Abstract
Long-term delivery of antibodies against the human immunodeficiency virus (HIV) using adeno-associated virus (AAV) vectors is a promising approach for the prevention or treatment of HIV infection. However, host antibody responses to the delivered antibody are a serious concern that could significantly limit the applicability of this approach. Here, we describe the dynamics and characteristics of the anti-antibody responses in monkeys that received either rhesus anti-simian immunodeficiency virus (SIV) antibodies (4L6 or 5L7) in prevention trials or a combination of rhesusized human anti-HIV antibodies (1NC9/8ANC195/3BNC117 or 10–1074/10E8/3BNC117) in therapy trials, all employing AAV1 delivery of IgG1. Eight out of eight monkeys that received the anti-HIV antibodies made persisting antibody responses to all three antibodies in the mix. Six out of six uninfected monkeys that received the anti-SIV antibody 4L6 and three out of six of those receiving anti-SIV antibody 5L7 also generated anti-antibodies. Both heavy and light chains were targeted, predominantly or exclusively to variable regions, and reactivity to complementarity-determining region (CDR)-H3 peptide could be demonstrated. There was a highly significant correlation of the magnitude of anti-antibody responses with the degree of sequence divergence of the delivered antibody from germline. Our results suggest the need for effective strategies to counteract the problem of antibody responses to AAV-delivered antibodies.
Introduction
In the classic approach to immunization, a microbial immunogen is delivered, there is an immune response to the immunogen, and it is hoped that the immune response is protective. The properties of human immunodeficiency virus (HIV), including its persistent replicative capacity and high variability, raise serious concerns whether such vaccine approaches in the classic sense could ever be sufficiently protective. A creative and promising strategy is to circumvent these difficulties by directly delivering potent and broadly neutralizing anti-HIV antibodies in a persisting fashion. An impressive array of such monoclonal antibodies have been isolated over the last several years.1,2,3,4,5,6 If satisfactory delivery methods that ensure long-lasting production in the recipient can be found, it becomes possible to envision a long-term sterilizing barrier to the vast majority of HIV-1 strains circulating in the population.
Adeno-associated virus (AAV) is ideal in many respects as a delivery vehicle since it has an outstanding safety record in clinical trials7 and, as long as the delivered protein is viewed as self, it can result in continuous durable expression of the transgene product for years.8,9,10,11,12 Studies in monkeys13,14,15,16 and in mice17 have already shown the extreme promise of this approach against HIV/ simian immunodeficiency virus (SIV). However, antibody responses to the delivered antibody can seriously limit the effectiveness of this strategy.13,14,15 In an initial study, rhesus macaques were inoculated with recombinant AAV coding for anti-SIV immunoadhesins (antibody-like molecules). Sterilizing protection against intravenous SIV challenge was observed in some recipients in that study but not among those that developed antibodies to the transgene product.13 At that time, it was not clear whether the antibody response raised against the delivered transgene product may have resulted from the use of immunoadhesins, which do not represent an authentic IgG structure.
Our group has recently used AAV to deliver mAbs from rhesus macaques in authentic, full-length IgG1 form to rhesus macaques.15 In those experiments, 9 of 12 rhesus monkeys had antibody responses to the delivered antibody that appeared to limit the concentration of the delivered antibody that could be achieved.15 Another group has also recently observed the appearance of anti-antibodies to AAV-delivered immunoglobulins.14 When a simianized form of bNAb VRC07 was delivered by AAV to rhesus monkeys, anti-antibody responses were consistently noted. Immune suppression with cyclosporine A was needed to achieve sustained expression; otherwise, the levels of delivered antibody became undetectable by week 9 after AAV inoculation.14
In the present study, we describe in detail the specificity and dynamics of anti-antibody responses in two prevention trials and two therapy trials in monkeys utilizing AAV delivery. Of seven different antibodies studied, anti-antibody responses were observed against all seven. The magnitude of the anti-antibody response correlated in a highly significant fashion with the sequence divergence of the delivered antibody from the germline.
Results
Host humoral responses against AAV-delivered anti-SIV antibodies mainly target the variable regions
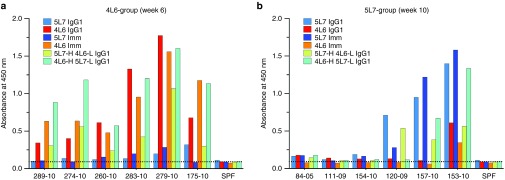
We have recently described the use of recombinant AAV1 in rhesus monkeys to deliver mAbs in IgG1 form.15 One group of six naive rhesus macaques received one anti-SIV mAb (4L6 IgG1) and another group of six received a different anti-SIV mAb (5L7 IgG1) in prevention experiments. These anti-SIV mAbs contained only authentic rhesus IgG sequences. At first, all the monkeys achieved good levels of delivered mAb in serum, but in several cases the levels subsequently dropped abruptly. All six animals in the 4L6 IgG1 group developed a clear anti-antibody response to the AAV-delivered mAb, as did three of the six that received 5L7 IgG1. While all the animals in the 4L6 IgG1 group became infected after challenge and did not show any protective effects from the mAb that they received, animals that received 5L7 IgG1 showed a lower viremia at peak, a delayed time to peak viremia and a lower viremia set-point. Moreover, one animal in the latter group that had the highest levels of delivered antibody showed apparent sterile protection after repeated challenges with neutralization-resistant and highly-pathogenic SIVmac239, including a challenge with 10 i.v. infectious doses. Interestingly, this monkey did not show detectable anti-antibodies. While the relevance of the anti-antibody response as a limiting factor for the protective effects was evident in our publication, the specifics of the anti-antibody responses were not further investigated.
