Abstract
Cryptococcus neoformans var. neoformans (serotype D) represents about 30% of the clinical isolates in Europe and is present less frequently in the other continents. It is the prevalent etiological agent in primary cutaneous cryptococcosis as well as in cryptococcal skin lesions of disseminated cryptococcosis. Very little is known about the genotypic diversity of this Cryptococcus subtype. The aim of this study was to investigate the genotypic diversity among a set of clinical and environmental C. neoformans var. neoformans isolates and to evaluate the relationship between genotypes, geographical origin and clinical manifestations. A total of 83 globally collected C. neoformans var. neoformans isolates from Italy, Germany, France, Belgium, Denmark, Greece, Turkey, Thailand, Japan, Colombia, and the USA, recovered from different sources (primary and secondary cutaneous cryptococcosis, disseminated cryptococcosis, the environment, and animals), were included in the study. All isolates were confirmed to belong to genotype VNIV by molecular typing and they were further investigated by MLST analysis. Maximum likelihood phylogenetic as well as network analysis strongly suggested the existence of a recombinant rather than a clonal population structure. Geographical origin and source of isolation were not correlated with a specific MLST genotype. The comparison with a set of outgroup C. neoformans var. grubii isolates provided clear evidence that the two varieties have different population structures.
Keywords: Cryptococcus, C. neoformans var. neoformans, C. neoformans var. grubii, MLST, Recombination
1. Introduction
Cryptococcus neoformans and C. gattii are two sibling yeast species responsible for cryptococcosis. This life-threatening disease is mainly associated with AIDS patients in the countries where the HIV infection burden is still high such as in sub-Saharan Africa and in South East Asia (Assogba et al., 2015; Park et al., 2009). In developed countries, however, the incidence of cryptococcosis in HIV-infected population is decreasing due to the introduction of high active antiretroviral therapy (HAART). In contrast, the disease is increasingly found in non-AIDS patients such as those with hematological neoplasms, recipients of organ transplantation, and victims of autoimmune diseases (Bratton et al., 2012; Henao-Martínez and Beckham, 2015; Sanchini et al., 2014).
C. neoformans is classified into two varieties, three serotypes and five molecular types. C. neoformans var. grubii, serotype A, is identified as the molecular types VNI, VNII and VNB, whereas C. neoformans var. neoformans, serotype D, belongs to molecular type VNIV. In addition, diploid or aneuploid intervarietal AD hybrids are identified as molecular type VNIII. Cryptococcus gattii has two serotypes, B and C, and four molecular types VGI, VGII, VGII, and VGIV (Heitman et al., 2010).
The recent C. gattii emergence from Vancouver Island (Canada) south to the Pacific Northwest of the United States, has contributed to the interest in C. gattii in different parts of the world and, at the same time, to highlight the differences from C. neoformans in both ecological distribution and clinical manifestations (Espinel-Ingroff and Kidd, 2015; Chen et al., 2014).
C. neoformans var. grubii is the prevalent agent of cryptococcosis and it is globally distributed. It is commonly isolated from pigeon and other bird excreta and soil as well as many species of trees (Cogliati, 2013). The main clinical manifestation and the cause of death is meningoencephalitis especially in immunocompromised patients (Kwon-Chung et al., 2014).
C. neoformans var. neoformans has not been extensively investigated and little is known about its ecology, distribution and clinical manifestations. The majority of clinical isolates were reported from Europe where it has a prevalence of 30% (Viviani et al., 2006), but it has also been found in North (Yan et al., 2002; Litvintseva et al., 2005) and South America (Pérez et al., 2008; Cortés et al., 2011; Meyer et al., 2003; Trilles et al., 2003) as well as in Asia (Sukroongreung et al., 1996; Feng et al., 2008; Capoor et al., 2008; Ikeda and Shinoda, 2000). Very few isolates have been recovered from the environment mainly from pigeon droppings. A recent large environmental survey carried out in Europe identified the association of this variety with different tree species (Cogliati et al., 2014). At present, little information about specific clinical manifestations is available although a skin tropism of this yeast has been shown. A study carried out on 108 Cryptococcus isolates recovered from patients with skin lesions clearly indicated that infection with serotype D isolates was one of the risk factors in cutaneous manifestations (Neuville et al., 2003).
The present study aims to investigate a large number of C. neoformans var. neoformans strains isolated from different geographical areas and from different sources in order to elucidate the genetic population structure of this variety.
2. Materials and methods
2.1. Isolates
Eighty-three C. neoformans var. neoformans isolates were investigated (Table 1). Twenty isolates were from Germany, 19 from Italy, 13 from Greece, 8 from Japan, 7 from France, 4 from Belgium, 4 from Denmark, 4 from Colombia, and 1 each from Australia, Thailand, Turkey and the USA. Twenty-six isolates were recovered from the environment (soil, trees, pigeon droppings and dust), 22 from cases of disseminated cryptococcosis, 17 from cases of documented primary cutaneous cryptococcosis, 4 from cases of probable primary cutaneous cryptococcosis, 9 from cases of secondary cutaneous cryptococcosis, and 5 from veterinary cases. All clinical cases were independent cases and no multiple isolates from the same patient were included in the study. In addition, data obtained from previous studies of 30 C. neoformans var. grubii isolates (11 VNB, 10 VNI, and 9 VNII) were included in the analysis as outgroup strains (Khayhan et al., 2013; Litvintseva et al., 2006; Sanchini et al., 2014; Cogliati et al., 2013; Meyer et al., 2009; Kaocharoen et al., 2013; Choi et al., 2010; Umeyama et al., 2013; Wiesner et al., 2012) (Table S1).
