Abstract
Lambda chromosomes are cut and packaged from concatemeric DNA by phage enzyme terminase. Terminase initiates DNA packaging by binding at a site called cosB and introducing staggered nicks at an adjacent site, cosN, to generate the left cohesive end of the DNA molecule to be packaged. After DNA packaging terminase recognizes and cuts the terminal cosN, an event that does not require a wild-type cosB. In this work a site, called cosQ, has been identified that is required for termination of DNA packaging. cosQ, defined by mutations in a sequence called R4, is located approximately 30 bp upstream from cosN. The order of sites is cosQ-cosN-cosB. Helper packaging of repressed, tandem prophage chromosomes demonstrated that a cosQ point mutation affects DNA packaging only when placed at the terminal cos site, whereas cosB mutations only affect packaging initiation. In vitro packaging studies confirmed that cosQ mutations do not affect packaging initiation. In vivo studies indicated that cosQ mutations do not affect cutting of initial cos sites but do cause a defect in packaging termination. cosQ mutants accumulated expanded phage heads, indicating that cosQ mutations affect a step that occurs after packaging of a substantial length of phage DNA. These results show that cosQ mutations define a site required for use of cos sites present at the ends of lambda chromosomes undergoing packaging. Available evidence suggests that other viruses, including phages T3 and T7 and the herpesviruses, may ultimately prove to use cosQ-like sites for packaging termination.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Marko M., Gold M. Early events in the in vitro packaging of bacteriophage lambda DNA. Virology. 1977 May 1;78(1):291–305. doi: 10.1016/0042-6822(77)90100-3. [DOI] [PubMed] [Google Scholar]
- Becker A., Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990 Jun;172(6):2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S., Daub E., Murialdo H. The overproduction of DNA terminase of coliphage lambda. Gene. 1987;60(2-3):277–289. doi: 10.1016/0378-1119(87)90236-8. [DOI] [PubMed] [Google Scholar]
- Chung Y. B., Hinkle D. C. Bacteriophage T7 DNA packaging. II. Analysis of the DNA sequences required for packaging using a plasmid transduction assay. J Mol Biol. 1990 Dec 20;216(4):927–938. doi: 10.1016/S0022-2836(99)80011-4. [DOI] [PubMed] [Google Scholar]
- Cue D., Feiss M. Genetic analysis of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda. J Mol Biol. 1992 Nov 5;228(1):58–71. doi: 10.1016/0022-2836(92)90491-2. [DOI] [PubMed] [Google Scholar]
- Cue D., Feiss M. Genetic analysis of mutations affecting terminase, the bacteriophage lambda DNA packaging enzyme, that suppress mutations in cosB, the terminase binding site. J Mol Biol. 1992 Nov 5;228(1):72–87. doi: 10.1016/0022-2836(92)90492-3. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J Mol Biol. 1974 Mar 15;83(4):511–525. doi: 10.1016/0022-2836(74)90511-7. [DOI] [PubMed] [Google Scholar]
- Feiss M., Fisher R. A., Siegele D. A., Nichols B. P., Donelson J. E. Packaging of the bacteriophage lambda chromosome: a role for base sequences outside cos. Virology. 1979 Jan 15;92(1):56–67. doi: 10.1016/0042-6822(79)90214-9. [DOI] [PubMed] [Google Scholar]
- Feiss M., Kobayashi I., Widner W. Separate sites for binding and nicking of bacteriophage lambda DNA by terminase. Proc Natl Acad Sci U S A. 1983 Feb;80(4):955–959. doi: 10.1073/pnas.80.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M., Sippy J., Miller G. Processive action of terminase during sequential packaging of bacteriophage lambda chromosomes. J Mol Biol. 1985 Dec 20;186(4):759–771. doi: 10.1016/0022-2836(85)90395-x. [DOI] [PubMed] [Google Scholar]