In order to characterize the anti-antibody responses in these two groups of animals, we tested enzyme-linked immunosorbent assay (ELISA) reactivity of serum samples from both groups against a set of purified proteins that included both 4L6 and 5L7 IgG1, immunoadhesin versions of these antibodies (IgG-like molecules that lack the constant domain of the light chain and the CH1 domain of the heavy chain) and hybrid versions: one corresponding to the heavy chain of 5L7 IgG1 with the light chain of 4L6 IgG1 (named 5L7-H 4L6-L IgG1), and another with the complementary combination (designated 4L6-H 5L7-L IgG1). Since all of these proteins had a κ-based light chain, we used an anti-lambda conjugate to detect antibodies with a lambda light chain reactive to the corresponding protein on an ELISA plate. Consistent with our previous results, all of the six monkeys in the 4L6 group showed readily-detected anti-antibodies to 4L6 IgG1 by week 6 after AAV-inoculation (Figure 1a, red bars), while in the 5L7 group three of the six animals showed clear anti-antibodies against 5L7 IgG1 (Figure 1b, blue bars). Monkey sera from the 4L6 IgG1 recipients reacted mostly against 4L6 IgG1 and had little or no reactivity against 5L7 IgG1 (Figure 1a); conversely, monkey sera from the 5L7 IgG1 recipients reacted mostly against 5L7 IgG1 and had little or no reactivity against 4L6 IgG1 (Figure 1b). Since both 4L6 IgG1 and 5L7 IgG1 share an identical constant region, these results indicate that anti-antibody responses in each group were mainly targeting the variable region of the corresponding antibody that the monkeys received. Only one monkey showed some degree of mixed reactivity: 153-10 (Figure 1b). These experiments not only allowed us to map the anti-antibody responses to the variable regions, they also allowed us to gauge relative reactivity to heavy versus light chains. For example, in all the animals of the 4L6 group (Figure 1a), a substantial portion of the anti-antibody response was directed against the variable region of the light chain of 4L6 IgG1 since little or no reactivity was detected to either 5L7 IgG1 or 5L7 Imm, but there was clear binding to 5L7-H 4L6-L IgG1. Only animal 120-09, the one with the lowest responses against 5L7 IgG1, did not show significant reactivity against the light chain. Overall, these results indicate that anti-antibodies are mainly targeting the variable regions of both the heavy and the light chain.
Figure 1.
Host anti-antibody responses to the adeno-associated virus (AAV)-delivered anti-SIV antibodies 5L7 IgG1 and 4L6 IgG1 mainly target the variable regions. Six naive rhesus monkeys received AAV-encoding anti-SIV mAb 5L7 IgG1 and another six received AAV encoding a different anti-SIV mAb, 4L6 IgG1. To dissect the anti-antibody responses to both antibodies, sera from the corresponding AAV recipients were tested against different versions of 5L7 IgG1 and 4L6 IgG1. Since both 4L6 and 5L7 had a κ-light chain, anti-antibodies were detected with a conjugated anti-lambda antibody. (a) Sera from the 4L6 IgG1 recipients and (b) sera from the 5L7 IgG1 recipients were tested against purified antibody 4L6 IgG1 and 5L7 IgG1, immunoadhesin versions of both, as well as hybrids. The immunoadhesin versions or antibody-like molecules (Imm in the graphs) consisted of only a single chain Fv, thus excluding the constant region of the light chain and the CH1 domain from the heavy chain. The hybrid versions of the antibodies consisted of the heavy chain from one antibody and the light chain from the other; therefore 5L7-H 4L6-L IgG1 consists of the heavy chain of 5L7 IgG1 with the light chain of 4L6 IgG1; conversely, 4L6-H 5L7-L IgG1 consists of the heavy chain of 4L6 IgG1 and the light chain of 5L7 IgG1. Since 4L6 IgG1 and 5L7 IgG1 shared the exact same constant region but differed in both heavy and light chain variable regions, antibodies that bound one but not the other were targeting the variable region. Serum from an SPF monkey was used as a negative control and to calculate the background level (dotted horizontal line). SPF, specific pathogen free.
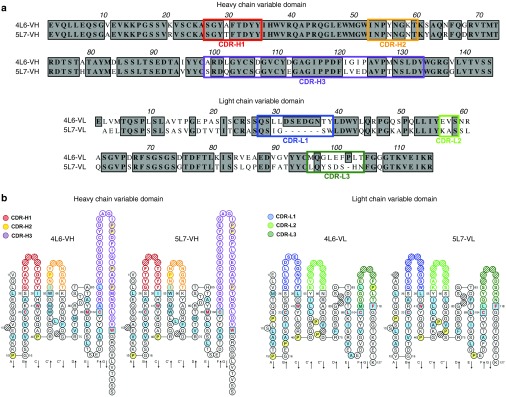
The 4L6 and 5L7 antibodies share a high degree of sequence similarity, particularly in the heavy chains (Figure 2a,b). Most of the sequence differences between the heavy chains were present in the complementarity-determining region (CDR)-H3 regions. The extensive sequence similarity between 4L6 and 5L7 IgG1 together with the limited cross-reactivity of the anti-antibody responses suggests that anti-antibodies to 4L6 and 5L7 are largely targeting regions of variable sequence differences between these two antibodies.
Figure 2.
Analysis of the amino acid sequence of the variable region of 4L6 IgG1 and 5L7 IgG1. 4L6 IgG1 and 5L7 IgG1 have similar sequences in the variable region of the heavy chain but differ more in the light chain. (a) Alignment of amino acid sequences of the variable region of both heavy and light chain of 4L6 IgG1 and 5L7 IgG1. Identical residues are boxed in gray, and complementarity-determining regions (CDRs) are indicated with colored frames. (b) Two-dimensional graphical representations of the variable domains or IMGT Collier de Perles, which were created by analyzing the corresponding nucleotide sequences with IMGT/V-QUEST.39,40 Numbering according to IMGT unique numbering for V-domain residues. CDRs for heavy chain (H1, H2, and H3) and light chain (L1, L2, and L3) are colored as indicated on the left; hatched-circle positions indicate gaps according to the IMGT numbering. Light blue indicates positions that are either hydrophobic amino acids (hydropathy index with positive value) or a tryptophan (W) as in 50% or more of analyzed variable domains, according to Pommié et al.42 Anchor positions are shown as squares and prolines in yellow. Amino acids with red and bold letters indicate the five conserved positions of a variable domain. Arrows indicate the β-strands and their direction. IMGT, international ImMunoGeneTics information system.