Table 1.
Clinical and molecular information of the 83 C. neoformans var. neoformans isolates investigated in the present study.
| Strain code | Category | Origin | Source | Date | Underlying disease | Molecular type |
Mating type |
Sequence type |
CAP59 | GPD1 | IGS1 | LAC1 | PLB1 | SOD1 | URA5 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WM629 | DC | Australia | Blood | 1987 | AIDS | VNIV | αD | 117 | 16 | 21 | 30 | 19 | 13 | 1 | 19 | Meyer |
| IUM 01-4729 | ENV | Belgium | Pigeon droppings | 2001 | – | VNIV | αD | 510 | 16 | 21 | 24 | 18 | 13 | 20 | 32 | Cogliati |
| IUM 01-4730 | ENV | Belgium | Dust | 2001 | – | VNIV | αD | 514 | 16 | 40 | 24 | 20 | 13 | 20 | 32 | Cogliati |
| IUM 97-4899 | PPCC | Belgium | Skin | 1997 | No risk factors | VNIV | αD | 507 | 16 | 3 | 26 | 39 | 14 | 20 | 34 | Cogliati |
| IUM 98-5036 | PPCC | Belgium | Skin | 1998 | Diabete | VNIV | αD | 506 | 16 | 3 | 24 | 20 | 13 | 20 | 32 | Cogliati |
| H0058-I-1406 | DC | Colombia | CSF | 2002 | AIDS | VNIV | αD | 335 | 27 | 28 | 30 | 19 | 14 | 17 | 41 | Escandon |
| H0058-I-2250 | DC | Colombia | CSF | 2004 | AIDS | VNIV | αD | 336 | 16 | 22 | 32 | 14 | 14 | 18 | 17 | Escandon |
| H0058-I-2291 | DC | Colombia | CSF | 2004 | AIDS | VNIV | αD | 160 | 16 | 21 | 30 | 19 | 13 | 17 | 19 | Escandon |
| H0058-I-2880 | DC | Colombia | CSF | 2007 | AIDS | VNIV | αD | 336 | 16 | 22 | 32 | 14 | 14 | 18 | 17 | Escandon |
| NIH-424 | ENV | Denmark | Pigeon nest | 1970 | – | VNIV | αD | 180 | 20 | 21 | 26 | 21 | 19 | 17 | 21 | Kwon-Chung |
| NIH-429 | ENV | Denmark | Pigeon nest | 1970 | – | VNIV | αD | 512 | 16 | 21 | 31 | 19 | 13 | 19 | 16 | Kwon-Chung |
| NIH-430 | ENV | Denmark | Pigeon nest | 1970 | – | VNIV | aD | 509 | 16 | 20 | 24 | 16 | 14 | 20 | 16 | Kwon-Chung |
| NIH-433 | ENV | Denmark | Pigeon nest | 1970 | – | VNIV | aD | 515 | 17 | 21 | 28 | 13 | 14 | 17 | 16 | Kwon-Chung |
| CNRMA00.330 | PCC | France | Skin | 2000 | No risk factor | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Dromer |
| CNRMA00.840 | SCC | France | Skin | 2000 | Hematological malignancy | VNIV | αD | 125 | 16 | 21 | 45 | 21 | 13 | 21 | 22 | Dromer |
| CNRMA07.1501 | SCC | France | CSF | 2007 | AIDS | VNIV | αD | 180 | 20 | 21 | 26 | 21 | 19 | 17 | 21 | Dromer |
| CNRMA97.697 | DC | France | CSF | 1997 | AIDS | VNIV | αD | 121 | 16 | 21 | 32 | 19 | 13 | 17 | 20 | Dromer |
| CNRMA98.480 | DC | France | CSF | 1998 | AIDS | VNIV | αD | 511 | 16 | 21 | 29 | 16 | 14 | 17 | 24 | Dromer |
| CNRMA99.1037 | PCC | France | Skin | 1999 | No risk factor | VNIV | αD | 122 | 16 | 21 | 32 | 24 | 13 | 17 | 20 | Dromer |
| MKT6301 | PCC | France | Skin | 2011 | No risk factor | VNIV | αD | 121 | 16 | 21 | 32 | 19 | 13 | 17 | 20 | Bienvenu |
| RKI 04-0061 | DC | Germany | – | 2004 | Liver disorder | VNIV | αD | 110 | 15 | 21 | 24 | 21 | 13 | 20 | 22 | Rickerts |
| RKI 04-0089 | PCC | Germany | Skin, hand | 2004 | Chronical asthma, corticosteroids | VNIV | αD | 486 | 16 | 22 | 32 | 19 | 13 | 17 | 22 | Rickerts |
| RKI 05-0151 | DC | Germany | Blood | 2005 | Diabetes | VNIV | αD | 531 | 16 | 29 | 24 | 20 | 13 | 20 | 18 | Rickerts |
| RKI 07-0173 | DC | Germany | – | 2007 | Liver disorder | VNIV | αD | 530 | 31 | 21 | 83 | 13 | 37 | 59 | 53 | Rickerts |
| RKI 08-0429 | DC | Germany | CSF | 2008 | AIDS | VNIV | αD | 487 | 16 | 21 | 32 | 24 | 13 | 17 | 32 | Rickerts |
| RKI 08-0572 | DC | Germany | – | 2008 | No risk factors | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 08-0591 | DC | Germany | – | 2008 | No risk factors | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 09-0102 | DC | Germany | – | 2009 | Sarcoidosis | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 09-0103 | DC | Germany | – | 2009 | Sarcoidosis | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 09-0388 | PCC | Germany | Skin | 2009 | No risk factor | VNIV | αD | 519 | 26 | 22 | 32 | 24 | 13 | 17 | 20 | Rickerts |