- Feiss M., Widner W. Bacteriophage lambda DNA packaging: scanning for the terminal cohesive end site during packaging. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3498–3502. doi: 10.1073/pnas.79.11.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
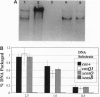
- Gold M., Becker A. The bacteriophage lambda terminase. Partial purification and preliminary characterization of properties. J Biol Chem. 1983 Dec 10;258(23):14619–14625. [PubMed] [Google Scholar]
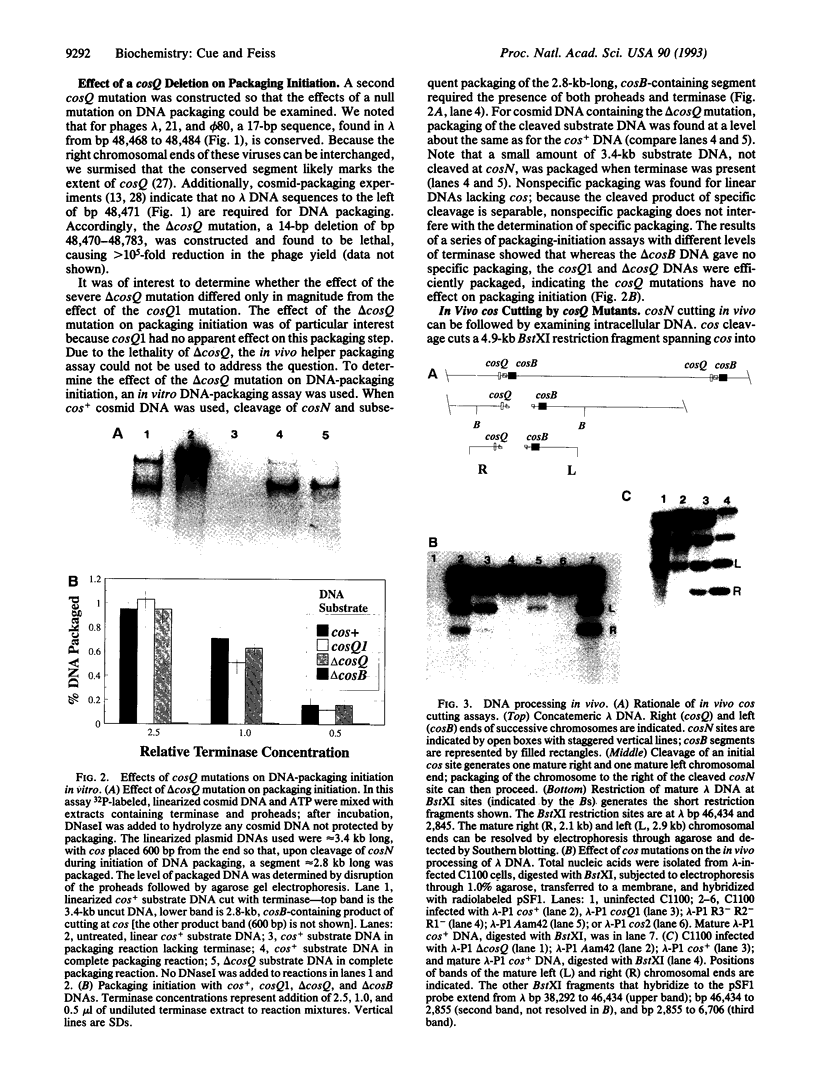
- Hashimoto C., Fujisawa H. DNA sequences necessary for packaging bacteriophage T3 DNA. Virology. 1992 Apr;187(2):788–795. doi: 10.1016/0042-6822(92)90480-d. [DOI] [PubMed] [Google Scholar]
- Hohn B. DNA sequences necessary for packaging of bacteriophage lambda DNA. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7456–7460. doi: 10.1073/pnas.80.24.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley G. H., Kemp C. L. Isolation and protein composition of normal and petit capsids of bacteriophage lambda. Virology. 1971 Nov;46(2):298–309. doi: 10.1016/0042-6822(71)90031-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi I., Stahl M. M., Stahl F. W. The mechanism of the chi-cos interaction in RecA-RecBC-mediated recombination in phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:497–506. doi: 10.1101/sqb.1984.049.01.056. [DOI] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T., Matsubara K. Lambda phage DNA sequences affecting the packaging process. Gene. 1983 Oct;24(2-3):199–206. doi: 10.1016/0378-1119(83)90080-x. [DOI] [PubMed] [Google Scholar]
- Murialdo H., Fife W. L. Synthesis of a trans-acting inhibitor of DNA maturation by prohead mutants of phage lambda. Genetics. 1987 Jan;115(1):3–10. doi: 10.1093/genetics/115.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinder G., Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J Virol. 1988 Feb;62(2):387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy J., Feiss M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage lambda terminase. J Bacteriol. 1992 Feb;174(3):850–856. doi: 10.1128/jb.174.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. P., Feiss M. Sites and gene products involved in lambdoid phage DNA packaging. J Bacteriol. 1993 Apr;175(8):2393–2399. doi: 10.1128/jb.175.8.2393-2399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Austin S. Isolation and characterization of P1 minireplicons, lambda-P1:5R and lambda-P1:5L. J Bacteriol. 1983 Feb;153(2):800–812. doi: 10.1128/jb.153.2.800-812.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Weisberg R. Packaging of prophage and host DNA by coliphage lambda. Nature. 1975 Jul 10;256(5513):97–103. doi: 10.1038/256097a0. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Xin W., Feiss M. Function of IHF in lambda DNA packaging. I. Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J Mol Biol. 1993 Mar 20;230(2):492–504. doi: 10.1006/jmbi.1993.1166. [DOI] [PubMed] [Google Scholar]
- Xu S. Y., Feiss M. Structure of the bacteriophage lambda cohesive end site. Genetic analysis of the site (cosN) at which nicks are introduced by terminase. J Mol Biol. 1991 Jul 20;220(2):281–292. doi: 10.1016/0022-2836(91)90013-v. [DOI] [PubMed] [Google Scholar]