Fine mapping of the rhesus anti-antibody responses to rhesus IgGs
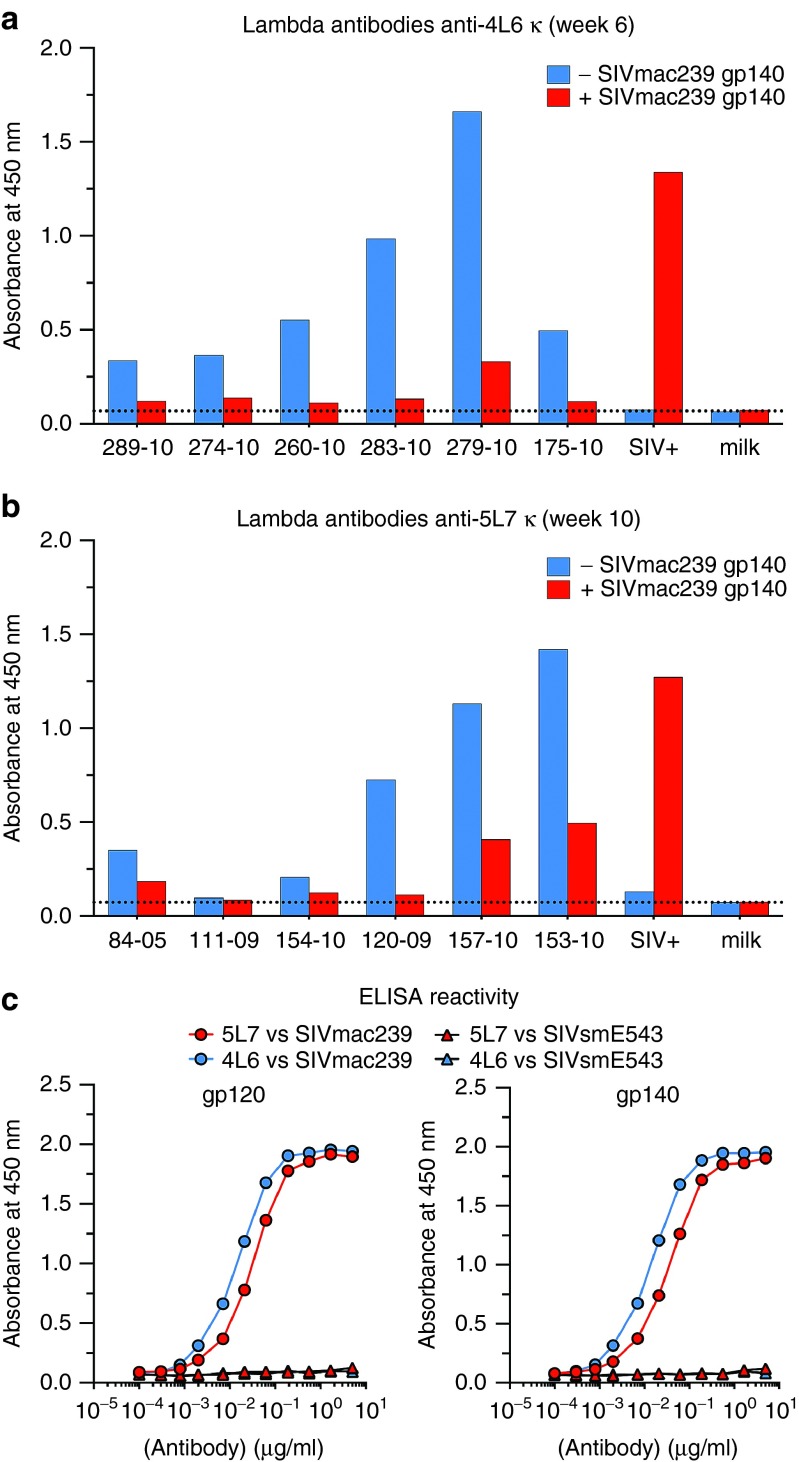
To further define the reactivity of the host anti-antibodies to 4L6 and 5L7 IgG1, we tested the ability of SIVmac239 gp140 to block the binding of anti-antibodies. Purified recombinant antibody was coated on ELISA plates and then preincubated with or without SIVmac239 gp140. Serum from each monkey was then tested for the detection of anti-antibodies against the antibody that each received. As shown in Figure 3a, preincubation with gp140 strongly blocked the binding of the anti-antibodies, and this is true for both 4L6 IgG1 and 5L7 IgG1 recipients. These results indicate that the antigen-binding site was being targeted and that binding of anti-antibodies to the delivered IgG blocked the ability of the delivered antibody to bind to its gp140 target. In the absence of preincubation with anti-antibody-containing monkey sera, purified 4L6 and 5L7 anti-SIV antibodies showed similar binding curves to SIVmac239 gp120 and gp140 by ELISA, but they did not bind detectably to SIVsmE543 gp120 or gp140 (Figure 3b).
Figure 3.
Preincubation with SIVmac239 gp140 results in decreased reactivity of sera against the delivered anti-SIV antibody. (a) Lambda anti-4L6 IgG1-κ antibodies were measured at the time of peak of anti-antibodies (week 6 after AAV administration) in sera from the 4L6 IgG1 recipients after preincubation with (red bars) or without (blue bars) SIVmac239 gp140. (b) Lambda anti-5L7 IgG1-κ antibodies were measured at time of peak (week 10 after AAV administration) in sera from the 5L7 IgG1 recipients after preincubation with (red bars) or without (blue bars) SIVmac239 gp140. A decrease in signal after preincubation with gp140 indicates that the presence of the antigen blocked the binding of a portion of the anti-antibodies. A SIV-positive serum was used as a positive control for the binding of gp140 to the antibody on the plate, and 5% milk in phosphate-buffered saline as a negative. (c) ELISA reactivity of 4L6 IgG1 and 5L7 IgG1 against SIVmac239 gp120 (left panel) and SIVmac239 gp140 (right panel). SIVsmE543 gp120 and gp140 were also correspondingly tested.
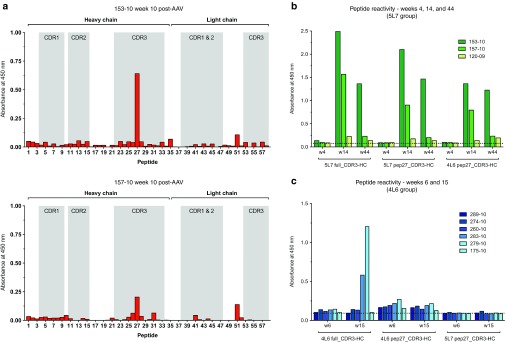
To further map and precisely define specific linear epitopes of the anti-antibody responses against the variable region of 5L7-IgG1, Pepscans were performed using overlapping peptides covering the entire variable regions of both the heavy chain and the light chain. The two monkeys with the highest levels of anti-5L7 IgG1 antibodies showed reactivity to peptide 27, as shown in Figure 4a. Peptide 27, which corresponds to the amino acid sequence SGGVCYEGAGIPPDF, is located in the midpoint of the CDR3 of 5L7, an area usually critical for epitope recognition and binding.18 In order to gain insight into this important reactivity, sera from the three monkeys with anti-antibodies in the 5L7 IgG1 group and from those that received 4L6 IgG1 were tested for reactivity against the full-length CDR3 peptide and peptide 27 matching the amino acid sequence of the antibody that they received, and to peptide 27 corresponding to the amino acid sequence that they did not receive, to test cross-reactivity (Figure 4b,c). In both groups of monkeys, reactivity against the corresponding full-length CDR3 peptide was detected in serum from the two animals that had shown the highest levels of anti-antibodies: 153-10 and 157-10 in the first group and 279-10 and 283-10 in the second. Reactivity to the corresponding peptides 27 was also detected in those samples, but was lower in sera from the monkeys of the 4L6 IgG1 group. Interestingly, no cross-reactivity was detected when peptide 27 containing the sequence that was not received was tested with the samples from the 4L6 group. In contrast, sera from both animals in the 5L7 group (153-10 and 157-10) did react with the peptide 27 sequence of the 4L6 antibody (Figure 4c). The amino acid sequences for the overlapping peptides used in the Pepscans from Figure 4a are shown in Supplementary Figure S1a, and alignment of the peptide sequences used in Figures 4b,c are shown in Supplementary Figure S1b.