| RKI 09-0393 | PCC | Germany | Skin | 2009 | No risk factor | VNIV | αD | 505 | 15 | 21 | 31 | 15 | 13 | 19 | 18 | Rickerts |
| RKI 09-0515 | DC | Germany | BAL | 2009 | Solid organ Tx | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Rickerts |
| RKI 09-0545 | DC | Germany | CSF | 2009 | AIDS | VNIV | αD | 160 | 16 | 21 | 30 | 19 | 13 | 17 | 19 | Rickerts |
| RKI 11-0047 | DC | Germany | CSF | 2010 | AIDS | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 11-0048 | DC | Germany | CSF | 2010 | AIDS | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 12-0155 | PCC | Germany | Skin | 2012 | No risk factor | VNIV | αD | 116 | 16 | 21 | 29 | 13 | 14 | 17 | 24 | Rickerts |
| RKI 12-0559 | PCC | Germany | Skin | 2012 | No risk factor | VNIV | αD | 513 | 16 | 21 | 43 | 21 | 13 | 20 | 18 | Rickerts |
| RKI 13-0490 | ENV | Germany | Pigeon droppings | 2013 | – | VNIV | aD | 522 | 27 | 21 | 53 | 19 | 14 | 19 | 16 | Rickerts |
| RKI 13-0491 | ENV | Germany | Pigeon droppings | 2013 | – | VNIV | aD | 523 | 29 | 21 | 83 | 21 | 14 | 58 | 53 | Rickerts |
| RKI 13-0492 | ENV | Germany | Pigeon droppings | 2013 | – | VNIV | aD | 524 | 29 | 21 | 83 | 40 | 14 | 58 | 53 | Rickerts |
| GRACA18BK1-3 | ENV | Greece | Eucalyptus tree | 2013 | – | VNIV | aD | 502 | 26 | 38 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRACP14BK1-1 | ENV | Greece | Pine tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRACP15SO1-1 | ENV | Greece | Pine tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRACP15SO1-2 | ENV | Greece | Pine tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRACP16HO1-1 | ENV | Greece | Pine tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRACP30BK1-1 | ENV | Greece | Plane tree | 2013 | – | VNIV | aD | 498 | 26 | 20 | 30 | 17 | 13 | 19 | 18 | Velegraki |
| GRAKI10SO1-1 | ENV | Greece | Olive tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRAKI11HO1-1 | ENV | Greece | Olive tree | 2013 | – | VNIV | aD | 499 | 26 | 21 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRAKI12SO1-1 | ENV | Greece | Plane tree | 2013 | – | VNIV | aD | 503 | 26 | 39 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRAKI13HO1-1 | ENV | Greece | Plane tree | 2013 | – | VNIV | aD | 503 | 26 | 39 | 30 | 22 | 13 | 19 | 18 | Velegraki |
| GRAKI28HO1-1 | ENV | Greece | Olive tree | 2013 | – | VNIV | aD | 500 | 26 | 22 | 44 | 17 | 14 | 23 | 23 | Velegraki |
| GRLMM26HO1-2 | ENV | Greece | Olive tree | 2013 | – | VNIV | aD | 489 | 1 | 1 | 1 | 1 | 1 | 27 | 1 | Velegraki |
| GRSAB18HO1-1 | ENV | Greece | Pine tree | 2013 | – | VNIV | aD | 501 | 26 | 37 | 30 | 38 | 13 | 19 | 18 | Velegraki |
| ITMPV22BK7-1 | ENV | Italy | Oak tree | 2014 | – | VNIV | αD | 496 | 16 | 3 | 30 | 19 | 13 | 17 | 19 | Cogliati |
| IUM 01-0956 | SCC | Italy | Skin, head | 2001 | AIDS | VNIV | αD | 521 | 27 | 13 | 12 | 6 | 9 | 8 | 13 | Cogliati |
| IUM 02-0826 | VET | Italy | Cat | 2002 | – | VNIV | aD | 517 | 22 | 21 | 30 | 24 | 21 | 17 | 20 | Cogliati |
| IUM 02-4295 | PPCC | Italy | Skin | 2002 | No risk factors | VNIV | aD | 516 | 22 | 3 | 31 | 24 | 14 | 17 | 20 | Cogliati |
| IUM 73-0017 | DC | Italy | CSF | 1973 | No risk factors | VNIV | αD | 508 | 16 | 3 | 31 | 24 | 14 | 17 | 16 | Cogliati |
