bdnf gene with behavioral detriments
Omega-3 fatty acids counteract the effects of the western diet.
Diets orchestrate key interactions between metabolic signals and Bdnf methylation.
Changes in Bdnf methylation may serve to build an “epigenetic memory”
Acknowledgements
This work was supported by National Institutes of Health Grant NS050465. We also acknowledge kind support of Letten Foundation and Professor Letten Saugstadt. There is no conflict of interest for any of the contributing authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceived and designed the experiments: ET FGP. Performed the experiments: ET YZ RA ZY. Analyzed the data: ET YZ. Contributed reagents/materials/analysis tools: ET FGP. Wrote the paper: ET FGP.
References
- Agrawal R, et al. Coupling energy homeostasis with a mechanism to support plasticity in brain trauma. Biochim Biophys Acta. 2014;1842:535–46. doi: 10.1016/j.bbadis.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Awad TA, et al. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–8. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- Boulle F, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17:584–96. doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- Burcin M, et al. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–8. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64:166–87. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, et al. Nicotinamide adenine dinucleotide (NAD)-regulated DNA methylation alters CCCTC-binding factor (CTCF)/cohesin binding and transcription at the BDNF locus. Proc Natl Acad Sci U S A. 2010;107:21836–41. doi: 10.1073/pnas.1002130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheng A, et al. Involvement of PGC-1α in the formation and maintenance of neuronal dendritic spines. Nat Commun. 2012;3:1250. doi: 10.1038/ncomms2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauncey MJ. Recent advances in nutrition, genes and brain health. Proc Nutr Soc. 2012;71:581–91. doi: 10.1017/S0029665112000237. [DOI] [PubMed] [Google Scholar]
- Farooqui AA, et al. Metabolic syndrome as a risk factor for neurological disorders. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-011-0840-1. 10.1007/s00018-011-0840-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, et al. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–40. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Filippova GN, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–13. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Cell biology: a clean energy programme. Nature. 2006;444:151–2. doi: 10.1038/444151a. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, et al. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F. Brain foods: the effects of nutrients on brain function. Nat Rev Neurosci. 2008;9:568–78. doi: 10.1038/nrn2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, et al. Brain-derived neurotrophic factor functions as a metabotrophin to mediate the effects of exercise on cognition. Eur J Neurosci. 2008;28:2278–87. doi: 10.1111/j.1460-9568.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, et al. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29:12764–7. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner RS, et al. Docosapentaenoic acid does not completely replace DHA in n-3 FA-deficient rats during early development. Lipids. 2003;38:431–5. doi: 10.1007/s11745-003-1080-2. [DOI] [PubMed] [Google Scholar]
- Hirayama T, et al. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–57. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Ikegame T, et al. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet. 2013;58:434–8. doi: 10.1038/jhg.2013.65. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb B, et al. Experience and the developing prefrontal cortex. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17186–93. doi: 10.1073/pnas.1121251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci. 2011;14:1001–8. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, et al. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–7. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–3. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Moore LD, et al. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, et al. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Oaks AW, et al. Age-dependent effects of A53T alpha-synuclein on behavior and dopaminergic function. PLoS ONE. 2013;8(4):e60378. doi: 10.1371/journal.pone.0060378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, et al. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–7. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Onishchenko N, et al. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106:1378–87. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- Pinto JG, et al. Comparing development of synaptic proteins in rat visual, somatosensory, and frontal cortex. Front Neural Circuits. 2013;7:97. doi: 10.3389/fncir.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, et al. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, et al. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–9. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, et al. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–26. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales AJ, et al. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br J Pharmacol. 2011;164:1711–21. doi: 10.1111/j.1476-5381.2011.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P. The Laboratory Rat: Relating Its Age With Human's. Int J Prev Med. 2013;4:624–30. [PMC free article] [PubMed] [Google Scholar]
- Simopoulos AP. Evolutionary aspects of diet, the omega-6/omega-3 ratio and genetic variation: nutritional implications for chronic diseases. Biomed Pharmacother. 2006;60:502–7. doi: 10.1016/j.biopha.2006.07.080. [DOI] [PubMed] [Google Scholar]
- Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–68. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Villegas A, et al. Dietary fat intake and the risk of depression: the SUN Project. PLoS One. 2011;6:e16268. doi: 10.1371/journal.pone.0016268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi E, et al. Vulnerability imposed by diet and brain trauma for anxiety-like phenotype: implications for post-traumatic stress disorders. PLoS One. 2013;8:e57945. doi: 10.1371/journal.pone.0057945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2:322–8. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MJ, et al. BDNF mRNA expression during postnatal development, maturation and aging of the human prefrontal cortex. Brain Res Dev Brain Res. 2002;139:139–50. doi: 10.1016/s0165-3806(02)00540-0. [DOI] [PubMed] [Google Scholar]
- Young G, Conquer J. Omega-3 fatty acids and neuropsychiatric disorders. Reprod Nutr Dev. 2005;45:1–28. doi: 10.1051/rnd:2005001. [DOI] [PubMed] [Google Scholar]
- Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–69. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–22. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.