Figure 4.
Analysis of antibody responses by Pepscan. (a) Sera from the recombinant AAV-5L7 recipients were tested by ELISA against overlapping peptides spanning both the heavy and the light chain variable regions of 5L7 IgG1. Heavy and light chain peptides are indicated by brackets. Shadows represent the areas corresponding to the CDRs. Pepscans for animals with the highest levels of anti-antibodies, 153-10 (upper panel) and 157-10 (lower panel) are shown. Sera samples corresponding to week 10 post-AAV administration, were tested at a 1/20 dilution. (b) Serum from the 5L7 IgG1 recipients was tested by ELISA against a peptide with the CDR3 sequence of the heavy chain of 5L7 (5L7 full_CDR3-HC), and against peptide 27 with the 5L7 sequence (5L7 pep27_CDR3-HC) and the peptide 27 sequence of the 4L6 antibody (4L6 pep27_CDR3-HC). Weeks 4, 14, and 44 after AAV administration were tested. (c) Same as in b but now serum from the 4L6 IgG1 recipients was tested against the corresponding versions of the peptides with 4L6 IgG1 sequences (4L6 full_CDR3-HC and 4L6 pep27_CDR3-HC) and, for cross-reactivity testing, against the peptide 27 with the sequence of 5L7 IgG1 (5L7 pep27_CDR3-HC). Weeks 6 and 15 after AAV administration were tested. Specific amino acid sequences for all these peptides can be seen in Supplementary Figure S1a.
Host humoral responses against AAV-delivered rhesusized anti-HIV antibodies also mainly target the variable regions
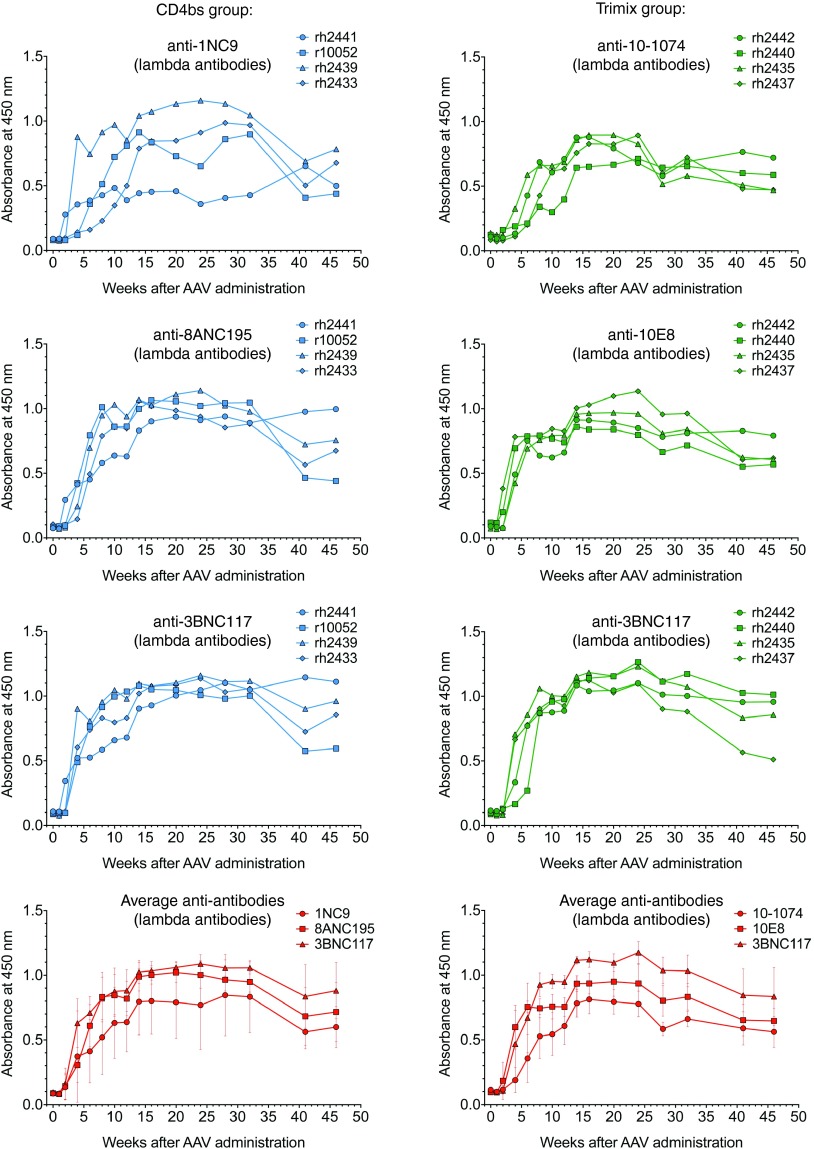
Two groups of four monkeys infected with SHIV-AD819 received different combinations of three different rhesusized anti-HIV mAbs 12 weeks postinfection in therapy trials. The mAbs that were used all had potent and broadly neutralizing activity. More specifically, one group got AAVs encoding rhesusized 1NC9, 8ANC195, and 3BNC117 while another group got AAVs encoding rhesusized 10–1074, 10E8, and 3BNC117. The first group was named “CD4bs” since 1NC9 and 3BNC117 have been reported to be CD4 binding site antibodies.2,20 Initially classified as a CD4bs antibody, 8ANC195 was subsequently shown to have a somewhat different specificity spanning both subunits of the envelope trimer.21 The second group was named “trimix” because of the three different specificities it received: 10–1074 targets a discontinuous epitope in the V3 loop22; 10E8 targets the membrane-proximal external region in gp4123 and 3BNC117 is a CD4bs mAb common to the previous group. Because of their human origin, antibody sequences of the anti-HIV antibodies were rhesusized prior to insertion into AAV, i.e., the human constant regions were substituted with equivalent rhesus constant region sequences. No change in the neutralization activity was observed after these antibodies were rhesusized (data not shown). Transient reductions in viral load were noted following the recombinant AAV administration, but viral loads quickly rebounded as the delivered mAbs disappeared from the serum of the treated monkeys. When sera from animals in both groups were tested for the presence of anti-antibodies, we observed that all the monkeys had mounted a humoral immune response against all three antibodies that each received (Figure 5). The anti-antibodies arose as soon as 2–4 weeks post-AAV administration and peaked around week 25, remaining quite stable for the duration of the study, almost 50 weeks. The CD4bs group data is shown in the left panels (in blue) and the trimix data in the right ones (green). Averages of the absorbance values were calculated for each antibody and represented in the two bottom panels (in red).