| IUM 77-0033 | SCC | Italy | Skin, right ear | 1977 | No risk factors | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 79-0801 | PCC | Italy | Skin, left leg | 1979 | Common variable immunodeficiency | VNIV | αD | 520 | 27 | 3 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 91-2588 | SCC | Italy | Skin, hand | 1991 | AIDS | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 92-0701 | DC | Italy | CSF | 1992 | AIDS | VNIV | αD | 508 | 16 | 3 | 31 | 24 | 14 | 17 | 16 | Cogliati |
| IUM 93-1543 | SCC | Italy | Skin | 1993 | AIDS | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 93-1656 | DC | Italy | CSF | 1993 | AIDS | VNIV | αD | 279 | 22 | 22 | 31 | 22 | 14 | 17 | 34 | Cogliati |
| IUM 97-4851 | SCC | Italy | Skin | 1997 | AIDS | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 98-0824 | SCC | Italy | Skin, right hand and arm | 1998 | Solid tumor prostate | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Cogliati |
| IUM 98-2742 | SCC | Italy | Skin, left hand | 1998 | AIDS | VNIV | αD | 112 | 16 | 22 | 31 | 24 | 14 | 17 | 16 | Cogliati |
| IUM 98-4987 | PPCC | Italy | Skin | 1998 | Solid tumor breast | VNIV | αD | 518 | 26 | 21 | 31 | 15 | 21 | 19 | 15 | Cogliati |
| NIH-530 | VET | Italy | Cow | 1972 | – | VNIV | αD | 112 | 16 | 22 | 31 | 24 | 14 | 17 | 16 | Kwon-Chung |
| PD1596 | VET | Italy | Cat | 2010 | – | VNIV | αD | 252 | 22 | 22 | 30 | 19 | 14 | 23 | 41 | Danesi |
| PD2270 | VET | Italy | Cat | 2011 | – | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Danesi |
| PD32 | VET | Italy | Cat | 2009 | – | VNIV | αD | 294 | 16 | 21 | 24 | 20 | 13 | 22 | 32 | Danesi |
| M9112 | PCC | Japan | Skin | 1985 | No risk factors | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9117 | PCC | Japan | Skin | 1988 | Severe cellular immunity deficiency | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9118 | PCC | Japan | Skin | 1988 | No risk factors | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9119 | PCC | Japan | Skin | 1988 | No risk factors | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9120 | PCC | Japan | Skin | 1983 | No risk factors | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9196 | PCC | Japan | CSF | 1989 | Malignant lymphoma | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9214 | PCC | Japan | Skin | 1989 | No risk factors | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| M9215 | PCC | Japan | Skin | 1993 | Systemic lupus erythematosus | VNIV | αD | 168 | 22 | 21 | 30 | 22 | 14 | 17 | 18 | Ikeda |
| CBS7816 | ENV | Thailand | Cuckoo droppings | 1997 | – | VNIV | αD | 126 | 17 | 21 | 28 | 19 | 14 | 1 | 20 | Kaochaoen |
| TRNCGB1H01-1 | ENV | Turkey | Pine tree | 2013 | – | VNIV | αD | 497 | 16 | 21 | 24 | 20 | 13 | 1 | 32 | Tore O |
| NIH-116 | ENV | USA, Virginia | Pigeon nest | 1960 | – | VNIV | αD | 135 | 27 | 22 | 43 | 24 | 13 | 17 | 20 | Kwon-Chung |
PCC = primary cutaneous cyrptococcosis; PPCC = probable primary cutaneous cryptococcosis; SCC = secondary cutaneous cryptococccosis; DC = disseminated cryptococcosis; ENV = environment; VET = veterinary; CSF = cerebrospinal fluid; BAL = Bronchoalveolar lavage.
2.2. Clinical case definitions
The cases of primary cutaneous cryptococcosis presented a single cutaneous lesion often on the arms or the legs which are primarily due to a traumatic injury. No Cryptococcus antigens from serum were detected and no isolates from other body sites were recovered. The cases that presented isolated cutaneous lesions but were not supported by the other clinical evidences were defined as probable.
Secondary cutaneous cryptococcosis patients presented multiple skin lesions with no specific body sites, positive Cryptococcus antigens or positive cultures from other clinical samples (blood, CSF, urine).
Disseminated cryptococcosis cases presented positive cultures from multiple body sites, positive Cryptococcus antigens, but no skin lesions.