Figure 5.
Dynamics of host anti-antibody responses to adeno-associated virus (AAV)-delivered broadly neutralizing anti-HIV antibodies in monkeys. Two groups of four SHIV-AD8-infected monkeys were delivered different combinations of three rhesusized anti-HIV mAbs after 12 weeks of infection in a therapeutic experiment using AAV. Purified rhesusized anti-HIV antibodies were used to coat ELISA plates and sera from the recipient monkeys were tested for the presence of anti-antibodies by probing with a horseradish-peroxidase-conjugated anti-lambda antibody. Engineered κ-bearing versions were used for those anti-HIV originally bearing a lambda light chain. Serum reactivity is shown for each individual monkey against each individual antibody that it received.
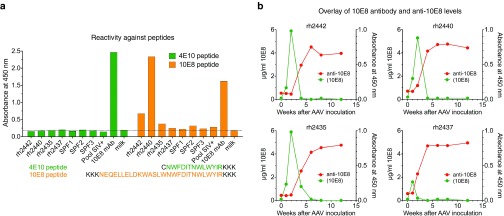
Since 10E8 is able to bind a linear epitope,23 we used two amino acid sequences, called the 4E10 peptide and the 10E8 peptide, as potential candidates for quantification purposes (Figure 6a). While both the shorter 4E10 peptide and the longer 10E8 peptide were capable of binding the 10E8 monoclonal antibody, only the shorter 4E10 peptide did not bind anti-Env antibodies present in the sera of the SHIV-AD8-infected monkeys (Figure 6a). Thus, the 4E10 peptide could be used to quantitate levels of AAV-delivered 10E8 monoclonal antibody in this study. Readily detectable levels of 10E8 were observed as early as 1 week post-AAV administration. The levels of 10E8 peaked at week 2 in three of the animals and at week 1 in one of the animals (Figure 6b). Peak levels of delivered 10E8 were in the range of 1.7–6 µg/ml. An abrupt, dramatic decline in levels of delivered 10E8 was coincident with the appearance of anti-10E8 antibodies (Figure 6b). In the monkey with the lowest levels of 10E8 in serum (rh2437), the anti-10E8 antibodies appeared earlier (week 2) while in the other three monkeys with higher levels of 10E8, anti-antibodies were seen later (mostly by week 4 post-AAV). Concentrations of delivered 10E8 mAb in serum dropped to below the limit of detection by 4–6 weeks following AAV administration (Figure 6b).
Figure 6.
The appearance of anti-10E8 antibodies is coincident in time with an abrupt and profound decrease in the levels of delivered 10E8. (a) Pre-AAV inoculation sera from the four monkeys infected with simian/human immunodeficiency virus (SHIV) that subsequently received 10E8 were tested for reactivity against two different peptides: 4E10 peptide with the minimum epitope sequence know to be bound by this antibody, and 10E8 peptide, a longer peptide that included the sequence of the 4E10 peptide.23 Lysines (K) and a cysteine (C) were added for improved solubility. Due to the lack of reactivity to the 4E10 peptide in sera from these SHIV-infected monkeys, the 4E10 peptide was used to quantitate the levels of delivered 10E8 in monkeys. (b) Levels of AAV-delivered 10E8 in sera from the four monkeys that received 10E8 in the trimix group (green) are plotted together with the corresponding levels of anti-10E8 (red).
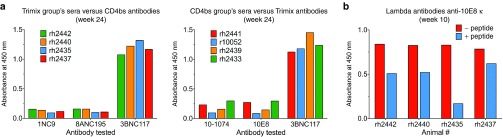
Next, sera from each group of animals were tested for anti-antibodies against the mAbs that the other group received. The reactivity was specific for the antibodies that each of the animals received (Figure 7a). Since all these mAbs shared an identical constant region, this indicates that the corresponding variable regions of each antibody were predominantly targeted in each group. As both groups got 3BNC117, this antibody worked as a positive control in these conditions.
Figure 7.
Host anti-antibody responses to adeno-associated virus (AAV)-delivered broadly neutralizing anti-HIV antibodies mainly target variable regions. (a) Sera from the group of monkeys that received the trimix combination of antibodies (10-1074, 10E8 and 3BNC117) were tested for the presence of anti-antibodies against the mAbs that the other group (CD4bs) received, i.e., 1NC9, 8ANC195, and 3BNC117 (upper panel) and the other way around (lower panel). Since the constant regions were all identical, the anti-antibodies were directed to the variable regions. 3BNC117 was used in both groups of monkeys. Sera from week 24 after AAV administration were tested. (b) Sera from the corresponding AAV recipients at week 10 (trimix group) was tested for reactivity against a κ version of 10E8 coated on an ELISA plate and preincubated with (blue bars) or without (red bars) 106 µmol/l of peptide corresponding to the minimum epitope sequence that 10E8 targets on the membrane-proximal external region.
We took further advantage of the fact that 10E8 could bind the 4E10 peptide for which reactivity was absent in pre-AAV serum (Figure 6a) to examine whether the peptide could block the reactivity of the anti-antibodies. Recombinant purified 10E8 coated on ELISA plates was preincubated with or without a 106 µmol/l solution of 4E10 peptide before sera from the corresponding group of monkeys were tested for anti-antibodies. As seen in Figure 7b, preincubation with this peptide partially blocked the binding of the anti-antibodies to 10E8 coated on the ELISA plates. This indicated that anti-antibodies targeted the antigen-binding site or a region very close to it, and that they could prevent the binding of 10E8 to its target.
Magnitude of host humoral responses to AAV-delivered rhesusized human antibodies correlates with extent of divergence from the germline
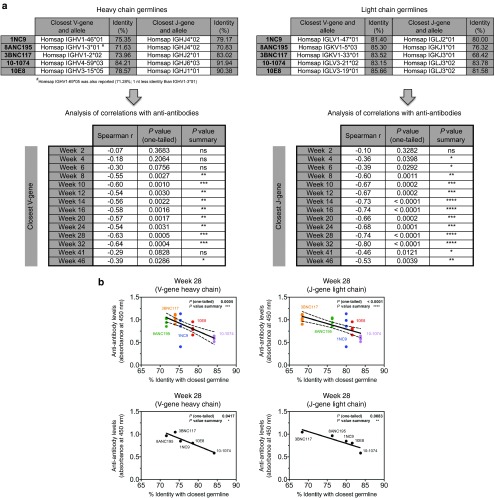
The nucleotide sequences of the human antibodies were analyzed with IMGT/V-QUEST to find the predicted closest human germline V- and J-genes and alleles (Figure 8a, top panels). The analysis was performed with the sequences of the variable regions of both heavy chains (left) and light chains (right). Percent identity with the predicted germline was also correspondingly obtained and correlations were calculated with the magnitude of the anti-antibody response (individual absorbance values for each antibody, obtained from Figure 5). Distance from the germline correlated strongly with the magnitude of the anti-antibody response (Figure 8a, panels below the arrows, and Figure 8b). Similar correlations were found when the closest germlines from rhesus monkeys were used for the analysis.
Figure 8.
Distance from germline correlates with the magnitude of the anti-antibody response. (a) Analysis of the corresponding nucleotide sequences was performed with the variable regions of both heavy chains (left) and light chains (right) of the adeno-associated virus (AAV)-delivered human antibodies using IMGT/V-QUEST. The closest V-genes and J-genes were obtained and identity percentages were calculated. Spearman correlations were then computed using the identity percentages and the levels of anti-antibody responses from Figure 5. Spearman correlation coefficients and P values with summary are shown, all calculated using GraphPad Prism 6 software. Code: ns (not significant) P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (b) Two correlations are shown: week 28 V-gene of heavy chain in the left panels and week 28 J-gene of the light chain in the right panels. The two top panels show calculations with individual values and the lower panels those with the averages.
Discussion
Individuals that become infected with HIV-1 typically mount a strain-specific neutralizing antibody response24,25; only a fraction of these individuals, however, go on to develop potent, broadly neutralizing activity over a prolonged period.6,26 The last 5–10 years has seen the accumulation of a number of remarkably potent, broadly-neutralizing mAbs from such individuals.2,20,22,23,26 At least five different specificities targeting conserved regions of the HIV envelope glycoprotein are represented in this collection.21 With respect to prevention scenarios, it is easy to envision how stable maintenance of protective concentrations of some combination of these monoclonal antibodies in high-risk individuals could provide a sterilizing barrier to the vast majority of HIV-1 strains circulating in the population. However, passive infusion of purified antibody is not a practical long-term solution because of the millions of people that need to be reached, the need for repeated administrations over a prolonged period, the massive amounts of purified antibody that would be needed, and the extremely high cost. We have been using monkey models to inform and guide development of the AAV delivery approach for use in people.
In our prevention trials (12 monkeys) and our therapy trials (8 monkeys), there were 36 opportunities for the generation of anti-antibody responses to AAV-delivered antibodies: 12 in the prevention trials since each monkey received only one antibody and 24 in the therapy trials since each monkey received a combination of three antibodies. Of the 36 opportunities, 33 of them had clear, readily-detectable anti-antibody responses. In the vast majority of cases (29/33), the anti-antiresponses persisted for as long as we followed the animals: >10 months to as long as 1.3 years. In all cases, the anti-antiresponses were directed predominantly or exclusively to the variable regions. The anti-antibody responses appeared to severely limit the concentration of delivered mAb that could be maintained. There is no way of knowing whether these results will be predictive of what will happen in human trials until such time that AAV-mAb delivery trials are carried out in humans; nonetheless, our findings raise serious concerns over this possibility.
It is reasonable to think that the immunogenicity of an AAV-delivered protein will be strongly influenced by the extent of divergence from endogenous protein, i.e., the degree of self versus nonself.27 Indeed, the strength of the immune response to factor IX in the setting of hemophilia correlates directly with the extent of divergence from endogenous coding information.28 Antibodies are inherently extremely variable in sequence and differ to varying degrees with the sequences present in the inherited germline DNA. Somatic mutation from germline sequences is a built-in process to give antibodies the specificity they need to fight the diverse array of microorganisms that the host may encounter. Furthermore, there is a complex “checkpoint” system to allow some antibodies that develop naturally, but not others, into the circulation of that individual.29 It is highly unlikely that an individual that receives a monoclonal antibody derived from another individual will have ever seen the divergent variable sequences present in that antibody.30
This problem with the “foreignness” of an externally delivered monoclonal antibody is compounded for applications to AIDS by the nature of the anti-HIV mAbs that one would like to deliver. The potent, broadly-neutralizing, anti-HIV mAbs are highly evolved over a prolonged period of time. They usually contain heavily hypermutated CDR and framework regions.2,20,22,23,26 Consistent with this line of reasoning, we report here for the first time a highly significant correlation of the magnitude of the host anti-antibody response with the distance from germline of the delivered antibody. Also along these lines, it is reasonable to think that the apparently diminished immunogenicity of AAV-delivered eCD4-Ig16 relates to its extensive identity with germline sequences and the minimal short stretch of nongermline sequences at its C-terminus.
Other factors could also possibly contribute to the immunogenicity of AAV-delivered antibodies. PERp1, a resident ER protein that is a component of the BiP (binding immunoglobulin protein) chaperon complex, is substantially upregulated during B to plasma cell differentiation and has a key role in ensuring correct folding and oligomerization of immunoglobulins.31,32 Muscle cells where the AAV is being administered are not B-cells and their controlling mechanisms may fall short in this regard, particularly at the very high levels of expression that are achieved with AAV vectors. It is also possible that some small percent of the inoculated AAV vector finds its way into professional antigen presenting cells such as dendritic cells and attempts to express the immunoglobulin in these cells could conceivably set off the immune response. Further work will be needed to understand the critical factors that underlie the anti-immunoglobulin responses to AAV-delivered antibodies and to develop practical strategies to circumvent these anti-antibody responses.