2.3. Molecular analysis
Molecular type and mating type were determined by multiplex PCRs as previously reported (Cogliati et al., 2000; Esposto et al., 2004). Multilocus sequence typing was performed according to the ISHAM consensus scheme (Meyer et al., 2009) and all sequences were deposited in the Cryptococcus MLST database (www.mycologylab.org).
The data of strains WM629, CBS7816, PD32, PD2270, PD1596, RKI 08-0429, RKI 04-0089, RKI 09-0388, RKI 09-0515, RKI 07-0173, RKI 05-0151, RKI 04-0061, RKI 09-0393, RKI 09-0545, RKI 09-0102, RKI 09-0103, RKI 11-0048, RKI 08-0591, RKI 08-0572, RKI 11-0047) were obtained from previous studies (Sanchini et al., 2014; Kaocharoen et al., 2013; Meyer et al., 2009; Danesi et al., 2014).
The concatenated sequences of the seven MLST genes (CAP59, GPD1, IGS1, LAC1, PLB1, SOD1, URA5) of the 83 C. neoformans var. neoformans and 30 C. neoformans var. grubii isolates were aligned by ClustalW algorithm (www.ebi.ac.uk) and the resulting file was converted in a Roehl data file by DnaSP software (Universitat de Barcelona, www.ub.edu/dnasp). Network analysis was performed using the median joining method included in the software Network v4.6 (Fluxus Technology Ltd., www.fluxus-engeneering.com).
Genetic population parameters and population comparisons were performed by DnaSP software whereas maximum likelihood phylogenetic analysis and average evolutionary divergence were calculated with the software Mega v6.0 (www.megasoftware.net).
The degree of recombination inside the population was also calculated using both the linkage disequilibrium test and the Watterson estimator (theta) method (DnaSP software). The linkage disequilibrium test is an extension of Fisher's exact probability test on contingency tables. The test consists in obtaining the probability of finding a table with the same marginal totals and which has a probability equal or less than the observed table. The null-hypothesis of non-random association between the two tested loci was confirmed if the probability was less than 0.05. The Watterson estimator (theta) method extrapolates two values that correspond to the expected theta value for a non-recombinant population and to the expected theta value for a free-recombinant population, and then it calculates the observed theta value in the investigated population.
3. Results
Molecular identification confirmed that all the C. neoformans var. neoformans isolates belonged to molecular type VNIV and that 63 were mating type α and 20 mating type a (Table 1).
The alignment of the 4092-bp sequences resulting by concatenating the seven MLST loci showed the presence of 425 polymorphic sites that identified 49 sequence types with a haplotype diversity (Hd) value of 0.965. LAC1 and URA5 were the most polymorphic loci discriminating 16 and 15 haplotypes, respectively. In contrast, the IGS1 locus was the least discriminatory locus with an Hd value of 0.241 (Table 2).
Table 2.
Genetic population parameters defining the population structure of the 83 var. neoformans isolates investigated.
| Locus | Sequence length (bp) | Polymorphic sites | Haplotypes | Haplotype diversity (Hd) | Recombining events | Linkage disequilibrium | Theta (no recombination; free recombination) |
|---|---|---|---|---|---|---|---|
| CAP59 | 560 | 41 | 9 | 0.761 | 4 | No | – |
| GPD1 | 558 | 58 | 5 | 0.241 | 1 | No | – |
| IGS1 | 781 | 57 | 11 | 0.772 | 1 | No | – |
| LAC1 | 481 | 149 | 15 | 0.866 | 20 | No | – |
| PLB1 | 534 | 40 | 7 | 0.584 | 0 | No | – |
| SOD1 | 537 | 73 | 11 | 0.628 | 2 | No | – |
| URA5 | 639 | 55 | 16 | 0.865 | 3 | No | – |
| All loci | 4092 | 425 | 49 | 0.965 | 31 | No | 87.1 (484.0; 17.4) |
Parameters were calculated by DnaSP v.5 software.
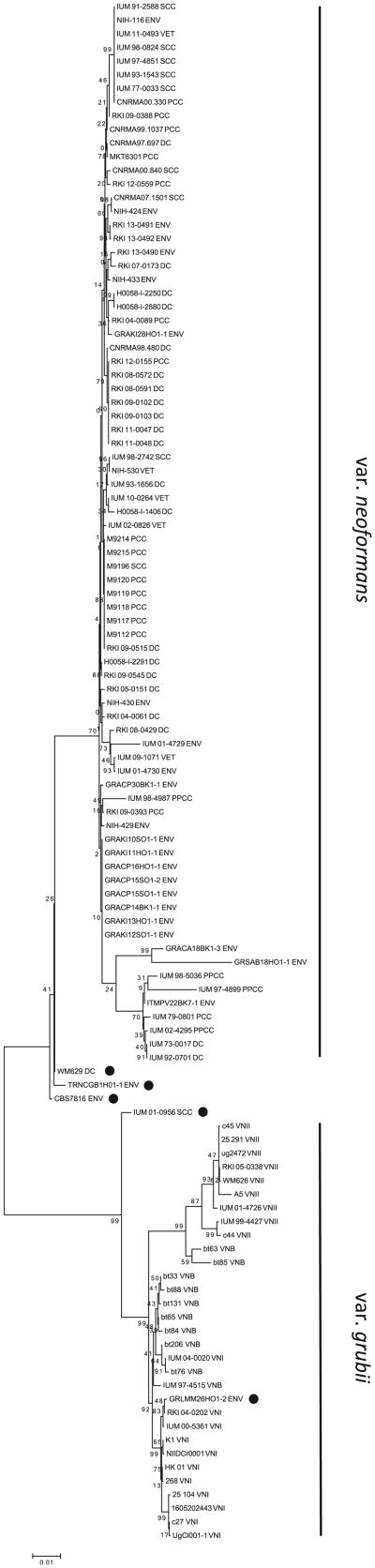
Maximum likelihood phylogenetic reconstruction showed that most of C. neoformans var. neoformans isolates were grouped in a unique cluster with an average evolutionary divergence of 0.008, twofold lower than that observed among C. neoformans var. grubii isolates. Four isolates (WM629, CBS7816, TRNCGB1HO1-1, IUM 01-0956) were situated between the two varieties with an ambiguous topology and one isolate (GRLMM26HO1-2) was unexpectedly more related to the VNI group (Fig. 1). A network analysis showed the same topology with a core region grouping the majority of C. neoformans var. neoformans isolates all linked by star-like branches. The topology of C. neoformans var. grubii population was very different with clear clusters and higher genetic distances. The two populations were linked by a long branch, along which were the five ambiguous isolates (Fig. 2A).
Fig. 1.
Maximun likelihood phylogenetic reconstruction including 83 C. neoformans var. neoformans (VNIV) and 30 C. neoformans var. grubii (VNI, VNII, VNB) isolates. Black dots indicate VNIV isolates with an ambiguous position. Numbers near the nodes represent the bootstrap values obtained for 1000 replications.
Fig. 2.
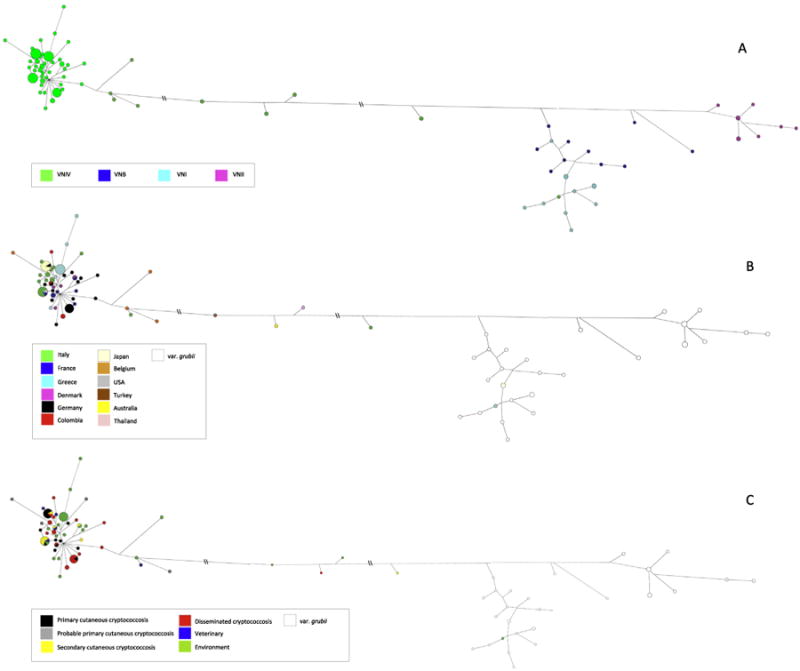
Network analysis performed by median joining algorithm. Panel A shows the different genotypes, panel B shows the different geographical origin, and panel C the different source of isolation. Double slash means that the branch has been shortened to fit the image.
The investigated isolates were therefore divided in three groups, C. neoformans var. neoformans, C. neoformans var. grubii and putative intervarietal recombinants, and average evolutionary divergence between the populations was calculated. The results showed that C. neoformans var. neoformans and C. neoformans var. grubii diverged 0.09 and both diverged from the putative recombinants around 0.05. The five putative recombinants were then checked to exclude mixed cultures and five single colonies from each of the original strains were collected and processed for molecular typing and ploidy determination by flow cytometry. The results confirmed that all strains from single colonies were VNIV aD or αD and that they were haploids (Fig. S1).
The linkage disequilibrium analysis of C. neoformans var. neoformans populations was also calculated for both the concatenated sequences and the single loci alignments. The results confirmed the absence of linkage disequilibrium in all the cases confirming that recombination in this population could not be excluded (Table 2). The Watterson estimator (theta) method results were in perfect agreement with that obtained in the linkage disequilibrium test strengthening for the hypothesis of a recombinant population (Table 2). In addition, the estimation of recombination events among the whole C. neoformans var. neoformans population reveals that at least 31 recombination events had occurred and that the most recombinant locus was LAC1 (20 events) (Table 2).