Materials and Methods
Antibody coding sequences and recombinant AAV. 4L6 and 5L7 immunoadhesin sequences13 served as a template and full-length antibodies were constructed by adding CH1 domain and CL domain of rhesus IgG to the already known immunoadhesin sequences as described elsewhere.15 Antibody sequences of the anti-HIV antibodies were kindly provided by Michel Nussenzweig (The Rockefeller University) and Mark Connors (National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIAID)). In order to rhesusize them, the variable domains of both heavy and light chain were identified using the IMGT/V-QUEST software (see below) and constant domains were swapped with the corresponding IgG1 rhesus sequences based on published sequences (GenBank: AAQ57550.1 for the heavy chain; AAD02577.1 for the κ chain, and ADX62855.1 for the lambda).33,34 Coding sequences were all designed in silico, codon-optimized and gene-synthesized (Genscript, Piscataway, NJ). Synthesized fragments were then cloned into NotI site of ssAAV vector plasmids. Production of recombinant AAV was conducted as described previously.35,36 In short, HEK 293 cells were transfected with recombinant AAV vector plasmid and two helper plasmids to allow generation of infectious AAV particles. After harvesting transfected cells and cell culture supernatant, AAV was purified by three sequential CsCl centrifugation steps. Vector genome number was assessed by real-time polymerase chain reaction, and the purity of the preparation was verified by electron microscopy and silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
For further details regarding our constructs, see ref. 15. Please note that the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) was used to enhance transgene expression.37 It has been suggested that the WPRE might encode a truncated version of the woodchuck hepatitis virus X protein.38 We have tested reactivity of sera from the AAV-administered monkeys to recombinant Protein X (Cusabio Biotech, Wuhan, China); this recombinant protein contains the amino acid sequence of the potentially expressed truncated protein. No significant reactivity was detected.
Identification of variable regions and closest germlines. In order to identify the variable regions of the corresponding antibodies, the nucleotide sequences were analyzed IMGT/V-QUEST (International ImMunoGeneTics information system/V-QUEry and STandardization; http://www.imgt.org/),39,40 an integrated software that analyzes rearranged immunoglobulin nucleotide sequences. Also with this tool, the V(D)J (variable(diversity)joining) genes and alleles were identified by alignment with the immunoglobulin germ line gene and allele sequences of the IMGT reference directory (Figures 2a and 8a). The CDRs and framework regions were identified on the basis of the IMGT unique numbering, and somatic mutations were identified. This tool was also used to create two-dimensional graphical representations of the variable regions, the IMGT Collier de Perles (Figure 2b).
Rhesus macaques and AAV immunization. The 20 animals in our study were Indian-origin rhesus macaques (Macaca mulatta). Twelve of them were housed at the New England Primate Research Center in a biosafety level 3 animal containment facility in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care and the Harvard Medical School Animal Care and Use Committee. Research was conducted according to the principles described in the Guide for the Care and Use of Laboratory Animals and was approved by the Harvard Medical School Animal Care and Use Committee.41 The remaining eight animals were housed at the Wisconsin National Primate Research Center and they were similarly cared for in accordance with the guidelines of the Weatherall Report under a protocol approved by the University of Wisconsin Graduate School Animal Care and Use Committee. All macaques tested negative for the presence of antibodies to SIV and AAV1 capsid prior to AAV administration. The weights of the animals ranged from 2.5 to 10.3 kg at the time of immunization. The 4L6 recipients received a total of 2.5 × 1013 AAV vector genomes per monkey and the 5L7 recipients received 1.6 × 1013 particles (see ref. 15 for details). The animals that received the rhesusized anti-HIV antibodies were infected with SHIV-AD8 and 12 weeks afterwards they were segregated into two equivalent groups based on viral loads. Then one group received AAVs encoding for 1NC9, 8ANC195 and 3BNC117 (total dose of 2.5 × 1013 AAV vector genomes per monkey) and the second received AAVs encoding for 10-1074, 10E8, and 3BNC117 (total dose of 3 × 1013 AAV vector genomes per monkey). AAV administration was conducted by deep intramuscular 0.5 ml injections. Each monkey received three different recombinant AAVs, one per antibody. Two injections of one recombinant AAV were given in the right quadriceps. Two injections of the second recombinant AAV were given in the left quadriceps. One injection of the third recombinant AAV was given in the right deltoid and one in the left deltoid.
Anti-antibody response. Measuring humoral responses to the AAV-delivered mAb is challenging since both antibody and anti-antibodies are of the same host species. To solve this problem, κ-bearing versions of the antibodies were engineered (for lambda bearer mAbs) and used to coat plates; then anti-antibodies were detected by means of an anti-lambda antibody (Southern Biotech, Birmingham, AL) in a regular ELISA. This way we could readily detect those anti-antibodies with a lambda light chain, which have been reported as the most prominent in our previous studies.15 To assess the target of the anti-antibody response we tested the reactivity of the serum samples against the corresponding purified mAb or mAbs. In some cases, different sets of peptides were also tested, including a whole library of overlapping peptides panning the entire variable region of both heavy and light chain of 5L7. Peptides were all synthesized by GenScript and specific sequences are indicated in Supplementary Figure S1.
Recombinant antibodies. HEK293T cells were expanded and then transfected with recombinant AAV vector plasmids. Cells were washed after 4 hours with prewarmed phosphate-buffered saline and then transferred to serum-free medium (Invitrogen, Carlsbad, CA). Afterwards, the antibody-containing medium was harvested, precleared by centrifugation, and filtered through 0.22-µm-pore-size membrane. Then, IgG was affinity purified using protein A Sepharose 4 Fast Flow (GE Healthcare, Uppsala, Sweden) followed by protein quantification with a Nanodrop UV spectrometer (Thermo Fisher Scientific, Wilmington, DE). Antibody purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent Coomassie blue staining (data not shown).
SUPPLEMENTARY MATERIAL Figure S1. Amino acid sequences of the overlapping peptides used in the Pepscans.
Acknowledgments
This project was supported by National Institutes of Health (NIH) grants P01 AI100263, R01 AI098446, and U19 AI095985 (to R.C.D.). We also acknowledge support from the Miami Center for AIDS Research (to J.M.M.-N.) at the University of Miami Miller School of Medicine funded by grant (P30AI073961) from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors would like to thank Li Zhou and William Lauer for technical assistance; the Gene Therapy Core at University of Massachusetts Medical School for AAV vector preparation and supportive advice, and the Wisconsin National Primate Research Center and the New England Primate Research Center veterinary staff for professional animal care. We would also like to thank Michel Nussenzweig (The Rockefeller University) and Mark Connors (NIH/NIAID) for providing the human antibody sequences. The authors wish to also acknowledge David Watkins (University of Miami), Matthew Gardner and Charles Bailey (The Scripps Research Institute) and Shaoyong Li (University of Massachusetts Medical School) for their insightful comments and critical reading of the manuscript.
Supplementary Material
Amino acid sequences of the overlapping peptides used in the Pepscans.
References
- Burton, DR and Mascola, JR (2015). Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16: 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F, Mouquet, H, Dosenovic, P, Scheid, JF, Scharf, L and Nussenzweig, MC (2013). Antibodies in HIV-1 vaccine development and therapy. Science 341: 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong, PD, Mascola, JR and Nabel, GJ (2013). Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol 13: 693–701. [DOI] [PubMed] [Google Scholar]
- Kwong, PD and Mascola, JR (2012). Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37: 412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, LM, Huber, M, Doores, KJ, Falkowska, E, Pejchal, R, Julien, JP et al.; Protocol G Principal Investigators. (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477: 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, DR, Desrosiers, RC, Doms, RW, Koff, WC, Kwong, PD, Moore, JP et al. (2004). HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5: 233–236. [DOI] [PubMed] [Google Scholar]
- Hastie, E and Samulski, RJ (2015). Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success–a personal perspective. Hum Gene Ther 26: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya, S and Berns, KI (2008). Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21: 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves, MA (2005). Adeno-associated virus: from defective virus to effective vector. Virol J 2: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, DM (2008). Self-complementary AAV vectors; advances and applications. Mol Ther 16: 1648–1656. [DOI] [PubMed] [Google Scholar]
- Schultz, BR and Chamberlain, JS (2008). Recombinant adeno-associated virus transduction and integration. Mol Ther 16: 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi, F and High, KA (2011). Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12: 341–355. [DOI] [PubMed] [Google Scholar]
- Johnson, PR, Schnepp, BC, Zhang, J, Connell, MJ, Greene, SM, Yuste, E et al. (2009). Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat Med 15: 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, KO, Wang, L, Joyce, MG, Yang, ZY, Balazs, AB, Cheng, C et al. (2015). Broadly neutralizing human immunodeficiency virus type 1 antibody gene transfer protects nonhuman primates from mucosal simian-human immunodeficiency virus infection. J Virol 89: 8334–8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, SP, Martinez-Navio, JM, Piatak, M Jr, Lifson, JD, Gao, G and Desrosiers, RC (2015). AAV-delivered antibody mediates significant protective effects against SIVmac239 challenge in the absence of neutralizing activity. PLoS Pathog 11: e1005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, MR, Kattenhorn, LM, Kondur, HR, von Schaewen, M, Dorfman, T, Chiang, JJ et al. (2015). AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs, AB, Chen, J, Hong, CM, Rao, DS, Yang, L and Baltimore, D (2012). Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, JL and Davis, MM (2000). Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13: 37–45. [DOI] [PubMed] [Google Scholar]
- Nishimura, Y, Shingai, M, Willey, R, Sadjadpour, R, Lee, WR, Brown, CR et al. (2010). Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J Virol 84: 4769–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid, JF, Mouquet, H, Ueberheide, B, Diskin, R, Klein, F, Oliveira, TY et al. (2011). Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333: 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, L, Scheid, JF, Lee, JH, West, AP Jr, Chen, C, Gao, H et al. (2014). Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep 7: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet, H, Scharf, L, Euler, Z, Liu, Y, Eden, C, Scheid, JF et al. (2012). Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA 109: E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J, Ofek, G, Laub, L, Louder, MK, Doria-Rose, NA, Longo, NS et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491: 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman, DD, Wrin, T, Little, SJ and Petropoulos, CJ (2003). Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA 100: 4144–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X, Decker, JM, Wang, S, Hui, H, Kappes, JC, Wu, X et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422: 307–312. [DOI] [PubMed] [Google Scholar]
- Walker, LM, Phogat, SK, Chan-Hui, PY, Wagner, D, Phung, P, Goss, JL et al.; Protocol G Principal Investigators. (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326: 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisgerault, F and Mingozzi, F (2015). The Skeletal Muscle Environment and Its Role in Immunity and Tolerance to AAV Vector-Mediated Gene Transfer. Curr Gene Ther 15: 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, O, Hoffman, BE, Moghimi, B, Nayak, S, Cooper, M, Zhou, S et al. (2009). Impact of the underlying mutation and the route of vector administration on immune responses to factor IX in gene therapy for hemophilia B. Mol Ther 17: 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann, H, Yurasov, S, Schaefer, A, Young, JW, Meffre, E and Nussenzweig, MC (2003). Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- Wang, C, Liu, Y, Cavanagh, MM, Le Saux, S, Qi, Q, Roskin, KM et al. (2015). B-cell repertoire responses to varicella-zoster vaccination in human identical twins. Proc Natl Acad Sci USA 112: 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y, Meunier, L and Hendershot, LM (2009). pERp1 is significantly up-regulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. Proc Natl Acad Sci USA 106: 17013–17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige, MJ, Hendershot, LM and Buchner, J (2010). How antibodies fold. Trends Biochem Sci 35: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scinicariello, F, Engleman, CN, Jayashankar, L, McClure, HM and Attanasio, R (2004). Rhesus macaque antibody molecules: sequences and heterogeneity of alpha and gamma constant regions. Immunology 111: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvas, P, Apoil, P, Fortenfant, F, Roubinet, F, Andris, J, Capra, D et al. (1999). Characterization of the three immunoglobulin G subclasses of macaques. Scand J Immunol 49: 595–610. [DOI] [PubMed] [Google Scholar]
- Zhang, H, Yang, B, Mu, X, Ahmed, SS, Su, Q, He, R et al. (2011). Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol Ther 19: 1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, SS, Li, H, Cao, C, Sikoglu, EM, Denninger, AR, Su, Q et al. (2013). A single intravenous rAAV injection as late as P20 achieves efficacious and sustained CNS Gene therapy in Canavan mice. Mol Ther 21: 2136–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey, R, Donello, JE, Trono, D and Hope, TJ (1999). Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 73: 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach, A, Bohne, J, Baum, C, Hermann, FG, Egerer, L, von Laer, D et al. (2006). Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther 13: 641–645. [DOI] [PubMed] [Google Scholar]
- Giudicelli, V, Brochet, X and Lefranc, MP (2011). IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011: 695–715. [DOI] [PubMed] [Google Scholar]
- Ruiz, M and Lefranc, MP (2002). IMGT gene identification and Colliers de Perles of human immunoglobulins with known 3D structures. Immunogenetics 53: 857–883. [DOI] [PubMed] [Google Scholar]
- National Research Council, IoLAR, Commission on Life Sciences. Guide for the Care and Use of Laboratory Animals. The National Academies Press: Washington, DC, 1996. [Google Scholar]
- Pommié, C, Levadoux, S, Sabatier, R, Lefranc, G and Lefranc, MP (2004). IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J Mol Recognit 17: 17–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences of the overlapping peptides used in the Pepscans.