Fig. 2B displays geographical origin of each isolate on the network tree. No correlation was observed between the country of isolation and the MLST profile. Only the eight Japanese isolates belonged all to the same genotype (ST168). Similarly, comparison between MLST profiles and source of isolation did not provide evidence of any specific correlation. ST135 and ST180 grouped together isolates from the environment with isolates from veterinary cases, from primary cutaneous cryptococcosis and from secondary cutaneous cryptococcosis whereas none of the environmental isolates shared the same ST with isolates from disseminated cryptococcosis (Fig. 2C).
4. Discussion
The present study investigated, for the first time by MLST, the genetic population structure of a large number of C. neoformans var. neoformans isolates from different geographical origin and from different sources. The results strongly support the hypothesis that this population is recombinant and are in agreement with the recent study carried out by other authors (Desnos-Ollivier et al., 2015) in a population of French clinical C. neoformans isolates. The high haplotype diversity as well as the low evolutionary divergence among C. neoformans var. neoformans population confirm that isolates are strictly correlated each to the other but they are characterized by high variability due to recombination. Statistical tests such as linkage disequilibrium analysis, Watterson estimator calculation, and recombining events evaluation also corroborate this hypothesis. Furthermore, the C. neoformans var. neoformans population investigated in the present study included 20 mating type a isolates recovered from both patients and the environment. This suggests that isolates of C. neoformans var. neoformans mating type a are more prevalent than those of C. neoformans var. grubii and therefore sexual reproduction may occur more frequently.
The high polymorphism observed for the LAC1 locus suggests that the laccase enzyme plays a crucial role for both the host infection and the environmental survival of this yeast. The survival in a particular ecological niche, such as a tree, could depend on the capacity of this yeast to degrade a wide range of phenolic compounds present in lignin of the tree trunk. In addition, since laccase is a key enzyme for melanin production this variety could gain an advantage for growth on surfaces exposed to light, for example the bark of the tree.
Both maximum likelihood phylogenetic analysis and network analysis identified a group of five isolates with an ambiguous position on the tree. The average evolutionary divergence between C. neoformans var. grubii and C. neoformans var. neoformans groups was twice than that observed between the two groups and the ambiguous isolates suggesting that the five isolates could represent haploid strains generated by intervarietal recombination (Xu et al., 2000; Kavanaugh et al., 2006). Laboratory isolated haploid intervarietal recombinant clones have been reported by crossing between H99, a strain of C. neoformans var. grubii VNI, and JEC20, a strain of C. neoformans var. neoformans (Kwon-Chung and Varma, 2006). This is in contrast with the recent published proposal (Hagen et al., 2015), advocating that C. neoformans var. neoformans and C. neoformans var. grubii are separate species. Further investigations and a larger number of C. neoformans var. neoformans isolates are needed to clarify this important issue.
The comparison between MLST profiles and geographical origin showed that isolates recovered from different countries and from different continents were grouped in the same cluster confirming the role of recombination in shortening the genetic divergence. However, clonal expansion of some genotypes could occur in geographical areas where physical barriers exist that prevent recombination. This could be true in Japan where all the isolates investigated belonged to the same ST even though they were isolated from different patients, at different time points and from different regions of Japan.
A similar analysis to this one, which compared the source of isolates, showed that the environmental genotypes could be the source of infections for animal, primary cutaneous cryptococcosis and secondary cutaneous cryptococcosis. In contrast, this was not observed for the isolates from disseminated cryptococcosis cases, which shared an identical ST but only with those from primary and secondary cutaneous cryptococcosis cases. Further studies are required to confirm this discrepancy.
In conclusion, our study showed that C. neoformans var. neoformans population is not evolving primarily by clonal expansion, as observed for C. neoformans var. grubii (Khayhan et al., 2013), and that intravarietal recombination is largely occurring and intervarietal recombination could not be excluded.
Supplementary Material
Acknowledgments
We thank Prof. F. Dromer and Prof. J. Kwon-Chung for providing some of the isolates and for her suggestions to improve this study, and Dr. T. Boekhout and Dr. B. Theelen for sequencing support.
Footnotes
Disclosures: The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Appendix A. Supplementary material: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fgb.2016.01.003.
References
- Assogba K, et al. Neuromeningeal cryptococcosis in sub-Saharan Africa: killer disease with sparse data. J Neurosci Rural Pract. 2015;6:221–224. doi: 10.4103/0976-3147.153231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton EW, et al. Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS ONE. 2012;7:e43582. doi: 10.1371/journal.pone.0043582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoor MR, et al. Current scenario of cryptococcosis and antifungal susceptibility pattern in India: a cause for reappraisal. Mycoses. 2008;51:258–265. doi: 10.1111/j.1439-0507.2007.01478.x. [DOI] [PubMed] [Google Scholar]
- Chen SC, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin Microbiol Rev. 2014;27:980–1024. doi: 10.1128/CMR.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 2010;10:769–778. doi: 10.1111/j.1567-1364.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, et al. Molecular epidemiology of Italian clinical Cryptococcus neoformans var. grubii isolates. Med Mycol. 2013;51:499–506. doi: 10.3109/13693786.2012.751642. [DOI] [PubMed] [Google Scholar]
- Cogliati M, et al. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med Mycol. 2000;38:97–103. doi: 10.1080/mmy.38.2.97.103. [DOI] [PubMed] [Google Scholar]
- Cogliati M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an Atlas of the molecular types. Scientifica. 2013;2013:675213. doi: 10.1155/2013/675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, et al. Environmental survey of Cryptococcus neoformans and C. gattii in European and Mediterranean area. Mycoses. 2014;57(Suppl. 1):30. [Google Scholar]
- Cortés JA, et al. Fungal bloodstream infections in tertiary care hospitals in Colombia. Rev Iberoam Micol. 2011;28:74–78. doi: 10.1016/j.riam.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Danesi P, et al. Multilocus sequence typing (MLST) and M13 PCR fingerprinting revealed heterogeneity amongst Cryptococcus species obtained from Italian veterinary isolates. FEMS Yeast Res. 2014;14:897–909. doi: 10.1111/1567-1364.12178. [DOI] [PubMed] [Google Scholar]
- Desnos-Ollivier M, et al. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. MBio. 2015;6:e00311. doi: 10.1128/mBio.00311-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A, Kidd SE. Current trends in the prevalence of Cryptococcus gattii in the United States and Canada. Infect Drug Resist. 2015;11:89–97. doi: 10.2147/IDR.S57686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposto MC, et al. Determination of Cryptococcus neoformans var. neoformans mating type by multiplex PCR. Clin Microbiol Infect. 2004;10:1092–1094. doi: 10.1111/j.1469-0691.2004.00972.x. [DOI] [PubMed] [Google Scholar]
- Feng X, et al. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 2008;8:930–938. doi: 10.1111/j.1567-1364.2008.00422.x. [DOI] [PubMed] [Google Scholar]
- Hagen F, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Heitman J, et al. Cryptococcus: From Human Pathogen to Model Yeast. ASM Press; Washington, DC, USA: 2010. [Google Scholar]
- Henao-Martínez AF, Beckham JD. Cryptococcosis in solid organ transplant recipients. Curr Opin Infect Dis. 2015;28:300–307. doi: 10.1097/QCO.0000000000000171. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Shinoda T. Mycological and serological diagnosis of cryptococcosis. Nihon Ishinkin Gakkai Zasshi. 2000;41:241–244. doi: 10.3314/jjmm.41.241. [DOI] [PubMed] [Google Scholar]
- Kaocharoen S, et al. Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl Trop Dis. 2013;7:e2297. doi: 10.1371/journal.pntd.0002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh LA, Fraser JA, Dietrich FS. Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol Biol Evol. 2006;23:1879–1890. doi: 10.1093/molbev/msl070. [DOI] [PubMed] [Google Scholar]
- Khayhan K, et al. Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE. 2013;8:e72222. doi: 10.1371/journal.pone.0072222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4:a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, et al. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, et al. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol. 2005;43:556–564. doi: 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, et al. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuville S, et al. Primary cutaneous cryptococcosis: a distinct clinical entity. Clin Infect Dis. 2003;36:337–347. doi: 10.1086/345956. [DOI] [PubMed] [Google Scholar]
- Park BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Pérez C, et al. Cryptococcus neoformans, Cryptococcus gattii: serotypes in Venezuela. Mycopathologia. 2008;166:149–153. doi: 10.1007/s11046-008-9132-1. [DOI] [PubMed] [Google Scholar]
- Sanchini A, et al. Molecular typing of clinical Cryptococcus neoformans isolates collected in Germany from 2004 to 2010. Med Microbiol Immunol. 2014;203:333–340. doi: 10.1007/s00430-014-0341-6. [DOI] [PubMed] [Google Scholar]
- Sukroongreung S, et al. Serotypes of Cryptococcus neoformans isolated from patients prior to and during the AIDS era in Thailand. Mycopathologia. 1996;135:75–78. doi: 10.1007/BF00436454. [DOI] [PubMed] [Google Scholar]
- Trilles L, et al. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med Mycol. 2003;41:383–390. doi: 10.1080/1369378031000137206. [DOI] [PubMed] [Google Scholar]
- Umeyama T, et al. Determination of epidemiology of clinically isolated Cryptococcus neoformans strains in Japan by multilocus sequence typing. Jpn J Infect Dis. 2013;66:51–55. doi: 10.7883/yoken.66.51. [DOI] [PubMed] [Google Scholar]
- Viviani MA, et al. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–619. doi: 10.1111/j.1567-1364.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- Wiesner DL, et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. MBio. 2012;3:e00196–12. doi: 10.1128/mBio.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Vilgalys R, Mitchell TG. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- Yan Z, Li X, Xu J. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 2002;40:965–972. doi: 10.1128/JCM.40.3.965-972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.